Abstract
Integration of hepadnavirus DNAs into host chromosomes can have oncogenic consequences. Analysis of host-viral DNA junctions of DHBV identified the terminally duplicated r region of the viral genome as a hotspot for integration. Since the r region is present on the 5′ and 3′ ends of double-stranded linear (DSL) hepadnavirus DNAs, these molecules have been implicated as integration precursors. We have produced a LMH chicken hepatoma cell line (LMH 66-1 DSL) which replicates exclusively DSL duck hepatitis B virus (DHBV) DNA. To test whether linear DHBV DNAs integrate more frequently than the wild type open circular DHBV DNAs, we have characterized the integration frequency in LMH 66-1 DSL cells by using a subcloning approach. This approach revealed that 83% of the LMH 66-1 DSL subclones contained new integrations, compared to only 16% of subclones from LMH-D2 cells replicating wild-type open circular DHBV DNA. Also, a higher percentage of the LMH 66-1 DSL subclones contained two or more new integrations. Mathematical analysis suggests that the DSL DHBV DNAs integrated stably once every three generations during subcloning whereas wild-type DHBV integrated only once every four to five generations. Cloning and sequencing of new integrations confirmed the r region as a preferred integration site for linear DHBV DNA molecules. One DHBV integrant was associated with a small deletion of chromosomal DNA, and another DHBV integrant occurred in a telomeric repeat sequence.
Hepadnaviruses infect the liver where they cause acute or persistent infection of hepatocytes, depending on the nature of the immune response mounted by the host (7). Infectious hepadnavirus virion particles contain open circular (OC) DNA formed in the cytoplasm (30). These nucleocapsids contain pregenomic RNA and the viral reverse transcriptase (P protein) plus additional chaperone molecules (14). The normal replication mechanism involves reverse transcription of the pregenomic RNA in nucleocapsids to form a full-length minus-strand DNA which contains a direct duplication of a nine-base sequence on its 5′ and 3′ ends. This sequence is called the terminally redundant r sequence (24). In the majority of cases, the viral DNA plus strand is initiated and synthesized from a specific position at the 5′ end of the minus strand (the DR2 site). This mechanism leads to the formation of OC viral DNA molecules in infectious virions (7, 16, 32, 33).
However, in approximately 5% of nucleocapsids, plus-strand synthesis is initiated from the 3′ end of the minus strand and this leads to the formation of a double-stranded linear (DSL) viral DNA molecule (29). DSL DHBV DNA can be circularized in hepatocytes which they infect and they replicate by a mechanism called illegitimate replication (38). This term was used for this type of replication because it leads to a very high frequency of mutant virus production, which amplifies itself through successive rounds of viral DNA replication (38).
The hepadnavirus replication mechanisms are unique for a virus replicating via reverse transcription because DHBV pregenomic RNAs are formed from a nuclear CCC DHBV DNA molecule and not an integrated provirus. Interestingly, one of the most striking sequelae of persistent infection with the mammalian hepadnaviruses is the occurrence of hepatocellular carcinoma (HCC) in the host liver (1, 22, 31). Molecular analysis of genomic DNA from such HCCs generally reveals the presence of clonally propagated viral DNA integrations (20, 22, 37). Therefore, while integration and provirus formation are not required for replication, integration does occur in host chromosomes during persistent infection (22, 37). Interestingly, molecular analysis of the integrations has shown that virtually all of them contain viral genomes with deletions and rearrangements. Thus, the integration process has been viewed as a pathway in which viral DNA normally destined for CCC DNA formation is diverted into nonfunctional integrations (8, 9, 20, 23). The presence of these integrations can have oncogenic consequences for the host since the integrations contain enhancers which can activate cellular promoter which are normally silent (5–7).
In the case of HCCs arising in woodchucks with persistent woodchuck hepatitis virus (WHV) infection, molecular analysis of cloned WHV DNA integrations has revealed a dramatic example of common activation of myc family proto-oncogenes (5–7, 11, 19). Specifically, when WHV DNA integrates near N-myc2, it generally activates the expression of a normally silent N-myc2 retroposon via an enhancer insertion mechanism (36). A second common integration site (the WIN locus) is located approximately 250 kb upstream from the N-myc2 gene (6). The mechanism by which integration at this site leads to activation of the N-myc proto-oncogene has yet to be described. In many other cases, integrations of hepatitis B virus (HBV) are implicated in cancer by their presence in or near growth regulatory genes. Altered expression of a number of genes by HBV DNA integrations have been reported, such as cyclin A (35), retinoic acid receptor (4), hst-1 oncogene (12), and mevalonate kinase (10). In the case of HBV, a commonly activated protooncogene has not yet been identified in human HCCs. However, the presence of many HBV DNA integrations at sites of chromosomal DNA deletions (23) and translocations (13) have implicated them as general mutagenic agents (8, 22).
Since molecular evidence clearly implicates hepadnavirus DNA integrations as potent carcinogenic agents, our aim has been to understand the natural history of integrations and the factors which either increase or decrease their frequency during persistent infections (21). Such an understanding may allow us to devise strategies to block or reduce their occurrence and reduce the risk of hepatocarcinogenesis in individuals with persistent infection. With this goal in mind, our laboratory has developed a single-cell cloning approach to study the natural history of integrations in growing cells. This approach detects new hepadnavirus integrations which are stable during the clonal growth of infected hepatoma cells (9).
Initially, we utilized the LMH-D2 cell line, which replicates circular (wild-type [WT]) duck hepatitis B virus (DHBV) (2, 15). These studies demonstrated that new DHBV DNA integrations could be detected in approximately 10 to 20% of the DNAs from LMH-D2 subclones (8). Cloning and sequencing of one of the DHBV DNA integrations (intb) revealed a structure strikingly similar to that of episomal DSL DHBV virion DNAs (9). To investigate whether linear episomal DHBV molecules might be more efficient integration substrates than circular DHBV DNAs, we produced a cell line (LMH 66-1 DSL) which produces only linear DHBV (17). We have investigated the frequency and natural history of DHBV DNA integrations in subclones of the above cell line and compared our data with previously reported data for integration of WT circular DHBV DNAs (8). Our calculations predict a frequency of one integration per three cell generations for linear DHBV versus one integration per four to five generations for the circular DHBV. Finally, cloning and sequencing of several new DHBV integrations has revealed some common features among the integrations and suggests mechanisms for DHBV DNA integration.
MATERIALS AND METHODS
Cell culture.
The LMH chicken hepatoma cell line (2, 15) was a generous gift from William Mason (Institute for Cancer Research, Philadelphia, Pa.). The plasmid 66-1 (17), which contains a 1.5× DHBV genome with five nucleotide mutations (29) in the 5′ DR1, was obtained from Dan Loeb (Madison, Wis.). The LMH cells were transfected with the plasmid 66-1 along with a selectable marker, pSV2Neo. G418-resistant cell clones were expanded and maintained in Dulbecco’s minimal essential medium-F12 with 10% fetal bovine serum and 200 μg of G418/ml. Single-cell subcloning of the LMH-DSL cell line was performed as previously described (8). Briefly, dilutions containing 100 to 200 single cells of the LMH-DSL cell line generated from the plasmid 66-1 were mixed with 1 × 105 G418-sensitive helper LMH cells and plated onto 100-mm dishes. The mixed culture was grown in medium without G418 for 2 days, and G418 (200 μg/ml) was then added to remove the helper cells. G418-resistant subclones were picked and transferred to 12-well culture plates and then to 100-mm dishes where they grew to 5 × 106 to 10 × 106 cells before being harvested for analysis. This represents approximately 23 generations of cell growth.
