Abstract
Heat stress (HS) adversely affects plant growth and productivity. The Class A1 HS transcription factors (HSFA1s) act as master regulators in the plant response to HS. However, how HSFA1-mediated transcriptional reprogramming is modulated during HS remains to be elucidated. Here, we report that a module formed by the microRNAs miR165 and miR166 and their target transcript, PHABULOSA (PHB), regulates HSFA1 at the transcriptional and translational levels to control plant HS responses. HS-triggered induction of MIR165/166 in Arabidopsis thaliana led to decreased expression of target genes including PHB. MIR165/166 overexpression lines and mutations in miR165/166 target genes enhanced HS tolerance, whereas miR165/166 knockdown lines and plants expressing a miR165/166-resistant form of PHB were sensitive to HS. PHB directly repressed the transcription of HSFA1s and globally modulated the expression of HS-responsive genes. PHB and HSFA1s share a common target gene, HSFA2, which is essential for activation of plant responses to HS. PHB physically interacted with HSFA1s and exerted an antagonistic effect on HSFA1 transcriptional activity. PHB and HSFA1s co-regulated transcriptome reprogramming upon HS. Together, these findings indicate that heat-triggered regulation of the miR165/166–PHB module controls HSFA1-mediated transcriptional reprogramming and plays a critical role during HS in Arabidopsis.
Heat-triggered regulation of the miR165/166–PHB module controls HSFA1-mediated transcriptional reprogramming and plays a critical role during heat stress in Arabidopsis.
Introduction
Plants are frequently challenged by extreme temperatures, which can adversely affect growth, development, distribution, and productivity. To acclimate to such stresses, plants have evolved strictly controlled regulatory mechanisms. When subjected to heat stress (HS), the expression of HS-responsive (HSR) genes is induced by HS transcription factors (HSFs). Plants contain more than 20 HSFs, which can be classified into 3 classes (A, B, and C) based on their oligomerization domains (Nover et al. 2001; Baniwal et al. 2004). In Arabidopsis (Arabidopsis thaliana), the 4 Class A1 HS transcription factors (HSFA1s), HSFA1a, HSFA1b, HSFA1d, and HSFA1e, act as master regulators of HS responses (Ohama et al. 2016).
With the exception of HSFB5, Class B HSFs contain the tetrapeptide motif LFGV and function as repressors in the regulation of HSR gene expression (Czarnecka-Verner et al. 2004; Ikeda and Ohme-Takagi 2009; Kumar et al. 2009; Ikeda et al. 2011). HSFA2, another HSF gene, serves as a direct target of HSFA1s and is crucial for the activation of HSR gene expression in plants (Charng et al. 2007). HSFs recognize and bind to heat shock elements (HSE: 5′-GAAnnTTC-3′) in the promoters of HSR genes to promote the accumulation of heat shock proteins (HSPs) (Busch et al. 2005). These HSPs are grouped into 5 classes according to their approximate molecular weight and include HSP100, HSP90, HSP70, HSP60, and small HSPs (sHSPs, 15–30 kD) (Vierling 1991; Trent 1996). HSPs function as molecular chaperones and maintain cellular proteostasis by preventing protein aggregation and denaturation.
In addition to protein factors, increasing evidence shows that microRNAs (miRNAs) are vital regulators of HS responses. miRNAs are noncoding RNAs that are processed from primary transcripts into ∼21-nucleotide-long molecules by the DICER-LIKE1 (DCL1)-associated processing complex and are essential for regulating plant growth and stress responses (Jones-Rhoades and Bartel 2004; Sunkar and Zhu 2004; Baker et al. 2005; Fujii et al. 2005; Lauter et al. 2005; Jones-Rhoades et al. 2006; Reyes and Chua 2007; Zhou et al. 2007; Khraiwesh et al. 2012; Rogers and Chen 2013). In Arabidopsis, miR398 strongly accumulates upon HS, while the transcript levels of its target genes such as COPPER/ZINC SUPEROXIDE DISMUTASE 1 (CSD1), CSD2, and COPPER CHAPERONE FOR CSD1 AND CSD2 (CCS) decrease (Guan et al. 2013). Single mutant plants lacking CSD1, CSD2, or CCS function are more tolerant to HS. By contrast, transgenic plants expressing forms of CSD1, CSD2, and CCS transcripts that are resistant to miR398-mediated cleavage are more vulnerable to HS than the wild type (WT) (Guan et al. 2013).
NAT398b/c are natural antisense transcripts of MIR398b/c genes and inhibit miR398 biogenesis, attenuating plant thermotolerance (Li et al. 2020). Additionally, heat-induced tocopherol (vitamin E) production promotes miR398 accumulation and improves heat tolerance (Fang et al. 2019). These results highlight the importance of miR398 abundance in plant thermotolerance. Other small RNAs, such as miR156, miR160, and trans-acting small interfering RNAs (ta-siRNAs), also play critical roles in the regulation of plant thermotolerance. miR156 regulates HS memory through SQUAMOSA-PROMOTER BINDING-LIKE (SPL) transcription factors (Stief et al. 2014). miR160 modulates the transcript levels of AUXIN RESPONSE FACTOR (ARF) genes to mediate heat tolerance (Lin et al. 2018). ta-siRNAs regulate their target genes HEAT-INDUCED TAS1 TARGET 1 (HTT1) and HTT2, to modulate plant responses to HS (Li et al. 2014).
The highly conserved miRNAs miR165/166, which differ by a single nucleotide, play essential roles in different biological processes by regulating members of the Class III HD-ZIP transcription factor family including PHABULOSA (PHB), PHAVOLUTA (PHV), REVOLUTA (REV), ATHB-8, and ATHB-15. Despite their close relationship, HD-ZIP III members also have antagonistic functions. HD-ZIP III genes are needed for patterning of different organs, as shown in previous studies (Mcconnell et al. 2001; Otsuga et al. 2001; Prigge et al. 2005; Carlsbecker et al. 2010; Smith and Long 2010; Lucas et al. 2013). Furthermore, the interconnection between HD-ZIPIII proteins and core components of the auxin, cytokinin, and abscisic acid network has also been uncovered (Jia et al. 2015; Yan et al. 2016; Sessa et al. 2018). Additionally, miR165 and miR166 are involved in abiotic stress responses (Yan et al. 2016; Zhang et al. 2018). In this study, we uncovered a function of miR165/166 in HS. We determined that the miR165/166–PHB module confers thermotolerance in Arabidopsis and revealed the underlying mechanism by which HSFA1-mediated transcriptional reprogramming is regulated by the miR165/166–PHB module to control plant responses to HS.
Results
HS triggers the accumulation of miR165/166, and overexpression of miR165/166 confers thermotolerance
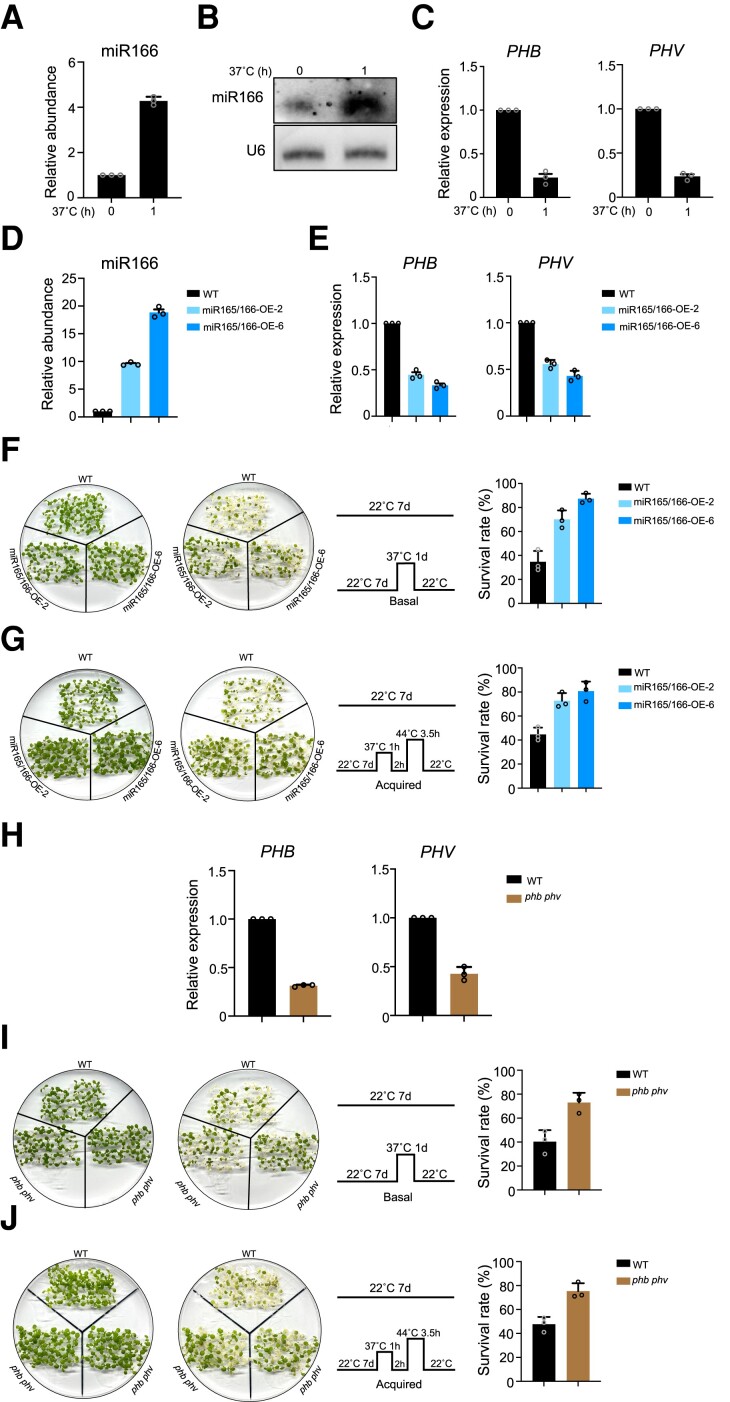
The highly conserved miRNAs miR165 and miR166 play critical roles in plant development and response to abiotic stress. However, it is unclear whether they are involved in HS. We observed that HS treatment dramatically induced miR165/166 accumulation (Fig. 1, A and B), leading to lower transcript levels for its target genes PHB and PHV (Fig. 1C). To explore the potential role of miR165/166 in HS, we generated and selected 2 independent transgenic lines overexpressing miR165/166 with higher miR165/166 levels and compared their thermotolerance phenotype to that of the WT. Mature miR165/166 markedly accumulated to levels 10 to 20 times greater than WT levels in these transgenic lines, whereas the transcript levels of miR165/166 target genes dramatically decreased (Fig. 1, D and E). When 7-d-old seedlings were subjected to HS at 37°C, we observed that miR165/166 overexpression lines were more tolerant to HS than the WT (Figs. 1, F and G, and S1A). To confirm our findings, we generated a phb phv double mutant (Fig. 1H) and analyzed its thermotolerance phenotype. Compared to the WT, the phb phv double mutant exhibited higher tolerance to HS (Figs. 1, I and J, and S1B).
Figure 1.
