Abstract
Retinoblastoma tumor suppressor protein (pRB) inhibition by tumor virus oncoproteins has been attributed to the need for these viruses to promote lytic viral nucleic acid synthesis by unscheduled entry into the S phase of the cell cycle. Kaposi’s sarcoma-associated herpesvirus (KSHV or HHV8) encodes a functional cyclin (vCYC) which is expressed during latency and can direct phosphorylation of pRB. We mapped the two major latent transcripts encoding vCYC, latent transcript 1 (LT1) and LT2, by cDNA sequencing, 5′ rapid amplification of cDNA ends, and primer extension analyses. Both LT1 and LT2 transcripts are spliced, originate from the same start site, and encode ORF K13 (vFLIP) as well as ORF72 (vCYC). The latency-associated nuclear antigen (LANA, ORF73) is encoded by LT1 but spliced from LT2. While differential expression of the two transcripts was not found, the promoter controlling LT1/LT2 transcription is regulated in a cell cycle-dependent manner. Activities of both KSHV LT1/LT2 and huCYC D1 luciferase promoter reporters transfected into NIH 3T3 cells increase 11- and 4-fold, respectively, after release from cell cycle arrest by serum starvation. Further, vCYC and huCYC D2 mRNA levels are low in naturally infected BCBL-1 cells arrested in late G1 with l-mimosine but increase in parallel during a 24-h period after release from cell cycle arrest. Cell cycle regulation of KSHV vCYC expression mimics cellular D cyclin regulation and may maintain infected cell cycling. This is consistent with an alternative hypothesis that tumor viruses have developed specific responses to innate cellular defenses against latent virus infection that include pRB-induced cell cycle arrest.
Kaposi’s sarcoma-associated herpesvirus (KSHV or human herpesvirus 8 [HHV8]) is a gammaherpesvirus (9, 30) etiologically linked to Kaposi’s sarcoma (KS), primary effusion lymphoma (PEL, also termed body cavity-based lymphoma [BCBL]), and a subset of multicentric Castleman’s disease (6, 9, 32, 42). Herpesviruses establish latent infections in their natural hosts characterized by persistence of the viral genome as a covalently closed, circular episome with limited viral gene expression (37). Most cells infected with KSHV demonstrate limited in situ viral gene expression consistent with virus latency (12, 33, 43). Tissue culture B-cell lines derived from PELs also have limited viral gene expression, although in some lines a minority population of cells can undergo spontaneous lytic replication (27, 35, 48). TPA (12-O-tetradecanoylphorbol-13-acetate) or sodium butyrate treatment of PEL cell lines substantially increases the proportion of KSHV-producing cells (27, 35).
Latently expressed viral genes have a particularly important role in maintaining the viral episome as well as in inducing immortalization of infected cells (23). KSHV transcription in PEL cell lines has been divided into three classes based on responsiveness to phorbol ester treatment (40). Class I transcripts are constitutively expressed and correspond to the presumed latency expression program, class II transcripts are expressed constitutively but are also induced by TPA treatment, and class III transcripts are only expressed after TPA induction corresponding to late lytic gene expression. A survey of KSHV gene transcription in BC-1 cells demonstrated that most KSHV genes are readily inducible by TPA treatment (class II or III transcripts). Two transcripts encoded on the right-hand end of the genome, however, show equal abundance in TPA-treated and untreated BC-1 cells, consistent with their designation as latent (class I) transcripts (40). Preliminary mapping of these transcripts indicated that the 6.0-kb transcript, designated latent transcript 1 (LT1), encodes ORF73, ORF72 (vCYC), and ORF K13 (vFLIP), while the 2.0-kb transcript, designated LT2, only encodes ORF72 and ORF K13 but not ORF73. Expression of ORF73-containing partial cDNA clones has demonstrated that this gene encodes the latency-associated nuclear antigen (LANA) (20, 33), an antigen target for several serological assays (14). In addition to the constitutively expressed latent transcripts LT1 and LT2, other class I KSHV transcripts in PEL cell lines include LT3 (40) and a transcript from the g block region of the KSHV genome that was not identified during the whole genome transcriptional mapping survey of Sarid et al. (9a, 40).
Characterization of LT1 and LT2 expression may provide important information on KSHV and its interaction with the host cell. vCYC, the protein product of the ORF72 gene, has been shown to be a functional cyclin capable of directing phosphorylation and inactivation of the retinoblastoma tumor suppressor protein (pRB) as well as phosphorylation of histone H1 (10, 15, 25). Mechanisms to inhibit pRB are a common feature of tumor viruses (for a review see reference 29). Expression of vCYC during latency suggests that KSHV has the capacity to independently regulate host cell transit through the pRB-controlled G1 checkpoint. Similarly, the vFLIP encoded by ORF K13 (sometimes referred to as ORF71) on the LT transcripts is likely to have anti-apoptotic activity that might affect survival of infected cells and contribute to the tumor phenotype (3). Thus, determining the expression patterns of the LT transcripts may delineate how KSHV affects cellular proliferation in tumors. Two considerations, however, are important for interpreting this data. First, posttranslational regulation is also likely to modulate expression of some latent proteins, such as vFLIP (32a). Second, KSHV shows evidence of tissue-specific transcriptional control such that results from studies of cultured cell lines may differ from tumors studied in situ. Cell culture studies, however, allow direct manipulation and experimentation of viral gene expression.
In this study, we examined the KSHV LT1 and LT2 transcripts by cDNA cloning, 5′ rapid amplification of cDNA ends (5′ RACE), and primer extension analyses. The promoter region for LT1 and LT2 (LP1/2) was examined with deletion reporter constructs to determine its transcriptional regulation. These studies also demonstrate that the LP1/2 promoter is activated in a cell cycle-dependent manner, which is one of the few examples of a viral gene regulated by the cell cycle. KSHV vCYC may supplement or substitute for downregulated cellular D cyclin activity to maintain cell cycle periodicity. Expression of the KSHV vCYC during latency and its regulation by the cell cycle is consistent with the virus attempting to reestablish cell cycle homeostasis in the setting of cellular activation of the G1 checkpoint to limit latent virus replication (29).
MATERIALS AND METHODS
Cells.
BC-1 (PEL-derived B-cell line coinfected with KSHV and Epstein-Barr virus [EBV]) (7), BCBL-1 (PEL-derived B-cell line infected with KSHV only) (35), P3HR1 (B-cell line infected with EBV), and BJAB (KSHV and EBV negative) were maintained at 37°C in RPMI 1640 medium (GibcoBRL, Gaithersburg, Md.) containing 2 mM l-glutamine and 10% fetal calf serum (FCS) (GibcoBRL) in the presence of 5% CO2. To induce lytic gene transcription, cells were exposed to 20 ng of TPA (Sigma Chemical Co., St. Louis, Mo.)/ml and harvested after 48 h (30). To inhibit virus DNA replication, phosphonoformic acid (PFA) (Sigma) was added at a concentration of 0.5 mM alone and in the presence of 20-ng/ml TPA for 48 h. NIH 3T3 and HeLa cell lines (American Type Culture Collection, Rockville, Md.) were maintained at 37°C in the presence of 5% CO2 in Dulbecco’s minimal essential medium (GibcoBRL) supplemented with 10% calf serum and FCS, respectively.
Cell cycle control of LT1/LT2 transcription.
