Abstract
Triple‐negative breast cancer (TNBC) is usually an aggressive disease with a poor prognosis and limited treatment options. The neurotrophic tyrosine receptor kinase (NTRK) gene fusions are cancer type‐agnostic emerging biomarkers approved by the Food and Drug Administration (FDA), USA, for the selection of patients for targeted therapy. The main aim of our study was to investigate the frequency of NTRK aberrations, i.e. fusions, gene copy number gain, and amplification, in a series of TNBC using different methods. A total of 83 TNBCs were analyzed using pan‐TRK immunohistochemistry (IHC), fluorescence in situ hybridization (FISH), real‐time polymerase chain reaction (RT‐PCR), and RNA‐based next‐generation sequencing (NGS). Of 83 cases, 16 showed pan‐TRK positivity although no cases had NTRK‐fusions. Indeed, FISH showed four cases carrying an atypical NTRK1 pattern consisting of one fusion signal and one/more single green signals, but all cases were negative for fusion by NGS and RT‐PCR testing. In addition, FISH analysis showed six cases with NTRK1 amplification, one case with NTRK2 copy number gain, and five cases with NTRK3 copy number gain, all negative for pan‐TRK IHC. Our data demonstrate that IHC has a high false‐positive rate for the detection of fusions and molecular testing is mandatory; there is no need to perform additional molecular tests in cases negativity for NTRK by IHC. In conclusion, the NTRK genes are not involved in fusions in TNBC, but both copy number gain and amplification are frequent events, suggesting a possible predictive role for other NTRK aberrations.
Keywords: triple‐negative breast carcinoma, neurotrophic tyrosine receptor kinase (NTRK), NTRK fusions, NTRK amplification, IHC, FISH, RT‐PCR, NGS
Introduction
Breast cancer is a clinically and genetically heterogeneous disease. Approximately 10–20% of breast cancers are triple‐negative breast cancer (TNBC), a subtype characterized by immunohistochemical lack of estrogen receptor (ER), progesterone receptor (PgR), and human epidermal growth factor receptor 2 (HER2) overexpression [1, 2, 3]. TNBCs have aggressive clinical behavior and poor prognosis since they are not responsive to either hormonal therapy or HER2‐specific inhibitors. The definition of new biomarkers with potential therapeutic implications represents one of the greatest challenges in the treatment of this subgroup of breast cancers.
In recent years, the neurotrophic tyrosine receptor kinase (NTRK) gene fusions have become new predictive biomarkers in several cancer types since the specific inhibitors have been developed and approved [4, 5]. Physiologically, the NTRK1, NTRK2, and NTRK3 genes encode for a family of receptor tyrosine kinases (Trk) that play a role in neuronal development, function, survival, and proliferation. NTRK gene fusions have been shown to be a driver mechanism in various tumors, although at a very low incidence [6]. NTRK fusions can be detected using different methods including immunohistochemistry (IHC), fluorescence in situ hybridization (FISH), real‐time‐polymerase chain reaction (RT‐PCR), and RNA‐based next‐generation sequencing (NGS) [6]. ESMO, the Japan Society of Clinical Oncology and the Japanese Society of Medical Oncology have proposed the recommendation of NTRK fusion identification, suggesting the use of IHC as a prescreening method in tumors with a low incidence with mandatory molecular confirmation of IHC positive cases [7, 8]. The prescreening test based on IHC has been suggested because of the high specificity and sensitivity of pan‐TRK IHC observed in several tumors [9]. On the other hand, several studies have shown discordant results between IHC and the other molecular methods of NTRK detection in various solid tumors [10, 11, 12, 13]. A previous study evaluated NTRK fusions in TNBC patient samples using IHC as a prescreening method and the other molecular methods to confirm the NTRK fusion [14]. NTRK IHC resulted in a high false‐positive rate of NTRK gene fusion suggesting that another molecular assay should be recommended for NTRK fusion detection.
Beyond NTRK fusions, Lee et al showed that the NTRK genes could be affected by amplification in various cancer types [10]. However, the possible predictive role of NTRK gene amplification is still unclear, and further studies are needed to confirm whether NTRK‐amplified tumors are suitable for treatment with specific inhibitors. The main aim of this study was to assess the frequency of NTRK gene aberrations, such as fusions, copy number gain, and amplification, in a TNBC series. The analysis was performed using different assays including IHC, FISH, RT‐PCR, and RNA‐based NGS.
Materials and methods
Case selection and tumor specimen collection
A series of 109 triple‐negative breast tumor tissue samples from surgical resections performed between 2019 and 2022 at the University of Campania ‘L. Vanvitelli’, the Ospedale Evangelico Betania, the University of Naples ‘Federico II’, the S.M. delle Grazie Hospital, and the IRCCS CROB were collected. We retrospectively recorded clinical and pathological parameters, including the age of the patient at initial diagnosis, the histological type, the grade, and the stage. We defined TNBC according to the current guidelines: negative for ER or PgR if <1% or 0% of tumor cell nuclei are immunoreactive and HER2 negativity defined as either IHC expression of 0–1+ or lack of gene amplification by FISH [15]. We used four cases of secretory breast cances harboring NTRK3 rearrangement as positive controls.
