Abstract
Objectives
How the local inflammatory environment regulates epigenetic changes in the context of inflammatory arthritis remains unclear. Here we assessed the transcriptional and active enhancer profile of monocytes derived from the inflamed joints of JIA patients, a model well-suited for studying inflammatory arthritis.
Methods
RNA sequencing and H3K27me3 chromatin immunoprecipitation sequencing (ChIP-seq) were used to analyse the transcriptional and epigenetic profile, respectively, of JIA synovial fluid-derived monocytes.
Results
Synovial-derived monocytes display an activated phenotype, which is regulated on the epigenetic level. IFN signalling-associated genes are increased and epigenetically altered in synovial monocytes, indicating a driving role for IFN in establishing the local inflammatory phenotype. Treatment of synovial monocytes with the Janus-associated kinase (JAK) inhibitor ruxolitinib, which inhibits IFN signalling, transformed the activated enhancer landscape and reduced disease-associated gene expression, thereby inhibiting the inflammatory phenotype.
Conclusion
This study provides novel insights into epigenetic regulation of inflammatory arthritis patient-derived monocytes and highlights the therapeutic potential of epigenetic modulation for the treatment of inflammatory rheumatic diseases.
Keywords: autoimmunity, monocytes, epigenetics, JIA, synovium
Rheumatology key messages.
Inflammatory phenotype of autoimmune monocytes is regulated on the epigenetic level.
IFN signaling is involved in the inflammatory phenotype of autoimmune monocytes.
JAK inhibitors alter the epigenetic landscape of autoimmune monocytes.
Introduction
Inflammatory arthritis is characterized by abnormal immune responses due to loss of immunological tolerance. The presence of autoreactive T and B cells and autoantibodies often links the pathogenesis of inflammatory arthritis to the adaptive immune system, but innate immune cells, such as monocytes, also play a key role. Monocytes can recruit adaptive immune cells to the site of inflammation by secreting chemokines and further perpetuate immune activation, through either direct cell–cell contact or abundant secretion of pro-inflammatory cytokines (i.e. IL-6, TNF-α). As such, inflammatory arthritis and many autoimmune diseases are associated with increased monocyte infiltration [1–5]. The exact mechanisms that regulate monocyte activity remain to be determined, but it is clear that environmental factors and epigenetic regulation are important in the pathogenesis of inflammatory arthritis [6]. Enhancers are cis-regulatory DNA regions, generally a few hundred base pairs in size, that are pivotal for the spatio-temporal regulation of gene expression by recruiting RNA polymerase II, transcription factors and co-factors, such as the histone acetyltransferase p300 or the mediator complex [7]. Enhancers regions are characterized by methylation of histone H3 at lysine 4 (H3K4me) and contain H3K27 acetylation in their active status [8, 9]. Around 60% of the single nucleotide polymorphisms (SNPs) associated with autoimmune disease are localized in enhancer regions and disease-associated variants preferentially map to enhancer regions specific for disease-relevant cell types [10–15]. For example, SNPs associated with multiple sclerosis (MS) and SSc have been demonstrated to map to monocyte enhancer regions, indicating that epigenetic regulation plays an important role in autoimmune disease pathogenesis [16]. In addition, alterations of the histone modification profile have been described in monocytes and other cell types derived from patients with inflammatory rheumatic diseases [17–19].
Information concerning epigenetic changes specifically at the site of inflammation is currently lacking. Reasons for this include the difficulty in obtaining enough immune cells from the local inflammatory site to perform in-depth analyses. For regulatory and effector T cells it has been demonstrated that the epigenetic landscape is altered at the site of autoinflammation and that these changes contribute to their pro-inflammatory phenotype [12, 20]. These studies indicate that the inflammatory environment can directly induce epigenetic changes that result in a pro-inflammatory phenotype that contributes to disease pathogenesis. The role of epigenetic regulation in monocytes at the site of inflammation has not been well studied. Here, we evaluated the role of active enhancers in monocytes in human inflammatory arthritis, by using monocytes derived from the inflamed joints of JIA patients. RNA-sequencing and chromatin immunoprecipitation (ChIP) sequencing for H3K27ac demonstrated that human monocytes from the local inflammatory site display an epigenetically-regulated activated phenotype. This phenotype was characterized by increased expression of IFN-responsive genes. Treatment of inflammatory site-derived monocytes with the JAK-1,2 inhibitor ruxolitinib reduced the active state of various enhancer regions, correlating with impaired expression of disease-associated genes. Taken together, these data demonstrate that IFN signalling induces epigenetic alterations and that these changes contribute to a pro-inflammatory phenotype, suggesting that modulating the epigenetic landscape might be a potential therapeutic approach for the treatment of inflammatory arthritis.
Methods
Collection of SF and peripheral blood (PB) samples
Twenty-six JIA patients were included in this study; 21 patients were diagnosed with oligoarticular JIA and five patients with polyarticular JIA [21]. Active disease was defined by physician global assessment of >1 active joint (swelling and/or limitation of movement) and inactive disease (remission) was defined as the absence thereof. PB and SF samples were obtained at the same moment, either via vein puncture or intravenous drip and therapeutic joint aspiration, respectively. Written informed consent was obtained from all patients either directly or from parents/guardians when the patients were younger than age 12 years. The study procedures were approved by the Institutional Review Board of the University Medical Center Utrecht (UMCU; METC nr: 11–499/C) and performed according to the principles expressed in the Helsinki Declaration. SF samples were treated with hyaluronidase (Sigma-Aldrich) for 30 min, 37°C. Synovial fluid mononuclear cells (SFMCs) and peripheral blood mononuclear cells (PBMCs) were isolated using Ficoll-Paque density gradient centrifugation (GE Healthcare) and were either used immediately or frozen and stored at –80°C until further use.
Monocyte isolation and culture
For RNA-sequencing of ex vivo HC and JA monocytes (Fig. 1), HC PBMCs and JIA SFMCs were thawed, CD3+ cells were depleted by human anti-CD3 microbeads (Miltenyi Biotec, Cologne, Germany) according to the manufacturer’s instructions and CD14+CD16+ cells were sorted by flow cytometry using a BD FACS Aria III (BD Biosciences, Franklin Lakes, New Jersey, US). For validation of ex vivo sequencing results, CD14+ were isolated from frozen HC PBMCs, JIA PBMCs and SFMCs by magnetic-activated cell sorting (MACS) using human CD14 microbeads (Miltenyi Biotec) according to the manufacturer’s instructions. For ChIP-sequencing of ex vivo HC and JA monocytes (Fig. 2), HC PBMCs, JIA PBMCs and JIA SFMCs CD14+ cells where isolated by flow cytometry using a BD FACS Aria II (BD Biosciences). For analysis of JQ1 sensitive genes, HC PBMCs were thawed and CD14+ cells were sorted by flow cytometry using a BD FACS Aria II (BD Biosciences). Subsequently, CD14+ cells were cultured o/n with 100 ng/ml LPS (Invivogen, Waltham, Massachusetts, US) and treated with 300 nM JQ1(–) or JQ1(+) (ApexBio, Houston Texas, US) in RPMI 1640 + GlutaMAX (Life Technologies, Carlsbad, California, US) supplemented with 100 U/ml penicillin (Gibco, Carlsbad, California, US), 100 mg/ml streptomycin (Gibco) and 10% heat-inactivated human AB-positive serum (Invitrogen) at 37°C in 5% CO2. For RNA- and ChIP-sequencing of Ruxolitinib-treated samples, CD14+ cells were isolated by magnetic-activated cell sorting (MACS) from JIA SFMCs and cultured for 4 h with or without 10 ng/ml IFNγ and 1 µM Ruxolitinib (Selleck Chemicals, Houston Texas, US).
