Abstract
Proteolytic processing is required for the activation of numerous viral glycoproteins. Here we show that the envelope glycoprotein from the Zaire strain of Ebola virus (Ebo-GP) is proteolytically processed into two subunits, GP1 and GP2, that are likely covalently associated through a disulfide linkage. Murine leukemia virions pseudotyped with Ebo-GP contain almost exclusively processed glycoprotein, indicating that this is the mature form of Ebo-GP. Mutational analysis identified a dibasic motif, reminiscent of furin-like protease processing sites, as the Ebo-GP cleavage site. However, analysis of Ebo-GP processing in LoVo cells that lack the proprotein convertase furin demonstrated that furin is not required for processing of Ebo-GP. In sharp contrast to other viral systems, we found that an uncleaved mutant of Ebo-GP was able to mediate infection of various cell lines as efficiently as the wild-type, proteolytically cleaved glycoprotein, indicating that cleavage is not required for the activation of Ebo-GP despite the conservation of a dibasic cleavage site in all filoviral envelope glycoproteins.
The glycoproteins of many enveloped viruses are initially synthesized as inactive precursors that, while able to bind to their cognate cellular receptors, are unable to mediate membrane fusion and, hence, viral entry. Proteolytic processing of the precursor polyprotein at specific cleavage sites is required to convert these glycoproteins to an active state and render the virus infectious. Examples of such viral glycoproteins include the envelope proteins of retroviruses such as human immunodeficiency virus type 1 (HIV-1) (27) and the avian leukosis and sarcoma viruses (ASLV) (8) as well as the hemagglutinin (HA) glycoprotein of the orthomyxovirus influenza A virus (24, 25) and the paramyxovirus Newcastle disease virus F protein (29, 35).
Endoproteolytic cleavage of the envelope glycoprotein is thus a critical step in the maturation of a virus, and the availability of cellular enzymes capable of processing the precursor polyprotein can be a major determinant of viral tropism and pathogenicity. For example, the HA glycoproteins of certain avirulent strains of influenza A viruses can be efficiently processed only by the endoproteases present within the cells of the respiratory tract (47). These viruses are therefore restricted to the respiratory tract and cannot cause a disseminating infection. In pathogenic viral strains, introduction of a polybasic cleavage site into HA renders the glycoprotein susceptible to proteolytic processing by a family of widely expressed cellular proteases, thereby expanding viral tropism (3, 23). It is believed that this expanded tropism is a pivotal determinant of the increased virulence of these viruses.
The envelope glycoproteins of the Ebola and Marburg viruses display significant homology to the oncoretroviral transmembrane (TM) glycoproteins (5, 45), especially those of ASLV (12). More striking than the strong amino acid similarities between these glycoproteins is the conservation of many putative functional domains such as a central CX6CC motif, the potential coiled coil, and the putative fusion peptide. Also conserved in all strains of Ebola virus is a stretch of basic residues that in ASLV constitute an endoproteolytic cleavage site (21, 32). Furthermore, the spacing between this basic residue-rich region and the adjacent presumptive fusion peptide is nearly identical between the Ebola virus and ASLV glycoproteins (1). This structural similarity suggests that the glycoproteins of Ebola virus and ASLV may utilize similar mechanisms to mediate membrane fusion and viral entry even though the triggers for these processes are clearly different: the ASLV envelope requires receptor-mediated activation, and the Ebola virus envelope glycoprotein (Ebo-GP) is pH dependent (6, 41, 48).
Since this dibasic motif is conserved in all strains of Ebola virus and is in a position analogous to the cleavage site of ASLV envelope, we hypothesized that Ebo-GP is endoproteolytically processed. Analysis of both wild-type and epitope-tagged forms of Ebo-GP revealed that this glycoprotein is proteolytically cleaved during maturation and that the two resulting subunits appear to be disulfide linked. Mutational analysis of the conserved dibasic motif identified this region as the Ebo-GP endoproteolytic processing site. Surprisingly, our results show that an uncleaved mutant of Ebo-GP is efficiently incorporated into murine leukemia virus (MLV) particles and is able to efficiently mediate viral entry, indicating that, in contrast to nearly all other viral systems where glycoprotein processing is observed, proteolytic cleavage is not essential for the membrane fusion activity of Ebo-GP.
MATERIALS AND METHODS
Cell lines and antibodies.
Human embryonic kidney 293T cells were maintained in Dulbecco’s modified Eagle medium supplemented with 10% bovine calf serum. Baby hamster kidney (BHK), murine NIH 3T3, African green monkey kidney (Vero and BSC-1), LoVo human colon carcinoma, Tb 1 lu bat lung, and bovine aorta endothelial cells were maintained in Dulbecco’s modified Eagle medium supplemented with 10% fetal calf serum and nonessential amino acids (0.1 mM). All cell lines were in addition supplemented with penicillin (100 U/ml) and streptomycin (100 mg/ml).
A rabbit polyclonal antibody recognizing the cytoplasmic tail of EnvA (anti-Rous sarcoma virus [RSV] tail serum) was generated as described previously (13, 14). Briefly, a peptide corresponding to the 23 carboxyl-terminal amino acids of EnvA was coupled to keyhole limpet hemocyanin (Pierce, Rockford, Ill.) and used to raise polyclonal sera in rabbits by Cocalico Biologicals, Inc. (Reamstown, Pa.). A rabbit polyclonal antibody recognizing the Zaire subtype of Ebo-GP was kindly provided by Anthony Sanchez (Centers for Disease Control and Prevention).
Plasmids and viruses.
pHIT60 and pHIT111 have been described previously (37). Plasmid pGEM7hFurin was kindly provided by Gary Thomas (Vollum Institute, Portland, Oreg.). The expression plasmid pCB6-Ebo-GP, encoding the envelope glycoprotein of the Mayinga strain of the Ebola virus Zaire subtype has been described previously (48), as has the EnvA expression plasmid, pCB6-EnvA (13). Plasmid pGEM-EMGP2, which contains the Ebo-GP cDNA is described in reference 36.
The Ebo-GP cDNA was excised from pGEM-EMGP2 by using the BamHI and KpnI restriction enzymes and cloned into plasmid pSP72 under the control of the T7 promoter. This plasmid was named pSP72-Ebo-GP.
An overlapping extension PCR protocol was used to produce an epitope-tagged version of Ebo-GP, termed Ebo-T, in which the 4 carboxyl-terminal amino acids of Ebo-GP were exchanged with the 23 carboxyl-terminal amino acids of EnvA. PCR was performed with pGEM-EMGP2 and pCB6-EnvA as templates. The flanking PCR primers used were OS 336 (5′-CGCTGAAGGTGTCGTTGC-3′), which is specific for a pGEM-EMGP2 sequence, and OS 168 (5′-CAGGGATCCGATGCTACTATATCC-3′), which is specific for a pCB6-EnvA sequence. The internal primers used were OS 444 (5′-TTATTCTGTATATGCAGAAAGATGATTAAT-3′) and its reverse complement, OS 345. This final PCR product was digested with restriction enzymes EcoRV and BamHI and cloned into pCB6-Ebo-GP to create plasmid pCB6-Ebo-T and into pSP72-Ebo-GP to create plasmid pSP72-Ebo-T.