Analysis of subclone DNA.
Total nuclear DNA of the LMH 66-1 cell line and its subclones was isolated as previously described (8). Briefly, the cells in culture dishes were trypsinized and lysed in the buffer containing 50 mM Tris HCl (pH 7.5), 1 mM EDTA, 5 mM MgCl2, and 0.5% Nonidet P-40. The pelleted nuclei were then lysed with 0.2% sodium dodecyl sulfate and treated with proteinase K (200 μg/ml) overnight, and the nucleic acids were extracted once with phenol and once with chloroform and were precipitated with enthanol. The cytoplasmic fraction was processed to isolate viral core particle DNA. It was treated with DNase I (100 μg/ml) for 1 h at 37°C. The reaction mixture was adjusted to contain 100 mM NaCl, 10 mM EDTA, 0.2% sodium dodecyl sulfate and proteinase K (200 μg/ml) and incubated overnight. The viral DNA was extracted as described above with phenol-chloroform. To isolate DHBV DNAs from the culture medium, DHBV virions secreted into the culture medium were concentrated with 15% polyethylene glycol and 1 M NaCl at 4°C. Virion DNA was isolated as described above for the cytoplasmic core particle DNA. For Southern blot analysis (27), nuclear or cytoplasmic DNA was digested with restriction enzymes overnight, electrophoresed through a 1% agarose gel, transferred to a Zetabind membrane, and hybridized with radiolabeled probes made by random priming (9).
Cloning and analysis of DHBV integrations.
To clone the DHBV integrations, total nuclear DNA isolated from LMH-DSL P1(5)-4 subclone was digested with SacI, ligated with lambda DASH II vector, and packaged with Gigapack extracts (Stratagene, La Jolla, Calif.). The genomic library was then screened with radiolabeled total DHBV DNA. The DHBV-positive phage clones were purified by two more cycles of plating and screening. Purified phage clones DNAs were digested with SacI to release the insert, and the DHBV DNA containing fragments were subcloned in the pBluescript II plasmid.
RESULTS
Establishment of a cell line that produces DSL DHBV DNA.
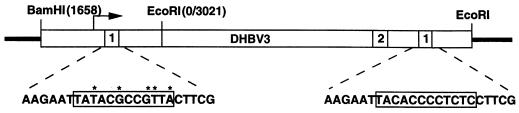
To investigate the frequency of integration of DSL DHBV molecules, we constructed a LMH cell line by using a previously characterized mutant DHBV, designated 5/12 (29), which was expected to produce DSL DHBV due to mutations in one copy of the DR1 sequence (five base substitutions in the 12-bp DR1). These mutations only allow virus DNA to be synthesized by the in situ priming mechanism (Fig. 1). The plasmid 66-1, which contained a 1.5× genome construct of the mutant 5/12 DHBV DNA was a gift from Dan Loeb. This plasmid was transfected into the LMH cells along with a selectable marker, PSV2neo (Fig. 1). A G418-resistant clone which secreted DHBV into the culture medium was expanded and was designated LMH 66-1 DSL.
FIG. 1.
Schematic diagram of the 66-1 plasmid. Plasmid 66-1 (17) was used for stable transfection of the LMH chicken hepatoma cells. It contains larger than genome size (3-kbp EcoRI fragment) DHBV3 (28) DNA for supporting viral replication. The BamHI-EcoRI (nucleotides 1658 to 3021) fragment was joined to the EcoRI monomer to create a “1 1/2mer” of DHBV DNA expression vector. The 5′ (left) DR1 (boxed) sequence contains 5-nucleotide substitutions from the WT DR1 located in the 3′ end (right). The pregenome RNA transcribed (arrow) from the vector will contain mutations in only the 5′ redundancy.
DHBV DNAs in LMH 66-1 DSL cells and secreted virions.
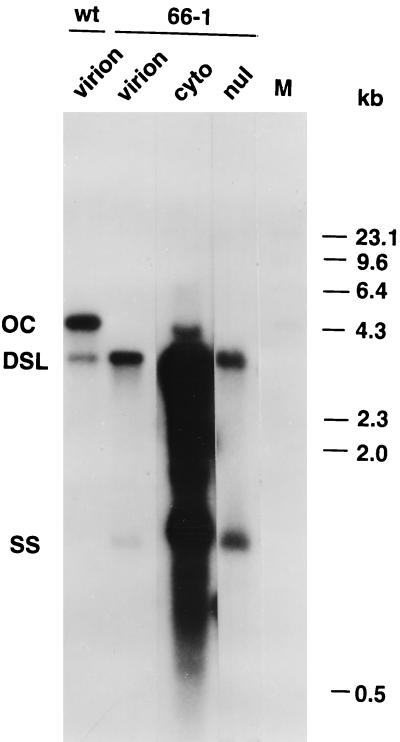
Southern blot analysis of DHBV virion DNAs secreted from LMH 66-1 DSL cells revealed two species of molecules. The major species was DSL DHBV DNA, and a very minor species migrated at the position of single-stranded (SS) DHBV DNA. No OC DHBV DNA molecules were detectable in the secreted virus preparations. In contrast, wild-type DHBV virions produced by LMH-D2 cells contained OC DHBV DNA molecules with a minor fraction of DSL molecules and little detectable SS DHBV DNA (Fig. 2).
FIG. 2.
Analysis of DHBV DNAs expressed from the LMH 66-1 DSL cell line. wt, WT DHBV produced by LMH-D2 cells; 66-1, virus present in LMH 66-1 DSL cells. Lanes: virion, viral DNA isolated from secreted virons; cyto, cytoplasmic DHBV DNAs; nul, nuclear DHBV DNAs; M, HindIII lambda phage marker fragments. The positions for open circular (OC), double-stranded linear (DSL), and single-stranded (SS) DHBV DNAs are indicated. Viral DNAs produced from the LMH 66-1 DSL and LMH-D2 (WT DHBV) cell lines were isolated and analyzed by Southern blotting as described in Materials and Methods. The cells were fractionated into nucleus (nul) and cytoplasm (cyto) fractions from which DHBV DNAs were extracted. The probe used was a random primed total DHBV DNA. Molecular weight standard (lane M) is a radiolabeled HindIII digest of lambda phage DNA.
In concordance with this picture, the cytoplasm of LMH 66-1 DSL cells contained an overwhelming majority of DSL and SS DHBV DNAs and replication intermediates (Fig. 2). Only a very small component of DHBV molecules migrating in the position of open circles was observed and these were observed only when the blot was highly overexposed for the cytoplasmic fraction, as shown in Fig. 2. Thus, the OC molecules are an extremely small fraction of the DHBV DNA molecules in the cytoplasm. This very small minority of OC DNAs could be synthesized by translocation or mismatch priming of the mutated plus-strand primer. Alternatively, these molecules could have arisen by circularization of DSL DHBV DNA molecules after their extraction. This could occur by end melting and hybridization of homologous r sequences at their ends. The specific structures of the very minor component of OC molecules was not determined.
