Abstract
The question of how best to protect the human population against a potential influenza pandemic has been raised by the recent outbreak caused by an avian H5N1 virus in Hong Kong. The likely strategy would be to vaccinate with a less virulent, laboratory-adapted H5N1 strain isolated previously from birds. Little attention has been given, however, to dissecting the consequences of sequential exposure to serologically related influenza A viruses using contemporary immunology techniques. Such experiments with the H5N1 viruses are limited by the potential risk to humans. An extremely virulent H3N8 avian influenza A virus has been used to infect both immunoglobulin-expressing (Ig+/+) and Ig−/− mice primed previously with a laboratory-adapted H3N2 virus. The cross-reactive antibody response was very protective, while the recall of CD8+ T-cell memory in the Ig−/− mice provided some small measure of resistance to a low-dose H3N8 challenge. The H3N8 virus also replicated in the respiratory tracts of the H3N2-primed Ig+/+ mice, generating secondary CD8+ and CD4+ T-cell responses that may contribute to recovery. The results indicate that the various components of immune memory operate together to provide optimal protection, and they support the idea that related viruses of nonhuman origin can be used as vaccines.
Any doubt that avian influenza A viruses can cross naturally into mammals and cause severe disease was removed by the recent outbreak in Hong Kong. A highly pathogenic H5N1 virus that circulates in domesticated birds infected at least 18 people and caused six deaths. Though the situation was controlled rapidly by the concerted efforts of viral epidemiologists and regulatory authorities, the experience served as a stark reminder that a human pandemic caused by a novel influenza A virus constitutes a very real danger. Experiments with a human isolate in laboratory mice have shown evidence of extreme virulence. Furthermore, this particular H5N1 strain kills chicken embryos so rapidly that there is little production of progeny virus. Since influenza virus vaccines are generally made from the infected allantoic fluid of hen eggs, developing appropriate strategies for dealing with such pathogens is a matter of some urgency (3, 4, 6, 14, 29, 33).
Both public health safety requirements and the lack of any preexisting human herd immunity impose major limitations on animal experiments with the H5N1 viruses. However, there is evidence that the human H3N2 viruses, which have been responsible for the recurring influenza epidemics over the past 30 years, also came originally from birds (27, 28). Furthermore, avian viruses carrying the H3 hemagglutinin (HA) molecule that provides the major determinants for the neutralizing antibody response (11, 30) are available for laboratory use (5). We have thus chosen to analyze the nature of protective immunity to an extremely pathogenic avian H3N8 influenza A virus (A/Duck/Hokkaido/8/80) that is conferred by prior priming with a mouse-adapted human H3N2 virus. This mimics the situation that would occur if a less virulent avian H5N1 virus were to be used to develop a vaccine intended for humans, a strategy that is currently under development (29).
MATERIALS AND METHODS
Viruses.
The analysis concentrated on the avian H3N8 virus A/Duck/Hokkaido/8/80. The isolate provided to us by Yoshihiro Kawaoka (St. Jude Children’s Research Hospital) had been passaged eight times through BALB/c mouse lungs and once in embryonated hen eggs. It was then passaged an additional four times in C57BL/6 (B6) mouse lungs, and graded doses of the final, frozen lung homogenate (MP12) were used to infect B6 mice. The MP12 virus was also passaged a further three times in chicken embryos (MP12EP3) to give a high-titer stock for infecting stimulators and target cells for the in vitro immunology analysis. The MP12EP3 virus was no less virulent for mice (data not shown) than the MP12 virus (Fig. 1). The studies described here used the H3N2 influenza A virus HKx31. HKx31, hereafter referred to as H3N2 virus, is a laboratory-generated reassortant between A/Aichi/68 (H3N2) and A/PR/8/34 (A/PR8; H1N1) which contains the surface HA and neuraminidase molecules of Aichi and the internal components of A/PR8 (17). Comparative sequence analysis of the H3N8 and H3N2 virus stocks used in the in vivo experiments showed differences in both the nucleoprotein (NP) and HA genes. The immunodominant epitope derived from the NP molecule (NP366–374 and presented in association with H-2Db was completely conserved between the H3N8 and H3N2 viruses, but residues flanking this epitope differed between the two (Table 1; GenBank entry nucleoprotein AF079571). Each of the five antibody domains of the HA1 component of the H3 glycoprotein contained amino acid changes (Table 1; GenBank entry HA1 chain AF079570). The unrelated B/Hong Kong/8/73 (B/HK) virus was used as a control. All viruses were titrated by allantoic inoculation into chicken embryos, and titers are expressed throughout as log10 50% egg infectious doses (EID50) (1). The H3N8 virus did not cause rapid death of the chicken embryos.
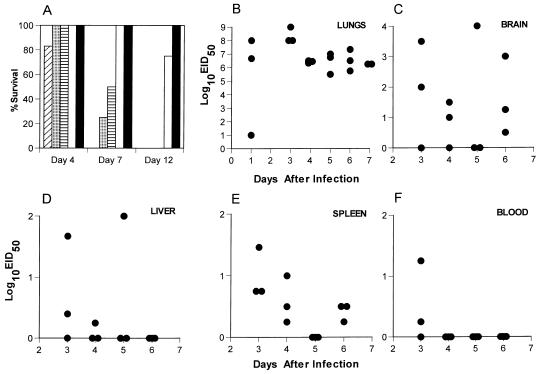
FIG. 1.
(A) Naive B6 mice were infected i.n. with 10-fold dilutions of the H3N8 virus and monitored daily for survival. Four mice per group were infected as follows: diagonal stripes, 105 EID50; dotted, 104 EID50; horizontal stripes, 103 EID50; open, 102 EID50; filled, 101 EID50. (B to F) Naive B6 mice were infected i.n. with 104 EID50 of H3N8 virus, and virus titers were determined for the lungs, brain, spleen, blood, and liver in embryonated chicken eggs.
TABLE 1.
