Abstract
The present research investigated whether accidental contact through stinging with honeybees, wasps, and hornets could represent a microbial hazard for humans. It has been previously suggested that such contact may transmit pathogens causing infections that could even be fatal for some susceptible individuals. Stinging simulation experiments were performed in the lab with live insects collected from the environment in Lemnos Island (north-eastern Greece), while different selective agar media targeting some clinically important bacteria (i.e., Staphylococcus aureus, Streptococcus pyogenes, Enterococcus faecalis/faecium, and Pseudomonas aeruginosa) were used as substrates for microbial recovery and identification. Results revealed none of the target pathogenic bacterial species in the honeybee samples, with bacilli, staphylococci, and micrococci dominating their surveyed microbiota. However, most of the suspect colonies isolated from wasps and hornets belonged to important hygienic indicators (i.e., enterococci, Proteus mirabilis, and coliforms), implying possible contact of these insects with fecal origin materials. To sum up, the microbiota that may be transmitted to humans through stinging appears to differ between honeybees and wasps/hornets, while the isolation from the latter samples of some other important opportunistic pathogens, such as Enterobacter spp. and Klebsiella spp., also known for multidrug resistance, could be an additional reason of concern.
Keywords: Hymenoptera stinging, Honeybees, Wasps, Microbiota, Bacterial pathogens, Infection
Introduction
Hymenoptera is a large order of insects, including members that are capable of stinging, such as bees, wasps, hornets, yellow jackets, and ants (Branstetter et al. 2018). Among them, bees are the dominant pollinators of angiosperms all over the world with more than 20,000 species described to date. These vary broadly in traits, such as nesting habitat, diet, and social behavior (Hedtke et al. 2013). More specifically, honeybees are social insects belonging to the Apis genus and live in well-organized communities. Besides their key beneficial role as pollinators in agriculture, they also provide several valuable products to humans, such as honey, beeswax, bee pollen, bee bread, royal jelly, and propolis (Martinello and Mutinelli 2021). These products are known for their plethora of functional (bioactive) properties and have been used by humans, mainly for nutrition and/or medicinal purposes, since ancient times (Cornara et al. 2017; Kurek-Górecka et al. 2020).
Although honeybees are beneficial to humans, they do still pose some danger due to their painful and venomous stings. Such exposure of humans to bee stinging dates back over 7000 years, when humans started to provide bee populations with artificial hives to harvest their honey and wax, or for pollination purposes (Pucca et al. 2019). However, most honeybees are not hostile toward humans or other animals and attack only when they are frightened and to defend their hives against intruders. Apis mellifera is the species that is mostly responsible for human envenoming in Europe (Pucca et al. 2019). Honeybees have barbed stingers that together with the venom sac are pulled out of the bee’s abdomen and remain in the victim’s skin following stinging; the insect dies shortly afterwards (Fitzgerald and Flood 2006). In contrast, wasps and hornets, both members of the Vespidae family, do not have barbed stingers and can sting multiple times without dying. They tend to be more aggressive than bees, stinging even without feeling threatened (Zirngibl and Burrows 2012).
Fortunately, most Hymenopteran stings are self-limiting (temporary) events, provoking only mild symptoms (such as erythema, edema, and pain at the sting site) which resolve without treatment in a few hours. However, massive envenomation may cause death depending on the victim’s age, body weight, number of stings, health, and immune status of the victim (Fitzgerald and Flood 2006). It has been roughly calculated that the European honeybee injects 147 μg of venom per sting, whereas most wasps inject about 17 μg of venom per sting, while the median lethal dose (LD50) of bee venom varies between 2.8 and 3.5 mg per kilogram of human body weight (Fitzgerald and Flood 2006). Therefore, a non-allergic person weighing 60–70 kg presents a 50% chance of death upon being stung by 1000–1500 bees, although deaths caused by many fewer stings have also been recorded (Pucca et al. 2019). Nevertheless, most deaths related to Hymenopteran stings are the result of immediate hypersensitivity reactions causing anaphylaxis. Such life-threatening anaphylactic reactions typically occur within 10 min of the sting and are not dose dependent or related to the number of stings (Adams et al. 2022).
Although bee venom is usually associated with pain resulting from the local inflammation in humans when stung by bees, during recent decades, venom has been applied for medicinal purposes because of its known antimicrobial and other therapeutical properties (Carpena et al. 2020; El-Seedi et al. 2020). Thus, bee venom through sting acupuncture is extensively used in apitherapy (Zhang et al. 2018). This alternate therapy relies on the usage of honeybee products, most importantly bee venom, for the treatment of several human diseases including neurodegenerative ones (such as Parkinson’s and Alzheimer’s), as well as some types of cancer (Oršolić 2012; Wehbe et al. 2019). For this reason, the venom is introduced into the human body by manual injection or directly via bee stings. However, with the latter, it is speculated that any bacteria or other microorganism found on either insects’ body or sting could be inoculated under the human epidermis during stinging. This may explain some reports of both local and disseminated bacterial infections after accidental bee stings (Anderson et al. 1995; Klug et al. 1982; Richardson and Schmitz 1997; Shahar and Frand 1979). Insect bites have also been reported to result in myocutaneous mycoses (Kontoyiannis et al. 2019). Viral infections following bee stings have also been reported (Can et al. 2021). Although all these infections are rarely fatal, there are also occasional reports of alarming deadly bacterial infections of susceptible human individuals following unintended honeybee stinging (Liang et al. 2021; Truskinovsky et al. 2001).
We therefore aimed to investigate whether accidental contact through stinging with honeybees could indeed represent a microbial hazard for humans. Wasps and hornets were also included in the experiments for comparative purposes. The latter are predaceous carnivores that live on other insects and sweet substances, such as sap and nectar, in contrast to honeybees that are herbivorous and live on nectar and pollen (Fitzgerald and Flood 2006). More specifically, our main goal was to detect whether some clinically important bacteria for human (i.e., Staphylococcus aureus, Streptococcus pyogenes, Enterococcus faecalis/faecium, and Pseudomonas aeruginosa) could be transmitted during the stinging procedure by these three important Hymenoptera insects (i.e., honeybees, wasps, and hornets). A series of four complementary stinging simulation experiments was performed.
Materials and methods
Collection of insects
A total of 288 honeybees (Apis mellifera), 252 wasps (Vespula germanica), and 36 hornets (Vespa orientalis) were collected from different environments, i.e., countryside, agricultural lands, and towns, in Lemnos Island, north-eastern Greece, during the period of April-May 2019. Special care was taken to increase the variety of the sampling sites (n = 9), by visiting various regions throughout the island, including both urban and rural areas. In addition, an equal number of insects of both categories (i.e., honeybees, vespids) were collected from each sampling site to minimize any unwanted bias that could arise due to the potential inhomogeneous environmental microbiota of each ecological niche. The numbers of each insect species inevitably differed due to the higher population density of honeybees and wasps compared to hornets at the sampling period (end of spring). The insects (n = 576) were caught and anesthetized by inhalation of carbon dioxide and directly transported to the Laboratory of Food Microbiology and Hygiene (LFMH; Department of Food Science and Nutrition, Lemnos), where they were individually placed in sterile Eppendorf tubes and temporarily stored at 4 °C for the stinging simulation experiments later the same day (see next section).