Immunohistochemistry
IHC was performed on 4 μm paraffin‐embedded whole tissue sections for each case. Pan‐TRK immunohistochemical staining for TrkA/B/C expression was performed using Ventana pan‐TRK antibody (clone EPR17341; #790‐7026, ready to use, Ventana Medical Systems, Tucson, AZ, USA), a rabbit recombinant monoclonal antibody reactive to a C‐terminal epitope conserved across TRK‐A, ‐B, and ‐C proteins and present in both wild type and chimeric proteins. All assays were performed on a fully automated BenchMark XT device (Ventana Medical Systems). Ganglia of the submucosal plexus of a normal vermiform appendix and an infantile fibrosarcoma with known ETV6‐NTRK3 fusion were used as positive controls and were run simultaneously with each sample. Lymphocytes served as internal negative controls. Tumors were considered positive if ≥1% of tumor cells exhibited positivity at any intensity above background. Different subcellular staining patterns were considered positive, as previously suggested (cytoplasmic, membranous, nuclear, and perinuclear) [8, 13, 14]. Signal intensity was expressed as a score, from 1 to 3, corresponding to weak, moderate, and strong signals. Two independent observers carried out immunohistochemical analysis, and both observers were blinded; in discordant cases, a consensus was reached by collegial discussion.
Fluorescence in situ hybridization
FISH was carried out on three 4‐μm‐thick sections cut from each formalin‐fixed paraffin‐embedded (FFPE) sample using the BOND FISH kit (Leica Biosystems, Newcastle Upon Tyne, UK) on the automated BOND system (Leica Biosystems) according to the manufacturer's instructions. This kit consists of a formamide mixture to reduce nonspecific hybridization of nucleic acid probes. NTRK1, NTRK2, and NTRK3 fusion detection was performed by three separate assays using specific break‐apart probes for each gene: ZytoLight SPEC NTRK1 Dual Color Break Apart Probe (ZytoVision, Bremerhaven, Germany); ZytoLight SPEC NTRK2 Dual Color Break Apart Probe (ZytoVision, Bremerhaven, Germany); ZytoLight SPEC NTRK3 Dual Color Break Apart Probe (ZytoVision, Bremerhaven, Germany). Slides were counterstained with 4′,6‐diamidino‐2‐phenylindole dihydrochloride (DAPI) in antifade solution and examined using an automated CytoVision platform (Leica Biosystems). FISH interpretation was performed with the automated fluorescence microscope Leica DM5500 B (Leica Biosystems) using the filter ET‐D/O/G for double Spectrum Green plus Spectrum Orange. FISH signals were counted in at least 100 nonoverlapping intact nuclei.
NTRK fusion interpretation
FISH was considered positive in relation to two different patterns: (1) a classic break‐apart pattern with one fusion signal and two separated 3′ orange and 5′ green signals (separation distance of at least two signal diameters between the green and orange signals); (2) an atypical pattern with one fusion signal and a single orange signal without a corresponding green signal. Tumors were considered positive if ≥15% of tumor cells exhibited gene rearrangements.
Interpretation of NTRK gene copy number gain and amplification
The mean copy numbers for NTRK1, NTRK2, and NTRK3 genes were evaluated. In order to exclude polyploidy, FISH for centromeric alpha‐satellite sequences specific for chromosomes 1, 9, and 15, where the NTRK1, NTRK2, and NTRK3 genes are located respectively, was performed on the cases showing copy number aberration of the genes. The FISH assay to evaluate polyploidy was performed using specific probes: Creative Bioarray CEN 1p FISH Probe Red; CytoCell Chromosome 9 Satellite III FISH Probe Aqua | OGT; CytoCell Chromosome 15 Alpha Satellite FISH Probe Red | OGT. In a normal interphase nucleus, two signals associated with the disomic status of chromosome 2 were expected; an increase in signals per nucleus indicated the polyploidy of the chromosome. The ratio between each NTRK gene mean copy number and the related CEP mean copy number was evaluated. Amplification of the NTRK genes was considered to occur when the NTRK locus‐specific probe/CEP ratio was ≥2; cases carrying NTRK copy number gain showed a NTRK locus‐specific probe/CEP ratio of <2 [15]. The criteria for copy number aberrations of NTRK genes were as follows: NTRK copy number gain with a mean copy number of 3–5 signals in ≥10% of cells and NTRK amplification with the presence of ≥6 copies of gene per cell in ≥10% of analyzed cells [16].
RNA extraction
The hematoxylin and eosin‐stained tumor slides of each case were reviewed by a pathologist to select representative areas of tumor suitable for molecular testing. RNA extraction from FFPE tumor samples was carried out according to the manufacturers' protocols utilizing the MagCore Total RNA FFPE One‐Step Kit (RBC Bioscience Corp., New Taipei City, Taiwan) on the automated extraction system MagCore Super (RBC Bioscience Corp.) based on magnetic bead extraction technology.
Real‐time PCR
The RNA isolated from FFPE tumor samples was used to analyze NTRK1, NTRK2, and NTRK3 fusions. Real‐time‐PCR (RT‐PCR) was performed using the EasyPGX ready NTRK fusion kit (Diatech Pharmacogenetics, Jesi, Italy), on the EasyPGX qPCR instrument 96 (Diatech Pharmacogenetics). The detectable, but not distinguishable, gene fusions detected with this test are listed in supplementary material, Table S1. A positive and a negative control were used. The detection was based on the use of the fluorescent probes marked as follow: probes marked with FAM for the targets and probes marked with HEX for the endogenous control genes. The data were analyzed by EasyPGX Analysis Software (Diatech Pharmacogenetics).