DNA extracted from the nuclei of LMH 66-1 DSL cells contained DSL and SS DHBV DNAs. Interestingly, there was no detectable CCC DHBV DNA in the nuclei of LMH 66-1 DSL cells. CCC DHBV DNAs are protein free and would migrate just ahead of the 2.0-kb DNA marker (Fig. 2), and we should have been able to detect as little as one CCC DHBV DNA molecule per nucleus. It is not known whether the DSL or SS DHBV DNAs in the nuclei of LMH 66-1 DSL cells are protein free as are the CCC DNAs in the LMH-D2 cells. These data demonstrate that DSL DHBV DNA molecules are not circularized in the nucleus of LMH hepatoma cells. This is in contrast to the circularization of DSL DHBV DNA, which occurs in primary duck hepatocytes (38). This property of LMH cells allowed us to carry out experiments without the complication of OC DHBV molecules being present in the nuclei of LMH 66-1 DSL cells. The pregenomic DHBV RNAs in these cells were produced from the transfected transgene DNA which was present in the cells. The transgene DNAs which had integrated into the chromosome are designated Tr and were detectable when the genomic DNA was digested with PstI, (see Fig. 4).
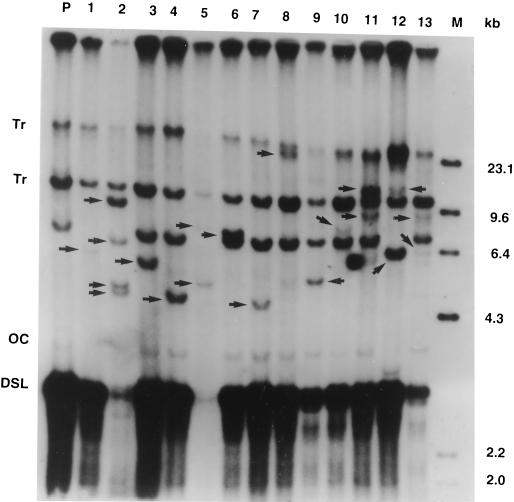
FIG. 4.
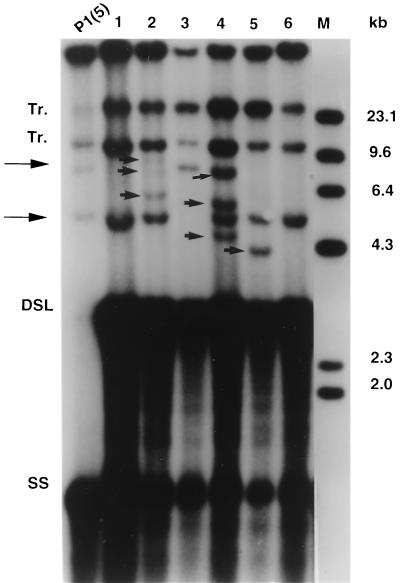
Southern blot analysis of DHBV DNA integrations in the first-generation subclones of the LMH 66-1 DSL cell line. Lanes: P, parental DNA; 1 to 13, DNAs from 13 first-cycle subclones. Tr, transgene bands. OC, DSL, and M are as defined in the legend for Fig. 2. Arrowheads indicate new DHBV DNA integrations in the subclones. The genomic DNAs were digested with PstI, a DHBV3 noncutter, and analyzed by Southern blotting with a total DHBV DNA probe. The LMH 66-1 DSL cell line contained two high-molecular-weight DNA bands (Tr) derived from integration of the plasmid 66-1 during stable transfection that are retained in all the subclones.
Single-cell subcloning of the LMH 66-1 DSL cell line revealed a high frequency of new DHBV integrations.
In order to estimate the frequency of stable DHBV integrations in the LMH 66-1 DSL cell line, we produced single cell subclones from the parental cells. A schematic of three cycles of subcloning protocol, illustrating the number of subclones in each cycle and the subclone numbers of lineages which were carried through sequential subcloning protocols, is illustrated in Fig. 3. All the nuclear DNAs were harvested from colonies which were allowed to grow through 23 to 24 cell divisions to reach approximately 4 × 106 to 8 × 106 cells. The genomic DNAs were digested with restriction enzyme PstI, whose recognition sequence is not present in the DHBV DNA of the mutant DHBV used in our experiments (28, 29). Therefore, each new band on a Southern blot, which was larger than DSL DHBV DNA, should represent a DHBV DNA integration.
FIG. 3.
Flow diagram of three cycles of single-cell subcloning of the LMH 66-1 DSL cell line. P, parental cell line used for initial subcloning. 1st, The first cycle of subcloning included 13 subclones, and subclones 5 and 8 were selected for a second round of subcloning; 2nd, six second-cycle subclones were derived from colony 5, and 12 second-cycle subclones were derived from colony 8; 3rd, third-cycle subclones were derived from second-cycle subclones P1(5)-5 and P1(5)-6, for a total of six third-cycle subclones. Open circles indicate subclones which were further subcloned, and solid circles denote subclones which were not further subcloned.
Southern blot analysis of genomic DNAs from the parental clone and all 13 first-generation subclones are shown in Fig. 4. The two bands in the Southern blot marked Tr were from the transfected DNA used to generate the cell line, and these were present in all the subclones. Another DHBV DNA band of approximately 7 kb was present in the parental DNA and also in 9 of 13 of the first-cycle subclones. Therefore, this integration occurred early in the selection of the LMH 66-1 DSL cell line and is present in most but not all of the cells. Alternatively, it could have been present in the initial cell used to generate the cell line and was subsequently lost from some cells. In either case, since it was present in the original parental DNA preparation and was common in the first cycle subclones, it was not counted as a new integration in our experiments.
Careful analysis of the new integration bands (above the DSL DHBV DNA band), revealed many bands with varied intensities. Previous subcloning data has shown that the variation in intensity is most often caused by integrations not present in all the cells in the colony (8, 9, 21). This means that the integration occurred after the first cell division initiating the colony. The total number of new integrations was 20, and at least one new integration was present in all of the subclones (Fig. 4). Eight subclones contained a single candidate new integration while four subclones contained two, and one subclone (no. 2) contained four candidate new integrations. The integrated transgene bands were present in the parental line and were stable throughout our experiments.
The LMH 66-1 DSL subcloning data were in sharp contrast to the data we had previously obtained for LMH-D2 cells which replicate WT DHBV. First-generation subcloning of that cell line revealed only two candidate new integrations in 12 subclones (8). Therefore, the new data strongly suggested that the integration frequency in our new LMH 66-1 DSL cell line was much higher than that of LMH-D2 cells.
To obtain a more accurate estimate of the integration frequency in LMH 66-1 DSL cells, it was necessary to carry out subsequent generations of subcloning, in which we grew the cells at the same rate for a known number of cell generations. We chose first-generation subclones 8, which contained one new integration band at approximately 23 kb (Fig. 3, and Fig. 4, lane 8), and subclone 5, which contained two new integrations at approximately 5 and 7 kb (Fig. 3, and Fig. 4, lane 5) for further lineage analysis.