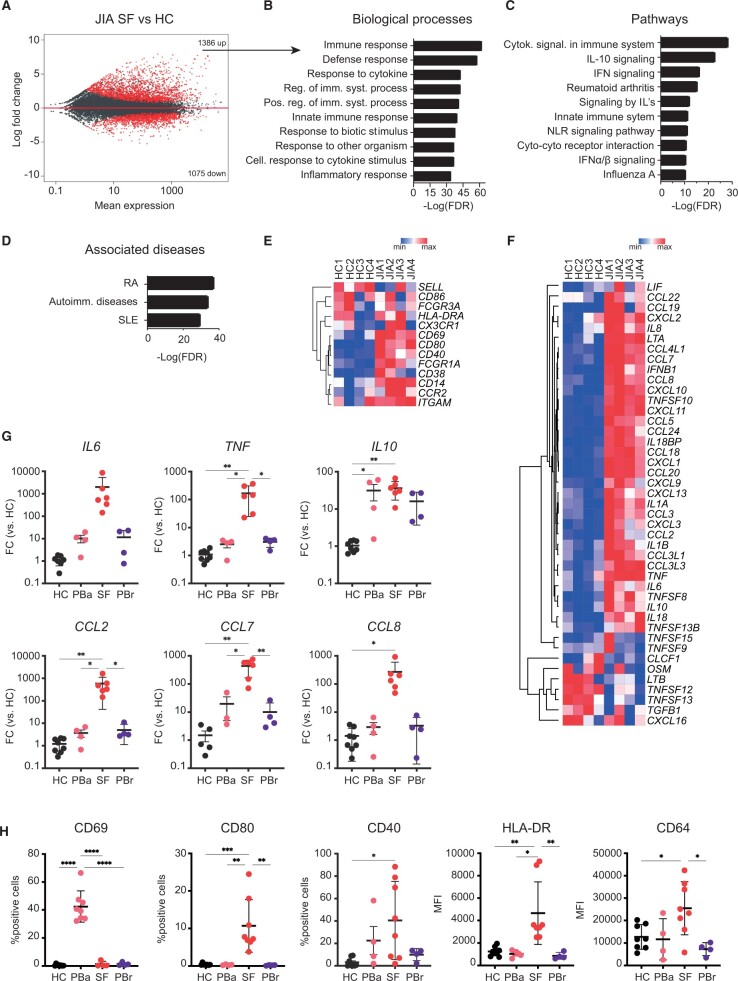
Figure 1.
Transcriptional analysis of JIA SF monocytes indicates an activated phenotype. (A) MA plot displaying genes differentially expressed between HC PB monocytes and JIA SF monocytes. Red dots indicate genes with FDR < 0.05. (B) Top 10 biological processes associated with genes that are significantly increased in JIA SF monocytes. (C) Top 10 pathways associated with genes that are significantly increased in JIA SF monocytes. (D) Top 3 diseases associated with genes that are significantly increased in JIA SF monocytes. (E, F) Heatmap demonstrating the expression of selected monocyte surface markers (E) and selected cytokines (F) and chemokines in HC PB and JIA SF monocytes. (G) Gene expression of selected cytokine and chemokines in monocytes derived from either the PB of HC, PB of JIA patients undergoing active disease (PBa), SF of JIA patients, and PB of JIA patients in remission (PBrem). (H) Protein levels of selected surface markers in monocytes derived from either the PB of HC, PB of JIA patients undergoing active disease (PBa), SF of JIA patients, and PB of JIA patients in remission (PBrem). P-values for F and G were calculated using an ordinary one-way ANOVA. *P < 0.05; **P < 0.01; ***P < 0.001. HC: healthy control; FDR: false discovery rate; PB: peripheral blood
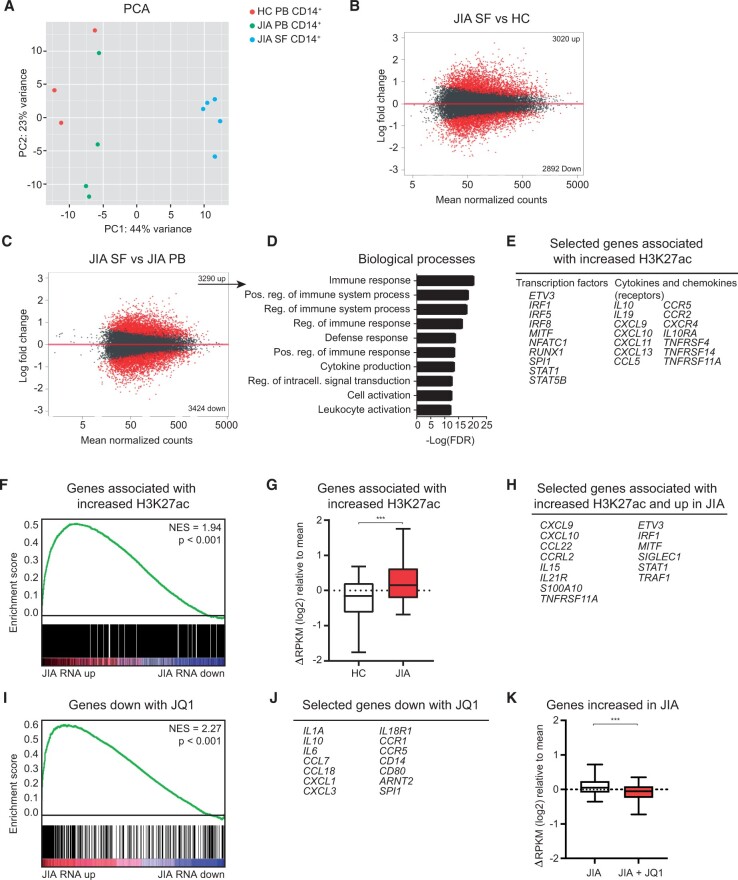
Figure 2.
