Abstract
Root nodules are major sources of nitrogen for soybean (Glycine max (L.) Merr.) growth, development, production, and seed quality. Symbiotic nitrogen fixation is time-limited, as the root nodule senesces during the reproductive stage of plant development, specifically during seed development. Nodule senescence is characterized by the induction of senescence-related genes, such as papain-like cysteine proteases (CYPs), which ultimately leads to the degradation of both bacteroids and plant cells. However, how nodule senescence–related genes are activated in soybean is unknown. Here, we identified 2 paralogous NAC transcription factors, GmNAC039 and GmNAC018, as master regulators of nodule senescence. Overexpression of either gene induced soybean nodule senescence with increased cell death as detected using a TUNEL assay, whereas their knockout delayed senescence and increased nitrogenase activity. Transcriptome analysis and nCUT&Tag-qPCR assays revealed that GmNAC039 directly binds to the core motif CAC(A)A and activates the expression of 4 GmCYP genes (GmCYP35, GmCYP37, GmCYP39, and GmCYP45). Similar to GmNAC039 and GmNAC018, overexpression or knockout of GmCYP genes in nodules resulted in precocious or delayed senescence, respectively. These data provide essential insights into the regulatory mechanisms of nodule senescence, in which GmNAC039 and GmNAC018 directly activate the expression of GmCYP genes to promote nodule senescence.
The senescence-induced NAC transcription factors GmNAC039 and GmNAC018 directly target and activate multiple cysteine protease genes to promote nodule senescence in soybean.
IN A NUTSHELL.
Background: Soybean (Glycine max) roots have nodules in which symbiotic bacteria reduce nitrogen gas into ammonium, which nourishes the plants. The termination of symbiotic nitrogen fixation, termed nodule senescence, is an active process and soybean nodules are usually severely senescent when the plant begins flowering, which appears counterproductive, as seed maturation requires a large amount of nitrogen. While much has been discovered about symbiotic signaling and nodule organogenesis over the past 2 decades, knowledge about nodule senescence is largely limited to decreased nitrogenase activity and leghemoglobin content, physiological changes, and gene expression profiles of senescent nodules. One molecular marker of senescent nodules is an increase in the transcript levels of genes encoding cysteine proteases that break down proteins and degrade nodule cells. However, how nodule senescence is activated at the molecular level and what components activate the expression of senescence-related genes are largely unknown.
Question: What are the key components regulating nodule senescence in soybean?
Findings: We identified 2 NAC transcription factors from soybean, GmNAC039 and its paralog GmNAC018, as regulators of nodule senescence. GmNAC039 directly binds to the promoters of at least 4 cysteine protease genes and activates their expression. Knocking out these 4 cysteine protease genes in soybean delayed nodule senescence and increased nitrogenase activity. Thus, GmNAC039 and GmNAC018 activate the expression of cysteine protease genes to promote nodule senescence in soybean.
Next steps: Nodule senescence is associated with extensive cell degradation, which leads to programed cell death in nodules. The expression of genes encoding proteins with proteolytic activities is required for nodule senescence. Given that cysteine proteases have key roles in soybean nodule senescence, identifying their target proteins is an important future research goal.
Introduction
Legume–rhizobia symbiosis leads to the development of nitrogen-fixing nodules, in which atmospheric N2 is reduced to ammonia and assimilated by the host plant. This process, known as symbiotic nitrogen fixation, could potentially be manipulated through biotechnology to achieve sustainable agriculture (Oldroyd and Dixon 2014). Soybean ( Glycine max (L.) Merr.) is a nutritious leguminous crop that is rich in protein and oil and used as a source of food and feed worldwide (Mus et al. 2016).
The life cycle of a nodule is much shorter than that of a soybean plant. Soybean nodules begin to have senescent cells about 4-wk post-inoculation. This process starts in the central cells of the nodule and gradually spreads outward, inducing complete degradation of the bacteroids and nodule necrosis (Puppo et al. 2005). Nodule senescence is accelerated when plants are subjected to biotic or abiotic stresses, including pathogen attack, darkness-induced starvation, high temperature, water deficit, drought, and nitrogen fertilizers, dramatically decreasing the symbiotic nitrogen-fixation efficiency.
Moreover, inadequate nitrogen supply is a major constraint for legume growth and yield under sterile soil conditions (Imsande 1986; Imsande and Schmidt 1998). For most leguminous plants, significant nodule senescence is observed during pod filling, when nutrient reallocation and remobilization occur to ensure yield and seed quality. Indeed, nitrogen fertilizers are typically applied during pod filling (reproductive stage R5) to improve soybean yield and seed protein content (Imsande 1986). Therefore, increasing active nitrogen fixation in nodules by delaying their senescence might be a powerful strategy to improve nitrogen-use efficiency during seed development.
Organ senescence is an active process that involves extensive cell degradation, resulting in cell death and organ abscission. Leaf senescence is a well-studied process, in which leaf color changes from green to yellow due to the degeneration of chloroplasts (Woo et al. 2019). Similarly, during nodule senescence, the symbiosomes (specialized organelles that contain the bacteroids) and the heme groups of leghemoglobin (a pigment involved in O2 delivery to the bacteroids) are degraded, causing a visible color change of nodules from pink to green or even light brown (Perez Guerra et al. 2010). A hallmark of nodule senescence is dramatically increased proteolysis, associated with activated transcription of protease genes, for degradation of the bacteria and plant cells in the nodules (i.e. the catabolism of proteins and other cellular components) (Puppo et al. 2005). Compared with the well-documented mechanisms governing leaf senescence (Diaz et al. 2008), the current understanding of nodule senescence is largely limited to the physiological changes and transcriptional reprograming that occur in nodules.
Research on nodule senescence dates back to at least the 1970s, when a study examined the degradation of nodule leghemoglobin in different legumes (Lehtovaara and Perttila 1978). As the heme groups of the oxygen-carrier protein leghemoglobin are degraded to biliverdin and bilirubin, the nodule color changes from red to green, one of the typical indicators of nodule senescence (Kazmierczak et al. 2020). The degradation of leghemoglobin means that the bacteroids are exposed to millimolar levels of free oxygen, leading to a sharp decline in nitrogenase activity. Hence, this redox change is also viewed as a typical feature of nodule senescence (Puppo et al. 2005).
Another well-studied feature of nodule senescence is seen in indeterminate nodules. Medicago ( Medicago truncatula ) indeterminate nodules have a visible senescence zone located at root proximal which is distinct from persistent apical meristem zone required for nodule development (Ferguson et al. 2010). The different locations of the senescence zone and meristem zone make it possible to identify specific cells involved in nodule senescence (Roy et al. 2020). However, this does not seem to be the case in determinant nodules. Due to the lack of active meristem cells in spherical nodule structure, the senescence in determinant nodules starts at the center and spreads outward leading to the necrosis of whole nodules (Ferguson et al. 2010). Thus, regulation mechanisms of senescence in determinate nodules might be different from that in indeterminate nodules.
Organ senescence could be activated prematurely by either environmental stresses or gene mutations or induced by developmental arrest. Senescence and developmental arrest are closely related. However, senescence could also be viewed as the last phase of the whole development and induce an irreversible arrest of growth (Woo et al. 2019). The continuation and aggravation of senescence will eventually induce a complete arrest of development leading to the necrosis and abscission of an organ (Woo et al. 2019). A typical example came from the study on M. truncatula nodules . Mutation at DNF2 (defective in nitrogen fixation), NAD1 (nodules with activated defense), or SymCRK (symbiotic cysteine-rich receptor-like kinase) could activate strong immune responses in nodules which finally induce nodule senescence and necrosis (Bourcy et al. 2013; Wang et al. 2016a; Domonkos et al. 2017 ; Berrabah et al. 2023). In either dnf2 , nad1 , or symCRK mutant plants, nodule senescence was only observed at the nitrogen-fixation zone in young nodules 7-d post inoculation (dpi) when senescence zone was not developed, but gradually expanded to full nodules at 4-wk post inoculation (wpi). Based on gene expression analysis in Medicago nad1 mutant nodules, expression of immune-related genes was significantly induced in young nodules, while senescence-related genes were induced in the mature nodules (Wang et al. 2016a). The results suggested that the activated immune responses might be the direct reason to promote senescence in nad1 mutant nodules, which finally leads to developmental arrest with small-size nodules developed in nad1 or dnf2 mutant plants.
The host plant seems to have a predominant role in regulating nodule senescence. Several studies have used transcriptome analysis to identify senescence-related plant genes in aged nodules (Maunoury et al. 2010; Chungopast et al. 2014; Wang et al. 2016a; Wang et al. 2019; Liu et al. 2021). Increased expression of a small family of genes encoding papain-like cysteine proteases (CYPs) and downregulation of genes encoding leghemoglobins have been used as molecular markers of nodule senescence (Kazmierczak et al. 2020; Yang et al. 2020). In soybean, cysteine protease (CYP) genes were identified to be highly induced in aged nodules (Alesandrini et al. 2003b; Van Wyk et al. 2014). However, it is unclear which GmCYP genes play key roles in regulating nodule senescence and how GmCYP transcription is activated. Based on the expression pattern of CYPs and their protease activities, it has been proposed that the function of CYPs is to degrade proteins or other key components required for nodule development (Sheokand and Brewin 2003; Pierre et al. 2014). However, the true function of CYPs in nodule senescence is unknown.
Because senescence could be viewed as a termination of plant development, the transcriptional switch from genes involved in growth to those involved in senescence must be tightly regulated in response to developmental signals and/or environmental stresses. Several transcription factor (TF) families have been identified as important regulators of leaf senescence–responsive gene expression, including NAC (NAM, ATAF1, and CUC2) (Balazadeh et al. 2010, 2011; Matallana-Ramirez et al. 2013; Fraga et al. 2021), MYB (Jaradat et al. 2013; Piao et al. 2019), AP2 (Phukan et al. 2017; Chen et al. 2022), and WRKY (Zhang et al. 2016; Yu et al. 2021).
NAC TFs are plant-specific and constitute one of the largest gene families in plants. NAC proteins were divided into different subgroups based on their C-terminal domain. Some NAC TFs function as transcriptional activators, while others possess an NAC repression domain (NARD) and function as transcriptional repressors (Hao et al. 2010). A great number of NAC TFs have been shown to play fundamental roles in organ senescence; for example, about one-third of the NAC TFs in Arabidopsis (Arabidopsis thaliana) are involved in plant senescence (Li et al. 2018). The Arabidopsis NAC TFs ANAC092, ANAC029, ANAC059, and ANAC016 promote leaf senescence, while ANAC082 and JUB1 (JUNGBRUNNEN1) are repressors of leaf senescence (Wu et al. 2012; Woo et al. 2019).
In soybean, an extensive genome-wide analysis identified more than 180 genes encoding NAC TFs (Melo et al. 2018). Soybean salt-induced NAC1 (GmSIN1) mediates the crosstalk between ABA (abscisic acid) signaling pathway and ROS (reactive oxygen species) production, to increase salt tolerance (Li et al. 2019). GmNAC06 is another NAC TF that promotes salt tolerance (Li et al. 2021). GmNAC081 and GmNAC030 form heterodimers to activate cell death program via upregulation of caspase1-like vacuolar processing enzyme ( VPE ) gene expression (Mendes et al. 2013). GmNAC020 promotes abiotic stress tolerance and lateral root formation in transgenic Arabidopsis plants (Hao et al. 2011). Soybean SHATTERING1-5 ( SHAT1-5 ) encodes a NAC TF that has been artificially selected for pod-shattering resistance during soybean domestication (Dong et al. 2014). Despite all these data, very few studies have focused on the involvement of NAC TFs in soybean developmental senescence.
Nodule senescence involves programed cell death, reactive oxygen species production, and increased expression of CYPs. However, the key factors that orchestrate nodule senescence have not been identified (Roy et al. 2020; Wang et al. 2022). In this study, we identified 2 NAC TFs, GmNAC039, and its paralog GmNAC018, as key regulators of nodule senescence through directly activating the expression of 4 GmCYP genes in soybean nodules. Knocking out these 4 genes delayed nodule senescence and increased the nitrogenase activity in soybean nodules. Our results demonstrate that the activation of CYP gene expression by 2 NAC transcription factors represents an important regulatory module mediating soybean nodule senescence.
Results
Nodule nitrogenase activity decreases during senescence
To investigate the molecular mechanisms underlying nodule senescence, we inoculated greenhouse-grown soybean ( G. max (L.) Merrill) cultivar Williams 82 (W82) plants with its symbiont, Bradyrhizobium diazoefficiens USDA 110, and monitored them over time (Fig. 1A). We evaluated the nitrogen-fixation ability of nodules at different developmental stages by examining their internal color. Accumulation of the pigment leghemoglobin results in a pink color and indicates that nitrogen fixation has started, while degradation of leghemoglobin results in a green or pale color and is associated with decreased nitrogen-fixation activity (Navascues et al. 2012). Because nodulation is not a homogenous process, we used the largest nodules at the base of the primary root for comparisons of senescence in different genotypes and at different plant developmental stages.
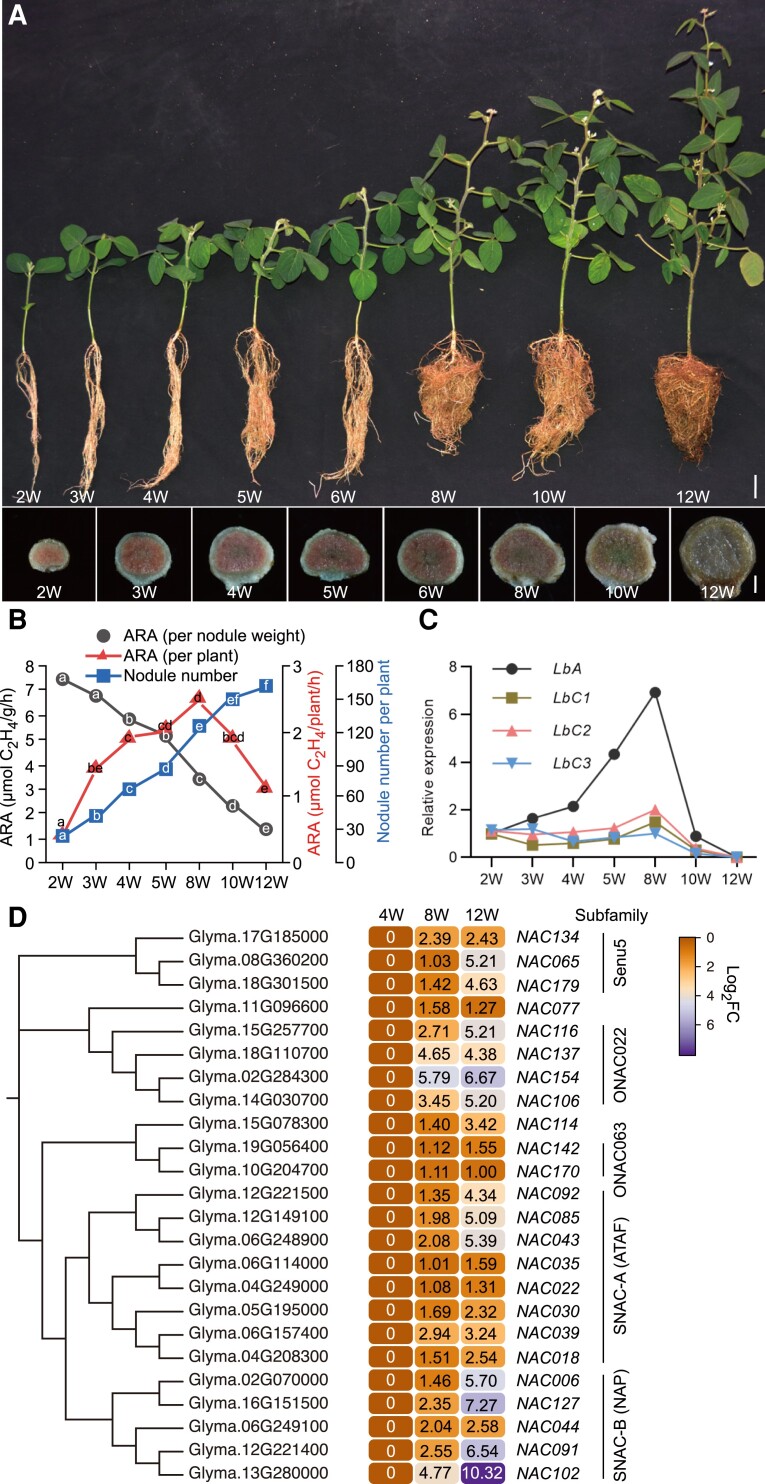
Figure 1.