HS triggers the accumulation of miR165/166, and miR165/166 overexpression enhances plants thermotolerance. A) RT-qPCR analysis of mature miR166 levels in WT with or without 37°C treatment. SnoR101 was used as an internal control. Values are means ± Se from 3 independent biological replicates, with each data point indicated. B) Northern blot analysis of the abundance of mature miR166 in WT grown at 22°C or exposed to HS treatment at 37°C for 1 h. C) RT-qPCR analysis of transcript levels for miR166 target genes in WT grown at 22°C or exposed to HS treatment at 37°C for 1 h. ACTIN was used as an internal control. Values are means ± Se from 3 independent biological replicates, with each data point indicated. D) RT-qPCR analysis of mature miR166 abundance in WT and miR165/166 overexpression lines (miR165/166-OE). SnoR101 was used as an internal control. Values are means ± Se from 3 independent biological replicates, with each data point indicated. E) RT-qPCR analysis of miR166 target genes in WT and miR165/166-OE. ACTIN mRNA was used as an internal control. Values are means ± Se from 3 independent biological replicates, with each data point indicated. F) Basal thermotolerance analysis and survival rate of WT and miR165/166-OE after HS. Seven-day-old seedlings were exposed to 37°C for 1 d and allowed to recover at 22°C for 3 d. About 200 seedlings for each biological replicate were used, and 3 independent biological replicates were performed. Each biological replicate includes 4 technical replicates of 50 seedlings. Values are means ± Se of 3 independent biological replicates, with each data point indicated. G) Acquired thermotolerance analysis and survival rate of WT and miR165/166-OE after HS. Seven-day-old seedlings were treated at 37°C for 1 h, returned to 22°C for 2 h, and then transferred to 44°C for 3.5 h and returned to 22°C for 3 d. About 200 seedlings for each biological replicate were used, and 3 independent biological replicates were performed. Each biological replicate includes 4 technical replicates. Values are means ± Se of 3 independent biological replicates, with each data point indicated. H) RT-qPCR analysis of transcript levels for miR166 target genes in WT and the phb phv double mutant. ACTIN was used as an internal control. Values are means ± Se from 3 independent biological replicates, with each data point indicated. I) Basal thermotolerance analysis and survival rate of WT and the phb phv double mutant after HS. Seven-day-old seedlings were exposed to 37°C for 1 d and allowed to recover at 22°C for 3 d. About 200 seedlings for each biological replicate were used, and 3 independent biological replicates were performed. Each biological replicate includes 4 technical replicates. Values are means ± Se of 3 independent biological replicates, with each data point indicated. J) Acquired thermotolerance analysis and survival rate of WT and the phb phv double mutant after HS. Seven-day-old seedlings were treated at 37°C for 1 h, returned to 22°C for 2 h, then transferred to 44°C for 3.5 h, and returned to 22°C for 3 d. About 200 seedlings for each biological replicate were used, and 3 independent biological replicates were performed. Each biological replicate includes 4 technical replicates. Values are means ± Se of 3 independent biological replicates, with each data point indicated.
We also examined the thermotolerance of 3-wk-old soil-grown WT plants and miR165/166 overexpression lines subjected to HS at 37°C for 4 d. We observed less damage in miR165/166 overexpression lines than in WT plants (Supplemental Fig. S2). Similar to miR165/166 overexpression transgenic plants, the phb phv double mutant subjected to 37°C displayed a more tolerant phenotype (Supplemental Fig. S3). These results demonstrate that as miR165/166 accumulation increases in miR165/166 overexpression lines, the transcript levels of miR165/166 target genes decrease, resulting in enhanced thermotolerance.
Knockdown of miR165/166 makes plants more sensitive to HS
To investigate the role of miR165/166 in thermotolerance, we generated 2 moderate miR165/166 knockdown lines, STTM165/166, using the short tandem target mimicry (STTM) approach (Yan et al. 2012). The accumulation of miR165/166 in the STTM165/166 lines dropped by 70% relative to the WT control (Fig. 2A), whereas transcript levels for miR165/166 target genes were ∼2 to 3.6 times higher than in WT (Fig. 2B). When 7-d-old WT and STTM165/166 seedlings were subjected to HS at 37°C, all STTM165/166 transgenic seedlings exhibited weaker thermotolerance than the WT control (Figs. 2, C and D, and S4A). These results suggest that lower levels of miR165/166 make seedlings more sensitive to HS.
Figure 2.
miR165/166 knockdown lines and PHB gain-of-function transgenic lines are more sensitive to HS. A) Analysis of mature miR166 abundance in WT and STTM165/166. SnoR101 was used as an internal control. Values are means ± Se from 3 independent biological replicates, with each data point indicated. B) Expression analysis of miR166 target genes in WT and STTM165/166. ACTIN mRNA was used as an internal control. Values are means ± Se from 3 independent biological replicates, with each data point indicated. C) Basal thermotolerance analysis and survival rate of WT and STTM165/166 after HS. Seven-day-old seedlings were exposed to 37°C for 1 d and allowed to recover at 22°C for 3 d. About 200 seedlings for each biological replicate were used, and 3 independent biological replicates were performed. Each biological replicate includes 4 technical replicates. Values are means ± Se from 3 independent biological replicates, with each data point indicated. D) Acquired thermotolerance analysis and survival rate of WT and STTM165/166 after HS. Seven-day-old seedlings were treated at 37°C for 1 h, returned to 22°C for 2 h, then transferred to 44°C for 3.5 h, and returned to 22°C for 3 d. About 200 seedlings for each biological replicate were used, and 3 independent biological replicates were performed. Each biological replicate includes 4 technical replicates. Values are means ± Se from 3 independent biological replicates, with each data point indicated. E) Expression analysis of transcript levels for miR166 target genes in WT and PHBpro:PHBm-FLAG (a miR165/166 cleavage-resistant version) transgenic lines. ACTIN was used as an internal control. Values are means ± Se from 3 independent biological replicates, with each data point indicated. F) Basal thermotolerance analysis and survival rate of WT and PHBpro:PHBm-FLAG transgenic lines after HS. Seven-day-old seedlings were exposed to 37°C for 1 d and allowed to recover at 22°C for 3 d. About 200 seedlings for each biological replicate were used, and 3 independent biological replicates were performed. Each biological replicate includes 4 technical replicates. Values are means ± Se from 3 independent biological replicates, with each data point indicated. G) Acquired thermotolerance analysis and survival rate of WT and PHBpro:PHBm-FLAG after HS. Seven-day-old seedlings were treated at 37°C for 1 h, returned to 22°C for 2 h, then transferred to 44°C for 3.5 h, and returned to 22°C for 3 d. About 200 seedlings for each biological replicate were used, and 3 independent biological replicates were performed. Each biological replicate includes 4 technical replicates. Values are means ± Se from 3 independent biological replicates, with each data point indicated.
We further explored this hypothesis using transgenic seedlings expressing a form of PHB that is resistant to miR165/166 cleavage, PHBpro:PHBm-FLAG. PHB transcript levels in these seedlings were much higher than those of WT (Fig. 2E), and PHBpro:PHBm-FLAG seedlings were more sensitive to HS compared to the WT (Fig. 2, F and G, and S4B). Consistent with these results, when 3-wk-old soil-grown WT and STTM165/166 and PHBpro:PHBm-FLAG plants were exposed to HS, the STTM165/166 and PHBpro:PHBm-FLAG plants displayed a severely damaged phenotype compared to the WT (Supplemental Figs. S5 and S6).
PHB represses the expression of HSFA1s
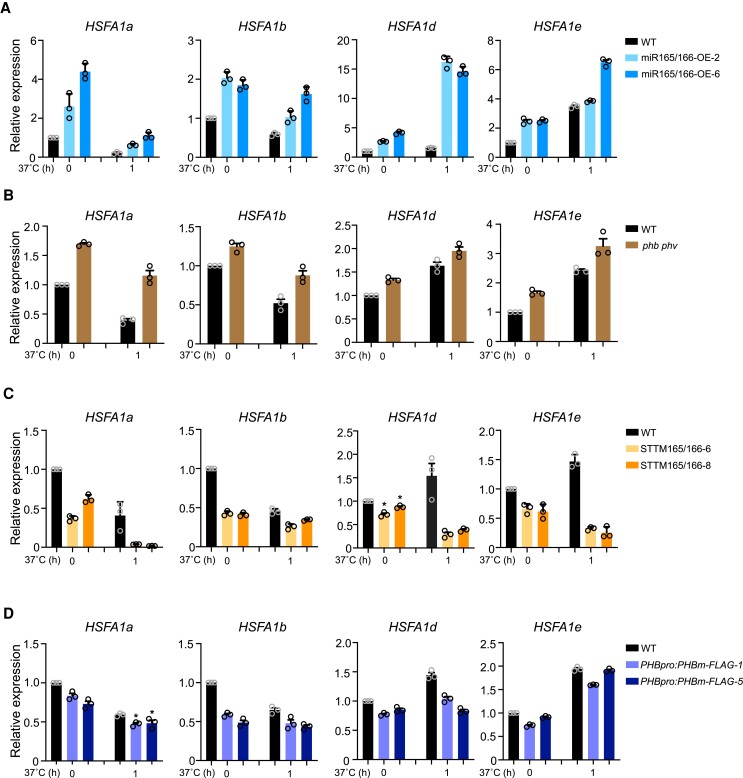
We investigated whether the miR165/166–PHB module regulates the expression of HSFA1s, which encode master regulators of HS responses. Surprisingly, expression analysis revealed that even at 22°C, HSFA1 transcript levels were higher in miR165/166-OE lines compared to the WT control (Fig. 3A). After HS treatment of 1 h at 37°C, HSFA1 transcript levels were still higher in miR165/166-OE lines compared to the WT (Fig. 3A). Similarly, in the phb phv double mutant, the expression of HSFA1s was higher relative to the WT regardless of HS treatment (Fig. 3B).
Figure 3.
The miR165/166–PHB module affects the expression of HSFA1s. A) Expression levels of HSFA1s in WT and miR165/166-OE grown at 22°C or exposed to HS treatment at 37°C for 1 h. ACTIN was used as an internal control. Values are means ± Se from 3 independent biological replicates, with each data point indicated. B) Expression levels of HSFA1s in WT and the phb phv double mutant grown at 22°C or exposed to HS treatment at 37°C for 1 h. ACTIN was used as an internal control. Values are means ± Se from 3 independent biological replicates, with each data point indicated. C) Expression levels of HSFA1s in WT and STTM165/166 grown at 22°C or exposed to HS treatment at 37°C for 1 h. ACTIN was used as an internal control. Values are means ± Se from 3 independent biological replicates, with each data point indicated. Significant differences were analyzed by ANOVA (*P < 0.05). D) Expression levels of HSFA1s in WT and PHBpro:PHBm-FLAG (a miR165/166 cleavage-resistant version) grown at 22°C or exposed to HS treatment at 37°C for 1 h. ACTIN was used as an internal control. Values are means ± Se from 3 independent biological replicates, with each data point indicated. Significant differences were analyzed by ANOVA (*P < 0.05).
We next assayed the expression of HSFA1s in STTM165/166 lines and observed that their transcript levels were significantly lower than in the WT again regardless of HS treatment (Fig. 3C). Similarly, HSFA1 transcript levels in the PHBpro:PHBm-FLAG transgenic lines were also lower relative to the WT grown at 22°C only or exposed to 37°C for 1 h (Fig. 3D). Together, these results demonstrate that the miR165/166–PHB module regulates the expression of HSFA1s.