Epigenetic regulation contributes to the activated phenotype of JIA SF monocytes. (A) PCA of H3K27ac signal in HC PB, JIA PB and JIA SF monocytes. (B and C) MA plot of H3K27ac regions different between JIA SF and HC PB and between JIA SF and JIA PB. Red dots indicate H3K27ac regions with FDR < 0.1. (D) Top 10 biological processes associated with genes significantly increased in JIA SF vs JIA PB. (E) Selected genes associated with increased H3K27ac signal in JIA SF vs JIA PB. (F) GSEA of genes associated with increased H3K27ac signal in JIA SF and genes differentially expressed within JIA SF. (G) Boxplot with 5%–95% whiskers displaying △RPKM (log2) values of genes associated with increased H3K27ac signal in HC and JIA. (H) Selected genes associated with increased H3K27ac signal and increased in JIA. (I) GSEA of genes decreased with JQ1 and genes differentially expressed within JIA SF. (J) Selected genes decreased with JQ1. (K) Boxplot with 5%–95% whiskers displaying △RPKM (log2) values of genes increased in JIA monocytes treated with or without JQ1. P-values for G and K were calculated using Wilcoxon-matched pairs signed rank test. *P < 0.05; **P < 0.01; ***P < 0.001. FDR: false discovery rate; GSEA: gene set enrichment analysis; HC: healthy control; PB: peripheral blood; PCA: principle component analysis; RPKM: reads per kilobase of exon per million reads mapped
ChIP-sequencing and analysis
ChIP-sequencing was performed as in [22]. Experimental and analysis details are described in the Supplementary Materials available at Rheumatology online.
RNA-sequencing and analysis
RNA-sequencing was performed as in [22]. Experimental and analysis details are described in the Supplementary Materials available at Rheumatology online.
Quantitative RT-PCR
Total RNA was extracted using the PicoPure RNA Isolation kit and cDNA synthesis was performed using the iScript cDNA synthesis kit (Bio-Rad, Hercules, California, US). cDNA samples were amplified with SYBR Select mastermix (Life Technologies) in a QuantStudio 12k flex (Thermo Fisher Scientific) according to the manufacturer’s protocol.
Multiplex immunoassay
Multiplex analysis (xMAP; Luminex, Austin, Texas, US) was performed as described previously on supernatant derived from HC and JIA-derived CD14+ cells either cultured o/n (to determined ex vivo cytokine production) or for 4 h in the presence of 1 µM ruxolitinib [23].
Results
Synovial-derived monocytes display an activated phenotype on the transcriptional level
To evaluate the transcriptional changes at the site of local autoimmune inflammation, monocytes were collected from inflamed joints of oligoarticular JIA patients. RNA-sequencing analysis of JIA CD14+CD16+ monocytes obtained from the synovial compartment (SF) and HC peripheral blood (PB)-derived CD14+CD16+ cells was performed. A total of 2443 genes were significantly differentially expressed (Fig. 1A). Genes that were upregulated in monocytes derived from the local inflammatory site are associated with regulation of the immune system and in particular with cytokine responses (Fig. 1B and C). Furthermore, the top three diseases associated with these genes are all autoimmune diseases (Fig. 1D). Analysis of monocyte surface markers revealed increased expression of several surface activation markers, including CD80, CD69, CCR2 and FCGR1A in monocytes (Fig. 1E). In addition, many cytokines and chemokines were expressed at a higher level by SF monocytes (Fig. 1F). To validate this, we analysed mRNA and protein levels of several cytokines, chemokines and surface markers in CD14+ monocytes derived from either the PB of HC, PB and SF of JIA patients undergoing active disease or in remission. We confirmed increased gene and protein expression in JIA SF-derived monocytes compared with HC monocytes and the majority of genes and proteins are also significantly higher expressed compared with JIA PB-derived monocytes (Fig. 1G, H; Supplementary Fig. S1A, available at Rheumatology online). In addition, mRNA and protein levels of these cytokines, chemokines, and surface markers are increased in polyarticular JIA patients (Supplementary Fig. S1B–D, available at Rheumatology online). Collectively, these data demonstrate that transcriptional changes in monocytes derived from the site of inflammation result in an activated phenotype associated with (auto-)inflammation.
H3K27ac regulation contributes to the activated phenotype of synovial monocytes
To determine whether the transcriptional changes in synovial monocytes are regulated on the epigenetic level, ChIP-sequencing for H3K27ac was performed on CD14+ cells from HC PB JIA PB- and JIA SF. Principal component analysis demonstrated that JIA SF monocytes clustered separately from HC and paired JIA PB-derived monocytes (PC1: 44%), indicating that their H3K27ac profile is distinct (Fig. 2A). Indeed, >3000 regions contain a significantly different H3K27ac signal in inflammation-derived monocytes compared with either HC or JIA PB monocytes, while JIA PB and HC PB monocytes are relatively similar regarding their H3K27ac profile (Fig. 2B, C; Supplementary Fig. S2A, available at Rheumatology online). Analysis of the genes associated with an increased H3K27ac signal in local inflammatory site-derived monocytes compared with JIA PB monocytes, indicated association with biological processes that are involved in regulation of the immune system and include many cytokine and chemokines of which the expression was upregulated in inflammatory monocytes (Fig. 2D and E). Indeed, gene set enrichment analysis demonstrated that genes associated with an increased H3K27ac signal are enriched within the mRNAs upregulated at the site of inflammation, indicating that an increased H3K27ac signal correlates with increased gene expression in synovial monocytes (Fig. 2F). In agreement with this, genes associated with an increased H3K27ac signal in JIA are more abundantly expressed in synovial monocytes compared with HC monocytes (Fig. 2G and H). To further assess whether changes in H3K27ac contribute to JIA gene expression, the bromodomain and extra-terminal domain (BET) inhibitor JQ1 was used to inhibit enhancer-mediated transcription [24]. Treatment of synovial monocytes with JQ1 preferentially reduced the expression of genes that were upregulated in JIA monocytes, indicating that expression of these genes depends on H3K27ac regulation (Fig. 2I–K; Supplementary Fig. S2B and S2C, available at Rheumatology online). Altogether, these data indicate that disease-associated gene expression in monocytes derived from the local inflammatory site is mediated by increased H3K27 acetylation.