Analysis of plant-specific NAC family during soybean nodule senescence. A) Plant and nodule development at different time points after inoculation with rhizobia. Five-day-old soybean seedlings grown in vermiculite were inoculated with rhizobia and grown for an additional 2, 3, 4, 5, 6, 8, 10, or 12 weeks (W). The upper panel illustrates plant growth at the different stages; scale bar = 5 cm. The lower panel shows longitudinal sections of nodules at the different stages; scale bar = 0.3 mm. The largest nodules at the base of the primary root were used for sections. B) Nitrogenase activity and nodule number at different developmental stages. Values represent the means ( 2W, n = 8;3W, n = 15;4W, n = 15;5W, n = 15;8W, n = 7;10W, n = 7;12W, n = 8 ). ARA, Acetylene reduction activity. Significant differences were determined by a two-tailed Student's t -test and are indicated by lower-case letters. C) Relative expression of leghemoglobin genes LbA, LbC1, LbC2, and LbC3 in nodules at different developmental stages determined using the comparative Ct method. Soybean ACT11 was used as an internal reference. Two samples for each time point were used for RT-qPCR. The largest nodules at the base of the primary root were harvested for each sample. D) Phylogenetic analysis of NAC proteins and gene expression patterns of senescence-induced NAC genes in soybean nodules. Expression data were obtained from RNA-Seq analysis of 4-, 8-, and 12 wpi (weeks post-inoculation) nodule samples. Number indicates the relative expression levels of genes.
Pink coloration was observed in longitudinal sections of the biggest nodule at the base of the primary root 2–6 wpi with rhizobia. By 4 wpi, a slightly brown area was observed in the center of the nodule, indicating that these cells had begun to senesce. By 8 wpi, the nodules had become pale pink, suggesting that senescence had begun to spread throughout the entire nodule. At 10 and 12 wpi, the nodules were partially green and light brown, respectively (Fig. 1A). The gradual changes in the color of the nodules between 8 and 12 wpi suggest that their nitrogenase activities gradually decreased during this time. The color change began at the center of nodules and gradually spread outward across the entire nodule (Fig. 1A). This finding is consistent with the conclusion that senescence in determinant nodules, such as those of soybean, begins at the center, and gradually spreads outward (Kazmierczak et al. 2020).
To further confirm the above conclusion, we performed acetylene reduction assays using all nodules from soybean plants at 2, 3, 4, 5, 8, 10, and 12 wpi with rhizobia. This assay is a direct measure of the relative reductase activity of nitrogenase in soybean nodules. The nitrogenase activity per plant at 2 wpi was relatively low but increased gradually as the number of mature nodules increased and peaked at 8 wpi (Fig. 1, A and B; Supplemental Fig. S1). By contrast, the nitrogenase activity per plant by 10 and 12 wpi had declined due to the increased number of senescent nodules on the whole plant (Fig. 1, A and B; Supplemental Fig. S1). The gradual reduction in nitrogenase activity per nodule fresh weight in plants from 2 to 12 wpi is consistent with a change in nodule color from pink to green and then to brown (Fig. 1, A and B). These data indicated that the combined nitrogen-fixation activity of all nodules from a soybean plant substantially declined after 8 wpi, with significant browning of the nodules, and that the color change from pink to brown reflects the progression of nodule development from maturity to senescence.
High nitrogenase activity depends on high levels of leghemoglobin, an oxygen-carrying protein that maintains the low-free-oxygen conditions needed for optimal nitrogenase activity in nodules (Wang et al. 2019). To determine the expression profiles of leghemoglobin genes in soybean nodules at different developmental stages, we collected several of the largest nodules at the base of primary roots for qPCR analysis. The soybean genome contains 4 major leghemoglobin genes: LbA, LbC1, LbC2, and LbC3. These Lb genes were expressed at high levels in the nodules from 2 to 8 wpi, but at lower levels at 12 wpi (Fig. 1C), consistent with the nitrogenase activities at these developmental stages. Together, these data show that the levels of nitrogenase activity and Lb gene expression decrease as soybean nodules senesce.
Transcription factors are expressed at high levels in senescent nodules
To study the molecular mechanisms of nodule senescence, we next performed a large-scale transcriptome analysis. Having established that nodules were severely senescent at 12 wpi (Fig. 1), we compared the gene expression profiles of nodules at 4, 8, and 12 wpi. Because nodulation is not a homogenous process, several bigger nodules at the base of each primary root were used for RNA-Seq analysis. We mined the soybean gene expression atlas (http://www.soybase.org) and compared the expression differences among 2,789 and 6,491 upregulated genes in 8 wpi vs. 4 wpi nodules and 12 wpi vs. 4 wpi nodules, respectively (Supplemental Data Set 1). We identified 1,710 genes (fold change (FC) > 2) that were significantly induced in nodules at both 8 and 12 wpi compared with those in nodules 4 wpi (Supplemental Fig. S2A).
To identify the TFs involved in regulating nodule senescence, we analyzed the upregulated genes encoding TFs among the 1,710 genes that were specifically expressed in senescing nodules (Supplemental Fig. S2B). Based on the plant TF database for soybean (http://planttfdb.gao-lab.org/), 210 genes encoding TFs from 5 of the main TF families (NAC, WRKY, MYB, ERF, and AGAMOUS-like) were identified as nodule senescence–induced (Supplemental Fig. S2C), suggesting that transcription factors might play important roles in activating the process of nodule senescence.
Two NAC transcription factors promote early senescence in soybean nodules
Abundant data indicate that a substantial number of NAC TFs play essential roles in regulating leaf senescence (reviewed by Kim et al. 2016), so we wondered whether any NAC TFs are involved in soybean nodule senescence. We compared the expression levels of soybean NAC genes in nodules at 8 and 12 wpi with those in nodules at 4 wpi. Expression profiles from the transcriptomic data and phylogenetic analysis showed that 24 NAC genes were specifically expressed in senescent nodules and clustered into 5 subgroups: SNAC-A (ATAF), SNAC-B (NAP), Senu5, ONAC022, and ONAC063 (Fig. 1D).
To investigate whether any of the senescence-induced NACs function in nodule senescence, we ectopically expressed them in soybean hairy roots using the leghemoglobin (Lb2) promoter and determined the nitrogenase activity and number of nodules per plant at 4 wpi. Due to gene duplication and high sequence similarity among the 24 NAC genes, we narrowed down the investigation to 10 NAC genes representing the 5 subfamilies (GmNAC018, GmNAC030, GmNAC039, GmNAC043, GmNAC065, GmNAC091, GmNAC114, GmNAC127, GmNAC142, and GmNAC154). Transgenic hairy roots overexpressing GmNAC114, GmNAC142, and GmNAC154 had similar nitrogenase activities and nodule numbers as the control (Supplemental Fig. S3, A to F), indicating that these NAC proteins might not substantially promote nodule early senescence.
Overexpression of GmNAC030, GmNAC065, GmNAC091, or GmNAC127 led to decreased nitrogenase activity but had no effect on the nodule numbers, except in the case of the GmNAC127 transgenic roots, which had slightly fewer nodules than the control transgens (Supplemental Fig. S3, A to D and J to L). Overexpression of GmNAC043 slightly increased the nitrogenase activity of nodules, but the nodule number was comparable to the control (Supplemental Fig. S3, G to I). Overexpression of GmNAC039 and GmNAC018 resulted in the most striking symbiotic phenotypes (Fig. 2). The shoots of plants with transgenic hairy roots overexpressing GmNAC039 or GmNAC018 displayed yellowish leaves, which is indicative of nitrogen deficiency (Fig. 2, A to D). In addition, at 4 wpi, GmNAC039-OE or GmNAC018-OE nodules were much smaller than control nodules and had light green or pale centers (Fig. 2, E and F).
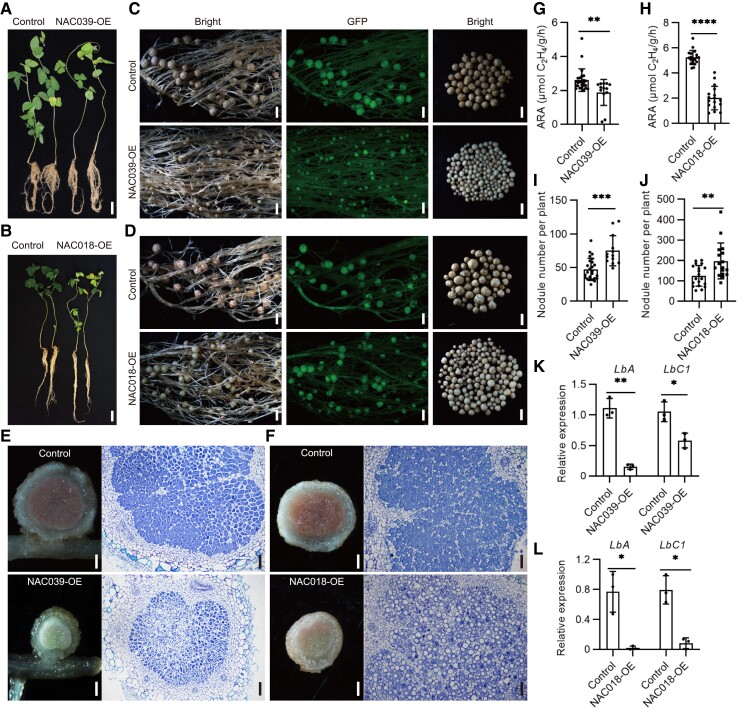
Figure 2.
Overexpression of GmNAC039 or GmNAC018 promotes nodule senescence in soybean. A and B) Comparison of growth phenotypes at 4 wpi of control soybean plants transformed with empty vector and plants overexpressing 3× Flag-tagged GmNAC039 or GmNAC018 (NAC039-OE or NAC018-OE). Scale bars = 5 cm. C and D) Nodule phenotypes of soybean plants harboring the empty vector (control) or overexpressing 3× Flag-tagged GmNAC039 or GmNAC018 (NAC039-OE or NAC018-OE). In each row, the two images at left show transgenic roots harboring the empty vector (Control) or overexpressing GmNAC039 or GmNAC018 (NAC039-OE or NAC018-OE) as seen under white light (first image) or fluorescent light (second image; green fluorescent protein (GFP) fluorescence). The third panels illustrate the size and color of nodules from one transgenic seedling. GFP served as a positive marker for transformed lines. Scale bars = 1.5 mm. E and F) Cell morphology and organization of nodules from transgenic hairy root plants of NAC039-OE (E) or NAC018-OE (F). The left panel illustrates the internal color of nodules from transgenic root, scale bars = 0.3 mm. The right panel represents longitudinal sections of nodules stained with toluidine blue dye. Scale bars = 100 μm. G and H) Nitrogenase activity of control and NAC039-OE (G) or NAC018-OE (H) nodules determined using an acetylene reduction assay. ARA, acetylene reduction activity. I and J) Nodule number of control, NAC039-OE (I), or NAC018-OE (J). Data shown in bar charts are mean ± SD. K and L) Relative expression of LbA and LbC1 in nodules from NAC039-OE (K), or NAC018-OE (L) from 3 transgenic roots per genotype, determined by qPCR. GmACT11 was used as the reference. Data shown in bar charts are means ± SD. Significance analyses were performed using GraphPad Prism version 9 based on paired Student's t-tests compared with the control (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001).
Toluidine blue staining of longitudinal cross sections in nodules revealed that GmNAC039 or GmNAC018 overexpression reduced the number of bacteroids and increased the size of vacuoles in infected cells compared to control nodules (Fig. 2, E and F). Additionally, nodules overexpressing GmNAC039 or GmNAC018 had significantly reduced nitrogenase activity compared with controls (Fig. 2, G and H). Interestingly, transgenic hairy roots expressing GmNAC039 or GmNAC018 formed more nodules than the controls (Fig. 2, I and J), perhaps as a result of a developmental regulatory mechanism that compensates for the drastically reduced nitrogenase activity of the nodules. In addition, leghemoglobin gene expression levels were reduced in the transgenic nodules compared with the controls (Fig. 2, K and L). All these results are consistent with the conclusion that nitrogenase activity was significantly reduced in the transgenic nodules. Therefore, these data suggest that among the 10 senescence-induced NAC genes tested, GmNAC039 and its paralog GmNAC018 were the strongest promoters of nodule early senescence.
To examine whether the defective nodules in GmNAC039-OE and GmNAC018-OE nodules are due to early senescence or developmental arrest, sections of nodules 2 wpi were made for cell morphology analysis. GmNAC039-OE or GmNAC018-OE transgenic roots generate similar-size nodules compared with those from control plants (Fig. 3A and Supplemental Fig. S4). An abundant, clear, and multi-vesicles or large-vacuolated cytoplasm with the reduced population of bacteroids in infected cells were observed in nodules of GmNAC039-OE or GmNAC018-OE relative to controls (Fig. 3A). Overexpression of GmNAC039 or GmNAC018 significantly reduced the nitrogenase activity in nodules (Fig. 3B), but did not significantly alter the nodule numbers compared with control nodules 2wpi (Fig. 3C). The data suggested that overexpression of GmNAC039 or GmNAC018 might promote early senescence in nodules at early stage.
Figure 3.
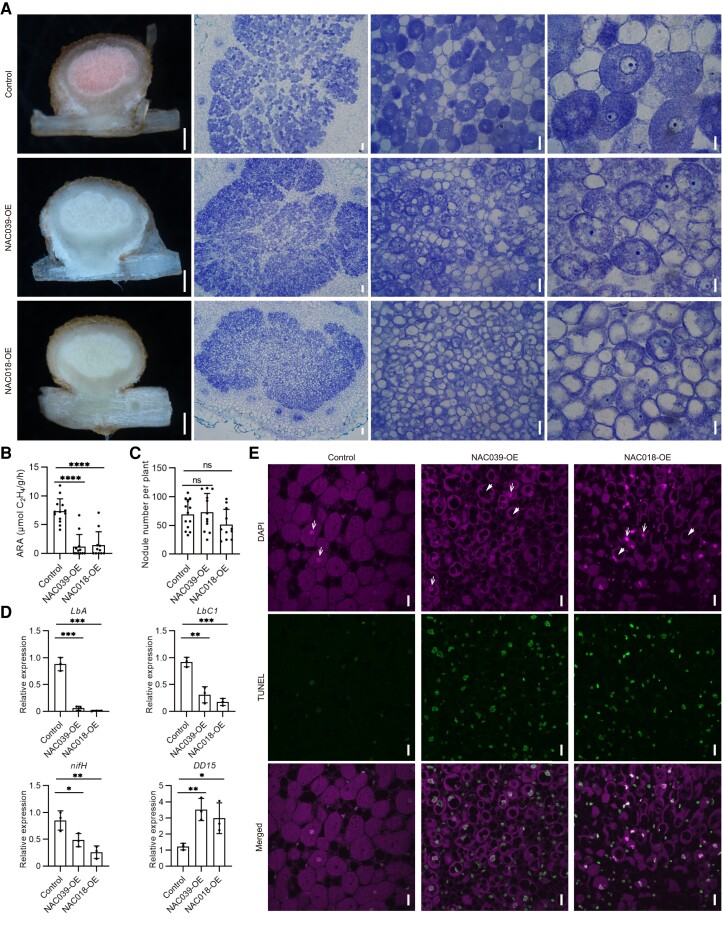
Cell structures of GmNAC039-OE/GmNAC018-OE transgenic nodules. A) Toluidine blue-stained transverse sections of transgenic nodules at 2 wpi. From left to right, scale bar = 0.5 mm, 50 μm, 25 μm, and 10 μm. B and C) Nitrogenase activity determined by acetylene reduction activity (ARA) (B) and nodule number (C) in transgenic roots at 2 wpi. D) RT-qPCR analyses of symbiotic genes and senescence-associated marker gene DD15 in transgenic nodules at 2 wpi. Data shown in bar charts are means ± SD from 3 biological replicates. E) Apoptosis analysis in NAC039-OE and NAC018-OE nodules 4 wpi using a TUNEL assay. The upper panel (DAPI) illustrates DAPI-stained nuclei and rhizobia. The middle panel (TUNEL) represents double-strand DNA breaks labeled using the TUNEL assay. The lower panel (Merged) represents merged images of the DAPI and TUNEL signals. Nuclei and vacuoles are indicated by white arrows and arrowheads, respectively. Scale bar = 20 μm. Significance analyses were performed using GraphPad Prism version 9 based on paired Student's t-tests compared with the control (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001).