Second-generation subcloning of LMH 66-1 DSL cells.
The second-generation subclones were grown for 23 to 24 cell divisions (4 × 106 to 8 × 106 cells) before DNA was prepared from each subcloned cell population for Southern blot analysis. New integrations which would be detected by this analysis would have occurred during the growth of the first-cycle subclone or during the first few cell divisions of the second-cycle subclone (as illustrated in Fig. 3). The results of the Southern blot analysis of second-cycle single-cell subclones are shown in Fig. 5 and 6.
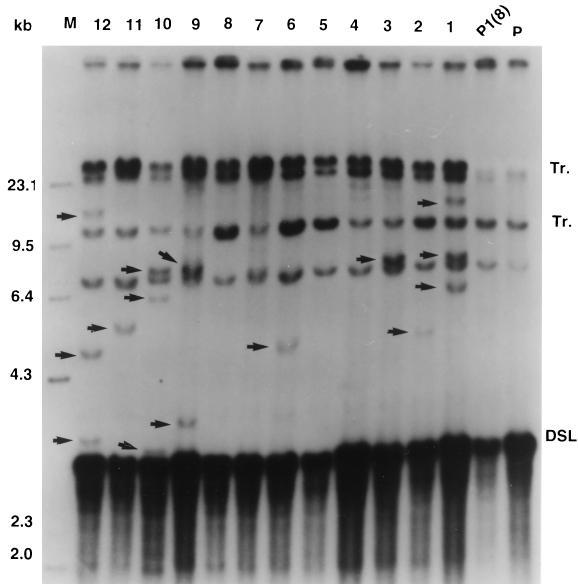
FIG. 5.
Southern blot analysis of the DHBV DNA integrations in the P1(8) lineage second-cycle subclones. Lanes: P, parental DNA; P1(8), nuclear DNA from first-cycle subclone P1(8); 1 to 12, nuclear DNAs from 12 second-cycle subclones derived from P1(8), digested with PstI, and analyzed by Southern blotting. Arrowheads indicate new DHBV DNA integrations detected in the second-cycle subclones. P1(8) had contained one new DHBV DNA integration at about 23 kb which was present in all the second-cycle subclones.
FIG. 6.
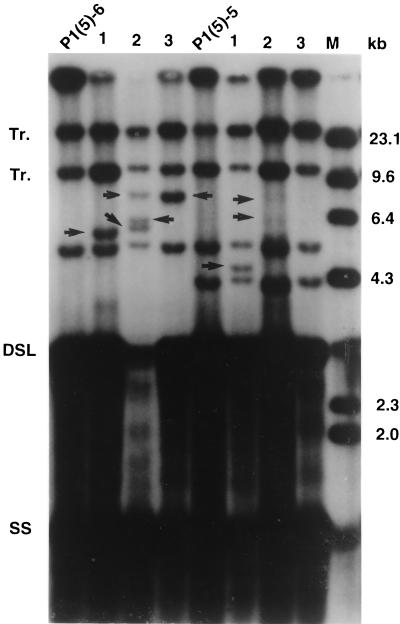
Southern blot analysis of the DHBV DNA integrations in the P1(5) lineage second-cycle subclones. Lanes: P1(5), nuclear DNA from first-cycle subclone P1(5); 1 to 6, nuclear DNAs from six second-cycle subclones derived from P1(5). Long arrows at left point to new integrations in first-cycle subclone P1(5) which segregate among second-cycle subclones. Smaller arrows within the blot point to new integrations in the second-cycle subclones. Abbreviations are as defined in the legend for Fig. 2.
First-generation subclone P1(8) had contained one new integration identified as a PstI fragment of about 23 kb. This integration was present in 100% of the second-generation subclones, which showed that it was present in the initial cell that produced the first-generation clone 8 (Fig. 5). Therefore, the 23-kb band was not included in the calculations which determined the number of integrations for this second-generation lineage. Southern blot analysis of the lineage 8 subclones revealed new integrations in 8 of 12 subclones (Fig. 5). Of the subclones with new integrations, one had two new integrations and remarkably, three had three new integrations. The total number of new integrations was 15 for 12 subclones (Table 1).
TABLE 1.
Frequencies of new stable DHBV DNA integrations in subclones replicating DSL versus WT OC DHBV DNAs
| Cell line | Specific lineage | No. of new DHBV integrations in subclone no.:
|
Ratio of clones with new integration to total clones | Ratio of clones without new integration to total clones | Ratio of new integrations to total clones | |||
|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | |||||
| LMH 66-1 DSL | Second generation | |||||||
| P1(8) | 4 | 4 | 1 | 3 | 8/12 | 4/12 | 15/12 | |
| P1(5) | 3 | 1 | 0 | 2 | 3/6 | 3/6 | 7/6 | |
| Third generation | ||||||||
| P1(5)-5 | 1 | 1 | 1 | 2/3 | 1/3 | 3/3 | ||
| P1(5)-6 | 2 | 1 | 3/3 | 0/3 | 5/3 | |||
| Combined second and third generations | 8 | 8 | 2 | 6 | 16/24 (66%) | 8/24 (33%) | 30/24 (125%) | |
| LMH-D2 (WT) | Combined second and third generationsa | 49 | 9 | 1 | 0 | 10/59 (16%) | 49/59 (84%) | 11/59 (18%) |
Data obtained from Gong et al. (8).
The first-generation P1(5) subclone contained two new DHBV integrations (Fig. 4, lane 5; Fig. 6). One of the second-generation subclones [P1(5)-3 (Fig. 6, lane 3)] contained the larger integration. This integration had been a “weak” integration in the first cycle and accordingly, the second-cycle subcloning revealed it was present in only one of six of the subclones. The other integration was present in the other five second-cycle subclones. This segregation illustrated that those integrations occurred in different progeny cells during the early clonal growth of the first-generation P1(5) population.
Three of the six P1(5) second-cycle subclones contained new DHBV integrations (Fig. 6). One subclone [P1(5)-5 (Fig. 6, lane 5)] contained one new integration and two subclones contained three [P1(5)-2 and -4 (Fig. 6, lanes 2 and 4)]. Thus, 50% of the second-generation subclones contained a new integration, yet the total number of new integrations was greater than the number of subclones (7 per 6 subclones) (Table 1). Summarizing the data for the clone 8 and 5 second-cycle lineages, 39% of the subclones did not contain a new integration. However, 21 new integrations were detected among the 18 second-generation subclones for an average of 1.2 new integrations per subclone (Table 1).
Third-generation subcloning of LMH 66-1 DSL cells.
To test whether our second-cycle integration data represented a true steady-state picture of the stable integration frequency in LMH 66-1 DSL cells, we conducted a third-cycle subcloning experiment. To do this, we used single cell subcloning of the second-cycle clone populations P1(5)-5 and P1(5)-6 (Fig. 6, lanes 5 and 6, respectively). As shown in Fig. 7, five of six third-cycle subclones contained new DHBV integrations that were not detected in the parental populations (Fig. 7). Three subclones contained one new integration, one subclone contained two new integrations, and one subclone contained three new integrations. A total of eight new integrations were detected in six subclones for an average of 1.3 integrations per subclone (Table 1).
