While prevalent in prokaryotes, physiological function and potential regulation of DNA 6mA modification in eukaryotes have yet to be fully explored. In a new study published in Cell Research, Ma et al. identified METL-9 as a DNA 6mA methyltransferase in Caenorhabditis elegans, and demonstrated that METL-9 could regulate innate immunity through 6mA-dependent and -independent pathways, suggesting new mechanisms for combating pathogen infection in eukaryotes.
N6-methyldeoxyadenosine (6mA) is well characterized as the most abundant DNA modification in many prokaryotes, regulating fundamental processes such as DNA transcription, replication, and repair.1 Since 2015, high levels of 6mA were detected in the genomes of several lower eukaryotes with unique distribution patterns, suggesting regulatory roles of 6mA as an additional DNA mark in these systems.2 In mammals, although the abundance tends to be low, 6mA has been proposed to play diverse functions in diseases, during animal development, and under stress conditions.3 However, recent studies questioned the presence and functions of 6mA in mammals, casting doubts on 6mA detection methods and suggesting that 6mA in mammalian genomic DNA (gDNA) could be products of misincorporation by DNA polymerases.4 Exploring the physiological role(s) of 6mA during different biological processes and characterizing 6mA effector proteins, such as methyltransferases and demethylases, are required to further support the existence of 6mA and understand its functions in eukaryotic systems.
Innate immunity is among first-line responders to defend against various pathogens. Epigenetic elements, including histone modifications and DNA 5-methylcytosine (5mC) modification, have been established to regulate innate immunity in eukaryotic cells.5 However, while 6mA has been well characterized within the restriction-modification systems as a defense mechanism in prokaryotes,1 its potential regulatory function in eukaryotic immune systems remain unexplored.
The study by Ma et al.6 in Cell Research reported that 6mA levels in C. elegans gDNA increased greatly when the animals were infected by bacteria (Fig. 1). The authors also observed a correlation between gDNA 6mA induction level and animal survival time after infection, suggesting a contribution of 6mA to innate immune responses. Ma et al. then applied biotin-labeled DNA oligos that bear previously reported 6mA-containing motifs to pull down proteins from the lysates of C. elegans. They identified METL-9 as a candidate of DNA 6mA methyltransferase. Overexpression of METL-9 increased 6mA levels in C. elegans gDNA, while knockout of metl-9 reduced basal 6mA levels and suppressed the induction of 6mA upon infection. The authors then performed in vitro DNA methylation assay using heterologously expressed C. elegans METL-9 and mouse METTL9 proteins, which validated the enzymatic activity of METL-9 as a 6mA methyltransferase. The in vitro assay revealed that METL-9 preferentially methylates single-stranded DNA, bubbled DNA, and stem-loop DNA, but not RNA and double-stranded DNA. The authors subsequently identified N172, L173, and D274 as important residues for METL9 enzymatic activity. C. elegans harboring N172K and D274G double mutations (metl-9mut) showed lower 6mA levels than wild-type animals under both normal and infected conditions. metl-9 KO and metl-9mut strains displayed diminished immunity and suppressed survival upon infection, suggesting that METL-9 could modulate innate immunity through its catalytic activity.
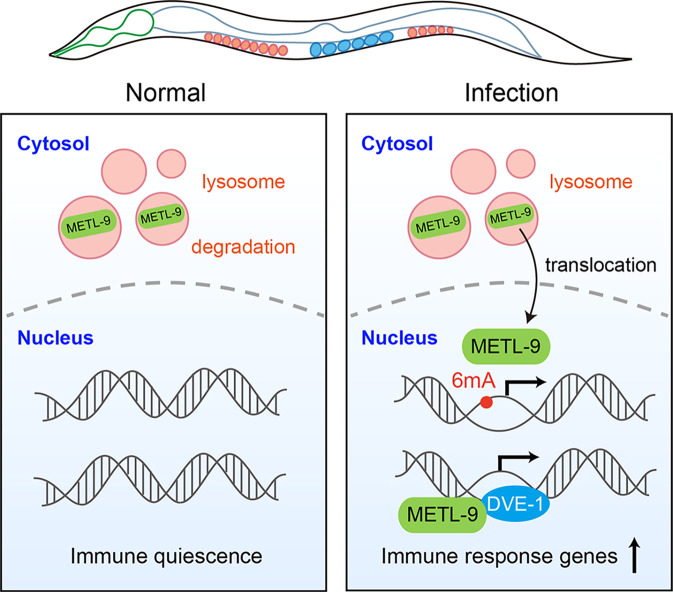
Fig. 1. METL9 modulates innate immune response in C. elegans through 6mA-dependent and -independent pathways.