Differences in amino acid residues for NP and HA molecules of H3N2 and H3N8 virusesa
| Viral component (reference) | Residue | Amino acid
|
|
|---|---|---|---|
| H3N2 | H3N8 | ||
| Flanking region of NP366–374b | 351 | K | R |
| 353 | L | V | |
| 357 | K | Q | |
| 375 | E | D | |
| HA1 antibody domains (9) | |||
| a | 144 | G | A |
| 226 | L | Q | |
| b | 193 | S | N |
| c | 92 | K | N |
| 278 | I | V | |
| d | 248 | K | Ec |
| e | 62 | I | R |
| 81 | N | D | |
The NP molecules of the H3N2 and H3N8 viruses are 94% homologous by identity, while the HA1 subunits are 96% homologous. There are a total of 12 amino acid differences in the HA1 subunits.
The Db-restricted CTL epitope ASNENMETM is completely conserved between the H3N2 and H3N8 viruses.
K in published sequence.
Mice, infection, and sampling.
The B6 mice were purchased from The Jackson Laboratory, Bar Harbor, Maine. A homozygous colony was established from μMT mice (18) backcrossed onto the B6 background that were obtained originally from The Jackson Laboratory. Apart from the exposure to influenza virus, the mice were maintained under specific-pathogen-free conditions throughout. Female mice aged 8 to 12 weeks were anesthetized with avertin (2,2,2-tribromoethanol) and then infected intranasally (i.n.) with 106.8 EID50 of the H3N2 virus, graded i.n. doses of the H3N8 virus, 105.6 EID50 of B/HK, or 103.0 EID50 of A/PR8. Some of the immunoglobulin-deficient (Ig−/−) μMT mice were also primed intraperitoneally (i.p.) with 105.2 EID50 of H3N8, as they succumbed to even a minimal i.n. challenge. Those mice used in survival studies were monitored daily and euthanized when severely clinically affected. Secondary challenge experiments were done with mice that had been primed for at least 4 weeks.
Inflammatory cells were obtained from anesthetized, infected mice by bronchoalveolar lavage (BAL). The BAL cells were first allowed to adhere on plastic petri dishes (Falcon, Lincoln Park, N.J.) for 1 h at 37°C to remove macrophages. Single-cell suspensions were made from the cervical lymph nodes (CLN), mediastinal lymph nodes (MLN), and spleens. The lungs, brain, liver, spleen, and blood (0.5 ml) were frozen (−70°C). Thawed samples were used for virus titration, with the solid tissues first being homogenized in 1 ml of Dulbecco’s phosphate-buffered saline (PBS; GibcoBRL, Grand Island, N.Y.). BAL was also done to sample mucosal Ig, and cells were removed by centrifugation.
Titration of H3N2-specific IgG in the lung.
The 96-well enzyme-linked immunosorbent assay (ELISA) plates (Nunc, Roskilde, Denmark) were coated with detergent-disrupted H3N2 virus at a concentration of 0.5 μg/well, washed with PBS–0.5% Tween 20 (Sigma, St. Louis, Mo.), and blocked with PBS–3% bovine serum albumin (Sigma). The plates were then incubated with threefold serial dilutions of the 1 ml of PBS used for BAL, followed by washing and incubation with anti-mouse IgG conjugated to alkaline phosphatase (Southern Biotechnology Associates, Birmingham, Ala.). The ELISAs were then developed with the substrate p-nitrophenyl phosphate, and optical density readings at 405 nm were done on a Bio-Rad Microplate Reader (model 3550; Bio-Rad, Richmond, Calif.).
Staining virus-specific CD8+ T cells.
Tetramers of major histocompatibility complex (MHC) class I glycoprotein plus viral peptide (2) were made from H-2Db complexed with influenza NP366–374 (ASNENMETM; NPP) or Sendai virus NP324–332 (FAPGNYPAL; SEV9) and avidin conjugated to phycoerythrin (PE) (10). The BAL cells were adhered, while the MHC class II+ and CD4+ populations were removed from the MLN and spleen cells by using Dynabeads (Dynal, Oslo, Norway) and a magnet (10). The Fc receptors were then blocked with purified anti-mouse CD16/CD32 (Fc-γRIII/II receptor; Pharmingen, San Diego, Calif.), and the lymphocytes were stained with either the NPP or SEV9 tetramer for 1 h at room temperature and then with fluorescein isothiocyanate-conjugated anti-CD8 (53-6.7; Pharmingen) for 30 min on ice. They were then washed and analyzed (2) on a FACScan using Cell Quest software (Becton Dickinson, Mountain View, Calif.).
Assaying functional CD8+ T cells.
Spleen or MLN cells were incubated under bulk culture conditions for 5 days in 12-well tissue culture plates (Costar, Cambridge, Mass.) at a responder-to-stimulator ratio of 2:1 in SMEM (GibcoBRL), 10% fetal calf serum (Atlanta Biologicals, Atlanta, Ga.), antibiotics, and 5 × 10−5 M 2-mercaptoethanol (Sigma) in a humidified 10% CO2 incubator (17). The split-well limiting-dilution analysis (LDA) used a 7-day culture period in round-bottomed 96-well tissue culture plates (Costar) in the presence of interleukin-2. Percent specific lysis was determined as [(experimental release − spontaneous release)/(maximum release − spontaneous release)] × 100. Levels of specific 51Cr release >3 times the standard deviation of the mean for the value in medium alone were considered to be positive for the LDA microcultures (21).
Assaying CD4+ T cells.
