SALT OVERLY SENSITIVE2 promotes shade avoidance by stabilizing phytochrome-interacting factors PIF4 and PIF5, and its kinase activity is enhanced by phytochromes A (phyA) and B (phyB) in the light.
Abstract
Sun-loving plants trigger the shade avoidance syndrome (SAS) to compete against their neighbors for sunlight. Phytochromes are plant red (R) and far-red (FR) light photoreceptors that play a major role in perceiving the shading signals and triggering SAS. Shade induces a reduction in the level of active phytochrome B (phyB), thus increasing the abundance of PHYTOCHROME-INTERACTING FACTORS (PIFs), a group of growth-promoting transcription factors. However, whether other factors are involved in modulating PIF activity in the shade remains largely obscure. Here, we show that SALT OVERLY SENSITIVE2 (SOS2), a protein kinase essential for salt tolerance, positively regulates SAS in Arabidopsis thaliana. SOS2 directly phosphorylates PIF4 and PIF5 at a serine residue close to their conserved motif for binding to active phyB. This phosphorylation thus decreases their interaction with phyB and posttranslationally promotes PIF4 and PIF5 protein accumulation. Notably, the role of SOS2 in regulating PIF4 and PIF5 protein abundance and SAS is more prominent under salt stress. Moreover, phyA and phyB physically interact with SOS2 and promote SOS2 kinase activity in the light. Collectively, our study uncovers an unexpected role of salt-activated SOS2 in promoting SAS by modulating the phyB-PIF module, providing insight into the coordinated response of plants to salt stress and shade.
IN A NUTSHELL.
Background: Sun-loving plants compete with their neighbors for sunlight by initiating shade avoidance syndrome (SAS). Phytochromes are plant photoreceptors that play a predominant role in perceiving the shaded environments and triggering SAS. PHYTOCHROME-INTERACTING FACTORS (PIFs) are a subset of basic helix–loop–helix (bHLH) family transcription factors that intrinsically promote plant growth. Upon light irradiation, photoactivated phytochrome B (phyB) interacts with PIFs via their active phytochrome B (APB)–binding motif and induces their rapid phosphorylation and degradation. Under shade, phyB is converted to the inactive form that cannot interact with PIFs; thus, the abundance of PIFs is increased, and plant growth is promoted.
Question: Are there any other factors that affect the phyB-PIF module in the shade?
Findings: We discovered that SALT OVERLY SENSITIVE2 (SOS2), a protein kinase essential for plant salt tolerance, positively regulates the response of Arabidopsis seedlings to shade. SOS2 directly phosphorylates PIF4 and PIF5 at a serine residue close to their conserved APB motif, thus decreasing the interactions of PIF4/PIF5 with phyB. Consequently, SOS2 posttranslationally promotes PIF4 and PIF5 protein accumulation, and the role of SOS2 in regulating PIF4/PIF5 protein abundance and SAS is more prominent under salt stress. Moreover, we showed that phyA and phyB physically interact with SOS2 and promote SOS2 kinase activity in the light.
Next steps: We aim to explore how phytochrome photoreceptors mediate the synergistic enhancement of SOS2 kinase activity by shade and salt stress. In addition, we will investigate whether SOS2 is also involved in plant responses to other environmental stresses such as drought, cold, or heat stress.
Introduction
Light is an important environmental cue that plays fundamental roles in regulating plant growth and development. In open stands or at the top of the canopy, plants sense full sunlight, in which the ratio of red (R) to far-red (FR) (R:FR) is high. However, in dense stands, a large portion of FR light is transmitted or reflected by plant tissues, leading to a decreased R:FR ratio in the shade (Casal 2012, 2013; Fiorucci and Fankhauser 2017; Yang and Li 2017). Shade-intolerant plants, such as Arabidopsis (Arabidopsis thaliana), sense the low R:FR light and rapidly trigger a set of responses named shade avoidance syndrome (SAS), displaying elongated hypocotyls and petioles, reduced branching and earlier flowering, thereby outgrowing their competitors (Casal 2013; Fiorucci and Fankhauser 2017; Fernández-Milmanda and Ballaré 2021).
Depending on plant density, SAS can be induced by 2 types of shade environments: neighbor detection and canopy shade. In neighbor detection, reflected FR light from neighboring plants leads to a low R/FR ratio but without a major drop in the amount of photosynthetically active radiation (PAR; R and blue [B] light). By contrast, in canopy shade, most R and B light is absorbed by upper leaves, thus leading to low PAR and low R:FR ratio underneath a canopy (Fiorucci and Fankhauser 2017; Fernández-Milmanda and Ballaré 2021). Therefore, a drop in the R:FR ratio serves as an early signal of a forming canopy (Ballaré et al. 1990), and plants interpret both types of shade environments as unfavorable conditions because they must compete with neighboring plants for sunlight.
The R and FR light spectra are perceived by the phytochrome (phy) family of photoreceptors in plants, and there are 5 phytochrome holoproteins in Arabidopsis: phytochromes A (phyA) to E (phyE) (Sharrock and Quail 1989; Franklin and Quail 2010; Fraser et al. 2016; Buti et al. 2020). Phytochromes exist in 2 photoconvertible forms: the active FR–absorbing Pfr form and the inactive R-absorbing Pr form (Bae and Choi 2008; Li et al. 2011; Legris et al. 2019; Cheng et al. 2021). Phytochromes are synthesized in the cytosol in the inactive Pr form; upon exposure to R light, phytochromes are converted to the Pfr form and translocated into the nucleus, thus leading to a set of changes in gene expression and photoresponses (Fankhauser and Chen 2008; Klose et al. 2015; Fraser et al. 2016; Buti et al. 2020; Favero 2020). Absorption of FR light converts the Pfr form back to the Pr form, and this photoconversion results in a dynamic photoequilibrium of Pr and Pfr form. Thus, low R:FR in shade displaces the photoequilibrium toward the inactive Pr form (Casal 2012, 2013; Fraser et al. 2016; Legris et al. 2019). Notably, phyB is deactivated under shade conditions, leading to a promotion of hypocotyl elongation, whereas phyA abundance is enhanced with decreasing R:FR ratios, preventing an excessive hypocotyl elongation of seedlings grown under deep shade (Casal 2012; Li et al. 2012; Martínez-García et al. 2014; Roig-Villanova and Martínez-García 2016; Yang et al. 2018; Pierik and Ballaré 2021).
Therefore, although both phyA and phyB play important roles in mediating SAS, their functional significance differs under different shade conditions: the deactivation of phyB under moderate shade (such as vegetation proximity) allows plants to compete for sunlight with their neighboring vegetation, whereas promotion of phyA abundance under deep shade (such as canopy shade) avoids unnecessary energy expense (Martínez-García et al. 2014; Fiorucci and Fankhauser 2017; Yang et al. 2018). Thus, the coordinated actions of phyA and phyB ensure an optimum hypocotyl elongation of seedlings under different shade conditions. In addition, it was shown that low B light, perceived by cryptochrome photoreceptors, can also trigger SAS and that low B light enhances the SAS induced by low R:FR light perceived by phytochromes (de Wit et al. 2016; Pedmale et al. 2016).
PHYTOCHROME-INTERACTING FACTORS (PIFs) belong to a group of basic helix–loop–helix (bHLH) transcription factors. All 8 PIFs identified so far in Arabidopsis contain a conserved active phytochrome B (APB)–binding motif which is necessary and sufficient for binding with phyB. However, only 2 PIF proteins, PIF1 and PIF3, have an additional active phytochrome A (APA)–binding motif that can mediate their interaction with phyA (Khanna et al. 2004; Duek and Fankhauser 2005; Leivar and Quail 2011; Leivar and Monte 2014; Lee and Choi 2017; Pham et al. 2018; Favero 2020; Pierik and Ballaré 2021). Among all PIFs found in Arabidopsis, PIF3-LIKE1 (PIL1, also known as PIF2), PIF4, PIF5, and PIF7 and, to a lesser extent, PIF1 and PIF3 have been implicated in SAS (Lorrain et al. 2008; Leivar and Quail 2011; Leivar et al. 2012; Li et al. 2012; Leivar and Monte 2014; Lee and Choi 2017; Buti et al. 2020). It has been well documented that upon R or white (W) light illumination, photoactivated phytochromes interact with PIFs and induce their rapid phosphorylation and proteasomal degradation. However, when plants are in shade, low R/FR favors the conversion of phyB into the inactive Pr form that no longer interacts with the PIF proteins, thus increasing their stability and activity (Lorrain et al. 2008; Leivar and Quail 2011; Casal 2013; Leivar and Monte 2014; Fraser et al. 2016; Fiorucci and Fankhauser 2017; Courbier and Pierik 2019; Buti et al. 2020; Favero 2020; Fernández-Milmanda and Ballaré 2021).
It was shown that PIF4 and PIF5 could rapidly reaccumulate in the shade, thus promoting hypocotyl growth by directly upregulating the expression of shade-induced genes (Lorrain et al. 2008; Hornitschek et al. 2009; Hersch et al. 2014). PIF7 plays a predominant role in mediating shade-induced growth responses, and PIF7 proteins are phosphorylated in W light but rapidly (within minutes) dephosphorylated in response to shade (Leivar et al. 2008; Li et al. 2012; de Wit et al. 2015; Mizuno et al. 2015). The expression of PIL1, a typical shade marker gene, displayed >100-fold induction within 30 min of shade treatment (Salter et al. 2003). LONG HYPOCOTYL IN FAR-RED 1 (HFR1), encoding an atypical bHLH protein, was shown to be a master negative regulator of SAS by forming non-DNA–binding heterodimers with PIF4 and PIF5 (Sessa et al. 2005; Hornitschek et al. 2009). Notably, HFR1 was shown to be rapidly induced by shade (Sessa et al. 2005).
Rapid phosphorylation of PIFs upon R light exposure suggests the existence of protein kinase(s) responsible for phosphorylating PIFs. Indeed, in the last decade, several kinases, belonging to different families, have been shown to phosphorylate PIF proteins. Casein kinase II (CK2), a ubiquitous Ser/Thr kinase, directly phosphorylates 7 Ser/Thr residues throughout the PIF1 protein. Ser/Thr to Ala mutations of these sites eliminated CK2 phosphorylation of PIF1 in vitro and significantly reduced light-induced degradation of PIF1 in vivo (Bu et al. 2011). BRASSINOSTE ROID INSENSITIVE2 (BIN2), a GLYCOGEN SYNTHASE KINASE3 (GSK3)–like kinase negatively regulating the brassinosteroid (BR) signaling pathway, was shown to phosphorylate the Ser/Thr residues in a conserved BIN2 phosphorylation consensus motif present in Arabidopsis PIF4/PIF5 proteins and their homologs in other plant species (Bernardo-García et al. 2014), thus facilitating their proteasomal degradation. In addition, BIN2 was also shown to induce PIF3 degradation in darkness by directly phosphorylating PIF3 (Ling et al. 2017). CONSTITUTIVELY PHOTOMORPHOGENIC1 (COP1), a key repressor of photomorphogenesis, interacts with SUPPRESSOR OF phyA-105 (SPA) proteins to form E3 ligase complexes, and it was shown that the COP1/SPA complexes promoted the accumulation of PIFs in the dark (Bauer et al. 2004; Ling et al. 2017; Pham et al. 2018). Interestingly, the COP1/SPA complexes interfere with the interaction between BIN2 and PIF3, thus inhibiting BIN2–mediated PIF3 phosphorylation to stabilize PIF3 in the dark (Ling et al. 2017). Photoregulatory protein kinases (PPKs), a family of nuclear protein kinases consisting of 4 members in Arabidopsis, interact with PIF3 and phyB in a light-dependent manner and mediate light-induced phosphorylation and degradation of PIF3 (Ni et al. 2017). A mitogen-activated protein kinase (MAPK) cascade, MKK10-MPK6, was recently shown to mediate R light-regulated cotyledon opening of Arabidopsis seedlings by phosphorylating PIF3 (Xin et al. 2018). Moreover, a recent study provided evidence showing that both oat (Avena sativa) and Arabidopsis phytochromes function as protein kinases that can directly phosphorylate PIFs in vitro, and unexpectedly, the protein kinase domain (KD) responsible for phosphorylating PIF3 is localized in the N-terminal photosensory domain (consisting of PAS-GAF-PHY tridomain) of recombinant oat phyA (Shin et al. 2016). Recently, the N-terminal serine/threonine KD of SPA proteins was shown to directly phosphorylate PIF1 and PIF4 (Paik et al. 2019; Lee et al. 2020), indicating that the SPA proteins also act as PIF protein kinases in addition to their roles in enhancing COP1 activity.
Under salt stress, a calcium-dependent protein kinase pathway, known as the SALT OVERLY SENSITIVE (SOS) pathway, is used by plants to transduce the salt stress signal and expel Na+ from cells (Zhu 2016; Yang and Guo 2018a, 2018b; Van Zelm et al. 2020; Zhao et al. 2020). Excess intracellular or extracellular Na+ elicits a cytosolic Ca2+ signal, which is sensed by SOS3 and SOS3-LIKE CALCIUM-BINDING PROTEIN8 (SCaBP8), 2 calcium-binding proteins belonging to the CALCINEURIN B-LIKE (CBL) protein family (Liu and Zhu 1997, 1998; Ishitani et al. 2000; Guo et al. 2001; Quan et al. 2007; Lin et al. 2009). SOS3 and SCaBP8 transduce the elevated Ca2+ signal by interacting with and activating SOS2, a serine/threonine protein kinase, and activated SOS2 phosphorylates and activates SOS1, a plasma membrane Na+/H+ antiporter, which extrudes Na+ from the cytosol to the apoplast (Halfter et al. 2000; Shi et al. 2000; Qiu et al. 2002; Quintero et al. 2002; Quan et al. 2007). Under nonstress conditions, 14-3-3, GIGANTEA (GI), and ABA INSENSITIVE 2 (ABI2) proteins interact with SOS2 and inhibit its kinase activity (Ohta et al. 2003; Kim et al. 2013; Zhou et al. 2014). However, 14-3-3 and GI proteins were shown to be degraded through the ubiquitin/26S proteasome pathway under salt stress, thereby releasing SOS2 kinase activity (Kim et al. 2013; Tan et al. 2016).
It was recently shown that low levels of NaCl in soil strongly inhibited shade-induced hypocotyl elongation through ABA-dependent signaling (Hayes et al. 2019). Here, we report an unexpected role of salt-activated SOS2 in promoting SAS by physically interacting with PIF4 and PIF5 to promote their protein abundance. Moreover, our data demonstrate that the kinase activity of SOS2 is enhanced by light through phyA and phyB and that SOS2 regulation of PIF4/PIF5 protein abundance and SAS is more prominent under salt stress. Together, our study demonstrates that SOS2 coordinately regulates plant responses to salt stress and shade, which seems to prevent exaggerated salt inhibition of shade-induced hypocotyl elongation.
Results
SOS2 positively regulates SAS in Arabidopsis
Salt treatment was recently shown to inhibit shade-induced hypocotyl elongation in Arabidopsis (Hayes et al. 2019). Considering the pivotal role of SOS2 in salt stress signaling, we asked whether SOS2 might be involved in regulating SAS. We obtained 2 T-DNA insertion lines of sos2, named sos2-T1 (SALK_016683) and sos2-T2 (SALK_056101), in which the T-DNA was inserted into the 11th intron and the promoter region of SOS2 in sos2-T1 and sos2-T2, respectively (Supplemental Fig. S1, A and B). Our reverse transcription-polymerase chain reaction (RT-PCR) analyses showed that SOS2 expression was notably knocked down in sos2-T2 and was not detectable in sos2-T1 mutant seedlings (Supplemental Fig. S1C). However, our immunoblots using polyclonal anti-SOS2 antibodies showed that SOS2 proteins were barely detectable in both sos2-T1 and sos2-T2 mutants (Supplemental Fig. S1D). We also included in this study a third mutant, sos2-2, which is in the gl1/gl1 Col background with a 2-bp deletion in the 8th exon of SOS2 (Liu et al. 2000; Supplemental Fig. S1, A and E).