To confirm this hypothesis, expression of genes involved in nodule development and senescence was analyzed. The transcript levels of symbiosis genes including GmLbA , GmLbC , and rhizobial nifH in the GmNAC039-OE or GmNAC018-OE nodules were detected at reduced levels (Fig. 3D). However, soybean DD15 (Glyma.02G021600), a nodule senescence-associated marker gene (Alesandrini et al. 2003a), was significantly induced in both GmNAC039-OE and GmNAC018-OE nodules compared with controls (Fig. 3D). These data indicated that GmNAC039 or GmNAC018 has a role in promoting nodule early senescence with decreased nitrogen-fixing ability during nodule development.
To further characterize the roles of GmNAC039 and GmNAC018 in nodule senescence, we quantified apoptosis in senescent nodules using a TUNEL assay, a method that detects double-strand breaks in the nuclear DNA, which increase during apoptosis (Xia 2018). Cells within longitudinally sectioned fresh nodules were stained both with 4′,6-diamidino-2-phenylindole (DAPI) and by fluorescently labeled terminal dUTP nick-end labeling. Transgenic nodules overexpressing either GmNAC039 or GmNAC018 displayed reduced numbers of bacteroids, large vacuole-like lytic structures, a typical marker of nodule senescence, and nuclear degradation (Fig. 3E). By contrast, the symbiotic cells of control nodules had fully developed symbiosomes that were packed in a radial pattern around an intact nucleus (Fig. 3E). These data further confirmed that GmNAC039 and GmNAC018 play essential roles in mediating soybean nodule senescence.
Knockout of GmNAC039 and GmNAC018 causes delayed nodule senescence
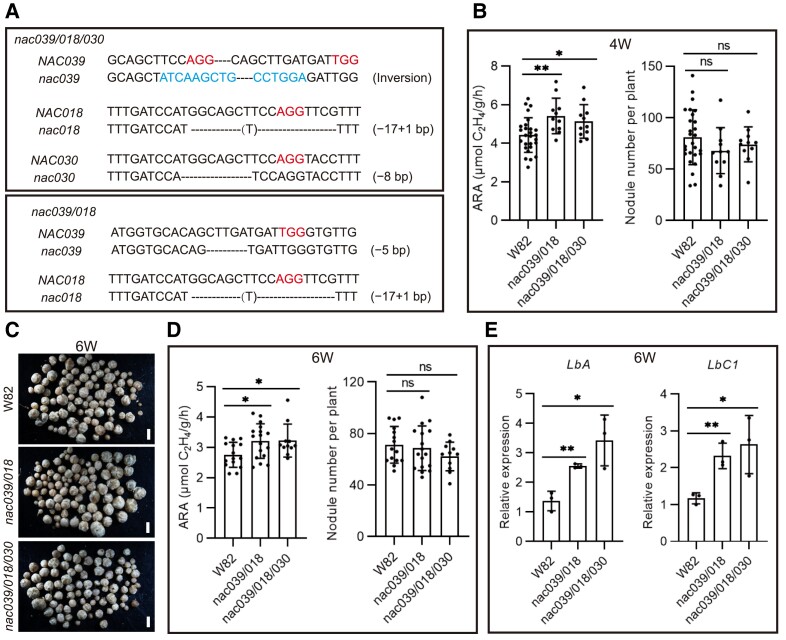
Having determined that overexpression of GmNAC039 or GmNAC018 accelerated nodule senescence in soybean, we hypothesized that knocking out these NAC genes would delay nodule senescence. GmNAC039, GmNAC018, and GmNAC030 belong to the same SNAC-A subgroup and share high sequence similarity (Supplemental Fig. S5A) and promote nodule senescence. Therefore, 4 gRNAs (guide RNAs) that simultaneously target all 3 NAC genes were designed and cloned into one plasmid using polycistronic-tRNA CRISPR/Cas9 technology (Xie et al. 2015) (Supplemental Fig. S5B). To verify the editing of these 3 NAC genes, the DNA sequences flanking each gRNA target site from 3 random individual transgenic hairy roots were amplified and sequenced (Supplemental Fig. S5, C and D). Sequencing of PCR products from independent biallelic mutant roots revealed a variety of mutations per transgenic root and mutation efficiencies with the 4 sgRNAs for GmNAC039 , GmNAC018, and GmNAC030 genes were approximately 100% (Supplemental Fig. S5, C and D).
Nitrogenase activity was measured in soybean plants at 6 wpi from the control (wild type transformed with empty vector) and GmNAC039/GmNAC018/GmNAC030 knockout (NACs-KO). The most of bigger nodules at the base of the primary roots of NACs-KO plants were slightly pinker in the center area than those formed by the control (Supplemental Fig. S5E). Consistent with this, the NACs-KO nodules had higher nitrogenase activity than the control nodules (Supplemental Fig. S5F), but there was no significant difference in nodule number (Supplemental Fig. S5G). The expression levels of LbA and LbC1 were higher in NACs-KO nodules than in control nodules (Supplemental Fig. S5, H and I), which is consistent with their higher nitrogenase activity.
To further confirm the role of GmNAC039, GmNAC018, and GmNAC030 in regulating nodule senescence, we generated stable transgenic soybean plants. Soybean nac039/nac018/nac030 triple knockout and nac039/nac018 double knockout homozygous mutants were obtained through gene editing (Fig. 4A). We observed no significant differences in growth rate between wild-type (WT) and stable NACs-KO plants during vegetative growth (Supplemental Fig. S6). Similar to the phenotype of the NACs-KO transgenic hairy roots, the nodules of both nac039/nac018/nac030 and nac039/nac018 homozygous mutant plants had higher nitrogenase activities than control nodules at 4 and 6 wpi (Fig. 4, B to D). The nodule numbers were similar for the control and the nac039/nac018/nac030 and nac039/nac018 homozygous mutant soybean plants (Fig. 4, B to D). Consistent with the elevated nitrogenase activity, significantly increased expression of the leghemoglobin genes LbA and LbC1 was detected in the mutant nodules compared to the WT nodules (Fig. 4E). Overall, these data strongly indicated that GmNAC039, GmNAC018, and GmNAC030 are essential components promoting nodule senescence in soybean.
Figure 4.
Nodule number and nitrogenase activity in nac039/nac018/nac030 knockout mutant plants. A) Gene editing types in NACs-KO (knockout) stably transformed plants. An inversion between gRNA1 and gRNA3 within NAC039, a 16 bp fragment deletion at gRNA1 within NAC018, and an 8-bp deletion at gRNA1 within NAC030 were found in the nac039/nac018/nac030 triple homozygous mutant plants. A 5-bp deletion at gRNA3 within NAC039 and 16 bp deletion at gRNA1 within NAC018 were found in the nac039/nac018 double homozygous mutant plants. B) Nitrogenase activity and nodule number of NACs-KO stably transformed plants at 4 wpi. Soybean wild-type W82 was used as the control. C) Nodule phenotype of NACs-KO stably transformed plants at 6 wpi. Scale bar = 1.5 mm. D) Nitrogenase activity and nodule number of NACs-KO stably transformed plants at 6 wpi. Data shown in bar charts are means ± SD. E) Relative expression of LbA and LbC1 in hairy root nodules at 6 wpi. Three individual nodule samples were analyzed by qPCR using GmACT11 as a reference. Data shown in bar charts are means ± SD. Asterisks show significant differences determined using GraphPad Prism version 9 through unpaired Student's t-tests of comparisons with the control (*P ≤ 0.05; **P ≤ 0.01; ns, no significant difference).
GmNAC039-mediated transcriptional activation is required for nodule senescence
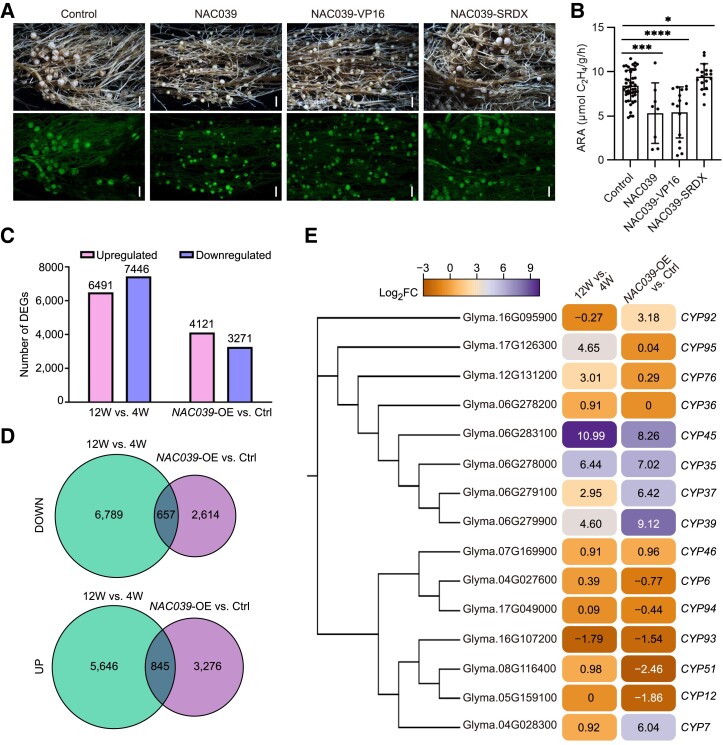
Previous studies have demonstrated that several NAC proteins contain a NARD and can function as either transcriptional activators or repressors (Hao et al. 2010; Nagahage et al. 2018). Amino acid sequence alignment showed that GmNAC039, GmNAC018, and GmNAC030 contain a conserved N-terminal DNA-binding domain with an NARD for transcriptional repression as well as a variable C-terminal transcriptional regulatory domain (Supplemental Fig. S5A). Therefore, we tested whether GmNAC039 functions as a transcriptional activator or repressor to promote nodule senescence. To test this, SRDX and VP16, protein domains known to effectively repress and activate gene expression, respectively, were fused with GmNAC039 for overexpression in soybean hairy roots. Transgenic GmNAC039 or GmNAC039-VP16 roots produced elevated numbers of nodules, which were small and white or light green, whereas nodules from control or GmNAC039-SDRX-expressing plants were pink (Fig. 5A), indicating that transcriptional activity of GmNAC039 is required for promoting nodule senescence. In agreement with their defective nodule development, transgenic hairy roots expressing GmNAC039-VP16 and GmNAC039 had nodules with significantly reduced nitrogenase activities compared with control nodules at 4 wpi, whereas nodules from GmNAC039-SDRX-expressing hairy roots had slightly higher nitrogenase activity than the controls (Fig. 5B). These results are in agreement with the observed nodule phenotypes of both transgenic hairy roots and stably transformed plants overexpressing or lacking GmNAC039 / GmNAC018 . These data indicate that GmNAC039-mediated transcriptional activation is required for accelerating nodule senescence in soybean.
Figure 5.
RNA-Seq analysis of gene expression patterns in NAC039-overexpression and naturally senescent nodules. A) Nodule phenotypes at 4 wpi of plants transformed with empty vector (Control), GmNAC039, GmNAC039-VP16, or GmNAC039-SRDX. Scale bars = 1.5 mm. B) Nitrogenase activity of nodules at 4 wpi. Asterisks show significant differences compared with the control determined by unpaired Student's t-test using GraphPad Prism version 9 (*P ≤ 0.05; ***P ≤ 0.001; ****P ≤ 0.0001). C) Comparison of the numbers of upregulated and downregulated differentially expressed genes (DEGs) in nodules at 12W vs. 4W and of NAC039-OE vs. Ctrl detected by RNA-Seq. D) Venn diagrams showing unique and common genes from different pairwise comparisons among DEG data sets. The 12W vs. 4W data set is shown in green, and the NAC039-OE vs. Ctrl. data set is shown in blue. E) Phylogenetic analysis and heatmap showing cysteine protease genes (CYPs) differentially expressed in naturally senescent nodules (12W vs. 4W) and NAC039-OE nodules (NAC039-OE vs. Ctrl). Significant DEGs had fold changes ≥ ± 1.5; P < 0.05. Number indicates the relative expression levels (log2 fold changes) of genes.
GmNAC039-overexpressing transgenic nodules have altered expression of senescence-related genes
To decipher the molecular mechanisms underlying the regulation of nodule senescence by GmNAC039, we aimed to identify its downstream target genes. To this end, we performed RNA-seq analysis to compare genome-wide transcript levels in GmNAC039-overexpressing and wild-type nodules at 12 wpi, a stage characterized by severe nodule senescence. Differentially expressed genes (DEGs) were identified (P-value < 0.05, FC ≥ 2) in comparisons between healthy nodules (at 4 wpi) and fully senescent nodules (at 12 wpi) of the wild type and between senescent GmNAC039-OE nodules and control transgenic nodules at 4 wpi (Supplemental Data Set 2). These DEGs included 6,491 upregulated and 7,446 downregulated genes in nodules at 12 wpi vs. 4 wpi and 4,121 upregulated and 3,271 downregulated genes in nodules of NAC039-OE vs. Control (Ctrl.) (Figure 5C).
We further classified DEGs from NAC039-OE vs. Ctrl. based on Gene Ontology (GO) and KEGG pathway enrichment analysis. Significant DEGs in GO analysis were related to cell wall biogenesis or responses to wounding, reactive oxygen species, and defense, indicative of plant–microbial interactions. Remarkably, genes involved in phenylpropanoid biosynthesis and in the metabolism of starch and amino acids were also highly enriched in the KEGG pathway analysis, indicative of plants undergoing successive developmental processes in response to stressful environments (Supplemental Fig. S7). These data indicate substantial transcriptional reprograming related to stress responses and immune responses during nodule senescence.
To determine whether GmNAC039 regulates the expression of senescence-related genes in nodules, DEGs common to NAC039-OE and 12-wpi nodules were identified by performing pairwise comparisons (NAC039-OE vs. Ctrl., 12 wpi vs. 4 wpi) of gene expression using the aligned reads. We identified 845 common upregulated genes and 657 common downregulated genes in the NAC039-OE vs. Ctrl. and 12 wpi vs. 4 wpi (Fig. 5D). These data indicated that GmNAC039 participates in controlling transcriptional reprograming of senescence-related genes during nodule senescence.
Given that GmNAC039-mediated transcriptional activation is required for nodule senescence, we focused on genes upregulated during nodule senescence as potential direct downstream targets of GmNAC039. Previously, strong expression of nodule-specific papain-like CYP genes was observed in senescent nodules in soybean (Van Wyk et al. 2014; Yuan et al. 2020), Chinese milk vetch ( Astragalus sinicus ) (Naito et al. 2000; Li et al. 2008), pea (Pisum sativum) (Kardailsky and Brewin 1996), and M. truncatula (Pierre et al. 2014; Deng et al. 2019) and is viewed as a hallmark of nodule senescence. Therefore, we analyzed the differentially expressed GmCYPs in both 12-wpi WT and GmNAC039-OE nodules. Among the upregulated DEGs common to GmNAC039-OE and 12 wpi nodules, GmCYP37, GmCYP35, GmCYP39, GmCYP45, and GmCYP7 were highly induced during nodule senescence (Fig. 5E). Phylogenetic analysis based on sequence similarities showed that all the nodule senescence–induced GmCYPs except for GmCYP7 are in the same subclade, have high sequence similarity, and are located adjacent to each other on chromosome 6 (Fig. 5E). The data indicate at least 4 GmCYP genes are nodule senescence marker genes and are highly induced in GmNAC039-OE and 12-wpi nodules.