In the normal condition (left), METL-9 is localized in lysosomes. After pathogen infection, METL-9 undergoes translocation to the cell nucleus and promotes transcription of genes related to immune response. METL-9 can deposit 6mA on DNA and interact with chromatin remodeler protein DVE-1, both contributing to the activation of genes related to immune response.
How could METL-9 and 6mA regulate innate immunity in C. elegans? Ma et al. performed RNA-seq in wild-type, metl-9 KO, and metl-9mut animals with and without pathogen infection. In the wild-type C. elegans, upregulated genes upon infection enrich pathways related to defense and immunity. The induction of these genes was compromised in metl-9 KO and metl-9mut strains. Applying a single-molecule real-time sequencing (SMRT-seq)-based method, the authors mapped 6mA on gDNA and confirmed the upregulation of 6mA in the genomes of the infected animals, especially at exons, promoters, and enhancers. Ma et al. then mutated promoter 6mA sites to guanine bases and found that mutations could suppress the activation of gene expression induced by infection, indicating a positive correlation between gDNA 6mA methylation and transcription. Interestingly, the authors observed that METL-9 was localized to the lysosome under normal conditions, while pathogen infection relocated METL-9 to the cell nucleus and increased the protein level of METL-9. These results suggest that infection may inhibit lysosome-mediated METL-9 degradation to elevate gDNA 6mA methylation and immune responses (Fig. 1).
Notably, Ma et al. found that metl-9mut C. elegans show partial resistance to pathogens compared to metl-9 KO animals, suggesting that METL-9 also regulates innate immunity through 6mA-independent mechanisms. Using protein pull-down followed by mass spectrometry, the authors revealed the DVE-1 protein that could interact with METL-9 upon pathogen infection (Fig. 1). DVE-1 is a known chromatin remodeler that regulates innate immune responses. Ma et al. further showed that the knockdown of dve-1 in the metl-9mut strain could repress pathogen resistance to the level observed in metl-9 KO animals, validating the contribution of the interaction between DVE-1 and METL-9 in modulating innate immunity.
Recent studies have suggested the involvement of 6mA in the response to heat shock and mitochondrial stresses.7–9 This study by Ma et al. provided an example that DNA 6mA can be installed by METL-9 in response to pathogen infection. These findings open up new questions for future investigations. An obvious question is how gDNA 6mA activates transcription and whether there are proteins that recognize the methylation. How cells regulate the translocation of METL-9 from lysosome to nucleus also remains unclear. Another interesting avenue is to explore similar pathways in mammalian systems.
References
- 1.Wion D, Casadesús J. Nat. Rev. Microbiol. 2006;4:183–192. doi: 10.1038/nrmicro1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luo GZ, et al. Nat. Rev. Mol. Cell Biol. 2015;16:705–710. doi: 10.1038/nrm4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boulias K, Greer EL. Nat. Rev. Genet. 2022;23:411–428. doi: 10.1038/s41576-022-00456-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng X, He C. Mol. Cell. 2023;83:343–351. doi: 10.1016/j.molcel.2023.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Q, Cao X. Nat. Rev. Immunol. 2019;19:417–432. doi: 10.1038/s41577-019-0151-6. [DOI] [PubMed] [Google Scholar]
- 6.Ma, C. et al. Cell Res. 10.1038/s41422-023-00826-y (2023).
- 7.Ma C, et al. Nat. Cell Biol. 2019;21:319–327. doi: 10.1038/s41556-018-0238-5. [DOI] [PubMed] [Google Scholar]
- 8.Hao Z, et al. Mol. Cell. 2020;78:382–395. doi: 10.1016/j.molcel.2020.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wan, Q. L. et al. Sci. Adv. 7, eabc3026 (2021).