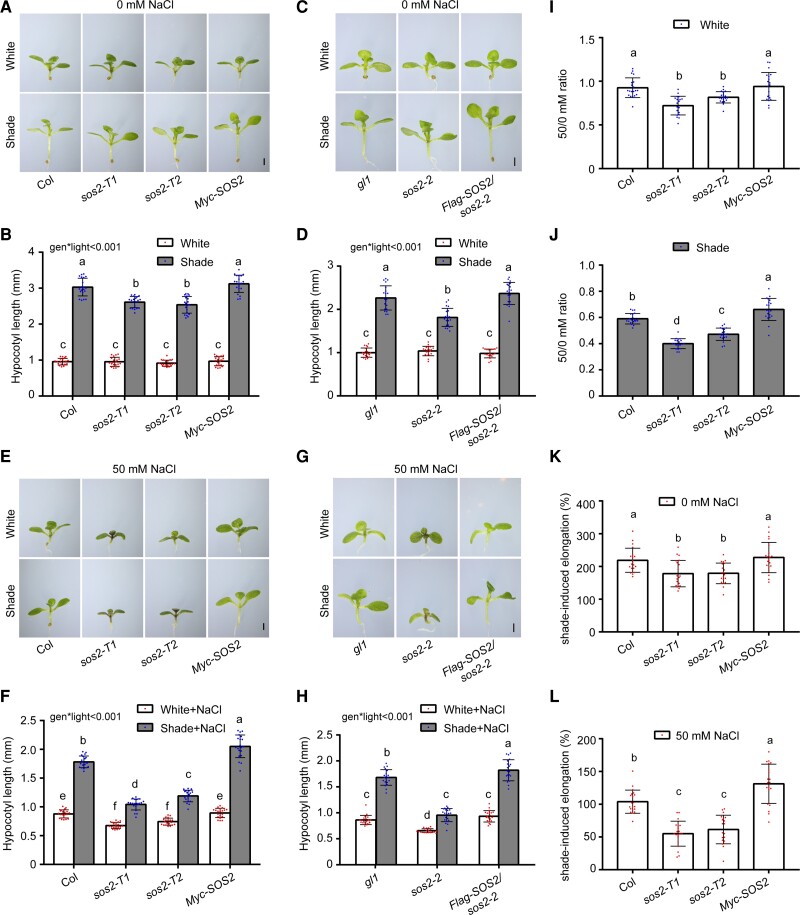
Next, we grew the 3 sos2 mutants together with their respective control plants under simulated W light (R/FR, 9; PAR, 56 μmol m−2 s−1) for 4 d and then either left them in simulated W light or transferred them to low R:FR (simulated shade [R/FR, 0.4; PAR, 56 μmol m−2 s−1]) for 5 more days. Interestingly, we observed that all 3 sos2 mutants exhibited similar phenotypes as their respective controls in simulated W light; however, they all developed significantly shorter hypocotyls in simulated shade (Fig. 1, A to D). Introduction of 35S-driven Flag-SOS2 into sos2-2 mutant seedlings efficiently rescued their short hypocotyl phenotype in the simulated shade (Fig. 1, C and D), indicating that the mutation of SOS2 was responsible for the short hypocotyl phenotype of sos2-2 mutant seedlings under shade.
Figure 1.
SOS2 positively regulates SAS in Arabidopsis. A and B) Phenotypes A) and hypocotyl lengths B) of Col, sos2-T1, sos2-T2, and Pro35S:Myc-SOS2 seedlings grown under simulated W light or shade (R/FR, 0.4). C and D) Phenotypes C) and hypocotyl lengths D) of gl1, sos2-2 (in gl1 background), and Pro35S:Flag-SOS2 sos2-2 seedlings grown under simulated W light or shade (R/FR, 0.4). E and F) Phenotypes E) and hypocotyl lengths F) of Col, sos2-T1, sos2-T2, and Pro35S:Myc-SOS2 seedlings grown under simulated W light or shade (R/FR, 0.4) with 50 mm NaCl treatment. G and H) Phenotypes G) and hypocotyl lengths H) of gl1, sos2-2 (in gl1 background), and Pro35S:Flag-SOS2 sos2-2 seedlings grown under simulated W light or shade (R/FR, 0.4) with 50 mm NaCl treatment. I and J) The ratios of hypocotyl lengths for the indicated seedlings grown on 50 mm NaCl versus 0 mm NaCl under simulated W light I) or shade (R/FR, 0.4) J). Error bars represent Sd from 20 seedlings. Different letters represent significant differences determined by 1-way ANOVA with Tukey's post hoc test (P < 0.05; Supplemental Data Set 1). K and L) The percentage of shade-induced elongation for the indicated seedlings grown without NaCl K) and on 50 mm NaCl L). Error bars represent Sd from 20 seedlings. Different letters represent significant differences determined by 1-way ANOVA with Tukey's post hoc test (P < 0.05; Supplemental Data Set 1). In A) to H), the seedlings were first grown under simulated W light for 4 d and then transferred to simulated shade (R/FR, 0.4) or remained under simulated W light for 5 more days. In A), C), E), and G), scale bar = 1 mm. In B), D), F), and H), error bars represent Sd from 20 seedlings; different letters represent significant differences determined by 2-way ANOVA with Tukey's post hoc test (P < 0.05; Supplemental Data Set 1). The interaction P value between genotypes and light conditions is shown inset (Supplemental Data Set 1).
Notably, under 3 tested shade conditions (R/FR, 0.8, 0.4, and 0.2; PAR, 56 μmol m−2 s−1), sos2 mutant seedlings always displayed defects in SAS (Supplemental Fig. S2). Moreover, when we treated Col seedlings with 50 mm NaCl, we observed that the hypocotyl growth of Col seedlings was not obviously affected by salt treatment under simulated W light but was significantly inhibited by salt treatment under shade conditions (Fig. 1, E to J). These observations were consistent with a recent study (Hayes et al. 2019). Moreover, we observed that sos2 mutant seedlings developed significantly shorter hypocotyls compared with the control plants under simulated W light when treated with 50 mm NaCl (Fig. 1, E to I). In addition, sos2 mutant seedlings displayed increased sensitivity, whereas the Pro35S:Myc-SOS2 seedlings exhibited decreased sensitivity to salt-inhibited hypocotyl growth under several shade conditions (Figs. 1, J to L, and S2). Together, our data indicated that SOS2 is involved in regulating SAS with or without salt stress, playing a more prominent role under salt stress.
To further substantiate the role of SOS2 in regulating SAS of the soil-grown plants, Col, sos2-T1, sos2-T2, and Pro35S:Myc-SOS2 seeds were sown onto wetted soil, stratified in darkness for 3 d, then moved to simulated W light with a long-day (16-h light/8-h dark) photoperiod, and watered with 0 or 50 mm NaCl. After 4 d, the seedlings were transferred to simulated shade (R/FR, 0.4) or remained under simulated W light for 5 more days, continuously watered with 0 or 50 mm NaCl. Notably, sos2 mutant seedlings developed shorter hypocotyls, whereas the Pro35S:Myc-SOS2 seedlings exhibited longer hypocotyls compared with Col in the shade under the treatment of 50 mm NaCl (Supplemental Fig. S3), thus reinforcing the notion that SOS2 coordinately regulates the responses of soil-grown plants to salt stress and shade.
We then asked whether the other SOS pathway components are also involved in regulating SAS. To this end, we compared the phenotypes of sos1, sos3, and scabp8 mutants with their respective control seedlings under simulated W light and shade conditions. We observed that whereas all 3 mutant seedlings were indistinguishable from their control plants under W light, sos1, but not sos3 and scabp8 mutant seedlings, developed shorter hypocotyls under shade (Supplemental Fig. S4, A to D). Notably, all 3 mutant seedlings developed shorter hypocotyls in the shade when treated with 50 mm NaCl (Supplemental Fig. S4, E to H). These observations indicated that the SOS pathway components are involved in SAS particularly under salt stress.
SOS2 posttranslationally promotes PIF4 and PIF5 protein accumulation especially in the shade
We next compared the expression of several early shade marker genes, such as PIL1 (Salter et al. 2003) and HFR1 in the sos2 mutant and control plants (Sessa et al. 2005). We first grew Col and sos2 mutant seedlings under simulated W light for 4 d and then transferred them to simulated shade for 1 h. Our RT-quantitative PCR (RT-qPCR) data showed that whereas the levels of PIL1 and HFR1 transcripts were not obviously regulated by SOS2 under W light, their expression was significantly downregulated in sos2 mutant seedlings in the shade compared to Col (Supplemental Fig. S5, A and B). We also examined the expression of several other shade-responsive genes, such as ARABIDOPSIS THALIANA HOMEOBOX PROTEIN2 (ATHB2), INDOLE-3-ACETIC ACID INDUCIBLE19 (IAA19), and YUCCA8 (YUC8) (Leivar et al. 2012; Li et al. 2012), and our RT-qPCR results indicated that SOS2 was also required for their shade-induced expression (Supplemental Fig. S5, C to E). Together, our data demonstrated that SOS2 positively regulates shade-responsive gene expression in Arabidopsis.
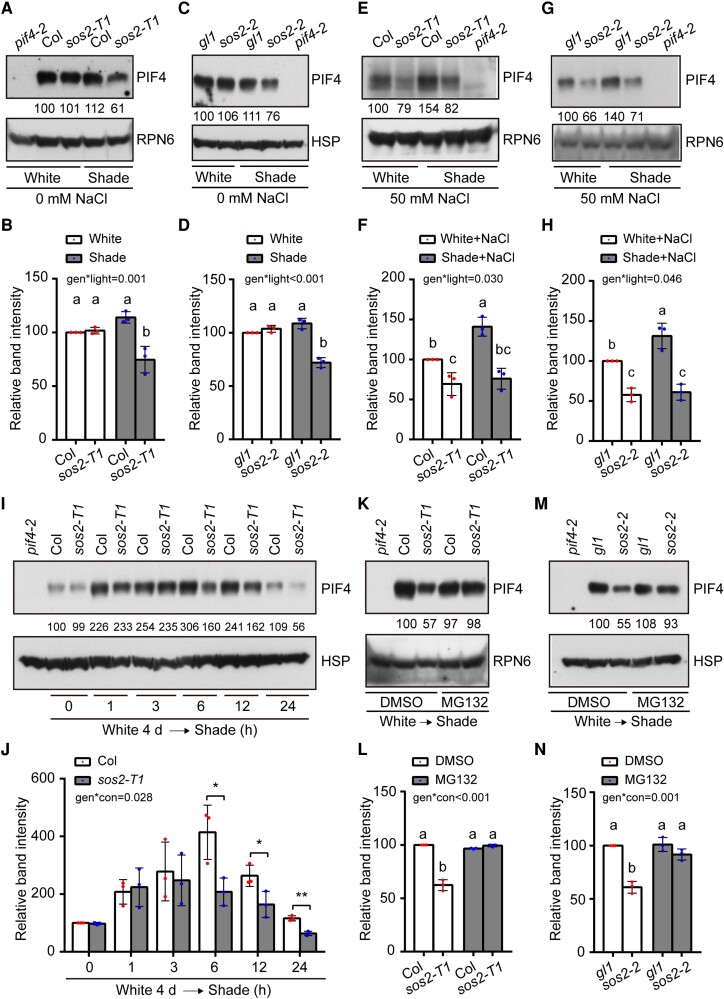
Since all these tested shade-induced genes were previously shown to be directly targeted by PIFs (Hornitschek et al. 2009; Leivar et al. 2012; Li et al. 2012; Zhang et al. 2013), we next asked whether SOS2 may regulate SAS by modulating the abundance of PIF proteins (Lorrain et al. 2008; Li et al. 2012). To test this, we first grew sos2 mutants and control seedlings under simulated W light for 4 d and then either transferred them to simulated shade or left them under simulated W light for 5 more days. Our immunoblot data indicated that the levels of PIF4 and PIF5 proteins in sos2 mutants were largely indistinguishable from those in the control seedlings in W light; however, we observed an obvious decrease in the levels of PIF4 and PIF5 proteins in sos2 mutants grown in the shade (Figs. 2, A to D, and S6, A and B).
Figure 2.
SOS2 promotes PIF4/PIF5 protein accumulation posttranslationally in the shade. A and B) Immunoblots showing the levels of PIF4 proteins in Col and sos2-T1 mutant seedlings grown under simulated W light or shade (R/FR, 0.4). Representative pictures are shown in A), and the relative levels of PIF4 proteins are shown in B). C and D) Immunoblots showing the levels of PIF4 proteins in gl1 and sos2-2 (in gl1 background) mutant seedlings grown under simulated W light or shade (R/FR, 0.4). Representative pictures are shown in C), and the relative levels of PIF4 proteins are shown in D). E and F) Immunoblots showing the levels of PIF4 proteins in Col and sos2-T1 mutant seedlings grown under simulated W light or shade (R/FR, 0.4) on 50 mm NaCl. Representative pictures are shown in E), and the relative levels of PIF4 proteins are shown in F). G and H) Immunoblots showing the levels of PIF4 proteins in gl1 and sos2-2 (in gl1 background) mutant seedlings grown under simulated W light or shade (R/FR, 0.4) on 50 mm NaCl. Representative pictures are shown in G), and the relative levels of PIF4 proteins are shown in H). I and J) Immunoblots showing the levels of PIF4 proteins in Col and sos2-T1 mutant seedlings grown under simulated W light for 4 d and then transferred to shade (R/FR, 0.4) for the indicated times. Representative pictures are shown in I), and the relative levels of PIF4 proteins are shown in J). In J), *P < 0.05 and **P < 0.01 (Student's t test; Supplemental Data Set 1) for the indicated pairs of samples. The interaction P value between genotypes and shade treatment time (conditions) was tested by 2-way ANOVA (Supplemental Data Set 1). K and L) Immunoblots showing the levels of PIF4 proteins in Col and sos2-T1 mutant seedlings grown under simulated W light for 4 d and then treated with mock (DMSO) or MG132 and transferred to shade (R/FR, 0.4) for 12 h. Representative pictures are shown in K) and the relative levels of PIF4 proteins are shown in L). M and N) Immunoblots showing the levels of PIF4 proteins in gl1 and sos2-2 (in gl1 background) mutant seedlings grown under simulated W light for 4 d and then treated with mock (DMSO) or MG132 and transferred to shade (R/FR, 0.4) for 12 h. Representative pictures are shown in M), and the relative levels of PIF4 proteins are shown in N). In A) to H), the seedlings were first grown under simulated W light for 4 d and then transferred to simulated shade (R/FR, 0.4) or remained under simulated W light for 5 more days. In A), E), G), and K), anti-RPN6 was used as a sample loading control; in C), I), and M), anti-HSP was used as a sample loading control. Numbers below the immunoblots in A), C), E), G), I), K), and M) indicate the relative band intensities of PIF4 normalized to the loading control. The ratio of the first clear band was set to 100. Error bars in B), D), F), H), J), L), and N) represent SD from 3 independent assays using 3 pools of seedlings. Different letters represent significant differences determined by 2-way ANOVA with Tukey's post hoc test (P < 0.05; Supplemental Data Set 1). The interaction P value between genotypes and light conditions is shown inset (Supplemental Data Set 1).
Notably, when treated with 50 mm NaCl, the PIF4 and PIF5 protein levels were both markedly decreased (∼80% and ∼40% relative to the wild-type control under shade and W light, respectively) in sos2 mutants compared to the control seedlings under both W light and shade (Figs. 2, E to H, and S6, C and D). Interestingly, we observed that whereas salt treatment did not significantly alter PIF4 protein abundance in Col seedlings grown under simulated W light, it notably increased PIF4 protein levels in Col seedlings grown under simulated shade (Supplemental Fig. S7). However, no significant changes in the levels of PIF3 and PIF7 proteins were observed in sos2 mutant seedlings in both simulated W light and shade conditions (Supplemental Fig. S8). Collectively, our data demonstrated that SOS2 regulates the abundance of PIF4 and PIF5 proteins in the shade particularly under salt stress.
To further investigate the role of SOS2 in modulating PIF4 protein accumulation in response to shade, Col and sos2 mutant seedlings were first grown in simulated W light for 4 d, then transferred to simulated shade for the indicated times ranging from 1 to 24 h, and then harvested and analyzed by immunoblotting. We observed that the levels of endogenous PIF4 proteins in Col seedlings remarkably increased in response to shade, peaking at 6 h after the transfer and then gradually decreasing after prolonged exposure to a low level at 24 h (Fig. 2, I and J), which may explain why the steady-state levels of endogenous PIF4 and PIF5 proteins were only marginally induced by the shade treatment (Figs. 2, A to D, and S6, A and B). However, although the levels of endogenous PIF4 proteins in sos2 mutant were largely similar to those of Col seedlings after 1 to 3 h of shade, they were clearly lower in sos2 mutant seedlings after 6 h of shade (Fig. 2, I and J). Together, these data indicated that the abundance of endogenous PIF4 proteins is strictly regulated in response to shade and that SOS2 is involved in controlling PIF4 protein abundance after a relatively long exposure to shade.
To explore the molecular mechanism underlying SOS2 promotion of PIF4 and PIF5 protein accumulation, we first asked whether SOS2 might regulate the transcript levels of PIF4 and PIF5. Our RT-qPCR analyses showed that the expression of PIF4 and PIF5 was not obviously decreased in sos2 mutants grown under simulated W light or shade with 0 or 50 mm NaCl (Supplemental Fig. S9, A and B). These results indicated that SOS2 might not promote PIF4 and PIF5 protein abundance in the shade and in response to salt stress through transcriptional upregulation of their expression. The transcript levels of PIF3 and PIF7 were also examined, and our data indicated that SOS2 also did not obviously upregulate PIF3 and PIF7 expression (Supplemental Fig. S9, C and D).