JAK inhibition alters the H3K27ac profile of synovial monocytes
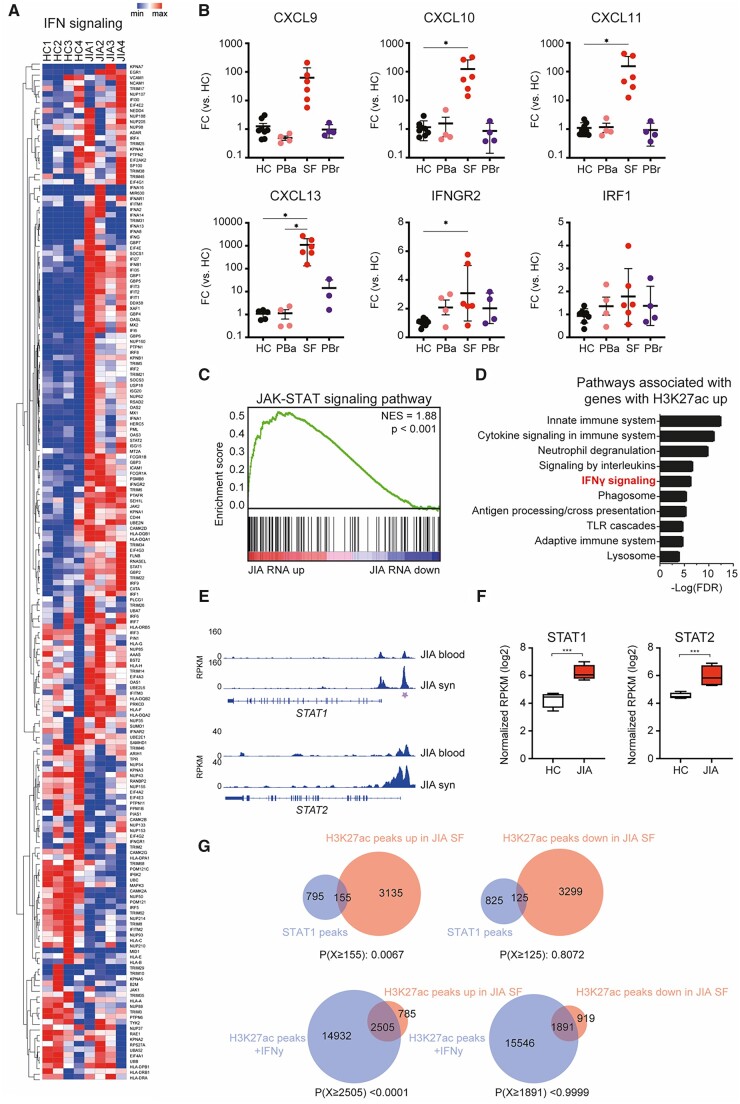
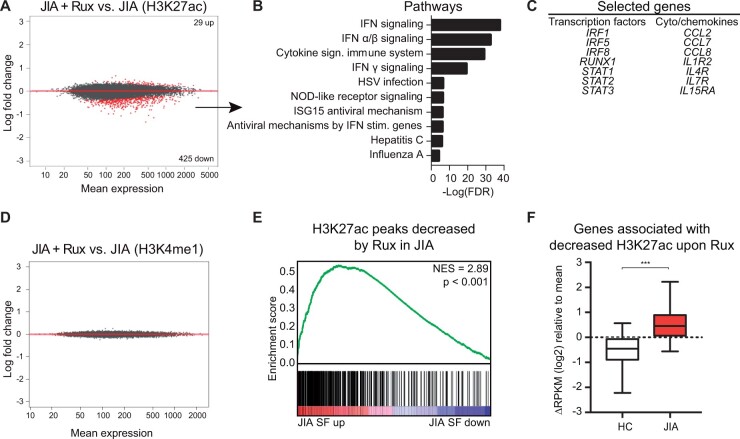
We observed that genes upregulated in inflammatory monocytes are associated with IFN signalling. Detailed analysis of IFN-signalling-associated genes (based on REACTOME IFN-signalling pathway) confirmed that a majority of these genes are indeed upregulated in inflammatory site-derived monocytes (Fig. 3A). RT-qPCR analysis validated the increased expression of several IFN-induced genes in JIA SF-derived CD14+ cells (Fig. 3B). The same trend was observed on the protein level and IFN-induced genes are also significantly increased in CD14+ monocytes derived from the SF of polyarticular JIA patients (Supplementary Fig. S3A and B, available at Rheumatology online). Pathway analysis of the genes associated with an increased H3K27ac signal in inflammatory monocytes indicated an association with IFNγ signalling (Fig. 3C). IFN signalling is mediated by the JAK and signal transducer and activator of transcription (STAT) pathway [25]. Gene set enrichment analysis demonstrated that genes belonging to the JAK-STAT signalling pathway are upregulated in synovial monocytes, suggesting that this pathway may regulate gene expression at the site of inflammation (Fig. 3D). Similarly, we observed increased expression of STAT1 and STAT2 and increased H3K27 acetylation of a STAT1 and STAT2-associated enhancer (Fig. 3E and F). To further define the role of the JAK-STAT signalling pathway, we assessed the H3K27ac and STAT1 ChIP-sequencing dataset of IFNγ-treated human primary monocytes generated by Qiao et al. (GSE43036 [26]). H3K27ac peaks increased in our inflammatory monocyte dataset were enriched for STAT1 binding and overlapped with H3K27ac signals mediated by IFNγ-treatment (Fig. 3G). To experimentally assess the involvement of IFN and JAK-STAT signalling in establishing the epigenetic profile within inflammatory monocytes, we treated JIA SF monocytes for 4 h with ruxolitinib, an inhibitor of JAK1 and JAK2, and performed ChIP-seq for H3K27ac and H3K4me1 [27]. While H3K27ac characterizes active enhancers, H3K4me1 is enriched at both active and inactive enhancer regions [9]. Ruxolitinib treatment induced ∼450 significant changes in the H3K27ac epigenome, of which the majority of changes comprised a reduced H3K27ac signal (Fig. 4A). Genes associated with a decrease in H3K27ac are involved in IFN signalling and included several components of the JAK-STAT signalling pathway, indicating an auto-regulatory feedback loop (Fig. 4B and C). Ruxolitinib did not alter the H3K4me1 profile of inflammatory monocytes (Fig. 4D). This is not surprising as histone methylation marks are less dynamically regulated than histone acetylation marks, which suggests that ruxolitinib impairs the activity of active enhancers [28]. Moreover, ruxolitinib treatment preferentially decreased the signal of H3K27ac peaks that were increased in inflammatory monocytes (Fig. 4E). Similarly, genes associated with a decreased H3K27ac signal upon ruxolitinib treatment were expressed to a higher extent in inflammation-derived monocytes compared with HC monocytes (Fig. 4F). Taken together, these data indicate that JAK inhibition preferentially reduces expression of genes associated with an increased H3K27ac signal in monocytes derived from the local inflammatory site.
Figure 3.