HSFA1s are direct targets of PHB
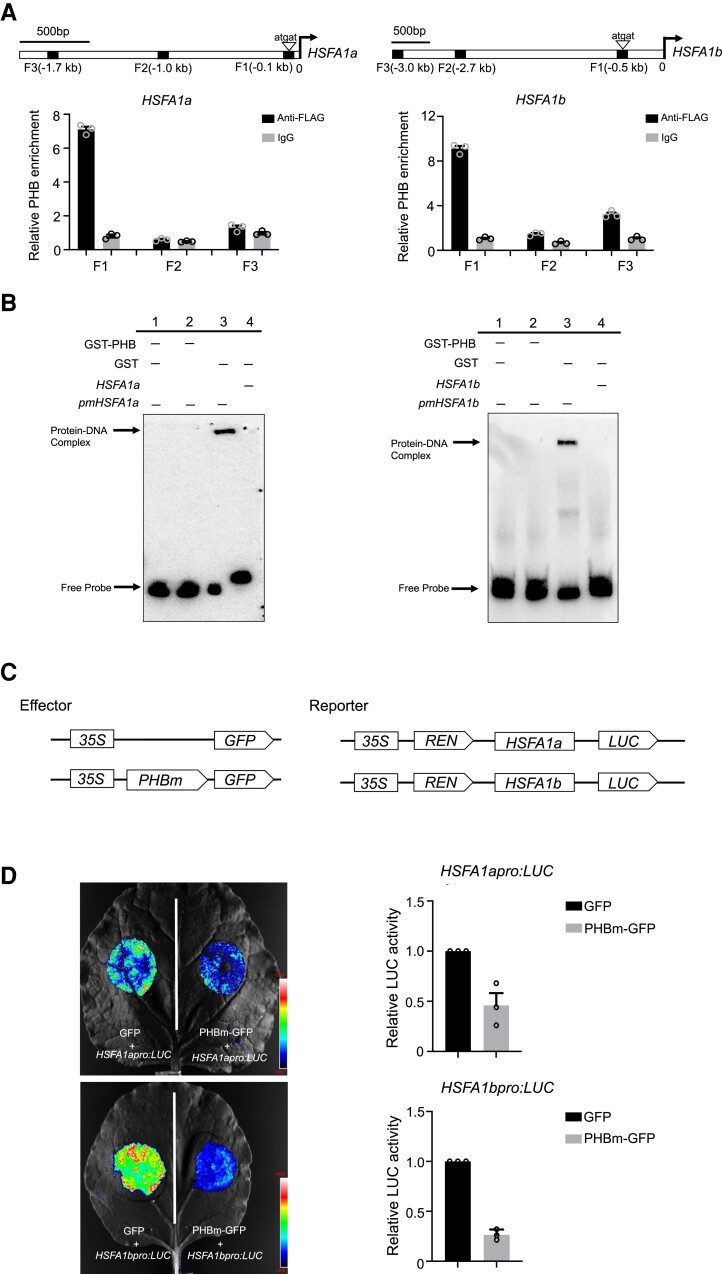
To investigate whether PHB can directly modulate the expression of HSFA1s, we identified potential PHB protein-binding sites in the promoter regions of HSFA1s (Fig. 4A). Utilizing PHBpro:PHBm-FLAG transgenic plants, we performed a chromatin immunoprecipitation (ChIP) assay to examine whether PHB associates with cis-regulatory sequences containing these binding motifs in vivo. As shown in Fig. 4A, we detected PHB occupancy in the gene promoter regions of HSFA1a and HSFA1b where PHB binding motifs were located. An electrophoretic mobility shift assay (EMSA) further revealed the direct PHB binding to the enriched PHB recognition motif fragment (Figs. 4B and S7), demonstrating that HSFA1a and HSFA1b are 2 direct targets of PHB.
Figure 4.
PHB directly binds to the promoters of HSFA1s and inhibits their transcription. A) Top: schematic diagrams of the promoter regions of HSFA1a and HSFA1b. Arrowheads indicate the positions of the core sequence (5′-atgat-3′) for the PHB recognition motif. Squares indicate the fragments amplified in ChIP-qPCR. Bottom: PHBpro:PHBm-FLAG seedlings were used for ChIP-qPCR, and IgG was used as the negative control. Values are means ± Se from 3 independent biological replicates, with each data point indicated. B) EMSA showing the direct binding of PHB to promoter regions of HSFA1a and HSFA1b. The positions of the DNA–protein complex and free probe are marked. The biotin-labeled probes containing the PHB recognition motif were incubated with GST-PHB recombinant proteins. The GST lane and mutated version of probes (pmHSFA1a and pmHSFA1b) lane were used as negative control. C) Schematic diagrams of effectors and reporters used in the transient dual-LUC reporter assay. Expression of PHB and REN was driven by the 35S promoter. The LUC reporter gene was driven by the HSFA1a or HSFA1b native promoter. D) Transient dual-LUC reporter assay in N. benthamiana leaves showing that PHB suppresses the promoter activity of HSFA1a and HSFA1b. Relative LUC activity was normalized to REN activity. Values are means ± Se from 3 independent biological replicates, with each data point indicated. For each replicate, at least 3 batches of plants were tested.
We performed a dual-luciferase (LUC) assay in Nicotiana benthamiana leaves to examine the effect of PHB on transcription from the HSFA1a and HSFA1b promoters, driving the firefly LUC reporter gene. Co-infiltration of the LUC reporters with 35S:PHB decreased relative LUC activity, indicating that PHB negatively regulates the transcription of HSFA1a and HSFA1b (Figs. 4, C and D, and S8). We also performed a ChIP assay to examine whether PHB associates with the promoter regions of the other 2 members of the HSFA1 family, HSFA1d and HSFA1e. As shown in Supplemental Fig. S9, we detected enrichment for the fragments containing the PHB recognition motif in the HSFA1d and HSFA1e promoter regions. These findings suggest that all 4 HSFA1 members are targets of PHB, which acts as a transcriptional repressor in modulating their transcription.
Global gene expression analysis reveals that PHB represses HSR gene expression
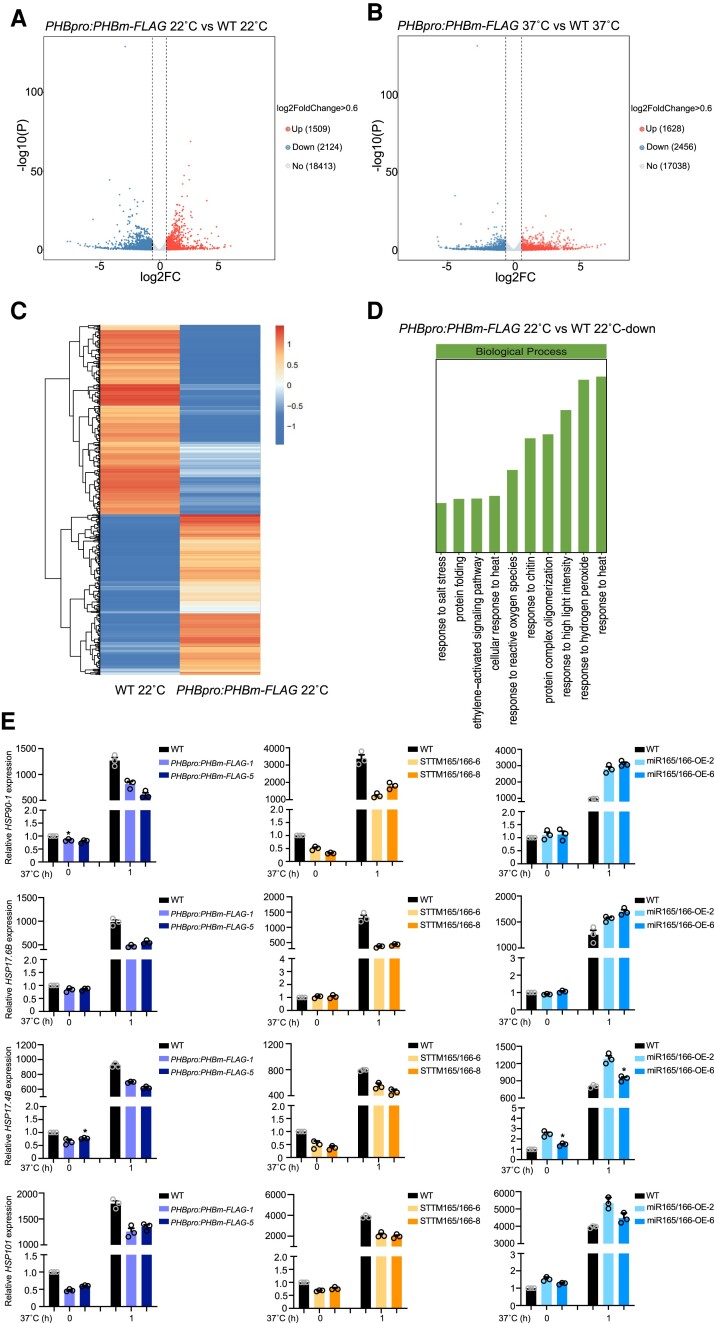
To decipher the mechanism of thermotolerance mediated by the miR165/166–PHB module, we performed transcriptome deep sequencing (RNA-seq) on 7-d-old WT and PHBpro:PHBm-FLAG seedlings grown at 22°C or exposed to 37°C for 1 h (Fig. 5). Without HS treatment, 1,509 genes were upregulated, and 2,124 genes were downregulated in PHBpro:PHBm-FLAG transgenic seedlings compared to WT controls (Fig. 5A). The transcriptome of PHBpro:PHBm-FLAG transgenic seedlings differed even more from that of the WT after HS treatment, with the number of upregulated genes increasing from 1,509 to 1,628 and the number of downregulated genes rising from 2,124 to 2,456 (Fig. 5B). Gene ontology (GO) term enrichment analysis showed that downregulated genes in PHBpro:PHBm-FLAG transgenic seedlings are enriched in terms related to response to HS (Fig. 5D).
Figure 5.
Transcriptome profiling of WT and PHBpro:PHBm-FLAG seedlings with or without HS. A) Volcano plot showing gene expression in WT and PHBpro:PHBm-FLAG grown at 22°C. Blue dots, downregulated genes; red dots, upregulated genes; gray dots, no change. B) Volcano plot showing gene expression in WT and PHBpro:PHBm-FLAG that were both exposed to 1 h of 37°C treatment. Blue dots, downregulated genes; red dots, upregulated genes; gray dots, no change. C) Heatmap of the transcriptome data from WT and PHBpro:PHBm-FLAG grown at 22°C. Red, upregulation; blue, downregulation; white, no change. The color scale represents the relative level of expression, which is normalized by the Z-score algorithm. Red, high Z-scores; blue, low Z-scores. D) GO enrichment analysis for downregulated genes in PHBpro:PHBm-FLAG compared to WT. E)HSP gene expression analysis in WT, PHBpro:PHBm-FLAG, STTM165/166, and miR165/166-OE grown at 22° or exposed to HS treatment at 37°C for 1 h. ACTIN was used as an internal control. Values are means ± Se from 3 independent biological replicates, with each data point indicated. Significant differences were analyzed by ANOVA (*P < 0.05).
To validate our RNA-seq data, we chose several HSP genes such as HSP90-1, HSP17.6B, HSP17.4B, HSP101, HSP17.4, and HSP17.6C for reverse transcription quantitative PCR (RT-qPCR) analysis. The expression levels of these genes were significantly lower in PHBpro:PHBm-FLAG seedlings relative to WT regardless of HS treatment (Figs. 5E and S10). We also examined the transcript levels of these genes in STTM165/166 and miR165/166-OE lines, which revealed their downregulation in STTM165/166 lines and their upregulation in miR165/166-OE lines compared to the WT control with or without HS treatment (Figs. 5E and S10). Together, these results indicate that PHB is involved in the transcriptional reprogramming of gene expression in response to HS.