To investigate whether SOS2 regulates PIF4 protein abundance posttranslationally, sos2 mutant and the control seedlings were first grown in simulated W light for 4 d, then treated with MG132 (an inhibitor of 26S proteasomes) or DMSO (the solvent for MG132), and transferred to simulated shade for 12 more hours. Our immunoblot data indicated that the levels of endogenous PIF4 proteins obviously decreased in DMSO–treated sos2 mutant seedlings in the shade; however, MG132 treatment effectively inhibited the decrease of PIF4 protein abundance in sos2 mutants (Fig. 2, K to N). These data indicated that SOS2 promoted PIF4 protein accumulation in the shade by inhibiting 26S proteasome-mediated PIF4 degradation. Collectively, our data demonstrated that SOS2 promotes PIF4/PIF5 protein accumulation posttranslationally in the shade.
Genetic relationship between SOS2 and PIF4/PIF5 in mediating SAS
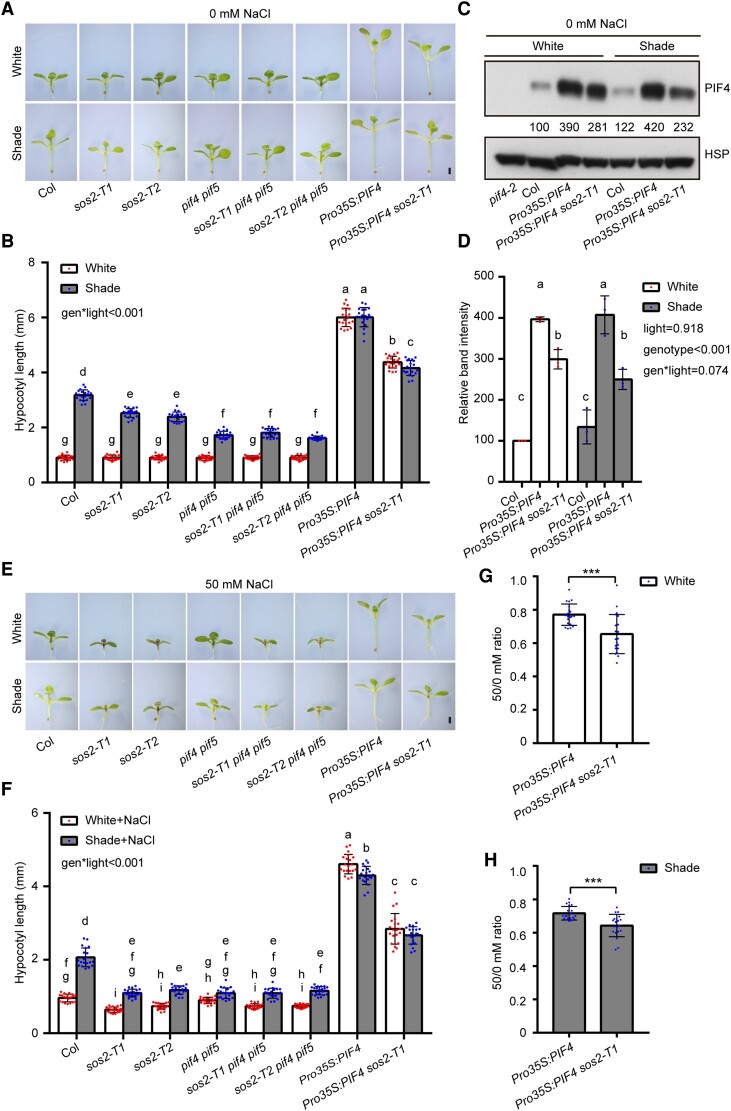
To determine the genetic relationships between SOS2 and PIF4/PIF5 in mediating SAS, we crossed sos2-T1 and sos2-T2 with pif4 pif5, respectively, and obtained homozygous sos2-T1 pif4 pif5 and sos2-T2 pif4 pif5 triple mutants (Supplemental Figs. S10, A and B). The 2 sos2 pif4 pif5 triple mutants were then grown together with Col, 2 sos2 single mutants, and pif4 pif5 double mutants under simulated W light and shade conditions. Interestingly, we observed that whereas the hypocotyl lengths of pif4 pif5 mutant seedlings were notably shorter than those of sos2 mutants in the shade, the hypocotyl lengths of sos2 pif4 pif5 mutants were indistinguishable from those of pif4 pif5 in the shade (Fig. 3, A and B), indicating that the pif4 pif5 mutations are epistatic to sos2 in regulating SAS.
Figure 3.
Genetic relationship between SOS2 and PIF4/PIF5 in mediating SAS. A and B) Phenotypes A) and hypocotyl lengths B) of Col, sos2-T1, sos2-T2, pif4 pif5, sos2-T1 pif4 pif5, sos2-T2 pif4 pif5, Pro35S:PIF4, and Pro35S:PIF4 sos2-T1 seedlings grown under simulated W light or shade (R/FR, 0.4). C and D) Immunoblots showing the levels of PIF4 proteins in Col, Pro35S:PIF4, and Pro35S:PIF4 sos2-T1 seedlings grown under simulated W light or shade (R/FR, 0.4). Anti-HSP was used as a sample loading control. Representative pictures are shown in C) and the relative levels of PIF4 proteins are shown in D). Numbers below the immunoblots in C) indicate the relative band intensities of PIF4 normalized to the loading control. The ratio of the first clear band was set to 100. E and F) Phenotypes E) and hypocotyl lengths F) of Col, sos2-T1, sos2-T2, pif4 pif5, sos2-T1 pif4 pif5, sos2-T2 pif4 pif5, Pro35S:PIF4, and Pro35S:PIF4 sos2-T1 seedlings grown under simulated W light or shade (R/FR, 0.4) on 50 mm NaCl. G and H) The ratios of hypocotyl lengths for the indicated seedlings grown on 50 mm NaCl versus 0 mm NaCl under simulated W light G) or shade (R/FR, 0.4) H). In A) to H), the seedlings were first grown under simulated W light for 4 d and then transferred to simulated shade (R/FR, 0.4) or remained under simulated W light for 5 more days. In A) and E), scale bar = 1 mm. Error bars in B), F), G), and H) represent Sd from 20 seedlings, and error bars in D) represent Sd from 3 independent assays using 3 pools of seedlings. Different letters in B), D), and F) represent significant differences determined by 2-way ANOVA with Tukey's post hoc test (P < 0.05; Supplemental Data Set 1). In G) and H), ***P < 0.001 (Student's t test; Supplemental Data Set 1) for the indicated pair of samples. The interaction P value between genotypes and light conditions is shown inset (Supplemental Data Set 1).
We also introduced the sos2-T1 mutation into Pro35S:PIF4 transgenic lines (Supplemental Fig. S10, C and D) and compared the phenotypes of Pro35S:PIF4 sos2-T1 and Pro35S:PIF4 seedlings grown in simulated W light and shade. Notably, we observed that Pro35S:PIF4 sos2-T1 developed significantly shorter hypocotyls than Pro35S:PIF4 seedlings in both conditions (Fig. 3, A and B). Consistent with this observation, PIF4 proteins accumulated to lower levels in Pro35S:PIF4 sos2-T1 than in Pro35S:PIF4 seedlings in both simulated W light and shade (Fig. 3, C and D), which was not due to decreased expression of the transgene after genetic crossing (Supplemental Fig. S10D). Together, our data demonstrated that pif4 pif5 mutations are epistatic to sos2 in regulating hypocotyl elongation in the shade and that SOS2 promotes PIF4 protein abundance posttranslationally under W light and shade conditions.
We then grew 2 sos2 pif4 pif5 triple mutants together with Col, 2 sos2 single mutants, and pif4 pif5 double mutant seedlings on 50 mm NaCl under simulated W light and shade conditions. Notably, we observed that in response to salt stress, 2 sos2 single mutants, 2 sos2 pif4 pif5 triple mutants, and pif4 pif5 double mutant seedlings all developed similar hypocotyl lengths in the shade (Fig. 3, E and F). Intriguingly, after measuring the ratios of hypocotyl lengths on 50 mm NaCl versus 0 mm NaCl, we found that compared with Pro35S:PIF4 seedlings, Pro35S:PIF4 sos2-T1 seedlings displayed increased sensitivity to salt-inhibited hypocotyl growth under both W light and shade conditions (Fig. 3, G and H). Together, our data demonstrated that the role of SOS2 in regulating shade-induced hypocotyl growth through PIF4/PIF5 is modulated by salt stress.
SOS2 physically interacts with PIF4 and PIF5
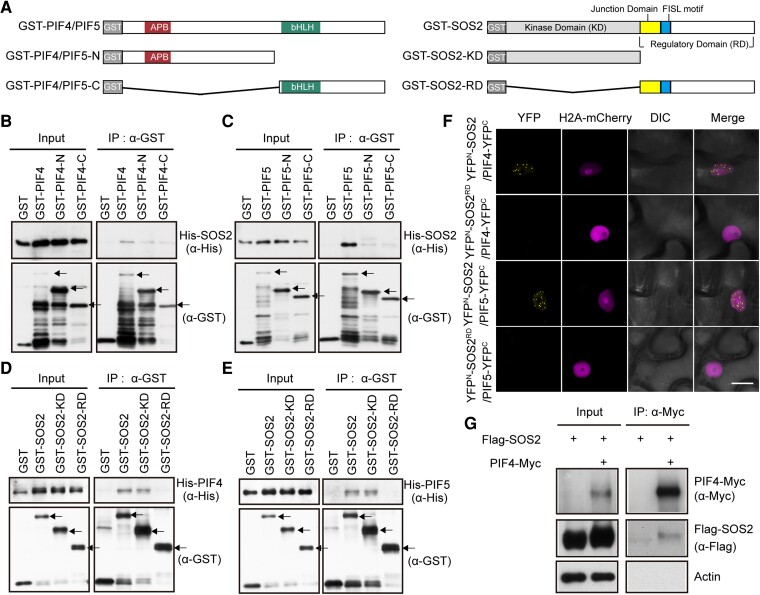
Next, we asked whether SOS2 could physically interact with PIF4 and PIF5. Since both PIF4 and PIF5 proteins harbor the conserved APB motif and bHLH domains (Duek and Fankhauser 2005; Leivar and Quail 2011), we first performed in vitro pull-down assays by expressing glutathione S-transferase (GST)-tagged full-length, N-terminal (containing the APB motif) or C-terminal (containing bHLH) domains of PIF4 and PIF5 (Fig. 4A) and histidine (His)-tagged full-length SOS2 in Escherichia coli. Our pull-down assays showed that GST-tagged full-length PIF4 and PIF5 proteins, but not GST alone, were able to pull down His-tagged SOS2 in vitro (Fig. 4, B and C). However, GST-tagged N-terminal and C-terminal domains of both PIF4 and PIF5 could only pull down markedly lower levels of His-SOS2 in vitro (Fig. 4, B and C), indicating that the full-length PIF4 and PIF5 proteins are required for interacting with SOS2.
Figure 4.
SOS2 physically interacts with PIF4 and PIF5. A) Schematic diagrams of GST-tagged PIF4/PIF5, PIF4/PIF5-N, PIF4/PIF5-C, SOS2, SOS2-KD, and SOS2-RD proteins. B and C) Pull-down assays showing that GST-tagged PIF4 B) and PIF5 C), but not GST alone, could pull down His-tagged SOS2 in vitro. The arrows indicate the GST-PIF4/5, GST-PIF4/5-N, and GST-PIF4/5-C proteins, respectively. D and E) Pull-down assays showing that GST-tagged SOS2 and SOS2-KD, but not GST alone, could pull down His-tagged PIF4 D) and PIF5 E) in vitro. The arrows indicate the GST-SOS2, GST-SOS2-KD, and GST-SOS2-RD proteins, respectively. F) BiFC assays showing the interactions between SOS2 and PIF4/PIF5 in N. benthamiana leaf cells. The indicated combinations of YFPN-SOS2, YFPN-SOS2RD PIF4-YFPC, and PIF5-YFPC constructs were co-transfected into N. benthamiana leaf cells, respectively. H2A-mCherry was the nuclear-localization marker. Scale bar = 20 μm. DIC, differential interference contrast. YFPN, N-terminal fragment of Yellow Fluorescent Protein; YFPC, C-terminal fragment of Yellow Fluorescent Protein. G) Co-IP assays showing that SOS2 associated with PIF4 in vivo. Flag-SOS2 and PIF4-Myc fusion proteins were transiently expressed in Arabidopsis (Col) protoplasts. Total proteins were extracted and incubated with Myc-trap agarose beads (AlpaLife). Total and precipitated proteins were examined by immunoblotting using antibodies against Myc, Flag and Actin, respectively.
We then investigated which domain of SOS2 mediates its interaction with PIF4 and PIF5. SOS2 consists of a conserved N-terminal KD and a C-terminal regulatory domain (RD) (Guo et al. 2001; Fig. 4A). Our in vitro pull-down assays showed that GST-tagged full-length and the N-terminal KD of SOS2, but not GST alone, were able to pull down His-tagged PIF4 and PIF5 proteins (Fig. 4, D and E). By contrast, the GST-tagged C-terminal RD of SOS2 was unable to pull down PIF4 and PIF5 proteins in vitro (Fig. 4, D and E). These data demonstrated that the KD of SOS2 mediates its interactions with PIF4 and PIF5.
To verify the physical interaction between SOS2 and PIF4/PIF5 in planta, bimolecular fluorescence complementation (BiFC) assays (Waadt et al. 2008) were performed by transiently expressing YFPN-SOS2 and PIF4/PIF5-YFPC fusions in Nicotiana benthamiana leaf cells. Our data showed that co-expression of YFPN-SOS2 with PIF4-YFPC or PIF5-YFPC led to strong YFP fluorescence (Figs. 4F and S11A). By contrast, YFPN-SOS2 co-transformed with GUS-YFPC or YFPN-tagged RD of SOS2 (YFPN-SOS2RD) co-transformed with PIF4-YFPC or PIF5-YFPC showed no detectable YFP fluorescence (Figs. 4F and S11A). Notably, we observed that SOS2 interacts with PIF4 and PIF5 in the nucleus, leading to the formation of nuclear bodies (Figs. 4F and S11A). These observations support the conclusion that SOS2 physically interacts with PIF4 and PIF5 in living plant cells.
To confirm the physical interaction between SOS2 and PIF4 in vivo, co-immunoprecipitation (co-IP) assays were performed by expressing PIF4-Myc and Flag-SOS2 in Arabidopsis protoplasts. Our immunoblot data showed that Flag-SOS2 was co-precipitated by the anti-Myc antibodies in the presence of PIF4-Myc (Fig. 4G), indicating that PIF4 associated with SOS2 in vivo. To test for physical interaction between SOS2 and PIF5 in vivo, Col, sos2-T1, and Pro35S:Myc-SOS2 seedlings were first grown in the dark for 4 d and then subjected to 30 min of shade treatment. Co-IP assays were then performed using the anti-Myc antibodies, and our immunoblot data showed that endogenous PIF5 proteins were co-precipitated by the anti-Myc antibodies in Pro35S:Myc-SOS2 seedlings, but not in Col and sos2-T1 mutant seedlings (Supplemental Fig. S11B), indicating that Myc-SOS2 associated with PIF5 in vivo. Collectively, our data demonstrated that SOS2 physically interacts with PIF4 and PIF5.