Stinging simulation experiments
For each insect, experiments were conducted in four different but complementary ways, trying to thoroughly simulate the stinging procedure: (i) live insects were forced to sting directly (Fig. 1A and B) each nutrient substrate (x4); (ii) live insects were forced to sting sterilized leather as a human skin simulation (Fig. 1C), and immediately afterward, their sting apparatus was transferred to each nutrient substrate (x4); (iii) the third pair of insects’ legs (back legs), which are always in contact with insects’ victims during stinging incidents, were aseptically removed and were attached to each nutrient substrate (x4); and (iv) the whole anesthetized insect was inserted in a tube with saline solution (1 mL for honeybees and wasps, 5 mL for hornets; quarter-strength Ringer’s solution; Lab M, Heywood, Lancashire, UK), vortexed for 1 min at medium intensity (using a mini vortex mixer, VXMNAL; OHAUS Europe GmbH, Nänikon, Switzerland), and then an aliquot (100 μL) of the resulting suspension was transferred to each nutrient substrate and spread (x5; including the four selective agar media plus a general purpose one; see next section). It should be noted that the second approach (concerning leather stinging) was followed only for the honeybees since the sting apparatus of wasps and hornets is not removed from their body after stinging. For each replicate of each stinging simulation experiment (x4), a different insect was always used. This is because a given honeybee cannot sting twice, while in addition, a second successive stinging by a wasp or hornet would hinder our purpose which was mostly to check potential bacterial transfer from an “unused” sting. Αll experimental procedures were applied under aseptic conditions, using sterile forceps, and working near the flame of a Bunsen burner.
Fig. 1.
Stinging simulation by wasps V. germanica (A), hornets V. orientalis (B), and honeybees A. mellifera (C)
Nutrient substrates used to isolate the target bacteria
Following each stinging simulation experiment (x4), four different types of selective agar media were used as the nutrient substrates to isolate the target bacteria, depending on the species. Thus, staphylococci were isolated on Baird Parker (BP) agar supplemented with Egg Yolk Tellurite (EYT), streptococci on blood agar supplemented with streptococcus selective supplement (COBA), enterococci on Kanamycin Aesculin Azide (KAA) agar, and pseudomonads on Pseudomonas Cephalothin, Fucidin, and Cetrimide (CFC) agar. BP, KAA, and CFC agars, together with their supplements, were all purchased from Lab M, while ready prepared streptococcal selective agar (COBA) plates (90 mm) were used that were provided by Oxoid (Thermo Fisher Scientific Inc, Microbiology, Perth, UK). Following inoculation of the plates, all the media were incubated aerobically at 37 °C for 24 h inside an incubator (Velp Scientifica™ FOC 215L; Fisher Scientific, Leicestershire, UK), except BP agar for which incubation was extended to 48 h (to obtain sufficiently grown colonies). Tryptone Soya Agar (TSA; Oxoid) was also surface inoculated as a general-purpose medium (i.e., in addition to the four selective agar media) in those stinging stimulation experiments where the whole anesthetized insects were individually subjected to vortexing (type iv; see “Stinging simulation experiments”). This was incubated at 30 °C for 72 h.
Recovery of microbial isolates and identification of bacterial species
Following incubation of the plates, some representative colonies were isolated from all those showing visible growth. To achieve maximum possible diversity, for plates harboring up to three colonies, all colonies were recovered, whereas for plates with more colonies, a selection was made about which to isolate based on morphology (i.e., shape, size, color, and texture). More specifically, from all the different types of stinging simulation experiments, a total of 77 colonies were recovered, of which 30 were isolated from the honeybee experiments and the other 47 from the vespids. All isolates were sub-cultured through streaking on TSA to confirm their purity, and a freshly prepared colony of each one was also subjected to Gram staining. Only those isolates that were shown to be bacteria following microscopic observation (n = 64) were subjected to catalase reaction and then proceeded to identification, using one of the following five approaches: (i) PCR amplification of 16S rRNA genes and sequencing using primer pairs either 63F/1542R or 27F/1518R and conditions as described by Galkiewicz and Kellogg (2008) and Giovannoni (1991), respectively (this was applied for 37 isolates from wasps and 5 isolates from hornets); (ii) PCR amplification of gyrase B (gyrB) gene and sequencing using primer pair UP-1/UP-2r and conditions as described by Yamamoto and Harayama (1995) (this was applied for 7 bacilli isolates from honeybees); (iii) PCR amplification of rpoB gene and sequencing using primer pair RpoB-F/RpoB-R and conditions as described by Hoffmann and Roggenkamp (2003) (this was applied for 5 Enterobacter spp. isolates from wasps); (iv) API® biochemical tests (using API®-Staph strips for 7 staphylococci/micrococci isolates and API®-20E strips for 1 Enterobacter sp. isolate, all from honeybees); and (v) matrix-assisted laser desorption-ionization time-of-flight mass spectrometry (MALDI-TOF MS) analysis as described by Jang and Kim (2018) (this was applied approach for 2 Paenibacillus spp. isolates from honeybees). These five different approaches were followed in our efforts to identify the species of each bacterial isolate since this was not possible for all the isolates through the 16S rRNA gene sequencing.
All PCRs were done using the Kapa Taq PCR Kit (KK1016, 500 U; Kapa Biosystems, Wilmington, MA, USA) on a FastGene® 96-well Ultracycler (FG-TC01 Gradient version; NIPPON Genetics EUROPE GmbH, Düren, Germany). All primer sequences that were here used are provided in Table 1. Before Sanger sequencing at the Institute of Applied Biosciences (CERTH, Thessaloniki, Greece), amplicons were purified using the NucleoSpin® Gel and PCR Clean-up Kit (MACHEREY-NAGEL GmbH & Co. KG, Düren, Germany). API® biochemical tests were done following the instructions of the manufacturer (bioMérieux SA, Marcy-l’Étoile, France), while MALDI-TOF MS analysis was executed on an external collaborating facility (Bioiatriki Healthcare Group, Athens, Greece). All identified isolates were long-term stored in the collection of LFMH at − 80 °C in cryovials containing Brain Heart Infusion (BHI) broth (Lab M), also supplied with 15% (v/v) glycerol.
Table 1.