GmNAC039 directly regulates the expression of CYP genes
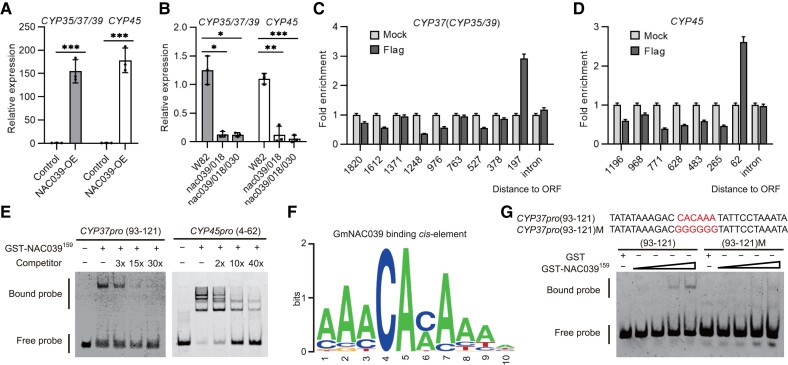
Due to their high expression levels and potential roles in senescent nodules, we hypothesized that GmCYP37, GmCYP35, GmCYP39, and GmCYP45 might be directly targeted and regulated by GmNAC039 and GmNAC018. To test this hypothesis, we analyzed the expression levels of GmCYP35, GmCYP37, GmCYP39, and GmCYP45 in GmNAC039-OE and nac039/nac018/nac030 mutant nodules. Due to their high sequence similarity, we used the same primers to amplify 3 CYP genes (GmCYP35, GmCYP37, and GmCYP39) and measured their expression levels using qPCR. A separate set of primers was used to amplify GmCYP45. As shown in Fig. 6, A and B, the expression of all 4 GmCYPs was highly induced in GmNAC039-OE nodules but suppressed in nac039/nac018 and nac039/nac018/nac030 mutant nodules compared with that in the control nodules. In addition, we observed a high sequence similarity in the promoter regions of GmCYP35, GmCYP37, and GmCYP39 (Supplemental Fig. S8A), which led us to hypothesize that the expression of GmCYP35, GmCYP37, and GmCYP39 might be regulated by similar TFs, including GmNAC039 and GmNAC018.
Figure 6.
NAC039 directly targets cysteine protease (CYP) genes. A) Gene expression of GmCYP35, GmCYP37, GmCYP39, and GmCYP45 in GmNAC039-OE nodules at 4 wpi determined by RT-qPCR. Data shown in bar charts are means ± SD of 3 biological repeats. B) Gene expression of GmCYP35, GmCYP37, GmCYP39, and GmCYP45 in 6-wpi nodules of NACs-KO stably transformed plants determined by RT-qPCR. Data shown in bar charts are means ± SE of 3 biological repeats. C and D) nCUT&Tag-qPCR analysis showing that GmNAC039 directly targets the GmCYP37 and GmCYP45 promoters. Anti-Flag antibody was used to pull down DNA fragments from GmNAC039-3×Flag transgenic nodules, followed by qPCR analysis using different primer pairs. The positions of the promoter segments are indicated relative to ATG. E) Confirmation of the binding of GmNAC039 to GmCYP37 and GmCYP45 promoters by EMSA. EMSA was performed using truncated GmNAC039 (residues 1–159) protein purified from E. coli. Different promoter fragments were labeled with fluorescein amidite (FAM), and unlabeled DNA was used as a competitor for GmNAC039 binding. The mobility shift represents GmNAC039 DNA binding activity. The positions of the promoter segments are indicated relative to ATG. F) GmNAC039 consensus binding sequence determined by a random DNA binding selection assay. Truncated glutathione S-transferase (GST)-NAC039 (residues 1–159) fused with GST tag was incubated with the random DNA fragments. 10-nt consensus sequences from the pulled-down sequences were analyzed online (http://weblogo.Berkeley.edu/). G) Identification of the conserved binding site of GmNAC039. Wild-type and mutant versions of nucleotides 93–121 of the GmCYP37 promoter were used for the EMSA. The DNA motif “CACAAA “in red font represents NAC039-binding sites, while “GGGGGG” indicates the mutant version [CYP37 pro (93-121)M].
To test this idea, we examined the ability of GmNAC039 to bind directly to the promoters of GmCYP35, GmCYP37, GmCYP39, and GmCYP45. The nodule is a specialized organ in which the majority of DNA is in the bacteroids, making it difficult to perform chromatin immunoprecipitation (ChIP) assays to identify the binding sites of GmNAC039. Because of this, we used a new method, a modified version of the nucleus CUT&Tag-qPCR (nCUT&Tag-qPCR) assay recently used in mammalian cells (Bartosovic et al. 2021). Soybean nodule cells were filtered using a special mesh to remove bacteroids before the assay. Filtered plant cells from GmNAC039-OE nodules were further confirmed using immunoblotting to detect FLAG-tagged GmNAC039 and stained with DAPI to check that the nuclei were intact and purified (Supplemental Fig. S9). Nuclei from GmNAC039-OE nodules were collected and used for the CUT&Tag assay. Genomic DNA fragments were then pulled down with or without an anti-FLAG antibody followed by qPCR analysis. Multiple primer pairs were designed to amplify DNA fragments from the 1,820- and 1,196-bp regions upstream of the start codons and introns of GmCYP37 and GmCYP45, respectively (Fig. 6, C and D). The GmCYP37 DNA region from −1 to −197 bp was detected at high levels by qPCR analysis in GmNAC039-OE nodule cells compared with the mock control (Fig. 6C). A single strong GmNAC039 binding peak was also identified in the GmCYP45 promoter region from −1 to −62 bp (Fig. 6D). These data suggest that GmNAC039 binds directly to the promoters of GmCYP37 and GmCYP45 near the start codon.
To confirm that GmNAC039 physically binds to the promoters of GmCYP37 and GmCYP45, an electrophoretic mobility shift assay (EMSA) was performed using the purified NAC domain of GmNAC039 and synthetic DNA fragments. To determine the specific binding sites of GmNAC039 on the GmCYP37 and GmCYP45 promoters, 5 different DNA fragments were synthesized: 4 between −76 and –205 bp of the GmCYP37 promoter and one from –4 to –62 bp of the GmCYP45 promoter. The NAC domain of GmNAC039 bound directly to the DNA fragments from −76 to −147 bp, −76 to −121 bp, and −93 to −121 bp, but not the fragment from −76 to −100 bp in the GmCYP37 promoter (Supplemental Fig. S8C and Fig. 6E). The NAC domain of GmNAC039 could also directly bind to the DNA fragment from −4 to −62 bp of the GmCYP45 promoter (Fig. 6E). We then wondered whether the GmNAC039 binding site is conserved among the promoters of GmCYP35/37/39/45. Sequence alignment analysis showed that the DNA fragment from −93 to −121 bp of GmCYP37 promoter is identical to sequences in the promoters of GmCYP35 and GmCYP39 (Supplemental Fig. S8A), but it is slightly different from the GmNAC039-bindng site (−4 to −62 bp) in the GmCYP45 promoter (Supplemental Figs. S8, A and B).
To precisely identify the cis-regulatory element targeted by GmNAC039, we conducted a random DNA-binding selection assay using random oligonucleotides. Analysis of the pulled-down sequences revealed a 10-nt consensus sequence that corresponded to a 23-bp sequence identified from 30 random oligonucleotides bound by the NAC domain of GmNAC039 (Supplemental Fig. S8D). Weblogo analysis showed that the core consensus sequence is CACA or CAAA, in which the flanking regions are A-rich (Fig. 6F). Furthermore, mutation of the core CAC(A)A motif in the region of −93 to −121 bp of the GmCYP37 promoter resulted in a dramatic reduction in GmNAC039 binding (Fig. 6G). Together, these data indicate that GmNAC039 directly targets the promoters of cysteine protease genes via the core consensus sequence CAC(A)A.
Overexpression of GmCYP genes promotes nodule senescence in soybean
Our results indicated that at least 4 GmCYP genes are direct targets of GmNAC039. We then sought to test whether these genes are involved in nodule senescence. Amino acid sequence alignment showed that GmCYP35, GmCYP37, GmCYP39, and GmCYP45 share a highly conserved amino acid sequence (Supplemental Fig. S10). Among these GmCYP proteins, GmCYP37 is paralogous to GmCYP35.
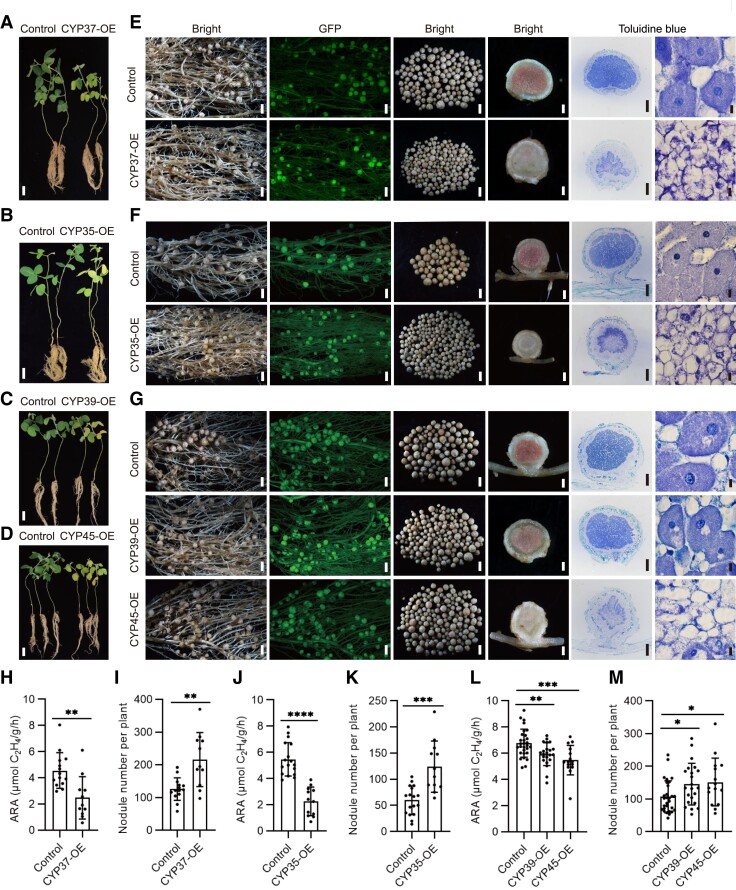
To test whether GmCYP35, GmCYP37, GmCYP39, GmCYP45, and GmCYP7 regulate nodule senescence, all 5 GmCYP genes were individually overexpressed in soybean roots using green fluorescent protein (GFP) as a selection marker, and the nodule phenotypes of the transgenic roots were examined. The leaves of GmCYP35, GmCYP37, GmCYP39, and GmCYP45 hairy root transformation plants displayed visible yellowing compared with the control plants, indicative of nitrogen deficiency (Fig. 7, A to D). Compared with control plants, transgenic hairy roots overexpressing GmCYP37, GmCYP35, GmCYP39, or GmCYP45 produced more nodules that were smaller in size compared with the control (Fig. 7, E to G). A toluidine blue staining assay showed that transgenic nodules overexpressing GmCYP35, GmCYP37, GmCYP39, or GmCYP45 had few symbiosomes and bacteroids (Fig. 7, E to G). At 4 wpi, these transgenic hairy roots produced more nodules, which were pale and had decreased nitrogenase activity compared to control nodules (Fig. 7, H to M). Although GmCYP7 is also the direct target of GmNAC039 (Supplemental Fig. S11, A and B), overexpression of GmCYP7 did not induce nodule senescence like the other GmCYPs tested (Supplemental Fig. S11, C to E). Together, these data indicated that GmCYP35, GmCYP37, GmCYP39, and GmCYP45 but not GmCYP7 play important roles in nodule senescence.
Figure 7.
The symbiotic phenotypes of plants overexpressing GmCYP37, GmCYP35, GmCYP39, or GmCYP45. A–D) Phenotypes of plants transformed with the empty vector (control), GmCYP37 (A), GmCYP35 (B), GmCYP39 (C), or GmCYP45 (D) at 4 wpi. Scale bars = 5 cm. E–G) Nodule phenotypes of transgenic roots at 4 wpi. GFP served as a positive marker for selection. The two left-most panels show nodulated transgenic roots under white light (first panel) and fluorescent light (second panel). The middle two panels show nodules picked from one transgenic root (third panel) and a longitudinal section of one of the nodules (fourth panel). The right-most two panels show nodule sections stained with toluidine blue dye and viewed at two different magnifications. The largest nodules at the base of the primary root were used for sections. Scale bars on the panels from left to right = 1.5 mm, 1.5 mm, 1.5 mm, 0.3 mm, 0.3, and 10 μm, respectively. H, J, and L) Nitrogenase activity of nodules at 4 wpi. I, K, and M) Nodule numbers of the overexpression hairy roots. Data are given as means ± SD. Significant differences were determined by unpaired Student's t-tests using GraphPad Prism version 9 (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001).
Knockout of GmCYP genes in soybean delays nodule senescence
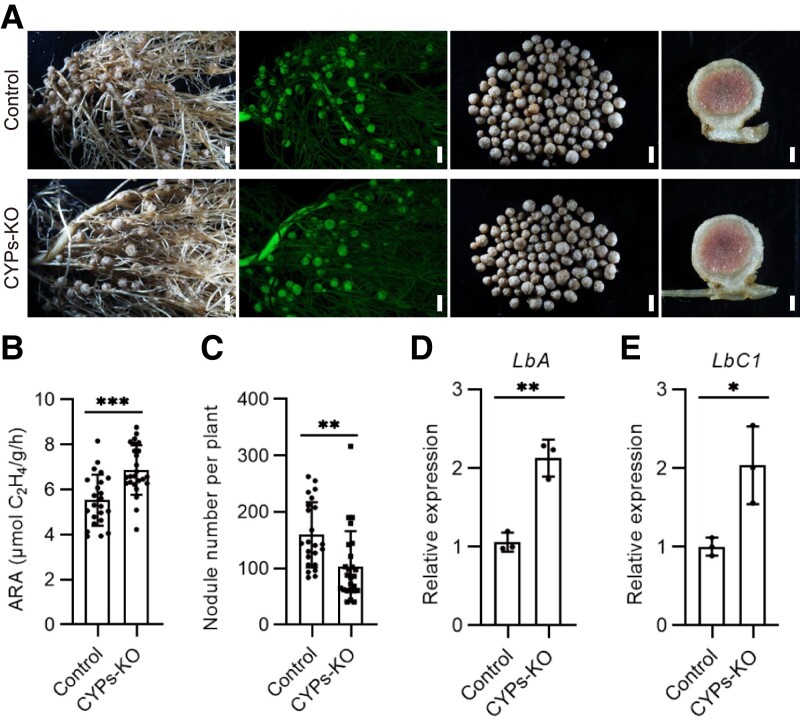
To further confirm the role of GmCYP35, GmCYP37, GmCYP39, and GmCYP45 in promoting nodule senescence, CRISPR–Cas9 technology was used to simultaneously target all 4 CYPs with conserved gRNAs (Supplemental Fig. S12A). The gene editing efficiency at the 4 different CYP loci in transgenic nodules was confirmed by sequencing DNA from 3 random individual transgenic hairy roots (Supplemental Fig. S12, B and C). Compared with the control, the nodules that formed at 6 wpi on CYP s-KO transgenic hairy roots were slightly pinker in the center and exhibited higher nitrogenase activity (Fig. 8, A and B). In contrast to the GmCYP overexpression lines, fewer nodules were generated on the CYPs-KO hairy roots compared with the control (Fig. 8C). Consistent with their increased nitrogenase activity, expression of the leghemoglobin genes LbA and LbC1 in CYPs-KO nodules was double that of the control nodules (Fig. 8, D and E). Together, these data confirm that, like GmNAC039 and GmNAC018, GmCYP35, GmCYP37, GmCYP39, and GmCYP45 play major roles in nodule senescence.