SOS2 promotes PIF4 protein stability by phosphorylating a serine residue close to the APB motif
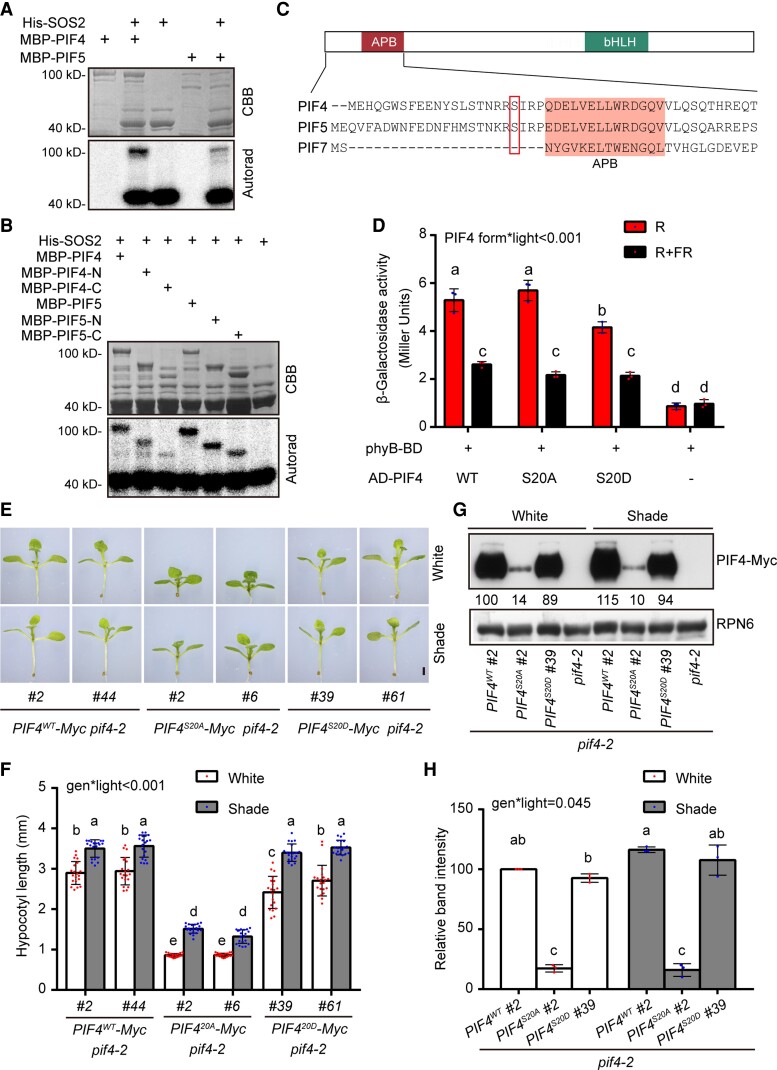
Since SOS2 is a well-characterized protein kinase, we next asked whether SOS2 could phosphorylate PIF4 and PIF5. We first performed in vitro phosphorylation assays using His-SOS2 and MBP-PIF4/PIF5 proteins expressed in and purified from E. coli. Our results showed that PIF4 and PIF5 were phosphorylated in the presence of SOS2 (Fig. 5A), indicating that they are substrates of SOS2 in vitro. Further analyses revealed that SOS2 preferentially phosphorylates the N-terminal regions than the C-terminal regions of both PIF4 and PIF5 (Fig. 5B). Mass spectrometry assays were then performed to identify the SOS2 phosphorylation sites in the PIF4 and PIF5 proteins. Interestingly, a conserved serine residue of PIF4 and PIF5, i.e. S20 of PIF4 and S22 of PIF5, were phosphorylated by SOS2 in vitro (Supplemental Fig. S12). Notably, this site is not conserved in PIF7 (Fig. 5C).
Figure 5.
SOS2 promotes PIF4 protein stability by phosphorylating a serine residue near to the APB motif. A and B) In vitro kinase assays showing that SOS2 directly phosphorylates full-length A) and truncated B) PIF4 and PIF5 proteins. In A) and B), top panels show CBB–stained SDS–PAGE gel containing His-SOS2 and MBP-PIF4/PIF5 proteins, and bottom panels show autoradiographs (Autorad) indicating SOS2 autophosphorylation (bottom bands) and MBP-PIF4/PIF5 phosphorylation (top bands). C) Schematic diagram of the domain structures of PIFs and the N-terminal sequences of PIF4, PIF5, and PIF7. The APB–binding motif (Khanna et al. 2004) is shaded, and the SOS2-phosphorylated serine residues in PIF4 and PIF5 are boxed. D) PIF4WT and PIF4S20A interacted with the Pfr form of phyB more strongly than PIF4S20D in yeast cells. Yeast cells transformed with the indicated plasmids were used for ONPG assays. The yeast cultures were irradiated with 5 min of R, or 5 min of R immediately followed by 5 min of FR, and then incubated for 2 h. The yeast cultures were then exposed to the same R or R + FR light treatments again and incubated for another 2 h. The β-galactosidase activities were then measured by liquid culture assays using ONPG as the substrate. Error bars represent Sd of 3 independent yeast cultures. Different letters represent significant differences determined by 2-way ANOVA with Tukey's post hoc test (P < 0.05; Supplemental Data Set 1). The interaction P value between PIF4 forms and light conditions is shown inset (Supplemental Data Set 1). E and F) Phenotypes E) and hypocotyl lengths F) of Super:PIF4WT-Myc pif4-2, Super:PIF4S20A-Myc pif4-2, and Super:PIF4S20D-Myc pif4-2 seedlings grown under simulated W light or shade (R/FR, 0.4). For each transgene, 2 independent homozygous lines with similar PIF4 expression levels were selected for further analyses. G and H) Immunoblots showing the levels of PIF4 proteins in Super:PIF4WT-Myc pif4-2, Super:PIF4S20A-Myc pif4-2, and Super:PIF4S20D-Myc pif4-2 seedlings grown under simulated W light or shade (R/FR, 0.4). Anti-RPN6 was used as a sample loading control. Representative pictures are shown in G), and the relative levels of PIF4 proteins are shown in H). Numbers below the immunoblots in G) indicate the relative band intensities of PIF4 normalized to the loading control. The ratio of the first band was set to 100. In E) to H), the seedlings were first grown under simulated W light for 4 d and then transferred to simulated shade (R/FR, 0.4) or remained under simulated W light for 5 more days. In E), scale bar = 1 mm. Error bars in F) represent Sd from 20 seedlings, and error bars in H) represent Sd from 3 independent assays using 3 pools of seedlings. Different letters in F) and H) represent significant differences determined by 2-way ANOVA with Tukey's post hoc test (P < 0.05; Supplemental Data Set 1). The interaction P value between genotypes and light conditions is shown inset (Supplemental Data Set 1).
The fact that this conserved serine residue of PIF4 and PIF5 is near the APB motif (Fig. 5C) prompted us to ask whether the phosphorylation status of this site might regulate phyB interactions with PIF4 and PIF5. To test this possibility, we employed a yeast 2-hybrid system (Shimizu-Sato et al. 2002) by adding phycocyanobilin (PCB) extracted from Spirulina into yeast media to allow phyB to form the Pfr and Pr forms, respectively, after R and FR irradiation. Our results showed that, consistence with previous reports (Khanna et al. 2004; Dong et al. 2020), wild-type PIF4 preferentially interacted with the Pfr form of phyB in yeast (Saccharomyces cerevisiae) cells (Fig. 5D).
However, whereas mutation of PIF4 S20 to phosphorylation-deficient alanine (S20A) did not obviously change PIF4 interaction with phyB Pfr, mutation of PIF4 S20 to phosphorylation-mimic aspartic acid (S20D) significantly decreased PIF4 interaction with phyB Pfr in yeast cells (Fig. 5D). Our immunoblot data showed that there were similar levels of AD-PIF4S20A, AD-PIF4S20D, and AD-PIF4WT proteins in the assayed yeast cells, indicating that these differences in interactions were not due to differences in protein abundance (Supplemental Fig. S13). Consistently, our in vivo plate assays showed that the expression level of the LacZ reporter gene was lower in R light-grown yeast transformants co-expressing phyB N621-LexA (N-terminal 621-amino acids of phyB fused with the LexA DNA-binding domain) and AD-PIF4S20D than those co-expressing phyB N621-LexA with AD-PIF4WT or AD-PIF4S20A (Supplemental Fig. S14). Together, these data demonstrate that phosphorylation of PIF4 at S20 decreased PIF4 interaction with phyB Pfr in yeast cells.
A recent study showed that the phosphorylation status of PIF7 could regulate its nuclear accumulation (Huang et al. 2018). To investigate whether SOS2-mediated phosphorylation could regulate the subcellular localization of PIF4, we transiently transfected Super:PIF4-GFP plasmid DNA into Arabidopsis protoplasts prepared from wild-type (Col) and sos2-T1 mutant seedlings, respectively. After transfection, the protoplasts were treated with simulated W light or shade for 3 h and then subjected to confocal microscopy. We observed that PIF4-GFP was constitutively localized in the nucleus of both Col and sos2 mutant protoplasts after W light or shade treatments (Supplemental Fig. S15). These observations indicated that the nuclear localization of PIF4 may not be regulated by SOS2-mediated phosphorylation.
To further investigate whether the phosphorylation status of PIF4 S20 could regulate PIF4 function in vivo, we generated transgenic Arabidopsis plants expressing Myc-tagged wild-type PIF4 (PIF4WT), PIF4S20A, or PIF4S20D, respectively, in the pif4-2 mutant background under the control of the constitutive Super promoter. Multiple independent transgenic lines were obtained for each construct, and 2 independent homozygous lines were selected for Super:PIF4WT-Myc, Super:PIF4S20A-Myc, and Super:PIF4S20D-Myc, respectively, based on the criterion that the expression levels of PIF4 were largely comparable in these lines (Supplemental Fig. S16A). Interestingly, we observed that whereas Super:PIF4S20D-Myc and Super:PIF4WT-Myc seedlings exhibited similar hypocotyl growth in the shade, Super:PIF4S20A-Myc seedlings developed substantially shorter hypocotyls in both simulated W light and shade (Fig. 5, E and F). Strikingly, our immunoblot data showed that whereas the S20D mutation did not obviously change the levels of transgenic PIF4 proteins, the S20A mutation led to a dramatically decreased stability of PIF4 in both simulated W light and shade (Fig. 5, G and H).
The expression levels of the shade-responsive genes HFR1, YUC8, and IAA19 in Super:PIF4WT-Myc, Super:PIF4S20A-Myc, and Super:PIF4S20D-Myc seedlings corresponded with the levels of transgenic PIF4 proteins in simulated shade (Supplemental Fig. S16, B to D). In addition, when we grew Super:PIF4WT-Myc, Super:PIF4S20A-Myc, and Super:PIF4S20D-Myc seedlings in simulated W light and shade on 50 mm NaCl, we observed similar phenotypes of Super:PIF4WT-Myc, Super:PIF4S20A-Myc, and Super:PIF4S20D-Myc seedlings and similar accumulation patterns of S20A and S20D PIF4 proteins with or without salt stress (Figs. 5, E to H, and S17). It should be noted that wild-type PIF4 and PIF4S20A mutant interacted similarly with phyB in yeast cells (Fig. 5D), whereas Super:PIF4WT-Myc and Super:PIF4S20D-Myc seedlings behaved similarly in simulated W light and shade (Fig. 5, E to H), which may be explained by the fact that SOS2, as well as other potential protein kinases that can phosphorylate S20 of PIF4, is absent in yeast cells but present in Arabidopsis. Collectively, our data suggest that the defect in phosphorylation of the S20 residue leads to enhanced destabilization of PIF4S20A under both simulated W light and shade, which is possibly due to increased interaction of PIF4S20A with phyB Pfr.
phyA and phyB physically interact with SOS2 to promote its kinase activity in the light
Next, we asked how SOS2 activity is regulated by light. We first grew wild-type (Col) seedlings in darkness, shade, or W, R, FR, and B light conditions and then examined the levels of SOS2 transcripts by RT-qPCR assays and the levels of endogenous SOS2 proteins by immunoblotting. Our results showed that both SOS2 transcript and protein levels were not markedly regulated by light (Supplemental Fig. S18).
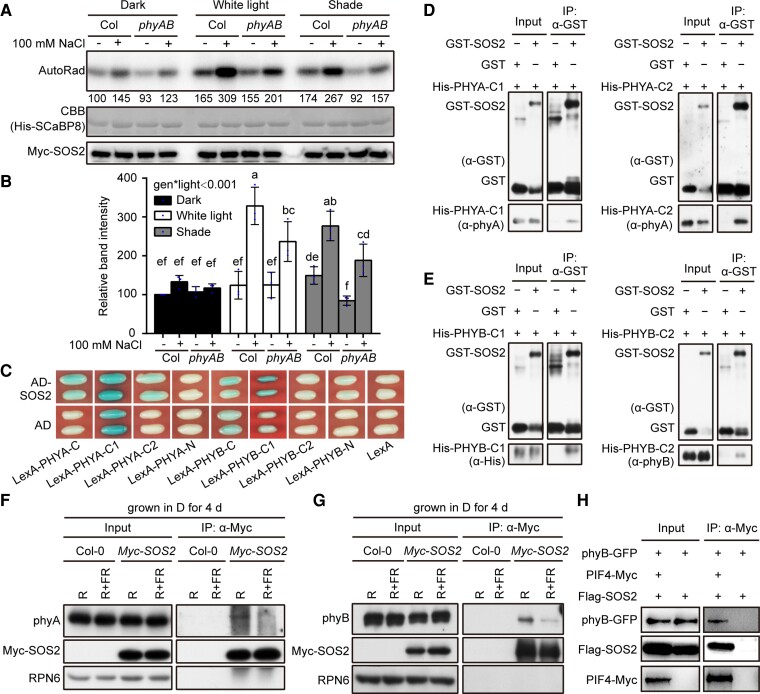
To investigate whether SOS2 kinase activity is regulated by light, we grew Pro35S:Myc-SOS2 transgenic seedlings in dark, W light, and shade conditions, respectively, and then performed semi-in vivo kinase assays to compare the kinase activities of SOS2 under different light conditions using immunoprecipitated Myc-SOS2 and recombinant His-SCaBP8 proteins (Lin et al. 2009). Interestingly, although it was well documented that NaCl treatment remarkably induced SOS2 kinase activity (Lin et al. 2009; Zhou et al. 2014; Ma et al. 2019), our data showed that salt stress only induced SOS2 kinase activities in the light, including both W light and shade, but not in the dark (Figs. 6, A and B, and S19), demonstrating that salt and light synergistically induce SOS2 kinase activity.
Figure 6.
SOS2 physically interacts with phyA and phyB. A, B) Semi-in vivo kinase assays showing SOS2 kinase activity in Pro35S:Myc-SOS2 and Pro35S:Myc-SOS2 phyA phyB seedlings grown under different light conditions. Dark, the seedlings were grown in darkness for 9 d; W light, the seedlings were grown in continuous W light (PAR, 50 μmol m−2 s−1) for 9 d; shade, the seedlings were first grown in continuous W light (PAR, 50 μmol m−2 s−1) for 5 d and transferred to simulated shade (R/FR, 0.6) for another 4 d and then treated with mock (-NaCl) or 100 mm NaCl for 12 h. Top panel shows autoradiograph indicating SOS2 kinase activity, middle panel shows CBB–stained SDS–PAGE gel containing His-SCaBP8 protein used as the SOS2 substrate, and bottom panel shows the immunoprecipitated Myc-SOS2 proteins detected by immunoblotting. Representative pictures are shown in A), and the relative levels of SOS2 kinase activity are shown in B). In A), numbers below the autoradiograph indicate the relative band intensities of SOS2 kinase activity normalized to those of the immunoprecipitated Myc-SOS2 proteins, respectively. The ratio of the first band was set to 100 for the gel. The results of the other 2 assays are shown in Supplemental Fig. S19. In B), error bars represent Sd from 3 independent assays using 3 pools of seedlings. Different letters represent significant differences by 2-way ANOVA with Duncan's post hoc test (P < 0.05; Supplemental Data Set 1). The interaction P value between genotypes and light conditions is shown inset (Supplemental Data Set 1). C) Yeast 2-hybrid assays showing that the HKRD domain (C2) of PHYA and the PRD domain (C1) of PHYB interact with SOS2 in yeast cells. D and E) Pull-down assays showing that GST-tagged SOS2, but not GST alone, could pull down His-tagged PRD domains (C1) and HKRD domains (C2) of PHYA and PHYB in vitro. F and G) Co-IP assays showing that SOS2 associated with phyA and phyB in vivo. Col and Pro35S:Myc-SOS2 seedlings were first grown in darkness for 4 d, then the total proteins were extracted and treated with 5 min of R light or with 5 min of R light followed by 5 min of FR light (R + FR) and then incubated with anti-Myc Affinity Gel (Sigma-Aldrich). The total and precipitated proteins were subjected to immunoblot analyses with antibodies against phyA, phyB, Myc, and RPN6, respectively. H) Co-IP assays showing that PIF4 associated with SOS2 and phyB in vivo. PIF4-Myc, phyB-GFP, and Flag-SOS2 proteins were first transiently expressed in Arabidopsis (Col-0) protoplasts, and then total proteins were extracted and incubated with Myc-trap agarose beads (AlpaLife). Total and precipitated proteins were examined by immunoblotting using antibodies against Myc, Flag, and GFP, respectively.
Since phyA and phyB are the 2 most important phytochrome photoreceptors in plants (Li et al. 2011; Legris et al. 2019), we generated homozygous Pro35S:Myc-SOS2 phyA phyB transgenic seedlings and performed semi-in vivo kinase assays to evaluate the role of phyA/phyB in regulating SOS2 kinase activity in the light. Intriguingly, our data showed that salt-induced SOS2 kinase activities were notably impaired in W light and shade conditions in the absence of phyA and phyB (Figs. 6, A and B, and S19), indicating that phyA and phyB promote salt-induced SOS2 kinase activity in the light.