Next‐generation sequencing
RNA input quantification was measured by RT‐PCR on the EasyPGX qPCR instrument 96 (Diatech Pharmacogenetics), detecting two highly conserved RNA regions of 105 and 175 bp through two probes labeled with FAM and HEX, respectively. The RNA concentration was assessed by quantification with a standard curve in the HEX channel. The ratio between the quantification (ng/μl) obtained in FAM and that obtained in the HEX allows evaluation of the DNA fragmentation. The data were analyzed by EasyPGX Analysis Software (Diatech Pharmacogenetics) in order to evaluate the concentration and degree of fragmentation of the samples. The RNA libraries were generated using the Myriapod NGS Cancer Panel RNA (Diatech Pharmacogenetics), according to the manufacturer's instructions. The kit allows the detection of the main gene fusions involving 10 recurrently rearranged cancer genes: ALK, ROS1, RET, NTRK1, NTRK2, NTRK3, FGFR2, FGFR3, PPARG, and the skipping of exon 14 of MET in total RNA isolated from tumor tissue. The RNA was retro‐transcribed into cDNA using random hexamers. Subsequently, cDNA was amplified by multiplex PCR using two primer mixtures to obtain fragments between 47 and 184 bases, including fusions of interest and endogenous control genes (PCR1). The amplification products were purified with magnetic beads to remove residual primers. An amplification‐based indexing reaction (PCR2) followed, which allowed a unique pair of two sample‐specific barcodes (indexes) and an Illumina platform‐specific adapter to be attached to each fragment. The libraries thus constituted were normalized in quantity by magnetic beads to guarantee a homogeneous coverage of the samples during sequencing. Finally, the normalized libraries were mixed (library pool) and sequenced in parallel on the Illumina MiSeq platform (Illumina Inc., San Diego, CA, USA) with MiSeq Reagent Kit v2 Micro (300 cycles) flow cell (Illumina Inc.). The data generated by the sequencer were analyzed locally with dedicated Myriapod NGS Data Analysis Software (v 4.0.2; Diatech Pharmacogenetics).
Statistical analysis
A Pearson chi‐square test was conducted using SPSS 20.0 for Mac (SPSS Inc., Chicago, IL, USA) to determine the association of NTRK aberrations with the clinical and pathological features.
Results
Clinical and pathological characteristics of patients
Overall, 109 TNBC patients were analyzed. The mean age of patients was 50 years (range 30–58 years); 84 (77%) were younger and 25 (23%) were older than 50 years of age. The cohort included 107 (98.2%) no special type carcinomas, 1 (0.9%) metaplastic carcinoma, and 1 (0.9%) pleomorphic lobular carcinoma; all cases were basal‐like. Of the 109 TNBC, 88 were grade (G)3 (80.7%) and 21 were grade G2 (19.3%). The postoperative pathological stages were I in 45 cases (41.2%), II in 45 cases (41.2%), and III in 19 cases (17.5%). The clinical and pathological features are summarized in Table 1.
Table 1.
NTRK aberrations and clinical–pathological features of the series analyzed
| NTRK1 FISH | NTRK2 FISH | NTRK3 FISH | NTRK‐fusion RNA‐based NGS | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. cases (%) | Pan‐TRK IHC positive | Isolated 5′ | CNG | Amp | CNG | CNG | Positive | Negative | NP | |
| Characteristics | 109 | 25 | 4 | 1 | 6 | 1 | 5 | 0 | 84 | 25 |
| Age | ||||||||||
| ≥50 | 25 (23%) | 11 (44%) | 2 (50%) | 1 (100%) | 2 (33.3%) | 0 | 1 (20%) | 0 | 18 (21.4%) | 7 (28%) |
| <50 | 84 (77%) | 14 (56%) | 2 (50%) | 0 | 4 (66.7%) | 1 (100%) | 4 (80%) | 0 | 66 (78.6%) | 18 (72%) |
| Histotype | ||||||||||
| NST | 107 (98.2%) | 25 (100%) | 4 (100%) | 1 (100%) | 6 (100%) | 1 (100%) | 5 (100%) | 0 | 84 (100%) | 25 (100%) |
| Metaplastic | 1 (0.9%) | 0 | 0 | 0 | 0 | 0 | 0 | |||
| Pleomorphic lobular | 1 (0.9%) | 0 | 0 | 0 | 0 | 0 | 0 | |||
| Grading | ||||||||||
| G2 | 21 (19.3%) | 5 (20%) | 0 | 1 (100%) | 1 (16.7%) | 0 | 2 (40%) | 0 | 11 (13.1%) | 10 (40%) |
| G3 | 88 (80.7%) | 20 (80%) | 4 (100%) | 0 | 5 (83.3%) | 1 (100%) | 3 (60%) | 0 | 73 (86.9%) | 15 (60%) |
| TNM stage | ||||||||||
| pT1, pN0 | 45 (41.2%) | 19 (76%) | 3 (75%) | 1 (100%) | 4 (66.7%) | 0 | 3 (60%) | 0 | 40 (47.6%) | 5 (20%) |
| pT2–pT3, pN0/pT1, pN1 | 45 (41.2%) | 6 (24%) | 0 | 0 | 2 (33.3%) | 1 (100%) | 1 (20%) | 0 | 32 (38.1%) | 13 (52%) |
| pT3, pN1/any T, pN2 | 19 (17.6%) | 0 | 1 (25%) | 0 | 0 | 0 | 1 (20%) | 0 | 12 (14.3%) | 7 (28%) |
| Neoadjuvant therapy | ||||||||||
| No | 66 (61%) | 21 (84%) | 4 (100%) | 1 (100%) | 5 (83.3%) | 1 (100%) | 4 (80%) | 0 | 58 (69%) | 8 (32%) |
| Yes | 43 (39%) | 4 (16%) | 0 | 0 | 1 (16.7%) | 0 | 1 (20%) | 0 | 26 (31%) | 17 (68%) |
| RCB | ||||||||||
| 0 | 0 | 0 | – | – | 0 | – | – | 0 | 0 | 0 |
| I | 4 (9.3%) | 0 | – | – | 0 | – | – | 0 | 3 (11.5%) | 1 (5.9%) |
| II | 31 (72.1%) | 3 (75%) | – | – | 1 (100%) | – | 1 (100%) | 0 | 18 (69.3%) | 13 (76.5%) |
| III | 8 (18.6%) | 1 (25%) | – | – | 0 | – | – | 0 | 5 (19.2%) | 3 (17.6%) |
Amp, gene amplification; CNG, copy number gain; NP, not performed; NST, no special type; RCB, residual cancer burden.