HSFA2 is a common target of both HSFA1s and PHB
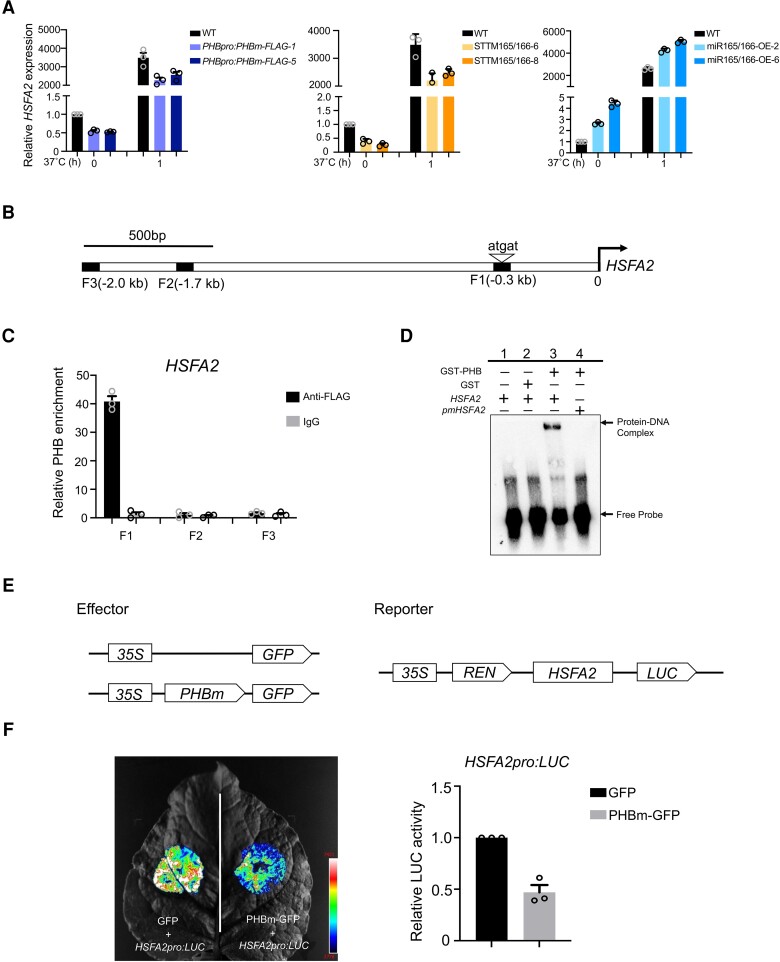
The above RNA-seq analysis revealed that the expression of the essential HSR gene regulator HSFA2 was lower in PHBpro:PHBm-FLAG transgenic seedlings compared to that of the WT with or without HS treatment. We conducted an RT-qPCR analysis to explore whether HSFA2 is regulated by the miR165/166–PHB module. HSFA2 mRNA abundance clearly decreased in PHBpro:PHBm-FLAG and STTM165/166 transgenic lines and dramatically increased in miR165/166-OE lines compared to the WT, regardless of HS treatment (Fig. 6A). Promoter sequence analysis indicated that the HSFA2 promoter contains a PHB recognition motif (Fig. 6B). To investigate whether PHB might directly regulate HSFA2 transcription, we performed a ChIP assay and EMSAs and determined that PHB directly binds to the HSFA2 promoter (Figs. 6, C and D, and S11). Given that HSFA1s can directly regulate HSFA2 transcription, these findings indicate that HSFA2 serves as a common target of both HSFA1s and PHB.
Figure 6.
HSFA2 is a direct target of PHB. A) Expression analysis of HSFA2 in WT, PHBpro:PHBm-FLAG, STTM165/166, and miR165/166-OE grown at 22°C or exposed to HS treatment at 37°C for 1 h. ACTIN was used as an internal control. Values are means ± Se from 3 independent biological replicates, with each data point indicated. B) Schematic diagram of the HSFA2 promoter region. The arrowhead indicates the position of the core sequence (5′-atgat-3′) for the PHB recognition motif. Squares indicate the fragments amplified in ChIP-qPCR. C) ChIP-qPCR analysis of PHB binding to the HSFA2 promoter. PHBpro:PHBm-FLAG seedlings were used for ChIP-qPCR, and IgG was used for the negative control. Values are means ± Se from 3 independent biological replicates, with each data point indicated. D) EMSA showing the direct binding of PHB to promoter regions of HSFA2. The positions of the DNA–protein complex and free probe are marked. The biotin-labeled probe containing the PHB recognition motif was incubated with recombinant GST-PHB. GST lane and mutated version of probes (pmHSFA2) lane were used for negative control. E) Schematic diagrams of effectors and reporters used in the transient dual-LUC reporter assay. Expression of PHB and REN was driven by the 35S promoter. The LUC reporter gene was driven by the HSFA2 native promoter. F) Transient dual-LUC reporter assay in N. benthamiana leaves showing that PHB suppresses the promoter activity of HSFA2. Relative LUC activity was normalized to REN activity. Values are means ± Se from 3 independent biological replicates, with each data point indicated. For each replicate, at least 3 batches of plants were determined.
We also conducted a dual-LUC assay in N. benthamiana leaves and found that the activity of the HSFA2pro:LUC reporter was substantially lower when co-infiltrating the effector construct 35S:PHB, supporting the negative role of PHB in the regulation of HSFA2 transcription (Figs. 6, E and F, and S12).
PHB physically interacts with HSFA1s
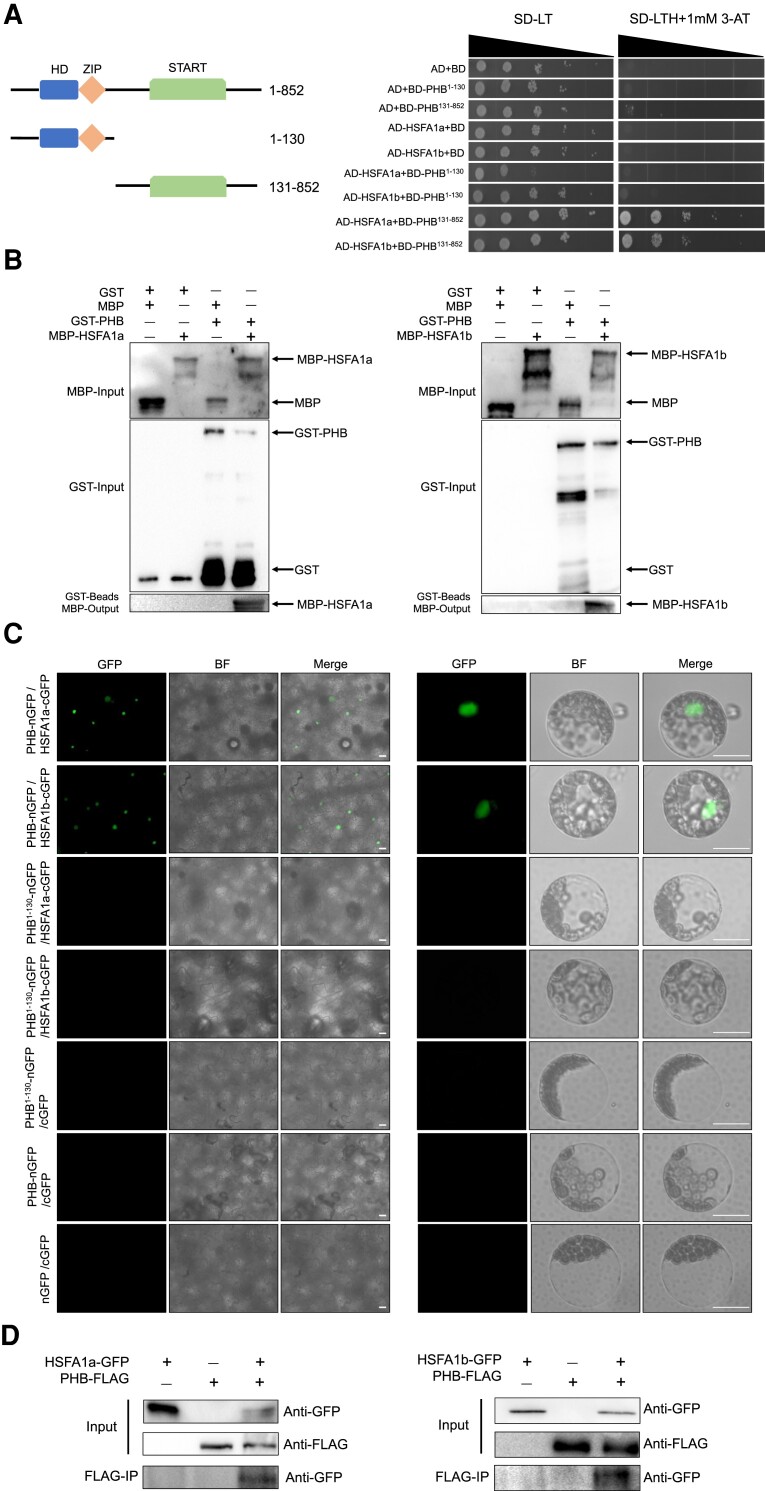
Given that the activity of HSFA1s is strictly controlled by protein–protein interactions, we investigated the possibility of direct interactions between PHB and the HSFA1 members HSFA1a and HSFA1b using a yeast 2-hybrid assay. Indeed, the C-terminal region, but not the N-terminal region of PHB, physically interacted with HSFA1a and HSFA1b (Fig. 7A). To validate this interaction, we conducted an in vitro pull-down assay using maltose-binding protein (MBP)-tagged HSFA1a or HSFA1b as a prey and glutathione S-transferase (GST)-tagged PHB as a bait. GST-tagged PHB, but not the control protein GST, interacted with HSFA1a and HSFA1b (Fig. 7B).
Figure 7.
PHB directly interacts with HSFA1a or HSFA1b. A) Yeast 2-hybrid assays showing the interaction between PHB and HSFA1a or HSFA1b. Left, schematic diagram showing the functional domains of PHB: START, steroidogenic acute regulatory protein-related lipid transfer domain; HD, homeodomain; ZIP, leucine zipper. –LT, synthetic defined (SD) medium lacking leucine and tryptophan; –LTH, SD medium lacking leucine, tryptophan, and histidine. 3-amino-1,2,4-triazole (3-AT) was added to the medium to reduce autoactivation. B) In vitro pull-down assays showing the interaction between PHB and HSFA1a or HSFA1b. Recombinant MBP-HSFA1a or MBP-HSFA1b was pulled down by GST-PHB and detected by anti-MBP antibody. C) BiFC assays showing that PHB physically interacts with HSFA1a or HSFA1b, whereas PHB1–130 lacking an interaction domain does not, in N. benthamiana leaves and Arabidopsis protoplasts. The fluorescence signal was captured by Zeiss Imager A2. Scale bars, 50 µm. D) Co-IP assays showing the interaction between PHB and HSFA1a or HSFA1b. PHB-FLAG was co-infiltrated with HSFA1a-GFP or HSFA1b-GFP in N. benthamiana leaves. PHB-FLAG protein was immunoprecipitated with anti-FLAG antibodies mixed with protein A/G agarose beads, and the immunoprecipitated HSFA1a-GFP or HSFA1b-GFP was then detected using an anti-GFP antibody.
We obtained further support for the interactions between PHB, HSFA1a, and HSFA1b by bimolecular fluorescence complementation (BiFC) assays. We generated constructs encoding PHB fused to the N-terminus of green fluorescent protein (GFP), while HSFA1a and HSFA1b were individually fused to the C-terminus of GFP. We then co-infiltrated the appropriate pairs of constructs into N. benthamiana leaves. Although we observed strong GFP fluorescence in the nucleus, we detected no signal in the negative controls (Fig. 7C). We confirmed this result in Arabidopsis protoplasts transfected with the same constructs (Fig. 7C).
To further validate the interaction between PHB and HSFA1s, we performed co-immunoprecipitation (co-IP) assays by co-expressing PHB-FLAG and HSFA1a-GFP or HSFA1b-GFP in N. benthamiana. We determined that HSFA1a or HSFA1b can be clearly pulled down by PHB (Fig. 7D). Together, these findings suggest that PHB can physically interact with HSFA1a and HSFA1b.