Primer sequences used in this study, together with their lengths (nt), GC contents (%), and references
| Name | Sequence (5′-->3′) | Length (nt) | GC (%) | Reference |
|---|---|---|---|---|
| 63F | CAG GCC TAA CAC ATG CAA GTC | 21 | 52.4 | Galkiewicz and Kellogg (2008) |
| 1542R | AAG GAG GTG ATC CAG CCG CA | 20 | 60.0 | |
| 27F | AGA GTT TGA TCM TGG CTC AG | 20 | 47.5 | Giovannoni (1991) |
| 1518R | AAG GAG GTG ATC CAN CCR CA | 20 | 55.0 | |
| UP-1 | GAA GTC ATC ATG ACC GTT CTG CAY GCN GGN GGN AAR TTY GA | 41 | 51.2 | Yamamoto and Harayama (1995) |
| UP-2r | AGC AGG GTA CGG ATG TGC GAG CCR TCN ACR TCN GCR TCN GTC AT | 44 | 59.1 | |
| RpoB-F | AAC CAG TTC CGC GTT GGC CTG G | 22 | 63.6 | Hoffmann and Roggenkamp (2003) |
| RpoB-R | CCT GAA CAA CAC GCT CGG A | 19 | 57.9 |
Results and discussion
It is generally acknowledged that the microbial ecology of the environment of an insect, together with its nutritional and social habits, are parameters that shape its microbiome, both externally and internally. That microbiome influences in turn the overall insect health and productivity (including nutrient acquisition, metabolism, growth and development, and immune performance), as well as its potential to contaminate other hosts with the microorganisms it carries (Khan et al. 2020; Kwong et al. 2017). In this research, we focused on that part of the culturable microbiota of honeybees and vespids (including both wasps and hornets) that could be transmitted during the stinging procedure. This was done to investigate whether that could harbor pathogenic bacteria for human and more specifically whether some clinically important bacterial species (i.e., S. aureus, S. pyogenes, E. faecalis/faecium, and P. aeruginosa) could be transmitted during stinging. The choice of the four target bacterial species was based on their known involvement in potentially serious human infections. Thus, both S. aureus and S. pyogenes are part of the human skin microbiota and are also found in the upper respiratory tract (Egert and Simmering 2016; Byrd et al. 2018). While S. aureus is a frequent member of skin microbiota, with 20–30% of humans being long-term carriers, S. pyogenes is an infrequent pathogenic species being still carried by the 1–5% of the healthy individuals. Both species can cause important human infections, ranging from mild superficial skin infections to life-threatening systemic diseases (Cheung et al. 2021; Walker et al. 2014). Enterococcus faecalis and E. faecium are both commensal bacteria inhabiting the gastrointestinal (GI) tract of healthy humans and other mammals but can also cause life-threatening infections in immunocompromised individuals (García-Solache and Rice 2019). P. aeruginosa is also an important opportunistic pathogen recognized for its widespread occurrence, multidrug resistance against many antibiotics, and its frequent involvement in many hospital-acquired infections (Horcajada et al. 2019).
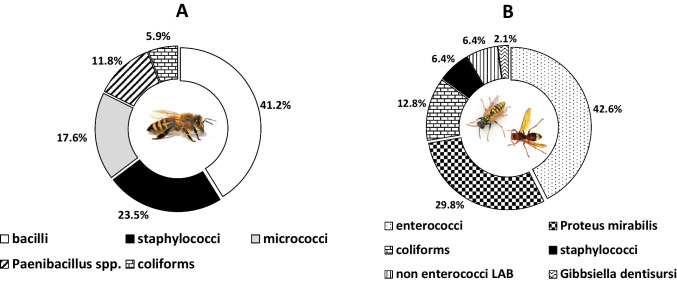
To stimulate the stinging procedure, four complementary approaches were followed. Thus, the surfaces of both sting apparatus and back legs, which are always in contact with insects’ victims during stinging incidents, as well as the whole insect, were surveyed for their potential carriage of the target bacterial species through a culture-based procedure using mainly selective agar media. A total of 77 microbial isolates were successfully recovered, of which 13 were identified by microscopic observation as yeasts. It is worth noting that all these yeast isolates were recovered from the honeybee experiments. Νo yeast isolate was recovered from the wasp and hornet experiments, although these sugar-demanding eukaryotic microorganisms may also colonize these insects (Jimenez et al. 2017; Madden et al. 2018). Thus, all 47 microbial isolates recovered from the vespids were bacteria (5 were isolated from hornets, 42 from wasps), together with the 17 bacterial isolates recovered from honeybees. The identification results of these 64 bacterial isolates are presented in Table 2, while their presence ratios for each insect type (honeybees/vespids) are recorded separately in Fig. 2. Interestingly, for honeybees, none of the identified bacterial species belonged to any of the target clinically important bacteria. Honeybees were found to harbor mainly bacilli, staphylococci, and micrococci on their surveyed microbiota, with these three microbial groups consisting of the 82.4% of bacteria isolated from these insects (n = 14/17). On the other hand, enterococci (of which 9 E. faecalis isolates), P. mirabilis, and coliforms constituted the 85.1% of bacteria isolated from wasps and hornets (n = 40/47), suggesting the contact of these insects with fecal origin materials.
Table 2.
Bacterial isolates recovered from honeybees (A. mellifera), wasps (V. germanica), and hornets (V. orientalis)
| s/n | Isolate code | Bacterial genus/species | Gram staining | Catalase reaction | Insect species (host) | Stinging simulation experimenta | Nutrient substrateb | Identif. methodc | NCBI accession numberd |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 3 | Bacillus cereus group | + | + | A. mellifera | iv | TSA | ii | OP009593 |
| 2 | 4 | Bacillus cereus group | + | + | A. mellifera | iv | TSA | ii | OP009594 |
| 3 | 5 | Bacillus cereus group | + | + | A. mellifera | iv | TSA | ii | OP009595 |
| 4 | 6 | Paenibacillus urinalis | − | + | A. mellifera | iv | TSA | v | - |
| 5 | 7 | Paenibacillus pabuli | − | + | A. mellifera | iv | TSA | v | - |
| 6 | 8Α | Staphylococcus capitis | + | + | A. mellifera | iv | TSA | iv | - |
| 7 | 8Β | Bacillus cereus group | + | + | A. mellifera | iv | TSA | ii | OP009596 |
| 8 | 9 | Bacillus cereus group | + | + | A. mellifera | iv | TSA | ii | OP009597 |
| 9 | 10 | Bacillus cereus group | + | + | A. mellifera | iv | TSA | ii | OP009598 |
| 10 | 14 | Enterobacter cloacae | − | + | A. mellifera | ii | CFC | iv | - |
| 11 | 15 | Micrococcus luteus | + | + | A. mellifera | ii | COBA | iv | - |
| 12 | 18 | Staphylococcus cohnii | + | + | A. mellifera | i | BP | iv | - |
| 13 | 22 | Staphylococcus warneri | + | + | A. mellifera | iii | BP | iv | - |
| 14 | 27 | Micrococcus luteus | + | + | A. mellifera | iii | COBA | iv | - |
| 15 | 28 | Bacillus cereus group | + | + | A. mellifera | iii | COBA | ii | OP009599 |
| 16 | 31 | Staphylococcus warneri | + | + | A. mellifera | i | ΒP | iv | - |
| 17 | 32 | Micrococcus luteus | + | + | A. mellifera | i | COBA | iv | - |
| 18 | 33 | Enterococcus sp. | + | − | V. germanica | iii | TSA | i | ON994583 |
| 19 | 34 | Enterococcus mundtii | + | − | V. germanica | iii | TSA | i | ON994584 |
| 20 | 35 | Enterococcus sp. | + | − | V. germanica | iii | TSA | i | ON994585 |