Pan‐TRK IHC
Overall, 25 of 109 cases showed positive pan‐TRK immunoreactivity; the remaining 84 cases were negative. In all IHC‐positive samples, a signal was present in more than 20% of tumor cells and the staining was observed exclusively in the cytoplasm. Signal intensity was strong in 2 cases (score 3), moderate in 9 cases (score 2), and weak in 14 cases (score 1) (Figure 1). NTRK IHC positivity was not statistically associated with clinical–pathological features. All 4 cases of secretory breast cancer selected as controls were pan‐TRK positive (supplementary material, Figure S1).
Figure 1.
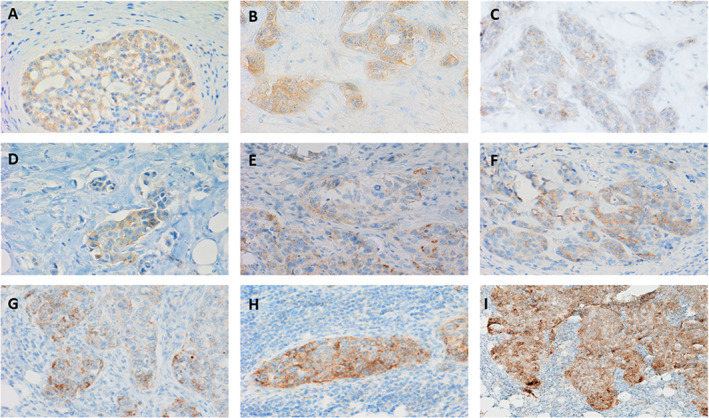
Representative pan‐TRK IHC results: (A–D) weak cytoplasmic immunoreactivity in neoplastic cells (original magnification ×40); (E–G) moderate cytoplasmic immunoreactivity in neoplastic cells (original magnification ×40); (H and I) strong cytoplasmic immunoreactivity in neoplastic cells (original magnification ×40).
Fluorescence in situ hybridization
FISH tests were performed on all cases to evaluate NTRK1, NTRK2, and NTRK3 fusions. In our series, no case was positive for NTRK fusions. Three cases showed NTRK1 rearrangement in approximately 7% of tumor cells analyzed, but this percentage was not sufficient to indicate positivity. Ten cases showed NTRK1 split‐apart signals that were less than two signal diameters apart in approximately 5–15% of tumor cells analyzed (Figure 2C). NTRK1 FISH showed 4 of 109 cases carrying an atypical pattern, particularly 2 cases with one fusion signal and a single green signal and 2 cases carrying an atypical pattern with three‐four fusion signals and a single green signal (Figure 2D). Moreover, six cases showed NTRK1 gene amplification and one case showed copy number gain; all cases showed disomy of chromosome 1 (Figure 2E,F). No atypical NTRK2 signal patterns were detected in our series; one case showed copy number gain, with disomy of chromosome 9 (Figure 2I). No atypical NTRK3 signal patterns were detected in our series; 5 of 109 cases showed copy number gain, with disomy of chromosome 15 (Figure 2L). NTRK FISH aberrations were not statistically associated with either clinical–pathological features or pan‐TRK expression. All 4 cases of secretory breast cancer selected as controls harbored rearranged NTRK3 (supplementary material, Figure S1).
Figure 2.
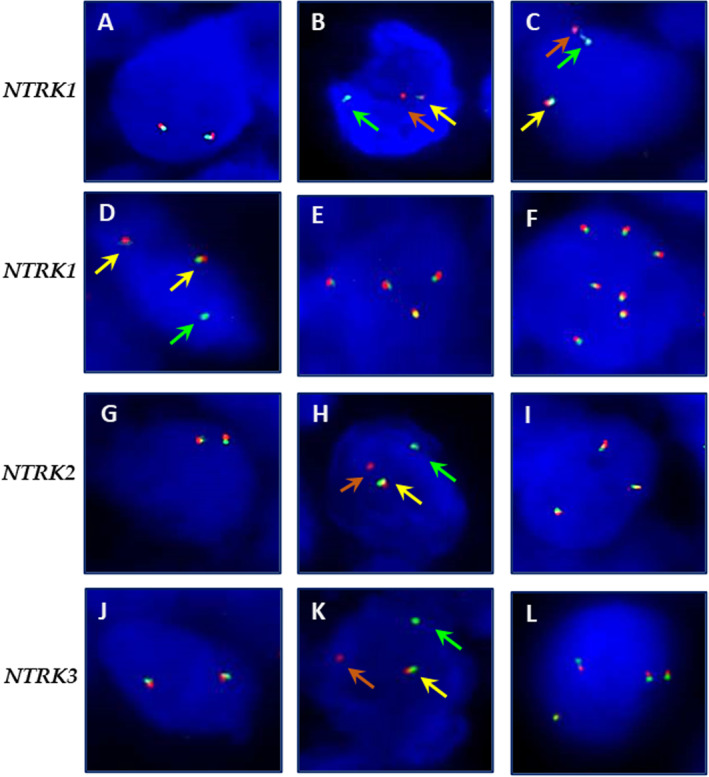
Representative NTRK1, NTRK2, and NTRK3 FISH results: (A) NTRK1 FISH showing the typical pattern wild type: two fusion signals (original magnification ×100); (B) NTRK1 FISH showing the typical pattern of rearrangement: one fusion signal (yellow arrow) and split 3′ (orange arrow) and 5′ (green arrow) signals, with a separation distance of at least two signal diameters between the green (green arrow) and orange (orange arrow) signals (original magnification ×100); (C) NTRK1 FISH showing one fusion signal (yellow arrow) and split 3′ (orange arrow) and 5′ (green arrow) signals, with a separation distance of less than two signal diameters between the green (green arrow) and orange signals (orange arrow) (original magnification ×100); (D) NTRK1 FISH showing an atypical pattern: two fusion signals (yellow arrows) and an isolated 5′ signal (green arrow) (original magnification ×100); (E) NTRK1 FISH showing copy number gain with four