PHB interferes with HSFA1 activity for binding to their targets
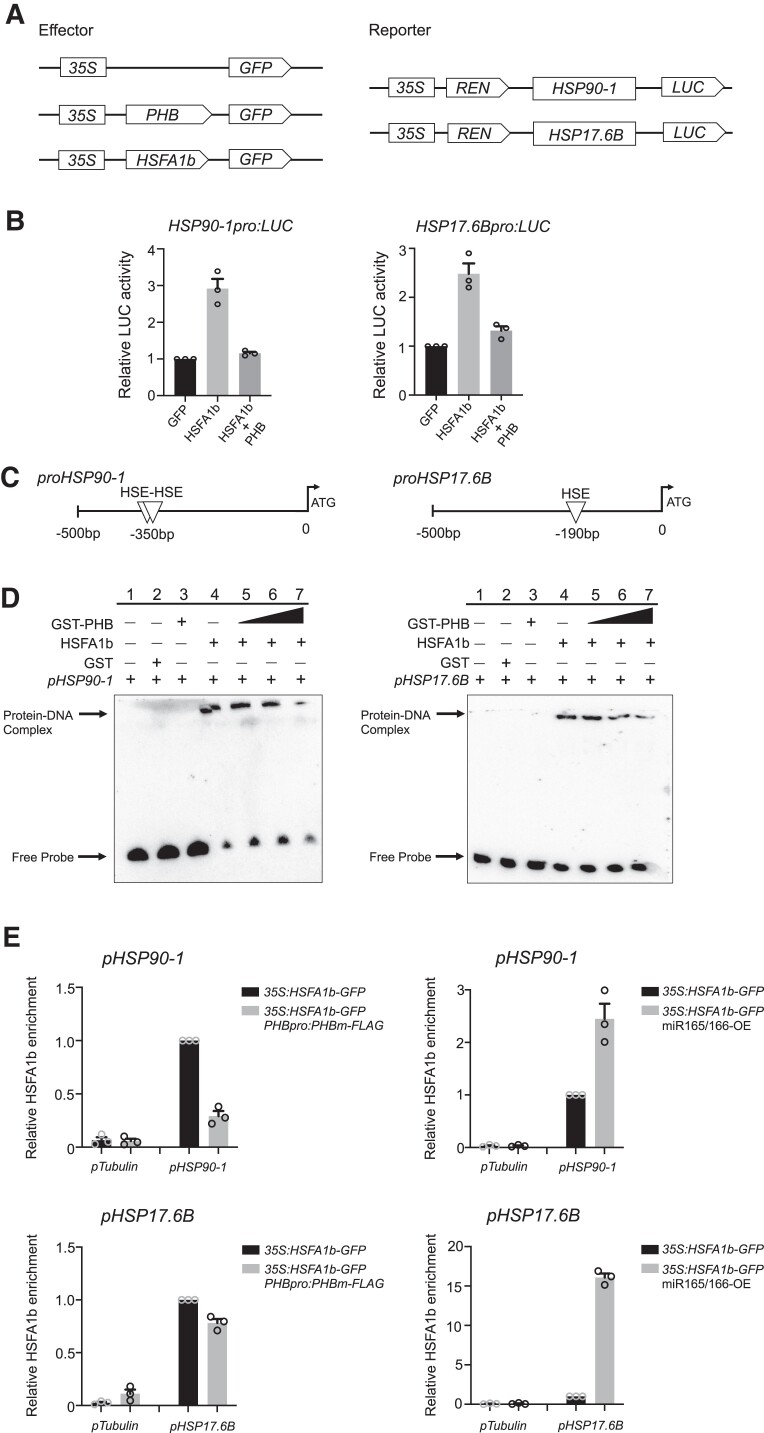
To determine whether direct interactions between PHB and HSFA1s affect transcriptional activity of the latter, we performed a dual-LUC assay where the expression of both HSFA1b and PHB was driven by the 35S promoter (Fig. 8A). For the assay, we selected the promoters of HSP90-1 and HSP17.6B, as they harbor only the binding sites for HSFA1s, but not the PHB recognition motif. LUC activity derived from the HSP90-1pro:LUC and HSP17.6Bpro:LUC reporter constructs was dramatically stimulated upon co-expression of HSFA1b (Fig. 8B). By contrast, the co-expression of HSFA1b and PHB repressed relative LUC activity compared to when HSFA1b was expressed (Fig. 8B). These findings indicate that PHB interferes with HSFA1b transcriptional function.
Figure 8.
PHB interferes with the transcriptional function of HSFA1b. A) Schematic diagrams of the effectors and reporters used in the transient dual-LUC reporter assay. The expression of PHB, HSFA1b, or REN was driven by the 35S promoter. The LUC reporter gene was driven by the native promoter of HSP90-1 or HSP17.6B. B) Transient dual-LUC reporter assays in N. benthamiana leaves showing that PHB has an antagonistic effect on HSFA1b for the regulation of HSP90-1 or HSP17.6B transcription. Relative LUC activity was normalized to REN activity. Values are means ± Se from 3 independent biological replicates, with each data point indicated. For each replicate, at least 3 batches of plants were determined. C) Schematic diagrams of the promoter regions of HSP90-1 and HSP17.6B. Triangles indicate the position of the HSE element for HSFA1s recognition. D) EMSA showing that PHB antagonizes HSFA1b binding to the promoters of HSP90-1 and HSP17.6B. The position of DNA–protein complex and free probe are marked. The biotin-labeled probe containing the HSE element was incubated with recombinant GST-PHB or MBP-HSFA1b. E) ChIP-qPCR analysis showing that the DNA binding of HSFA1b to the HSE element at the promoters of HSP90-1 and HSP17.6B is inhibited by PHB. Seven-day-old seedlings of 35S:HSFA1b-GFP, 35S:HSFA1b-GFP PHBpro:PHBm-FLAG, and 35S:HSFA1b-GFP miR165/166-OE were used. qPCR data with the TUBULIN promoter (TUBpro) as a negative control. Values are means ± Se from 3 independent biological replicates, with each data point indicated.
We performed an EMSA to test whether PHB affects the binding activity of HSFA1b to an oligonucleotide containing the HSE motif. As shown in Figs. 8, C and D, and S13, HSFA1b did bind to the cis-element; however, when recombinant PHB was added, the binding of HSFA1b to the cis-element dramatically decreased. We performed a ChIP assay to examine the effects of PHB on the binding of HSFA1b to the promoters of HSP90-1 and HSP17.6B in vivo. Compared to 35S:HSFA1b-GFP seedlings, the enrichment of HSFA1b at the promoters of HSP90-1 and HSP17.6B was lower in 35S:HSFA1b-GFP PHBpro:PHBm-FLAG seedlings (Fig. 8E), which we generated by crossing 35S:HSFA1b-GFP and PHBpro:PHBm-FLAG transgenic lines, therefore maintaining similar expression levels for HSFA1b (Supplemental Fig. S14, A and B). However, in the 35S:HSFA1b-GFP miR165/166-OE lines, which we obtained by crossing 35S:HSFA1b-GFP and miR165/166-OE lines and thus maintain similar HSFA1b expression levels as the single transgenic lines (Supplemental Fig. S14, C and D), the enrichment of HSFA1b at the promoters of HSP90-1 and HSP17.6B was enhanced compared to 35S:HSFA1b-GFP seedlings (Fig. 8E). These findings indicate that PHB disrupts the DNA binding of HSFA1b and has an antagonistic effect on HSFA1b in the regulation of HSR gene expression.
PHB mediates the HSFA1-triggered transcriptional reprogramming upon HS
HSFA1s function as master regulators in the activation of HSR transcriptional networks, whereas PHB has an antagonistic effect on the regulation by HSFA1s. We hypothesized that the transcriptional reprogramming triggered by HSFA1s upon HS might be mediated by PHB. Accordingly, we compared the transcriptome profile between HS-treated PHBpro:PHBm-FLAG seedlings and the HS-treated hsfa1a hsfa1b hsfa1d hsfa1e quadruple mutant, which was generated in the Col-0 genetic background (Fig. 9A). After HS treatment, we identified 3,989 genes regulated by HSFA1s (quadruple mutant compared to WT) and 4,084 genes regulated by PHB (PHBpro:PHBm-FLAG relative to WT) (Fig. 9B). Almost half of these genes (1,945 genes) were co-regulated by HSFA1s and PHB. In all, 733 genes and 809 genes were upregulated or downregulated, respectively, in both the hsfa1 quadruple mutant and PHBpro:PHBm-FLAG seedlings (Fig. 9B). Importantly, for the subset of 1,945 genes co-regulated by HSFA1s and PHB, most (1,517 of 1,945) including many HSPs displayed HS-triggered alterations in expression (WT HS-treated compared to WT untreated) (Fig. 9, C and D). Collectively, these data indicate that PHB plays a crucial role in mediating the HSFA1-triggered transcriptome reprogramming during HS.
Figure 9.
The miR165/166–PHB module plays an important role in plant HS pathway. A) Left: structures of HSFA1 loci and the position of the T-DNA insertions marked by triangles. Boxes indicate exons. Right, RT-qPCR analysis of the transcript levels of HSFA1 genes. Values are means ± Se from 3 independent biological replicates, with each data point indicated. B) Venn diagrams showing the extent of overlap between the sets of DEGs regulated by PHB (PHBpro:PHBm-FLAG 37°C relative to WT 37°C) compared to DEGs regulated by HSFA1s (quadruple mutant 37°C relative to WT 37°C). In total, 1,945 genes were co-regulated by HSFA1s and PHB. Among these genes, the changes of 1,542 genes are in the same trend in the hsfa1 quadruple mutant and PHBpro:PHBm-FLAG seedlings. A total of 733 genes and 809 genes were upregulated or downregulated, respectively, in both the hsfa1 quadruple mutant and PHBpro:PHBm-FLAG seedlings. The changes of the remaining 403 genes are in opposite trend in the hsfa1 quadruple mutant and PHBpro:PHBm-FLAG seedlings. C) Percentage of heat-regulated genes among co-regulated genes in PHBpro:PHBm-FLAG and hsfa1a hsfa1b hsfa1d hsfa1e after 37°C treatment. Of the 1,945 co-regulated genes, ∼78% (1517 of 1945) are associated with the heat response. D) Heatmap of HSP genes in WT, hsfa1a hsfa1b hsfa1d hsfa1e, and PHBpro:PHBm-FLAG with 37°C treatment. Red, upregulation; blue, downregulation; white, no change. The color scale represents the relative level of expression, which is normalized by the Z-score algorithm. Red, high Z-scores; blue, low Z-scores.
The miR165/166–PHB module acts upstream of HSFA1s genetically in the control of thermotolerance
Given that the activity of HSFA1s can be regulated by the miR165/166–PHB module, we investigated the genetic relationship between HSFA1s and the miR165/166–PHB module in the regulation of HS responses. To this end, we compared the thermotolerance phenotype of miR165/166-OE lines and miR165/166-OE lines in the hsfa1a hsfa1b hsfa1d mutant background. Although the expression level of miR165/166 target genes was similar in miR165/166-OE and hsfa1a hsfa1b hsfa1d miR165/166-OE (Supplemental Fig. S15A), we observed that the stronger thermotolerance phenotype of miR165/166-OE lines is suppressed when assessed in the hsfa1a hsfa1b hsfa1d mutant background (Supplemental Fig. S15B). Thus, the miR165/166–PHB module acts upstream of HSFA1s genetically in the regulation of thermotolerance.
Discussion
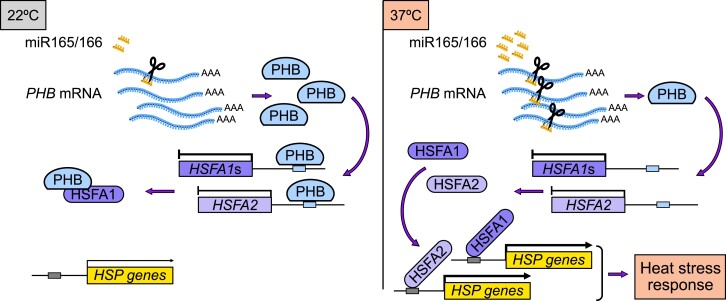
In this study, we identified miR165/166 as essential to the HS response and provided evidence that the miR165/166–PHB module contributes greatly to HS response in Arabidopsis (Fig. 10). The miR165/166 knockdown transgenic lines, STTM165/166, and PHBpro:PHBm-FLAG plants are hypersensitive to HS (Fig. 2, C and D and F and G). By contrast, the miR165/166 overexpression lines and the phb phv double mutant are more tolerant of HS (Fig. 1, F and G and I and J). We determined that the miR165/166–PHB module influences the expression of HSFA1 genes and that PHB can bind to the promoter regions of HSFA1s to inhibit their transcription (Figs. 3 and 4). RNA-seq analysis revealed that PHB-mediated thermotolerance involves suppression of HSR gene expression (Fig. 5, D and E). In addition, we identified HSFA2 as a common target of PHB and HSFA1s (Fig. 6). Our work revealed that PHB directly interacts with HSFA1s (Fig. 7) and has an antagonistic effect on their binding to the promoters of HSR genes, thereby interfering with the transcriptional function of HSFA1s (Fig. 8). Transcriptome profile comparison between the hsfa1 quadruple mutant and PHBpro:PHBm-FLAG seedlings upon HS further reinforced the idea that PHB is involved in global transcriptional reprogramming by HSFA1s during HS (Fig. 9). Our data thus reveal the miR165/166–PHB module as an important positive regulator for thermotolerance and show how the miR165/166–PHB module controls plant response to HS by regulating HSFA1-mediated transcriptional reprogramming during HS (Fig. 10).