The CD4+ T-cell population was enriched by incubating MLN or spleen populations with anti-CD8 (53-6.72; American Type Culture Collection) and anti-MHC class II (TIB 120; American Type Culture Collection), followed by anti-rat Ig- and anti-mouse Ig-coated Dynabeads and depletion with a magnet (25). The final population contained >85% CD4+ T cells. The gamma interferon (IFN-γ) ELISPOT assay used 96-well filtration plates (Millipore, Bedford, Mass.) that were coated with purified rat anti-mouse IFN-γ (Pharmingen) at 4 μg/ml, washed with PBS, and blocked with SMEM (GibcoBRL) containing 10% fetal calf serum (Atlanta Biologicals) for 1 h at room temperature. The CD4+ T cells were plated at a maximum concentration of 4 × 105 per well and serially diluted twofold. These responders were then cultured for 68 h with either uninfected or H3N2-infected irradiated (2,500 rads) splenocytes dispensed at a final concentration of 5 × 105 per well. The plates were washed four times with PBS–0.05% Tween 20 (Sigma), stained overnight at 4°C with 2 μg of biotin anti-mouse IFN-γ (Pharmingen) per ml, then washed again, and incubated with peroxidase-labeled goat antibiotin (Vector Laboratories, Burlingame, Calif.) at 5 μg/ml for 1 h at room temperature. After a further washing, the plates were incubated with the developing substrate (3-amino-9-ethylcarbazole; Sigma) for 15 min at room temperature and then washed with distilled H2O to stop the reaction. The peroxidase-positive spots were then counted microscopically, and the data were used to determine the virus-specific CD4+ T-cell frequency.
In vivo T-cell depletion.
Mice were injected with ascites fluid containing the CD4-specific monoclonal antibody (MAb) GK1.5, the CD8-specific MAb 2.43, or a control rat Ig, commencing 5 days before infection and continuing at 2- to 3-day intervals thereafter (24). The efficacy of the protocol was checked at time of sampling, with flow cytometric analysis (anti-CD4-PE antibody RM4-4 and anti-CD8-PE antibody 53-5.8; Pharmingen) always showing <1% of the respective population remaining.
RESULTS
Infection in immunologically naive mice.
The first step was to analyze the nature of the infectious process caused by the H3N8 virus. The mean survival time following respiratory challenge of naive, adult B6 mice with a uniformly lethal dose (104.0 EID50) of the mouse-passaged H3N8 influenza A virus was 5.8 ± 1.7 days (Fig. 1A). Those remaining alive on day 7 had lung titers of >106.0 EID50 (Fig. 1B). Evidence of significant systemic spread, a characteristic observed only with extremely virulent influenza A viruses (15), was detected in samples of liver, spleen, and brain (Fig. 1C to E). The titers were variable in all sites other than the lung (Fig. 1B to E) and could not be attributed to the concurrent presence of infected blood (Fig. 1F), though there was early evidence of minimal viremia. This H3N8 virus is clearly very pathogenic for laboratory mice.
Protection conferred by H3N2 priming.
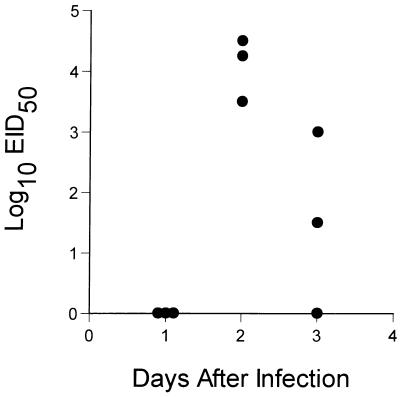
The next question was whether mice can be protected against this fatal influenza virus infection by prior exposure to a related but much less virulent virus. The HA molecule of the HKx31 influenza A virus, which has been analyzed extensively in mouse model systems, is 96% homologous to that of the H3N8 virus, with sequence differences in each of the five antibody-binding domains (Table 1; references 13a and 30). Mice that had recovered from respiratory exposure to the HKx31 virus, hereafter referred to as H3N2 virus, were challenged i.n. with 104.0 EID50 of the H3N8 virus. No virus was recovered from the lung at 24 h after infection, but titers as high as 104.5 EID50 were detected in mice sampled at 48 h and were obviously declining by the next day (Fig. 2). None of the H3N2-primed mice showed any obvious signs of clinical impairment, and all were clearly protected from a dose of the H3N8 virus that achieved titers of >108.0 EID50 in the lungs of naive mice and was uniformly lethal (Fig. 1A and B).
FIG. 2.
Naive B6 mice were primed i.n. with 106.8 EID50 of the H3N2 virus and rested for 1 month prior to challenge i.n. with 104 EID50 of the H3N8 virus. Virus titers in the lung were determined on days 1 to 3 after infection.
Role of antibody.
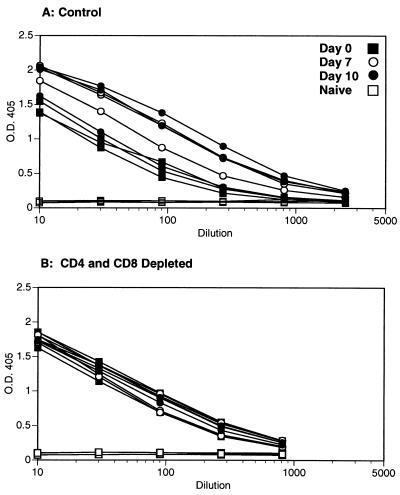
The protective effect of prior H3N2 priming (Fig. 2) could be mediated by humoral immunity, by the recall of T-cell memory, or by both sets of mechanisms. In terms of the humoral response, specific antibodies to the H3N2 and H3N8 viruses are cross-reactive in the standard hemagglutination inhibition test; preincubating the H3N8 virus in 10% mouse serum from H3N2-primed mice prior to i.n. challenge completely prevented the development of symptoms, whereas preincubation with 10% normal mouse serum resulted in 100% mortality (data not shown). The next step was to determine the susceptibility profile of immune mice that lack one or more components of the specific host response. Challenge with a lethal dose of the H3N8 virus (104 EID50) was thus repeated in B6 mice and congenic, Ig−/− μMT mice which had been primed 2 to 4 months previously with either the H3N2 or the H3N8 virus. The immune B6 (Ig+/+) mice were protected and showed 100% survival, while the immune μMT (Ig−/−) mice were completely susceptible to the secondary challenge (Table 2). Eliminating both the CD4+ and the CD8+ T cells by treating the Ig+/+ B6 mice with lymphocyte subset-specific MAbs commencing prior to challenge with the H3N8 virus still resulted in 100% survival (Table 2). Levels of H3N2-specific IgG in lung washes of mock-treated and T-cell-depleted mice were determined on days 0, 7, and 10 after secondary infection with the H3N8 virus. Significant virus-specific antibody titers were found when both mock-treated and T-cell-depleted (Fig. 3) mice were compared to naive mice. It seems that although the levels of H3N2-specific antibody at the mucosal surface were not sufficient to prevent the H3N8 virus from infecting at least some respiratory epithelial cells, the Ig response generated by H3N2 priming protected against the development of lethal pneumonia (compare Fig. 1 and 2).