fusion signals (original magnification ×100); (F) NTRK1 FISH showing gene amplification with seven fusion signals (original magnification ×100); (G) NTRK2 FISH showing the typical wild type pattern: two fusion signals (original magnification ×100); (H) NTRK2 FISH showing the typical pattern of rearrangement: one fusion signal (yellow arrow) and split 3′ (orange arrow) and 5′ (green arrow) signals, with a separation distance of at least two signal diameters between the green (green arrow) and orange signals (orange arrow) (original magnification ×100); (I) NTRK2 FISH showing gene copy number gain with three fusion signals(original magnification ×100); (J) NTRK3 FISH showing the typical wild type pattern: two fusion signals (original magnification ×100); (K) NTRK3 FISH showing the typical pattern of rearrangement: one fusion signal (yellow arrow) and split 3′ (orange arrow) and 5′ (green arrow) signals, with a separation distance of at least two signal diameters between the green (green arrow) and orange signals (orange arrow)(original magnification ×100); (L) NTRK3 FISH showing copy number gain with four fusion signals (original magnification ×100).
Real‐time RT‐PCR
All cases were analyzed by RT‐PCR analysis. No NTRK fusions were detected.
RNA‐based NGS
The NGS analysis was performed on 84 of 109 cases; unfortunately, 25 cases were not tested with NGS as the RNA quantity and the quality were not adequate for this assay. No NTRK fusions were identified in the cases analyzed. Among 25 cases that were pan‐TRK IHC positive, 23 were NTRK1‐3 wild type and 2 cases were not analyzed by NGS (Table 2). Among four cases with an atypical NTRK1 FISH pattern, three cases were NTRK1 wild type and one case was not analyzed by NGS (Table 2).
Table 2.
Comparison of different assays in cases carrying NTRK aberrations
| NTRK FISH | ||||||
|---|---|---|---|---|---|---|
| Case | Pan‐TRK IHC score | NTRK1 | NTRK2 | NTRK3 | RT‐PCR | RNA‐NGS |
| 1 | 3 | ap (4F 1G) | NR | NR | NR | wt |
| 2 | 3 | CNG | CNG | CNG | NR | wt |
| 3 | 2 | ap (1F 1G) | NR | CNG | NR | NP |
| 4 | 2 | NR | NR | CNG | NR | wt |
| 5 | 2 | NR | NR | NR | NR | wt |
| 6 | 2 | NR | NR | NR | NR | wt |
| 7 | 2 | NR | NR | NR | NR | wt |
| 8 | 2 | NR | NR | NR | NR | wt |
| 9 | 2 | NR | NR | NR | NR | wt |
| 10 | 2 | NR | NR | NR | NR | wt |
| 11 | 2 | NR | NR | NR | NR | wt |
| 12 | 1 | NR | NR | NR | NR | wt |
| 13 | 1 | NR | NR | NR | NR | wt |
| 14 | 1 | NR | NR | NR | NR | NP |
| 15 | 1 | NR | NR | NR | NR | wt |
| 16 | 1 | NR | NR | NR | NR | wt |
| 17 | 1 | NR | NR | NR | NR | wt |
| 18 | 1 | NR | NR | NR | NR | wt |
| 19 | 1 | NR | NR | NR | NR | wt |
| 20 | 1 | NR | NR | NR | NR | wt |
| 21 | 1 | NR | NR | NR | NR | wt |
| 22 | 1 | NR | NR | NR | NR | wt |
| 23 | 1 | NR | NR | NR | NR | wt |
| 24 | 1 | NR | NR | NR | NR | wt |
| 25 | 1 | NR | NR | NR | NR | wt |
| 26 | 0 | ap (1F 1G) | NR | NR | NR | wt |
| 27 | 0 | ap (3F 1G) | NR | NR | NR | wt |
| 28 | 0 | Amp | NR | CNG | NR | NP |
| 29 | 0 | Amp | NR | CNG | NR | NP |
| 30 | 0 | Amp | NR | NR | NR | NP |
| 31 | 0 | Amp | NR | NR | NR | NP |
| 32 | 0 | Amp | NR | NR | NR | NP |
| 33 | 0 | Amp | NR | NR | NR | NP |
Amp: gene amplification; ap, atypical pattern (F, fused signals; G, green signals); CNG, copy number gain; NP, not performed; NR, not rearranged; wt, wild type.