Figure 10.
A working model for the transcriptional reprogramming mediated by the miR165/166–PHB module to regulate plant thermotolerance. HS triggers the accumulation of miR165/166, leading to the downregulation of its target genes. The miR165/166–PHB module regulates thermotolerance through at least 2 pathways. One is through direct transcriptional regulation of HSFs. PHB can modulate the transcription of HSRs in an HSFA1-dependent manner. PHB can also regulate the transcription of HSRs in an HSFA1-independent manner by directly regulating the transcription of heat-inducible HSFA2. In addition, PHB physically interacts with HSFA1s and interferes with their transcription function.
The highly conserved HSFs act as central regulators of HSR genes in both animals and plants. In plants, HSFA1s are the master regulator and activate the HSR gene expression upon HS (Jones-Rhoades et al. 2006; Reyes and Chua 2007; Ohama et al. 2017). Notably, unlike HSFA2, which is 1 of the HS-inducible HSFAs, the effect of overexpressing HSFA1s is relatively limited, because HSFA1s activity is strictly controlled at the posttranslational level. Protein–protein interactions are important for regulation of and by HSFA1s. Previous studies have shown that HSP70 and HSP90 physically interact with HSFA1s and negatively regulate their activity by modulating their nuclear localization and transactivation activity (Fujii et al. 2009; Smith and Long 2010; Ohama et al. 2017). In this work, we revealed that PHB physically interacts with HSFA1s and has an antagonistic effect on their regulation of HSR gene expression at the posttranslational level. In Arabidopsis, the HSFA1 members HSFA1a, HSFA1b, and HSFA1d can physically interact with one another and appear to function as homocomplexes or heterocomplexes (Yoshida et al. 2011). The interaction of PHB with HSFA1s might interfere with the formation of such functional HSFA1s complexes, and the competition between PHB and HSFA1s will affect the binding ability of HSFA1s to the promoters of HSR genes. Under normal conditions, the physical interaction of PHB and HSFA1s inhibits the transcriptional activity of HSFA1s. Upon HS, the production of miR165/166 is dramatically induced, which in turn represses PHB expression. Consequently, the inhibition of PHB on regulation by HSFA1s activity is limited. Our results also show that the miR165/166–PHB module can regulate HSFA1 at the transcriptional level and that PHB represses the expression of HSFA1s by binding to their promoter regions. Thus, under nonstress conditions, PHB not only represses the transcription of HSFA1s but also inhibits their transcriptional activity. Under HS conditions, the miR165/166-mediated cleavage of PHB mRNA is dramatically enhanced, and the negative effect of PHB on the expression of HSFA1s and activity of HSFA1s was reduced, leading to increased tolerance to HS.
Transcriptome-wide reprogramming of gene expression is critical for plant responses to HS. HSFA1-mediated transcriptional reprogramming contributes greatly to thermotolerance. Transcriptome profiling analysis of the hsfa1 quadruple mutant and PHB gain-of-function transgenic plants revealed that almost half of the genes regulated by HSFA1 are also controlled by PHB under HS (Fig. 9B). Combined with the evidence that PHB controls the expression of HSFA1s and activity of HSFA1s, we hypothesize that PHB can regulate the transcriptional cascade in an HSFA1-dependent manner. Notably, HS-inducible HSFA2 is also directly regulated by PHB and serves as a common target of HSFA1 and PHB (Fig. 6). We predict that PHB might also orchestrate HS-triggered transcriptional reprogramming in an HSFA1-independent manner, possibly via its direct regulation of HSFA2 or some other factors.
Our work focused here on the downstream core signaling components underlying thermotolerance and showed that HS-induced accumulation of miR165/166 reduces the transcript levels of miR165/166 target genes (Fig. 1, A to C). How the abundance of miR165/166 is controlled and the underlying mechanism mediating the modulation of miR165/166 under HS is still unknown. Identification of regulatory factors upstream of the miR165/166–PHB module will further our understanding of how it regulates HS responses.
HS affects plant growth and development. When confronted with HS, plants utilize different regulators to coordinate their developmental regulatory program and stress responses modulation networks to survive. miR165/166 is an important conserved miRNA and plays a vital role in plant development and HS responses. Like miR165/166, the developmental regulators miR156 and miR160 are also essential HS components (Stief et al. 2014; Lin et al. 2018). These findings suggest that miRNA might mediate environmental regulation of growth and development in plant response to HS. The use of miRNAs to counterbalance the adverse effects of HS on plant growth and development could be a good strategy for plants to respond to HS. Therefore, it is worth identifying more miRNAs involved in HS and deciphering the connection between miRNAs and different HS components. This analysis will provide a deep insight into the mechanism of miRNAs in HS response and eventually facilitate the improvement of crop stress tolerance and yield stability in the future.
Materials and methods
Plant materials and growth conditions
The WT and mutant Arabidopsis (A. thaliana) plants used in this study were in the Col-0 background. STTM165/166 lines, miR165/166 overexpression lines, PHBpro:PHBm-FLAG (a miR165/166 cleavage-resistant version) lines, and phb phv double mutant were described previously (Yan et al. 2012). For the generation of 35S:HSFA1b-GFP transgenic plants, the plasmid was transformed into WT Arabidopsis plants by the floral dip method (Clough and Bent 1998). More than 20 independent lines resistance to Basta were isolated and characterized, and T3 transgenic plants were used for analysis. The Arabidopsis T-DNA insertion mutants hsfa1a (SALK_068042) and hsfa1d (SALK_022404) and the hsfa1b hsfa1e double mutant (CS2103365) were obtained from the Nottingham Arabidopsis Stock Centre (http://arabidopsis.info/). The triple mutant hsfa1a hsfa1b hsfa1e was obtained by crossing the hsfa1a single mutant and the hsfa1b hsfa1e double mutant. Then, the hsfa1a hsfa1b hsfa1d hsfa1e quadruple mutant was generated by crossing the hsfa1a hsfa1b hsfa1e triple mutant and the hsfa1d single mutant. The hsfa1a hsfa1b hsfa1d triple mutant and hsfa1a hsfa1b hsfa1d hsfa1e quadruple mutant were identified from the F2 segregating progeny of F1 heterozygous plants. Homozygous plants were isolated by genotyping using the PCR primers listed in Supplemental Table S1. Seeds were surface sterilized and sown on solid culture medium (Murashige–Skoog [MS] salts, 1% [w/v] sucrose, and 0.6% [w/v] agar). After 2 d of stratification at 4°C in darkness, seeds were placed into a growth incubator (PERCIVAL) at 22°C in a 16-h light (100 µmol m−2s−1, white LED lamps)/8-h dark photoperiod.
Thermotolerance assays
Thermotolerance assays were performed as described (Guan et al. 2013; Han et al. 2020). Surface-sterilized seeds were sown on MS medium with 1% (w/v) sucrose and placed in a growth incubator at 22°C in a 16-h light/8-h dark photoperiod. The plates were then transferred to a growth chamber after 7 d for 1 h at 37°C before collecting samples for RT-qPCR analysis.
For basal thermotolerance analysis, 7-d-old seedlings were incubated at 37°C for 24 h and allowed to recover at 22°C for 3 d. For acquired thermotolerance analysis, 7-d-old seedlings were first incubated at 37°C for 1 h and then transferred at 22°C for 2 h. After 2 h at 22°C, seedlings were treated at 44°C for 3.5 h and returned to 22°C.
Three-week-old soil-grown plants were treated at 37°C for 4 d and then allowed to recover at 22°C for 3 d. Photographs of the damage were taken, and survival rates were measured after 3 d of recovery.
Plasmid construction
To obtain the 35S:HSFA1b-GFP plasmid, the full-length coding sequence (CDS) of HSFA1b was amplified by PCR from WT (Col-0) cDNA and inserted into the BamHI-Smal sites of the pENTR-TOPO vector. The insert was recombined into the pEarlygate 103 vector containing the sequence encoding GFP using the Gateway LR Clonase II Enzyme system (Thermo Fisher).
For in vitro pull-down assays, the full-length CDS of PHB was inserted into SmaI-SalI sites of the pGEX4T-1 vector to generate GST-PHB, and the CDS of HSFA1a and HSFA1b was individually cloned into the BamHI-XbaI and XbaI-PstI sites of pMAL-c2x to generate MBP-HSFA1a and MBP-HSFA1b, respectively.
Two fragments of the PHB CDS (from 1 to 390 bp or 391 to 2,556 bp) for yeast 2-hybrid assays were individually cloned into the NdeI-EcoRI sites of pGBKT7 (GAL4 DNA-binding domain, Clontech) bait vector, and the full-length CDS of HSFA1a or HSFA1b was cloned into the Smal-BamHI or NdeI-EcoRI sites, respectively, of the pGADT7 (GAL4 activation domain, Clontech) prey vector.
For BiFC assays, the full-length or truncated CDS of PHB was inserted into the SacI-SmaI sites or EcoRI-Smal sites of the pENTR-TOPO vector respectively and then cloned in-frame with the sequence encoding the N-terminal half of GFP using the pGWB6 to generate the 35S:PHB-nGFP construct. Similarly, the full-length CDS of HSFA1a or HSFA1b was first inserted into the BamHI-SmaI sites of the pENTR-TOPO vector and then cloned in-frame with the sequence encoding the C-terminal half of GFP using the pGWB5 to obtain the 35S:HSFA1a-cGFP or 35S:HSFA1b-cGFP construct, respectively.
To perform the dual-LUC reporter assay, putative promoter sequences (∼2.0 kb) of HSFA1a, HSFA1b, HSFA2, HSP90-1, and HSP17.6B were amplified by PCR from Col-0 genomic DNA and cloned individually into the pGreenII 0800-LUC vector to drive the firefly LUC gene, with the Renilla luciferase (REN) gene controlled by the 35S promoter. The primer sequences are listed in Supplemental Data Set 1.
RT-qPCR analysis
Total RNA was extracted from 7-d-old seedlings that were ground in liquid nitrogen using TRIzol reagent (Invitrogen) and treated with DNase I (New England Biolabs) to remove genomic DNA contamination. Approximately 4 µg of total RNA was used for first-strand cDNA synthesis with a ReverTra Ace qPCR RT Master Mix (Toyobo). qPCR reactions were performed using a 2× SYBR Green Real-Time PCR Master Mix (Toyobo) on a CFX96 Real-Time PCR system (BioRad). ACTIN mRNA was used as an internal control. Three independent biological replicates were conducted, with 3 technical replicates for each biological replicate. The primers used are listed in Supplemental Data Set 1.
Mature miRNA RT-qPCR analysis
Mature miRNA abundance analysis was conducted as previously described (Chen et al. 2005). According to the manufacturer's protocol, total RNA was isolated using an RNeasy mini kit (Qiagen) from 7-d-old seedlings, accompanied by genomic DNA digestion with an RNase-free DNase set (Qiagen). A TaqMan microRNA Reverse Transcription Kit (Invitrogen) was used for miRNA reverse transcription and TaqMan Universal Master Mix II (Thermo Fisher) for qPCR. Arabidopsis SnoR101 was used as an internal control. At least 3 independent biological replicates were conducted, with 3 technical replicates for each biological replicate. Primer sequences are listed in Supplemental Data Set 1.
Northern blotting analysis
The northern blotting analysis was conducted as described by Guan et al. (2013). Small RNAs were extracted from 7-d-old WT seedlings with a miRcute miRNA Isolation Kit (TIANGEN) following the manufacturer's protocol. Approximately 30 µg of small RNAs were resolved on 15% polyacrylamide denaturing gels (8 m urea) and transferred onto a Hybond N+ membrane (Amersham Biosciences, GE Healthcare). The membrane was cross-linked by UV irradiation for 3 min and hybridized with biotin-labeled oligonucleotides at 42°C for 12 h. The membrane signals were detected with a Chemiluminescent Nucleic Acid Detection Module Kit (Thermo Fisher). The U6 probe was used as the control. Three biological replicates were performed, and the sequences of the oligonucleotides used as probes are listed in Supplemental Data Set 1.