IFNγ-associated genes are increased in JIA SF monocytes. (A) Heatmap demonstrating the expression of IFNγ signalling-associated genes in HC PB and JIA SF monocytes. (B) Gene expression of selected IFNγ signalling-associated genes in monocytes derived from either the PB of HC, PB of JIA patients undergoing active disease (PBa), SF of JIA patients, and PB of JIA patients in remission (PBrem). (C) Top 10 pathways associated with genes associated with an increased H3K27ac signal. (D) Gene set enrichment of JAK-STAT signalling pathway-associated genes and genes differentially expressed within JIA SF monocytes. (E) Gene track for STAT1 and STAT2 displaying H3K27ac signals for JIA PB and JIA SF. Purple asterisk indicates H3K27ac regions that are significantly different. (F) Normalized RPKM (log2) values of STAT1 and STAT2 expression in HC and JIA monocytes. (G) Venn diagrams displaying the overlap of H3K27ac peaks upregulated or downregulated in JIA SF monocytes with STAT1 or H3K27ac peaks of IFNγ-treated monocytes. STAT1 and H3K27ac data was obtained from GSE43036 (GSM1057010 and GSM1057016, respectively). P-values for B were calculated using an ordinary one-way ANOVA. P-values for E were calculated using a hypergeometric test. *P < 0.05; **P < 0.01; ***P < 0.001. HC: healthy control; PB: peripheral blood; RPKM: reads per kilobase of exon per million reads mapped
Figure 4.
Ruxolitinib alters JIA SF monocytes at the epigenetic level. (A) MA plot of H3K27ac regions different within JIA SF monocytes after ruxolitinib treatment. Red dots indicate genes with FDR < 0.1. (B) Top 10 pathways associated with genes significantly decreased by ruxolitinib. (C) Selected genes significantly decreased by ruxolitinib. (D) MA plot of H3K4me1 regions different within JIA SF monocytes after ruxolitinib treatment. Red dots indicate genes with an FDR < 0.1. (E) Gene set enrichment analysis of H3K27ac peaks decreased by ruxolitinib in JIA and H3K27ac peaks different in JIA SF. (F) Boxplot with 5%–95% whiskers displaying △RPKM (log2) values of genes associated with a decreased H3K27ac signal in JIA monocytes after ruxolitinib treatment. P-value for E was calculated using a Wilcoxon-matched pairs signed rank test. *P < 0.05; **P < 0.01; ***P < 0.001. JIA: juvenile idiopathic arthritis; RPKM: reads per kilobase of exon per million reads mapped; SF: synovial fluid
Ruxolitinib inhibits the activated phenotype of JIA SF monocytes
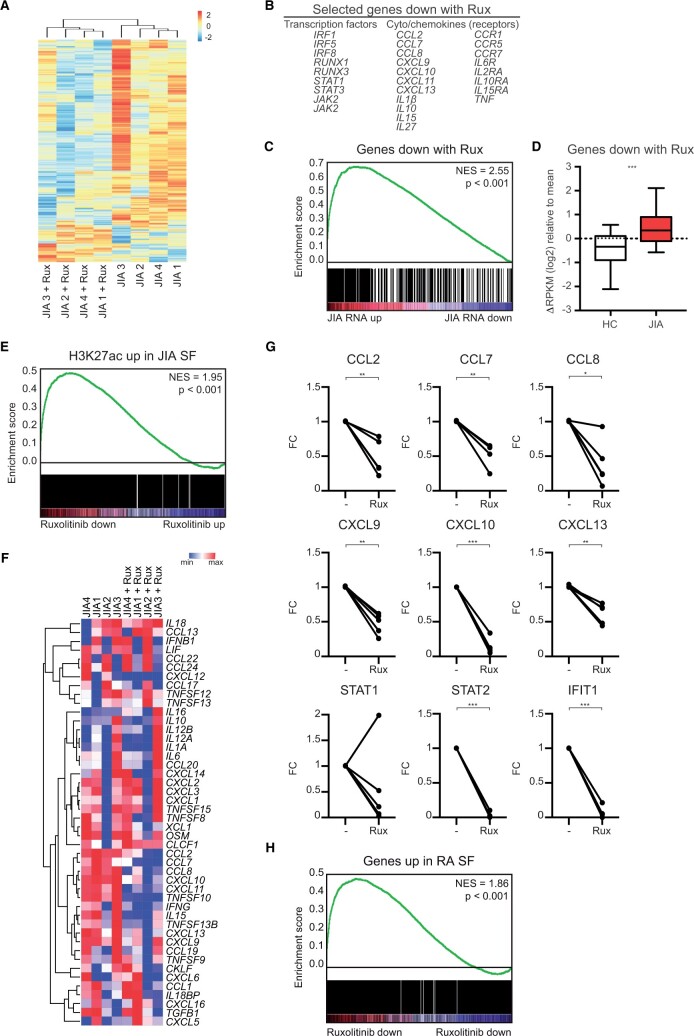
Alteration of the epigenetic landscape by ruxolitinib affected the activated phenotype of synovial monocytes, with the most predominant effect on IFN and cytokine signalling (Fig. 5A and B; Supplementary Fig. S4A–C, available at Rheumatology online). Genes decreased by ruxolitinib are enriched within the genes that are upregulated in JIA, indicating that ruxolitinib leads to reduced expression of disease-associated genes (Fig. 5C and D). Furthermore, genes associated with an increased H3K27ac signal in inflammatory monocytes were preferentially decreased by ruxolitinib, indicating that ruxolitinib is targeting H3K27ac-regulated genes (Fig. 5E). We also analysed the effect of ruxolitinib on JIA SF monocytes that were treated in vitro with IFNγ. IFNγ treatment itself did not have a strong impact on the transcriptome of JIA SF monocytes (Supplementary Fig. S4D and E, available at Rheumatology online). We speculate that this is because the epigenetic and transcriptional changes induced by IFNγ have already taken place within the synovial compartment and/or that the cultured monocytes secrete IFNγ which can activate themselves, and that adding exogenous IFNγ consequently does not have much of an impact anymore. Therefore, the effect of ruxolitinib on IFNγ-treated monocytes is also similar to the effect of ruxolitinib on untreated monocytes (Supplementary Fig. S4F and S4G, available at Rheumatology online). Expression of many cytokines and chemokines was increased in synovial monocytes compared with HC monocytes. Ruxolitinib inhibits the expression of many of these cytokines, as demonstrated by RNA-sequencing and qPCR validation experiments (Fig. 5F and G). To assess whether these observations could be extended to other local inflammatory site-derived monocytes, we assessed monocytes derived from the SF of RA patients. This revealed that RA-associated genes are also preferentially decreased by ruxolitinib (Fig. 5H), indicating that our observations are not specific for JIA but represent local autoinflammation as a whole. Taken together, these data demonstrate that ruxolitinib-mediated inhibition of enhancer activity results in repression of the expression of genes associated with (auto-)inflammation in monocytes.