Comparison between the different methods
All 25 pan‐TRK IHC positive cases showed no fusions by FISH, RT‐PCR, and NGS testing. All six cases carrying NTRK1 gene amplification and all cases with copy number gain of NTRK1, NTRK2, or NTRK3 were pan‐TRK IHC negative. All cases showing NTRK1 rearrangement in <15% of tumor cells analyzed were negative by RT‐PCR and NGS. In particular, all three cases harboring the NTRK1 rearrangement in approximately 7% of the scattered neoplastic cells by FISH showed 3–10 on‐target reads by NGS, which is considered negligible for possible gene rearrangement. All cases that showed NTRK1 split‐apart signals less than two signals in diameter in approximately 5–15% of tumor cells were negative by RT‐PCR and NGS. All cases that showed an atypical NTRK1 FISH pattern were negative by RT‐PCR and NGS. Complete agreement was found between RT‐PCR and NGS results (Table 2).
Discussion
NTRK gene fusions have been described in various tumor types with variable frequency. Tumors harboring NTRK fusions are sensitive to treatment with specific inhibitors regardless of histological features. To date, NTRK target inhibitors have been approved by the Food and Drug Administration (FDA), USA, for the treatment of TRK fusion‐positive cancers in a tumor agnostic way. Previous studies have reported that the incidence of NTRK gene fusions in breast carcinoma, not otherwise specified, is very low, ranging from 0 to 0.08% [17, 18, 19], whereas secretory breast carcinoma is characterized by NTRK3‐ETV6 fusion [20]. Moreover, clinical trials have demonstrated that secretory breast carcinoma patients carrying NTRK fusions have a high response to TRK inhibitor therapy [21]. To the best of our knowledge, only two reports have evaluated NTRK fusions in TNBC and no clinical trial results are available to date [14, 17]. The ESMO guidelines recommend NTRK IHC as a screening method to select cases for additional molecular testing to evaluate the fusions [7]. Pan‐TRK IHC represents a useful screening test since it is feasible in most pathology laboratories at moderate cost allowing the inclusion of all potential positive cases in triage for molecular testing. Several studies have analyzed the sensitivity and specificity of NTRK IHC, with very different results. In the literature, some studies have reported a sensitivity of pan‐TRK IHC of approximately of 100% [22, 23]. Other data showed lower sensitivity to detect NTRK fusions by IHC, particularly in the studies reported by Solomon et al (88%) [9], Hondelink et al (82%) [24], Koopman et al (79%) [12], and Gatalica et al (75%) [13]. A false‐negative IHC result was observed in particular for cases carrying NTRK3 rearrangements, regardless of the tumor type [9, 12, 13, 24]. In our TNBC series, pan‐TRK IHC was not sensitive, since all 25 pan‐TRK positive cases were wild type by molecular testing. Similarly, different percentages of specificity have been reported frequently according to the type of tumor. Solomon et al showed a specificity of 100% for carcinomas of the colon, lung, thyroid, pancreas, and biliary tract, with a lower specificity in breast and salivary gland carcinomas, 82 and 52%, respectively [9]. Since pan‐TRK IHC negative patients lose the possibility of further molecular investigation in clinical practice, good technical and interpretative practices for pan‐TRK IHC are recommended to avoid false negative results [25]. Our results confirm that pan‐TRK IHC has high specificity and negative predictive value for the detection of NTRK gene fusions. In our series, 14 of 25 pan‐TRK positive cases (56%) displayed only weak immunoreaction and were wild type by molecular analysis, suggesting that the cases showing weak pan‐TRK expression could be disregarded, significantly reducing the need for, and costs of, further molecular tests.
Molecular testing remains the gold standard for NTRK fusion detection. However, several critical pitfalls must be considered in the choice and interpretation of the available tests. Historically, FISH has been used to identify gene fusion detection in clinical practice, as in the identification of ALK, ROS1, and RET rearrangements in lung cancer. Regarding NTRK FISH, no guidelines for interpretation have yet been provided, thus comparison studies with NGS and RT‐PCR could clarify the evaluation criteria. Several interpretive doubts are currently unresolved in the interpretation of NTRK FISH, i.e. the cutoff for defining a rearranged case, the atypical patterns, the distance of separation of the split‐apart signals [7, 17, 26, 27]. In our study, some cases with dubious FISH have been solved by comparison with NGS and RT‐PCR; in particular, all cases showing separation of the split‐apart signals by less than two signal diameters did not show fusions by the orthogonal assays suggesting the strict use of at least two signal diameters to define a true rearrangement. Regarding the FISH threshold, the two cases with pan‐TRK IHC positive (intensity score 1+) and NTRK1 FISH showing about 7% of tumor cells rearranged were negative by NGS. Our and other previous findings related to NTRK FISH reveal the need for consensus criteria in the evaluation of this test in order to avoid false results. In our series, we identified four cases with an isolated 5′ signal pattern by NTRK1 FISH. Furthermore, two of these four cases showed pan‐TRK IHC positive staining, while no fusion was documented by NGS. Similarly to ALK FISH interpretation, the loss of 3′ signal (isolated 5′ signal) is routinely considered negative in the interpretation of NTRK FISH. Although isolated 5′ signal ALK FISH is still currently considered negative, two patients carrying this atypical pattern have tested positive with NGS and showed complete response to crizotinib, a specific ALK inhibitor [28]. In our series, the cases with isolated 5′ signal NTRK1 were negative by both RT‐PCR and NGS; however, the lesson learnt from ALK rearrangement suggests that the cases harboring atypical NTRK patterns in FISH should be better characterized through other orthogonal assays. In this context, the NGS approach provides high‐throughput data and the analysis of multiple genetic loci simultaneously. However, some samples are not suitable for this test related to the quantity and the quality of the RNA [29].