FIG. 7.
Southern blot analysis of DHBV DNA integrations in the third-cycle subclones of the P1(5) lineage. Lanes: P1(5)-5 and P1(5)-6, DNAs from second-cycle subclones used to derive third-cycle subclones; 1 to 3, three third-cycle subclones each derived from the respective second-cycles subclones. Abbreviations are as defined in the legend for Fig. 2.
Combining the integration frequencies for the second- and third-generation subclones, we observed that 33% (8/24) of the subclones did not contain a new integration. However, the total number of new integrations, (30 per 24 subclones) averaged 1.25 integrations per subclone (Table 1). This was due to the presence of greater than one new integration in 33% (8/24) of the subclones. This was a dramatic increase over the previously observed integration of LMH-D2 subclones, in which an average of 84% (49/59) of the second- and third-generation subclones did not contain a new integration, and in which there were only 0.18 new integrations observed per subclone (Table 1).
The structures of the DHBV DNA integrations in LMH 66-1 cells resemble linear DHBV DNA molecules but often with a few nucleotides deleted from either terminus.
Having established a cell line that integrated DHBV DNA at a high frequency, we wanted to study the integration mechanism by comparing the structure of the newly integrated DHBV DNA with that of episomal DSL DHBV molecules. To do this, we cloned three new integrations from subclone P1(5)-4 (Fig. 6, lane 4). We generated a SacI genomic library in Lambda phage DASH II vector from genomic DNA of subclone P1(5)-4. Once the clones were plaque purified, DNA was prepared, and the complete integrated DHBV DNA molecule in each integration was sequenced along with immediate flanking cellular DNA. Analyses of the three viral DNA integrations in the LMH P1(5)-4 clone are shown in Fig. 8 in comparison to the structure of DSL DHBV produced by LMH 66-1 cells.
FIG. 8.
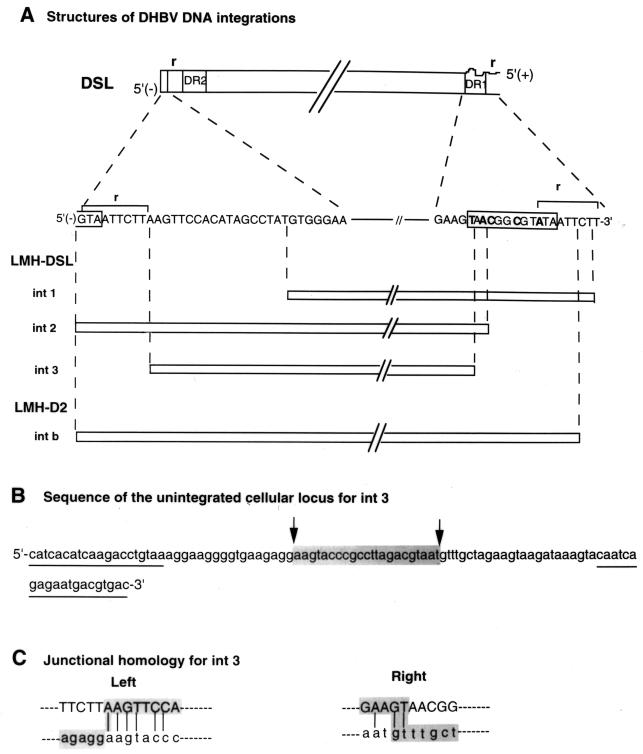
Structure of three DHBV DNA integrations from the LMH 66-1 DSL cell line. (A) Top section labeled DSL is a schematic map of a DSL DHBV virion DNA produced from LMH 66-1 DSL cell line and the specific minus-strand DNA sequences present at each end of the DSL molecule. Curvy line shows the 5′ end of the plus strand (+) of DSL DHBV DNA, which contains an 18-nucleotide RNA primer including the r-terminal redundancy region. Nucleotides in bold in DR1 indicate the mutated nucleotide changes to produce DSL DNAs. Section labeled LMH-DSL (int1, int2, and int3) is a map of three new integrations cloned from LMH-DSL subclone P1(5)-4 (integration bands seen in Fig. 6, lane 4). Vertical dashed lines denote left and right viral junctions with cellular DNA. LMH-D2 intb is a previously reported WT DHBV integration (9) with a structure similar to that of DSL DNA. Complete DHBV integrations and their immediate cellular flanking DNA were sequenced manually or by an automatic sequencer using oligonucleotides derived from the DHBV genome and cellular DNA. Slashes (//) in the diagram represent the uninterrupted DHBV genome. (B) The unintegrated cellular locus for LMH 66-1 DSL int3 was isolated by PCR from the untransfected LMH cell line using oligonucleotides derived each from the left and right flanking cellular DNA of int3. One specific fragment was amplified by PCR from the LMH cell line from the oligonucleotide primers (underlined) and was sequenced. The shaded sequence represents region of the unintegrated locus DNA that was not present in the int3 flanking cellular DNA. The two arrows indicate possible sites of deletion of cellular DNA during int3 integration. (C) DHBV DNA at the junction site of int3 (upper-case letters) and cellular genomic DNA sequences across the left and right junctions of int3 (lower-case letters) were aligned for comparison. Vertical lines denote the homology between the viral and cellular DNA sequences at the junctions. The shaded sequences are int3 junctional sequences.
Each of the DHBV DNA integrations contained one copy of the DHBV genome that was generally colinear with a DSL DHBV DNA. However, the viral DNA sequences at the junctions with cellular DNA differed in each clone, yet at the same time, they were all closely clustered within 30 nucleotides of either terminus of the minus strand. The viral junctions at the 3′ terminus were all localized in the end 18-nucleotide region that would contain an RNA primer following synthesis of the DSL DNA by the so-called in situ priming mechanism (Fig. 8A). The three DHBV DNA integrations isolated from the LMH 66-1 subclones contained all or part of the DR1 sequence and each had retained the mutations constructed in the DHBV expression plasmid.
Integration at DHBV int3 resulted in a small deletion of chromosomal DNA.
We next isolated the unintegrated locus of chromosomal DNA corresponding to the chromosomal integration site for int3 (Fig. 8A). This was done by PCR using oligonucleotide primers derived from the left and right flanking cellular DNA. The complete sequence of the normal cellular integration site is shown in Fig. 8B. Sequence analysis of the intact site versus the left and right junction sequences of int3 revealed that a deletion of 22 bp of the chromosomal DNA occurred at the DHBV integration locus (Fig. 8B). Alignment of the unintegrated locus and DHBV sequences across the junctions of int3 further revealed a 2- to 4-bp homology at each of the viral-cellular DNA junctions (Fig. 8C). This short junctional homology between the hepadnaviral and cellular DNAs has previously been observed and most likely has a role in the integration mechanism.
DHBV integration int1 is associated with telomere repeat sequences.