We then asked whether SOS2 could physically interact with phyA and phyB. To this end, we first employed a yeast 2-hybrid system using bait vectors expressing the N-terminal, C-terminal, PAS-related (designated as C1), or His kinase–related (designated as C2) domains of PHYA and PHYB apoproteins fused to the LexA DNA binding domain (Supplemental Fig. S20) and the prey vector expressing the full-length SOS2 protein fused to the activation domain (AD). Our results showed that the C-terminal domains of both PHYA and PHYB, particularly C2 of PHYA and C1 of PHYB, interacted with SOS2 in yeast cells (Fig. 6C). To further verify the interactions between SOS2 and the C-terminal domains of PHYA and PHYB, we performed in vitro pull-down assays using GST-tagged full-length SOS2 and His-tagged C1 and C2 domains of PHYA or PHYB. Our pull-down assays showed that GST-SOS2, but not GST alone, was able to pull down His-tagged C1 and C2 domains of both PHYA and PHYB in vitro (Fig. 6, D and E). Collectively, these data indicated that SOS2 physically interacted with the C-terminal domains of both PHYA and PHYB in yeast cells and in vitro.
To confirm the physical interactions between SOS2 and phyA/phyB in vivo, we conducted co-IP assays using Col and Pro35S:Myc-SOS2 transgenic seedlings grown in the dark for 4 d. To determine which form (Pr or Pfr) of phyA and phyB associated with Myc-SOS2 more strongly, total proteins were extracted in the darkroom and exposed to 5 min of R light or 5 min of R light immediately followed by 5 min of FR light. Our immunoblot data showed that both phyA and phyB were co-precipitated with anti-Myc antibodies in Pro35S:Myc-SOS2 but not in Col seedlings (Fig. 6, F and G). Moreover, larger amounts of phyA and phyB were co-precipitated with Myc-SOS2 after R light exposure than R plus FR light irradiation (Fig. 6, F and G), indicating that SOS2 preferentially interacts with the Pfr forms of phyA and phyB in vivo.
To further substantiate the associations of phyB, SOS2, and PIF4 in vivo, we performed co-IP assays by expressing Flag-SOS2, phyB-GFP, and PIF4-Myc proteins in Arabidopsis protoplasts. Our immunoblot data showed that phyB-GFP and Flag-SOS2 were co-precipitated by the anti-Myc antibodies in the presence of PIF4-Myc (Fig. 6H), indicating that PIF4 associated with phyB and SOS2 in vivo. Collectively, our data demonstrated that SOS2 is tightly associated with the phyB-PIF4/5 module and that photoactivated phyA and phyB physically interact with SOS2 to promote its kinase activity in the light.
SOS2 genetically interacts with phyA/phyB in mediating SAS
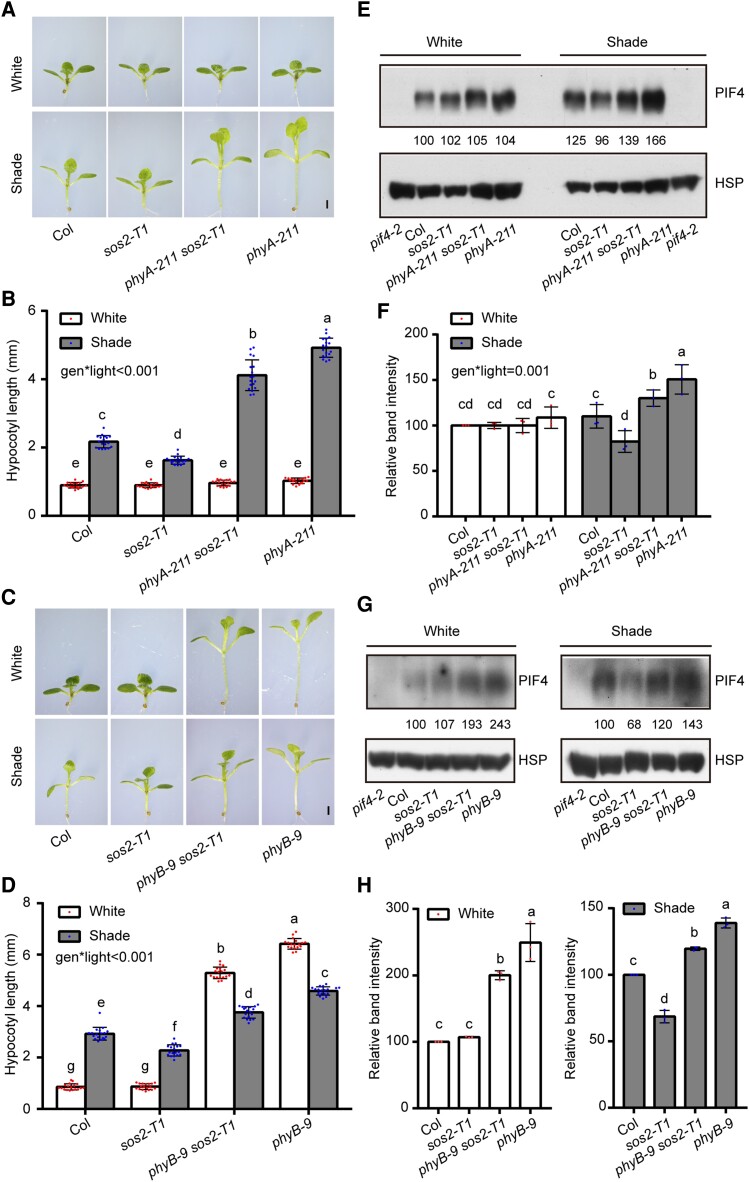
Since both phyA and phyB play important roles in mediating SAS (Martínez-García et al. 2014; Roig-Villanova and Martínez-García 2016; Yang et al. 2018), we finally investigated the genetic relationships between SOS2 and phyA/phyB in SAS. We generated phyA-211 sos2-T1 and phyB-9 sos2-T1 double mutants by genetic crossing and grew them together with Col and their respective single mutant seedlings in simulated W light and shade. Interestingly, we observed that under simulated W light, phyA-211 sos2-T1 seedlings were indistinguishable from Col and their single mutants, whereas the hypocotyls of phyB-9 sos2-T1 double mutant seedlings were of intermediate lengths compared with Col and phyB mutant seedlings (Fig. 7, A to D). Moreover, under simulated shade, both phyA-211 sos2-T1 and phyB-9 sos2-T1 double mutant seedlings developed hypocotyls that were longer than Col but shorter than phyA-211 or phyB-9 mutants (Fig. 7, A to D). These observations indicate that SOS2 contributes significantly to the long hypocotyl phenotypes of phyA-211 and phyB-9 mutant seedlings in the shade.
Figure 7.
SOS2 genetically interacts with phyA/phyB in mediating SAS. A and B) Phenotypes A) and hypocotyl lengths B) of Col, sos2-T1, phyA-211 sos2-T1, and phyA-211 seedlings first grown under simulated W light for 4 d and then transferred to simulated shade (R/FR, 0.4) or remained under simulated W light for 5 more days. In A), scale bar = 1 mm. In B), error bars represent Sd from 18 seedlings. Different letters represent significant differences by 2-way ANOVA with Tukey's post hoc test (P < 0.05; Supplemental Data Set 1). The interaction P value between genotypes and light conditions is shown inset (Supplemental Data Set 1). C and D) Phenotypes C) and hypocotyl lengths D) of Col, sos2-T1, phyB-9 sos2-T1, and phyB-9 seedlings first grown under simulated W light for 4 d and then transferred to simulated shade (R/FR, 0.8) or remained under simulated W light for 5 more days. In C), scale bar = 1 mm. In D), error bars represent Sd from 18 seedlings. Different letters represent significant differences by 2-way ANOVA with Tukey's post hoc test (P < 0.05; Supplemental Data Set 1). The interaction P value between genotypes and light conditions is shown inset (Supplemental Data Set 1). E and F) Immunoblots showing the levels of PIF4 proteins in Col, sos2-T1, phyA-211 sos2-T1, and phyA-211 seedlings first grown under simulated W light for 4 d and then transferred to simulated shade (R/FR, 0.4) or remained under simulated W light for 5 more days. Anti-HSP was used as sample loading control. Representative pictures are shown in E), and the relative levels of PIF4 proteins are shown in F). The interaction P value between genotypes and light conditions is shown inset (Supplemental Data Set 1). G and H) Immunoblots showing the levels of PIF4 proteins in Col, sos2-T1, phyB-9 sos2-T1, and phyB-9 seedlings first grown under simulated W light for 4 d and then transferred to simulated shade (R/FR, 0.8) or remained under simulated W light for 5 more days. Anti-HSP was used as sample loading control. Representative pictures are shown in G), and the relative levels of PIF4 proteins are shown in H). In E) and G), numbers below the immunoblots indicate the relative band intensities of PIF4 normalized to the loading control. The ratio of the first clear band was set to 100. Error bars in F) and H) represent Sd from 3 independent assays using 3 pools of seedlings. Different letters represent significant differences by 2-way F) and 1-way ANOVA H) with Duncan's post hoc test (P < 0.05; Supplemental Data Set 1).
We also compared the levels of PIF4 proteins in phyA-211 sos2-T1 and phyB-9 sos2-T1 double mutants with those in Col and their respective single mutant seedlings. Our immunoblot data (Fig. 7, E to H) showed that the steady-state levels of PIF4 corresponded well with the hypocotyl lengths of the examined seedlings of different genotypes in both simulated W light and shade (Fig. 7). Therefore, SOS2-regulated PIF4 protein abundance might explain, at least in part, the hypocotyl phenotypes of phyA-211 sos2-T1 and phyB-9 sos2-T1 double mutant seedlings grown under simulated W light and shade conditions. Together, our data indicated that the SOS2-PIF4 module plays a pivotal role in the regulation of SAS in Arabidopsis.
Discussion
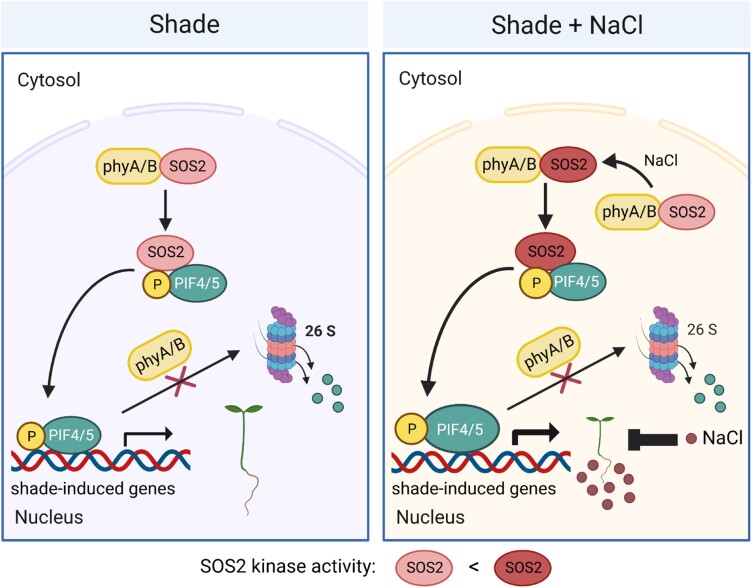
In this study, we demonstrated that SOS2, a protein kinase essential for plant salt tolerance, positively regulates plant SAS (Fig. 1). We further showed that SOS2 physically interacts with PIF4 and PIF5 and directly phosphorylates a conserved serine residue close to their APB motifs, thus decreasing their interactions with active phyB and posttranslationally promoting PIF4/PIF5 protein accumulation (Figs. 2 to 5). Notably, our data indicated that the role of SOS2 in regulating PIF4/PIF5 protein abundance and SAS is more prominent under salt stress (Figs. 1 and 2). Moreover, our data revealed that both phyA and phyB interact with SOS2 and enhance salt-induced SOS2 kinase activity in both W light and shade conditions (Fig. 6). Together, although salt stress exerts an overall suppressive effect on shade-induced plant growth (Hayes et al. 2019), our study uncovers an unexpected role of salt-activated SOS2 in promoting SAS (Fig. 8). Thus, SOS2 serves as a key integrator of external light environment and internal salt stress signaling pathways by modulating the phyB-PIF signaling module.
Figure 8.
A working model depicting that SOS2 positively regulates SAS by promoting PIF4 and PIF5 protein accumulation in the shade. In the shade, phyA and phyB interact with SOS2 and promote its kinase activity. SOS2 directly phosphorylates PIF4 and PIF5 at a serine residue close to their APB motif, thus decreasing their interactions with phyB. Therefore, SOS2 inhibits 26S proteasome-mediated degradation of PIF4/PIF5 and posttranslationally promotes their protein accumulation in the shade. Increased levels of PIF4/PIF5 proteins promote hypocotyl elongation in the shade by modulating the expression of shade-responsive genes. Under both shade and salt stress, shade-induced hypocotyl growth of Arabidopsis seedlings is overall inhibited by salt stress (Hayes et al. 2019), but salt-activated SOS2 more robustly promotes PIF4 and PIF5 protein accumulation.
Salt stress hampers plant growth and development, due to the reduction of water availability caused by high concentrations of salts in the soil and the toxic effects of high concentrations of Na+ and Cl− on plants (Munns and Tester 2008; van Zelm et al. 2020). Thus, the recent observation that salt stress strongly inhibits shade-induced hypocotyl elongation in plants (Hayes et al. 2019) is reasonable, because plants experiencing water limitation due to salinity would suffer even more if they activate SAS to enhance their exposure to sunlight, which at the same time also generates increased water demand (Pierik and Testerink 2014; Pierik and Ballaré 2021). However, the short-hypocotyl phenotype of sos2 mutants under shade may not be due to the toxic effects of Na+ on plant growth, because our data showed that the Na+ contents were similar in Col and sos2 mutant seedlings grown under shade without salt stress (Supplemental Fig. S21). In addition, although we did observe an increased accumulation of Na+ in sos2 mutant seedlings grown under shade and treated with 50 mm NaCl, similar levels of Na+ accumulated in Pro35S:PIF4 sos2-T1 seedlings which developed much longer hypocotyls than sos2 mutant seedlings (Figs. 3 and S21). Collectively, our results suggest that the decrease in PIF4 protein abundance is responsible for the short-hypocotyl phenotype of sos2 mutant seedlings grown in the shade, although we could not exclude the possibility that the increased Na+ content in the absence of functional SOS2 may also affect hypocotyl growth of seedlings grown under shade and salt stress.
It is both interesting and unexpected that SOS2, whose kinase activity is greatly activated by salt stress (Figs. 6, A and B, and S19; Lin et al. 2009; Zhou et al. 2014; Ma et al. 2019), promotes plant SAS particularly under salt stress. Thus, our uncovered role of SOS2 in regulating SAS seems contradictory to the overall effect of salt stress on SAS. However, it should be noted that plants under salt stress also respond, although to a lesser extent, to shade (Hayes et al. 2019), while our results indicated that SOS2 plays a prominent role in plant response to shade under salt stress (Fig. 1). Therefore, our data demonstrate that when plants suffer from both salt stress and shade, salt- and light-activated SOS2 not only increases plant tolerance to salt but also promotes plant response to shade.
Similar observations were also made in our recent study on how plants respond to low temperatures. Hypocotyl growth of Arabidopsis seedlings is inhibited at low ambient temperatures (e.g. 17 °C) compared with that at 22 °C; however, low ambient temperatures induce the expression of C-repeat binding factor/dehydration-responsive element-binding protein1 (CBF1) whose product acts to promote hypocotyl growth by increasing PIF4 and PIF5 protein accumulation (Dong et al. 2020). Collectively, our study provides a scenario in which plants facing both shade and salt stress respond to both stresses through the action of SOS2, which seems to prevent an exaggerated response of salt inhibition of shade-induced hypocotyl elongation.
Shaded environments are unfavorable conditions for plants because they must compete with neighboring plants for sunlight. Based on this understanding, it is not surprising that plants grown under shade exhibit characteristics similar to those of plants under other stress conditions. For example, early flowering is a characteristic phenotype of SAS (Casal 2012, 2013; Fiorucci and Fankhauser 2017; Yang and Li 2017) and is also observed in plants responding to abiotic stresses, such as drought, high or low temperatures, high-intensity light, and nutrient stress (Kolár and Senková 2008; Castro Marín et al. 2011; Takeno 2012, 2016; Kazan and Lyons 2016). Nevertheless, it should be noted that different types of stresses induce completely opposite morphological responses in plants: whereas shade and high ambient temperatures usually promote cell elongation in hypocotyls, stems, and petioles, drought and soil salinity inhibit growth and elongation in general (Pierik and Testerink 2014; Qi et al. 2022). Since plants grown under natural conditions often deal with multiple abiotic stresses simultaneously, it is important and urgent to investigate how the different stress signaling pathways are integrated in plants at different levels because the combinatorial effects of different stresses on plant performance and yield could not be predicted based on the knowledge of single stresses (Pierik and Testerink 2014).
Together, our data demonstrate that SOS2 plays a pivotal role when plants are faced with both salt stress and shade. Our study thus broadens our understanding of how plants coordinately respond to multiple environmental stresses.