Beyond NTRK fusions, our findings show an increase in the copy number of NTRK genes in our TNBC series. Copy number gain was present for NTRK1 and NTRK2 with a frequency of 1.2% and for NTRK3 in 6% of cases analyzed. Moreover, NTRK1 gene amplification was present in 6 of 83 cases (7.2%) but none of these cases showed pan‐TRK positive staining. The amplification of ‘driver’ genes represents a frequent mechanism related to tumorigenesis through the deregulation of both normal growth and survival pathways in cancer [30]. NTRK gene amplification could play a role in cancer, regardless of the presence of fusions. NTRK gene amplification was previously described in other solid tumors such melanoma, uterine leiomyosarcoma, squamous cell lung cancer and gastric adenocarcinoma [10]. Lee et al have analyzed a total of 1,250 tumor specimens and detected NTRK amplification in 28 cases of various types of cancer. Among these cases, only four were positive for pan‐TRK IHC, including one melanoma, one sarcoma, one lung cancer, and one gastric cancer [10]. These data demonstrated that NTRK gene amplification does not necessarily result in protein overexpression, as found in all our TNBCs. Similarly to our findings, Lee et al showed that the amplification mainly affects NTRK1 rather than NTRK2 and NTRK3. However, they used a different threshold for defining gene amplification; specifically, we used a cutoff of mean copy number gene/cell ≥6.0 copies, while Lee et al used a mean copy number of gene/cell of ≥4.0 copies. Beyond the mean number of gene copies per cell it is mandatory for the correct analysis of gene amplification to evaluate the ratio between the gene copies and the relative centromere to exclude polysomy. From this point of view, we have demonstrated disomy of chromosome 1 in our NTRK1 amplified cases. Until now, few data have been reported regarding the sensitivity to NTRK‐specific inhibitors of patients harboring NTRK amplification regardless of fusions [31]. Currently, the phase II clinical trial NCT04879121 is recruiting patients with NTRK gene amplification solid tumors that are locally advanced or metastatic to evaluate the effect of larotrectinib in terms of overall response rate [32]. Further data are needed about NTRK amplification to understand its possible clinical significance, it is being understood that to date only tumors carrying NTRK fusions have been shown to benefit from treatment with specific inhibitors.
In conclusion, different from other tumors, pan‐TRK IHC showed a high false‐positive rate in TNBC for the detection of NTRK fusions, as determined by molecular testing. Our findings show that the NTRK genes are not involved in fusions in TNBC, but both copy number gain and amplification of NTRK genes are frequent events with an unknown potential predictive role. The role of NTRK gene amplification as an oncogenic driver will be clarified by the results of future clinical trial [32]. Moreover, our data provide useful guidelines for NTRK FISH interpretation suggesting the use of a stringent cut‐off for positivity of at least >15% of the cells analyzed and a mandatory separation distance of the split‐apart signals of two signal diameters to define a real gene fusion. Finally, the finding of an isolated 5′ signal remains an open issue, thus great attention must be paid in clinical practice to these cases.
Author contributions statement
FZM, MA and RF conceived the study. SB, RG, FM, GA, GC, GV, AS, LI, FF and MO collected samples. MA, MM, RG, GV and RF evaluated the pathology. SB and FZM carried out experiments. MA, MM, RG, GV and RF performed analyses. FZM, MA and RF interpreted data. FZM and RF drafted the first version of the manuscript. FZM, MA and RF reviewed and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Informed consent statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of University of Campania ‘L. Vanvitelli’‐AORN ‘Ospedale dei Colli’ Naples, Italy with approval number 21 of 17 January 2020.
Supporting information
Figure S1. Representative results from a secretory breast carcinoma used as a positive control
Table S1. Panel of gene fusions detected but not distinguishable by RT‐PCR
Acknowledgements
The authors thank Diatech Pharmacogenetics, Jesi, Italy for support in the interpretation of NGS data.
The authors disclose the receipt of financial support for the research from Roche Diagnostic SPA; project ‘Validazione del pan‐TRK (EPR17341) Assay nella selezione dei casi NTRK positive’ N.32523 (approved 18 February 2020). The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
No conflicts of interest were declared.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1. Wang X, Guda C. Integrative exploration of genomic profiles for triple negative breast cancer identifies potential drug targets. Medicine 2016; 95: e4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Garrido‐Castro AC, Lin NU, Polyak K. Insights into molecular classifications of triple‐negative breast cancer: improving patient selection for treatment. Cancer Discov 2019; 9: 176–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Prado‐Vázquez G, Gámez‐Pozo A, Trilla‐Fuertes L, et al. A novel approach to triple‐negative breast cancer molecular classification reveals a luminal immune‐positive subgroup with good prognoses. Sci Rep 2019; 9: 1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cocco E, Scaltriti M, Drilon A. NTRK fusion‐positive cancers and TRK inhibitor therapy. Nat Rev Clin Oncol 2018; 15: 731–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lange AM, Lo HW. Inhibiting TRK proteins in clinical cancer therapy. Cancer 2018; 10: 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zito Marino F, Pagliuca F, Ronchi A, et al. NTRK fusions, from the diagnostic algorithm to innovative treatment in the era of precision medicine. Int J Mol Sci 2020; 21: 3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Marchiò C, Scaltriti M, Ladanyi M, et al. ESMO recommendations on the standard methods to detect NTRK fusions in daily practice and clinical research. Ann Oncol 2019; 30: 1417–1427. [DOI] [PubMed] [Google Scholar]