A similar PCR strategy was used to produce the proteolytic processing site mutants CL-1 and CL(−). In this case, the DNA templates used were pGEM-EMGP2 and pCB6-Ebo-T so as to produce the mutants in both the wild-type (Ebo-GP) and epitope-tagged (Ebo-T) forms of Ebo-GP. The flanking primers used were OS 336 and either OS 7 (5′-AATACGACTCACTATAG-3′), which is specific for the T7 primer in pGEM-EMGP2, or OS 168, which is specific for pCB6-EnvA and thus pCB6-Ebo-T. The internal primers used were either OS 446 (5′-AGAAGAACTGCAGCAGAAGCAATTGTCAATGCT-3′) and OS 447 (5′-TGCTGCAGTTCTTCTCCCGCCTGTGATCAG-3′) for production of the CL-1 mutant, OS 503 (5′-AGAGCAACTGCAAGAGAAGCAATTGTCAATGCT-3′) and OS 504 (5′-TCTTGCAGTTGCTCTCCCGCCTGTGATCAG-3′) for production of the CL-2 mutant, OS 505 (5′-AGAGCAACTGCTGCAGAAGCAATTGTCAATGCT-3′) and OS 506 (5′-TGCAGCAGTTGCTCTCCCGCCTGTGATCAG-3′) for production of the CL-3 mutant, OS 437 (5′-GCAGGTACCGCAGCAGAAGCAATTGTCAATGCT-3′) and OS438 (5′-GAAGTAGGTACCTGCCCCGCCTGTGATCAGTCC-3′) for production of the CL-7 mutant, or OS 435 (5′-GCAGGTACCGCAGCAGAAGCAATTGTCAATGCT-3′) and OS 436 (5′-TGCTGCGGTACCTGCCCCGCCTGTGATCAGTCC-3′) for production of the CL(−) mutant. These PCR products were either digested with the EcoRV and EcoRI restriction enzymes and cloned into pCB6-Ebo-GP to produce plasmids pCB6-Ebo-GP.CL-1 and pCB6-Ebo-GP.CL(−) or digested with EcoRV and BamHI and cloned into pBC6-Ebo-T to produce plasmids pCB6-Ebo-T.CL-1 and pCB6-Ebo-T.CL(−).
The vaccinia virus vTF-7, encoding the T7 polymerase, was obtained from Bernie Moss, National Institutes of Health.
Sanchez et al. (36) have described a putative signal peptide at the amino terminus of Ebo-GP. The predicted cleavage site for this signal peptide removes the amino-terminal 32 amino acids of Ebo-GP and produces a mature form of the protein which has an Ile in the amino-terminal position. All numbering of the Ebo-GP amino acid sequence will therefore start with Ile1.
Production of MLV pseudotypes.
MLV particles pseudotyped with the various forms of Ebo-GP described above were produced through modification of a transient MLV packaging system as previously described (37). Briefly, 20 μg of either pCB6-Ebo-GP, pCB6-Ebo-T, pCB6-Ebo-GP.CL-1, pCB6-Ebo-GP.CL(−), pCB6-Ebo-T.CL-1, or pCB6-Ebo-T.CL(−) was mixed with 20 μg of a plasmid encoding the MLV Gag-Pol (pHIT60) and 20 μg of a plasmid containing a packageable genome encoding the β-galactosidase reporter gene (pHIT111). 293T cells were transfected with these DNA mixtures by a standard CaPO4 procedure (48) to produce MLV(Ebo-GP), MLV(Ebo-T), MLV(Ebo-GP.CL-1), MLV(Ebo-GP.CL(−), MLV(Ebo-T.CL-1), or MLV(Ebo-T.CL(−)) respectively. Approximately 48 h posttransfection, the cellular supernatants containing these viruses were collected and clarified by filtration through 0.45-μm-pore-size syringe filters. These supernatants were stored at either 4 or −80°C as viral stocks.
All experiments involving the production or functional analysis of these replication-incompetent MLV pseudotypes were performed under biosafety level 2 containment as approved by the University of Pennsylvania Institutional Biosafety Committee.
To analyze the incorporation into MLV virions of the various forms of Ebo-GP described above, 3.5-ml aliquots of clarified viral stocks were layered onto 2 ml of 20% sucrose in phosphate-buffered saline (PBS) and centrifuged at 55,000 rpm in an SW55 rotor for 15 min. Pelleted virions were lysed in radioimmunoprecipitation assay (RIPA) buffer (140 mM NaCl, 10 mM Tris [pH 8.0], 5 mM EDTA, 1% sodium deoxycholate, 1% Triton X-100, 0.1% sodium dodecyl sulfate [SDS]) and resolved by SDS-polyacrylamide gel electrophoresis (PAGE). Expression of the various forms of Ebo-GP was detected by Western blotting as previously described (48), using either the anti-Ebo-GP (1:1,000 dilution) or anti-RSV-tail (1:1,500 dilution) serum described above. A horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (Pierce) was used at a 1:20,000 dilution.
Dissociation of GP1 and GP2.
MLV(Ebo-GP), MLV(Ebo-T), and MLV(Ebo-GP.CL(−)) were produced as described above and centrifuged at 25,000 rpm in an SW28 rotor for 90 min. The supernatants were decanted, and the viral pellets resuspended in TNE buffer (50 mM Tris [pH 8], 130 mM NaCl, 1 mM EDTA) overnight at 4°C. Urea (8 M in PBS) was added to these viral stocks to a final concentration of 0, 2, 4, or 6 M in the presence or absence of dithiothreitol (DTT) at a final concentration of 100 mM. These solutions were incubated at 37°C for 30 min and then layered onto 20% sucrose in PBS and centrifuged at 55,000 rpm in an SW55 rotor for 15 min. The resulting viral pellets were lysed in RIPA buffer and resolved by SDS-PAGE. Ebo-GP expression was detected by Western blotting with the anti-Ebo-GP and anti-RSV tail sera, as described above.
Expression of Ebo-GP and Ebo-T in LoVo cells.
LoVo cells were seeded at 3 × 105 cells/well of a six-well dish the day before transfection. Mixtures containing either no DNA, 5 μg of pSP72-Ebo-T, or 5 μg of pSP72-Ebo-T and 5 μg of pGEM7hFurin were prepared and used to transfect the LoVo cells by a standard CaPO4 method as described above. Two hours posttransfection, the cells were refed and infected with the vaccinia virus vTF-7 at a multiplicity of infection of approximately 10. One hour postinfection, the cells were again refed. Eighteen hours postinfection, the cells were lysed in Triton lysis buffer (50 mM Tris [pH 8], 5 mM EDTA, 150 mM NaCl, 1% Triton X-100). Cellular lysates were resolved by SDS-PAGE, and expression of the different forms of Ebo-T was examined by Western blot analysis with the anti-RSV tail serum as described above.
Analysis of Ebo-GP processing requirement for viral entry.