Figure 5:
Ruxolitinib decreases the activated phenotype of JIA SF monocytes. (A) Supervised clustering analysis based on differences in gene expression in JIA SF monocytes after treatment with ruxolitinib. (B) Selected genes downregulated by ruxolitinib. (C) Gene set enrichment analysis of genes downregulated by ruxolitinib, and genes differentially expressed in JIA SF monocytes vs HC monocytes. (D) Boxplot with 5%–95% whiskers displaying △RPKM (log2) values of genes decreased by ruxolitinib in signal in HC and JIA monocytes. (E) Gene set enrichment analysis of genes associated with an increased H3K27ac signal in JIA SF monocytes and genes affected by ruxolitinib treatment in JIA SF monocytes. (F) Heatmap demonstrating the expression of selected cytokines and chemokines in JIA SF monocytes with and without ruxolitinib treatment, PB and JIA SF monocytes. (G) Gene expression of selected genes in JIA SF monocytes affected by ruxolitinib treatment. P-values for D and G were calculated using a Wilcoxon-matched pairs signed rank test. (H) Gene set enrichment analysis of genes increased in RA SF monocytes and genes affected by ruxolitinib treatment in JIA SF monocytes. *P < 0.05; **P < 0.01; ***P < 0.001. HC: healthy control; PB: peripheral blood; RA: rheumatoid arthritis
Discussion
Here, we assessed the transcriptional and H3K27ac profile of synovial monocytes isolated from JIA patients. We observed that these inflammatory monocytes display an activated phenotype, illustrated by increased gene expression of surface activation markers and inflammatory cytokines and chemokines. This transcriptional phenotype was mirrored in the H3K27ac landscape, as expression of genes that are associated with an increased H3K27ac signal was increased. This suggests that epigenetic regulation contributes to disease-associated gene expression and thus disease pathogenesis.
For several inflammatory rheumatic diseases, such as RA and SLE, an increased frequency in intermediate monocytes (CD14+CD16+) has been described [29, 30]. Furthermore, intermediate monocytes have been described to be increased in the PB and SF of enthesitis‐related arthritis, a specific subset of JIA [31]. The JIA patients used in this study do not have elevated levels of CD14+CD16+ in the PB, but the frequency of intermediate monocytes in the SF is significantly increased compared with HC and JIA PB, which is in agreement with previous reports [32]. Therefore, we sorted CD14+CD16+ from JIA SF as well as HC PB and compared those for our transcriptional profiling. Unfortunately, this was not possible for the ChIP-sequencing experiments as there were insufficient numbers of intermediate monocytes from the PB of HC and JIA patients and therefore isolated CD14+ cells were used. Some of the epigenetic differences observed between inflammation-derived monocytes and PB monocytes might therefore be the result of a different ratio of classical (CD14+CD16-) monocytes vs intermediate monocytes. However, because we did observe a strong correlation between the H3K27ac profile and gene expression of inflammatory monocytes, we assume that these differences do not drastically affect the epigenetic profile on a global level.
Our data demonstrated that JIA synovial monocytes are epigenetically distinct, regarding their H3K27ac status, from JIA PB and HC monocytes, while the latter are relatively similar. This raises the question whether the epigenetic alterations in inflammatory monocytes are cell-intrinsic or a reflection of the highly inflammatory synovial environment. IFNγ is one of the pro-inflammatory mediators present within the SF of oligoarticular, polyarticular as well as systemic JIA patients [33]. Natural type I IFN producing cells have been detected in the synovial tissue of JIA patients, although IFNα levels in SF are reported to be low [34]. There are some indications that IFNγ might play an important role within JIA pathogenesis. For example, increased responsiveness of PB-derived classical monocytes as well as naive CD4+ T cells towards IFNγ stimulation, as indicated by increased phospho-STAT3 and pospho-STAT1 levels, respectively, has been described for polyarticular JIA [35]. In addition, gene expression analysis of SFMCs of extended oligoarticular JIA patients revealed increased IFNγ levels compared with persistent oligoarticular JIA SFMCs, which corresponded with a trend of more IFNγ in the SF of these patients [36]. Furthermore, IFNγ has been described to be produced by both CD4+ and CD8+ SF T cells and contributes to the resistance of SF CD8+ T cells to suppression [37]. Our data indicated that transcriptional and epigenetic changes in inflammation-derived monocytes are associated with the IFN-signalling pathway. Together with the lack of H3K27ac differences between JIA PB and HC PB-derived monocytes, this suggests that monocytes are epigenetically altered at the site of inflammation due to cell-extrinsic factors, leading to altered gene expression which results in increased activation and cytokine production and thereby creating an auto-inflammatory loop. This is in agreement with our in vitro experiments using ruxolitinib, were we cultured synovial monocytes in the presence and absence of exogenous IFNγ and noticed that the effect of ruxolitinib on the transcriptome was very similar.
An epigenetic role for IFNγ and type I IFNs has been described in macrophages. IFNγ primes existing regulatory elements by increasing histone acetylation and chromatin remodeling, which leads to increased responsiveness of macrophages to pro-inflammatory stimuli, such as LPS or type I interferons [26, 38]. This increased responsiveness is illustrated by increased transcription of TNF and IL6, pro-inflammatory cytokines which we found to be highly expressed by inflammatory monocytes. IFNα has a similar role and primes chromatin to enable robust transcriptional responses to weak signals [39]. For example, IFNα enhances TNF inflammatory function by preventing the silencing of inflammatory genes. IFNγ can also induce de novo or latent enhancer formation by inducing transcription factors, such as STATs and interferon regulatory factors (IRFs), that cooperate with lineage-determining transcription factors to open chromatin and deposit enhancer-associated histone marks in mouse macrophages [40]. We observed that inhibiting IFN signalling in JIA synovial monocytes using the JAK inhibitor ruxolitinib profoundly affected the H3K27ac landscape, while the H3K4me1 profile was unaffected. This indicates that in our experimental setting, ruxolitinib predominantly affects existing regulatory elements by reducing H3K27ac deposition. This is in line with observations in T cells, where STATs have been demonstrated to have a major effect on enhancers bound by the histone acetyltransferase p300, while the impact of STATs on H3K4me1-positive enhancers was minimal [41]. However, JAK inhibition prior to IFNγ stimulation has been demonstrated to affect H3K27ac as well as H3K4me1 at stimuli-specific enhancers [40]. This suggests that, in patients, next to altering the established epigenetic landscape of monocytes at the site of inflammation, JAK inhibition might also affect formation of de novo enhancers. JAK inhibitors are currently being used for the treatment of RA, ulcerative colitis and psoriasis and recently the JAK inhibitor tofacitinib has been approved for the treatment or patients with active polyarticular course JIA [42, 43]. The latter is based on a phase 3 study evaluating the efficacy and safety of tofacitinib which demonstrated that the occurrence of disease flares was significantly decreased in tofacitinib-treated patients compared with placebo-treated patients [44–46]. Our data indicate that the therapeutic effect of JAK inhibition in JIA is, at least partly, established by altering the epigenetic landscape of inflammatory monocytes, thereby reducing the expression of disease-associated genes. Gene set enrichment analysis demonstrated that genes upregulated in RA synovial-derived monocytes are enriched within the genes downregulated by ruxolitinib in JIA, indicating that these genes in RA might also be JAK-dependent and regulated by H3K27ac [47]. It will be interesting to study if JAK inhibitors have a similar effect in other autoimmune diseases as well. Furthermore, the development of next-generation JAK-inhibitors that selectively inhibit JAK2 will allow for more precise control of disease-associated gene expression, as JAK2 is upregulated in inflammatory monocytes [43]. Altogether our data provide further rationale for the treatment of inflammatory arthritis patients with JAK inhibitors and indicate that other molecules that affect the epigenetic landscape of inflammatory monocytes, or other immune cells, can have a therapeutic effect as well.