- 8. Naito Y, Mishima S, Akagi K, et al. Japan Society of Clinical Oncology/Japanese Society of Medical Oncology‐led clinical recommendations on the diagnosis and use of tropomyosin receptor kinase inhibitors in adult and pediatric patients with neurotrophic receptor tyrosine kinase fusion‐positive advanced solid tumors, cooperated by the Japanese Society of Pediatric Hematology/Oncology. Int J Clin Oncol 2020; 25: 403–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Solomon JP, Linkov I, Rosado A, et al. NTRK fusion detection across multiple assays and 33,997 cases: diagnostic implications and pitfalls. Mod Pathol 2020; 33: 38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee SJ, Kim NKD, Lee S, et al. NTRK gene amplification in patients with metastatic cancer. Precis Future Med 2017; 1: 129–137. [Google Scholar]
- 11. Cappellesso R, Nozzoli F, Zito Marino F, et al. NTRK gene fusion detection in atypical Spitz tumors. Int J Mol Sci 2021; 22: 12332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Koopman B, Kuijpers C, Groen H, et al. Detection of NTRK fusions and TRK expression and performance of pan‐TRK immunohistochemistry in routine diagnostics: results from a nationwide community‐based cohort. Diagnostics 2022; 12: 668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gatalica Z, Xiu J, Swensen J, et al. Molecular characterization of cancers with NTRK gene fusions. Mod Pathol 2019; 32: 147–153. [DOI] [PubMed] [Google Scholar]
- 14. Wu S, Shi X, Ren X, et al. Evaluation of NTRK gene fusion by five different platforms in triple‐negative breast carcinoma. Front Mol Biosci 2021; 8: 654387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hammond ME, Hayes DF, Dowsett M, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol 2010; 28: 2784–2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zito Marino F, Botti G, Aquino G, et al. Unproductive effects of ALK gene amplification and copy number gain in non‐small‐cell lung cancer. ALK gene amplification and copy gain in NSCLC. Int J Mol Sci 2020; 21: 4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Remoué A, Conan‐Charlet V, Bourhis A, et al. Non‐secretory breast carcinomas lack NTRK rearrangements and TRK protein expression. Pathol Int 2019; 69: 94–96. [DOI] [PubMed] [Google Scholar]
- 18. Vranic S, Palazzo J, Sanati S, et al. Potential novel therapy targets in neuroendocrine carcinomas of the breast. Clin Breast Cancer 2019; 19: 131–136. [DOI] [PubMed] [Google Scholar]
- 19. Rosen EY, Goldman DA, Hechtman JF, et al. TRK fusions are enriched in cancers with uncommon histologies and the absence of canonical driver mutations. Clin Cancer Res 2020; 26: 1624–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vasudev P, Onuma K. Secretory breast carcinoma: unique, triplenegative carcinoma with a favorable prognosis and characteristic molecular expression. Arch Pathol Lab Med 2011; 135: 1606–1610. [DOI] [PubMed] [Google Scholar]
- 21. Scott LJ. Larotrectinib: first global approval. Drugs 2019; 79: 201–206. [DOI] [PubMed] [Google Scholar]
- 22. Berrino E, Bragoni A, Annaratone L, et al. Pursuit of gene fusions in daily practice: evidence from real‐world data in wild‐type and microsatellite instable patients. Cancer 2021; 13: 3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Murphy DA, Ely HA, Shoemaker R, et al. Detecting gene rearrangements in patient populations through a 2‐step diagnostic test comprised of rapid IHC enrichment followed by sensitive next‐generation sequencing. Appl Immunohistochem Mol Morphol 2017; 25: 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hondelink LM, Schrader A, Asri Aghmuni G, et al. The sensitivity of pan‐TRK immunohistochemistry in solid tumours: a meta‐analysis. Eur J Cancer 2022; 173: 229–237. [DOI] [PubMed] [Google Scholar]
- 25. Conde E, Hernandez S, Sanchez E, et al. Pan‐TRK immunohistochemistry: an example‐based practical approach to efficiently identify patients with NTRK fusion cancer. Arch Pathol Lab Med 2021; 145: 1031–1040. [DOI] [PubMed] [Google Scholar]
- 26. George J, Walter V, Peifer M, et al. Integrative genomic profiling of large‐cell neuroendocrine carcinomas reveals distinct subtypes of high‐grade neuroendocrine lung tumors. Nat Commun 2018; 9: 1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Del Castillo M, Chibon F, Arnould L, et al. Secretory breast carcinoma: a histopathologic and genomic Spectrum characterized by a joint specific ETV6‐NTRK3 gene fusion. Am J Surg Pathol 2015; 39: 1458–1467. [DOI] [PubMed] [Google Scholar]
- 28. Guyard A, Charpy C, Théou‐Anton N, et al. Isolated 5' signals are an atypical pattern to Be considered as positive for ALK rearrangement: a brief report of three cases and review of the literature. Transl Oncol 2019; 12: 784–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Teixidó C, Giménez‐Capitán A, Molina‐Vila MÁ, et al. RNA analysis as a tool to determine clinically relevant gene fusions and splice variants. Arch Pathol Lab Med 2018; 142: 474–479. [DOI] [PubMed] [Google Scholar]
- 30. Zito Marino F, Rocco G, Morabito A, et al. A new look at the ALK gene in cancer: copy number gain and amplification. Expert Rev Anticancer Ther 2016; 16: 493–502. [DOI] [PubMed] [Google Scholar]
- 31. Hempel D, Wieland T, Solfrank B, et al. Antitumor activity of larotrectinib in esophageal carcinoma with NTRK gene amplification. Oncologist 2020; 25: e881–e886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larotrectinib for the Treatment of NTRK Amplification Positive, Locally Advanced or Metastatic Solid Tumors. [Accessed 4 August 2022]. Available from: https://clinicaltrials.gov/ct2/show/NCT04879121
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Representative results from a secretory breast carcinoma used as a positive control
Table S1. Panel of gene fusions detected but not distinguishable by RT‐PCR
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