TABLE 2.
Susceptibility of primed mice to challenge with the H3N8 virusa
| Mouse strain | T-cell depletionb | Mean survival time (days) ± SD
|
|
|---|---|---|---|
| Naive mice | Immune mice | ||
| B6 | Nil | 5.6 ± 0.6 | >21 |
| CD4 | 4.3 ± 0.5 | >21 | |
| CD8 | 4.8 ± 0.5 | >21 | |
| CD4 + CD8 | 5.3 ± 1.3 | >21 | |
| μMT | Nil | 7.5 ± 2.9 | 6.1 ± 1.0, 4.7 ± 2.1c |
B6 and μMT mice were infected i.n. with 106.8 EID50 of the H3N2 virus and rested for 2 to 4 months. Groups of four mice were challenged i.n. with 104.0 EID50 of the H3N8 virus. The data are cumulated from several experiments and are for a total of 8 to 12 mice for each treatment.
The mice were treated with MAb GK1.5 to CD4, MAb 2.43 to CD8, or rat Ig as a control (Nil), commencing 5 days prior to challenge and continuing every 2 or 3 days thereafter (20).
This group of μMT mice was primed i.p. with 105.2 EID50 of the H3N8 virus and rested for 7 weeks prior to i.n. challenge with 104.0 EID50 of the H3N8 virus.
FIG. 3.
Naive B6 mice were infected i.n. with 106.8 EID50 of the H3N2 virus and rested for 4 months. Rat Ig-treated control mice (A) or mice depleted of both CD4+ and CD8+ T cells (B) were infected i.n. with 104 EID50 of the H3N8 virus. BAL was done on days 0, 7, and 10 after secondary infection. The BAL cells were removed by centrifugation, and H3N2-specific IgG titers were determined by ELISA. Each curve represents a single animal, and three animals are shown per time point. O.D. 405, optical density at 405 nm.
Secondary CD4+ and CD8+ T-cell responses in primed Ig+/+ mice.
The H3N2-specific antibody levels in the lung increased between days 0 and 7 after challenge with the H3N8 virus in control mice but remained constant in mice depleted of both CD4+ and CD8+ T cells (Fig. 3). Furthermore, viral lung titers of these same animals showed that the control group had completely cleared virus by day 7 after infection with the H3N8 virus (data not shown). In contrast, virus was still present in lung samples from two of three T-cell-depleted mice (101.5 EID50 and 102.5 EID50, respectively) at day 10 after secondary challenge (data not shown). Despite the apparent delay in clearance, all of nine T-cell-depleted animals survived for >45 days (data not shown). Taken together, these data suggest that although antibody-mediated protective mechanisms are sufficient to ensure survival following a potentially lethal secondary challenge, the CD4+ and CD8+ T-cell subsets also play a role in the secondary responses of Ig+/+ mice. Thus, secondary CD4+ and CD8+ T-cell responses were examined as follows. Mice that had been given the H3N2 virus 8 months previously were infected i.n. with (i) the lethal H3N8 virus, which shares HA-specific CD4+ T-cell epitopes with the H3N2 virus, (ii) the serologically different A/PR8 (H1N1) virus, which shares the NP366–374 epitope recognized by the majority of the responding CD8+ T cells, and (iii) an influenza B virus (B/HK) that is not known to cross-react in any way with the influenza A viruses but causes a similar inflammatory pathology in the murine lung (10).
We have previously shown that staining of lymph node, spleen, and BAL populations with the NPP tetramer (tetrameric complex of Db bound to NP peptide) allows enumeration of virus-specific CD8+ T cells during an influenza virus infection (10). The recruitment of tetramer-positive (CD8+ NPP+) T cells to the lung was comparable for the mice challenged with the H3N8 and H1N1 viruses (Table 3), which is intriguing since the extent of replication (and thus antigen load) for the H1N1 virus would be expected to be much greater (reference 16 and Fig. 2). The value for the B/HK challenge presumably reflects the background associated with the nonspecific recruitment of memory cytotoxic T lymphocytes (mCTL) to the pneumonic lung (26). Both of the influenza A viruses, but not the B/HK virus, stimulated the development of effector CTL (eCTL) in the lymph nodes. The greatest increase in the prevalence of the virus-specific CD4+ T-helper precursor (Thp) population was seen for the mice challenged with the H3N8 virus (Table 3). This is not surprising, as many of the epitopes recognized by CD4+ T cells in H-2b mice are derived from the HA molecule (21a). Clearly, even in the presence of neutralizing antibody, the antigen load is sufficient to restimulate both the CD4+ and CD8+ T-cell responses. This, in turn, may account for a more vigorous humoral response and accelerated viral clearance in animals not depleted of T cells.
TABLE 3.
CD4+ and CD8+ T-cell responses in H3N8-challenged, H3N2-primed B6 micea
| Challenge virus | % CD8+ NPP+ T cellsb
|
% Specific lysisc
|
Reciprocal Thp frequencyd
|
||||
|---|---|---|---|---|---|---|---|
| BAL | MLN | Spleen | MLN | Spleen | MLN | Spleen | |
| B/HK | 3.8 | 0.1 | 0.4 | 0 | 0 | 20,000 | 33,333 |
| H3N8 | 9.6 | 2.3 | 0.8 | 23 | 0 | 523 | 1,785 |
| H1N1 | 7.3 | 1.8 | 1.4 | 32 | 9 | 1,754 | 8,333 |
B6 mice were infected i.n. with 106.8 EID50 of H3N2 virus and rested for 7 to 8 months; they were then challenged i.n. with 105.6 EID50 of the B/HK virus, 103 EID50 of the A/PR8 (H1N1) virus, or 104 EID50 of H3N8 virus and sampled 5 days later.