All cell lines described above were seeded at 3 × 105 to 5 × 105 cells/well of a six-well dish the day before infection. Various dilutions [1:1, 1:10, or 1:100, depending on the previously observed MLV (vesicular stomatitis virus) pseudotype titer on the various cell types (48)] of either MLV(Ebo-GP) or MLV(Ebo-GP.CL(−)) were made in 1 ml of maintenance medium and used to challenge these cells as described previously (48). Equal amounts of either MLV(Ebo-GP) or MLV(Ebo-GP.CL(−)) were used in each infection as judged by Western blot analysis of lysed virions with the anti-Ebo-GP serum and by visualization of MLV Gag proteins by Ponceau S staining of nitrocellulose membranes to which viral lysates resolved by SDS-PAGE had been transferred. Forty eight hours postchallenge, the cells were fixed with 2% paraformaldehyde and stained for β-galactosidase activity as described previously (48). Viral titers were determined by microscopic examination of stained cells and the enumeration of β-galactosidase-positive cells. These titers were expressed as the number of β-galactosidase-positive cells per milliliter of viral stock used in the infection (infectious units [IU] per milliliter). The multiplicity of infection in these experiments was always less than 0.1.
RESULTS
Production of an epitope-tagged form of Ebo-GP.
To facilitate analysis of the Ebola virus glycoproteins, we introduced an epitope tag at the carboxyl terminus of Ebo-GP. An overlapping extension PCR protocol was used to exchange the nucleotides encoding the 4 carboxyl-terminal amino acids of Ebo-GP with those encoding the 12 carboxyl-terminal amino acids of EnvA. The resulting PCR product was cloned into the expression plasmid pCB6 under the control of the cytomegalovirus immediate-early promoter, to create plasmid pCB6-Ebo-T. Plasmid pCB6-Ebo-GP, which encodes the wild-type form of Ebo-GP, has been described previously (48). A schematic diagram of the predicted protein products of both pCB6-Ebo-GP (Ebo-GP) and pCB6-Ebo-T (Ebo-T) is shown in Fig. 1A.
FIG. 1.
(A) Schematic diagram of the predicted protein products of pCB6-EboGP and pCB6-Ebo-T. Speckled boxes represent leader peptides; hatched boxes represent TM domains. Gray shading indicates the central region of Ebo-GP that is divergent between the different strains of Ebola virus. Arrows indicate the putative endoproteolytic processing sites, as well as the Ebo-GP cytoplasmic tail. The RSV cytoplasmic tail, used as an epitope tag in Ebo-T, is indicated by an arrow; potential glycosylation sites are shown (|∨). To evaluate the incorporation of Ebo-GP and Ebo-T into MLV virions, 293T cells were transfected with MLV Gag-Pol and genome constructs with either pCB6-Ebo-GP (Ebo-GP) or pCB6-Ebo-T (Ebo-T) or without a glycoprotein construct (Mock). Viral supernatants were collected and partially purified by pelleting through 20% sucrose. Cell lysates (B) and lysed pellets (C and D) were analysed by SDS-PAGE and Western blotting with either the anti-Ebo-GP serum (B and C) or the anti-RSV tail serum (D). Arrows indicate the GP1, GP2, and GP0 proteins; positions of molecular mass markers (in kilodaltons) are shown to the left of each gel.
293T cells were transiently transfected with either pCB6-Ebo-GP or pCB6-Ebo-T. Forty-eight hours posttransfection, cellular lysates were prepared, resolved by SDS-PAGE, and analyzed by Western blotting with an anti-Ebo-GP serum. Lysates from cells transfected with either the wild-type or epitope-tagged construct demonstrated a pair of bands with apparent molecular masses of approximately 110 to 140 kDa (Fig. 1B), matching those previously described for Ebo-GP (9, 44, 48). This result suggests that the epitope tag did not affect the processing of Ebo-GP.
Production and analysis of MLV(Ebo-GP) and MLV(Ebo-T) pseudotypes.
We have previously demonstrated efficient incorporation of Ebo-GP into MLV particles (48). To examine the incorporation of the epitope-tagged Ebo-T into MLV particles, 293T cells were transiently transfected with plasmids encoding MLV gag/pol (pHIT 60), an MLV genome containing a β-galactosidase reporter gene (pHIT111), and either pCB6-Ebo-GP or pCB6-Ebo-T. Forty-eight hours posttransfection, cellular supernatants, containing MLV(Ebo-GP) or MLV(Ebo-T), respectively, were collected, clarified by filtration, and stored as viral stocks at either 4 or −80°C.
These viral stocks were partially purified by centrifugation through 20% sucrose, lysed in RIPA buffer, and analyzed by SDS-PAGE and Western blotting with an anti-Ebo-GP serum. This analysis detected a single band, with a mobility similar to that previously described for the mature form of Ebo-GP (44, 48), in the viral lysates of both MLV(Ebo-GP) and MLV(Ebo-T) (Fig. 1C; compare lanes 2 and 3). In contrast, Western blot analysis of these viral lysates with the polyclonal anti-RSV tail serum demonstrated instead a predominant 28-kDa band in the lysates of MLV(Ebo-T) virions and not the 140-kDa band seen with the anti-Ebo-GP serum (Fig. 1D, lane 3). These data suggest that Ebo-GP is proteolytically processed into two subunits during maturation. Moreover, the detection of a band of approximately 160 kDa in the lysates of MLV(Ebo-T) virions that was weakly reactive with the anti-RSV tail serum (Fig. 1D, lane 3) implies that the processing of Ebo-GP may not be complete and that unprocessed forms of Ebo-GP are incorporated into virions. By analogy to the nomenclature for influenza virus HA, we propose to name the two subunits of Ebo-GP GP1 (the larger subunit recognized by the anti-Ebo-GP serum) and GP2 (the smaller subunit recognized by the anti-RSV-tail serum) (Fig. 1C and D); the uncleaved form of Ebo-GP we propose to name GP0 (Fig. 1D).
Dissociation of GP1 from MLV(Ebo-T) by reduction and denaturation.
The detection of two different Ebo-GP-specific bands in the lysates of pseudotyped virions strongly suggested that Ebo-GP is a cleaved glycoprotein. We reasoned that if Ebo-GP on the virion was indeed cleaved, then we should be able to separate the two subunits by mild denaturation and/or reduction.
To examine the potential proteolytic processing of the Ebola virus glycoproteins, MLV(Ebo-GP) and MLV(Ebo-T) pseudotypes were produced as described above and concentrated by ultracentrifugation, and the viral pellets were resuspended overnight in an isotonic buffer. These virions were then treated with various concentrations of urea in the presence or absence of 100 mM DTT for 30 min at 37°C. Following this incubation, the virions were centrifuged through 20% sucrose to separate the virion-associated proteins from those dissociated from the particles by denaturation and/or reduction. The resulting viral pellet was lysed in RIPA buffer, and protein composition was analyzed by SDS-PAGE and Western blotting with either the anti-Ebo-GP or anti-RSV tail serum.
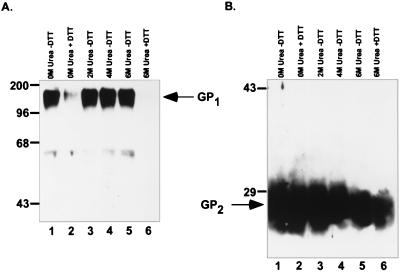
Lysates of untreated MLV(Ebo-T) virions demonstrated both GP1- and GP2-specific bands when analyzed with the anti-Ebo-GP and anti-RSV tail sera (Fig. 2, lanes 1). Treatment of the MLV(Ebo-T) pseudotypes with up to 6 M urea in the absence of DTT did not alter the protein composition of the virions from that seen in untreated virions (Fig. 2; compare lanes 1, 3, 4, and 5). However, treatment of these virions with 100 mM DTT either alone or in combination with 6 M urea dramatically reduced the intensity of the GP1-specific band, while GP2 remained associated with the virion (Fig. 2; lanes 2 and 6). This result indicates that these treatments were sufficient to dissociate the two subunits from one another. Similar analysis of virions carrying the Ebo-GP also demonstrated a specific loss of GP1 with either 100 mM DTT or 6 M urea–100 mM DTT treatment (data not shown). These data strongly support the hypothesis that Ebo-GP is endoproteolytically processed into two subunits during maturation. Furthermore, it suggests that the two subunits are covalently associated by disulfide bonds.