Figure 8.
Phenotypes caused by CRISPR/Cas9-mediated CYP35/CYP37/CYP39/CYP45 knockout in soybean hairy roots. A) Nodulation phenotypes at 6 wpi of plants transformed with the empty vector (control) or the CYPs CRISPR vector (CYPs-KO). The first panel in each row shows the nodulated roots under white light. The second panel shows GFP fluorescence signals representing positive transgenic nodules. The third panel shows the nodules produced by a single plant. The fourth panel shows a longitudinal section of a nodule. The largest nodules at the base of the primary root were used for the sections. From left to right, scale bars = 1.5, 1.5, 1.5, and 0.3 mm. B and C) Nitrogenase activity (B) and nodule number (C) of the control and CYPs-KO hairy roots. Asterisks show significant differences determined by unpaired Student's t-tests using GraphPad Prism version 9 (**P ≤ 0.01; ***P ≤ 0.001). D and E) Relative expression of LbA and LbC1 in control and CYPs-KO hairy root nodules. Data shown in bar charts are means ± SD of 3 biological repeats.
Discussion
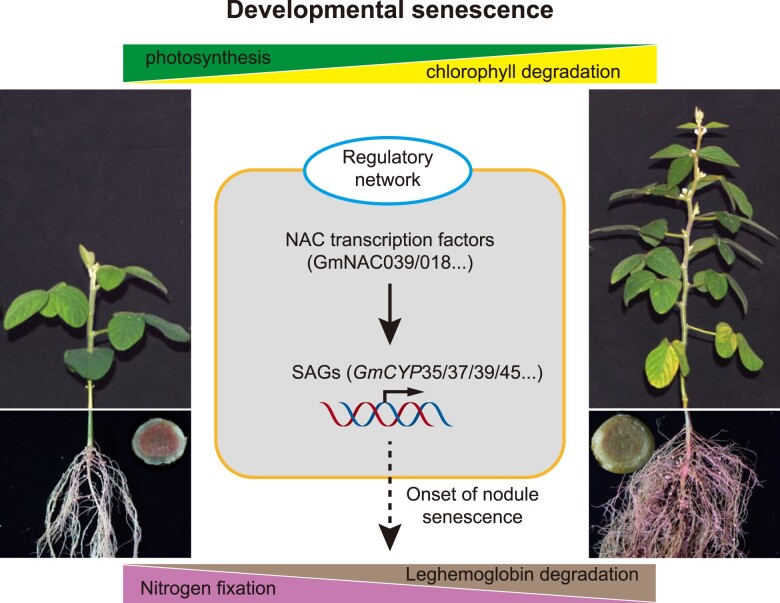
Nodule senescence, marked by high levels of cysteine proteases (CYPs), is an active process programed by the whole plant development. However, the key components regulating nodule senescence remain largely unknown, limiting our understanding of the regulatory mechanisms underlying this process. In this study, we explored the molecular basis of nodule senescence in the soybean-rhizobia symbiosis. Two NAC TFs, GmNAC039 and its paralog GmNAC018, were identified as essential regulators of soybean nodule senescence. GmNAC039 directly binds to the promoters of 4 GmCYP genes and enhances their expression. In turn, GmCYPs promote nodule senescence in soybean. Our data provide clear evidence for a regulatory pathway in which the activation of GmCYPs expression by GmNAC TFs regulates nodule senescence in soybean (Fig. 9).
Figure 9.
Model of GmNAC–GmCYP regulation of nodule senescence. Chlorophyll degradation and decreased photosynthesis are hallmarks of leaf senescence. In nodules, the amount of leghemoglobin is an indicator of nitrogen-fixation ability. When soybean is at the full bloom stage, the nodules undergo extensive senescence along with a reduction in nitrogen-fixation capacity. Some NAC transcription factors, including GmNAC039 and GmNAC018, are significantly induced during nodule senescence. This model proposes that GmNAC039/018 may activate downstream SAG expression to promote nodule senescence in soybean.
Nodules are specialized plant organs that provide large amounts of nitrogenous nutrients to the host. However, the life cycle of a nodule, from initiation to complete senescence, is usually shorter than that of its host plant. For example, most soybean plant nodules senesce during the bloom stage, causing a rapid decrease in nitrogenase activity. Nodule senescence seems to be poorly timed in soybean plants, as this process reduces nitrogen fixation when the plant has an urgent need for nitrogen to support seed development. Therefore, external N fertilizer must be applied to meet the requirements of seed development. The termination of a nodule development may result from the nutrient remobilization that occurs at this stage of plant development to ensure seed quality (Imsande 1986; Imsande and Schmidt 1998; Schiltz et al. 2005). In any case, delaying the senescence of symbiotic nodules could prolong active nitrogen fixation, resulting in an increased overall nitrogen supply for seed development during seed filling.
The NAC family is one of the largest TF gene families in plants, and its members play a central role in organ senescence. Among ∼180 genes encoding NAC TFs identified in the soybean genome (Melo et al. 2018), 79 are differentially expressed and involved in leaf senescence (Melo et al. 2018; Fraga et al. 2021). The key roles of NAC TFs in leaf senescence have been well studied in Arabidopsis. For example, ATAF1 (ARABIDOPSIS TRANSCRIPTION ACTIVATOR FACTOR 1) and ATAF2, 2 SNAC-A subfamily NAC TFs and representative senescence-associated genes, promote age-dependent and dark-induced leaf senescence (Garapati et al. 2015; Nagahage et al. 2020). A SNAC-A septuple mutant in Arabidopsis displayed delayed ABA-induced leaf senescence (Takasaki et al. 2015), and ANAC07 regulates both age- and dark-induced leaf senescence (Li et al. 2016). Through evolutionary analysis of nodule senescence–induced NACs, we established that 8 of 11 NAC genes in the SNAC-A subfamily and 5 of 23 NAC genes in the SNAC-B subfamily are induced in senescent nodules.
Based on the transcriptional changes that occur during leaf and nodule senescence, the SNAC-A and SNAC-B subfamily proteins are important components in the regulation of senescence. In soybean, GmNAC039, its paralog GmNAC018, and GmNAC030 belong to the SNAC-A subfamily and have identical expression patterns in senescent nodules. The essential roles of GmNAC039, GmNAC018, and GmNAC030 in mediating nodule senescence were confirmed by a variety of transgenic studies in transgenic hairy roots and stably transformed plants. Given that GmNAC030 was also shown to regulate soybean leaf senescence (Fraga et al. 2021), this gene might have dual roles in regulating both leaf and nodule senescence. Overall, these data support the roles of SNAC-A subfamily members not only in leaf senescence but also in nodule senescence during plant development.
In addition to SNAC-A TFs, 5 SNAC-B subfamily TFs (GmNAC127 and its paralog GmNAC006, GmNAC102 and its paralog GmNAC091, and GmNAC044) were also highly expressed in senescent nodules. However, their abilities to promote nodule senescence were not as strong as that of GmNAC039 or GmNAC018. Instead, GmNAC127, GmNAC006, and GmNAC102 play roles in soybean abiotic stress responses to salinity and drought (Tran et al. 2009; Hussain et al. 2017). The importance of SNAC-B TFs in response to environmental stresses has also been studied in Arabidopsis. ANAC047, the Arabidopsis ortholog of GmNAC127, increased plant salt tolerance when it was fused with the chimeric repressor SRDX (Mito et al. 2011). ANAC029, another SNAC-B TF, plays a positive role in salt-induced senescence in Arabidopsis (Guo and Gan 2006). Similarly, the rice (Oryza sativa) SNAC-B TF ONAC058 (OsNAP) is induced during the onset of leaf senescence and improves drought and salt tolerance when overexpressed in rice (Liang et al. 2014). In soybean, overexpression of 2 SNAC-B TFs, GmNAC127, and GmNAC091, also led to early senescence during nodule development, but the senescence phenotype was much weaker than that caused by overexpression of GmNAC039 and GmNAC018. Thus, although both SNAC-A TFs and SNAC-B TFs subfamilies function in soybean nodule senescence, SNAC-A TFs other than SNAC-B TFs seem to play prominent roles in this process.
NAC TFs are involved in various growth and developmental processes, as well as in environmental stress responses (Nuruzzaman et al. 2013). In general, many TFs function as activators or repressors of gene expression in different processes, and NAC TFs are certainly no exception (Fujita et al. 2004; Tran et al. 2004; Mendes et al. 2013). For example, ATAF1 serves as a core transcriptional activator of senescence by inducing the senescence-related gene ORESARA1 (ORE1) and repressing the photosynthesis-associated gene GOLDEN2-LIKE1 (GLK1) during developmental and stress-induced leaf senescence in Arabidopsis (Garapati et al. 2015). OsNAC6, a rice ortholog of Arabidopsis ATAF1, positively regulates disease resistance and salinity tolerance, whereas ATAF2 represses pathogenesis-related (PR) genes in Arabidopsis (Delessert et al. 2005; Nakashima et al. 2007).
Some TFs have dual functions as transcriptional activators and repressors (Sridhar et al. 2006; Vaquerizas et al. 2009; Mendes et al. 2013). GmNAC81 and GmNAC30 form homo- and heterodimers to function either as transcriptional activators or repressors in response to abiotic stresses (Mendes et al. 2013). These 2 GmNACs regulate programed cell death by transactivating the expression of the VPE gene (Mendes et al. 2013). Our data revealed that ectopic expression of GmNAC039 and GmNAC039-VP16 in nodules contributed to nodule senescence, whereas GmNAC039-SRDX did not. This result suggests that GmNAC039 functions as a transcriptional activator in accelerating nodule senescence.
Leaf senescence is often accompanied by a series of metabolic changes that allow nutrient recycling and remobilization (Diaz et al. 2008). Cysteine proteases are the most abundantly expressed gene family during leaf senescence in Arabidopsis (Guo et al. 2004). Cysteine protease genes are often markers for senescence, such as SAG12 (SENESCENCE-ASSOCIATED GENE 12), which is involved in Rubisco degradation during leaf senescence (James et al. 2018). A genome-wide survey identified 106 putative GmCYP genes in soybean (Yuan et al. 2020). There is increasing evidence suggesting that CYP expression is dramatically induced during nodule senescence in different legumes (Kardailsky and Brewin 1996; Pierre et al. 2014; Yuan et al. 2020). For example, a cysteine protease gene from M. truncatula, MtCP77, positively regulates nodule senescence by promoting plant programmed cell death and ROS accumulation (Deng et al. 2019).
Our RNA-seq results showed that several GmCYP genes from the same phylogenetic clade, GmCYP37, GmCYP35, GmCYP39, and GmCYP45, are strongly induced in GmNAC039-overexpressing nodules and naturally senescent nodules. The upregulation of these GmCYPs in GmNAC039-overexpressing nodules suggests that they likely play important roles in soybean nodule senescence. However, the degradation targets of the upregulated CYPs remain unknown, and their identification would be of great interest. Our in vivo and in vitro assay results showed that GmNAC039 specifically binds to a segment of the GmCYP37 promoter that is highly conserved in the promoters of GmCYP37, GmCYP35, and GmCYP39. The 29-bp region identified as the GmNAC039 binding site is an A/T-rich, CAC(A)A-containing motif, similar to the binding sites for Arabidopsis ATAF2 and soybean GmNAC030 (Wang and Culver 2012; Mendes et al. 2013). A conserved CAC(A)A motif was also identified in the GmCYP45 promoter, but not at the same position as in the GmCYP37 promoter, which is consistent with our results showing that GmNAC039 binds to different regions of the GmCYP37 and GmCYP45 promoters in vivo. Therefore, GmCYP37, GmCYP35, GmCYP39, and GmCYP45 are direct targets of GmNAC039 via binding to their conserved CAC(A)A motifs.
To distinguish between early senescence and developmental arrest in GmNAC039-OE or GmNAC018-OE nodules, analysis of the symbiosis genes and nodule senescence-associated genes is performed. GmNAC039-OE induced the expression of nodule senescence-associated genes DD15 and GmCYPs and suppressed the expression of Lbs genes and rhizobial nifH genes, suggesting that GmNAC039 has a role in promoting early senescence in soybean nodules. Senescence has been viewed as the last phase of development and induces developmental arrest. Thus, it was not surprising to see that nodules 4wpi generated from GmNAC039-OE and GmNAC018-OE plants were at smaller sizes than control nodules. Thus, we propose that GmNAC039 or GmNAC018 have primary roles in activating early senescence in nodules which then inevitably induces developmental arrest leading to smaller-size nodule formation. But, how senescence mediates developmental arrest is unclear.
Legume CYPs are putative hydrolytic enzymes that function as executors of cell death during nodule senescence, but their precise functions remain unknown. In M. truncatula , the nodule senescence-induced cysteine protease gene MtCP6 is exclusively expressed in the symbiosomes of infected cells (Pierre et al. 2014), indicating that this cysteine protease may directly participate in symbiosome degradation, leading to the formation of the central lytic vacuole in infected cells. It is predicted that a putative N-terminal endoplasmic reticulum- and/or vacuole-targeting signal peptide might direct CYP proteins to the large lytic vacuoles or to the bacteroid-containing symbiosomes to contribute to protein proteolysis and degradation (Vincent and Brewin 2000; Pierre et al. 2014).
In cells of senescent nodules, vacuoles and symbiosomes collapse to form a large lytic vacuolar compartment through membrane fusion (Limpens et al. 2009; Gavrin et al. 2014). The large lytic compartment formed during symbiosome degradation harbors typical vacuolar enzymes for lipid, nucleotide, and protein degradation in the peribacteroid space (Vincent and Brewin 2000; Van de Velde et al. 2006). Symbiosome degradation is similar to chloroplast degradation, during which the chloroplasts become encapsulated in cytoplasmic vesicles and are delivered to the vacuole/lysosome to be broken down by proteolytic enzymes (Dominguez and Cejudo 2021). Whether GmCYP-containing vesicles are involved in symbiosome degradation is currently unknown. Further experiments are required to identify GmCYP-interacting partners and target substrates to understand how CYPs mediate symbiosome degradation within host plant cells during nodule senescence.
Overall, our data provide valuable insights into the molecular mechanisms of nodule senescence in soybean. We identified 2 key NAC TFs, GmNAC039, and GmNAC018, which are essential for soybean nodule senescence through their transcriptional activation of multiple GmCYP genes. Thus, genetic modification of GmNAC–GmCYP module might be an efficient way to prolong active nitrogen fixation in nodules during soybean developmental stages with the potential to improve soybean production or seed quality. However, how GmNAC039 and GmNAC018 are programed by whole plant development and what are the targets of CYPs are 2 opening questions remaining to be answered in the future.
Materials and methods
Plant materials and growth conditions
Soybean ( G. max (L.) Merrill) cultivar Williams 82 (W82) was used in this study. For the W82 nodulation assay, soybean seeds were sterilized with chlorine gas for 14–18 h and germinated in pots filled with sterilized vermiculite in a greenhouse at 26 °C for 5 d under a day/night cycle of 16/8 h. Then, the seedlings were watered with Fahraeus medium (FM) containing 0.5 mm KNO3 and inoculated with 20 ml Bradyrhizobium diazoefficiens strain USDA 110 (OD600 = 0.01) for each pot. Soybean seedlings were grown at 400–710 nm, 200 µmol m −2 s −1 light condition (16-h light/8-h dark) in a growth room at 26 °C. The largest nodules at the base of the primary roots were collected at different time points post inoculation for symbiotic phenotyping and RNA isolation.