Materials and methods
Plant materials and growth conditions
The wild-type A. thaliana used in this study is the Columbia (Col) accession, unless otherwise indicated. The Pro35S:Myc-SOS2 (Lin et al. 2009), scabp8 (Quan et al. 2007), pif4-2 (Leivar et al. 2008), pif5-3 (Khanna et al. 2008), pif4-101 pif5-1 (pif4 pif5) (de Lucas et al. 2008), Pro35S:PIF4 (Huq and Quail 2002), pif7-2 (Leivar et al. 2008), phyB-9 (Reed et al. 1993), and phyA-211 (Reed et al. 1994) were in the Col background, and sos1 (Zhu et al. 1998), sos2-2 (Zhu et al. 1998; Liu et al. 2000), sos3 (Zhu et al. 1998), and Pro35S:Flag-SOS2 sos2-2 (Zhou et al. 2014) were in the gl1/gl1 Col background and had been described previously. The sos2-T1 (SALK_016683) and sos2-T2 (SALK_056101) mutants were obtained from the Arabidopsis Biological Resource Center (ABRC). The phyA phyB (phyA-211 phyB-9), sos2-T1 pif4 pif5, sos2-T2 pif4 pif5, Pro35S:PIF4 sos2-T1, phyA-211 sos2-T1, and phyB-9 sos2-T1 mutants were generated by genetic crossing.
After sterilizing, seeds were stratified at 4 °C for 3 d and then sown on half-strength Murashige and Skoog (MS) medium with 0 or 50 mm NaCl. The petri dishes were incubated in growth chambers (Percival Scientific) under simulated W light (R/FR, 9; PAR, 56 μmol m−2 s−1) (Supplemental Fig. S22) for 4 d and then were either left in simulated W light or transferred to simulated shade (R/FR, 0.8, 0.4, or 0.2; PAR, 56 μmol m−2 s−1) (Supplemental Fig. S22) for 5 more days before hypocotyl measurements were made. Simulated W light and shade were provided by Snap-Lite LED modules (Quantum Devices).
To examine the phenotypes of plants grown in the soil, seeds were sown onto wetted soil, stratified in darkness for 3 d and then moved to simulated W light (R/FR, 9; PAR, 56 μmol m−2 s−1) with a long-day (16-h light/8-h dark) photoperiod and watered with 0 or 50 mm NaCl. After 4 d, the seedlings were transferred to simulated shade (R/FR, 0.4; PAR, 56 μmol m−2 s−1) with a long-day photoperiod or remained under simulated W light for 5 more days and continuously watered with 0 or 50 mm NaCl before the phenotypes were analyzed. The 7- to 10-d seedlings of tobacco (N. benthamiana) were transferred to soil and grown in the greenhouse with a long-day (16-h light/8-h dark) photoperiod at 28 °C.
Plasmid construction and generation of transgenic Arabidopsis plants
The PHYB-BD and AD-PIF4 constructs were described previously (Zhang et al. 2018; Dong et al. 2020). To generate the AD-PIF4S20A and AD-PIF4S20D constructs, the Mut Express II Fast Mutagenesis Kit V2 (Vazyme) was used with the AD-PIF4 plasmid as the template and the primers shown in Supplemental Table S1 according to the manufacturer's instructions. The LexA-PHYA-N, LexA-PHYA-C, LexA-PHYA-C1, LexA-PHYA-C2, LexA-PHYB-N, LexA-PHYB-C, LexA-PHYB-C1, LexA-PHYB-C2, phyB N621-LexA, and AD-PIF4 constructs were described previously (Li et al. 2021; Zhang et al. 2018). To generate the AD-SOS2 construct, the full-length coding sequence of SOS2 was cloned into the EcoRI-SalI sites of the pB42AD vector (Clontech) using the primers shown in Supplemental Table S1.
The GST-PIF4, GST-PIF5, His-PIF4, His-PIF5, GST-SOS2, GST-SOS2-KD, GST-SOS2-RD, His-SOS2, His-PHYA-C1, His-PHYA-C2, His-PHYB-C1, and His-PHYB-C2 constructs were described previously (Guo et al. 2001; Dong et al. 2020; Yan et al. 2020). To generate the GST-PIF4-N, GST-PIF4-C, GST-PIF5-N, and GST-PIF5-C constructs, the respective coding sequences were amplified by PCR using the primers shown in Supplemental Table S1 and then cloned into the EcoRI-XhoI sites of the pGEX-4T-1 vector (Amersham Biosciences), respectively. To generate the MBP-PIF4, MBP-PIF4-N, MBP-PIF4-C, MBP-PIF5, MBP-PIF5-N, and MBP-PIF5-C constructs, the respective coding sequences were amplified by PCR using the primers shown in Supplemental Table S1 and then cloned into the SalI-EcoRI sites of the pMal-c5x vector (NEB), respectively. To generate the GST-SOS2-C (a.a. 202-447) and His-PIF7 constructs used for generating anti-SOS2 and anti-PIF7 antibodies, the indicated coding sequences were amplified by PCR using the primers shown in Supplemental Table S1 and then cloned into the EcoRI-SalI sites of the pGEX-4T-1 vector (Amersham Biosciences) or EcoRI-XhoI sites of the pET28a vector (Novagen), respectively.
The YFPN-SOS2 and GUS-YFPC constructs were described previously (Ma et al. 2019; Li et al. 2020). To generate the YFPN-SOS2RD construct, the coding sequence of SOS2RD (a.a. 268-446) was cloned into the BamHI-KpnI sites of the pSPYNE vector (Waadt et al. 2008) using the primers shown in Supplemental Table S1. To generate the PIF4-YFPC and PIF5-YFPC constructs, the full-length coding sequences of PIF4 and PIF5 were cloned into the SalI-KpnI sites of the pSPYCE (MR) vector (Waadt et al. 2008), respectively, using the primers shown in Supplemental Table S1.
The Flag-SOS2 construct was described previously (Zhou et al. 2014). To generate the Super:PIF4WT-Myc construct, the full-length coding sequence of PIF4 was cloned into the SalI-KpnI sites and fused with Myc in the pSuper1300 vector (Liu et al. 2017). To generate the Super:PIF4S20A-Myc and Super:PIF4S20D-Myc constructs, the mutated coding sequences of PIF4 were amplified by PCR using the AD-PIF4S20A and AD-PIF4S20D constructs as the templates and the primers shown in Supplemental Table S1 and then cloned into the SalI-KpnI sites and fused with Myc, respectively, in the pSuper1300 vector (Liu et al. 2017).
To generate the PIF4-GFP construct, the full-length coding sequence of PIF4 was amplified by PCR using the primers shown in Supplemental Table S1 and then cloned into the SalI-KpnI sites and fused with GFP in the pSuper1300 vector (Liu et al. 2017; Song et al. 2023). To generate the phyB-GFP construct, the full-length coding sequence of phyB was cloned into the XbaI-KpnI sites fused with GFP in the pSuper1300 vector (Liu et al. 2017; Song et al. 2023).
To generate the Super:PIF4WT-Myc, Super:PIF4S20A-Myc, Super:PIF4S20D-Myc, and Pro35S:Myc-SOS2 transgenic plants, the corresponding constructs were transformed into Agrobacterium tumefaciens (strain GV3101) and then transformed into pif4-2 or phyA phyB (phyA-211 phyB-9) mutants, respectively, by the floral dip method (Clough and Bent 1998).
All of the primers used to generate the above-mentioned constructs are listed in Supplemental Table S1, and all of the constructs were confirmed by sequencing prior to usage in various assays. The transgenic plants were selected on MS medium with hygromycin B. T2 plants showing 3:1 segregation for hygromycin B resistance were considered single insertion lines and were selected for isolation of homozygous lines for further studies.
Yeast 2-hybrid assays
Yeast 2-hybrid assays using the LexA-based system were performed as described previously (Qi et al. 2020). The indicated combinations of LexA-PHYA/B-N, LexA-PHYA/B-C, LexA-PHYA/B-C1, LexA-PHYA/B-C2, phyB N621-LexA, AD-SOS2, AD-PIF4WT, AD-PIF420A, and AD-PIF420D were co-transformed into the yeast strain EGY48, respectively. The yeast (S. cerevisiae) transformants were selected on SD/-Trp-Ura-His agar plates at 30 °C and then grown on SD/Gal/Raf/-Trp-Ura-His agar plates containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) together with or without PCB (10 μmol L−1) in darkness or continuous R (30 μmol m−2 s−1) or FR (50 μmol m−2 s−1) light for B color development.
Yeast 2-hybrid assays using the GAL4-based system were performed as described previously (Zhou et al. 2018; Dong et al. 2020; Yan et al. 2020). The indicated combinations of PHYB-BD and AD-PIF4, AD-PIF4S20A, or AD-PIF4S20D were transformed into the yeast strain Y190, respectively. The yeast transformants were cultivated in SD/-Trp-Leu liquid medium for 12 h at 30 °C and then cultivated in SD/-Trp-Leu liquid medium supplemented with 20 µm PCB for another 12 h in darkness at 30 °C. The yeast cultures were then irradiated with 5 min of R alone or with 5 min of R immediately followed by 5 min of FR irradiation, and cultures were then incubated for 2 h at 30 °C. Then, the yeast cultures were exposed to R or R + FR light treatments again and incubated for another 2 h. β-Galactosidase activities were measured by liquid culture assays using o-nitrophenyl-β-d-galactopyranoside (ONPG) as the substrates as described previously (Sheerin et al 2015; Zhou et al 2018).
RT-qPCR assays
RNA extraction, reverse transcription, and real-time PCR assays were performed as described previously (Wang et al. 2019). Briefly, total RNA was extracted from Arabidopsis seedlings using the RNeasy Plant Mini kit (Tiangen). The cDNAs were synthesized from 1 µg of total RNA using RevertAid First Strand cDNA Synthesis Kits (Thermo Fisher Scientific). Real-time PCR assays were performed using Power Up SYBR Green PCR Master Mix (Thermo Fisher Scientific) and gene specific primers listed in Supplemental Table S1. RT-qPCR was performed in 3 technical replicates for each sample, and the relative expression levels were normalized to that of the TUBULIN3 gene.
Immunoblotting
Total proteins were extracted as described previously (Qiu et al. 2017; Dong et al. 2020; Yan et al. 2020). Briefly, Arabidopsis seedlings were homogenized in extraction buffer (150 μL per 50-mg sample) consisting of 100 mm Tris–HCl (pH 7.5), 100 mm NaCl, 5 mm EDTA (pH 8.0), 20% [w/v] glycerol, 5% [w/v] SDS, 40 mm β-mercaptoethanol, 20 mm DTT, 10 mm N-ethylmaleimide, 2 mm PMSF, 80 μm MG132, 80 μm MG115, 1× EDTA-free protease inhibitor cocktail, and 1% [v/v] phosphatase inhibitor cocktail. Samples were immediately boiled 10 min in a dark room under dim green light and then centrifuged 10 min at 13,000 × g at room temperature. Proteins from the supernatants were used in the subsequent immunoblot assays, run on SDS–PAGE gels, and blotted onto polyvinylidene difluoride membranes. Primary antibodies used in this study include anti-PIF4 (1:1,000 [v/v], catalog no. AS163955; Agrisera), anti-PIF5 (1:1,000 [v/v], catalog no. AS122112; Agrisera), anti-GST (1:1,000 [v/v], catalog no. G7781; Sigma-Aldrich), anti-His (1:1,000 [v/v], catalog no. H1029; Sigma-Aldrich), anti-HSP (1:1,000 [v/v], catalog no. AbM51099-31-PU; Beijing Protein Innovation), anti-Myc (1:1,000 [v/v], catalog no. MF083; Mei5 Biotechnology), anti-GFP (1:3,000 [v/v], catalog no. 11814460001; Roche), anti-Flag (1:3,000 [v/v], catalog no. F3165; Sigma-Aldrich), anti-GAPDH (1:1,000 [v/v], catalog no. AC033; ABclonal), anti-RPN6 (1:1,000 [v/v]; Zhou et al. 2018), anti-phyA (1:1,000 [v/v]; Zhang et al. 2018), and anti-phyB (1:1,000 [v/v]; Dong et al. 2020; Yan et al. 2020). Secondary antibodies used in this study include goat antirabbit IgG (whole molecule), HRP (1:8,000 [v/v], catalog no. A-9169; Sigma-Aldrich), goat antimouse IgG (whole molecule), HRP (1:8,000 [v/v], catalog no. A9044; Sigma-Aldrich), rabbit antigoat IgG (whole molecule), and HRP (1:8,000 [v/v], catalog no. A5420; Sigma-Aldrich).
The anti-SOS2 and anti-PIF7 polyclonal antibodies were made by Beijing Protein Innovation (BPI). Briefly, GST-SOS2-C (a.a. 202-447) and His-PIF7 proteins were first expressed in E. coli and then purified and used as antigens to immunize rabbits for production of polyclonal antisera. The anti-SOS2 antibody is also used in the companion study by Ma et al. (2023). Antigen affinity purified anti-SOS2 and anti-PIF7 antibodies were used in immunoblots (1:500 [v/v]).
Co-IP assays
To test the in vivo associations of SOS2 with phyA and phyB, Col and Pro35S:Myc-SOS2 seedlings were grown in darkness for 4 d and then harvested. To test the in vivo association of SOS2 with PIF5, Col, sos2-T1, and Pro35S:Myc-SOS2 seedlings grown in darkness for 4 d were transferred to simulated shade for 0.5 h and then harvested. The seedlings were homogenized in an extraction buffer containing 150 mm NaCl, 10 mm MgCl2, 50 mm Tris–HCl (pH 7.5), 1 mm EDTA, 0.1% [v/v] Nonidet P-40, 1 mm PMSF, 1× MG132, 1× EDTA-free protease inhibitor cocktail, and 1× EDTA-free phosphatase inhibitor cocktail. After centrifugation twice at 12,000 × g for 15 min, the proteins were treated with the indicated combinations of R/FR light pulses (for SOS2 association with phyA/phyB) and then incubated with anti-Myc Affinity Gel (Sigma-Aldrich). For co-IP assays to test the SOS2-PIF4 and PIF4-SOS2-phyB associations in vivo, Flag-SOS2, PIF4-Myc, and phyB-GFP proteins were transiently expressed as indicated in Arabidopsis (Col) protoplasts. After extraction, proteins were incubated with Myc-trap agarose beads (AlpaLife). The beads were then gently washed 4 times (10 min each time) with protein extraction buffer at 4 °C, and the immunoprecipitated proteins were eluted in 2× SDS loading buffer at 95 °C for 15 min and analyzed by immunoblotting.
BiFC assays
The BiFC assays were performed as described previously (Waadt et al. 2008). Briefly, the indicated combinations of constructs were transfected into N. benthamiana leaves for transient expression by Agrobacterium leaf infiltration (Schweiger and Schwenkert 2014). After infiltration, plants were grown under a 16-h-light/8-h-dark cycle for 3 d. The YFP fluorescence signal was detected and visualized by a confocal laser scanning microscope (ZEISS LSM 880) (lasers: 488 nm, 15%; 561 nm, 15%; gains: 600; pinhole: 100 μm).
Confocal microscopy
To detect the subcellular localization of PIF4-GFP, Super:PIF4-GFP plasmid DNA was transiently transfected into Arabidopsis (Col or sos2-T1 mutant) protoplasts as described previously (Xu et al. 2014). After transfection, the protoplasts were incubated in the dark for 16 h and then treated with simulated W light or shade for 3 h, and then the images were captured with a ZEISS LSM 880 confocal laser scanning microscope (lasers: 488 nm, 15%; 561 nm, 15%; gains: 600; pinhole: 100 μm).
Preparation of recombinant proteins
All constructs were transformed into E. coli BL21 (DE3) cells, and the expression of fusion proteins was induced by 0.5 mm isopropyl-β-d-thiogalactoside (IPTG). After ultrasonication, the GST-fusion proteins were purified with Glutathione Sepharose 4B beads (GE Healthcare), the His-fusion proteins were purified with nickel nitrilotriacetic acid beads (Qiagen), and the MBP-fusion proteins were purified with Amylose Resin (New England Biolabs).
In vitro pull-down assays
For in vitro pull-down assays, 2 µg of recombinant bait proteins (GST-PIF4, GST-PIF4-N, GST-PIF4-C, GST-PIF5, GST-PIF5-N, GST-PIF5-C, GST-SOS2, GST-SOS2-KD, GST-SOS2-RD, or GST) and 2 µg of prey proteins (His-SOS2, His-PHYA/B-C1, His-PHYA/B-C2, His-PIF4, or His-PIF5) were added into 200-μL binding buffer containing 50 mm Tris–HCl (pH 7.5), 100 mm NaCl, 0.2% [w/v] glycerol, and 0.6% [v/v] Triton X-100. After incubation for 2 h at 4 °C, the mixed proteins were incubated with Glutathione Sepharose 4B beads (GE Healthcare) for another 2 h at 4 °C, and then the beads were washed 6 times with the binding buffer. The pulled-down proteins were eluted in 2× SDS loading buffer at 100 °C for 15 min and then detected by immunoblotting.