RNA-seq analysis
Total RNA was extracted from 7-d-old WT, PHBpro:PHBm-FLAG and hsfa1a hsfa1b hsfa1d hsfa1e seedlings grown under normal conditions and 7-d-old WT, PHBpro:PHBm-FLAG and hsfa1a hsfa1b hsfa1d hsfa1e seedlings with heat treatment (37°C for 1 h) using the TRIzol reagent (Invitrogen). The purity and quantification of RNA were evaluated with a NanoDrop 2000 spectrophotometer (Thermo Fisher), while RNA integrity was assessed with an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara). For library construction, a VAHTS Universal V6 RNA-seq Library Prep Kit was used.
Transcriptome sequencing and subsequent analysis were performed by OE Biotech Co., Ltd. (Shanghai, China) using an Illumina Novaseq 6000 platform. After removal of low-quality reads with Q20 <85%, clean reads were mapped to the Arabidopsis reference genome (TAIR10.1-NCBI) using HISAT2 (Kim et al. 2015). Principal component analysis (PCA) was conducted to evaluate the biological replication of samples using R (v 3.2.0). Differential expression analysis was conducted using DESeq2 R package (Love et al. 2014). GO enrichment analysis of differentially expressed genes (DEGs) was conducted using clusterprofile (v4.6.0) to look for significantly enriched terms. P-values were adjusted using the Benjamini and Hochberg approach. In each pair-wise comparison, genes with |log2 fold-change| > 0.6 and a P-value < 0.05 found by DEseq2 were considered as differentially expressed.
ChIP assays
ChIP assays were conducted as previously described (Guan et al. 2013). Briefly, 7-d-old PHBpro:PHBm-FLAG, 35S:HSFA1b-GFP, 35S:HSFA1b-GFP PHBpro:PHBm-FLAG, and 35S:HSFA1b-GFP miR165/166-OE seedlings were used. Approximately 3 g of seedlings was fixed for 20 min with 1% (w/v) formaldehyde. Glycine (125 mm) was added to terminate cross-linking, and the samples were ground to powder. Chromatin extracts were isolated with 25 mL of isolation buffer and sheared into ∼500-bp fragments with a sonicator (Diagenode) and were then precleared using protein A/G agarose beads (Santa Cruz). DNA precipitated by 5 µL of anti-GFP (Abmart, P30010L) or anti-FLAG (Abmart, M20008L) antibodies was washed for 10 min using 4 different buffers (low salt buffer; high salt buffer; LiCl buffer; and TE buffer) at 4°C and then reverse cross-linked in a 65°C incubator for 12 h. Proteinase K (1 mg/mL) was used to remove proteins at 45°C for 1 h. qPCR was performed with the purified DNA. IgG (Abmart, B30011M) was used as the negative control. Three independent biological replicates were conducted. Primers used in the ChIP-qPCR assays are listed in Supplemental Data Set 1.
EMSAs
EMSAs were conducted essentially as described previously (Li et al. 2014). Recombinant GST-PHB and MBP-HSFA1b were produced in the BL21 strain of Escherichia coli. Oligonucleotide probes (∼65 bp) were synthesized with their 3′ end labeled with biotin by Sangon Biotech Co., Ltd. (Shanghai, China). To generate double-stranded probes, sense and antisense oligonucleotide probes were heated for 10 min at 95°C and cooled down to room temperature. EMSA was carried out using a Light Shift Chemiluminescent EMSA Kit (Thermo Fisher). The reaction mixtures were incubated at room temperature for ∼25 min and electrophoresed on 6% native polyacrylamide gels in 0.5× Tris borate EDTA (TBE) at 100 V for 100 min. Then, the DNA was electrophoretically transferred to nylon membranes (GE Healthcare) at 400 mm for 30 min. The resulting membranes were exposed to UV light for ∼5 min to cross-link the transferred free probes and bound probes to the membrane and then washed with buffers. Signals were detected by a Tanon-5500 Chemiluminescent Imaging System (Tanon, China). The relevant oligonucleotide sequences are listed in Supplemental Data Set 1.
Yeast 2-hybrid assays
Yeast 2-hybrid assays were carried out as previously described (Li et al. 2014). The constructs were transformed into the yeast strain AH109 (Clontech). Yeast cells were plated onto selective medium SD/–Leu/–Trp/–His (−LTH). 3-amino-1,2,4-triazole (3-AT) was added to the selection medium to reduce the autoactivation of reporter genes. The relevant primer sequences are listed in Supplemental Data Set 1.
In vitro pull-down assays
Pull-down analysis was performed according to Friedrich et al. (2021). Recombinant GST and MBP fusion proteins were produced in the BL21 strain of E. coli and then were purified using Mag-Beads GST (Sangon, China) or Amylose Resin (New England Biolabs). The purified proteins were immunoblotted with anti-GST (GenScript, 1:2,000) or anti-MBP (New England Biolabs, 1:5,000) antibodies. The relevant primer sequences are listed in Supplemental Data Set 1.
BiFC assays
BiFC assays were conducted as previously described (Li et al. 2014). The constructs were transformed into Agrobacterium (Agrobacterium tumefaciens) strain GV3101. Constructs harboring sequences encoding the C- or N-terminal halves of GFP were co-infiltrated in young leaves of N. benthamiana. After 48 h, infiltrated leaves were harvested to detect GFP fluorescence signals. Additionally, plasmids were also co-transfected into Arabidopsis protoplasts using the polyethylene glycol-mediated (PEG-mediated) transient transformation method (Yoo et al. 2007), and GFP fluorescence signals were observed after 12 h of incubation. GFP fluorescence signals were detected using a microscope (Zeiss Imager A2) with the GFP filter (excitation band EX 470/40 nm, emission band EM530/40 nm). The Arabidopsis protoplast assay was performed as previously described (Marion et al. 2008). At least 3 independent experiments were conducted using different batches of plants. More than 6 plants were infiltrated, and over 500 protoplasts were examined for each independent biological replicate. The relevant primer sequences are listed in Supplemental Data Set 1.
Dual-LUC assays
Dual-LUC assays and transient expression assays were carried out as previously described by Hu et al. (2019). In brief, the vectors, 35S:GFP, 35S:PHB-GFP, and 35S:HSFA1b-GFP, were used as effectors. The vectors, HSFA1apro:LUC, HSFA1bpro:LUC, HSFA2pro:LUC, HSP90-1pro:LUC, and HSP17.6Bpro:LUC, were used as reporters. Effector and reporter constructs were co-infiltrated as appropriate pairs into N. benthamiana leaves via Agrobacterium, and leaves were collected 48 h after infiltration. The LUC and REN activity values were determined with a Dual-LUC Reporter Assay System (Promega) on a Synergy NEO2 luminometer (BioTek), and then LUC activity was normalized to REN. The relevant prime sequences are listed in Supplemental Data Set 1.
Co-IP assays
Co-immunoprecipitation assays were performed as previously described (Friedrich et al. 2021). Briefly, the 35S:PHB-FLAG, 35S:HSFA1a-GFP, and 35S:HSFA1b-GFP plasmids were transformed into Agrobacterium strain GV3101 and co-infiltrated into young N. benthamiana leaves. After 48 h, total proteins were extracted from leaves using protein extraction kit (CWBIO, CW0885) and incubated overnight with anti-FLAG antibody (Abmart, M20008L, 1:5,000). The immunocomplexes were then collected by protein A/G agarose beads (Santa Cruz) and washed with immunoprecipitation buffer. The eluted proteins were subsequently detected by immunoblotting using an anti-GFP antibody (Abmart, P30010L, 1:5,000).
Statistical analysis
Statistical analyses were performed as described in each figure legend. Statistical data are provided in Supplemental Data Set 2.
Accession numbers
Raw RNA-seq reads were deposited at the National Center for Biotechnology Information (NCBI) under the accession number PRJNA911441. The sequence data for genes in this article can be found in The Arabidopsis Information Resource under the following accessions numbers: PHB (At2g34710), PHV (At1g30490), HSFA1a (At4g17750), HSFA1b (At5g16820), HSFA1d (At1g32330), HSFA1e (At3g02990), HSFA2 (At2g26150), HSP90-1 (At5g52640), HSP17.6B (At2g29500), HSP101 (At1g74310), HSP17.4B (At1g54050), HSP17.4 (At3g46230), and HSP17.6C (At1g53540).
Supplementary Material
Acknowledgments
We are grateful to Drs. Xianzhong Feng and Zhong Zhao for their assistance or helpful discussions. We also thank Drs. Zhongnan Yang and Jirong Huang for generously providing the pGreenII-0800 vector and the BiFC vectors.
Contributor Information
Jie Li, School of Life Sciences, East China Normal University, Shanghai 200241, People’s Republic of China.
Yiming Cao, School of Life Sciences, East China Normal University, Shanghai 200241, People’s Republic of China.
Jiaxin Zhang, School of Life Sciences, East China Normal University, Shanghai 200241, People’s Republic of China.
Cuijing Zhu, School of Life Sciences, East China Normal University, Shanghai 200241, People’s Republic of China.
Guiliang Tang, Department of Biological Sciences, Michigan Technological University, Houghton, MI 49931, USA.
Jun Yan, School of Life Sciences, East China Normal University, Shanghai 200241, People’s Republic of China.
Author contributions
J.Y. designed the research. J.L., Y.C., J.Z., and C.Z. conducted the research. J.L., Y.C., J.Z., C.Z., G.T., and J.Y. analyzed the data. J.L., G.T., and J.Y. wrote and edited the manuscript.
Supplemental data
The following materials are available in the online version of this article.
The following materials are available in the online version of this article.
Supplemental Figure S1. Thermotolerance analysis of 7-d-old WT, miR165/166-OE and phb phv seedlings.
Supplemental Figure S2. Thermotolerance phenotype and survival rate of soil-grown WT and miR165/166-OE.
Supplemental Figure S3. Thermotolerance phenotype and survival rate of soil-grown WT and the phb phv double mutant.
Supplemental Figure S4. Thermotolerance analysis of 7-d-old WT, STTM165/166 and PHBpro:PHBm-FLAG seedlings.
Supplemental Figure S5. Thermotolerance phenotype and survival rate of soil-grown WT and STTM165/166.
Supplemental Figure S6. Thermotolerance phenotype and survival rate of soil-grown WT and PHBpro:PHBm-FLAG.
Supplemental Figure S7. EMSA showing the direct binding of PHB to the promoter regions of HSFA1a or HSFA1b.
Supplemental Figure S8. Transient dual-LUC reporter assay in N. benthamiana leaves showing that PHB suppresses the promoter activity of HSFA1a and HSFA1b, whereas PHB131–852 lacking the DNA-binding domain does not.
Supplemental Figure S9. ChIP assay showing that PHB is enriched on the promoter region of HSFA1d or HSFA1e.
Supplemental Figure S10. Expression analysis of HSP genes in WT, PHBpro:PHBm-FLAG, STTM165/166, and miR165/166-OE grown at 22°C or exposed to 37°C for 1 h.
Supplemental Figure S11. EMSA showing the direct binding of PHB to promoter regions of HSFA2.
Supplemental Figure S12. Transient dual-LUC reporter assay in N. benthamiana leaves showing that PHB suppresses the promoter activity of HSFA2, whereas PHB131–852 lacking of DNA-binding domain does not.
Supplemental Figure S13. EMSA assay showing that PHB antagonizes HSFA1b to bind to the promoters of HSP90-1 and HSP17.6B.
Supplemental Figure S14. Analysis of 35S:HSFA1b-GFP, 35S:HSFA1b-GFP PHBpro:PHBm-FLAG, and 35S:HSFA1b-GFP miR165/166-OE transgenic plants.