To investigate whether the LP1/2 promoter is dependent on the cell cycle, the full-length pGL3.6 promoter was transfected into NIH 3T3 cells. These cells were then arrested in G0 by serum deprivation for 60 h. Following growth arrest, the cells were released by addition of medium containing 20% FCS for 14 h. Comparisons were made to similarly treated cells transfected with the human cyclin D1 promoter (cell cycle dependent) in pA3-luc, a kind gift of Richard G. Pestell (1), and a constitutively active cytomegalovirus (CMV) promoter (cell cycle independent) cloned into pGL3-luc. Cells were harvested 0 and 14 h after serum stimulation and split into fractions for luciferase and β-galactosidase assays and for fluorescence-activated cell sorting (FACS).
To directly assess the cell cycle dependence of KSHV gene expression, l-mimosine {β-[N-(3-hydroxy-4-pyridone)]-α-aminopropionic acid} (Calbiochem, La Jolla, Calif.) was used to arrest BCBL-1 cells in late G1 phase of the cell cycle. l-mimosine solution was made fresh in phosphate-buffered saline (PBS) for each experiment and added to FCS-containing medium. Cells were plated at approximately 3 × 106 cell/ml, and l-mimosine was added at 200 mM final concentration. After 20 h of incubation, cells were collected by centrifugation and prepared for flow cytometry and RNA extraction. Release from cell cycle arrest was achieved by resuspending cells in fresh medium without l-mimosine. Cells were harvested for flow cytometry and RNA extraction at 0, 3, 6, 10, and 24 h after release. Cell viability was evaluated by light microscopy after trypan blue staining.
RNA preparation and Northern analysis.
Total RNA from TPA-treated and untreated BC-1 cells was extracted by the RNAzol method (Tel-Test, Friendswood, Tex.). mRNA was selected by using the PolyATract mRNA isolation kit (Promega, Madison, Wis.). Northern blotting was performed on mRNA (300 ng per lane) by using a 1% formaldehyde-agarose gel and transfer onto nylon membranes (GeneScreen, NEN Research Products, Boston, Mass.). DNA probes for the LT1- and LT2-encoded genes ORF72, ORF73, and ORF K13 were synthesized by PCR and 32P-labeled by random priming (RediPrime DNA labeling system, Amersham, Buckinghamshire, England). The primers used were as follows: ORF72 (vCYC), sense, 5′-CTCCAATGGCAACTGCCAAT-3′; antisense, 5′-TTAATAGCTGTCCAGAATGCG-3′; ORF73 (LANA), sense, 5′-TATGCAGCAGGAGCAGGAGACGGTGGAA-3′; antisense, 5′-TGTCATTTCCTGTGGAGAGTCCCCA-3′; ORF K13 (vFLIP), sense, 5′-CAAGCCGTCGACATGGCCACT-3′; antisense, 5′-CAGCTTGTAAGCTTTGGTGTATGG-3′. A probe from the late lytic ORF25 gene encoding the major capsid protein (MCP) was used as a comparative control: ORF25 (MCP), sense, 5′-GGCGACATTCATCAACCTCAGG-3′; antisense 5′-ATATCATCCTGTGCGTTCACGAC-3′. Probes for human cyclins D1, D2, and D3 were a kind gift from I. Bernard Weinstein. Hybridization was performed at 42°C in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–50% formamide–5× Denhardt’s solution–2% sodium dodecyl sulfate (SDS), 10% dextran sulfate–100 mg of denatured sheared salmon sperm DNA/ml. β-Actin and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probes were used to standardize loading.
Construction and screening of BC-1 cell line cDNA library.
A cDNA phage library of BC-1 cell line was constructed in the ZAP ExpressTM vector (Stratagene, La Jolla, Calif.). Clones identified with the ORF72 (vCYC) probe were plaque purified; positive phages were converted into phagemids by employing the ExAssist helper phage (Stratagene), and inserts were sequenced by automated DNA sequencing (ABI 377 Sequenator; Perkin-Elmer, Foster City, Calif.) with specific internal primers.
5′ RACE.
5′ rapid amplification of cDNA ends (5′ RACE) was performed by using the MarathonTM cDNA amplification kit (Clontech, Palo Alto, Calif.). To overcome difficulties in reverse transcription because of high GC content and repeat regions, we synthesized the first strand cDNA by employing oligo(dT) primer and avian myeloblastosis virus reverse transcriptase (Promega) at 55°C. The RACE primers RP1 (nucleotide [nt] 123,598 to 123,691), 5′-AGAGCCTGGAGTTTCAGGGTGCTC-3′, and RP2 (nt 123830 to 123849), 5′-TCCCCAGGACCTTGGTTTTC-3′) and an adapter primer were used for specific cDNA PCR amplification.
Primer extension.
Primers PE1 (5′-ATAAGTCAGCCGGACCAAGC-3′), PE2 (5′-AAATGCAAGTGCGGAGCGGCGA-3′), and PE3 (5′-GACCTCAGGCGCATTCCCGG-3′) were end-labeled with [γ-32P]ATP and hybridized to 10 mg of RNA in 0.15 M KCl–10 mM Tris-HCl (pH 8.3)–1 mM EDTA at 65°C for 90 min, followed by equilibration to room temperature. Reverse transcription was performed with Superscript II RNase H reverse transcriptase (GibcoBRL). Reaction products were precipitated by the addition of 2.5 volumes of ethanol and resolved on a 6% polyacrylamide–7 M urea gel in Tris-borate-EDTA. Parallel DNA sequencing reactions were performed with Sequenase 2.0 (United States Biochemical Corporation, Cleveland, Ohio). pGEM template sequenced with the M13 forward primer was used as a size marker.
Plasmids.
Reporter gene plasmids were constructed by ligating LP1/2 fragments into the EcoRI and XhoI sites of pGL3-basic vector, which contains a promoterless luciferase gene downstream of the cloning site (Promega). The various LP1/2 fragments upstream from nt +30 relative to the start site were synthesized by PCR (30 PCR cycles of 94°C, 2 min; 60°C, 1 min; 72°C, 1 min; and final extension, 72°C, 7 min) with antisense primer (+9 to +30) 5′-AGCTGCCTCCAAATGATACACA-3′ and sense primers pGL3.1 (5′-CT GGGGCACCAATCAGAAAGTA-3′), pGL3.2 (5′-ACCACTGAACCGGTGCCAGCAA-3′), pGL3.3 (5′-TAGCCAGATGAACGCCACCCAA-3′), pGL3.4 (5′-CTTTTTTGCCAGGTAACGCTAA-3′), and pGL3.6 (5′-ACGGTCCTCCACGCAACTGTAA-3′) with BC-1 DNA as template.
DNA transfection and reporter assays.
DNA transfections into HeLa and NIH 3T3 cells were carried out with the calcium phosphate CellPhect Transfection kit (Pharmacia Biotech, Piscataway, N.J.). For each transfection into BJAB, 107 cells were pelleted, resuspended in 0.4 ml of 1640 RPMI, and mixed with luciferase reporter plasmid DNA. Cells were electroporated at 250 V and 960 mF with a Gene Pulser (Bio-Rad, Hercules, Calif.). Transfection efficiency was normalized by cotransfection of a reporter plasmid, pcDNA3.1/HisB/lacZ (Invitrogen, Carlsbad, Calif.). Cell lysates were prepared 48 h after transfection by using Reporter Lysis Buffer (Promega). In all cases, three or more separate transfections were performed, and results shown are the averages of the experiments. In the case of transcriptional activation studies with TPA, we could not use an internal standard for transfection variation by using a second reporter gene since TPA activates the normalization promoter. Instead, we used multiple repeats to compensate for variation in standard transfection conditions.