| 21 | 36 | Enterococcus sp. | + | − | V. germanica | iii | TSA | i | ON994586 |
| 22 | 37 | Enterococcus sp. | + | − | V. germanica | iii | TSA | i | ON994587 |
| 23 | 38 | Staphylococcus epidermidis | + | + | V. germanica | iv | ΒP | i | ON994588 |
| 24 | 39 | Enterococcus sp. | + | − | V. germanica | iii | KAA | i | ON994589 |
| 25 | 40 | Enterococcus sp. | + | − | V. germanica | iii | KAA | i | ON994590 |
| 26 | 41 | Enterococcus mundtii | + | − | V. germanica | iii | COBA | i | ON994591 |
| 27 | 42 | Enterococcus mundtii | + | − | V. germanica | iii | COBA | i | ON994592 |
| 28 | 44 | Proteus mirabilis | − | + | V. orientalis | iv | TSA | i | ON994593 |
| 29 | 46 | Proteus mirabilis | − | + | V. orientalis | iv | TSA | i | ON994594 |
| 30 | 47 | Gibbsiella dentisursi | − | + | V. orientalis | iv | TSA | i | ON994595 |
| 31 | 48 | Enterococcus faecalis | + | − | V. orientalis | iv | KAA | i | ON994596 |
| 32 | 49 | Staphylococcus sp. | + | + | V. orientalis | i | KAA | i | ON994597 |
| 33 | 50 | Staphylococcus sp. | + | + | V. germanica | iv | TSA | i | ON994598 |
| 34 | 53 | Enterobacter cancerogenus | − | + | V. germanica | iv | TSA | iii | OP009588 |
| 35 | 54 | Enterococcus faecalis | + | − | V. germanica | iv | TSA | i | ON994599 |
| 36 | 55 | Enterobacter cancerogenus | − | + | V. germanica | iv | CFC | iii | OP009589 |
| 37 | 56 | Enterobacter cancerogenus | − | + | V. germanica | iv | CFC | iii | OP009590 |
| 38 | 57 | Enterobacter cancerogenus | − | + | V. germanica | iv | CFC | iii | OP009591 |
| 39 | 58 | Enterobacter sp. | − | + | V. germanica | i | CFC | iii | OP009592 |
| 40 | 59Α | Enterococcus faecalis | + | − | V. germanica | i | KAA | i | ON994600 |
| 41 | 59Β | Klebsiella sp. | − | + | V. germanica | i | KAA | i | ON994601 |
| 42 | 60 | Proteus mirabilis | − | + | V. germanica | iii | CFC | i | ON994602 |
| 43 | 61 | Proteus mirabilis | − | + | V. germanica | iii | CFC | i | ON994603 |
| 44 | 62 | Proteus mirabilis | − | + | V. germanica | iii | CFC | i | ON994604 |
| 45 | 63 | Proteus mirabilis | − | + | V. germanica | iii | CFC | i | ON994605 |
| 46 | 64 | Proteus mirabilis | − | + | V. germanica | iii | CFC | i | ON994606 |
| 47 | 65 | Proteus mirabilis | − | + | V. germanica | iii | CFC | i | ON994607 |
| 48 | 66 | Proteus mirabilis | − | + | V. germanica | iii | CFC | i | ON994608 |
| 49 | 67 | Proteus mirabilis | − | + | V. germanica | iii | CFC | i | ON994609 |
| 50 | 68 | Proteus mirabilis | − | + | V. germanica | iii | COBA | i | ON994610 |
| 51 | 69 | Enterococcus gallinarum† | − | + | V. germanica | iii | COBA | i | ON994611 |
| 52 | 70 | Lactococcus lactis† | + | − | V. germanica | iii | COBA | i | ON994612 |
| 53 | 71 | Enterococcus faecalis | + | − | V. germanica | iii | COBA | i | ON994613 |
| 54 | 72 | Proteus mirabilis | − | + | V. germanica | iii | COBA | i | ON994614 |
| 55 | 73 | Enterococcus gallinarum† | + | − | V. germanica | iii | COBA | i | ON994615 |
| 56 | 74 | Lactococcus lactis | + | − | V. germanica | iii | COBA | i | ON994616 |
| 57 | 75 | Enterococcus faecalis | + | − | V. germanica | iii | COBA | i | ON994617 |
| 58 | 76 | Enterococcus faecalis | + | − | V. germanica | iii | KAA | i | ON994618 |
| 59 | 77 | Enterococcus faecalis | + | − | V. germanica | iii | KAA | i | ON994619 |
| 60 | 78 | Lactiplantibacillus plantarum | + | − | V. germanica | iii | KAA | i | ON994620 |
| 61 | 79 | Proteus mirabilis | − | + | V. germanica | iii | KAA | i | ON994621 |
| 62 | 81 | Enterococcus faecalis | + | − | V. germanica | iii | KAA | i | ON994622 |
| 63 | 82 | Proteus mirabilis | − | + | V. germanica | iii | KAA | i | ON994623 |
| 64 | 83 | Enterococcus faecalis | + | − | V. germanica | iii | KAA | i | ON994624 |
aSee “Stinging simulation experiments”
bsee “Nutrient substrates used to isolate the target bacteria”
csee “Recovery of microbial isolates and identification of bacterial species”
daccession numbers of nucleotide sequences at National Center for Biotechnology Information (NCBI) databases (https://www.ncbi.nlm.nih.gov/)
†The primer pair 27F/1518R was used for the amplification and sequencing of the 16S rRNA genes
Fig. 2.
Ratios of bacterial isolates recovered from honeybees (A) and wasps and hornets (B)
Regarding honeybee’s microbiota, species belonging to the Bacillus genus are ubiquitous in nature and especially in soil and dust where they can differentiate themselves into oval endospores, being able to remain in this metabolically dormant state for years (Checinska et al. 2015). The species Bacillus anthracis, B. cereus, and Bacillus thuringiensis are phylogenetically closely related and together with some others that are less well studied (e.g., Bacillus mycoides, Bacillus pseudomycoides, Bacillus cytotoxicus etc.) belong to the so-called B. cereus group which contains species with pathogenic potential (Ehling-Schulz et al. 2019). Thus, B. anthracis is the etiologic agent of anthrax, a disease with a severity that varies depending on the host and route of infection. This can infect all mammals, some birds, and possibly even reptiles, but it primarily affects ruminants because of their frequent environmental exposure to that pathogen (Pilo and Frey 2018). Humans may be infected by ingesting spores found in foods (such as meat from infected animals), but today, most naturally acquired human cases of anthrax are cutaneous infections which are provoked following contact with infected animals or their products. B. cereus is commonly recognized as a foodborne pathogen, being able to produce toxins both upon its growth in foods and also inside the human gut, while strains of this species can also cause localized wound and eye infections, as well as systemic disease particularly in immunosuppressed individuals, intravenous drug users, and neonates, due to their ability to produce several tissue-destructive exoenzymes (Enosi Tuipulotu et al. 2021). B. thuringiensis strains are commonly known as entomopathogens, being able to produce potent species-specific insecticidal toxins, and for this reason have been commercialized for use as biopesticides in agriculture and for developing genetically modified cultivars (Palma et al. 2014). However, there are also reports documenting the ability of this microorganism to cause various infectious diseases in immunocompromised human individuals due to its ability to produce various virulence factors acting against mammalian cells (Celandroni et al. 2014). It should be noted that the differentiation of the species of the B. cereus group is really challenging (Ehling-Schulz et al. 2019), while in our case, this was not possible by either of the two approaches we followed (full sequencing of both the 16S rRNA and gyrB genes).