FIG. 2.
Dissociation of GP1 from GP2. MLV(Ebo-GP) and MLV(Ebo-T) were produced by transient transfection of 293T cells. Viral supernatants were collected from transfected cells and partially purified by pelleting through 20% sucrose. The resulting viral pellet was resuspended in PBS and treated with the indicated concentrations of urea with (+) or without (−) 100 mM DTT. Treated virions were again partially purified by pelleting through 20% sucrose. Lysed viral pellets were analyzed by SDS-PAGE and Western blotting with either (A) the anti-Ebo-GP serum (A) or the anti-RSV tail serum (B). Arrows indicate the GP1 and GP2 proteins; positions of molecular mass markers (in kilodaltons) are shown to the left of each gel.
Production of site-directed mutations within the proposed Ebo-GP proteolytic processing site.
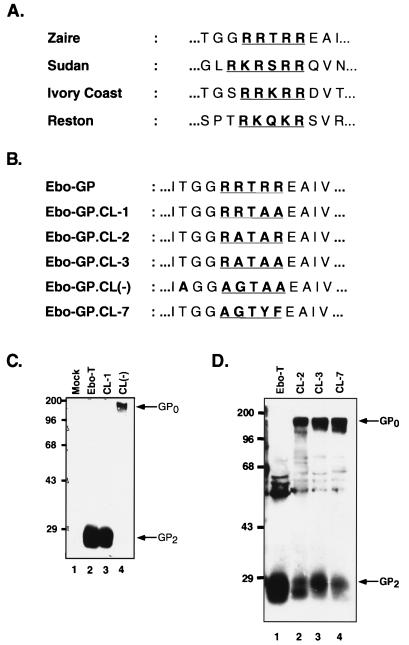
Conserved within the glycoproteins of all known filoviruses is a consensus dibasic proteolytic processing site similar to that found in other viral glycoproteins such as ASLV EnvA and the HIV-1 envelope glycoprotein (8, 27) (Fig. 3A). In the envelope glycoprotein from the Mayinga strain of Ebola virus Zaire subtype used for these studies, this putative dibasic processing site comprises Arg465, Arg466, Thr467, Arg468, and Arg 469 (RRTRR). To determine whether this sequence comprised the endoproteolytic processing site, we introduced two mutations into both the Ebo-GP and Ebo-T cDNAs. These mutations were designed to replace the basic arginine residues of this putative processing site with apolar amino acids. In the mutant CL-1, Arg468 and Arg469 were changed to Ala to produce the sequence RRTAA; in the mutant CL(−), this sequence was changed to AGTAA (Fig. 3B). To analyze the effects of these mutations on processing, MLV particles pseudotyped with either Ebo-T, Ebo-T.CL-1, or Ebo-T.CL(−) were produced as described above, partially purified by centrifugation through 20% sucrose, and analyzed by SDS-PAGE and Western blotting with the anti-RSV tail serum. These results demonstrated that like Ebo-T, the Ebo-T.CL-1 mutant was incorporated efficiently into virions and was processed efficiently into two subunits, as Western blot analysis revealed the presence of GP2, and not GP0, in the MLV(Ebo-GP.CL-1) virion lysates (Fig. 3C; compare lanes 2 and 3). Analysis of virions produced with Ebo-GP.CL(−) also indicated efficient incorporation of this mutant glycoprotein, however, in this instance, Western blot analysis with the anti-RSV tail serum revealed the specific loss of GP2 and the gain of an approximately 160-kDa band (Fig. 3C, lane 4). This larger form of Ebo-GP was likely representative of the unprocessed form of Ebo-GP, GP0, suggesting that this mutation blocked the endoproteolytic processing of Ebo-T. In support of this hypothesis, Western blot analysis of MLV particles pseudotyped either Ebo-GP, Ebo-GP.CL-1, or Ebo-GP.CL(−) with the anti-Ebo-GP serum also demonstrated a specific loss of GP1 in the lysates of virions pseudotyped with the Ebo-GP.CL(−) glycoprotein and the gain of an approximately 160-kDa band (data not shown). Together, these data strongly suggest that the identified dibasic motif in the envelope glycoprotein of Ebola Zaire is the endoproteolytic processing site.
FIG. 3.
Identification of the Ebo-GP endoproteolytic processing site. (A) Comparison of the conserved dibasic motifs in the envelope glycoproteins of all known strains of Ebola virus. (B) Comparison of the Ebo-GP endoproteolytic processing site with those of five mutant glycoproteins. (C) Western blot analysis of Ebo-T, Ebo-T.CL-1, and Ebo-T.CL(−). 293T cells were transfected with either pCB6-Ebo-T (Ebo-T), pCB6-Ebo-T.CL-1 (CL-1), or pCB6-Ebo-T.CL(−) [CL(−)] or without a glycoprotein construct (Mock). Viral lysates prepared 48 h posttransfection were analyzed by SDS-PAGE and Western blotting with the anti-RSV tail serum. Arrows indicate the GP2 and GP0 proteins. Positions of molecular mass markers (in kilodaltons) are shown to the left of each gel. (D) Western blot analysis of Ebo-T, Ebo-T.CL-2, Ebo-T.CL-3, and Ebo-T.CL-7. Viral preparation and analysis were performed as for panel C. Arrows indicate the GP2 and GP0 proteins; positions of molecular mass markers (in kilodaltons) are shown to the left of each gel.
To further define the processing site requirements of Ebo-GP, we produced three other mutant glycoproteins in which the cleavage site was changed to RATAR (Ebo-GP.CL-2), RATAA (Ebo-GP.CL-3), and AGTYF (Ebo-GP.CL-7), respectively. The CL-7 mutant contains a consensus chymotrypsin cleavage site so as to allow in vitro processing of the glycoprotein. Western blotting of the lysates of MLV pseudotypes containing these glycoproteins with the anti-RSV tail serum demonstrated that all three were still endoproteolytically processed (Fig. 3D), demonstrating that the Ebo-GP cleavage site is relatively insensitive to mutagenesis. Furthermore, the glycoprotein is cleaved even when the consensus dibasic site is replaced by a chymotrypsin cleavage site, suggesting that in the native Ebo-GP structure, this region is highly exposed and likely accessible to a broad array of proteases.
GP0 is not dissociated from MLV(Ebo-GP.CL(−)) by reduction and denaturation.