FACS analysis.
Cells (2 × 106) were fixed with 80% ethanol, washed, and incubated with 1 mg of RNase (Sigma)/ml and 500 mg of propidium iodide/ml in PBS containing 0.3% Nonidet P-40 for 30 min. Samples were analyzed for DNA content by standard methods with a FACstar Plus flow cytometer (Becton Dickinson, Franklin Lakes, N.J.). ModFit LT software (Becton Dickinson) was used for data analysis.
RESULTS
Northern blot analysis.
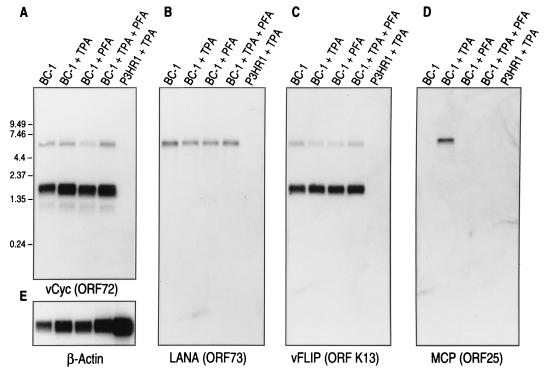
Probes internal to ORF72 (vCYC) (Fig. 1A) and ORF K13 (vFLIP) (Fig. 1C) detect 6.0- and 2.0-kb bands corresponding to LT1 and LT2 transcripts, respectively, as previously reported (40). In contrast, the ORF73 (LANA) probe only detects the 6.0-kb LT1 transcript. Prolonged exposure with the ORF72 probe also demonstrates the presence of a low-abundance transcript approximately 1 kb in size that does not hybridize with either the ORF K13 or ORF73 probe (Fig. 1A). We were unable to further characterize this small transcript due to its low abundance. As previously demonstrated (13, 40), the LT1 and LT2 transcripts are not induced by TPA treatment nor are they inhibited by treatment with the DNA polymerase inhibitor PFA (Fig. 1A to C). In comparison, expression of the 7.0-kb ORF25 transcript is both induced by TPA and inhibited by PFA treatment, which is consistent with its designation as a late lytic-phase gene (Fig. 1D).
FIG. 1.
Northern hybridization of BC-1 mRNA with ORF72 (A), ORF73 (B), ORF K13 (C), and ORF 25 (D) probes. Probe hybridizations for mRNA from uninduced BC-1 cells (lane 1), BC-1 cells treated with 20-ng/ml TPA for 48 h (lane 2), BC-1 cells treated with 0.5 mM PFA (lane 3), BC-1 cells treated with both 20-ng/ml TPA and 0.5 mM PFA (lane 4), and KSHV-negative, EBV-infected P3HR1 cells treated with 20-ng/ml TPA (lane 5) are shown. ORF72 (vCYC) (panel A) and ORF K13 (vFLIP) (panel C) probes hybridize to both the 6.0-kb LT1 and the 2.0-kb LT2 bands while only the 6.0-kb LT1 band hybridizes with the ORF73 probe (panel B). A low-abundance 1-kb band is detected with the ORF72 probe alone (panel A). LT1 and LT2 bands are not induced by TPA and are not inhibited by PFA treatment, consistent with their designation as class I or latent viral transcripts (40). P3HR1 mRNA does not cross-hybridize to any of the KSHV probes. (E) The same as the blot shown in panel A except it has been stripped and reprobed with a β-actin probe to control for equal loading.
Transcript mapping.
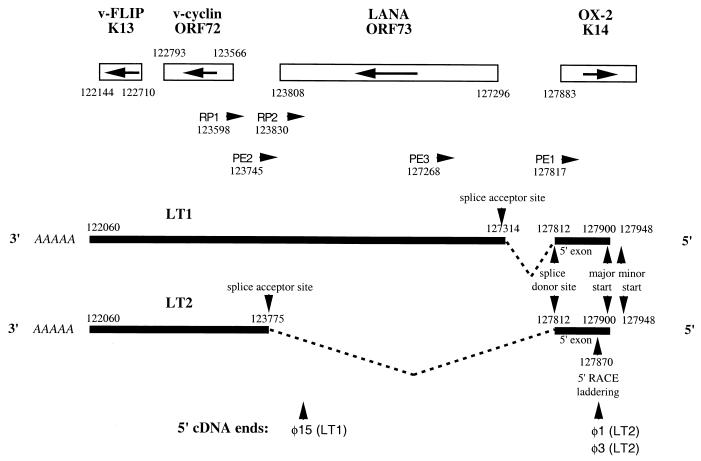
To characterize LT1 and LT2, we screened approximately 2 × 105 clones from a BC-1 cDNA library with a radiolabeled ORF72 PCR probe. Approximately 30 positive clones were identified, and 8 were isolated for plasmid excision and sequencing. cDNA inserts ranged from 1.4 to 1.8 kb, and each possessed a poly(A) tail with a conserved polyadenylation signal, AAUAAA (24 to 33 bp from the 3′ terminus), at a position corresponding to nt 122,093 in the BC-1 sequence (39). While most of the cDNA inserts were found to be prematurely 5′ truncated, three cDNAs provided information on transcript splicing patterns. Two cDNAs, φ1 and φ3, are full-length (or near full-length, see below) cDNAs of the LT2 transcript (Fig. 2). Both are spliced and bicistronic, encoding ORF K13, ORF72, and a short, noncoding 58-bp exon 5′ to the ORF73 gene. The intervening region (nt 123,775 to 127,812), including ORF73, between ORF72 and the 5′ exon, is spliced out from these two inserts at conserved splice donor (5′-ATAAACA∧GTGAGTA-3′) and acceptor (5′-TCCCTAG∧AAGCCAC-3′) sequences (4). Both φ1 and φ3 have the same 5′ ends corresponding to nt 127,870. A third cDNA clone, φ15, was the only cDNA from the eight cDNAs isolated which corresponded to the LT1 transcript (Fig. 2). It is a truncated cDNA which is colinear to genomic DNA throughout its length (i.e., it does not possess the splice site present in LT2) and contains ORF K13 and ORF72 as well as a portion of the carboxy-terminal coding sequence of ORF73, indicating that all three genes are expressed on the LT1 transcript.
FIG. 2.
Depiction of the transcript mapping results for LT1 and LT2 based on cDNA sequencing, 5′ RACE, and primer extension studies. The arrangement and orientation of genes for this region of the KSHV BC-1 genome are shown with nucleotide positions designated by Russo et al. (39). Black bars represent the LT1 and LT2 transcripts, introns are represented as dashed lines, and nucleotide positions for polyadenylation sites, splice junctions, and start sites are indicated. The positions of 5′ truncation for cDNA phages φ1, φ3, and φ15 are shown. Primer positions and orientations used for 5′ RACE (RP1, RP2, and RP3) and primer extension (PE1, PE2, and PE3) analyses are shown.