Plasmid construction
For gene overexpression, a 1261-bp promoter fragment upstream of the start codon of the M. truncatula leghemoglobin gene Lb2 (Medtr5g066070) was amplified and inserted between the PstI/XbaI restriction sites of the pUB-GFP-3xFlag plasmid (Yu et al. 2018) to replace the ubiquitin promoter, resulting pUB-GFP-pLb2-3xFlag. The full-length coding sequences of NAC genes and genomic DNA of CYPs genes were amplified and cloned into pUB-GFP-pLb2-3xFlag through XbaI/StuI double digestion. These constructs were introduced into Agrobacterium rhizogenes K599 for hairy root transformation. For gene knockout experiments, the CRISPR/Cas9 vector was constructed as previously reported (Yu et al. 2018). In brief, the ubiquitin promoter was amplified from pUB-GFP-3xFLAG and introduced into the KpnI/XhoI restriction sites of pBluescript sk(+)−2 × 35S–Cas9 to replace the 2 × 35S promoter (Wang et al. 2016b). Then, LjUbipro-Cas9 was introduced into the KpnI/EcoRI restriction sites of pCAMBIA1300-sGFP. The LjU6-gRNA fragments between the KpnI and XbaI sites of pBluescript SK(+)−LjU6-tRNA–gRNA vector were cloned into the pCAMBIA1300−sGFP LjUbipro-Cas9 vector, which was then used for gene knockouts via soybean hairy root transformation. The CRISPR/Cas9 target sequences were designed using the web tool CRISPR-P 2.0 (http://crispr.hzau.edu.cn/CRISPR2/). NAC039 (residues 1–159) was introduced into the EcoRI/SalI restriction sites of pGEX–4T1 (GenBank: U13853.1) to express glutathione S-transferase (GST)-tagged protein in Escherichia coli. All primers and vectors used for vector construction are listed in Supplemental Data Set 3.
Hairy root transformation
Hairy root transformation was performed as described previously with some modifications (Kereszt et al. 2007; Toth et al. 2016). In brief, soybean seeds were sterilized with chlorine gas for 14–18 h and germinated in sterilized vermiculite for 5 d. A double-edged blade was then used to cut the seedling about 1 cm above the hypocotyl to remove the root part. The cut ends of the shoots were inoculated with A. rhizogenes strain K599 carrying the gene construct and then placed in sterilized vermiculite medium under high humidity for 1–2 d in the dark followed by 3–5 d in the light. After removing the untransformed roots, plants were transferred to a 178 × 98 × 69-mm box with liquid FM and cultured for 8–10 d under 16-h light/8-h dark conditions at 26 °C. Successfully transformed roots were identified by the presence of GFP fluorescence, observed using a stereoscopic fluorescence microscope (Nikon) or portable dual fluorescent protein flashlights (LUYOR-3415RG). One healthy GFP-positive transgenic root was selected, and all other roots were removed. The base of the shoot was covered with a thin film to inhibit the formation of untransformed roots after planting. The transformed plants were transferred to vermiculite pots and inoculated with B. diazoefficiens USDA 110 (OD600 = 0.01, 20 ml) in liquid FM containing 0.5 mm KNO3 3 d later. The symbiotic phenotypes of the transformed plants were assessed 4 or 6 wpi. The largest nodules (3–5) on the base of the primary roots were used for sections and reverse transcription quantitative PCR (RT-qPCR). At least 20 transgenic hairy roots were generated and used for analysis in each experiment. For gene overexpression, nodule samples were harvested at 4 wpi. For gene knockout experiments, nodule samples were harvested at 6 wpi. At least 3 transgenic hairy roots were randomly selected to extract the genome DNA for determining the targeted DNA insertions/deletions in CRISPR/Cas9-based knockout experiments. The primer sequences used for generating vectors are shown in Supplemental Data Set 3. Toluidine blue staining and TUNEL (terminal deoxynucleotidyl transferase dUTP nick-end labeling) assay of soybean nodule sections from 20 to 30 samples were performed as described previously (Wang et al. 2016a; Xia 2018).
Stable transgenic soybean
The DNA fragments containing pUbi:Cas9 and pLjU6:gRNAs were digested with Eco RI and Xba I and ligated into pZY101 for stable soybean transformation mediated by Agrobacterium tumefaciens strain EHA105 (Zeng et al. 2004). Stably transformed soybean plants were generated using a previously reported Agrobacterium-mediated method (Chen et al. 2018).
Nitrogenase activity
An acetylene reduction activity (ARA) assay was used to measure the nitrogenase activity of root nodules. Acetylene gas was produced by reacting CaC2 (Sigma–Aldrich CAS No.: 75-20-7) with H2O and purified by filtering through a saturated CuSO4 solution. The nodulated roots from each plant were placed inside a sealed 40-ml vial. To begin the assay, 3 ml of the air inside the vial was removed and then replaced with 3 ml acetylene gas. The vials were placed upside down in a tray of water to keep them sealed and incubated at 28 °C for 2 h. Then, the vials were cooled with ice to stop the acetylene reduction reaction. One hundred microliters of the gas in each vial was used for the measurement of ethylene content (the product of acetylene reduction) using a GC–4000A gas chromatograph (Dongxi, Beijing, China). Nitrogenase activity was calculated by normalizing nodule fresh weight and/or per plant (Yu et al. 2018). For each experiment, root nodules from at least 20 soybean plants were used for analysis. Statistical analysis was carried out using GraphPad Prism version 9 based on a paired t-test; a probability value of less than 0.05 was regarded as statistically significant.
Reverse transcription quantitative PCR analysis
Total RNA was extracted using an EasyPure Plant RNA Kit (Yeasen, Shanghai, China) according to the manufacturer's instructions. The RNA samples were quantified using a NANODROP 2000 (Thermo–Fisher, Waltham, MA, USA). First-strand cDNA was synthesized from 1 μg of total RNA using a Primescript RT Reagent Kit (ABclonal, Wuhan, China). RT-qPCR was performed using SYBR Select Master Mix reagent (ABclonal, Wuhan, China) on an Applied Biosystems ViiA 7 Real-Time PCR System under the standard cycling conditions: 2 min at 50 °C, 10 min at 95 °C, followed by 40 cycles of 15 s at 94 °C and 1 min at 60 °C. The soybean Actin gene ( GmACT11 ) and bacterial 16S ribosomal RNA gene (16S) were used as an internal control. All the qPCR data were generated from 3 biological replicates with 3 technical repeats for each biological replicate. The primers are listed in Supplemental Data Set 3.
Electrophoretic mobility shift assay
EMSAs were performed as described previously (Xiao et al. 2020). The truncated NAC039 (residues 1–159) was fused with a GST tag. The GST-tagged fusion product was purified using GST MagBeads (GenScript, Nanjing, China) according to the manufacturer's instructions. The binding buffer contained 20 mm Tris–HCl (pH 8.0), 50 mm KCl, 1 mm MgCl2, 1 mm dithiothreitol (DTT), 0.5 mm EDTA, 0.05% Triton X–100 (v/v), and 5% glycerol (v/v). The oligonucleotide probes were labeled with FAM at the 5′ end. Unlabeled oligonucleotides of the same sequence were used as competitors. The oligonucleotides used are listed in Supplemental Data Set 3.
Multiple alignment and phylogenetic analysis
The relevant protein and promotor sequences of G. max are available at Phytozome (https://phytozome-next.jgi.doe.gov/). The protein alignment was conducted using Clustal Omega on the EMBL-EBI website (https://www.ebi.ac.uk/Tools/msa/clustalo/). The promoter alignment was conducted using MUSCLE on the EMBL-EBI website (https://www.ebi.ac.uk/Tools/msa/muscle/). The analyzed amino acid and nucleotide sequences were edited using ESPript 3.0 (https://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi). The upregulated NAC genes in the 12 wpi/4 wpi and 8 wpi/4 wpi comparisons and the upregulated CYPs common to GmNAC039-OE and 12-wpi nodules were used for phylogenetic analyses. The protein sequences were matched using MAFFT (GitHub–GSLBiotech/mafft: Align multiple amino acid or nucleotide sequences), and the phylogenetic tree was constructed using FastTree (FastTree 2.1: approximately–Maximum–Likelihood Trees for Large Alignments (microbesonline.org)) based on the maximum likelihood method.
Transcriptome analysis
Several of the largest nodules at the base of the primary roots were collected at 4, 8, and 12 wpi. Nodules overexpressing GmNAC039, which were green in color, were harvested at 4 wpi. Three individual samples were sequenced using Illumina HiseqPE150. Raw sequence files from the Illumina pipeline were evaluated using FastQC software (GitHub–s-andrews/FastQC). Next, we used STAR (GitHub–alexdobin/STAR: RNA-seq aligner) to generate the soybean (Wm82.a4.v1, SoyBase.org) genome index file, specifying a length of 149 (MaxReadLength-1) as the genomic sequence around the annotated junction to be used in constructing the splice junction database (Dobin et al. 2013). RSEM (deweylab/RSEM (github.com)) was used to extract reference transcripts from the soybean genome with gene annotations in a general transfer format file to calculate expression values (Li and Dewey 2011). The STAR aligner was used to obtain RSEM alignments of reads to reference transcripts. Abundance was determined as the estimated fraction of transcripts aligned to a given isform or gene. Subsequently, the differential expression analysis was performed using the R package DESeq2, and the differentially expressed genes were screened with Log2 fold change ≥ 1, E–value < 0.05, followed by GO annotation and KEGG annotation (Love et al. 2014).
Nucleus CUT&Tag-qPCR (nCUT&Tag-qPCR)
GmNAC039–3×FLAG overexpression nodules were harvested at 3 wpi for nCUT&Tag-qPCR assays, which were performed as described previously with modifications (Kaya-Okur et al. 2020; Bartosovic et al. 2021; Ouyang et al. 2021). About 0.1 g of fresh nodules was chopped thoroughly to complete homogeneity in a plastic petri dish with 1 ml phosphate-buffered saline (1× PBS, pH 7.4) containing protease inhibitor (P9599, Sigma–Aldrich) on ice. The homogenate was filtered through 2 layers of Miracloth to remove large debris, followed by separation of plant cell nuclei from the rhizobia using 5 μm polyethersulfone (PES) film. The PES film was washed twice with 1 ml 1× PBS to remove residual rhizobia and rinsed with 1 ml 1× PBS to obtain the nuclei. The nuclei were isolated by centrifuging the filtrate in a swinging bucket rotor at 1000 g for 10 min at 4 °C. The nuclei pellet was then washed twice with 500 µl ice-cold wash buffer: 20 mm HEPES buffer (pH 7.5), 150 mm NaCl, 0.5 mm spermidine, and protease inhibitor. The nuclei were resuspended with 100 µl wash buffer. The collected nuclei were stained with 10 μg/ml DAPI and observed using a fluorescence microscope (Nikon ECLIPSE 80i, Tokyo, Japan). The following procedures were performed according to the instructions of the CUT&Tag kit (Yeasen, Shanghai, China). In brief: (i) the nuclei were bound to ConA beads, (ii) the bound nuclei were incubated with primary antibody (anti-FLAG: F1804, Sigma–Aldrich), (iii) the bound nuclei were incubated with secondary antibody (Goat Anti-Mouse IgG H&L: ab6708 Abcam), (iv) the bound nuclei were incubated with pA/G-Transposome adapter complex, and the transposase was activated, (v) the genomic DNA was released by Proteinase K digestion and recovered, and (vi) the genomic DNA was subjected to PCR amplification and purification. DNA library pooling without the addition of primary antibody was used as the negative control. qPCR conditions were as follows: initial denaturation, 50 °C for 2 min followed by 95 °C for 10 min; amplification, 40 cycles of 95 °C for 15 s, 60 °C for 30 s, and 72 °C for 20 s. The reaction mix with SYBR was prepared according to the manufacturer's instructions (ABclonal, Wuhan, China). CUT&Tag-qPCR experiments were performed with 2 biological replicates with 3 technical repeats for each assay. Primers used for the real-time PCR are listed in Supplemental Data Set 3.
Random DNA-binding selection assay
The Random DNA-Binding Selection Assay (RDSA) was performed according to previously described procedures with some modifications (Wang and Luo 2015). The double-stranded DNA fragments used for RDSA consisted of 23-bp random sequence regions flanked by adaptors for PCR amplification using RDSA-F and RDSA-R primers. The reaction conditions were as follows: 5 min at 94 °C; 30 cycles of 30 s at 94 °C, 30 s at 50 °C, and 30 s at 72 °C; 10 min at 72 °C. Truncated GST-NAC039 (residues 1–159) bound to anti-GST magnetic beads was incubated with the RDSA random DNA fragments (5–10 μg) in 500 μl RDSA buffer (5 mm Tris–HCl (pH 8.0), 75 mm NaCl, 2.5 mm MgCl2, 0.5 mm EDTA, 5% glycerol (v/v), 1% Tween (v/v), and 1 mm DTT) for at least 2 h at 4 °C with gentle rolling. The bound DNA was purified with an equal volume of 24:1 (chloroform:isoamyl alcohol). The purified DNA was used for the next round of PCR amplification. The PCR products were purified using SiO2. The DNA templates and amplification cycles require proper adjustment for each RDSA cycle. After 8 or more rounds of binding and amplification, the eluted DNA was cloned and ligated into the vector pGEM-T (Cat#A1360, Promega). Individual clones were then randomly selected for sequencing. The consensus sequence was analyzed online (http://weblogo.berkeley.edu/). In each RDSA cycle, BSA and increasing amounts of the nonspecific competitor poly (dIdC) (0, 50, 100, 150, 200, 250, 300, and 350 ng) were added to increase the stringency.
Statistical analysis
Statistical tests used are listed in material methods or figure legends. Statistical data are provided in Supplemental Data Set 4.
Accession numbers
Sequence data from this article can be found in the Phytozome database (https://phytozome-next.jgi.doe.gov/) under the following accession numbers: LbA (Glyma.10G199100), LbC1 (Glyma.10G199000), LbC2 (Glyma.20G191200), LbC3 (Glyma.10G198800), NAC134 (Glyma.17G185000), NAC065 (Glyma.08G360200), NAC179 (Glyma.18G301500), NAC077 (Glyma.11G096600), NAC116 (Glyma.15G257700), NAC137 (Glyma.18G110700), NAC154 (Glyma.02G284300), NAC106 (Glyma.14G030700), NAC114 (Glyma.15G078300), NAC142 (Glyma.19G056400), NAC170 (Glyma.10G204700), NAC092 (Glyma.12G221500), NAC085 (Glyma.12G149100), NAC043 (Glyma.06G248900), NAC035 (Glyma.06G114000), NAC022 (Glyma.04G249000), NAC030 (Glyma.05G195000), NAC039 (Glyma.06G157400), NAC018 (Glyma.04G208300), NAC006 (Glyma.02G070000), NAC127 (Glyma.16G151500), NAC044 (Glyma.06G249100), NAC091 (Glyma.12G221400), and NAC102 (Glyma.13G280000). The sequence alignments and tree file for the above NAC proteins are shown in Supplemental Files 1 and 2, respectively. CYP92 (Glyma.16G095900), CYP95 (Glyma.17G126300), CYP76 (Glyma.12G131200), CYP36 (Glyma.06G278200), CYP45 (Glyma.06G283100), CYP35 (Glyma.06G278000), CYP37 (Glyma.06G279100), CYP39 (Glyma.06G279900), CYP46 (Glyma.07G169900), CYP6 (Glyma.04G027600), CYP94 (Glyma.17G049000), CYP93 (Glyma.16G107200), CYP51 (Glyma.08G116400), CYP12 (Glyma.05G159100), and CYP7 (Glyma.04G028300). The sequence alignments and tree file for the above CYP proteins are shown in Supplemental Data Files 3 and 4, respectively. The RNA-seq data generated in this study have been deposited at the National Center for Biotechnology Information Sequence Read Archive (SRA) database under accession number: PRJNA843914.