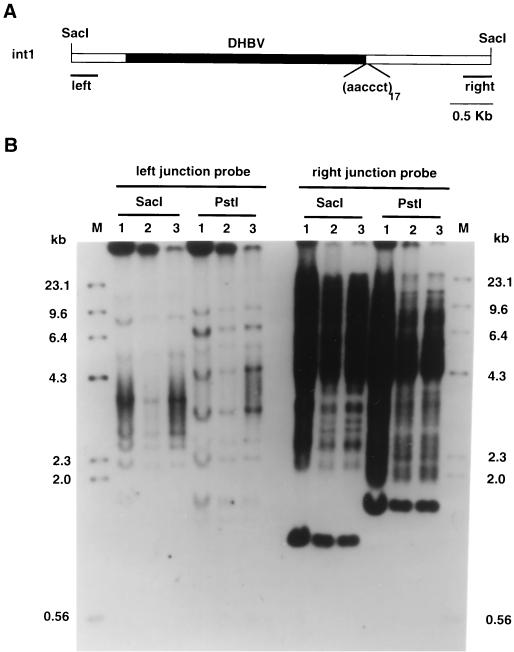
Sequence analysis of the right-hand flanking cellular DNA for the DHBV int1 revealed 17 copies of the sequence aaccct, which is a telomere-associated repeat sequence conserved in vertebrates (Fig. 9A) (18). While hepadnaviral DNA integrations have previously been found to be associated with the cellular repetitive DNA (25), no report has shown them to be directly linked to telomere repeat sequences.
FIG. 9.
Integration of int1 in the telomere repeat sequence and repetitive DNA. (A) Schematic diagram of the genomic SacI fragment containing DHBV DNA integration int1 isolated from subclone P1(5)-4. Filled rectangle indicates the DHBV DNA in the integration (junctions shown in Fig. 8). aaccct indicates the 17 copies of the telomere repeat sequence located at the right junction of the integration. Open rectangle represents flanking cellular DNA. Left and right denote the size and location of the flanking probes used in the Southern blot in panel B. (B) Analysis of the integration site of int1. Left and right junction probes: Southern blots of subclone DNAs digested with either SacI or PstI and hybridized with either the left or right junction probes as denoted. Lanes: 1, untransfected LMH cells; 2, Parental LMH 66-1 DSL DNA; 3, P1(5)-4 subclone DNA. The probes were made from left or right flanking cellular DNA of int3 regions indicated in panel A. The probe regions were amplified by PCR from the cloned genomic SacI fragment, subcloned into plasmid vectors, and isolated for use. For appropriate analysis of the banding patterns, refer to the text. M, marker DNA.
Since telomeres are often at the end of chromosomes, this raised the possibility that the integration had linked the end of one chromosome with another chromosome fragment in a translocation mechanism. To investigate this, we mapped the DHBV integration site using flanking sequence probes from each side of DHBV int1. One problem we had was that both probes hybridized to repetitive DNA elements in the chicken genome (LMH cells are chicken hepatoma cells). As shown in Fig. 9B, we detected numerous distinct bands with both probes in the untransfected LMH cell line (Fig. 9B, lane 1) and also in the LMH 66-1 parental populations (Fig. 9B, lane 2), and the subclone P1(5)-4 that contained int1 (Fig. 9B, lane 3). The results indicated that both sides of the flanking cellular DNA of the integration int1 contained moderately repetitive sequences. Due to the presence of the repeated sequences in each flanking probe, we could not determine conclusively whether a chromosome rearrangement had occurred at the integration site. However, the right cellular flanking DNA detected stronger hybridizing bands than the left flanking DNA.
DISCUSSION
Previous work had suggested that DSL molecules of DHBV might serve as precursors of new DHBV integrations which occur in LMH cells (9, 29). We reasoned that a cell line which produced DSL molecules might exhibit a higher frequency of DHBV DNA integration than a cell line which produced WT OC DHBV DNA. To test this hypothesis, we generated an LMH cell line which produced exclusively DHBV virions containing DSL DHBV DNA molecules (17). Interestingly, the successful establishment of the LMH 66-1 DSL cell line demonstrated that LMH chicken hepatoma cells lack a mechanism for efficient circularization of DSL molecules. In contrast, primary duck hepatocytes circularize DSL DHBV DNAs very efficiently (38). Such circularization would be expected to reduce the pool of linear DHBV DNA integration precursors. This in turn could explain why integration is a rare event in duck liver and why congenitally infected ducks generally do not develop liver cancer.
We used a previously established single-cell subcloning approach to measure the frequency of stable new DHBV DNA integrations which occurred in the cells. In our Southern blot assay, integrations which occurred after the third cell division of colony growth would be undetectable since they would be present in the subclone in less than one of eight of the cells in the final colony. In order to be able to normalize our data between subclones, we grew each subclone to approximately 8 × 106 cells or 24 cell generations.
Subcloning of the parental cell population revealed a 100% frequency of new integrations in the first-cycle subclones. To normalize our data to a defined number of cell generations (8, 9), second- and third-cycle subclones were grown for approximately 24 cell divisions (8 × 106 cells) before harvesting and preparation of genomic DNAs for Southern blot analysis. These procedures matched those carried out earlier for WT DHBV subcloning and allowed direct comparison with our previously published data.
The combined second- and third-generation integration data revealed that 66% of the LMH 66-1 DSL subclones contained new integrations compared to only 16% of comparable LMH-D2 (WT) subclones. This difference was significant at the P = 0.0001 level using a chi-square test. In addition, 33% of the LMH 66-1 DSL subclones contained more than one new integration compared to only 1 to 2% of the LMH-D2 (WT) subclones. As a consequence, the ratio of new integrations per subclone for the LMH 66-1 DSL cell line was 1.25 and for LMH-D2 cells it was only 0.18.
The Southern blot approach we used can detect only the subset of DHBV integrations which are stable in host chromosomes. Although we have observed the loss of specific integrations in LMH lineages, such losses (of previously stable integrations) are rare. Therefore, in our experiments, when we followed cell lineages through three subcloning cycles, we observed a continuous accumulation of stable new integrations. These data suggest that the presence of one integration does not block the acquisition of additional new integrations in the same cell. Furthermore, the percentage of cells which do not contain an integration should decrease with every integration cycle.
Therefore, the percentage of subclones which do not contain a new integration should steadily decrease as a colony goes through successive cell generations. For example, if after three generations (23 cells) one of eight cells in the colony would acquire an integration, that would leave seven-eighths of the cells without a new integration. After three more generations, seven-eighths × seven-eighths (or 7/82), would not have an integration. A mathematical formula to describe the steady decrease in the percentage of cells without an integration is X = (1 − 1/2k)t where X = the percentage of subclones which do not contain a new integration (from Table 1), t = the number of integration cycles needed to arrive at the percentage of clones which do not contain an integration, and k = the number of generations per integration cycle.
We measured X = 33% of the DSL subclones without a new integration. If we hypothesize that k = 3, solving for t we get t = 8.3. Therefore, our calculations predict that it should take tk or 24.9 generations of cell growth to yield a population in which 33% of the cells do not contain an integration. This prediction fits closely with our data, since we grew our subclones approximately 24 generations before harvest. Thus, the data and mathematical model fit a frequency of one stable integration per three generations for DSL DHBV DNAs in LMH 66-1 DSL cells.
In contrast, for WT DHBV, we measured X = 84% of the subclones without a stable new integration. If we hypothesize that k = 5, solving for t we get t = 5.5. Therefore, it should take 5.5 × 5, or 27.5 generations to reach the integration frequency we observed in the LMH-D2 subclones. This is slightly greater than the 24 generations we grew the LMH-D2 subclones. Therefore, we estimate that the integration frequency for WT DHBV in LMH-D2 cells is approximately one integration per four to five generations.