The BAL cells were processed, stained with the tetramer and anti-CD8, and analyzed as described in Materials and Methods to determine the prevalence of virus-specific (CD8+ NPP+) T cells. The MLN and spleen cells were enriched for CD8+ T cells and stained with the tetramer and anti-CD8 prior to analysis. The level of background staining with the SEV9 tetramer was <0.1%.
Freshly isolated MLN and spleen cells were used directly as effectors in a 6-h 51Cr release assay with H3N2-infected MC57G targets at an effector-to-target ratio of 100:1. Lysis of uninfected targets at this ratio was <4.0%.
Enriched CD4+ T cells from the MLN and spleen were incubated for 68 h with either uninfected or H3N2-infected stimulators and assayed for IFN-γ production by ELISPOT as described in Materials and Methods. The results are expressed as reciprocal virus-specific CD4+ Thp frequencies.
Limited protection of the Ig−/− mice by CD8+ T-cell-mediated immunity.
The Ig−/− μMT mice were primed i.n. with the H3N2 virus or i.p. with the H3N8 virus and then challenged i.n. with the H3N8 virus to analyze the protective efficacy of the secondary CD8+ T-cell response (Fig. 4). A similar protocol involving the challenge of H3N2-immune μMT mice with the homologous H3N2 virus resulted in a rapid clearance of virus from the lung. This protective effect was greatly diminished by the elimination of the CD8+ T-cell subset (24). The immunodominant NP366–374 peptide (5) recognized in association with H-2Db is present and completely conserved in both the H3N2 and the H3N8 viruses. However, there is variation in the flanking regions (Table 1) which might potentially influence the processing and presentation of the epitope (7) and thus the magnitude of the CD8+ T-cell response. Restimulating the immune T-cell populations with virus under conditions of bulk culture followed by testing in a standard 51Cr release assay showed that cross-reactive eCTL were generated in both the H3N2- and H3N8-primed μMT mice (Table 4). Similarly, determining mCTL frequencies by LDA established that the spectra of T-cell priming were comparable following i.n. (H3N2) or i.p. (H3N8) exposure to these two influenza viruses (Table 4).
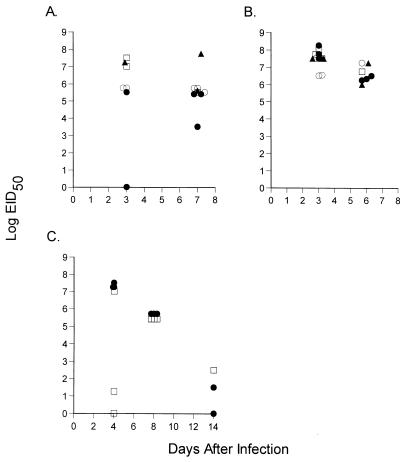
FIG. 4.
(A and B) μMT mice were primed i.n. with 106.8 EID50 of the H3N2 virus (A) or i.p. with 105.2 EID50 of the H3N8 virus (B) and rested for 6 weeks. They were then either mock (rat Ig)-treated (•) or depleted of CD4+ T cells (□), CD8+ T cells (▴), or both (▵) as described in Materials and Methods and then challenged i.n. with 104.0 EID50 of the H3N8 virus. Virus titers in the lungs were determined. (C) μMT mice were infected i.n. with 106.8 EID50 of H3N2 virus, rested for 10 weeks, and then challenged i.n. with 103 EID50 of the H3N8 virus. Virus titers in the lungs were determined.
TABLE 4.
Cross-reactivity of the CTL response for the H3N2 and H3N8 virusesa
| Immunizing virus | In vitro restimulation | % Specific 51Cr releaseb
|
Reciprocal CTLp frequencyc
|
|||
|---|---|---|---|---|---|---|
| Nil | H3N2 | H3N8 | H3N2 | H3N8 | ||
| H3N2 | H3N2 | 1 | 58 | 59 | 1,722 | 1,615 |
| H3N8 | 16 | 50 | 53 | 2,684 | 2,091 | |
| H3N8 | H3N2 | 7 | 63 | 56 | 2,013 | 2,065 |
| H3N8 | 4 | 49 | 45 | 3,914 | 2,781 | |
Ig−/− μMT mice were uninfected (Nil) or infected i.n. with 106.8 EID50 of the H3N2 virus or i.p. with 105.2 EID50 of the H3N8 virus and rested for 4 weeks prior to harvesting of CLN, MLN, and spleen cells for bulk culture or LDA assays.
Cells were restimulated in vitro with either H3N2- or H3N8-infected stimulators as indicated; values for bulk cultures are shown at an effector-to-target ratio of 25:1 in a 6-h assay using MC57G target cells.
LDA cultures were assayed for levels of specific 51Cr release >3 times the standard deviation to calculate reciprocal precursor CTL (CTLp) frequencies (21).
What are the characteristics of the secondary CD8+ T-cell response in these primed mice? Recruitment of substantial numbers of CD8+ NPP+ T cells to the pneumonic lung was apparent at 6 days after i.n. challenge of the H3N2- and H3N8-immune, but not naive, μMT mice with 104.0 EID50 of the H3N8 virus (Fig. 5A to C). However, the lung titers were still high (Fig. 4A and B), and this dose of virus is uniformly lethal in the absence of antibody (Table 2). Furthermore, any control mediated by the CD8+ T cells was more apparent for the H3N2-primed mice than for the H3N8-primed mice (see below and Table 5), reflecting the greater eCTL numbers recovered from the virus-infected lung (Fig. 5B and C). The extreme virulence of the H3N8 virus (Fig. 1) is clearly the key determinant of susceptibility, as a secondary response of similar magnitude was previously shown to accelerate the rapid CD8+ T-cell-mediated clearance of the less pathogenic A/PR8 (H1N1) influenza virus from H3N2-primed B6 mice (10).