The CL(−) mutation (AGTAA) appears to block endoproteolytic processing of the Ebola virus glycoprotein. To confirm this observation, we attempted to determine if the presumably unprocessed glycoprotein in virions carrying this mutant glycoprotein were resistant to dissociation by reduction and denaturation. Wild-type and CL(−)-pseudotyped virions were produced as described above and gently ultracentrifuged, and the resulting viral pellets were resuspended in an isotonic buffer overnight at 4°C. These concentrated virions were then treated with 100 mM DTT, alone or with 6 M urea, for 30 min at 37°C. Virion-associated proteins were partially purified by centrifugation through 20% sucrose. After lysis with RIPA buffer, the protein composition of the pelleted virions was analyzed by SDS-PAGE and Western blotting with the polyclonal anti-Ebo-GP antibody.
As demonstrated previously, treatment of virus containing wild-type Ebo-GP with DTT or urea and DTT efficiently dissociated GP1 from the virions (Fig. 4A; compare lanes 1, 2, and 3). In contrast, incubation of CL(−)-pseudotyped virions under either of these conditions did not result in a significant release of the glycoprotein from the virions (Fig. 4B). These results strongly support the hypothesis that the Ebo-GP.CL(−) protein represents an uncleaved form of Ebo-GP.
FIG. 4.
GP0 cannot be dissociated from virions by denaturation or reduction. MLV(Ebo-GP) (A) and MLV(Ebo-GP.CL(−) (B) were produced by transient transfection of 293T cells. Forty-eight hours posttransfection, virion-containing cellular supernatants were collected and partially purified through 20% sucrose. The resulting viral pellets were resuspended in PBS and treated with the indicated concentrations of urea, with (+) or without (−) 100 mM DTT. These treated virions were then partially purified by pelleting through 20% sucrose, lysed, and analyzed by SDS-PAGE and Western blotting with the anti-Ebo-GP serum. Arrows indicate GP1 and GP0; positions of molecular mass markers (in kilodaltons) are shown to the left of each gel.
Proteolytic processing of Ebo-GP does not require furin.
Furin is a widely expressed cellular protease that cleaves proteins at paired basic amino acid sites (20). Moreover, it has been shown to be responsible for the cleavage of a number of viral glycoproteins, including the human parainfluenza virus type 3 F protein (31) and the HAs of various strains of pathogenic influenza A viruses (19, 40). To address whether furin is the cellular enzyme responsible for the endoproteolytic processing of Ebo-GP, we attempted to express this envelope glycoprotein in a human colon carcinoma cell line (LoVo) that specifically lacks furin activity (42, 43).
Ebo-T was expressed in LoVo cells with or without the coexpression of human furin. Cellular lysates were prepared, resolved by SDS-PAGE and examined by Western blot analysis with both the anti-Ebo-GP and anti-RSV tail sera. When these cellular lysates were examined with the anti-Ebo-GP polyclonal serum, it was apparent that Ebo-T produced in LoVo cells was of a greater apparent molecular mass than when furin was coexpressed (Fig. 5A; compare lanes 1 and 2). This larger form of Ebo-GP, found specifically in the lysates of LoVo cells expressing Ebo-T alone, appears to be similar in size to GP0, suggesting that in the absence of furin, Ebo-T is not efficiently cleaved. Similar results were obtained when the wild-type form of Ebo-GP was expressed in LoVo cells (data not shown). However, when these same cellular lysates were examined with the anti-RSV tail serum, it became apparent that there was significant processing of Ebo-T in these furin-deficient LoVo cells. In addition, an increased amount of GP2 was observed when human furin was coexpressed in the LoVo cells with Ebo-T (Fig. 5B, lanes 1 and 2). The presence of GP2 in the LoVo cell lysates indicates that Ebo-T is endoproteolytically processed in the absence of furin and implies that furin is likely not the only cellular enzyme responsible for the endoproteolytic processing of Ebo-GP.
FIG. 5.
Expression of Ebo-T in LoVo cells. LoVo cells were transiently transfected with either no DNA (Mock), pCB6-Ebo-T (Ebo-T), or pCB6-Ebo-T and pGEM7hFurin (Ebo-T + Furin). Two hours posttransfection, the cells were infected with the vaccinia virus vTF-7. Eighteen hours postinfection, the cells were lysed and cellular protein expression was examined by SDS-PAGE and Western blotting with either the anti-Ebo-GP serum (A) or the anti-RSV tail serum (B). Positions of molecular mass markers (in kilodaltons) are shown to the left of each gel.
Analysis of entry mediated by Ebo-GP.CL(−).
The conservation of an endoproteolytic processing site in the glycoproteins of all known strains of Ebola virus strongly suggests that cleavage may be important for glycoprotein function. To investigate the importance of processing for Ebo-GP-mediated viral entry, we produced MLV particles pseudotyped with either the wild-type glycoprotein or the cleavage-deficient mutant, Ebo-GP.CL(−). These viral pseudotypes were normalized with respect to Ebo-GP by Western blot analysis with the anti-Ebo-GP antiserum and then used to challenge a variety of target cell lines as described in Materials and Methods.
As shown previously (48), MLV virions pseudotyped with Ebo-GP (MLV(Ebo-GP)) were able to mediate efficient infection of a wide variety of cell lines (Table 1), with titers ranging from 2.3 × 105 IU/ml on the human embryonic kidney 293T cell line to 1.2 × 102 IU/ml on the Tb 1 lu bat lung cell line. Surprisingly, MLV particles pseudotyped with the uncleaved mutant of Ebo-GP were also able to infect these cell lines efficiently, with the exception of the African green monkey kidney cell line BSC-1, which was consistently 3- to 10-fold less susceptible to MLV particles carrying the CL(−) glycoprotein than wild-type Ebo-GP. However, there was no difference in titer between MLV pseudotyped with either of these two glycoproteins on Vero cells which, like the BSC-1 cells, are derived from African green monkey kidney. It seems unlikely that the small difference observed on BSC-1 cells represents a major effect of processing upon glycoprotein function. Therefore, it appears that, in contrast to the glycoproteins of the oncoretroviruses, to which the Ebola virus glycoproteins bear a striking homology, endoproteolytic processing of Ebo-GP is not required for glycoprotein function; this suggests a more subtle reason for the conservation of this dibasic endoproteolytic processing site in the Ebola virus glycoproteins.
TABLE 1.
Efficient processing of Ebo-GP is not required for infectivitya
| Cell line | Species | Titer (IU/ml)
|
% of wild-type value | |
|---|---|---|---|---|
| MLV (Ebola) | MLV (Ebola CL(−)) | |||
| 293T | Human | 2.1 × 105 | 2.3 × 105 | 110 |
| HeLa | Human | 3.5 × 102 | 5.3 × 102 | 151 |
| Vero | Simian | 1.7 × 104 | 1.7 × 104 | 100 |
| Cos 7 | Simian | 8.0 × 103 | 9.8 × 103 | 123 |
| BSC-1 | Simian | 1.0 × 104 | 8.4 × 102 | 8 |
| NIH 3T3 | Murine | 2.6 × 104 | 2.1 × 104 | 81 |
| BHK | Hamster | 1.6 × 104 | 1.4 × 104 | 88 |
| MDBK | Bovine | 2.4 × 103 | 3.6 × 103 | 150 |
| BAECb | Bovine | 2.3 × 103 | 1.8 × 103 | 78 |
| Tb 1 lu | Bat | 1.2 × 102 | 2.0 × 102 | 167 |
Data from representative experiments are shown. Similar results have been obtained in replicate experiments.