Once we estimated the integration frequency (k = 3 for DSL and approximately 5 for WT), we wanted to predict the total number of integrations which would occur in a colony after t cycles of integration where one integration occurs every k generations. We let St equal the total number of integrations at the tth cycle, where one cycle is equivalent to k generations. Also, we let Nt be the total number of cells at the tth cycle, (i.e., kt generations). Thus, St = t(2k)t−1; t = 1,2, … ; and Nt = 2kt. Thus, the ratio of number of integrations per cell at the tth cycle can be derived as follows: St/Nt = t(2k)t−1/2kt = t2kt−k/2kt = t/2k where k = 1, 2, 3 and t = 1, 2, 3.
According to the formula, for colonies grown 24 generations, the following ratios are predicted. When k = 2, 3, 4, or 5, then St/Nt = 3.0, 1.0, 0.375, or 0.15, respectively. The experimentally determined ratio of new integrations per clone was 1.25 for LMH 66-1 DSL clones, fitting a k = 3 frequency, and the ratio for LMH D2 clones was 0.18, fitting a k = 5 frequency. Therefore, using two different mathematical approaches, i.e., the calculation of the percentage of subclones without an integration and the calculation of the total number of integrations per subclone, the data analysis suggest a frequency of one integration per three generation for DSL DHBV DNAs versus one integration per four to five generation frequency for WT DHBV DNAs.
DHBV integration mechanisms.
The structures of the three DHBV integrations isolated from the LMH 66-1 subclones bear a striking resemblance to a complete DSL DHBV DNA molecule and may be derived directly from the virion DSL DNA. The structures of LMH 66-1 DSL cell line integrations were also comparable to intb, that we previously cloned from LMH-D2 cells (Fig. 8A). intb contains a complete DHBV genome with only three nucleotides deleted from the 3′ terminus of the minus strand. The heterogeneity of the viral junctions of DHBV integrations in LMH 66-1 DSL cells is very similar to integrations in LMH-D2 cells (9a). An overwhelming majority of the viral junctions are localized in the 70-bp region bracketed by the DR sequences. Similar preference of the viral junction sites has been observed for hepadnaviral DNA integrations isolated from tumors (3, 26, 34). One explanation for the highly preferred DHBV DNA junctions in LMH cells is that the majority of the DNA integrations are derived from linearized DHBV DNAs or from the minority linear forms present in the WT DHBV population.
The most frequent viral junctions of DHBV DNA integrations in LMH cells map in the 18-nucleotide region at the 3′ terminus of the minus strand that would contain an RNA:DNA duplex in the DSL DNA (Fig. 8A). The viral junctions near the 5′ terminus of the minus strand are located in the 70-bp cohesive overlap and are more scattered than at the 3′ terminus of the minus strand. This can be explained if the integration substrate DSL DNAs had contained an incompletely elongated DHBV plus strand and therefore had an SS region toward the 5′ end of the minus strand. An SS region may be more susceptible than double-stranded DNA to nuclease digestions prior to or during the integration which would lead to greater heterogeneity at the 5′ viral junction sites.
The significance of 17-copy of the aaccct telomere repeat DNA directly linked to DHBV int1 is not known. Stretches of the aaccct sequence repeat are found primarily in the telomeres of chromosomes (18). The opposite cellular flanking DNA of int1 does not contain additional telomeric repeat sequence. One possibility is that int1 was integrated near the telomeric region of the chromosome at the borderline of telomere repeat sequence and other repetitive DNA. Another possibility is that int1 was integrated into other regions of the chromosomes, e.g., centromeres, that also contain some telomeric repeat sequence. Still another possibility involves modification of the ends of the linear DHBV DNA or the broken ends of chromosomes by the telomerase complex during integration. It is known that telomerase can also synthesize telomeres de novo onto nontelomeric DNA termini in addition to elongating preexisting telomere tracts (18). Telomerase activity can be detected in transformed cell lines, including LMH cells (34a).
ACKNOWLEDGMENTS
We thank Dan Loeb for graciously providing the 5/12 DHBV DSL clone which was used to generate the LMH 66-1 DSL cell line. We also thank Jesse Summers for his thoughts on approaches to calculating integration frequencies and to thank William Mason for his critical reading of the manuscript.
This work was supported by U.S. Public Health Service grant CA37232 from the National Cancer Institute (to C.E.R.), grant DK-17702 from the Digestive Disease Center Grant Program, Cancer Center grant P30CA13330, and NIH training grant CA09060 (to S.S.G.).
REFERENCES
- 1.Buendia M A. Hepatitis B virus and hepatocellular carcinoma. Adv Cancer Res. 1992;59:167–226. doi: 10.1016/s0065-230x(08)60306-1. [DOI] [PubMed] [Google Scholar]
- 2.Condreay L, Aldrich C, Coates L, Mason W, Wu T-T. Efficient duck hepatitis B virus production by an avian tumor cell line. J Virol. 1990;64:3249–3258. doi: 10.1128/jvi.64.7.3249-3258.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dejean A, Sonigo P, Wain-Hobson S, Tiollais P. Specific hepatitis B virus integration in hepatocellular DNA through a viral 11-base-pair direct repeat. Proc Natl Acad Sci USA. 1984;81:5350–5354. doi: 10.1073/pnas.81.17.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de The H, Marchio A, Tiollais P, Dejean A. A novel thyroid hormone receptor-related gene inappropriately expressed in human hepatocellular carcinoma. Nature (London) 1987;347:294–298. doi: 10.1038/330667a0. [DOI] [PubMed] [Google Scholar]
- 5.Fourel G, Trepo C, Bougueleret L, Hengelein B, Ponzetto A, Tiollais P, Buendia M A. Frequent activation of N-myc genes by hepadnavirus insertion in woodchuck liver tumors. Nature. 1990;347:294–298. doi: 10.1038/347294a0. [DOI] [PubMed] [Google Scholar]
- 6.Fourel G, Conturier J, Wei Y, Apiou F, Tiollais P, Buendia M A. Evidence for long-range oncogene activation by hepadnavirus insertion. EMBO J. 1994;13:2526–2534. doi: 10.1002/j.1460-2075.1994.tb06542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ganem D. Hepadnaviridae and their replication. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2703–2737. [Google Scholar]
- 8.Gong S S, Jensen A D, Rogler C E. Loss and acquisition of duck hepatitis B virus integrations in lineages of LMH-D2 chicken hepatoma cells. J Virol. 1996;70:2000–2007. doi: 10.1128/jvi.70.3.2000-2007.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gong S S, Jensen A D, Wang H, Rogler C E. Duck hepatitis B virus integrations in LMH chicken hepatoma cells: identification and characterization of new episomally derived integrations. J Virol. 1995;69:8102–8108. doi: 10.1128/jvi.69.12.8102-8108.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Gong, S., and C. Rogler. Unpublished data.