The three Staphylococcus species recovered from honeybees (S. capitis, S. cohnii, and S. warneri) are all coagulase-negative (CoN) that are not normally dangerous to healthy people as skin commensals. However, all these have been implicated in infective endocarditis and bacteremia, both consisting of serious infections in immunocompromised individuals (Diaconu et al. 2019; Soldera et al. 2013; Thakker et al. 2021), while in neonates, they are included in the most common endemic nosocomial pathogens (Michels et al. 2021). This is in accordance with the general belief that CoN could be dangerous to sensitive population groups, such as the neonates, elderly, and immunocompromised, when these bacteria succeed to enter the body (Natsis and Cohen 2018). The same is also true for Micrococcus luteus, an obligate aerobic catalase-positive Gram-positive bacterium that can also cause infections in susceptible people (Martín Guerra et al. 2019; Seifert et al. 1995) and has been shown to survive in oligotrophic environments for extended periods of time (Greenblatt et al. 2004).
The two Paenibacillus isolates recovered from honeybees (i.e., P. urinalis and P. pabuli) belong to a genus with species that were formerly included in Bacillus. Like the latter, this contains spore-former species that are isolated from a variety of environments, most of them being found in soil and often associated with plant roots where they can promote plant growth via nitrogen fixation, phosphate utilization, iron acquisition, and phytohormone production (Grady et al. 2016). Some species (such as Paenibacillus larvae, Paenibacillus apiarius, and Paenibacillus glabratella) are pathogens to honeybees and other invertebrates; others are opportunistic pathogens for humans, rarely tending to cause infections to immunocompromised people, with P. urinalis being the first time isolated from a human urine sample in 2008 (Roux et al. 2008).
Bacteria of the Enterobacter genus belonging to the Enterobacteriaceae family are facultative anaerobic, Gram-negative, and widely distributed in nature and encountered in various environmental habitats. To date, that genus comprises more than twenty species that are found in soil, water, sewage, and plants (as either endophytic or phytopathogens for various plant species) and are also commensals of the animal and human gut (Davin-Regli et al. 2019). Alarmingly, in recent decades, Enterobacter spp. have emerged as important nosocomial pathogens mainly for immunocompromised patients hosted in intensive care units (ICUs). Indeed, these pathogens are included in the so-called ESKAPE group (E. faecium, S. aureus, Klebsiella pneumoniae, Acinetobacter baumannii, P. aeruginosa, and Enterobacter species), which comprises bacterial species that are leading etiological agents of resistant and life-threatening nosocomial infections (Chang et al. 2022). E. cloacae was the sole species isolated from the honeybees (one isolate), while five other Enterobacter isolates were recovered from wasps, of which the four belonged to E. cancerogenus species. Like other Enterobacter, E. cloacae is ubiquitous in terrestrial and aquatic environments and is also an important nosocomial (opportunistic) pathogen that can be responsible for bacteremia, endocarditis, septic arthritis, osteomyelitis, and several other infections. The skin and the GI tract are the sites through which this bacterium is most commonly acquired, while it is worth noting that Enterobacter spp. in general are responsible for approximately 5% of nosocomial bacteremia cases (Mezzatesta et al. 2012).
Twenty enterococci isolates were recovered from vespids comprising the 42.6% of bacteria isolated from these insects, of which nine were E. faecalis, three E. mundtii, and two E. gallinarum. It was not possible to identify the other six Enterococcus spp. isolates to species level (through the 16S rRNA gene sequencing approach here followed). In general, enterococci are Gram-positive bacteria that can survive in conditions unfavorable to other bacteria (García-Solache and Rice 2019) and are frequently employed as hygienic indicators, with their presence in foods and water denoting fecal contamination (Boehm and Sassoubre 2014). These are commensals of the GI tract of many metazoans, from insects to humans. Although they normally do not cause disease once found in that habitat, they can become pathogenic when they infect sites outside the intestine (Yuen and Ausubel 2014). Recently, enterococci have gained attention as important nosocomial pathogens, with E. faecalis and E. faecium being the two species causing the majority of human enterococcal infections (Yuen and Ausubel 2014). Alarmingly, both species present intrinsic resistance to several common antibiotics (such as cephalosporins and aminoglycosides) but can also readily acquire further resistance to other antibiotics, due to plasticity in their genomes (García-Solache and Rice 2019). More specifically, E. faecalis is well adapted to persist in multiple niches within the mammalian host and can quickly adjust its metabolism to survive in new environments, something which, together with the arsenal of strategies it possesses to confront innate and adaptive immune mechanisms, enable it to cause infections in many sites within the host including the GI tract, urinary tract, wounded epithelium, heart, and blood (Kao and Kline 2019).
Proteus mirabilis was the most abundant species that was isolated from vespids, with a total of 14 isolates recovered from wasps and hornets (consisting of the 29.8% of the isolates). It is a Gram-negative rod-shaped proteolytic bacterium belonging to the Enterobacteriaceae family and in humans is an opportunistic pathogen well-known for its swarming motility and ability to produce urease, often causing catheter-associated urinary tract infections (UTIs) (Armbruster et al. 2018). P. mirabilis can be encountered in various environments, including soil, water, and sewage, but is generally a commensal of the GI tracts of humans and animals. However, although commensal, P. mirabilis is equipped with a shocking arsenal of virulence factors and is thus capable of causing a variety of human infections, with bacteremia and sepsis provoked by this bacterium presenting a high mortality rate. Alarmingly, the existence of multidrug-resistant isolates further complicates the therapeutic treatment options (Girlich et al. 2020). In general, Proteus spp. bacteria, like enterococci and coliforms, are often regarded as indicators of fecal pollution of the environments, with human and animal feces being probably their source in those environments (Drzewiecka 2016).
The third most isolated bacterial group from vespids was coliforms which are also considered strong indicators of fecal contamination (Mishra et al. 2018). Thus, five Enterobacter spp. isolates and one Klebsiella sp. isolate were recovered from wasps (12.8% of the isolates). Like most other coliforms, Klebsiella species belong to the Enterobacteriaceae family. They are found everywhere in nature and considered opportunistic pathogens able to cause a wide range of diseases, particularly among those with weakened immune systems, such as neonates, elderly, and immunocompromised individuals, including pneumonia, UTIs, bloodstream infections, and sepsis (Bengoechea and Sa Pessoa 2019). Alarmingly, the increasing isolation of multidrug-resistant strains, such as those belonging to the K. pneumoniae species, significantly hampers the therapeutic options for the treatment of these infections (Navon-Venezia et al. 2017).