5′ RACE was performed on cDNA reverse-transcribed poly(A)-selected BC-1 RNA to further delineate splicing patterns for LT1 and LT2. Two primers were designed to preferentially amplify either the LT1 or LT2 cDNA. The 5′ RACE primer 1 (RP1) (Fig. 2), located immediately 3′ to the splice acceptor site, at nt 123,775, exclusively amplified a 235-bp PCR product corresponding to the LT2 transcript (data not shown). The RP1 PCR product had the same 5′ end (nt 127,870) and splice junction (nt 123,775 to 127,812) as that found in the cDNA φ1 and φ3 LT2 inserts. This product is consistent with preferential amplification of only the spliced LT2 cDNA by the RP1 primer. The RP2 primer, located immediately 5′ to the splice acceptor site at nt 123,775 (within the LT2 intron), amplified a 3,542-bp product corresponding to the LT1 transcript. The sequence of the RP2 5′ RACE product demonstrated that LT1 also has a spliced intron sequence that uses the same splice donor site as LT2 (nt 127,812) but the splice acceptor site is at nt 127,314, corresponding to a second conserved splice acceptor sequence (5′-TTGTCAG∧ACCAGAT-3′). The RP2 5′ RACE product also had the same 5′ end at nt 127,870 as the RP1 product.
To confirm the authenticity of the transcription start sites determined by 5′ RACE and cDNA sequencing, we performed primer extension analyses. Primer extension primer 1 (PE1) was designed to anneal to a 3′ site (nt 127,817) located 54 bp 3′ to the putative transcriptional start site determined by 5′ RACE at nt 127,870. The PE1 primer (Fig. 2) synthesized two oligonucleotide products (Fig. 3), which extends the presumed start site(s) beyond the site determined by 5′ RACE and cDNA sequencing. The major transcriptional start site for both LT1 and LT2 transcripts is a unique adenine at nt 127,900 located 30 bp beyond the start site determined by cDNA sequencing and 5′ RACE. A second minor start site is present at nt 127,948, which is usually less intense than the nt 127,900 product on repeated analysis. A laddering pattern (Fig. 3) is present 47 to 54 bp from the PE1 primer which corresponds to the nt 127,870 “start” site found by cDNA and 5′ RACE analyses. This is consistent with premature termination of the reverse transcriptase reaction and is the likely explanation for the shorter products found by these methods. The primer extension results for PE1 shown in Fig. 3 are atypical in that the minor start site at nt 127,948 (Fig. 3, 132 bp upstream of the PE1 primer) is more intense than the major start site at nt 127,900 (Fig. 3, 84 bp upstream of the PE1 primer) but the figure is shown to highlight the laddering pattern occurring at nt 127,870 (Fig. 3, 47 to 54 bp upstream of the PE1 primer). These start sites were confirmed for the LT1 transcript by using a primer (PE3) located within the LT2 intron at nt 127,268 and for the LT2 transcript by using a primer (PE2) located 3′ to the splice acceptor site at nt 123,745. For both transcripts, transcription was predominantly initiated at nt 127,900.
FIG. 3.

Primer extension results obtained with the PE1 primer. Two start sites are present at 84 bp (nt 127,900) and 132 bp (nt 127,948) from the PE1 primer. A laddering pattern consistent with premature reverse transcription termination is present at 47 to 54 bp from the PE1 primer (nt 127,863 to 127,870), corresponding to the start sites determined by cDNA sequencing and 5′ RACE analyses.
Taken together, the cDNA sequencing, 5′ RACE, and primer extension analyses indicate that LT1 and LT2 are overlapping, alternatively spliced, and polycistronic transcripts (Fig. 2). The LT1 transcript (i) corresponds to the 6.0-kb mRNA, (ii) encodes ORF K13, ORF72, and ORF73, (iii) has a major transcriptional start site at nt 127,900 and a minor transcriptional start site at nt 127,948, and (iv) uses a splice junction with the splice donor site at nt 127,812 (5′-ATAAACA∧GTGAGTA-3′) and a splice acceptor site at nt 127,314 (5′-TTGTCAG∧ACCAGAT-3′). In contrast, the LT2 transcript (i) corresponds to the 2.0-kb mRNA, (ii) encodes ORF K13 and ORF72 but not ORF73, (iii) has an identical 3′ end to LT1 and shares the same major 5′ start site, (iv) uses the same splice donor site as LT1 at nt 127,812 but has a splice acceptor sequence at nt 123,775 (5′-TCCCTAG∧AAGCCAC-3′), which splices the ORF73 gene out of the LT2 transcript.
Analysis of the promoter sequence.
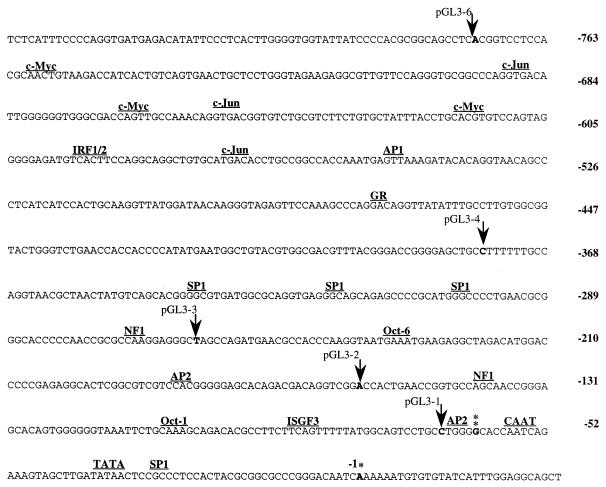
An 842-bp sequence encompassing the LP region for LT1 and LT2 (LP1/2) was examined for potential regulatory sites (Fig. 4). LP1/2 possesses a noncanonical TATA box 34 bp upstream of the nt 127,900 mRNA transcription start site and a CAAT element 15 bp upstream of the putative TATA box. Additionally, two conserved initiator (Inr) elements, TATCATTT (+13 to +20 bp) and CTCCACTA (−20 to −28 bp), flank the major transcription start site. Inr elements are present in TATA-containing promoters as well as those that lack a confirmed TATA box (38). Inr elements bind the transcription factor TFIID and can replace a TATA box as a site to initiate RNA polymerase II-dependent transcription. In this case, transcriptional initiation is less precisely positioned and occurs at a cluster of start points (5, 26).
FIG. 4.
Nucleotide sequence of the latent promoter region (LP1/2) for LT1-2. The first nucleotide of the presumed major transcription start site at nt 127,900 is marked by a single asterisk and is indicated in boldface. The second start site is marked by a double asterisk, and potential transcription factor binding sites in the promoter region are indicated. The locations of promoter deletions used for reporter studies indicated by arrows.
A search of the LP1/2 region for potential transcription factor binding sites identified an SP1 binding site located 3′ of the TATA box and immediately 5′ to one of the Inr sites. Several more conserved SP1 binding sites are clustered around bp −350. Conserved Oct-1 and Oct-6 binding sites were also found (Fig. 4). Oct-1 is a ubiquitous transcription factor that recognizes an octamer sequence also recognized by Oct-2, a lymphoid-specific transcription factor. Two conserved interferon regulatory factor (IRF1/2) binding sites and a semiconserved interferon-stimulated response element (ISGF3) are present, which suggests the promoter may be regulated by interferon signal transduction pathways as has been shown for the EBV EBNA-1 Qp promoter (31, 46). Conserved c-Jun, c-myc, and NF1 binding sites are also present in this region. One AP-1 conserved binding site is present, although LT1-2 transcription has not been found to be responsive to TPA (13, 14, 40).
LP1/2-luciferase reporter activity.