Supplementary Material
Acknowledgments
The authors thank Prof. Kabin Xie for kindly providing the plasmid pGTR, Prof. Zhongming Zhang for providing valuable suggestions on the research, Dr. Yong Feng for providing help on transcriptomic data analysis, and Xianpeng Zhang and Zeyuan Xu for providing some materials of soybean nodulation assay. We thank other undergraduates and graduates in the Cao's Lab for providing help on soybean nodulation assay.
Contributor Information
Haixiang Yu, National Key Laboratory of Agricultural Microbiology, Hubei Hongshan Laboratory, College of Life Science and Technology, Huazhong Agricultural University, Wuhan, Hubei 430070, China.
Aifang Xiao, National Key Laboratory of Agricultural Microbiology, Hubei Hongshan Laboratory, College of Life Science and Technology, Huazhong Agricultural University, Wuhan, Hubei 430070, China.
Jiashan Wu, National Key Laboratory of Agricultural Microbiology, Hubei Hongshan Laboratory, College of Life Science and Technology, Huazhong Agricultural University, Wuhan, Hubei 430070, China.
Haoxing Li, National Key Laboratory of Agricultural Microbiology, Hubei Hongshan Laboratory, College of Life Science and Technology, Huazhong Agricultural University, Wuhan, Hubei 430070, China.
Yan Duan, National Key Laboratory of Agricultural Microbiology, Hubei Hongshan Laboratory, College of Life Science and Technology, Huazhong Agricultural University, Wuhan, Hubei 430070, China.
Qingshan Chen, Key Laboratory of Soybean Biology of Chinese Ministry of Education, Key Laboratory of Soybean Biology and Breeding/Genetics of Chinese Agriculture Ministry, Northeast Agricultural University, Harbin, Heilongjiang 150038, China.
Hui Zhu, National Key Laboratory of Agricultural Microbiology, Hubei Hongshan Laboratory, College of Life Science and Technology, Huazhong Agricultural University, Wuhan, Hubei 430070, China.
Yangrong Cao, National Key Laboratory of Agricultural Microbiology, Hubei Hongshan Laboratory, College of Life Science and Technology, Huazhong Agricultural University, Wuhan, Hubei 430070, China.
Author contributions
H.Y., A.X., and Y.C. conceived the idea and designed the research; H.Y., A.X., and J.W. performed most of the experiments; Y.D. performed some experiments on vector construction; HL analyzed the sequencing data; H.Y. and A.X. analyzed the data; H.Z. and Q.C. provided valuable suggestions for the study; H.Y., A.X., and Y.C. wrote the paper.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Nodulation phenotype of W82 at different growth stages.
Supplemental Figure S2. RNA-Seq analysis of soybean nodules at 4, 8, and 12 wpi.
Supplemental Figure S3. Phenotypes of transgenic nodules expressing NAC family genes.
Supplemental Figure S4. Nodule phenotypes of transgenic roots expressing GmNAC039 and GmNAC018 at 2 wpi.
Supplemental Figure S5. Protein sequence alignment and nodule phenotypes of CRISPR/Cas9-mediated GmNAC039/GmNAC018/GmNAC030 knockout hairy roots.
Supplemental Figure S6. Growth phenotype of NACs-KO mutant soybean plants.
Supplemental Figure S7. GO and KEGG analysis of DEGs in GmNAC039-OE nodules.
Supplemental Figure S8. The consensus binding sequence of GmNAC039
Supplemental Figure S9. Isolation of nodule nuclei from GmNAC039-3×FLAG transgenic nodules.
Supplemental Figure S10. GmCYP35, GmCYP37, GmCYP39, and GmCYP45 protein sequence alignment.
Supplemental Figure S11. Binding of GmNAC039 to the GmCYP7 promoter and phenotypes of GmCYP7-overexpressing roots.
Supplemental Figure S12. CYP gene editing by CRISPR/Cas9 in transgenic nodules.
Supplemental Data Set 1. RNA-Seq analysis of soybean nodules at 4, 8, and 12 wpi.
Supplemental Data Set 2. RNA-Seq analyses of GmNAC039-OE nodules vs. control nodules and 12W vs. 4W nodules.
Supplemental Data Set 3. Primers used in this study.
Supplemental Data Set 4. Statistical data.
Supplemental File 1. The sequence alignments for NAC proteins shown in Fig. 1 .
Supplemental File 2. The tree file for NAC proteins shown in Fig. 1 .
Supplemental File 3. The sequence alignments for CYP proteins shown in Fig. 5 .
Supplemental File 4. The tree file for NAC proteins shown in Fig. 5 .
Supplemental File 5. The sequence alignments for transcription factors shown in Supplemental Fig. S2.
Supplemental File 6. The tree file for transcription factors shown in Supplemental Fig. S2.
Funding
This work was supported by the National Key R&D Program of China (2021YFF1001200), the National Natural Science Foundation of China (32090063, 32001438, and 32000191), the Natural Science Foundation of Hubei Province (2020CFA008 and 2020CFB289). H.Y., A.X., and Y.C. were supported by BaiChuan and Longyun Programs from the College of Life Science and Technology at the HZAU.
References
- Alesandrini F, Frendo P, Puppo A, Hérouart D. Isolation of a molecular marker of soybean nodule senescence. Plant Physiol Biochem. 2003a:41(8):727–732. 10.1016/S0981-9428(03)00110-4 [DOI] [Google Scholar]
- Alesandrini F, Mathis R, Van de Sype G, Herouart D, Puppo A. Possible roles for a cysteine protease and hydrogen peroxide in soybean nodule development and senescence. New Phytol. 2003b:158(1):131–138. 10.1046/j.1469-8137.2003.00720.x [DOI] [Google Scholar]
- Balazadeh S, Kwasniewski M, Caldana C, Mehrnia M, Zanor MI, Xue GP, Mueller-Roeber B. ORS1, an H2O2-responsive NAC transcription factor, controls senescence in Arabidopsis thaliana. Mol Plant. 2011:4(2):346–360. 10.1093/mp/ssq080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazadeh S, Siddiqui H, Allu AD, Matallana-Ramirez LP, Caldana C, Mehrnia M, Zanor MI, Kohler B, Mueller-Roeber B. A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J. 2010:62(2):250–264. 10.1111/j.1365-313X.2010.04151.x [DOI] [PubMed] [Google Scholar]
- Bartosovic M, Kabbe M, Castelo-Branco G. Single-cell CUT&tag profiles histone modifications and transcription factors in complex tissues. Nat Biotechnol. 2021:39(7):825–835. 10.1038/s41587-021-00869-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrabah F, Bernal G, Elhosseyn AS, Kassis CE, L'Horset R, Benaceur F, Wen J, Mysore KS, Garmier M, Gourion B, et al. . Insight into the control of nodule immunity and senescence during Medicago truncatula symbiosis. Plant Phyiol. 2023:191(1):729–746. 10.1093/plphys/kiac505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourcy M, Brocard L, Pislariu CI, Cosson V, Mergaert P, Tadege M, Mysore KS, Udvardi MK, Gourion B, Ratet P. Medicago truncatula DNF2 is a PI-PLC-XD-containing protein required for bacteroid persistence and prevention of nodule early senescence and defense-like reactions. New Phytol. 2013:197(4):1250–1261. 10.1111/nph.12091 [DOI] [PubMed] [Google Scholar]
- Chen L, Cai Y, Liu X, Yao W, Guo C, Sun S, Wu C, Jiang B, Han T, Hou W. Improvement of soybean agrobacterium-mediated transformation efficiency by adding glutamine and asparagine into the culture media. Int J Mol Sci. 2018:19(10):3039. 10.3390/ijms19103039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Feng P, Tang B, Hu Z, Xie Q, Zhou S, Chen G. The AP2/ERF transcription factor SlERF.F5 functions in leaf senescence in tomato. Plant Cell Rep. 2022:41(5):1181–1195. 10.1007/s00299-022-02846-1 [DOI] [PubMed] [Google Scholar]
- Chungopast S, Hirakawa H, Sato S, Handa Y, Saito K, Kawaguchi M, Tajima S, Nomura M. Transcriptomic profiles of nodule senescence in Lotus japonicus and Mesorhizobium loti symbiosis. Plant Biotechnol. 2014:31(4):345–349. 10.5511/plantbiotechnology.14.1021a [DOI] [Google Scholar]
- Delessert C, Kazan K, Wilson IW, Van Der Straeten D, Manners J, Dennis ES, Dolferus R. The transcription factor ATAF2 represses the expression of pathogenesis-related genes in Arabidopsis. Plant J. 2005:43(5):745–757. 10.1111/j.1365-313X.2005.02488.x [DOI] [PubMed] [Google Scholar]
- Deng J, Zhu F, Liu J, Zhao Y, Wen J, Wang T, Dong J. Transcription factor bHLH2 represses CYSTEINE PROTEASE 77 to negatively regulate nodule senescence. Plant Physiol. 2019:181(4):1683–1703. 10.1104/pp.19.00574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz C, Lemaitre T, Christ A, Azzopardi M, Kato Y, Sato F, Morot-Gaudry JF, Le Dily F, Masclaux-Daubresse C. Nitrogen recycling and remobilization are differentially controlled by leaf senescence and development stage in Arabidopsis under low nitrogen nutrition. Plant Physiol. 2008:147(3):1437–1449. 10.1104/pp.108.119040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-Seq aligner. Bioinformatics. 2013:29(1):15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez F, Cejudo FJ. Chloroplast dismantling in leaf senescence. J Exp Bot. 2021:72(16):5905–5918. 10.1093/jxb/erab200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domonkos Á, Kovács S, Gombár A, Kiss E, Horváth B, Kováts GZ, Farkas A, Tóth MT, Ayaydin F, Bóka K, et al. . NAD1 Controls defense-like responses in Medicago truncatula symbiotic nitrogen fixing nodules following rhizobial colonization in a BacA-independent manner. Genes (Basel). 2017:8(12):387. 10.3390/genes8120387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Yang X, Liu J, Wang BH, Liu BL, Wang YZ. Pod shattering resistance associated with domestication is mediated by a NAC gene in soybean. Nat Commun. 2014:5(1):3352. 10.1038/ncomms4352 [DOI] [PubMed] [Google Scholar]
- Ferguson BJ, Indrasumunar A, Hayashi S, Lin MH, Lin YH, Reid DE, Gresshoff PM. Molecular analysis of legume nodule development and autoregulation. J Integr Plant Biol. 2010:52(1):61–76. 10.1111/j.1744-7909.2010.00899.x [DOI] [PubMed] [Google Scholar]
- Fraga OT, de Melo BP, Quadros IPS, Reis PAB, Fontes EPB. Senescence-associated Glycine max (Gm)NAC genes: integration of natural and stress-induced leaf senescence. Int J Mol Sci. 2021:22(15):8287. 10.3390/ijms22158287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M, Tran LS, Yamaguchi-Shinozaki K, Shinozaki K. A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 2004:39(6):863–876. 10.1111/j.1365-313X.2004.02171.x [DOI] [PubMed] [Google Scholar]
- Garapati P, Xue GP, Munne-Bosch S, Balazadeh S. Transcription factor ATAF1 in Arabidopsis promotes senescence by direct regulation of key chloroplast maintenance and senescence transcriptional cascades. Plant Physiol. 2015:168(3):1122–1139. 10.1104/pp.15.00567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrin A, Kaiser BN, Geiger D, Tyerman SD, Wen ZY, Bisseling T, Fedorova EE. Adjustment of host cells for accommodation of symbiotic bacteria: vacuole defunctionalization, HOPS suppression, and TIP1g retargeting in Medicago. Plant Cell. 2014:26(9):3809–3822. 10.1105/tpc.114.128736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Cai Z, Gan S. Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ. 2004:27(5):521–549. 10.1111/j.1365-3040.2003.01158.x [DOI] [Google Scholar]
- Guo Y, Gan S. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 2006:46(4):601–612. 10.1111/j.1365-313X.2006.02723.x [DOI] [PubMed] [Google Scholar]
- Hao YJ, Song QX, Chen HW, Zou HF, Wei W, Kang XS, Ma B, Zhang WK, Zhang JS, Chen SY. Plant NAC-type transcription factor proteins contain a NARD domain for repression of transcriptional activation. Planta. 2010:232(5):1033–1043. 10.1007/s00425-010-1238-2 [DOI] [PubMed] [Google Scholar]
- Hao YJ, Wei W, Song QX, Chen HW, Zhang YQ, Wang F, Zou HF, Lei G, Tian AG, Zhang WK, et al. . Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J. 2011:68(2):302–313. 10.1111/j.1365-313X.2011.04687.x [DOI] [PubMed] [Google Scholar]
- Hussain RM, Ali M, Feng X, Li X. The essence of NAC gene family to the cultivation of drought-resistant soybean (Glycine max L. Merr.) cultivars. BMC Plant Biol. 2017:17(1):55. 10.1186/s12870-017-1001-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imsande J. Ineffective utilization of nitrate by soybean during pod fill. Physiol Plant. 1986:68(4):689–694. 10.1111/j.1399-3054.1986.tb03419.x [DOI] [Google Scholar]
- Imsande J, Schmidt JM. Effect of N source during soybean pod filling on nitrogen and sulfur assimilation and remobilization. Plant Soil. 1998:202(1):41–47. 10.1023/A:1004313326745 [DOI] [Google Scholar]
- James M, Poret M, Masclaux-Daubresse C, Marmagne A, Coquet L, Jouenne T, Chan P, Trouverie J, Etienne P. SAG12, A major cysteine protease involved in nitrogen allocation during senescence for seed production in Arabidopsis thaliana. Plant Cell Physiol. 2018:59(10):2052–2063. 10.1093/pcp/pcy125 [DOI] [PubMed] [Google Scholar]
- Jaradat MR, Feurtado JA, Huang D, Lu Y, Cutler AJ. Multiple roles of the transcription factor AtMYBR1/AtMYB44 in ABA signaling, stress responses, and leaf senescence. BMC Plant Biol. 2013:13(1):192. 10.1186/1471-2229-13-192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky IV, Brewin NJ. Expression of cysteine protease genes in pea nodule development and senescence. Mol Plant Microbe Interact. 1996:9(8):689–695. 10.1094/MPMI-9-0689 [DOI] [PubMed] [Google Scholar]
- Kaya-Okur HS, Janssens DH, Henikoff JG, Ahmad K, Henikoff S. Efficient low-cost chromatin profiling with CUT&tag. Nat Protoc. 2020:15(10):3264–3283. 10.1038/s41596-020-0373-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmierczak T, Yang L, Boncompagni E, Meilhoc E, Frugier F, Frendo P, Bruand C, Gruber V, Brouquisse R. Legume nodule senescence: a coordinated death mechanism between bacteria and plant cells. Adv Bot Res. 2020:94:181–212. 10.1016/bs.abr.2019.09.013 [DOI] [Google Scholar]
- Kereszt A, Li D, Indrasumunar A, Nguyen CD, Nontachaiyapoom S, Kinkema M, Gresshoff PM. Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat Protoc. 2007:2(4):948–952. 10.1038/nprot.2007.141 [DOI] [PubMed] [Google Scholar]
- Kim HJ, Nam HG, Lim PO. Regulatory network of NAC transcription factors in leaf senescence. Curr Opin Plant Biol. 2016:33:48–56. 10.1016/j.pbi.2016.06.002 [DOI] [PubMed] [Google Scholar]
- Lehtovaara P, Perttila U. Bile-pigment formation from different leghaemoglobins. Methine-bridge specificity of coupled oxidation. Biochem J. 1978:176(2):359–364. 10.1042/bj1760359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Chen R, Jiang Q, Sun X, Zhang H, Hu Z. GmNAC06, a NAC domain transcription factor enhances salt stress tolerance in soybean. Plant Mol Biol. 2021:105(3):333–345. 10.1007/s11103-020-01091-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011:12(1):323. 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Gao J, Yao LY, Ren GD, Zhu XY, Gao S, Qiu K, Zhou X, Kuai BK. The role of ANAC072 in the regulation of chlorophyll degradation during age- and dark-induced leaf senescence. Plant Cell Rep. 2016:35(8):1729–1741. 10.1007/s00299-016-1991-1 [DOI] [PubMed] [Google Scholar]