Supplementary Material
Acknowledgements
We thank the Luminex core facility in the University Medical Center Utrecht, the Netherlands, for determining cytokine concentrations.
Contributor Information
Janneke G C Peeters, Center for Translational Immunology, University Medical Center Utrecht, Utrecht University, Utrecht, The Netherlands.
Arjan Boltjes, Center for Translational Immunology, University Medical Center Utrecht, Utrecht University, Utrecht, The Netherlands.
Rianne C Scholman, Center for Translational Immunology, University Medical Center Utrecht, Utrecht University, Utrecht, The Netherlands.
Stephin J Vervoort, Center for Molecular Medicine, University Medical Center Utrecht, Utrecht University, Utrecht, The Netherlands.
Paul J Coffer, Center for Molecular Medicine, University Medical Center Utrecht, Utrecht University, Utrecht, The Netherlands.
Michal Mokry, Laboratory of Experimental Cardiology, UMC Utrecht, Utrecht University, Utrecht, The Netherlands; Department of Pediatric Gastroenterology, Wilhelmina Children's Hospital, University Medical Center Utrecht, Utrecht University, Utrecht, The Netherlands.
Sebastiaan J Vastert, Center for Translational Immunology, University Medical Center Utrecht, Utrecht University, Utrecht, The Netherlands; Department of Pediatric Rheumatology and Immunology, Division of Pediatrics, Wilhelmina Children’s Hospital, University Medical Center Utrecht, Utrecht University Utrecht, The Netherlands.
Femke van Wijk, Center for Translational Immunology, University Medical Center Utrecht, Utrecht University, Utrecht, The Netherlands.
Jorg van Loosdregt, Center for Translational Immunology, University Medical Center Utrecht, Utrecht University, Utrecht, The Netherlands.
Supplementary material
Supplementary material is available at Rheumatology online.
Data availability
The sequencing data have been deposited online. Not all raw RNA-sequencing and ChIP-sequencing data from the HC and JIA patients used in this study are publicly available due to research participant privacy/consent. Gene expression data of ex vivo HC and JIA monocytes have been deposited in the Gene Expression Omnibus database (GSE220264). Data from RNA-seq and ChIP-seq experiments with Ruxolitinib- and IFNγ-stimulated JIA monocytes, RNA-seq experiments with JQ1-treated JIA monocytes and ChIP-seq experiments with ex-vivo HC and JIA monocytes are accessible at 10.5281/zenodo.7487852. All other data are available in the manuscript and its supplementary materials available at Rheumatology online.
Contribution statement
Conceptualization: J.G.C.P., F.vW., J.vL. Performed experiments: J.G.C.P., A.B., R.C.C., S.J.Ve. Data analysis: J.G.C.P., A.B., S.J.Ve., M.M. Writing—original draft: J.G.C.P. Writing—review and editing: J.G.C.P., A.B., P.J.C., S.J.Va., F.vW., J.vL. Supervision: J.vL.
Funding
This project was supported by the Netherlands Organisation for Scientific Research.
Disclosure statement: The authors have declared no conflict of interests.
References
- 1. Ishikawa O, Ishikawa H. Macrophage infiltration in the skin of patients with systemic sclerosis. J Rheumatol 1992;19:1202–6. [PubMed] [Google Scholar]
- 2. Carlsen H, Baekkevold E, Morton H, Haraldsen G, Brandtzaeg P. Monocyte-like and mature macrophages produce CXCL13 (B cell-attracting chemokine 1) in inflammatory lesions with lymphoid neogenesis. Blood 2004;104:3021–7. [DOI] [PubMed] [Google Scholar]
- 3. Misharin AV, Cuda CM, Saber R et al. Nonclassical Ly6C(-) monocytes drive the development of inflammatory arthritis in mice. Cell Rep 2014;9:591–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Magnusson M, Brynjólfsson S, Dige A et al. Macrophage and dendritic cell subsets in IBD: ALDH+ cells are reduced in colon tissue of patients with ulcerative colitis regardless of inflammation. Mucosal Immunol 2016;9:171–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sack U, Stiehl P, Geiler G. Distribution of macrophages in rheumatoid synovial membrane and its association with basic activity. Rheumatol Int 1994;13:181–6. [DOI] [PubMed] [Google Scholar]
- 6. Cho JH, Gregersen PK. Genomics and the multifactorial nature of human autoimmune disease. N Engl J Med 2011;365:1612–23. [DOI] [PubMed] [Google Scholar]
- 7. Alberts B, Johnson A, Lewis J et al. Molecular biology of the cell. 4th edn. New York: Garl. Sci., 2002. [Google Scholar]
- 8. Heintzman ND, Stuart RK, Hon G et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 2007;39:311–8. [DOI] [PubMed] [Google Scholar]
- 9. Creyghton MP, Cheng AW, Welstead GG et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA 2010;107:21931–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Farh KK-H, Marson A, Zhu J et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 2015;518:337–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ernst J, Kheradpour P, Mikkelsen TS et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 2011;473:43–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peeters JGC, Vervoort SJ, Tan SC et al. Inhibition of super-enhancer activity in autoinflammatory site-derived T cells reduces disease-associated gene expression. Cell Rep 2015;12:1986–96. [DOI] [PubMed] [Google Scholar]
- 13. Parker SCJ, Stitzel ML, Taylor DL et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc Natl Acad Sci USA 2013;110:17921–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vahedi G, Kanno Y, Furumoto Y et al. Super-enhancers delineate disease-associated regulatory nodes in T cells. Nature 2015;520:558–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Seumois G, Chavez L, Gerasimova A et al. Epigenomic analysis of primary human T cells reveals enhancers associated with TH2 memory cell differentiation and asthma susceptibility. Nat Immunol 2014;15:777–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hnisz D, Abraham BJ, Lee TI et al. Super-enhancers in the control of cell identity and disease. Cell 2013;155:934–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ballestar E, Li T. New insights into the epigenetics of inflammatory rheumatic diseases. Nat Rev Rheumatol 2017;13:593–605. [DOI] [PubMed] [Google Scholar]
- 18. Van Der Kroef M, Castellucci M, Mokry M et al. Histone modifications underlie monocyte dysregulation in patients with systemic sclerosis, underlining the treatment potential of epigenetic targeting. Ann Rheum Dis 2019;78:529–38. [DOI] [PubMed] [Google Scholar]
- 19. Shi L, Zhang Z, Song L et al. Monocyte enhancers are highly altered in systemic lupus erythematosus. Epigenomics 2015;7:921–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mijnheer G, Lutter L, Mokry M et al. Conserved human effector Treg cell transcriptomic and epigenetic signature in arthritic joint inflammation. Nat Commun 2021;12:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Petty RE, Southwood TR, Manners P et al. International League of Associations for Rheumatology classification of juvenile idiopathic arthritis: second revision. J Rheumatol 2004;31:390–2. [PubMed] [Google Scholar]