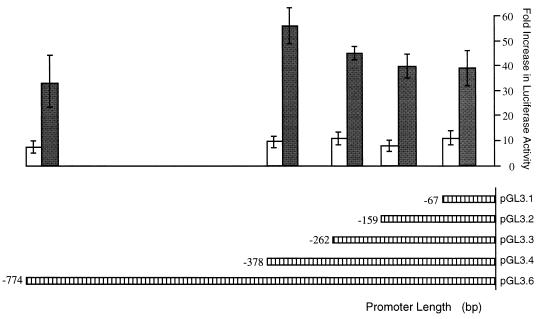
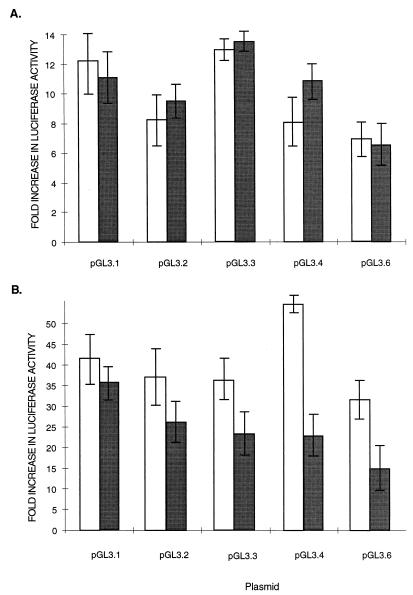
To confirm that the 5′ flanking region functions as a promoter, we assessed its ability to drive expression of a reporter gene in transiently transfected cells. Promoter-reporter gene recombinants were constructed in which various lengths of the LP1/2 promoter were cloned in front of a luciferase reporter gene (pGL3.1-6) (Fig. 4). The sites of fusion were in the 5′ untranslated leader sequence of LT1-2 corresponding to nt 127,870. Results from three independent experiments are shown in Fig. 5. When transfected into human epithelial HeLa cells, a construct containing sequences −774 to +30 (pGL3.6) demonstrated 10-fold greater luciferase activity over that of the basal activity of the pGL3-basic luciferase construct. Truncation of promoter length from the 5′ end had little effect on promoter activity, and a minimal promoter containing 67 bp of the promoter sequence was able to initiate reporter transcription efficiently. Similar results were found when reporter plasmids were transfected into the EBV-negative B-cell line BJAB; however, luciferase activity was much higher for each promoter construct. In contrast to the experiments with HeLa cells, luciferase activity in the BJAB cell line was 32-fold higher for pGL3.6 (−774 to +30) and 58-fold higher for pGL3.4 (−378 to +30) than for pGL3-basic. The higher transcriptional activity of the reporter in BJAB compared to HeLa cells may represent lymphoid-specific transcription, while the decreased activity of pGL3.6 compared to that of pGL3.4 in BJAB cells suggests a possible lymphoid-specific repressor element in this region.
FIG. 5.
Luciferase expression levels for LP1/2-luciferase reporter constructs in HeLa (white bars) and BJAB (grey bars) cells. HeLa cells were transfected with 1 mg of reporter gene and 1 mg of control plasmid pcDNA3.1/lacZ. BJAB cells were transfected with 10 mg of reporter gene and 10 μg of control plasmid pcDNA3.1/lacZ. Mean promoter activities obtained from three transfections after normalization for β-galactosidase expression are indicated relative to the pGL3-luc basic reporter plasmid lacking a promoter sequence.
Although the presence of a conserved AP-1 (−550) suggests responsiveness to TPA, we found no evidence for enhanced reporter activity for any of the five LP1/2 luciferase constructs in HeLa (Fig. 6A) and BJAB (Fig. 6B) cells treated with 20-ng/ml TPA. TPA treatment of BJAB cells resulted in minimal inhibition of reporter activity for the pGL3.4 and pGL3.6 constructs but this may have been due to cellular toxicity rather than a direct effect of TPA-induced signaling on the AP1 promoter site. Similarly, assays with LP1/2 luciferase-reporter constructs showed no response to interferon treatment or transfection with either human IRF-1 or KSHV vIRF expression plasmids in either HeLa and BJAB cells despite the presence of potential interferon responsive elements in the promoter (not shown).
FIG. 6.
Effect of TPA treatment on LP1/2-luciferase promoter reporter constructs. HeLa (A) and BJAB (B) cells cultured in the presence (grey bars) or the absence (white bars) of 20-ng/ml TPA were transfected with LP1/2-luciferase constructs as described in the legend for Fig. 4 and assayed for luciferase activity. No evidence for TPA inducibility was found for either cell type.
Cell cycle control of LT1-2 transcription.
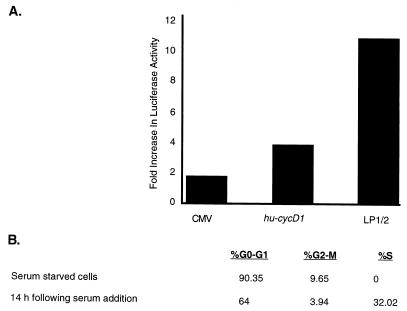
To examine cell cycle regulation of the LP1/2 promoter, the pGL3.6 luciferase reporter activity was compared to those of a cellular cyclin D1 (huCYC D1) promoter and a constitutively active CMV promoter in NIH 3T3 cells. Promoter-reporter constructs were transfected into NIH 3T3 cells which were then serum arrested in 0.1% FCS for 60 h. Release from serum starvation by serum stimulation for 14 h resulted in a 4-fold induction for the human cyclin D1 promoter and an 11-fold induction of the KSHV LP1/2 promoter compared to only a 1.7-fold induction for the CMV promoter (Fig. 7A). Parallel flow cytometry determinations (Fig. 7B) demonstrate that serum starvation for 60 h effectively arrests mitogenesis of NIH 3T3 cells (0% in S phase) and addition of serum for 14 h initiates cell cycle progression (32% in S phase).
FIG. 7.
(A) Cell cycle-dependent induction of the pGL3.6 LP1/2-luciferase reporter. NIH 3T3 cells were transfected with the indicated constructs and cultured in low serum (0.1% FCS) for 60 h. Bars indicate the levels of induction of luciferase activity 14 h after readdition of 20% FCS compared to pretreatment luciferase activity for the constitutively active CMV, the human cyclin D1, and the KSHV LP1/2 promoters. (B) Cell cycle distributions as determined by FACS analyses at 0 and 14 h after serum readdition. Serum starvation for 60 h arrested 90% of NIH 3T3 cells in G0/G1, whereas 32% of cells were entering S phase 14 h after serum stimulation.