Bovine aorta endothelial cells.
DISCUSSION
In this study, we demonstrated that Ebo-GP is endoproteolytically processed during maturation into two subunits that we have designated GP1 and GP2, thus confirming other recently described results (46). It is the smaller, membrane-anchored subunit, GP2, which is homologous to the TM glycoproteins of the oncoretroviruses and contains many of the putative structures believed to be important for glycoprotein-mediated membrane fusion. When the epitope-tagged form Ebo-T was pseudotyped into MLV particles, we observed that the cleaved form of the glycoprotein was preferentially incorporated into the virions. The peripheral GP1 subunit could be specifically stripped from these pseudotyped particles by treatment with the reducing agent DTT but not with the denaturant urea, indicating that the two subunits are likely disulfide linked. Furthermore, mutational analysis of this glycoprotein identified a dibasic motif, conserved in all membrane-anchored filoviral glycoproteins, as the Ebo-GP endoproteolytic cleavage site.
Furin is a proprotein convertase that has been implicated in the endoproteolytic processing of numerous cellular proteins (39) as well as the proteolytic activation of many viral glycoproteins, including HA of certain strains of influenza A virus (19, 40) and the prM glycoprotein of tick-borne encephalitis virus (38). Furin is expressed in a wide variety of cell types and cleaves proteins after dibasic motifs of the general consensus Arg-X-Lys/Arg-Arg (20). To determine whether furin was required for the processing of Ebo-GP, we analyzed Ebo-T expression in the furin-deficient LoVo cell line. We found that the processing of Ebo-T, although incomplete, was still observed in these cells, indicating that other endoproteases that may be present in the LoVo cells, such as PC2 or PACE4 (39), are competent to cleave the Ebo-GP. Contrary to our results, Volchkov and others (46) have recently reported that Ebo-GP is not cleaved in LoVo cells, leading them to hypothesize that furin is required for the endoproteolytic processing of this glycoprotein. The presence of an epitope tag at the carboxyl terminus of Ebo-T provides us with an extremely powerful assay with which to detect Ebo-GP processing, and it is likely the lack of such a sensitive assay that is the cause for the discrepancy between our results. Moreover, the hypothesis that furin is not required for Ebo-GP processing is strengthened by the fact that the Ebo-GP cleavage mutants Ebo-GP.CL-1, Ebo-GP.CL-2, and Ebo-GP.CL-3, in which the consensus furin recognition site has been disrupted, are still processed, albeit less efficiently than the wild type. Indeed, in the Ebo-GP.CL-3 mutant, only one arginine residue remains at the cleavage site.
Our result demonstrating processing in LoVo cells is not entirely surprising, as it has been shown that the HIV-1 glycoprotein, which is also processed at a consensus furin-like protease recognition motif, is efficiently cleaved in the absence of furin (7, 16, 22, 30). These data strongly suggest that Ebo-GP may be cleaved by a variety of proprotein convertases in different cell types and not by furin alone. Moreover, Ebo-GP was cleaved even when the dibasic motif was replaced with a consensus chymotrypsin protease recognition motif (Ebo-GP.CL-7), suggesting that the cleavage site of this glycoprotein may be highly exposed in the native structure and thus accessible to a great number of different proteases.
Endoproteolytic processing is a requirement for the fusogenic activity of many viral glycoproteins, including the HIV-1 envelope glycoprotein, the MLV envelope glycoprotein, and the influenza A virus HA (11, 24, 25, 27). Site-directed mutagenesis of both the HIV-1 and ASLV glycoprotein cleavage sites has shown that in the absence of processing, these glycoproteins are unable to mediate viral entry (8, 10, 17, 32). Similarly, many apathogenic strains of influenza A virus are restricted to replicating in the cells of the respiratory tract due to the localized expression of proteases capable of cleaving the HA glycoprotein (39). Acquisition of a furin-like endoprotease recognition site in the HA of these viruses, and the consequent expanded cellular tropism, is a pivotal determinant of the increased pathogenesis of virulent influenza A virus strains (3, 4, 39). It was therefore surprising, especially in view of the close relationship between the Ebola virus and retrovirus TM sequences, to discover that the uncleaved Ebo-GP mutant, Ebo-GP.CL(−) was able to mediate viral entry into a variety of cell types as efficiently as the wild-type glycoprotein, indicating that, unlike most other cleaved viral glycoproteins, endoproteolytic processing is not required for Ebo-GP function. A recent report suggests that the coronavirus mouse hepatitis virus (MHV) A59 does not require glycoprotein cleavage either in vivo or in vitro (2, 18). In contrast to the filoviruses, however, many coronavirus glycoproteins have no obvious conserved cleavage site and are not generally found as cleaved proteins, and thus the relevance of this result with MHV A59 for Ebola virus is unclear.
The lack of a requirement for endoproteolytic processing is puzzling, since the dibasic motif is highly conserved in the envelope glycoproteins of all known strains of Ebola virus and the closely related Marburg virus (1, 46). It is possible that processing of Ebo-GP is required for viral replication only in certain cell types. The Sindbis virus E2 glycoprotein is normally endoproteolytically processed during maturation (26). However, cleavage is required only for viral growth in invertebrate cells, while the virus retains the ability to grow in the cells of vertebrates, albeit with significantly reduced efficiency (33). A similar situation may exist for Ebola virus, where glycoprotein processing may be important for the infection of certain species or tissue types. However, our preliminary survey of eight cell lines of different tissues and species revealed only a modest 3- to 10-fold reduction in one cell line. The absolute conservation of this dibasic cleavage site in all filoviruses might suggest that endoproteolytic processing of Ebo-GP is critical for some stage of the viral life cycle. We may discover that glycoprotein processing is important for Ebola virus replication in the species that is the natural reservoir for this virus. If this is the case, and endoproteolytic processing of Ebo-GP is required only for the infection of certain cell types, then the analysis of the ability of MLV(Ebo-GP) and MLV(Ebo-GP.CL(−)) to infect a very wide selection of different cell types from diverse species might provide important clues for the identification of the elusive Ebola virus reservoir.
Another possible explanation for the highly conserved cleavage site is that cleavage confers an advantage to Ebo-GP-mediated viral entry that cannot be resolved in our one-step infection assay. Over many rounds of viral replication during an infection in vivo, a small advantage might provide the selective pressure required for retention of this motif. It has been shown for MHV A59 that although cleavage is not required for viral infectivity, the uncleaved glycoprotein mediates cell-to-cell fusion with delayed kinetics (1a, 15). Similarly, while proteolytic processing of the Sindbis virus E2 glycoprotein is not required for viral infectivity, virions containing mutations that abrogate E2 processing are avirulent in vivo and produce small plaques in vitro (34). Techniques for reverse genetics are not available for filoviruses but do exist for the closely related rhabdoviruses (28). When such techniques become available for the filoviruses, it will be of great interest to determine whether viruses containing an uncleavable form of Ebo-GP can replicate in vitro and if such viruses are pathogenic in animal models. A recent report described the use of peptides to inhibit the endoproteolytic processing of Ebo-GP (46). If, as with Sindbis virus, proteolytic processing of the glycoprotein is important in pathogenesis, then the administration of such peptides might represent a novel approach to treating individuals infected with this deadly virus.