Three staphylococci, of which one is S. epidermidis (also included in CoN species previously reported); three lactic acid bacteria (LAB), including two Lactococcus lactis and one Lactiplantibacillus plantarum; and Gibbsiella dentisursi were the remaining bacterial species isolated from wasps and hornets. In general, LAB comprises a group of generally recognized as safe (GRAS) bacteria that are found in decomposing plants, human GI, and vaginal microflora and used for centuries in food fermentations (Raman et al. 2022). A few members of this group are also included in the core gut microbiota of social bees (Kwong and Moran 2016). G. dentisursi is a largely unknown, rod-shaped, Gram-negative species of the Enterobacteriaceae family that was first isolated from the oral cavity of a bear in Japan (Saito et al. 2012).
Conclusions
Although the target pathogenic bacterial species (i.e., S. aureus, S. pyogenes, E. faecalis/faecium, and P. aeruginosa) were not recovered from honeybees, the isolation from these insects of some important opportunistic pathogens (such as E. cloacae and CoN staphylococci), together with potentially pathogenic species of the B. cereus group, does not in theory exclude the possibility of a honeybee sting being fatal to someone with a weakened immune system. However, to the best of our knowledge, such opportunistic pathogens have never been described as etiological agents of infections caused following bee stinging. In addition, it would be probably rather impossible to not isolate at all opportunistic bacterial pathogens from an organism living free in the environment, such as the honeybee, also considering that most bacteria, if not all microorganisms, may cause infections under certain (opportunistic) conditions. On the other hand, most of the suspect colonies isolated from wasps and hornets belonged to important hygienic indicators (i.e., enterococci, P. mirabilis, and coliforms), suggesting that these insects were in contact with fecal origin materials. The simultaneous isolation from these insects of some other important opportunistic pathogenic bacterial species, such as Enterobacter spp. (10.6% of isolates) and Klebsiella spp., also known for multidrug resistance, might cause serious concern. Future studies could be conducted employing a larger sample of insects and preferably metagenomic approaches to try to fully unravel the total (i.e., not only the culturable) sting microbiota of the most important Hymenoptera species, such as the ones already performed on the gut microbiota of some of them.
Acknowledgements
We thank the President of the Agricultural and Apicultural Cooperative of Lemnos Mr. Dimitrios Palaiologos, who helped us during insects’ collection. We thank the personnel from Bioiatriki Healthcare Group (Athens, Greece) who identified the two Paenibacillus spp. isolates through MALDI-TOF MS analysis. We also thank Dr. Stephen Fleming for editing the manuscript. Alexandros Papachristoforou’s research is supported by Vita Bee Health.
Author contribution
Ioanna Gkitsaki: methodology, validation, formal analysis, and investigation. Alexandros Papachristoforou: conceptualization, methodology, investigation, resources, wring, review, editing, and visualization. Sofia Michailidou: methodology, validation, investigation, data curation, wring, review, and editing. Nikolaos Karamvalis: investigation. Ioannis Iliadis: investigation. Dimitra Graikini: investigation. Christina Sakarikou: methodology, investigation, wring, review, and editing. Evangelos Tsoukis: methodology, investigation, wring, review, and editing. Anagnostis Argyriou: methodology, resources, wring, review, and editing. Efstathios Giaouris: methodology, formal analysis, investigation, resources, writing original draft, wring, review, editing, visualization, supervision, and project administration.
Funding
Open access funding provided by HEAL-Link Greece.
Data availability
The data presented in this study are available on reasonable request from the corresponding author.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors have read and agreed to the published version of the manuscript.
Conflict of interest
The authors declare no conflict of interest.
Footnotes
The original online version of this article was revised: The authors regret that an unnecessary sentence** was embedded at the beginning of the results and discussion section.
** “We refer to 3 different parameters:
(i) microbial ecology of the environment
(ii) nutritional habits
(iii) social habits”
The original article has been corrected.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
2/24/2023
A Correction to this paper has been published: 10.1007/s10123-023-00342-4
Contributor Information
Alexandros Papachristoforou, Email: alpapach@agro.auth.gr.
Efstathios Giaouris, Email: stagiaouris@aegean.gr.
References
- Adams KE, Tracy JM, Golden DBK. Anaphylaxis to stinging insect venom. Immunol Allergy Clin North Am. 2022;42:161–173. doi: 10.1016/j.iac.2021.09.003. [DOI] [PubMed] [Google Scholar]
- Anderson CR, Jenkins D, Tron V, Prendiville JS. Wells’ syndrome in childhood: case report and review of the literature. J Am Acad Dermatol. 1995;33:857–864. doi: 10.1016/0190-9622(95)90423-9. [DOI] [PubMed] [Google Scholar]
- Armbruster CE, Mobley HLT, Pearson MM. Pathogenesis of Proteus mirabilis infection. Eco Sal Plus. 2018;8:10.1128/ecosalplus.ESP-0009-2017. doi: 10.1128/ecosalplus.ESP-0009-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengoechea JA, Sa Pessoa J. Klebsiella pneumoniae infection biology: living to counteract host defences. FEMS Microbiol Rev. 2019;43:123–144. doi: 10.1093/femsre/fuy043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm AB, Sassoubre LM. Enterococci as indicators of environmental fecal contamination. In: Gilmore MS, Clewell DB, Ike Y, Shankar N, editors. Enterococci: from commensals to leading causes of drug resistant infection [Internet] Boston: Massachusetts Eye and Ear Infirmary; 2014. [PubMed] [Google Scholar]
- Branstetter MG, Childers AK, Cox-Foster D, Hopper KR, Kapheim KM, Toth AL, Worley KC. Genomes of the Hymenoptera. Curr Opin Insect Sci. 2018;25:65–75. doi: 10.1016/j.cois.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd AL, Belkaid Y, Segre JA. The human skin microbiome. Nat Rev Microbiol. 2018;16:143–155. doi: 10.1038/nrmicro.2017.157. [DOI] [PubMed] [Google Scholar]
- Can C, Yazicioglu M, Gokalp S, Ozkayin N. Parvovirus infection in a child presenting with erythema multiforme and vasculitis after a yellow jacket bee sting. J Trop Pediatr. 2021;67:fmaa043. doi: 10.1093/tropej/fmaa043. [DOI] [PubMed] [Google Scholar]