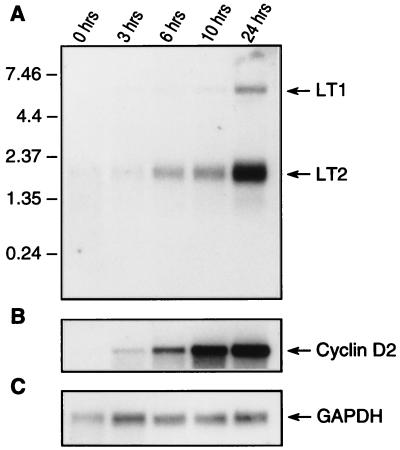
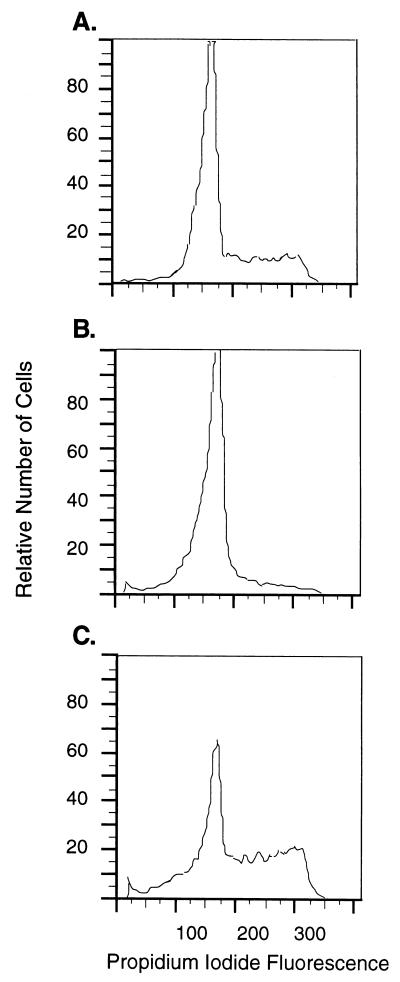
Direct confirmation of KSHV LP1/2 dependence on the cell cycle was performed by Northern analysis of LT1 and LT2 expression in BCBL-1 cells (Fig. 8). Unlike NIH 3T3 cells, PEL cells are resistant to arrest by serum starvation and continue to proliferate in the absence of serum but undergo marked loss of viability, an effect similar to that seen for EBV-immortalized cells (11). Growth arrest of PEL cells, however, was achieved by 200 mM l-mimosine treatment for 20 h, which arrests cells in late G1 phase (16). l-mimosine-induced arrest is reversible by washing BCBL-1 cells with fresh medium (Fig. 9). For comparison, expression of the human cyclin D2 gene was examined because preliminary Northern analyses showed that only cyclin D2, and not cyclin D1 or D3, was appreciably expressed in BCBL-1 cell lines. KSHV LT1 and LT2 (Fig. 8A) and human cyclin D2 (Fig. 8B) expression was nearly absent in l-mimosine-arrested cells at time zero (87% G0/G1) (Fig. 9B). Expression of these transcripts progressively increased in parallel after release from l-mimosine arrest. l-mimosine arrest had no effect on GAPDH transcription used to confirm RNA loading (Fig. 8C).
FIG. 8.
Time course of KSHV vCYC (A) and cellular huCYC D2 (B) mRNA expression in BCBL-1 cells after release from l-mimosine G1 arrest. GAPDH mRNA expression (C) was used as a control for equal mRNA loading. Cells were arrested for 20 h by treatment with 200 mM l-mimosine and released from cell cycle arrest by washing in fresh medium. Cell cultures were harvested at 0, 3, 6, 10, and 24 h after l-mimosine washout and prepared for mRNA extraction. Expression of KSHV LT1 and LT2 mRNA mirrors expression of cellular CYC D2, which is expressed early in G1 phase of the cell cycle.
FIG. 9.
FACS analysis demonstrating that BCBL-1 cells are reversibly arrested by l-mimosine treatment. (A) Cell cycle distribution of exponentially growing BCBL-1 prior to l-mimosine arrest. (B) G1 arrest of BCBL-1 cells at 0 h as shown in Fig. 8, after treatment with 200 nM l-mimosine for 20 h. (C) Cell cycle progression of BCBL-1 cells 24 h after l-mimosine washout, as shown in Fig. 8.
DISCUSSION
Like other herpesviruses, KSHV gene transcription is limited during presumed virus latency (40, 48). KSHV latent (or class I) gene expression has been functionally defined as being constitutive in PEL tissue culture cells (40) in that it is not induced by phorbol esters nor inhibited by DNA polymerase inhibitors. In contrast, class III genes such as ORF25, which encodes the MCP, are expressed in a manner consistent with late lytic cycle genes in that they are not expressed without TPA treatment in some highly restricted cell lines such as BC-1 (40).
In this study we defined the nucleotide sequences and gene organization of two KSHV latent transcripts, LT1 and LT2, that were found during a survey for KSHV gene transcription (40). The constitutive transcription of LT1-2 transcripts is consistent with transcriptional studies of vCYC mRNA performed with BC-1 and BC-3 cell lines (13) and for LANA protein expression with BC-1 cells (14). Both LT1 and LT2 are spliced transcripts which have the same transcriptional start site and a splice junction beginning after a short untranslated 5′ leader sequence. The LT1 intron is a short noncoding region, whereas the LT2 intron splices out the entire ORF73 gene. Our primer extension analysis suggests that an additional transcription start site is present; however, we found no evidence for differential regulation of LT1 and LT2, and a more comprehensive examination of study of transcriptional factor activation may reveal important differences in expression of these transcripts. The reasons why the KSHV ORF73 gene encoding LANA is spliced from LT2 and why the virus possesses these two overlapping transcripts can only remain speculative since little is currently known about the functions of the LANA protein.
The promoter region for these transcripts, LP1/2, was transcriptionally active in both epithelial and lymphoid cell lines by using a luciferase reporter construct. The higher activity in BJAB cells suggests transcription might be specifically enhanced in lymphoid cell lines. Although a 67-bp minimal LP1/2 promoter was sufficient to drive efficient transcription of the reporter gene, the putative promoter region contains a number of potential transcriptional factor binding sites that might regulate transcription. While examination of promoter-reporter constructs in tissue culture provides important information on LT1-2 transcriptional regulation, an important caveat is that cell type-specific transcriptional programs exist for KSHV (28, 34). Direct examination of tissue-specific transcription and translation is necessary to extend our results directly to tumors in situ.
At least two nuclear proteins, LANA (ORF73) and vCYC (ORF72) are expressed during viral latency (12, 30). LANA possesses a long acidic repeat domain which is reminiscent of a similar domain in the latent EBV EBNA-1 protein that inhibits cytotoxic T-lymphocyte recognition of this antigen (24). At present it is unclear whether LANA functions like EBNA-1 in maintaining viral episomal replication or has other functions during viral latency. No functional data are available to determine if ORF73 plays a role in cell cycle regulation or transformation; however, NIH 3T3 cells stably expressing ORF73 alone are not transformed (40a). KSHV vCYC has a high degree of sequence homology with cellular D-type cyclins (8) and is functionally similar to these cell cycle control proteins (10). vCYC can overcome the G1-S block mediated by pRb in SAOS-2 cells (10) and induce pRb and histone H1 phosphorylation via CDK6 activation (15, 25). Further, the viral cyclin, unlike cellular D-type cyclins, is resistant to p16, p21, and p27 CDK inhibitors (44). A third protein encoded by these latent transcripts, vFLIP (ORF K13), is a dominant negative inhibitor of Fas/APO1-activated apoptosis. Preliminary studies suggest that this protein may be under posttranscriptional regulation and is not expressed during latency in PEL cells despite active transcription of the gene on LT1 and LT2 (32a).
Cellular D-type cyclin expression is required for progression through the G1 cell cycle checkpoint controlled by the pRB tumor suppressor protein. This checkpoint is therefore a critical target for proliferative signals in G1. Several tumor viruses, such as simian virus 40, human papillomavirus 16, and adenovirus, have evolved mechanisms to subvert the host cell cycle through direct binding and inactivation of the pRB tumor suppressor protein (18). Other tumor viruses, including EBV and human T-cell lymphotropic virus type 1, have evolved mechanisms to indirectly inactivate pRB (29). The EBV latent immortalizing genes EBNA-LP and EBNA-2, for example, induce cyclin D2 expression (17, 21, 41). Although EBNA-LP protein levels are stable throughout the cell cycle, this protein is phosphorylated in a cell cycle stage-specific manner, suggesting possible functional regulation (22). KSHV also indirectly inactivates pRB through pRB phosphorylation in conjunction with cyclin-dependent kinase 6 (CDK 6). This is an intriguing correlation since many of the cellular genes induced by EBV are also encoded by the KSHV genome (28, 29). In vitro overexpression of cyclin D induces apoptosis (36), presumably through deregulated overexpression of E2F-1 and induction of p14ARF (2). Similarly, overexpression of vCYC in NIH 3T3 cells induces cell death (40b).