Supplemental Figure S15. The enhanced thermotolerance phenotype of miR165/166-OE is suppressed when they in the hsfa1a hsfa1b hsfa1d mutant background.
Supplemental Data Set 1. Oligonucleotides used in this study.
Supplemental Data Set 2. Summary of statistical analyses.
Funding
This work was supported by grants from Science and Technology Commission of Shanghai Municipality, China (No.18PJ1402800, 20ZR1417900, and 22N11900400).
References
- Baker CC, Sieber P, Wellmer F, Meyerowitz EM. The early extra petals1 mutant uncovers a role for microRNA miR164c in regulating petal number in Arabidopsis. Curr Biol. 2005:15(4):303–315. 10.1016/j.cub.2005.02.017 [DOI] [PubMed] [Google Scholar]
- Baniwal SK, Bharti K, Chan KY, Fauth M, Ganguli A, Kotak S, Mishra SK, Nover L, Port M, Scharf K-D, et al. Heat stress response in plants: a complex game with chaperones and more than twenty heat stress transcription factors. J Biosci. 2004:29(4):471–487. 10.1007/BF02712120 [DOI] [PubMed] [Google Scholar]
- Busch W, Wunderlich M, Schöffl F. Identification of novel heat shock factor-dependent genes and biochemical pathways in Arabidopsis thaliana. Plant J. 2005:41(1):1–14. 10.1111/j.1365-313X.2004.02272.x [DOI] [PubMed] [Google Scholar]
- Carlsbecker A, Lee JY, Roberts CJ, Dettmer J, Lehesranta S, Zhou J, Lindgren O, Moreno-Risueno MA, Vatén A, Thitamadee S, et al. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 2010:465(7296):316–321. 10.1038/nature08977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charng YY, Liu HC, Liu NY, Chi WT, Wang CN, Chang SH, Wang TT. A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol. 2007:143(1):251–262. 10.1104/pp.106.091322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Real-time quantification of microRNA by stem-loop RT-PCR. Nucleic Acids Res. 2005:33(20):e179. 10.1093/nar/gni178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998:16(6):735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Czarnecka-Verner E, Pan S, Salem T, Gurley WB. Plant class B HSFs inhibit transcription and exhibit affinity for TFIIB and TBP. Plant Mol Biol. 2004:56(1):57–75. 10.1007/s11103-004-2307-3 [DOI] [PubMed] [Google Scholar]
- Fang X, Zhao G, Zhang S, Li Y, Gu H, Li Y, Zhao Q, Qi Y. Chloroplast-to-nucleus signaling regulates microRNA biogenesis in Arabidopsis. Dev Cell. 2019:48(3):371–382.e4. 10.1016/j.devcel.2018.11.046 [DOI] [PubMed] [Google Scholar]
- Friedrich T, Oberkofler V, Trindade I, Altmann S, Brzezinka K, Lämke J, Gorka M, Kappel C, Sokolowska E, Skirycz A. , et al. Heteromeric HSFA2/HSFA3 complexes drive transcriptional memory after heat stress in Arabidopsis. Nat Commun. 2021:(1):3426–3441. 10.1038/s41467-021-23786-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK. In vitro reconstitution of an abscisic acid signalling pathway. Nature 2009:462(7273):660–664. 10.1038/nature08599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK. A miRNA involved in phosphate-starvation response in Arabidopsis. Curr Biol. 2005:15(22):2038–2043. 10.1016/j.cub.2005.10.016 [DOI] [PubMed] [Google Scholar]
- Guan Q, Lu X, Zeng H, Zhang Y, Zhu J. Heat stress induction of miR398 triggers a regulatory loop that is critical for thermotolerance in Arabidopsis. Plant J. 2013:74(5):840–851. 10.1111/tpj.12169 [DOI] [PubMed] [Google Scholar]
- Han SH, Park YJ, Park CM. HOS1 activates DNA repair system to enhance plant thermotolerance. Nat Plants 2020:6(12):1439–1446. 10.1038/s41477-020-00809-6 [DOI] [PubMed] [Google Scholar]
- Hu Y, Han X, Yang M, Zhang M, Pan J, Yu D. The transcription factor INDUCER OF CBF EXPRESSION1 interacts with ABSCISIC ACID INSENSITIVE5 and DELLA proteins to fine-tune abscisic acid signaling during seed germination in Arabidopsis. Plant Cell 2019:31(7):1520–1538. 10.1105/tpc.18.00825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Mitsuda N, Ohme-Takagi M. Arabidopsis HsfB1 and HsfB2b act as repressors of the expression of heat-inducible HSFs but positively regulate the acquired thermotolerance. Plant Physiol. 2011:157(3):1243–1254. 10.1104/pp.111.179036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Ohme-Takagi M. A novel group of transcriptional repressors in Arabidopsis. Plant Cell Physiol. 2009:50(5):970–975. 10.1093/pcp/pcp048 [DOI] [PubMed] [Google Scholar]
- Jia X, Ding N, Fan W, Yan J, Gu Y, Tang X, Li R, Tang G. Functional plasticity of miR165/166 in plant development revealed by small tandem target mimic. Plant Sci. 2015:233:11–21. 10.1016/j.plantsci.2014.12.020 [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell. 2004:14(6):787–799. 10.1016/j.molcel.2004.05.027 [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAS and their regulatory roles in plants. Annu Rev Plant Biol. 2006:57(1):19–53. 10.1146/annurev.arplant.57.032905.105218 [DOI] [PubMed] [Google Scholar]
- Khraiwesh B, Zhu JK, Zhu J. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim Biophys Acta. 2012:1819(2):137–148. 10.1016/j.bbagrm.2011.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015:12(4):357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Busch W, Birke H, Kemmerling B, Nurnberger T, Schoffl F. Heat shock factors HsfB1 and HsfB2b are involved in the regulation of Pdf1.2 expression and pathogen resistance in Arabidopsis. Mol Plant. 2009:2(1):152–165. 10.1093/mp/ssn095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauter N, Kampani A, Carlson S, Goebel M, Moose S. MicroRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc Natl Acad Sci USA. 2005:102(26):9412–9417. 10.1073/pnas.0503927102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li X, Yang J, He Y. Natural antisense transcripts of MIR398 genes suppress microR398 processing and attenuate plant thermotolerance. Nat Commun. 2020:11(1):5351. 10.1038/s41467-020-19186-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Liu J, Liu Z, Li X, Wu F, He Y. HEAT-INDUCED TAS1 TARGET1 mediates thermotolerance via HEAT STRESS TRANSCRIPTION FACTOR A1a-directed pathways in Arabidopsis. Plant Cell 2014:26(4):1764–1780. 10.1105/tpc.114.124883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JS, Kuo CC, Yang IC, Tsai WA, Shen YH, Lin CC, Liang YC, Li YC, Kuo YW, King YC, et al. MicroRNA160 modulates plant development and heat shock protein gene expression to mediate heat tolerance in Arabidopsis. Front Plant Sci. 2018:9:68. 10.3389/fpls.2018.00068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014:15(12):550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas WJ, Groover A, Lichtenberger R, Furuta K, Yadav SR, Helariutta Y, He XQ, Fukuda H, Kang J, Brady SM, et al. The plant vascular system: evolution, development and functions. J Integr Plant Biol. 2013:55(4):294–388. 10.1111/jipb.12041 [DOI] [PubMed] [Google Scholar]
- Marion J, Bach L, Bellec Y, Meyer C, Gissot L, Faure JD. Systematic analysis of protein subcellular localization and interaction using high-throughput transient transformation of Arabidopsis seedlings. Plant J. 2008:56(1):169–179. 10.1111/j.1365-313X.2008.03596.x [DOI] [PubMed] [Google Scholar]
- Mcconnell JR, Emery JF, Eshed Y, Bao N, Bowman J, Barton MK. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 2001:411(6838):709–713. 10.1038/35079635 [DOI] [PubMed] [Google Scholar]
- Nover L, Bharti K, Döring P, Mishra SK, Ganguli A, Scharf KD. Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones 2001:6(3):177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohama N, Kusakabe K, Mizoi J, Zhao H, Kidokoro S, Koizumi S, Takahashi F, Ishida T, Yanagisawa S, Shinozaki K, et al. The transcriptional cascade in the heat stress response of Arabidopsis is strictly regulated at the level of transcription factor expression. Plant Cell 2016:28(1):181–201. 10.1105/tpc.15.00435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohama N, Sato H, Shinozaki K, Yamaguchi-Shinozaki K. Transcriptional regulation network of plant heat stress response. Trends in Plant Sci. 2017:22(1):53–65. 10.1016/j.tplants.2016.08.015 [DOI] [PubMed] [Google Scholar]
- Otsuga D, De Guzman B, Prigge MJ, Drews GN, Clark SE. REVOLUTA regulates meristem initiation at lateral positions. Plant J. 2001:25(2):223–236. 10.1046/j.1365-313x.2001.00959.x [DOI] [PubMed] [Google Scholar]
- Prigge MJ, Otsuga D, Alonso JM, Ecker JR, Drews GN, Clark SE. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 2005:17(1):61–76. 10.1105/tpc.104.026161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes JL, Chua NH. ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J. 2007:49(4):592–606. 10.1111/j.1365-313X.2006.02980.x [DOI] [PubMed] [Google Scholar]
- Rogers K, Chen X. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 2013:25(7):2383–2399. 10.1105/tpc.113.113159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa G, Carabelli M, Possenti M, Morelli G, Ruberti I. Multiple links between HD-Zip proteins and hormone networks. Int J Mol Sci. 2018:19(12):4047. -. 10.3390/ijms19124047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZR, Long JA. Control of Arabidopsis apical-basal embryo polarity by antagonistic transcription factors. Nature 2010:464(7287):423–426. 10.1038/nature08843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stief A, Altmann S, Hoffmann K, Pant BD, Scheible WR, Baurle I. Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell 2014:26(4):1792–1807. 10.1105/tpc.114.123851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Zhu JK. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 2004:16(8):2001–2019. 10.1105/tpc.104.022830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trent JD. A review of acquired thermotolerance, heat-shock proteins, and molecular chaperones in archaea. FEMS Microbiol Rev. 1996:18(2–3):249–258. 10.1111/j.1574-6976.1996.tb00241.x [DOI] [Google Scholar]
- Vierling E. The roles of heat shock proteins in plants. Annu Rev Plant Physiol Plant Mol Biol. 1991:42(1):579–620. 10.1146/annurev.pp.42.060191.003051 [DOI] [Google Scholar]
- Yan J, Gu Y, Jia X, Kang W, Pan S, Tang X, Chen X, Tang G. Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell 2012:24(2):415–427. 10.1105/tpc.111.094144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Zhao C, Zhou J, Yang Y, Wang P, Zhu X, Tang G, Bressan RA, Zhu JK. The miR165/166 mediated regulatory module plays critical roles in ABA homeostasis and response in Arabidopsis thaliana. PLoS Genet. 2016:12(11):e1006416. 10.1371/journal.pgen.1006416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc. 2007:2(7):1565–1572. 10.1038/nprot.2007.199 [DOI] [PubMed] [Google Scholar]
- Yoshida T, Ohama N, Nakajima J, Kidokoro S, Mizoi J, Nakashima K, Maruyama K, Kim J, Seki M, Todaka D, et al. Arabidopsis HsfA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Mol Genet Genomics. 2011:286(5–6):321–332. 10.1007/s00438-011-0647-7 [DOI] [PubMed] [Google Scholar]
- Zhang S, Dou Y, Li S, Ren G, Chevalier D, Zhang C, Yu B. DAWDLE interacts with DICER-LIKE proteins to mediate small RNA biogenesis. Plant Physiol. 2018:177(3):1142–1151. 10.1104/pp.18.00354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou GK, Kubo M, Zhong R, Demura T, Ye ZH. Overexpression of miR165 affects apical meristem formation, organ polarity establishment and vascular development in Arabidopsis. Plant Cell Physiol. 2007:48(3):391–404. 10.1093/pcp/pcm008 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.