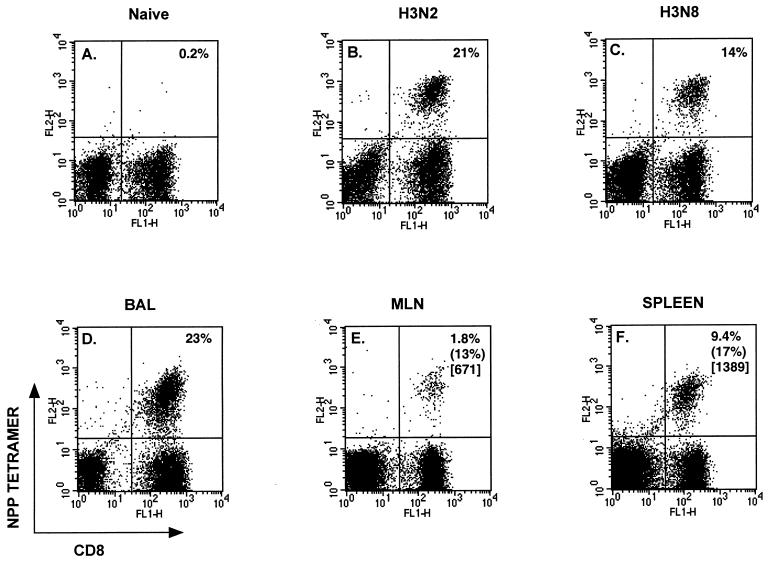
FIG. 5.
(A to C) Staining profiles for CD8+ NPP+ lymphocytes obtained for BAL cells harvested from naive (A), H3N2-immune (B), or H3N8-immune (C) μMT mice on day 6 after i.n. challenge with 104.0 EID50 of the H3N8 virus. (D to F) Prevalence of virus-specific CD8+ T cells for BAL (D), MLN (E), and spleen (F) cells harvested from μMT mice on day 12 after i.n. challenge with 250 EID50 of the H3N8 virus. Staining with the control SEV9 tetramer was always <0.1%. Percentages in parentheses denote levels of specific 51Cr release (see footnotes to Table 4) after direct assay of freshly isolated lymphocyte populations; numbers in brackets are reciprocal Thp frequencies for IFN-γ-producing CD4+ T cells (see footnotes to Table 3).
TABLE 5.
CD8+ T-cell-mediated protection in Ig−/− micea
| Immunizing virus | T-cell depletionb | Group size | Mean survival time (days) ± SD | % Survival >21 days |
|---|---|---|---|---|
| None | Nil | 5 | 10.4 ± 2.2 | 0 |
| CD8 | 5 | 5.8 ± 1.5 | 0 | |
| H3N2 | Nil | 9 | 14 ± 8.4 | 78 |
| CD8 | 10 | 12.9 ± 1.9 | 20 | |
| H3N8 | Nil | 10 | 9.7 ± 2.6 | 30 |
| CD8 | 6 | 7.7 ± 1.8 | 0 |
μMT animals were primed either i.n. with 106.8 EID50 of H3N2 or i.p. with 105.2 EID50 of H3N8 and rested for 14 to 16 weeks prior to i.n. challenge with 250 EID50 of the H3N8 virus.
The mice were given rat Ig (Nil) or were CD8 depleted as described for Table 2.
Decreasing the magnitude of the H3N8 challenge to 103.0 EID50 resulted in the long-term survival of 10% of the primed μMT mice, with evidence that the infection was being controlled with time (Fig. 4C). The early stage of the infectious process was much more variable following exposure to this lower dose, though lung titers were uniformly high by day 8 after infection (Fig. 4C). Reducing the virus challenge a further fourfold (250 EID50) increased both the interval to the development of severe symptoms and the numbers of mice that recovered (Table 5). Depleting the CD8+ subset in mice secondarily challenged with the 250 EID50 dose of H3N8 virus showed that this protective effect was mediated largely by the secondary eCTL response (Table 5). The eCTL were present at high frequency in the BAL (Fig. 5D) of mice challenged with the 250 EID50 dose and could also be detected in the MLN and spleen by staining with the NPP tetramer (Fig. 5E and F). Furthermore, the lymphoid tissue contained both eCTL and virus-specific CD4+ T cells that could be stimulated by in vitro culture (Fig. 5E and F). Taken together, the results in Fig. 4, Fig. 5, and Table 5 indicate that the CD8+ T-cell response is capable of handling a very low dose of the H3N8 influenza virus in the absence of antibody, but not in a way that is uniformly protective (Table 5).
DISCUSSION
The extreme susceptibility of laboratory mice to respiratory challenge with the H3N8 influenza A virus demonstrates very clearly that clinical outcome depends on a race between the growth characteristics of the pathogen and the development of the specific host response. The extent of virus-induced damage to lung epithelium in immunologically naive mice is simply too great by the time that virus-specific antibody and eCTL populations become available at the site of pathology. Previous analysis utilizing an H3N2 challenge in H1N1-primed B6 mice has shown that the secondary CTL response develops in the MLN and takes at least 4 to 5 days before the effectors are available in the infected respiratory tract (10). Our studies (24, 25) and others (8, 12) using Ig−/− μMT mice indicate that primed CD8+ T cells can provide at least some protection against novel influenza A viruses, which may be the reason that not all those infected during the recent H5N1 outbreak in Hong Kong succumbed (29). However, although priming the CD8+ T-cell compartment can protect Ig−/− μMT mice against homologous challenge with high titers of the HKx31 virus (24), the recall of the mCTL to eCTL function is still too slow to protect from all but a very low dose of the H3N8 virus.