The convergent evolution of pRB inhibitory mechanisms by tumor viruses illustrates the importance of the pRB checkpoint to the life cycles of both DNA and RNA tumor viruses. pRB inhibition has been attributed to the need for a virus to induce S-phase DNA synthesis in order to expand virion production (18). This is likely to be the case for at least some tumor viruses during lytic virus replication. EBV, for example, encodes lytic replication transactivator proteins (BRLF1 and BZLF1) which directly inhibit pRB and p53 proteins (45, 47). However, pRB inactivation during virus latency does not preferentially replicate viral genome over host cell genome. An alternative explanation for pRB inhibition during virus latency is that cell cycle arrest serves as an antiviral mechanism to limit latent virus replication (29). Our data is consistent with this latter hypothesis in that the KSHV vCYC is expressed during virus latency. Rather than being constitutively overexpressed, vCYC transcription appears to be closely regulated by cell cycle-specific signaling events. Swanton and colleagues have demonstrated that the KSHV vCYC is resistant to cellular CDK inhibitors that control D cyclin activities (44). This suggests that KSHV replaces cellular D cyclin activity with a viral cyclin that is resistant to cellular control mechanisms. In this case, vCYC expression may reestablish cell cycle homeostasis in the presence of active cellular antiviral responses that would otherwise induce cell cycle arrest.
ACKNOWLEDGMENTS
This work was supported by grants CA67391 and CA73564 from the National Institutes of Health.
ADDENDUM
After submitting this article we learned of the publication by Dittmer et al. (12a) of transcriptional mapping results for the KSHV LT1 and LT2 transcripts which are similar to our own results reported here.
REFERENCES
- 1.Albanese C, Johnson J, Watanabe G, Eklund N, Vu D, Arnold A, Pestell R G. Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J Biol Chem. 1995;270:23589–23597. doi: 10.1074/jbc.270.40.23589. [DOI] [PubMed] [Google Scholar]
- 2.Bates S, Phillips A C, Clark P A, Stott F, Peters G, Ludwig R L, Vousden K H. p14ARF links the tumour suppressors RB and p53. Nature. 1998;395:124–125. doi: 10.1038/25867. [DOI] [PubMed] [Google Scholar]
- 3.Bertin J, Armstrong R C, Ottilie S, Martin D A, Wang Y, Banks S, Wang G H, Senkevich T G, Alnemri E S, Moss B, Lenardo M J, Tomaselli K J, Cohen J I. Death effector domain-containing herpesvirus and poxvirus proteins inhibit both Fas- and TNFR1-induced apoptosis. Proc Natl Acad Sci USA. 1997;94:1172–1176. doi: 10.1073/pnas.94.4.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bodescot M, Perricaudet M. Epstein-Barr virus mRNAs produced by alternative splicing. Nucleic Acids Res. 1986;14:7103–7114. doi: 10.1093/nar/14.17.7103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bulfone-Paus S, Dempsey L A, Maizels N. Host factors LR1 and Sp1 regulate the Fp promoter of Epstein-Barr virus. Proc Natl Acad Sci USA. 1995;92:8293–8297. doi: 10.1073/pnas.92.18.8293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences are present in AIDS-related body cavity based lymphomas. New Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 7.Cesarman E, Moore P S, Rao P H, Inghirami G, Knowles D M, Chang Y. In vitro establishment and characterization of two AIDS-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi’s sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood. 1995;86:2708–2714. [PubMed] [Google Scholar]
- 8.Cesarman E, Nador R G, Bai F, Bohenzky R A, Russo J J, Moore P S, Chang Y, Knowles D M. Kaposi’s sarcoma-associated herpesvirus contains G protein-coupled receptor and cyclin D homologs which are expressed in Kaposi’s sarcoma and malignant lymphoma. J Virol. 1996;70:8218–8223. doi: 10.1128/jvi.70.11.8218-8223.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;265:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 9a.Chang, Y. Unpublished observations.
- 10.Chang Y, Moore P S, Talbot S J, Boshoff C H, Zarkowska T, Godden-Kent D, Paterson H, Weiss R A, Mittnacht S. Cyclin encoded by KS herpesvirus. Nature. 1996;382:410. doi: 10.1038/382410a0. [DOI] [PubMed] [Google Scholar]
- 11.Cherney B W, Bhatia K, Tosato G. A role for deregulated c-Myc expression in apoptosis of Epstein-Barr virus-immortalized B cells. Proc Natl Acad Sci USA. 1994;91:12967–12971. doi: 10.1073/pnas.91.26.12967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis M A, Sturzl M A, Blasig C, Schreier A, Guo H G, Reitz M, Opalenik S R, Browning P J. Expression of human herpesvirus 8-encoded cyclin D in Kaposi’s sarcoma spindle cells. J Natl Cancer Inst. 1997;89:1868–1874. doi: 10.1093/jnci/89.24.1868. [DOI] [PubMed] [Google Scholar]
- 12a.Dittmer D, Lagunoff M, Renne R, Staskus K, Haase A, Ganem D. A cluster of latently expressed genes in Kaposi’s sarcoma-associated herpesvirus. J Virol. 1998;72:8309–8315. doi: 10.1128/jvi.72.10.8309-8315.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flore O, Gao S J. Effect of DNA synthesis inhibitors on Kaposi’s sarcoma-associated herpesvirus cyclin and major capsid protein gene expression. AIDS Res Hum Retroviruses. 1997;13:1229–1233. doi: 10.1089/aid.1997.13.1229. [DOI] [PubMed] [Google Scholar]
- 14.Gao S-J, Kingsley L, Hoover D R, Spira T J, Rinaldo C R, Saah A, Phair J, Detels R, Parry P, Chang Y, Moore P S. Seroconversion to antibodies against Kaposi’s sarcoma-associated herpesvirus-related latent nuclear antigens before the development of Kaposi’s sarcoma. New Engl J Med. 1996;335:233–241. doi: 10.1056/NEJM199607253350403. [DOI] [PubMed] [Google Scholar]
- 15.Godden-Kent D, Talbot S J, Boshoff C, Chang Y, Moore P S, Weiss R A, Mittnacht S. The cyclin encoded by Kaposi’s sarcoma associated herpesvirus (KSHV) stimulates cdk6 to phosphorylate the retinoblastoma protein and histone H1. J Virol. 1997;71:4193–4198. doi: 10.1128/jvi.71.6.4193-4198.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffman B D, Hanauske-Abel H M, Flint A, Lalande M. A new class of reversible cell cycle inhibitors. Cytometry. 1991;12:26–32. doi: 10.1002/cyto.990120105. [DOI] [PubMed] [Google Scholar]
- 17.Hollyoake M, Stuhler A, Farrell P, Gordon J, Sinclair A. The normal cell cycle activation program is exploited during the infection of quiescent B lymphocytes by Epstein-Barr virus. Cancer Res. 1995;55:4784–4787. [PubMed] [Google Scholar]
- 18.Jansen-Dürr P. How viral oncogenes make the cell cycle. Trends Genet. 1996;12:270–275. doi: 10.1016/0168-9525(96)81455-7. [DOI] [PubMed] [Google Scholar]