Kinase assays
For semi-in vivo kinase assays, Pro35S:Myc-SOS2 and Pro35S:Myc-SOS2 phyA phyB seedlings grown in dark, W light, and shade conditions were treated with mock (1/2 MS liquid medium) or 100 mm NaCl for 12 h. After harvesting, the plants were ground to powder in liquid nitrogen and then homogenized in precooled IP buffer (10 mm Tris–HCl, pH7.6, 0.5% [v/v] Nonidet-P40, 2 mm EDTA, 150 mm NaCl, and 1× EDTA-free protease inhibitor cocktail). The proteins were centrifuged at 12,000 × g for 10 min and incubated with anti-Myc Affinity Gel (Sigma-Aldrich) for 2 h at 4 °C. Equal amounts of immunoprecipitated Myc-SOS2 proteins were used for the further kinase assays with His-SCaBP8 proteins as the substrates.
The in vitro kinase assays were performed as described previously (Lin et al. 2009). Briefly, the indicated recombinant proteins (His-SOS2 and MBP-PIF4/PIF5 proteins) were added into 15 µL of the kinase buffer (20 mm Tris–HCl, pH 8.0, 5 mm MgCl2, 1 mm CaCl2, 10 µm ATP, and 1 mm DTT) containing 0.1 μL [γ-32P] ATP (1 μCi). After incubation at 30 °C for 30 min, the reactions were terminated by the addition of 6× SDS loading and were boiled at 100 °C for 8 min. The samples were separated by 10% [w/v] SDS–PAGE and stained with Coomassie Brilliant Blue (CBB) R 250, and then the gels were exposed to a phosphor screen (Amersham Biosciences). The autoradiographic signals (Autorad) were detected by a Typhoon 9410 phosphor imager (Amersham Biosciences).
Na+ content measurement
To measure the contents of Na+ in different seedlings, Col, sos2-T1, and Pro35S:PIF4 sos2-T1 seeds were sown on half-strength MS medium with 0 or 50 mm NaCl, then incubated under simulated W light (R/FR, 9; PAR, 56 μmol m−2 s−1) (Supplemental Fig. S22) for 4 d, and then transferred to simulated shade (R/FR, 0.8, 0.4, or 0.2; PAR, 56 μmol m−2 s−1) (Supplemental Fig. 22) for 5 more days before measurement. The seedlings were harvested separately, with each pool of seedlings containing >60 individual plants. The seedlings were first oven-dried at 80 °C for at least 48 h. After weighing, samples were digested with 68% [w/v] HNO3 and then created a dilution series using 1% [v/v] hydrochloric acid. The Na+ contents were then determined using a 4100 MP-AES spectrometer (Agilent, Santa Clara, CA, USA).
Mass spectrometry assays
Liquid chromatography–tandem mass spectrometry (LC–MS/MS) assays were performed to identify the phosphorylation sites of PIF4 and PIF5 by SOS2 in vitro. Briefly, the indicated recombinant proteins (His-SOS2 and MBP-PIF4/PIF5 proteins) were added into 60 µL of the kinase buffer (20 mm Tris–HCl, pH 8.0, 5 mm MgCl2, 1 mm CaCl2, 10 µm ATP, and 1 mm DTT) and incubated at 30 °C for 30 min. The proteins were reduced with DTT, alkylated with iodoacetamide, and digested by trypsin (1:50) overnight at 37 °C. The resulting peptides were diluted with 0.1% [v/v] formic acid and centrifuged at 12,000 × g for 20 min. The supernatant was collected for LC–MS/MS assays using the nano-Acquity nano HPLC (Waters, Milford, MA, USA) coupled with a Thermo Q-Exactive high resolution mass spectrometer (Thermo Scientific, Waltham, MA, USA).
Quantification and statistical analysis
Protein quantification was performed with ImageJ. One-way ANOVA and 2-way ANOVA were performed with SPSS statistical software, and Student's t tests were performed in Microsoft Excel. Different letters represent significant differences at P < 0.05, and levels that are not significantly different are indicated with the same letter. Values are represented as means ± Sd. The results of all statistical analyses were shown in Supplemental Data Set 1.
Accession numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: PIF4 (At2g43010), PIF5 (At3g59060), PIF3 (At1g09530), PIF7 (At5g61270), PHYB (At2g18790), PHYA (At1g09570), SOS2 (At5g35410), SOS1 (At2g01980), SOS3 (At5g24270), SCABP8 (At4g33000), PIL1 (At2g46970), HFR1 (At1g02340), IAA19 (At3g15540), ATHB2 (At4g16780), and YUC8 (At4g28720).
Supplementary Material
Acknowledgments
We thank Peter Quail and Chuanyou Li for PIF-related seeds and Lin Li for pif7 mutant seeds and helpful suggestions.
Contributor Information
Run Han, State Key Laboratory of Plant Environmental Resilience, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Liang Ma, State Key Laboratory of Plant Environmental Resilience, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Yang Lv, State Key Laboratory of Plant Environmental Resilience, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Lijuan Qi, State Key Laboratory of Plant Environmental Resilience, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Jing Peng, State Key Laboratory of Plant Environmental Resilience, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Hong Li, State Key Laboratory of Plant Environmental Resilience, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Yangyang Zhou, State Key Laboratory of Plant Environmental Resilience, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Pengyu Song, State Key Laboratory of Plant Environmental Resilience, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Jie Duan, State Key Laboratory of Plant Environmental Resilience, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Jianfang Li, State Key Laboratory of Plant Environmental Resilience, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Zhen Li, State Key Laboratory of Plant Environmental Resilience, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
William Terzaghi, Department of Biology, Wilkes University, Wilkes-Barre, PA 18766, USA.
Yan Guo, State Key Laboratory of Plant Environmental Resilience, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Jigang Li, State Key Laboratory of Plant Environmental Resilience, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Author contributions
Jig.L., Y.G., R.H., and L.M. designed the research. R.H., L.M., Y.L., L.Q., J.P., H.L., Y.Z., P.S., J.D., and Jia.L. performed the research. Jig.L., Y.G., and Z.L. discussed and interpreted the data. Jig.L., W.T., R.H., and L.M. wrote the paper.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Genotyping of the 3 sos2 mutants.
Supplemental Figure S2. SOS2 positively regulates SAS in Arabidopsis.
Supplemental Figure S3. SOS2 positively regulates SAS of soil-grown plants with or without salt stress.
Supplemental Figure S4. The SOS pathway components are involved in regulating SAS particularly under salt stress.
Supplemental Figure S5. The expression of shade-induced genes is decreased in sos2 mutant seedlings.
Supplemental Figure S6. The protein abundance of PIF5 is regulated by SOS2 in the shade.
Supplemental Figure S7. SOS2 promotes PIF4 protein accumulation in the shade under 50 mm NaCl.
Supplemental Figure S8. The levels of PIF3 and PIF7 proteins are not obviously regulated by SOS2 in the shade.
Supplemental Figure S9. The expression levels of PIF3, PIF4, PIF5, and PIF7 in sos2 mutant seedlings grown under W light and shade conditions.
Supplemental Figure S10. Genotyping of sos2 pif4 pif5 and Pro35S:PIF4 sos2-T1 seedlings.
Supplemental Figure S11. SOS2 physically interacts with PIF4 and PIF5.
Supplemental Figure S12. SOS2 directly phosphorylates PIF4 and PIF5 in vitro at a serine residue near to their APB motif.
Supplemental Figure S13. The levels of phyB-BD and AD-PIF4 proteins in yeast cells used for yeast 2-hybrid assays.
Supplemental Figure S14. PIF4WT and PIF4S20A interacted more strongly with the Pfr form of phyB than PIF4S20D in yeast cells.
Supplemental Figure S15. Nuclear localization of PIF4 is not regulated by SOS2.
Supplemental Figure S16. The transcript levels of PIF4 and shade-responsive genes in Super:PIF4WT-Myc pif4-2, Super:PIF4S20A-Myc pif4-2, and Super:PIF4S20D-Myc pif4-2 seedlings.
Supplemental Figure S17. Phenotypes and the levels of PIF4 proteins of Super:PIF4WT-Myc pif4-2, Super:PIF4S20A-Myc pif4-2, and Super:PIF4S20D-Myc pif4-2 seedlings under salt stress.
Supplemental Figure S18. SOS2 transcript and protein levels are not significantly regulated by light.
Supplemental Figure S19. SOS2 kinase activity is synergistically induced by salt and light.
Supplemental Figure S20. Schematic diagrams of domain structures of PHYA and PHYB used for yeast 2-hybrid and in vitro pull-down assays.
Supplemental Figure S21. Contents of Na+ in seedlings grown under simulated shade and treated with 0 or 50 mm NaCl.
Supplemental Figure S22. Light conditions used in this study.
Supplemental Table S1. Summary of primers used in this study.
Supplemental Data Set 1. Statistical results tables.
Funding
This work was supported by grants from the National Natural Science Foundation of China (32225006 to J.L. (Jigang Li), and 32200245 to L.Q.), the China Postdoctoral Science Foundation (2022M723410 to J.P.), the China National Postdoctoral Program for Innovative Talents (BX20200371 to L.Q.), and the Beijing Outstanding University Discipline Program.
References
- Bae G, Choi G. Decoding of light signals by plant phytochromes and their interacting proteins. Annu Rev Plant Biol. 2008:59(1):281–311. 10.1146/annurev.arplant.59.032607.092859 [DOI] [PubMed] [Google Scholar]
- Ballaré CL, Scopel AL, Sánchez RA. Far-red radiation reflected from adjacent leaves: an early signal of competition in plant canopies. Science 1990:247(4940):329–332. 10.1126/science.247.4940.329 [DOI] [PubMed] [Google Scholar]
- Bauer D, Viczián A, Kircher S, Nobis T, Nitschke R, Kunkel T, Panigrahi KC, Adám E, Fejes E, Schäfer E, et al. . Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 2004:16(6):1433–1445. 10.1105/tpc.021568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo-García S, de Lucas M, Martínez C, Espinosa-Ruiz A, Davière J-M, Prat S. BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes Dev. 2014:28(15):1681–1694. 10.1101/gad.243675.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu Q, Zhu L, Dennis MD, Yu L, Lu SX, Person MD, Tobin EM, Browning KS, Huq E. Phosphorylation by CK2 enhances the rapid light-induced degradation of phytochrome interacting factor 1 in Arabidopsis. J Biol Chem. 2011:286(14):12066–12074. 10.1074/jbc.M110.186882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buti S, Hayes S, Pierik R. The bHLH network underlying plant shade-avoidance. Physiol Plant. 2020:169(3):312–324. 10.1111/ppl.13074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ. Shade avoidance. Arabidopsis Book 2012:10:e0157. 10.1199/tab.0157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ. Photoreceptor signaling networks in plant responses to shade. Annu Rev Plant Biol. 2013:64(1):403–427. 10.1146/annurev-arplant-050312-120221 [DOI] [PubMed] [Google Scholar]
- Castro Marín I, Loef I, Bartetzko L, Searle I, Coupland G, Stitt M, Osuna D. Nitrate regulates floral induction in Arabidopsis, acting independently of light, gibberellin and autonomous pathways. Planta 2011:233(3):539–552. 10.1007/s00425-010-1316-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M-C, Kathare PK, Paik I, Huq E. Phytochrome signaling networks. Annu Rev Plant Biol. 2021:72(1):217–244. 10.1146/annurev-arplant-080620-024221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998:16(6):735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Courbier S, Pierik R. Canopy light quality modulates stress responses in plants. iScience 2019:22:441–452. 10.1016/j.isci.2019.11.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M, Davière J-M, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S. A molecular framework for light and gibberellin control of cell elongation. Nature 2008:451(7177):480–484. 10.1038/nature06520 [DOI] [PubMed] [Google Scholar]
- de Wit M, Keuskamp DH, Bongers FJ, Hornitschek P, Gommers CMM, Reinen E, Martínez-Cerón C, Fankhauser C, Pierik R. Integration of phytochrome and cryptochrome signals determines plant growth during competition for light. Curr Biol. 2016:26(24):3320–3326. 10.1016/j.cub.2016.10.031 [DOI] [PubMed] [Google Scholar]
- de Wit M, Ljung K, Fankhauser C. Contrasting growth responses in lamina and petiole during neighbor detection depend on differential auxin responsiveness rather than different auxin levels. New Phytol. 2015:208(1):198–209. 10.1111/nph.13449 [DOI] [PubMed] [Google Scholar]
- Dong X, Yan Y, Jiang B, Shi Y, Jia Y, Cheng J, Shi Y, Kang J, Li H, Zhang D, et al. . The cold response regulator CBF1 promotes Arabidopsis hypocotyl growth at ambient temperatures. EMBO J. 2020:39(13):e103630. 10.15252/embj.2019103630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duek PD, Fankhauser C. bHLH class transcription factors take centre stage in phytochrome signalling. Trends Plant Sci. 2005:10(2):51–54. 10.1016/j.tplants.2004.12.005 [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Chen M. Transposing phytochrome into the nucleus. Trends Plant Sci. 2008:13(11):596–601. 10.1016/j.tplants.2008.08.007 [DOI] [PubMed] [Google Scholar]
- Favero DS. Mechanisms regulating PIF transcription factor activity at the protein level. Physiol Plant. 2020:169(3):325–335. 10.1111/ppl.13075 [DOI] [PubMed] [Google Scholar]
- Fernández-Milmanda GL, Ballaré CL. Shade avoidance: expanding the color and hormone palette. Trends Plant Sci. 2021:26(5):509–523. 10.1016/j.tplants.2020.12.006 [DOI] [PubMed] [Google Scholar]
- Fiorucci A-S, Fankhauser C. Plant strategies for enhancing access to sunlight. Curr Biol. 2017:27(17):R931–R940. 10.1016/j.cub.2017.05.085 [DOI] [PubMed] [Google Scholar]
- Franklin KA, Quail PH. Phytochrome functions in Arabidopsis development. J Exp Bot. 2010:61(1):11–24. 10.1093/jxb/erp304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser DP, Hayes S, Franklin KA. Photoreceptor crosstalk in shade avoidance. Curr Opin Plant Biol. 2016:33:1–7. 10.1016/j.pbi.2016.03.008 [DOI] [PubMed] [Google Scholar]
- Guo Y, Halfter U, Ishitani M, Zhu J-K. Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell 2001:13(6):1383–1400. 10.1105/TPC.010021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfter U, Ishitani M, Zhu J-K. The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc Natl Acad Sci U S A. 2000:97(7):3735–3740. 10.1073/pnas.97.7.3735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes S, Pantazopoulou CK, van Gelderen K, Reinen E, Tween AL, Sharma A, de Vries M, Prat S, Schuurink RC, Testerink C, et al. . Soil salinity limits plant shade avoidance. Curr Biol. 2019:29(10):1669–1676.e4. 10.1016/j.cub.2019.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersch M, Lorrain S, de Wit M, Trevisan M, Ljung K, Bergmann S, Fankhauser C. Light intensity modulates the regulatory network of the shade avoidance response in Arabidopsis. Proc Natl Acad Sci U S A. 2014:111(17):6515–6520. 10.1073/pnas.1320355111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P, Lorrain S, Zoete V, Michielin O, Fankhauser C. Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 2009:28(24):3893–3902. 10.1038/emboj.2009.306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Zhang Q, Jiang Y, Yang C, Wang Q, Li L. Shade-induced nuclear localization of PIF7 is regulated by phosphorylation and 14-3-3 proteins in Arabidopsis. eLife. 2018:7:E31636. 10.7554/eLife.31636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E, Quail PH. PIF4, A phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 2002:21(10):2441–2450. 10.1093/emboj/21.10.2441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani M, Liu J, Halfter U, Kim C-S, Shi W, Zhu J-K. SOS3 Function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell 2000:12(9):1667–1677. 10.1105/tpc.12.9.1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K, Lyons R. The link between flowering time and stress tolerance. J Exp Bot. 2016:67(1):47–60. 10.1093/jxb/erv441 [DOI] [PubMed] [Google Scholar]
- Khanna R, Huq E, Kikis EA, Al-Sady B, Lanzatella C, Quail PH. A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix–loop–helix transcription factors. Plant Cell 2004:16(11):3033–3044. 10.1105/tpc.104.025643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R, Shen Y, Marion CM, Tsuchisaka A, Theologis A, Schäfer E, Quail PH. The basic helix–loop–helix transcription factor PIF5 acts on ethylene biosynthesis and phytochrome signaling by distinct mechanisms. Plant Cell 2008:19(12):3915–3929. 10.1105/tpc.107.051508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W-Y, Ali Z, Park HJ, Park SJ, Cha J-Y, Perez-Hormaeche J, Quintero FJ, Shin G, Kim MR, Qiang Z, et al. . Release of SOS2 kinase from sequestration with GIGANTEA determines salt tolerance in Arabidopsis. Nat Commun. 2013:4(1):1352. 10.1038/ncomms2357 [DOI] [PubMed] [Google Scholar]