- 22. Peeters JGC, Vervoort SJ, Tan SC et al. Inhibition of super-enhancer activity in autoinflammatory site-derived T cells reduces disease-associated gene expression. Cell Rep 2015;12:1986–96. [DOI] [PubMed] [Google Scholar]
- 23. de Jager W, Prakken BJ, Bijlsma JWJ, Kuis W, Rijkers GT. Improved multiplex immunoassay performance in human plasma and synovial fluid following removal of interfering heterophilic antibodies. J Immunol Methods 2005;300:124–35. [DOI] [PubMed] [Google Scholar]
- 24. Filippakopoulos P, Qi J, Picaud S et al. Selective inhibition of BET bromodomains. Nature 2010;468:1067–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stark GR, Darnell JE. The JAK-STAT pathway at twenty. Immunity 2012;36:503–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Qiao Y, Giannopoulou EG, Chan CH et al. Synergistic activation of inflammatory cytokine genes by interferon-γ-induced chromatin remodeling and toll-like receptor signaling. Immunity 2013;39:454–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lin Q, Meloni D, Pan Y et al. Enantioselective synthesis of Janus Kinase inhibitor INCB018424 via an organocatalytic aza-michael reaction. Org Lett 2009;11:1999–2002. [DOI] [PubMed] [Google Scholar]
- 28. Mews P, Zee BM, Liu S et al. Histone methylation has dynamics distinct from those of histone acetylation in cell cycle reentry from quiescence. Mol Cell Biol 2014;34:3968–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kawanaka N, Yamamura M, Aita T et al. CD14+,CD16+ blood monocytes and joint inflammation in rheumatoid arthritis. Arthritis Rheum 2002;46:2578–86. [DOI] [PubMed] [Google Scholar]
- 30. Henriques A, Inês L, Carvalheiro T et al. Functional characterization of peripheral blood dendritic cells and monocytes in systemic lupus erythematosus. Rheumatol Int 2012;32:863–9. [DOI] [PubMed] [Google Scholar]
- 31. Gaur P, Myles A, Misra R, Aggarwal A. Intermediate monocytes are increased in enthesitis-related arthritis, a category of juvenile idiopathic arthritis. Clin Exp Immunol 2017;187:234–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schmidt T, Berthold E, Arve-Butler S et al. Children with oligoarticular juvenile idiopathic arthritis have skewed synovial monocyte polarization pattern with functional impairment – a distinct inflammatory pattern for oligoarticular juvenile arthritis. Arthritis Res Ther 2020;22:186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. de Jager W, Hoppenreijs EPAH, Wulffraat NM et al. Blood and synovial fluid cytokine signatures in patients with juvenile idiopathic arthritis: a cross-sectional study. Ann Rheum Dis 2007;66:589–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gattorno M, Chicha L, Gregorio A et al. Distinct expression pattern of IFN-α and TNF-α in juvenile idiopathic arthritis synovial tissue. Rheumatology 2007;46:657–65. [DOI] [PubMed] [Google Scholar]
- 35. Throm AA, Moncrieffe H, Orandi AB et al. Identification of enhanced IFN-γ signaling in polyarticular juvenile idiopathic arthritis with mass cytometry. JCI Insight 2018;3:e121544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hunter PJ, Nistala K, Jina N et al. Biologic predictors of extension of oligoarticular juvenile idiopathic arthritis as determined from synovial fluid cellular composition and gene expression. Arthritis Rheum 2010;62:896–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Petrelli A, Wehrens E, Scholman R et al. Self-sustained resistance to suppression of CD8+ teff cells at the site of autoimmune inflammation can be reversed by tumor necrosis factor and interferon-γ blockade. Arthritis Rheumatol 2016;68:229–36. [DOI] [PubMed] [Google Scholar]
- 38. Ivashkiv LB. IFNγ: signalling, epigenetics and roles in immunity, metabolism, disease and cancer immunotherapy. Nat Rev Immunol 2018;18:545–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Park SH, Kang K, Giannopoulou E et al. Type I IFNs and TNF cooperatively reprogram the macrophage epigenometo promote inflammatory activation. Nat Immunol 2017;18:1104–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ostuni R, Piccolo V, Barozzi I et al. Latent enhancers activated by stimulation in differentiated cells. Cell 2013;152:157–71. [DOI] [PubMed] [Google Scholar]
- 41. Vahedi G, Takahashi H, Nakayamada S et al. STATs shape the active enhancer landscape of T cell populations. Cell 2012;151:981–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drugs.com. Xeljanz (tofacitinib) FDA Approval History - Drugs.com. https://www.drugs.com/history/xeljanz.html (20 April 2020, date last accessed).
- 43. Schwartz DM, Kanno Y, Villarino A et al. JAK inhibition as a therapeutic strategy for immune and inflammatory diseases. Nat Rev Drug Discov 2017;16:843–62. [DOI] [PubMed] [Google Scholar]
- 44. Ruperto N, Brunner HI, Zuber Z et al. Pharmacokinetic and safety profile of tofacitinib in children with polyarticular course juvenile idiopathic arthritis: results of a phase 1, open-label, multicenter study. Pediatr Rheumatol 2017;15:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ruperto N, Synoverska O, Ting T et al. OP0291 tofacitinib for the treatment of polyarticular course juvenile idiopathic arthritis: results of a phase 3, randomised, double-blind, placebo-controlled withdrawal study. Ann Rheum Dis 2020;79:180.2–1. [Google Scholar]
- 46. Ruperto N, Brunner H, Synoverska O et al. Tofacitinib in juvenile idiopathic arthritis: a double-blind, placebo-controlled, withdrawal phase 3 randomised trial. Lancet 2021;398:1984–96. [DOI] [PubMed] [Google Scholar]
- 47. Rajasekhar M, Olsson AM, Steel KJA et al. MicroRNA-155 contributes to enhanced resistance to apoptosis in monocytes from patients with rheumatoid arthritis. J Autoimmun 2017;79:53–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data have been deposited online. Not all raw RNA-sequencing and ChIP-sequencing data from the HC and JIA patients used in this study are publicly available due to research participant privacy/consent. Gene expression data of ex vivo HC and JIA monocytes have been deposited in the Gene Expression Omnibus database (GSE220264). Data from RNA-seq and ChIP-seq experiments with Ruxolitinib- and IFNγ-stimulated JIA monocytes, RNA-seq experiments with JQ1-treated JIA monocytes and ChIP-seq experiments with ex-vivo HC and JIA monocytes are accessible at 10.5281/zenodo.7487852. All other data are available in the manuscript and its supplementary materials available at Rheumatology online.