- Li S, Wang N, Ji DD, Zhang WX, Wang Y, Yu YC, Zhao SZ, Lyu MH, You JJ, Zhang YY, et al. . A GmSIN1/GmNCED3s/GmRbohBs feed-forward loop acts as a signal amplifier that regulates root growth in soybean exposed to salt stress. Plant Cell. 2019:31(9):2107–2130. 10.1105/tpc.18.00662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Woo HR, Guo H. Genetic redundancy of senescence-associated transcription factors in Arabidopsis. J Exp Bot. 2018:69(4):811–823. 10.1093/jxb/erx345 [DOI] [PubMed] [Google Scholar]
- Li Y, Zhou L, Li Y, Chen D, Tan X, Lei L, Zhou J. A nodule-specific plant cysteine proteinase, AsNODF32, is involved in nodule senescence and nitrogen fixation activity of the green manure legume Astragalus sinicus. New Phytol. 2008:180(1):185–192. 10.1111/j.1469-8137.2008.02562.x [DOI] [PubMed] [Google Scholar]
- Liang C, Wang Y, Zhu Y, Tang J, Hu B, Liu L, Ou S, Wu H, Sun X, Chu J, et al. . OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc Natl Acad Sci USA. 2014:111(27):10013–10018. 10.1073/pnas.1321568111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens E, Ivanov S, van Esse W, Voets G, Fedorova E, Bisseling T. Medicago N2-fixing symbiosomes acquire the endocytic identity marker rab7 but delay the acquisition of vacuolar identity. Plant Cell. 2009:21(9):2811–2828. 10.1105/tpc.108.064410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Rasing M, Zeng T, Klein J, Kulikova O, Bisseling T. NIN Is essential for development of symbiosomes, suppression of defence and premature senescence in Medicago truncatula nodules. New Phytol. 2021:230(1):290–303. 10.1111/nph.17215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 2014:15(12):550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matallana-Ramirez LP, Rauf M, Farage-Barhom S, Dortay H, Xue GP, Droge-Laser W, Lers A, Balazadeh S, Mueller-Roeber B. NAC Transcription factor ORE1 and senescence-induced BIFUNCTIONAL NUCLEASE1 (BFN1) constitute a regulatory cascade in Arabidopsis. Mol Plant. 2013:6(5):1438–1452. 10.1093/mp/sst012 [DOI] [PubMed] [Google Scholar]
- Maunoury N, Redondo-Nieto M, Bourcy M, Van de Velde W, Alunni B, Laporte P, Durand P, Agier N, Marisa L, Vaubert D, et al. . Differentiation of symbiotic cells and endosymbionts in Medicago truncatula nodulation are coupled to two transcriptome-switches. PLoS One. 2010:5(3):e9519. 10.1371/journal.pone.0009519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo BP, Fraga OT, Silva JCF, Ferreira DO, Brustolini OJB, Carpinetti PA, Machado JPB, Reis PAB, Fontes EPB. Revisiting the soybean GmNAC superfamily. Front Plant Sci. 2018:18(9):1864. 10.3389/fpls.2018.01864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes GC, Reis PA, Calil IP, Carvalho HH, Aragao FJ, Fontes EP. GmNAC30 and GmNAC81 integrate the endoplasmic reticulum stress- and osmotic stress-induced cell death responses through a vacuolar processing enzyme. Proc Natl Acad Sci USA. 2013:110(48):19627–19632. 10.1073/pnas.1311729110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito T, Seki M, Shinizaki K, Ohme-Takagi M, Matsui K. Generation of chimeric repressors that confer salt tolerance in Arabidopsis and rice. Plant Biotechnol J. 2011:9(7):736–746. 10.1111/j.1467-7652.2010.00578.x [DOI] [PubMed] [Google Scholar]
- Mus F, Crook MB, Garcia K, Costas AG, Geddes BA, Kouri ED, Paramasivan P, Ryu MH, Oldroyd GED, Poole PS, et al. . Symbiotic nitrogen fixation and the challenges to its extension to nonlegumes. Appl Environ Microbiol. 2016:82(13):3698–3710. 10.1128/AEM.01055-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahage ISP, Sakamoto S, Nagano M, Ishikawa T, Kawai-Yamada M, Mitsuda N, Yamaguchi M. An NAC domain transcription factor ATAF2 acts as transcriptional activator or repressor dependent on promoter context. Plant Biotechnol. 2018:35(3):285–289. 10.5511/plantbiotechnology.18.0507a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahage ISP, Sakamoto S, Nagano M, Ishikawa T, Mitsuda N, Kawai-Yamada M, Yamaguchi M. An Arabidopsis NAC domain transcription factor, ATAF2, promotes age-dependent and dark-induced leaf senescence. Physiol Plant. 2020:170(2):299–308. 10.1111/ppl.13156 [DOI] [PubMed] [Google Scholar]
- Naito Y, Fujie M, Usami S, Murooka Y, Yamada T. The involvement of a cysteine proteinase in the nodule development in Chinese milk vetch infected with Mesorhizobium huakuii subsp. rengei. Plant Physiol. 2000:124(3):1087–1096. 10.1104/pp.124.3.1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Tran LS, Van Nguyen D, Fujita M, Maruyama K, Todaka D, Ito Y, Hayashi N, Shinozaki K, Yamaguchi-Shinozaki K. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 2007:51(4):617–630. 10.1111/j.1365-313X.2007.03168.x [DOI] [PubMed] [Google Scholar]
- Navascues J, Perez-Rontome C, Gay M, Marcos M, Yang F, Walker FA, Desbois A, Abian J, Becana M. Leghemoglobin green derivatives with nitrated hemes evidence production of highly reactive nitrogen species during aging of legume nodules. Proc Natl Acad Sci USA. 2012:109(7):2660–2665. 10.1073/pnas.1116559109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuruzzaman M, Sharoni AM, Kikuchi S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front Microbiol. 2013:3(4):248. 10.3389/fmicb.2013.00248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GE, Dixon R. Biotechnological solutions to the nitrogen problem. Curr Opin Biotechnol. 2014:26:19–24. 10.1016/j.copbio.2013.08.006 [DOI] [PubMed] [Google Scholar]
- Ouyang W, Zhang X, Peng Y, Zhang Q, Cao Z, Li G, Li X. Rapid and low-input profiling of histone marks in plants using nucleus CUT&tag. Front Plant Sci. 2021:12(12):634679. 10.3389/fpls.2021.634679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez Guerra JC, Coussens G, De Keyser A, De Rycke R, De Bodt S, Van De Velde W, Goormachtig S, Holsters M. Comparison of developmental and stress-induced nodule senescence in Medicago truncatula. Plant Physiol. 2010:152(3):1574–1584. 10.1104/pp.109.151399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phukan UJ, Jeena GS, Tripathi V, Shukla RK. Regulation of Apetala2/ethylene response factors in plants. Front Plant Sci. 2017:21(8):150. 10.3389/fpls.2017.00150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao W, Kim SH, Lee BD, An G, Sakuraba Y, Paek NC. Rice transcription factor OsMYB102 delays leaf senescence by down-regulating abscisic acid accumulation and signaling. J Exp Bot. 2019:70(10):2699–2715. 10.1093/jxb/erz095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre O, Hopkins J, Combier M, Baldacci F, Engler G, Brouquisse R, Herouart D, Boncompagni E. Involvement of papain and legumain proteinase in the senescence process of Medicago truncatula nodules. New Phytol. 2014:202(3):849–863. 10.1111/nph.12717 [DOI] [PubMed] [Google Scholar]
- Puppo A, Groten K, Bastian F, Carzaniga R, Soussi M, Lucas MM, de Felipe MR, Harrison J, Vanacker H, Foyer CH. Legume nodule senescence: roles for redox and hormone signalling in the orchestration of the natural aging process. New Phytol. 2005:165(3):683–701. 10.1111/j.1469-8137.2004.01285.x [DOI] [PubMed] [Google Scholar]
- Roy S, Liu W, Nandety RS, Crook A, Mysore KS, Pislariu CI, Frugoli J, Dickstein R, Udvardi MK. Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation. Plant Cell. 2020:32(1):15–41. 10.1105/tpc.19.00279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiltz S, Munier-Jolain N, Jeudy C, Burstin J, Salon C. Dynamics of exogenous nitrogen partitioning and nitrogen remobilization from vegetative organs in pea revealed by N-15 in vivo labeling throughout seed filling. Plant Physiol. 2005:137(4):1463–1473. 10.1104/pp.104.056713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheokand S, Brewin NJ. Cysteine proteases in nodulation and nitrogen fixation. Indian J Exp Biol. 2003:41:1124–1132. [PubMed] [Google Scholar]
- Sridhar VV, Surendrarao A, Liu Z. APETALA1 And SEPALLATA3 interact with SEUSS to mediate transcription repression during flower development. Development. 2006:133(16):3159–3166. 10.1242/dev.02498 [DOI] [PubMed] [Google Scholar]
- Takasaki H, Maruyama K, Takahashi F, Fujita M, Yoshida T, Nakashima K, Myouga F, Toyooka K, Yamaguchi-Shinozaki K, Shinozaki K. SNAC-as, stress-responsive NAC transcription factors, mediate ABA-inducible leaf senescence. Plant J. 2015:84(6):1114–1123. 10.1111/tpj.13067 [DOI] [PubMed] [Google Scholar]
- Toth K, Batek J, Stacey G. Generation of soybean (Glycine max) transient transgenic roots. Curr Protoc Plant Biol. 2016:1(1):1–13. 10.1002/cppb.20017 [DOI] [PubMed] [Google Scholar]
- Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell. 2004:16(9):2481–2498. 10.1105/tpc.104.022699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran LS, Quach TN, Guttikonda SK, Aldrich DL, Kumar R, Neelakandan A, Valliyodan B, Nguyen HT. Molecular characterization of stress-inducible GmNAC genes in soybean. Mol Genet Genomics. 2009:281(6):647–664. 10.1007/s00438-009-0436-8 [DOI] [PubMed] [Google Scholar]
- Van de Velde W, Guerra JC, De Keyser A, De Rycke R, Rombauts S, Maunoury N, Mergaert P, Kondorosi E, Holsters M, Goormachtig S. Aging in legume symbiosis. A molecular view on nodule senescence in Medicago truncatula. Plant Physiol. 2006:141(2):711–720. 10.1104/pp.106.078691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wyk SG, Du Plessis M, Cullis CA, Kunert KJ, Vorster BJ. Cysteine protease and cystatin expression and activity during soybean nodule development and senescence. BMC Plant Biol. 2014:14(1):294. 10.1186/s12870-014-0294-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat Rev Genet. 2009:10(4):252–263. 10.1038/nrg2538 [DOI] [PubMed] [Google Scholar]
- Vincent JL, Brewin NJ. Immunolocalization of a cysteine protease in vacuoles, vesicles, and symbiosomes of pea nodule cells. Plant Physiol. 2000:123(2):521–530. 10.1104/pp.123.2.521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Culver JN. DNA Binding specificity of ATAF2, a NAC domain transcription factor targeted for degradation by tobacco mosaic virus. BMC Plant Biol. 2012:12(1):1471–2229. 10.1186/1471-2229-12-157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Dong W, Murray J, Wang E. Innovation and appropriation in mycorrhizal and rhizobial symbioses. Plant Cell. 2022:34(5):1573–1599. 10.1093/plcell/koac039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Luo M. Random DNA binding selection assay (RDSA). Bio Protoc. 2015:5(8):e1452. 10.21769/BioProtoc.1452 [DOI] [Google Scholar]
- Wang L, Rubio MC, Xin X, Zhang B, Fan Q, Wang Q, Ning G, Becana M, Duanmu D. CRISPR/Cas9 knockout of leghemoglobin genes in Lotus japonicus uncovers their synergistic roles in symbiotic nitrogen fixation. New Phytol. 2019:224(2):818–832. 10.1111/nph.16077 [DOI] [PubMed] [Google Scholar]
- Wang L, Wang L, Tan Q, Fan Q, Zhu H, Hong Z, Zhang Z, Duanmu D. Efficient inactivation of symbiotic nitrogen fixation related genes in Lotus japonicus using CRISPR-cas9. Front Plant Sci. 2016b:31(7):1333. 10.3389/fpls.2016.01333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Yu H, Luo L, Duan L, Cai L, He X, Wen J, Mysore KS, Li G, Xiao A, et al. . NODULES WITH ACTIVATED DEFENSE 1 is required for maintenance of rhizobial endosymbiosis in Medicago truncatula. New Phytol. 2016a:212(1):176–191. 10.1111/nph.14017 [DOI] [PubMed] [Google Scholar]
- Woo HR, Kim HJ, Lim PO, Nam HG. Leaf senescence: systems and dynamics aspects. Annu Rev Plant Biol. 2019:70(1):347–376. 10.1146/annurev-arplant-050718-095859 [DOI] [PubMed] [Google Scholar]
- Wu A, Allu AD, Garapati P, Siddiqui H, Dortay H, Zanor MI, Asensi-Fabado MA, Munné-Bosch S, Antonio C, Tohge T, et al. . JUNGBRUNNEN1, A reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell. 2012:24(2):482–506. 10.1105/tpc.111.090894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X. Phenotypic analysis and molecular markers of plant nodule senescence. Methods Mol Biol. 2018:1744:65–80. 10.1007/978-1-4939-7672-0_5 [DOI] [PubMed] [Google Scholar]
- Xiao A, Yu H, Fan Y, Kang H, Ren Y, Huang X, Gao X, Wang C, Zhang Z, Zhu H, et al. . Transcriptional regulation of NIN expression by IPN2 is required for root nodule symbiosis in Lotus japonicus. New Phytol. 2020:227(2):513–528. 10.1111/nph.16553 [DOI] [PubMed] [Google Scholar]
- Xie K, Minkenberg B, Yang Y. Boosting CRISPR/cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci USA. 2015:112(11):3570–3575. 10.1073/pnas.1420294112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Syska C, Garcia I, Frendo P, Boncompagni E. Involvement of proteases during nodule senescence in leguminous plants. In: Bruijn F, editor. The model legume Medicago truncatula. Hoboken (NJ): John Wiley & Sons, Inc; 2020. p. 683–693. 10.1002/9781119409144.ch85 [DOI] [Google Scholar]
- Yu Y, Qi Y, Xu J, Dai X, Chen J, Dong CH, Xiang F. Arabidopsis WRKY71 regulates ethylene-mediated leaf senescence by directly activating EIN2, ORE1 and ACS2 genes. Plant J. 2021:107(6):1819–1836. 10.1111/tpj.15433 [DOI] [PubMed] [Google Scholar]
- Yu H, Xiao A, Dong R, Fan Y, Zhang X, Liu C, Wang C, Zhu H, Duanmu D, Cao Y, et al. . Suppression of innate immunity mediated by the CDPK-Rboh complex is required for rhizobial colonization in Medicago truncatula nodules. New Phytol. 2018:220(2):425–434. 10.1111/nph.15410 [DOI] [PubMed] [Google Scholar]
- Yuan S, Ke D, Li R, Li X, Wang L, Chen H, Zhang C, Huang Y, Chen L, Hao Q, et al. . Genome-wide survey of soybean papain-like cysteine proteases and their expression analysis in root nodule symbiosis. BMC Plant Biol. 2020:20(1):517. 10.1186/s12870-020-02725-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng P, Vadnais DA, Zhang Z, Polacco JC. Refined glufosinate selection in Agrobacterium-mediated transformation of soybean [Glycine max (L.) Merrill]. Plant Cell Rep. 2004:22(7):478–482. 10.1007/s00299-003-0712-8 [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhao M, Song Q, Zhao L, Wang G, Zhou C. Identification and function analyses of senescence-associated WRKYs in wheat. Biochem Biophys Res Commun. 2016:474(4):761–767. 10.1016/j.bbrc.2016.05.034 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.