- 19.Jung J U, Stager M, Desrosiers R C. Virus-encoded cyclin. Mol Cell Biol. 1994;14:7235–7244. doi: 10.1128/mcb.14.11.7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kedes D H, Lagunoff M, Renne R, Ganem D. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi’s sarcoma-associated herpesvirus. J Clin Investig. 1997;100:2606–2610. doi: 10.1172/JCI119804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kempkes B, Spitkovsky D, Jansen-Dürr P, Ellwart J W, Kremmer E, Rottenberger C, Bornkamm G W, Hammerschmidt W. B-cell proliferation and induction of early G1-regulating proteins by Epstein-Barr virus mutants conditional for EBNA2. EMBO J. 1995;14:88–96. doi: 10.1002/j.1460-2075.1995.tb06978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kitay M K, Rowe D T. Cell cycle stage-specific phosphorylation of the Epstein-Barr virus immortalization protein EBNA-LP. J Virol. 1996;70:7885–7893. doi: 10.1128/jvi.70.11.7885-7893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klein G. Epstein-Barr virus strategy in normal and neoplastic B cells. Cell. 1994;77:791–793. doi: 10.1016/0092-8674(94)90125-2. [DOI] [PubMed] [Google Scholar]
- 24.Levitskaya J, Coram M, Levitsky V, Imreh S, Stigerwald-Mullen P M, Klein G, Kurilla M G, Masucci M G. Inhibition of antigen processing by the internal repeat region of the Epstein-Barr virus nuclear antigen-1. Nature. 1995;375:685–688. doi: 10.1038/375685a0. [DOI] [PubMed] [Google Scholar]
- 25.Li M, Lee H, Yoon D W, Albrecht J C, Fleckenstein B, Neipel F, Jung J U. Kaposi’s sarcoma-associated herpesvirus encodes a functional cyclin. J Virol. 1997;71:1984–1991. doi: 10.1128/jvi.71.3.1984-1991.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manzano-Winkler B, Novina C D, Roy A L. TFII is required for transcription of the naturally TATA-less but initiator-containing Vbeta promoter. J Biol Chem. 1996;271:12076–12081. doi: 10.1074/jbc.271.20.12076. [DOI] [PubMed] [Google Scholar]
- 27.Miller G, Heston L, Grogan E, Gradoville L, Rigsby M, Sun R, Shedd D, Kushnaryov V M, Grossberg S, Chang Y. Selective switch between latency and lytic replication of Kaposi’s sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J Virol. 1997;71:314–324. doi: 10.1128/jvi.71.1.314-324.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore P S, Boshoff C, Weiss R A, Chang Y. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science. 1996;274:1739–1744. doi: 10.1126/science.274.5293.1739. [DOI] [PubMed] [Google Scholar]
- 29.Moore P S, Chang Y. Antiviral activity of tumor-suppressor pathways: clues from molecular piracy by KSHV. Trends Genet. 1998;14:144–150. doi: 10.1016/s0168-9525(98)01408-5. [DOI] [PubMed] [Google Scholar]
- 30.Moore P S, Gao S J, Dominguez G, Cesarman E, Lungu O, Knowles D M, Garber R, Pellett P E, McGeoch D J, Chang Y. Primary characterization of a herpesvirus agent associated with Kaposi’s sarcoma. J Virol. 1996;70:549–558. doi: 10.1128/jvi.70.1.549-558.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nonkwelo C, Ruf I K, Sample J. Interferon-independent and -induced regulation of Epstein-Barr virus EBNA-1 gene transcription in Burkitt lymphoma. J Virol. 1997;71:6887–6897. doi: 10.1128/jvi.71.9.6887-6897.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parravicini C, Corbellino M, Paulli M, Magrini U, Lazzarino M, Moore P S, Chang Y. Expression of a virus-derived cytokine, KSHV vIL-6, in HIV seronegative Castleman’s disease. Am J Pathol. 1997;151:1517–1521. [PMC free article] [PubMed] [Google Scholar]
- 32a.Parravicini, C., and Y. Chang. Unpublished observations.
- 33.Rainbow L, Platt G M, Simpson G R, Sarid R, Gao S J, Stoiber H, Herrington C S, Moore P S, Schulz T F. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J Virol. 1997;71:5915–5921. doi: 10.1128/jvi.71.8.5915-5921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Renne R, Blackbourn D, Whitby D, Levy J, Ganem D. Limited transmission of Kaposi’s sarcoma-associated herpesvirus in cultured cells. J Virol. 1998;72:5182–5188. doi: 10.1128/jvi.72.6.5182-5188.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. Lytic growth of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 36.Resnitzky D, Gossen M, Bujard H, Reed S I. Acceleration of the G1/S phase transition by expression of cyclins D1 and E with an inducible system. Mol Cell Biol. 1994;14:1669–1679. doi: 10.1128/mcb.14.3.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roizman B. The family Herpesviridae. In: Roizman B, Whitley R J, Lopez C, editors. The human herpesviruses. New York, N.Y: Raven Press, Ltd.; 1993. pp. 1–9. [Google Scholar]
- 38.Roy A L, Carruthers C, Gutjahr T, Roeder R G. Direct role for Myc in transcription initiation mediated by interactions with TFII-I. Nature. 1993;365:359–361. doi: 10.1038/365359a0. [DOI] [PubMed] [Google Scholar]
- 39.Russo J J, Bohenzky R A, Chien M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarid R, Flore O, Bohenzky R A, Chang Y, Moore P S. Transcription mapping of the Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1) J Virol. 1998;72:1005–1012. doi: 10.1128/jvi.72.2.1005-1012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40a.Sarid, R. Unpublished observation.
- 40b.Sarid, R., and C. Boshoff. Unpublished observation.
- 41.Sinclair A J, Palmero I, Peters G, Farrell P J. EBNA-2 and EBNA-LP cooperate to cause G0 to G1 transition during immortalization of resting human B lymphocytes by Epstein-Barr virus. EMBO J. 1994;13:3321–3328. doi: 10.1002/j.1460-2075.1994.tb06634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soulier J, Grollet L, Oskenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d’Agay M-F, Clauvel J-P, Raphael M, Degos L, Sigaux F. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 43.Staskus K A, Zhong W, Gebhard K, Herndier B, Wang H, Renne R, Beneke J, Pudney J, Anderson D J, Ganem D, Haase A T. Kaposi’s sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J Virol. 1997;71:715–719. doi: 10.1128/jvi.71.1.715-719.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swanton C, Mann D J, Fleckenstein B, Neipel F, Peters G, Jones N. Herpes viral cyclin/Cdk6 complexes evade inhibition by CDK inhibitor proteins. Nature. 1997;390:184–187. doi: 10.1038/36606. [DOI] [PubMed] [Google Scholar]
- 45.Zacny V L, Wilson J, Pagano J S. The Epstein-Barr virus immediate-early gene product, BRLF1, interacts with the retinoblastoma protein during the viral lytic cycle. J Virol. 1998;72:8043–8051. doi: 10.1128/jvi.72.10.8043-8051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang L, Pagano J S. IRF-7, a new interferon regulatory factor associated with Epstein-Barr virus latency. Mol Cell Biol. 1997;17:5748–5757. doi: 10.1128/mcb.17.10.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Q, Gutsch D, Kenney S. Functional and physical interaction between p53 and BZLF1: implications for Epstein-Barr latency. Mol Cell Biol. 1994;14:1929–1938. doi: 10.1128/mcb.14.3.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhong W, Wang H, Herndier B, Ganem D. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc Natl Acad Sci USA. 1996;93:6641–6646. doi: 10.1073/pnas.93.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]