- Carpena M, Nuñez-Estevez B, Soria-Lopez A, Simal-Gandara J. Bee venom: an updating review of its bioactive molecules and its health applications. Nutrients. 2020;12:3360. doi: 10.3390/nu12113360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celandroni F, Salvetti S, Senesi S, Ghelardi E. Bacillus thuringiensis membrane-damaging toxins acting on mammalian cells. FEMS Microbiol Lett. 2014;361:95–103. doi: 10.1111/1574-6968.12615. [DOI] [PubMed] [Google Scholar]
- Chang RYK, Nang SC, Chan HK, Li J. Novel antimicrobial agents for combating antibiotic-resistant bacteria. Adv Drug Deliv Rev. 2022;187:114378. doi: 10.1016/j.addr.2022.114378. [DOI] [PubMed] [Google Scholar]
- Checinska A, Paszczynski A, Burbank M. Bacillus and other spore-forming genera: variations in responses and mechanisms for survival. Annu Rev Food Sci Technol. 2015;6:351–369. doi: 10.1146/annurev-food-030713-092332. [DOI] [PubMed] [Google Scholar]
- Cheung GYC, Bae JS, Otto M. Pathogenicity and virulence of Staphylococcus aureus. Virulence. 2021;12:547–569. doi: 10.1080/21505594.2021.1878688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornara L, Biagi M, Xiao J, Burlando B. Therapeutic properties of bioactive compounds from different honeybee products. Front Pharmacol. 2017;8:412. doi: 10.3389/fphar.2017.00412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davin-Regli A, Lavigne JP, Pagès JM. Enterobacter spp.: update on taxonomy, clinical aspects, and emerging antimicrobial resistance. Clin Microbiol Rev. 2019;32:e00002–e00019. doi: 10.1128/CMR.00002-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaconu R, Golumbeanu E, Constantin A, Donoiu I. Native valve endocarditis with Staphylococcus warneri. BMJ Case Rep. 2019;12:e229546. doi: 10.1136/bcr-2019-229546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drzewiecka D. Significance and roles of Proteus spp. bacteria in natural environments. Microb Ecol. 2016;72:741–758. doi: 10.1007/s00248-015-0720-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egert M, Simmering R. The microbiota of the human skin. Adv Exp Med Biol. 2016;902:61–81. doi: 10.1007/978-3-319-31248-4_5. [DOI] [PubMed] [Google Scholar]
- Ehling-Schulz M, Lereclus D, Koehler TM. The Bacillus cereus group: Bacillus species with pathogenic potential. Microbiol Spectr. 2019;7:10.1128/microbiolspec.GPP3-0032-2018. doi: 10.1128/microbiolspec.GPP3-0032-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Seedi H, Abd El-Wahed A, Yosri N, Musharraf SG, Chen L, Moustafa M, Zou X, Al-Mousawi S, Guo Z, Khatib A, Khalifa S. Antimicrobial properties of Apis mellifera’s bee venom. Toxins. 2020;12:451. doi: 10.3390/toxins12070451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enosi Tuipulotu D, Mathur A, Ngo C, Man SM. Bacillus cereus: epidemiology, virulence factors, and host-pathogen interactions. Trends Microbiol. 2021;29:458–471. doi: 10.1016/j.tim.2020.09.003. [DOI] [PubMed] [Google Scholar]
- Fitzgerald KT, Flood AA. Hymenoptera stings. Clin Tech Small Anim Pract. 2006;21:194–204. doi: 10.1053/j.ctsap.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Galkiewicz JP, Kellogg CA. Cross-kingdom amplification using bacteria-specific primers: complications for studies of coral microbial ecology. Appl Environ Microbiol. 2008;74:7828–7831. doi: 10.1128/AEM.01303-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Solache M, Rice LB. The Enterococcus: a model of adaptability to its environment. Clin Microbiol Rev. 2019;32:e00058–e00018. doi: 10.1128/CMR.00058-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni SJ. The polymerase chain reaction. In: Stackebrandt E, Goodfellow M, editors. Sequencing and hybridization techniques in bacterial systematics. New York, NY: John Wiley & Sons; 1991. pp. 177–201. [Google Scholar]
- Girlich D, Bonnin RA, Dortet L, Naas T. Genetics of acquired antibiotic resistance genes in Proteus spp. Front Microbiol. 2020;11:256. doi: 10.3389/fmicb.2020.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady EN, MacDonald J, Liu L, Richman A, Yuan ZC. Current knowledge and perspectives of Paenibacillus: a review. Microb Cell Fact. 2016;15:203. doi: 10.1186/s12934-016-0603-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt CL, Baum J, Klein BY, Nachshon S, Koltunov V, Cano RJ. Micrococcus luteus - survival in amber. Microb Ecol. 2004;48:120–127. doi: 10.1007/s00248-003-2016-5. [DOI] [PubMed] [Google Scholar]
- Hedtke SM, Patiny S, Danforth BN. The bee tree of life: a supermatrix approach to apoid phylogeny and biogeography. BMC Evol Biol. 2013;13:138. doi: 10.1186/1471-2148-13-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann H, Roggenkamp A. Population genetics of the nomenspecies Enterobacter cloacae. Appl Environ Microbiol. 2003;69:5306–5318. doi: 10.1128/AEM.69.9.5306-5318.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horcajada JP, Montero M, Oliver A, Sorlí L, Luque S, Gómez-Zorrilla S, Benito N, Grau S. Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin Microbiol Rev. 2019;32:e00031–e00019. doi: 10.1128/CMR.00031-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang KS, Kim YH. Rapid and robust MALDI-TOF MS techniques for microbial identification: a brief overview of their diverse applications. J Microbiol. 2018;56:209–216. doi: 10.1007/s12275-018-7457-0. [DOI] [PubMed] [Google Scholar]
- Jimenez SI, Carroll C, Babcock T, Derstine N, Hadwin A, Moore M, Gries G. Yeasts harbored by vespine wasps in the pacific northwest. Environ Entomol. 2017;46:217–225. doi: 10.1093/ee/nvw173. [DOI] [PubMed] [Google Scholar]
- Kao PHN, Kline KA. Dr. Jekyll and Mr. Hide: how Enterococcus faecalis subverts the host immune response to cause infection. J Mol Biol. 2019;431:2932–2945. doi: 10.1016/j.jmb.2019.05.030. [DOI] [PubMed] [Google Scholar]
- Khan KA, Al-Ghamdi AA, Ghramh HA, Ansari MJ, Ali H, Alamri SA, Al-Kahtani SN, Adgaba N, Qasim M, Hafeez M. Structural diversity and functional variability of gut microbial communities associated with honeybees. Microb Pathog. 2020;138:103793. doi: 10.1016/j.micpath.2019.103793. [DOI] [PubMed] [Google Scholar]
- Klug R, Immerman R, Giron JA. Bee bite and the toxic shock syndrome. Ann Intern Med. 1982;96:382. doi: 10.7326/0003-4819-96-3-382_1. [DOI] [PubMed] [Google Scholar]