ACKNOWLEDGMENTS
We thank Gary Thomas (Vollum Institute, Portland, Oreg.) for the human furin expression plasmid, George Prendergast for the LoVo cell line, and Bernie Moss (National Institutes of Health) for vaccinia virus vTF-7. We thank John Balliet for critical reading of the manuscript and other members of the Bates laboratory for useful discussions.
This work was supported by grant CA63531 to P.B. from the National Institutes of Health.
REFERENCES
- 1.Bates, P. Unpublished observations.
- 1a.Bos E C, Heijnen L, Luytjes W, Spaan W J. Mutational analysis of the murine coronavirus spike protein: effect on cell-to-cell fusion. Virology. 1995;214:453–463. doi: 10.1006/viro.1995.0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bos E C, Luytjes W, Spaan W J. The function of the spike protein of mouse hepatitis virus strain A59 can be studied on virus-like particles: cleavage is not required for infectivity. J Virol. 1997;71:9427–9433. doi: 10.1128/jvi.71.12.9427-9433.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bosch F X, Garten W, Klenk H D, Rott R. Proteolytic cleavage of influenza virus hemagglutinins: primary structure of the connecting peptide between HA1 and HA2 determines proteolytic cleavability and pathogenicity of avian influenza viruses. Virology. 1981;113:725–735. doi: 10.1016/0042-6822(81)90201-4. [DOI] [PubMed] [Google Scholar]
- 4.Bosch F X, Orlich M, Klenk H D, Rott R. The structure of the hemagglutinin, a determinant for the pathogenicity of influenza viruses. Virology. 1979;95:197–207. doi: 10.1016/0042-6822(79)90414-8. [DOI] [PubMed] [Google Scholar]
- 5.Bukreyev A, Volchkov V E, Blinov V M, Netesov S V. The GP-protein of Marburg virus contains the region similar to the ‘immunosuppressive domain’ of oncogenic retrovirus P15E proteins. FEBS Lett. 1993;323:183–187. doi: 10.1016/0014-5793(93)81476-g. [DOI] [PubMed] [Google Scholar]
- 6.Damico R L, Crane J, Bates P. Receptor-triggered membrane association of a model retroviral glycoprotein. Proc Natl Acad Sci USA. 1998;95:2580–2585. doi: 10.1073/pnas.95.5.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Decroly E, Vandenbranden M, Ruysschaert J M, Cogniaux J, Jacob G S, Howard S C, Marshall G, Kompelli A, Basak A, Jean F, et al. The convertases furin and PC1 can both cleave the human immunodeficiency virus (HIV)-1 envelope glycoprotein gp160 into gp120 (HIV-1 SU) and gp41 (HIV-I TM) J Biol Chem. 1994;269:12240–12247. [PubMed] [Google Scholar]
- 8.Dong J Y, Dubay J W, Perez L G, Hunter E. Mutations within the proteolytic cleavage site of the Rous sarcoma virus glycoprotein define a requirement for dibasic residues for intracellular cleavage. J Virol. 1992;66:865–874. doi: 10.1128/jvi.66.2.865-874.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elliott L H, Kiley M P, McCormick J B. Descriptive analysis of Ebola virus proteins. Virology. 1985;147:169–176. doi: 10.1016/0042-6822(85)90236-3. [DOI] [PubMed] [Google Scholar]
- 10.Freed E O, Myers D J, Risser R. Mutational analysis of the cleavage sequence of the human immunodeficiency virus type 1 envelope glycoprotein precursor gp160. J Virol. 1989;63:4670–4675. doi: 10.1128/jvi.63.11.4670-4675.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freed E O, Risser R. The role of envelope glycoprotein processing in murine leukemia virus infection. J Virol. 1987;61:2852–2856. doi: 10.1128/jvi.61.9.2852-2856.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallaher W R, Ball J M, Garry R F, Griffin M C, Montelaro R C. A general model for the transmembrane proteins of HIV and other retroviruses. AIDS Res Hum Retroviruses. 1989;5:431–440. doi: 10.1089/aid.1989.5.431. [DOI] [PubMed] [Google Scholar]
- 13.Gilbert J M, Bates P, Varmus H E, White J M. The receptor for the subgroup A avian leukosis-sarcoma viruses binds to subgroup A but not to subgroup C envelope glycoprotein. J Virol. 1994;68:5623–5628. doi: 10.1128/jvi.68.9.5623-5628.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert J M, Hernandez L D, Balliet J W, Bates P, White J M. Receptor-induced conformational changes in the subgroup A avian leukosis and sarcoma virus envelope glycoprotein. J Virol. 1995;69:7410–7415. doi: 10.1128/jvi.69.12.7410-7415.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gombold J L, Hingley S T, Weiss S R. Fusion-defective mutants of mouse hepatitis virus A59 contain a mutation in the spike protein cleavage signal. J Virol. 1993;67:4504–4512. doi: 10.1128/jvi.67.8.4504-4512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu M, Rappaport J, Leppla S H. Furin is important but not essential for the proteolytic maturation of gp160 of HIV-1. FEBS Lett. 1995;365:95–97. doi: 10.1016/0014-5793(95)00447-h. [DOI] [PubMed] [Google Scholar]
- 17.Guo H G, Veronese F M, Tschachler E, Pal R, Kalyanaraman V S, Gallo R C, Reitz M S., Jr Characterization of an HIV-1 point mutant blocked in envelope glycoprotein cleavage. Virology. 1990;174:217–224. doi: 10.1016/0042-6822(90)90070-8. [DOI] [PubMed] [Google Scholar]
- 18.Hingley S T, Leparc-Goffart I, Weiss S R. The spike protein of murine coronavirus mouse hepatitis virus strain A59 is not cleaved in primary glial cells and primary hepatocytes. J Virol. 1998;72:1606–1609. doi: 10.1128/jvi.72.2.1606-1609.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horimoto T, Nakayama K, Smeekens S P, Kawaoka Y. Proprotein-processing endoproteases PC6 and furin both activate hemagglutinin of virulent avian influenza viruses. J Virol. 1994;68:6074–6078. doi: 10.1128/jvi.68.9.6074-6078.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hosaka M, Nagahama M, Kim W S, Watanabe T, Hatsuzawa K, Ikemizu J, Murakami K, Nakayama K. Arg-X-Lys/Arg-Arg motif as a signal for precursor cleavage catalyzed by furin within the constitutive secretory pathway. J Biol Chem. 1991;266:12127–12130. [PubMed] [Google Scholar]
- 21.Hunter E, Hill E, Hardwick M, Bhown A, Schwartz D E, Tizard R. Complete sequence of the Rous sarcoma virus env gene: identification of structural and functional regions of its product. J Virol. 1983;46:920–936. doi: 10.1128/jvi.46.3.920-936.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inocencio N M, Sucic J F, Moehring J M, Spence M J, Moehring T J. Endoprotease activities other than furin and PACE4 with a role in processing of HIV-I gp160 glycoproteins in CHO-K1 cells. J Biol Chem. 1997;272:1344–1348. doi: 10.1074/jbc.272.2.1344. [DOI] [PubMed] [Google Scholar]