- Klose C, Viczián A, Kircher S, Schäfer E, Nagy F. Molecular mechanisms for mediating light-dependent nucleo/cytoplasmic partitioning of phytochrome photoreceptors. New Phytol. 2015:206(3):965–971. 10.1111/nph.13207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolár J, Senková J. Reduction of mineral nutrient availability accelerates flowering of Arabidopsis thaliana. Plant Physiol. 2008:165(15):1601–1609. 10.1016/j.jplph.2007.11.010 [DOI] [PubMed] [Google Scholar]
- Lee N, Choi G. Phytochrome-interacting factor from Arabidopsis to liverwort. Curr Opin Plant Biol. 2017:35:54–60. 10.1016/j.pbi.2016.11.004 [DOI] [PubMed] [Google Scholar]
- Lee S, Paik I, Huq E. SPAs promote thermomorphogenesis by regulating the phyB-PIF4 module in Arabidopsis. Development 2020:147(19):dev189233. 10.1242/dev.189233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legris M, Ince Y, Fankhauser C. Molecular mechanisms underlying phytochrome-controlled morphogenesis in plants. Nat Commun. 2019:10(1):5219. 10.1038/s41467-019-13045-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E. PIFs: systems integrators in plant development. Plant Cell 2014:26(1):56–78. 10.1105/tpc.113.120857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Al-Sady B, Carle C, Storer A, Alonso JM, Ecker JR, Quail PH. The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 2008:20(2):337–352. 10.1105/tpc.107.052142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Quail PH. PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 2011:16(1):19–28. 10.1016/j.tplants.2010.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Tepperman JM, Cohn MM, Monte E, Al-Sady B, Erickson E, Quail PH. Dynamic antagonism between phytochromes and PIF family basic helix-loop-helix factors induces selective reciprocal responses to light and shade in a rapidly responsive transcriptional network in Arabidopsis. Plant Cell 2012:24(4):1398–1419. 10.1105/tpc.112.095711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li G, Wang H, Wang Deng X. Phytochrome signaling mechanisms. Arabidopsis Book 2011:9:e0148. 10.1199/tab.0148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ljung K, Breton G, Schmitz RJ, Pruneda-Paz J, Cowing-Zitron C, Cole BJ, Ivans LJ, Pedmale UV, Jung H-S, et al. . Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 2012:26(8):785–790. 10.1101/gad.187849.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Qin X, Song P, Han R, Li J. A LexA-based yeast two-hybrid system for studying light-switchable interactions of phytochromes with their interacting partners. aBIOTECH. 2021:2(2):105–116. 10.1007/s42994-021-00034-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zhou H, Zhang Y, Li Z, Yang Y, Guo Y. The GSK3-like kinase BIN2 is a molecular switch between the salt stress response and growth recovery in Arabidopsis thaliana. Dev Cell. 2020:55(3):367–380.e6. 10.1016/j.devcel.2020.08.005 [DOI] [PubMed] [Google Scholar]
- Lin H, Yang Y, Quan R, Mendoza I, Wu Y, Du W, Zhao S, Schumaker KS, Pardo JM, Guo Y. Phosphorylation of SOS3-LIKE CALCIUM BINDING PROTEIN8 by SOS2 protein kinase stabilizes their protein complex and regulates salt tolerance in Arabidopsis. Plant Cell 2009:21(5):1607–1619. 10.1105/tpc.109.066217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling J-J, Li J, Zhu D, Deng XW. Noncanonical role of Arabidopsis COP1/SPA complex in repressing BIN2-mediated PIF3 phosphorylation and degradation in darkness. Proc Natl Acad Sci U S A. 2017:114(13):3539–3544. 10.1073/pnas.1700850114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Ishitani M, Halfter U, Kim CS, Zhu JK. The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc Natl Acad Sci U S A. 2000:97(7):3730–3734. 10.1073/pnas.97.7.3730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhu J-K. An Arabidopsis mutant that requires increased calcium for potassium nutrition and salt tolerance. Proc Natl Acad Sci U S A. 1997:94(26):14960–14964. 10.1073/pnas.94.26.14960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhu J-K. A calcium sensor homolog required for plant salt tolerance. Science. 1998:280(5371):1943–1945. 10.1126/science.280.5371.1943 [DOI] [PubMed] [Google Scholar]
- Liu Z, Jia Y, Ding Y, Shi Y, Li Z, Guo Y, Gong Z, Yang S. Plasma membrane CRPK1-mediated phosphorylation of 14-3-3 proteins induces their nuclear import to fine-tune CBF signaling during cold response. Mol Cell. 2017:66(1):117–128.e5. 10.1016/j.molcel.2017.02.016 [DOI] [PubMed] [Google Scholar]
- Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 2008:53(2):312–323. 10.1111/j.1365-313X.2007.03341.x [DOI] [PubMed] [Google Scholar]
- Ma L, Han R, Yang Y, Liu X, Li H, Zhao X, Jian L, Fu H, Huo Y, Sun L, et al. . Phytochromes enhance SOS2-mediated PIF1 and PIF3 phosphorylation and degradation to promote Arabidopsis salt tolerance. Plant Cell 2023;35(8):2997–3020. 10.1093/plcell/koad117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Ye J, Yang Y, Lin H, Yue L, Luo J, Long Y, Fu H, Liu X, Zhang Y, et al. . The SOS2-SCaBP8 complex generates and fine-tunes an AtANN4-dependent calcium signature under salt stress. Dev Cell. 2019:48(5):697–709.e5. 10.1016/j.devcel.2019.02.010 [DOI] [PubMed] [Google Scholar]
- Martínez-García JF, Gallemí M, Molina-Contreras MJ, Llorente B, Bevilaqua MR, Quail PH. The shade avoidance syndrome in Arabidopsis: the antagonistic role of phytochrome A and B differentiates vegetation proximity and canopy shade. PLoS One 2014:9(10):e109275. 10.1371/journal.pone.0109275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T, Oka H, Yoshimura F, Ishida K, Yamashino T. Insight into the mechanism of end-of-day far-red light (EODFR)-induced shade avoidance responses in Arabidopsis thaliana. Biosci Biotechnol Biochem. 2015:79(12):1987–1994. 10.1080/09168451.2015.1065171 [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008:59(1):651–681. 10.1146/annurev.arplant.59.032607.092911 [DOI] [PubMed] [Google Scholar]
- Ni W, Xu S-L, González-Grandío E, Chalkley RJ, Huhmer AFR, Burlingame AL, Wang Z-Y, Quail PH. PPKs mediate direct signal transfer from phytochrome photoreceptors to transcription factor PIF3. Nat Commun. 2017:8(1):15236. 10.1038/ncomms15236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Guo Y, Halfter U, Zhu J-K. A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc Natl Acad Sci U S A. 2003:100(20):11771–11776. 10.1073/pnas.2034853100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik I, Chen F, Ngoc Pham V, Zhu L, Kim J-I, Huq E. A phyB-PIF1-SPA1 kinase regulatory complex promotes photomorphogenesis in Arabidopsis. Nat Commun. 2019:10(1):4216. 10.1038/s41467-019-12110-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedmale UV, Huang S-C, Zander M, Cole BJ, Hetzel J, Ljung K, Reis PAB, Sridevi P, Nito K, Nery JR, et al. . Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell 2016:164(1-2):233–245. 10.1016/j.cell.2015.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham VN, Kathare PK, Huq E. Phytochromes and phytochrome interacting factors. Plant Physiol. 2018:176(2):1025–1038. 10.1104/pp.17.01384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierik R, Ballaré CL. Control of plant growth and defense by photoreceptors: from mechanisms to opportunities in agriculture. Mol Plant. 2021:14(1):61–76. 10.1016/j.molp.2020.11.021 [DOI] [PubMed] [Google Scholar]
- Pierik R, Testerink C. The art of being flexible: how to escape from shade, salt, and drought. Plant Physiol. 2014:166(1):5–22. 10.1104/pp.114.239160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L, Liu S, Li C, Fu J, Jing Y, Cheng J, Li H, Zhang D, Wang X, Dong X, et al. . PHYTOCHROME-INTERACTING FACTORS interact with the ABA receptors PYL8 and PYL9 to orchestrate ABA signaling in darkness. Mol Plant. 2020:13(3):414–430. 10.1016/j.molp.2020.02.001 [DOI] [PubMed] [Google Scholar]
- Qi L, Shi Y, Terzaghi W, Yang S, Li J. Integration of light and temperature signaling pathways in plants. J Integr Plant Biol. 2022:64(2):393–411. 10.1111/jipb.13216 [DOI] [PubMed] [Google Scholar]
- Qiu Q-S, Guo Y, Dietrich MA, Schumaker KS, Zhu J-K. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci U S A. 2002:99(12):8436–8441. 10.1073/pnas.122224699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Pasoreck EK, Reddy AK, Nagatani A, Ma W, Chory J, Chen M. Mechanism of early light signaling by the carboxy-terminal output module of Arabidopsis phytochrome B. Nat Commun. 2017:8(1):1905. 10.1038/s41467-017-02062-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan R, Lin H, Mendoza I, Zhang Y, Cao W, Yang Y, Shang M, Chen S, Pardo JM, Guo Y. SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 2007:19(4):1415–1431. 10.1105/tpc.106.042291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero FJ, Ohta M, Shi H, Zhu J-K, Pardo JM. Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc Natl Acad Sci U S A. 2002:99(13):9061–9066. 10.1073/pnas.132092099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Nagatani A, Elich TD, Fagan M, Chory J. Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 1994:104(4):1139–1149. 10.1104/pp.104.4.1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Poole DS, Furuya M, Chory J. Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 1993:5(2):147–157. 10.1105/tpc.5.2.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig-Villanova I, Martínez-García JF. Plant responses to vegetation proximity: a whole life avoiding shade. Front Plant Sci. 2016:7:236. 10.3389/fpls.2016.00236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter MG, Franklin KA, Whitelam GC. Gating of the rapid shade-avoidance response by the circadian clock in plants. Nature 2003:426(6967):680–683. 10.1038/nature02174 [DOI] [PubMed] [Google Scholar]
- Schweiger R, Schwenkert S. Protein–protein interactions visualized by bimolecular fluorescence complementation in tobacco protoplasts and leaves. J Vis Exp. 2014:(85):51327. 10.3791/51327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa G, Carabelli M, Sassi M, Ciolfi A, Possenti M, Mittempergher F, Becker J, Morelli G, Ruberti I. A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. Genes Dev. 2005:19(23):2811–2815. 10.1101/gad.364005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock RA, Quail PH. Novel phytochrome sequences in Arabidopsis thaliana: structure, evolution, and differential expression of a plant regulatory photoreceptor family. Gene Dev. 1989:3(11):1745–1757. 10.1101/gad.3.11.1745 [DOI] [PubMed] [Google Scholar]
- Sheerin DJ, Menon C, zur Oven-Krockhaus S, Enderle B, Zhu L, Johnen P, Schleifenbaum F, Stierhof Y-D, Huq E, Hiltbrunner A. Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 2015:27(1):189–201. 10.1105/tpc.114.134775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Ishitani M, Kim C, Zhu J-K. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci U S A. 2000:97(12):6896–6901. 10.1073/pnas.120170197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu-Sato S, Huq E, Tepperman JM, Quail PH. A light-switchable gene promoter system. Nat Biotechnol. 2002:20(10):1041–1044. 10.1038/nbt734 [DOI] [PubMed] [Google Scholar]
- Shin A-Y, Han Y-J, Baek A, Ahn T, Kim SY, Nguyen TS, Son M, Lee KW, Shen Y, Song P-S, et al. . Evidence that phytochrome functions as a protein kinase in plant light signalling. Nat Commun. 2016:7(1):11545. 10.1038/ncomms11545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song P, Yang Z, Guo C, Han R, Wang H, Dong J, Kang D, Guo Y, Yang S, Li J. 14-3-3 Proteins regulate photomorphogenesis by facilitating light-induced degradation of PIF3. New Phytol. 2023:237(1):140–159. 10.1111/nph.18494 [DOI] [PubMed] [Google Scholar]
- Takeno K. Stress-induced flowering. In: Ahmad P, Prasad M, editors. Abiotic stress responses in plants. New York: (NY: ): Springer; 2012. p. 331–345. [Google Scholar]
- Takeno K. Stress-induced flowering: the third category of flowering response. J Exp Bot. 2016:67(17):4925–4934. 10.1093/jxb/erw272 [DOI] [PubMed] [Google Scholar]
- Tan T, Cai J, Zhan E, Yang Y, Zhao J, Guo Y, Zhou H. Stability and localization of 14-3-3 proteins are involved in salt tolerance in Arabidopsis. Plant Mol Biol. 2016:92(3):391–400. 10.1007/s11103-016-0520-5 [DOI] [PubMed] [Google Scholar]
- van Zelm E, Zhang Y, Testerink C. Salt tolerance mechanisms of plants. Annu Rev Plant Biol. 2020:71(1):403–433. 10.1146/annurev-arplant-050718-100005 [DOI] [PubMed] [Google Scholar]
- Waadt R, Schmidt LK, Lohse M, Hashimoto K, Bock R, Kudla J. Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 2008:56(3):505–516. 10.1111/j.1365-313X.2008.03612.x [DOI] [PubMed] [Google Scholar]
- Wang X, Guo C, Peng J, Li C, Wan F, Zhang S, Zhou Y, Yan Y, Qi L, Sun K, et al. . ABRE-BINDING FACTORS play a role in the feedback regulation of ABA signaling by mediating rapid ABA induction of ABA co-receptor genes. New Phytol. 2019:221(1):341–355. 10.1111/nph.15345 [DOI] [PubMed] [Google Scholar]
- Xin X, Chen W, Wang B, Zhu F, Li Y, Yang H, Li J, Ren D. Arabidopsis MKK10-MPK6 mediates red-light-regulated opening of seedling cotyledons through phosphorylation of PIF3. J Exp Bot. 2018:69(3):423–439. 10.1093/jxb/erx418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Li J, Gangappa SN, Hettiarachchi C, Lin F, Andersson MX, Jiang Y, Deng XW, Holm M. Convergence of light and ABA signaling on the ABI5 promoter. PLoS Genet. 2014:10(2):e1004197. 10.1371/journal.pgen.1004197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Li C, Dong X, Li H, Zhang D, Zhou Y, Jiang B, Peng J, Qin X, Cheng J, et al. . MYB30 Is a key negative regulator of Arabidopsis photomorphogenic development that promotes PIF4 and PIF5 protein accumulation in the light. Plant Cell 2020:32(7):2196–2215. 10.1105/tpc.19.00645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Guo Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018a:217(2):523–539. 10.1111/nph.14920 [DOI] [PubMed] [Google Scholar]
- Yang Y, Guo Y. Unraveling salt stress signaling in plants. J Integr Plant Biol. 2018b:60(9):796–804. 10.1111/jipb.12689 [DOI] [PubMed] [Google Scholar]
- Yang C, Li L. Hormonal regulation in shade avoidance. Front Plant Sci. 2017:8:1527. 10.3389/fpls.2017.01527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Xie F, Jiang Y, Li Z, Huang X, Li L. Phytochrome A negatively regulates the shade avoidance response by increasing auxin/indole acidic acid protein stability. Dev Cell. 2018:44(1):29–41.e4. 10.1016/j.devcel.2017.11.017 [DOI] [PubMed] [Google Scholar]
- Zhang S, Li C, Zhou Y, Wang X, Li H, Feng Z, Chen H, Qin G, Jin D, Terzaghi W, et al. . TANDEM ZINC-FINGER/PLUS3 is a key component of phytochrome A signaling. Plant Cell 2018:30(4):835–852. 10.1105/tpc.17.00677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Mayba O, Pfeiffer A, Shi H, Tepperman JM, Speed TP, Quail PH. A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet. 2013:9(1):e1003244. 10.1371/journal.pgen.1003244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Zhang H, Song C, Zhu J-K, Shabala S. Mechanisms of plant responses and adaptation to soil salinity. The Innovation 2020:1(1):100017. 10.1016/j.xinn.2020.100017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Lin H, Chen S, Becker K, Yang Y, Zhao J, Kudla J, Schumaker KS, Guo Y. Inhibition of the Arabidopsis SALT OVERLY SENSITIVE pathway by 14-3-3 proteins. Plant Cell 2014:26(3):1166–1182. 10.1105/tpc.113.117069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Yang L, Duan J, Cheng J, Shen Y, Wang X, Han R, Li H, Li Z, Wang L, et al. . Hinge region of Arabidopsis phyA plays an important role in regulating phyA function. Proc Natl Acad Sci U S A. 2018:115(50):E11864–E11873. 10.1073/pnas.1813162115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J-K. Abiotic stress signaling and responses in plants. Cell 2016:167(2):313–324. 10.1016/j.cell.2016.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J-K, Liu J, Xiong L. Genetic analysis of salt tolerance in Arabidopsis. Evidence for a critical role of potassium nutrition. Plant Cell 1998:10(7):1181–1191. 10.1105/tpc.10.7.1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.