- Kontoyiannis PD, Koons GL, Hicklen RS, Mikos AG, Kontoyiannis DP. Insect bite-associated invasive fungal infections. Open Forum Infect Dis. 2019;6:ofz385. doi: 10.1093/ofid/ofz385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurek-Górecka A, Górecki M, Rzepecka-Stojko A, Balwierz R, Stojko J. Bee products in dermatology and skin care. Molecules. 2020;25:556. doi: 10.3390/molecules25030556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong WK, Medina LA, Koch H, Sing KW, Soh EJY, Ascher JS, Jaffé R, Moran NA. Dynamic microbiome evolution in social bees. Sci Adv. 2017;3:e1600513. doi: 10.1126/sciadv.1600513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong WK, Moran NA. Gut microbial communities of social bees. Nat Rev Microbiol. 2016;14:374–384. doi: 10.1038/nrmicro.2016.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang JH, Liang WH, Deng YQ, Fu ZG, Deng JL, Chen YH. Vibrio vulnificus infection attributed to bee sting: a case report. Emerg Microbes Infect. 2021;10:1890–1895. doi: 10.1080/22221751.2021.1977589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden AA, Epps MJ, Fukami T, Irwin RE, Sheppard J, Sorger DM, Dunn RR. The ecology of insect-yeast relationships and its relevance to human industry. Proc Biol Sci. 2018;285:20172733. doi: 10.1098/rspb.2017.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín Guerra JM, Martín Asenjo M, Rodríguez Martín C. Bacteraemia by Micrococcus luteus in an inmunocompromised patient. Med Clin. 2019;152:469–470. doi: 10.1016/j.medcli.2018.09.011. [DOI] [PubMed] [Google Scholar]
- Martinello M, Mutinelli F. Antioxidant activity in bee products: a review. Antioxidants. 2021;10:71. doi: 10.3390/antiox10010071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezzatesta ML, Gona F, Stefani S. Enterobacter cloacae complex: clinical impact and emerging antibiotic resistance. Future Microbiol. 2012;7:887–902. doi: 10.2217/fmb.12.61. [DOI] [PubMed] [Google Scholar]
- Michels R, Last K, Becker SL, Papan C. Update on coagulase-negative staphylococci-what the clinician should know. Microorganisms. 2021;9:830. doi: 10.3390/microorganisms9040830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra M, Arukha AP, Patel AK, Behera N, Mohanta TK, Yadav D. Multi-drug resistant coliform: water sanitary standards and health hazards. Front Pharmacol. 2018;9:311. doi: 10.3389/fphar.2018.00311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsis NE, Cohen PR. Coagulase-negative Staphylococcus skin and soft tissue infections. Am J Clin Dermatol. 2018;19:671–677. doi: 10.1007/s40257-018-0362-9. [DOI] [PubMed] [Google Scholar]
- Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev. 2017;41:252–275. doi: 10.1093/femsre/fux013. [DOI] [PubMed] [Google Scholar]
- Oršolić N. Bee venom in cancer therapy. Cancer Metastasis Rev. 2012;31:173–194. doi: 10.1007/s10555-011-9339-3. [DOI] [PubMed] [Google Scholar]
- Palma L, Muñoz D, Berry C, Murillo J, Caballero P. Bacillus thuringiensis toxins: an overview of their biocidal activity. Toxins. 2014;6:3296–3325. doi: 10.3390/toxins6123296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilo P, Frey J. Pathogenicity, population genetics and dissemination of Bacillus anthracis. Infect Genet Evol. 2018;64:115–125. doi: 10.1016/j.meegid.2018.06.024. [DOI] [PubMed] [Google Scholar]
- Pucca MB, Cerni FA, Oliveira IS, Jenkins TP, Argemí L, Sørensen CV, Ahmadi S, Barbosa JE, Laustsen AH. Bee updated: current knowledge on bee venom and bee envenoming therapy. Front Immunol. 2019;10:2090. doi: 10.3389/fimmu.2019.02090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman J, Kim JS, Choi KR, Eun H, Yang D, Ko YJ, Kim SJ. Application of lactic acid bacteria (LAB) in sustainable agriculture: advantages and limitations. Int J Mol Sci. 2022;23:7784. doi: 10.3390/ijms23147784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson D, Schmitz JP. Chronic relapsing cervicofacial necrotizing fasciitis: case report. J Oral Maxillofac Surg. 1997;55:403–408. doi: 10.1016/s0278-2391(97)90136-1. [DOI] [PubMed] [Google Scholar]
- Roux V, Fenner L, Raoult D. Paenibacillus provencensis sp. nov., isolated from human cerebrospinal fluid, and Paenibacillus urinalis sp. nov., isolated from human urine. Int J Syst Evol Microbiol. 2008;58:682–687. doi: 10.1099/ijs.0.65228-0. [DOI] [PubMed] [Google Scholar]
- Saito M, Shinozaki-Kuwahara N, Takada K. Gibbsiella dentisursi sp. nov., isolated from the bear oral cavity. Microbiol Immunol. 2012;56:506–512. doi: 10.1111/j.1348-0421.2012.00464.x. [DOI] [PubMed] [Google Scholar]
- Seifert H, Kaltheuner M, Perdreau-Remington F. Micrococcus luteus endocarditis: case report and review of the literature. Zentralbl Bakteriol. 1995;282:431–435. doi: 10.1016/s0934-8840(11)80715-2. [DOI] [PubMed] [Google Scholar]
- Shahar E, Frand M. Pseudomonas arthritis following puncture wounds of the foot. J Pediatr. 1979;95:671. doi: 10.1016/s0022-3476(79)80793-3. [DOI] [PubMed] [Google Scholar]
- Soldera J, Nedel WL, Cardoso PR, d’Azevedo PA. Bacteremia due to Staphylococcus cohnii ssp. urealyticus caused by infected pressure ulcer: case report and review of the literature. Sao Paulo Med J. 2013;131:59–61. doi: 10.1590/s1516-31802013000100010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakker RA, Chatila K, Reynoso D, Karnath B. Native and prosthetic valve Staphylococcus capitis endocarditis: a review of the literature. Cardiol Res. 2021;12:140–145. doi: 10.14740/cr1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truskinovsky AM, Dick JD, Hutchins GM. Fatal infection after a bee sting. Clin Infect Dis. 2001;32:E36–E38. doi: 10.1086/318451. [DOI] [PubMed] [Google Scholar]
- Walker MJ, Barnett TC, McArthur JD, Cole JN, Gillen CM, Henningham A, Sriprakash KS, Sanderson-Smith ML, Nizet V. Disease manifestations and pathogenic mechanisms of Group A Streptococcus. Clin Microbiol Rev. 2014;27:264–301. doi: 10.1128/CMR.00101-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehbe R, Frangieh J, Rima M, El Obeid D, Sabatier JM, Fajloun Z. Bee venom: overview of main compounds and bioactivities for therapeutic interests. Molecules. 2019;24:2997. doi: 10.3390/molecules24162997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Harayama S. PCR amplification and direct sequencing of gyrB genes with universal primers and their application to the detection and taxonomic analysis of Pseudomonas putida strains. Appl Environ Microbiol. 1995;61:1104–1109. doi: 10.1128/aem.61.3.1104-1109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen GJ, Ausubel FM. Enterococcus infection biology: lessons from invertebrate host models. J Microbiol. 2014;52:200–210. doi: 10.1007/s12275-014-4011-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Liu Y, Ye Y, Wang XR, Lin LT, Xiao LY, Zhou P, Shi GX, Liu CZ. Bee venom therapy: potential mechanisms and therapeutic applications. Toxicon. 2018;148:64–73. doi: 10.1016/j.toxicon.2018.04.012. [DOI] [PubMed] [Google Scholar]
- Zirngibl G, Burrows HL. Hymenoptera stings. Pediatr Rev. 2012;33:534–535. doi: 10.1542/pir.33-11-534. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available on reasonable request from the corresponding author.