- 23.Kawaoka Y, Webster R G. Sequence requirements for cleavage activation of influenza virus hemagglutinin expressed in mammalian cells. Proc Natl Acad Sci USA. 1988;85:324–328. doi: 10.1073/pnas.85.2.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klenk H D, Rott R, Orlich M, Blodorn J. Activation of influenza A viruses by trypsin treatment. Virology. 1975;68:426–439. doi: 10.1016/0042-6822(75)90284-6. [DOI] [PubMed] [Google Scholar]
- 25.Lazarowitz S G, Choppin P W. Enhancement of the infectivity of influenza A and B viruses by proteolytic cleavage of the hemagglutinin polypeptide. Virology. 1975;68:440–454. doi: 10.1016/0042-6822(75)90285-8. [DOI] [PubMed] [Google Scholar]
- 26.Mayne J T, Rice C M, Strauss E G, Hunkapiller M W, Strauss J H. Biochemical studies of the maturation of the small Sindbis virus glycoprotein E3. Virology. 1984;134:338–357. doi: 10.1016/0042-6822(84)90302-7. [DOI] [PubMed] [Google Scholar]
- 27.McCune J M, Rabin L B, Feinberg M B, Lieberman M, Kosek J C, Reyes G R, Weissman I L. Endoproteolytic cleavage of gp160 is required for the activation of human immunodeficiency virus. Cell. 1988;53:55–67. doi: 10.1016/0092-8674(88)90487-4. [DOI] [PubMed] [Google Scholar]
- 28.Mebatsion T, Schnell M J, Cox J H, Finke S, Conzelmann K K. Highly stable expression of a foreign gene from rabies virus vectors. Proc Natl Acad Sci USA. 1996;93:7310–4731. doi: 10.1073/pnas.93.14.7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagai Y, Ogura H, Klenk H. Studies on the assembly of the envelope of Newcastle disease virus. Virology. 1976;69:523–538. doi: 10.1016/0042-6822(76)90482-7. [DOI] [PubMed] [Google Scholar]
- 30.Ohnishi Y, Shioda T, Nakayama K, Iwata S, Gotoh B, Hamaguchi M, Nagai Y. A furin-defective cell line is able to process correctly the gp160 of human immunodeficiency virus type 1. J Virol. 1994;68:4075–4079. doi: 10.1128/jvi.68.6.4075-4079.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ortmann D, Ohuchi M, Angliker H, Shaw E, Garten W, Klenk H D. Proteolytic cleavage of wild type and mutants of the F protein of human parainfluenza virus type 3 by two subtilisin-like endoproteases, furin and Kex2. J Virol. 1994;68:2772–2776. doi: 10.1128/jvi.68.4.2772-2776.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perez L G, Hunter E. Mutations within the proteolytic cleavage site of the Rous sarcoma virus glycoprotein that block processing to gp85 and gp37. J Virol. 1987;61:1609–1614. doi: 10.1128/jvi.61.5.1609-1614.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Presley J F, Polo J M, Johnston R E, Brown D T. Proteolytic processing of the Sindbis virus membrane protein precursor PE2 is nonessential for growth in vertebrate cells but is required for efficient growth in invertebrate cells. J Virol. 1991;65:1905–1909. doi: 10.1128/jvi.65.4.1905-1909.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Russell D L, Dalrymple J M, Johnston R E. Sindbis virus mutations which coordinately affect glycoprotein processing, penetration, and virulence in mice. J Virol. 1989;63:1619–1629. doi: 10.1128/jvi.63.4.1619-1629.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Samson A C, Fox C F. Precursor protein for Newcastle disease virus. J Virol. 1973;12:579–587. doi: 10.1128/jvi.12.3.579-587.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanchez A, Kiley M P, Holloway B P, Auperin D D. Sequence analysis of the Ebola virus genome: organization, genetic elements, and comparison with the genome of Marburg virus. Virus Res. 1993;29:215–240. doi: 10.1016/0168-1702(93)90063-s. [DOI] [PubMed] [Google Scholar]
- 37.Soneoka Y, Cannon P M, Ramsdale E E, Griffiths J C, Romano G, Kingsman S M, Kingsman A J. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 1995;23:628–633. doi: 10.1093/nar/23.4.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stadler K, Allison S L, Schalich J, Heinz F X. Proteolytic activation of tick-borne encephalitis virus by furin. J Virol. 1997;71:8475–8481. doi: 10.1128/jvi.71.11.8475-8481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steiner D F, Smeekens S P, Ohagi S, Chan S J. The new enzymology of precursor processing endoproteases. J Biol Chem. 1992;267:23435–23438. [PubMed] [Google Scholar]
- 40.Stieneke-Grober A, Vey M, Angliker H, Shaw E, Thomas G, Roberts C, Klenk H D, Garten W. Influenza virus hemagglutinin with multibasic cleavage site is activated by furin, a subtilisin-like endoprotease. EMBO J. 1992;11:2407–2414. doi: 10.1002/j.1460-2075.1992.tb05305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takada A, Robison C, Goto H, Sanchez A, Murti K G, Whitt M A, Kawaoka Y. A system for functional analysis of Ebola virus glycoprotein. Proc Natl Acad Sci USA. 1997;94:14764–14769. doi: 10.1073/pnas.94.26.14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takahashi S, Kasai K, Hatsuzawa K, Kitamura N, Misumi Y, Ikehara Y, Murakami K, Nakayama K. A mutation of furin causes the lack of precursor-processing activity in human colon carcinoma LoVo cells. Biochem Biophys Res Commun. 1993;195:1019–1026. doi: 10.1006/bbrc.1993.2146. [DOI] [PubMed] [Google Scholar]
- 43.Takahashi S, Nakagawa T, Kasai K, Banno T, Duguay S J, Van de Ven W J, Murakami K, Nakayama K. A second mutant allele of furin in the processing-incompetent cell line, LoVo. Evidence for involvement of the homo B domain in autocatalytic activation. J Biol Chem. 1995;270:26565–26569. doi: 10.1074/jbc.270.44.26565. [DOI] [PubMed] [Google Scholar]
- 44.Volchkov V E, Becker S, Volchkova V A, Ternovoj V A, Kotov A N, Netesov S V, Klenk H D. GP mRNA of Ebola virus is edited by the Ebola virus polymerase and by T7 and vaccinia virus polymerases. Virology. 1995;214:421–430. doi: 10.1006/viro.1995.0052. [DOI] [PubMed] [Google Scholar]
- 45.Volchkov V E, Blinov V M, Netesov S V. The envelope glycoprotein of Ebola virus contains an immunosuppressive-like domain similar to oncogenic retroviruses. FEBS Lett. 1992;305:181–184. doi: 10.1016/0014-5793(92)80662-z. [DOI] [PubMed] [Google Scholar]
- 46.Volchkov V E, Feldmann H, Volchkova V A, Klenk H D. Processing the Ebola virus glycoprotein by the proprotein convertase furin. Proc Natl Acad Sci USA. 1998;95:5762–5767. doi: 10.1073/pnas.95.10.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Webster R G, Rott R. Influenza virus A pathogenicity: the pivotal role of hemagglutinin. Cell. 1987;50:665–666. doi: 10.1016/0092-8674(87)90321-7. [DOI] [PubMed] [Google Scholar]
- 48.Wool-Lewis R J, Bates P. Characterization of Ebola virus entry by using pseudotyped viruses: identification of receptor-deficient cell lines. J Virol. 1998;72:3155–3160. doi: 10.1128/jvi.72.4.3155-3160.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]