Though the H3N8 virus has a variable capacity for systemic spread, naive μMT mice suffered no obvious consequences following i.p. challenge with a dose 1,000 times higher than that causing uniformly lethal disease following i.n. exposure. Thus, the requirement for enzymatic cleavage of the viral HA molecule that substantially limits the production of infectious influenza A viruses to the murine lung mucosa still determines the pathogenesis of the disease (13). The secondary localization of the H3N8 virus to the brain detected following i.n. infection of B6 mice is probably a consequence of the much greater viral load resulting from the continued replication in the respiratory tract. The present experiments show that this can result in measurable viremia, although of very limited duration. The analysis also makes the point that it may be possible to protect mammals against a novel influenza virus infection by injecting fully virulent virus subcutaneously or intramuscularly, though such a strategy would obviously be too risky to consider using in humans. However, an attenuated, live virus vaccine might be given safely via this route to people that lack cross-reactive neutralizing antibody, with much less risk than for administration via a respiratory route. Such an approach could be considered for limiting a rapidly spreading pandemic caused by an extremely virulent avian influenza A virus, provided that an attenuated variant is available.
The H3-specific antibody response in the B6 mice clearly prevents the development of lethal pneumonia following respiratory challenge with the H3N8 virus, though there is still some replication in the lung. In general, preexisting antibody has been shown to be the major mechanism of protection against secondary influenza virus infection (11). This may reflect direct neutralization of the inoculum (reviewed in reference 11) by virus-specific IgG or IgA already present at the surface of the lung mucosa. Also, the virus-specific Ig could act to prevent further spread in the respiratory tract by the neutralization of free virions or by opsonizing the virus for uptake by macrophages. Ig may also enable macrophages to mediate antibody-dependent cellular cytotoxicity (11).
The rapid control of the H3N8 infection in the H3N2-immune B6 mice by neutralizing antibody in no way inhibited the development of the secondary CD4+ and CD8+ T-cell responses. Previous studies have shown that virus-specific CD8+ T-cell responses are generated in the presence of neutralizing antibody (23). The present analysis extends this observation to show that both CD4+ and CD8+ T-cell responses develop in the presence of neutralizing antibody. Furthermore, though the recall of CD4+ T-cell memory has been shown to have relatively little protective effect in Ig−/− μMT mice depleted of the CD8+ T-cell subset (24), such primed T-cell help could obviously be a significant factor in the Ig+/+ group. Overall, the results indicate that cross-reactive CD8+ eCTL (20) and the neutralizing antibody response may both play a part in recovery from a virulent influenza virus infection, with the latter mechanism being much more important.
The laboratory-adapted viruses used in these experiments express H3 molecules derived from viruses circulating in humans (H3N2) and birds (H3N8) in 1968 and 1980, respectively. Sequencing the HA1 subunits showed at least one amino acid difference for each of the five antibody-binding domains. The 96% sequence homology for these two HA1 regions is fairly representative of that seen for many of the “drifted” H3N2 viruses that cause sequential pandemics (16, 19, 22, 31, 32). Such antibody-mediated selection pressure was shown to result in a 9.2% change in the HA1 region of human H3N2 viruses isolated over a 10-year period. The avian viruses do not show much evidence of such drift in their natural, maintaining host (16). The present results indicate that any recent incursion of an avian virus into the human population could be dealt with by using a virus that is likely to be already available as a nonpathogenic laboratory strain adapted for growth in embryonated hen eggs, the basic starting point for the currently prevalent vaccine technology. The same is true for the H7N7 viruses, which also loom as possible human pathogens (28). The use of contemporary molecular approaches to develop attenuated vaccine candidates for the influenza A viruses that seem most likely to cross from domestic animals into humans is worth considering.
ACKNOWLEDGMENTS
We thank Vicki Henderson for help in preparing the manuscript, and we thank Kristin Branum for expert technical assistance.
This work was supported by Public Health Service grants AI08831, AI29579, AI38359, and CA21765 and by the American Lebanese-Syrian Associated Charities.
REFERENCES
- 1.Allan W, Tabi Z, Clearly A, Doherty P C. Cellular events in the lymph node and lung of mice with influenza: consequences of depleting CD4+ T cells. J Immunol. 1990;144:3980–3986. [PubMed] [Google Scholar]
- 2.Altman J D, Moss P A H, Goulder P J R, Barouch D H, McHeyzer-Williams M G, Bell J I, McMichael A J, Davis M M. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- 3.Claas E C J, Osterhaus A D M E, van Beek R, De Jong J C, Rimmelzwaan G F, Senne D A, Karauss S, Shortridge K F, Webster R G. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet. 1998;351:472–477. doi: 10.1016/S0140-6736(97)11212-0. [DOI] [PubMed] [Google Scholar]
- 4.Cohen J. The flu pandemic that might have been. Science. 1997;277:1600–1601. doi: 10.1126/science.277.5332.1600. [DOI] [PubMed] [Google Scholar]
- 5.Deckhut A M, Allan W, McMickle A, Eichelberger M, Blackman M A, Doherty P C, Woodland D L. Prominent use of Vβ8.3 T cells in H-2Db nucleoprotein epitope. J Immunol. 1993;151:2658–2666. [PubMed] [Google Scholar]
- 6.De Jong J C, Osterhaus A D M E, Webster R G, Lim W L. A pandemic warning? Nature. 1997;389:554. doi: 10.1038/39218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eisenlohr L C, Yewdell J W, Bennink J R. Flanking sequences influence the presentation of an endogenously synthesized peptide to cytotoxic T lymphocytes. J Exp Med. 1992;175:481–487. doi: 10.1084/jem.175.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Epstein S L, Lo C, Misplon J A, Bennink J R. Mechanisms of protective immunity against influenza virus infection in mice without antibodies. J Immunol. 1998;160:322–327. [PubMed] [Google Scholar]
- 9.Flynn K, Müllbacher A. The generation of memory antigen-specific cytotoxic T cell responses by CD28/CD80 interactions in the absence of antigen. Eur J Immunol. 1997;27:456–462. doi: 10.1002/eji.1830270216. [DOI] [PubMed] [Google Scholar]
- 10.Flynn K J, Belz G T, Altman J D, Ahmed R, Woodland D L, Doherty P C. Virus-specific CD8+ T cells in primary and secondary influenza pneumonia. Immunity. 1998;8:683–691. doi: 10.1016/s1074-7613(00)80573-7. [DOI] [PubMed] [Google Scholar]
- 11.Gerhard W, Mozdzanowska K, Furchner M, Washko G, Maiese K. Role of the B-cell response in recovery of mice from primary influenza virus infection. Immunol Rev. 1997;159:95–103. doi: 10.1111/j.1600-065x.1997.tb01009.x. [DOI] [PubMed] [Google Scholar]
- 12.Graham M B, Braciale T J. Resistance to and recovery from lethal influenza virus infection in B lymphocyte-deficient mice. J Exp Med. 1997;186:2063–2068. doi: 10.1084/jem.186.12.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horimoto T, Kawaoka Y. The hemagglutinin cleavability of a virulent avian influenza virus by subtilisin-like endoproteases is influenced by the amino acid immediately downstream of the cleavage site. Virology. 1995;210:466–470. doi: 10.1006/viro.1995.1363. [DOI] [PubMed] [Google Scholar]
- 13a.Stech, J. Unpublished data.