- 10.Gracef E, Caselmann W H, Wells J, Koshy R. Insertional activation of mevalonate kinase by hepatitis B virus DNA in a human hepatoma cell line. Oncogene. 1994;9:81–87. [PubMed] [Google Scholar]
- 11.Hansen L J, Tennant B C, Seeger C, Ganem D. Differential activation of myc family members in hepatic carcinogenesis by closely related hepatitis B viruses. Mol Cell Biol. 1993;13:659–667. doi: 10.1128/mcb.13.1.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hatada I, Tokino T, Ochiya T, Matsubara K. Co-amplification of integrated hepatitis B virus DNA and transforming gene hst-1 in hepatocellular carcinoma. Oncogene. 1988;3:537–540. [PubMed] [Google Scholar]
- 13.Hino O, Shows T B, Rogler C E. Hepatitis B virus integration site in hepatocellular carcinoma at chromosome 17:18 translocation. Proc Natl Acad Sci USA. 1986;83:8338–8342. doi: 10.1073/pnas.83.21.8338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu J, Seeger C. hsp90 is required for the activity of a hepatitis B virus reverse transcriptase. Proc Natl Acad Sci USA. 1996;93:1060–1064. doi: 10.1073/pnas.93.3.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawaguchi T, Nomura K, Hirayama Y, Kitagawa T. Establishment and characterization of a chicken hepatocellular carcinoma cell line LMH. Cancer Res. 1987;47:4460–4464. [PubMed] [Google Scholar]
- 16.Koeck J, Schlicht H-J. Analysis of the earliest steps of hepadnaviral replication: genome repair after infectious entry into hepatocytes does not depend on viral polymerase activity. J Virol. 1993;67:4867–4874. doi: 10.1128/jvi.67.8.4867-4874.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loeb D D, Hirsch R C, Ganem D. Sequence-independent RNA cleavages generate the primers for plus strand DNA synthesis in hepatitis B virus: implication for other reverse transcribing elements. EMBO J. 1991;10:3533–3540. doi: 10.1002/j.1460-2075.1991.tb04917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Melek M, Greene E C, Shippen D E. Processing of nontelomeric 3′ ends by telomerase: default template alignment and endonucleolytic cleavage. Mol Cell Biol. 1996;16:3437–3445. doi: 10.1128/mcb.16.7.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moroy T, Marchio A, Etiemble J, Trepo C, Tiollais P, Buendia M A. Rearrangement and enhanced expression of C-myc in hepatocellular carcinoma of hepatitis virus infected woodchucks. Nature (London) 1986;324:276–279. doi: 10.1038/324276a0. [DOI] [PubMed] [Google Scholar]
- 20.Nagaya T, Nakamua T, Tokina T, Tsurimoto T, Imai M, Mayumi T, Kamino K, Yamamura K, Matsubara K. The mode of hepatitis B virus DNA integration in chromosomes of human hepatocellular carcinoma. Genes Dev. 1987;1:773–782. doi: 10.1101/gad.1.8.773. [DOI] [PubMed] [Google Scholar]
- 21.Peterson J, Dandri M, Burkle A, Zhang L, Rogler C. Increase in the frequency of hepadnavirus DNA integrations of oxidative DNA damage and inhibition of DNA repair. J Virol. 1997;71:5455–5463. doi: 10.1128/jvi.71.7.5455-5463.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rogler C E. Cellular and molecular mechanisms of hepatocarcinogenesis associated with hepadnavirus infection. Curr Top Microbiol Immunol. 1991;168:103–141. doi: 10.1007/978-3-642-76015-0_6. [DOI] [PubMed] [Google Scholar]
- 23.Rogler C E, Sherman M, Su C Y, Shafritz D A. Deletion in chromosome 11p associated with a hepatitis B integration site in hepatocellular carcinoma. Science. 1985;230:319–322. doi: 10.1126/science.2996131. [DOI] [PubMed] [Google Scholar]
- 24.Seeger C, Summers J, Mason W S. Viral DNA synthesis. Curr Top Microbiol Immunol. 1991;168:41–61. doi: 10.1007/978-3-642-76015-0_3. [DOI] [PubMed] [Google Scholar]
- 25.Shaul Y, Garcia P D, Schonberg S, Rutter W J. Integration of hepatitis B virus DNA in chromosome-specific satellite sequences. J Virol. 1986;59:731–734. doi: 10.1128/jvi.59.3.731-734.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shih C, Burke K, Chou M-J, Zeldis J B, Yang C-S, Lee C-S, Isselbacher K J, Wands J R, Goodman H M. Tight clustering of human hepatitis B virus integration sites in hepatomas near a triple-stranded region. J Virol. 1987;61:3491–3498. doi: 10.1128/jvi.61.11.3491-3498.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 28.Sprengel R, Kuhn C, Will H, Schaller H. Comparative sequence analysis of duck and human hepatitis B virus genome. J Med Virol. 1985;15:323–333. doi: 10.1002/jmv.1890150402. [DOI] [PubMed] [Google Scholar]
- 29.Staprans S, Loeb D D, Ganem D. Mutations affecting hepadnaviral plus-strand synthesis dissociate primer cleavage from translocation and reveal the origin of linear viral DNA. J Virol. 1991;65:1255–1262. doi: 10.1128/jvi.65.3.1255-1262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Summers J, Mason W. Replication of the genome of hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell. 1982;29:403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- 31.Szmuness W. Hepatocellular carcinoma and the hepatitis B virus: evidence for a causal association. Prog Med Virol. 1978;24:40–69. [PubMed] [Google Scholar]
- 32.Tuttleman J, Pourcel C, Summers J. Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell. 1986;47:451–460. doi: 10.1016/0092-8674(86)90602-1. [DOI] [PubMed] [Google Scholar]
- 33.Wang G, Seeger C. Novel mechanism for reverse transcription in hepatitis B viruses. J Virol. 1993;67:6507–6512. doi: 10.1128/jvi.67.11.6507-6512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang H-P, Rogler C E. Topoisomerase I-mediated integration of hepadnavirus DNA in vitro. J Virol. 1991;65:2381–2392. doi: 10.1128/jvi.65.5.2381-2392.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34a.Wang, H. P., and C. E. Rogler. Unpublished data.
- 35.Wang J, Zindy F, Chenivesse X, Lamas E, Henglein B, Brechot C. Modification of cyclin A expression by hepatitis B virus DNA integration in a hepatocellular carcinoma. Oncogene. 1992;7:1653–1656. [PubMed] [Google Scholar]
- 36.Wei Y, Fourel G, Ponzetto A, Silvestro M, Tiollais P, Buendia M-A. Hepadnavirus integration: mechanism of activation of the N-myc2 retrotransposon in woodchuck liver tumors. J Virol. 1992;66:5265–5276. doi: 10.1128/jvi.66.9.5265-5276.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yaninuma K, Kobayashi H, Morishima T, Matsuyama K, Koike K. Multiple integration sites of hepatitis B virus DNA in hepatocellular carcinoma and chronic active hepatitis tissues from children. J Virol. 1987;61:1808–1813. doi: 10.1128/jvi.61.6.1808-1813.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang W, Summers J. Illegitimate replication of linear hepadnaviral DNA through nonhomologous recombination. J Virol. 1995;69:4029–4036. doi: 10.1128/jvi.69.7.4029-4036.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]