- 14.Subbarao K, Klimov A, Katz J, Regnery H, Lim W, Hall H, Perdue M, Swayne D, Bender C, Huang J, Hemphill M, Rowe T, Shaw M, Xu X, Fukuda K, Cox N. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science. 1998;279:393–396. doi: 10.1126/science.279.5349.393. [DOI] [PubMed] [Google Scholar]
- 15.Kawaoka Y. Equine H7N7 influenza A viruses are highly pathogenic in mice without adaptation: potential use as an animal model. J Virol. 1991;65:3891–3894. doi: 10.1128/jvi.65.7.3891-3894.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kida H, Kawaoka Y, Naeve C W, Webster R G. Antigenic and genetic conservation of H3 influenza virus in wild ducks. Virology. 1987;159:109–119. doi: 10.1016/0042-6822(87)90353-9. [DOI] [PubMed] [Google Scholar]
- 17.Kilbourne E D. Future influenza vaccines and the use of genetic recombinants. Bull WHO. 1969;41:643–606. [PMC free article] [PubMed] [Google Scholar]
- 18.Kitamura D, Roes J, Kuhn R, Rajewsky K. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin μ chain gene. Nature. 1991;350:423–426. doi: 10.1038/350423a0. [DOI] [PubMed] [Google Scholar]
- 19.Krystal M, Young J F, Palese P, Wilson I A, Skehel J J, Wiley D C. Sequential mutations in hemagglutinins of influenza B virus isolates: definition of antigenic domains. Proc Natl Acad Sci USA. 1983;80:4527–4531. doi: 10.1073/pnas.80.14.4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lalvani A, Brooks R, Hambleton S, McMichael W J. Rapid effector function in CD8+ memory T cells. J Exp Med. 1997;186:859–865. doi: 10.1084/jem.186.6.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Müllbacher A, Hill A B, Blanden R V, Cowden W B, King N J, Hla R T. Alloreactive cytotoxic T cells recognize MHC class I antigen without peptide specificity. J Immunol. 1991;147:1765–1772. [PubMed] [Google Scholar]
- 21a.Riberdy, J. Unpublished data.
- 22.Rogers G N, Paulson J C, Daniels R S, Skehel J J, Wilson I A, Wiley D C. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature. 1983;304:76–78. doi: 10.1038/304076a0. [DOI] [PubMed] [Google Scholar]
- 23.Seiler P, Bründler M A, Zimmermann C, Weibel D, Bruns M, Hengartner H, Zinkernagel R M. Induction of protective cytotoxic T cell responses in the presence of high titers of virus-neutralizing antibodies: implications for passive and active immunization. J Exp Med. 1998;187:649–654. doi: 10.1084/jem.187.4.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Topham D J, Doherty P C. Clearance of an influenza A virus by CD4+ T cells is inefficient in the absence of B cells. J Virol. 1998;76:882–885. doi: 10.1128/jvi.72.1.882-885.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Topham D J, Tripp R A, Hamilton-Easton A M, Sarawar S R, Doherty P C. Quantitative analysis of the influenza virus-specific CD4+ T cell memory in the absence of B cells and Ig. J Immunol. 1996;157:2949–2952. [PubMed] [Google Scholar]
- 26.Tripp R A, Hou S, McMickle A, Houston J, Doherty P C. Recruitment and proliferation of CD8+ T cells in respiratory virus infections. J Immunol. 1995;154:6013–6021. [PubMed] [Google Scholar]
- 27.Webster R G. Influenza virus: transmission between species and relevance to emergence of the next human pandemic. Arch Virol. 1997;13:105–113. doi: 10.1007/978-3-7091-6534-8_11. [DOI] [PubMed] [Google Scholar]
- 28.Webster R G. Predictions for future human influenza pandemics. J Infect Dis. 1997;176:514–519. doi: 10.1086/514168. [DOI] [PubMed] [Google Scholar]
- 29.Webster R G, Hay A J. In: Textbook of influenza. Nicholson K G, Webster R G, Hay A J, editors. Oxford, England: Blackwell Science Ltd.; 1998. pp. 561–565. [Google Scholar]
- 30.Wiley D, Skehel J J. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu Rev Biochem. 1987;56:365–394. doi: 10.1146/annurev.bi.56.070187.002053. [DOI] [PubMed] [Google Scholar]
- 31.Wiley D C, Wilson I A, Skehel J J. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature. 1981;289:373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- 32.Wilson I A, Skehel J J, Wiley D C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 33.Yuen K Y, Chan P K S, Peiris M, Tsang D N C, Que T L, Shortridge K F, Cheung P Y, To W K, Ho E T F, Sung R, Cheng A F B members of the H5N1 study group. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet. 1998;351:467–471. doi: 10.1016/s0140-6736(98)01182-9. [DOI] [PubMed] [Google Scholar]