ABSTRACT.
In leprosy, early diagnosis is crucial to prevent transmission and onset of disabilities of the disease. The purpose of this study was to determine usefulness of quantitative real-time polymerase chain reaction (PCR) in clinically diagnosed cases of leprosy. Thirty-two leprosy cases were included. The real-time PCR was performed using commercial kit targeting Mycobacterium leprae–specific insertion sequence element. The slit skin smear was positive in two (22.2%) borderline tuberculoid (BT) patients, five (83.3%) borderline lepromatous (BL) patients, and seven (50%) lepromatous leprosy (LL). The positivity of quantitative real-time PCR in BT, BL, LL, and pure neuritic leprosy were 77.8%, 83.3%, 100%, and 33.3%, respectively. Using histopathology as the gold standard, sensitivity of quantitative real-time PCR was 93.1%, and specificity was 100%. The DNA load was higher in LL (3,854.29/106 cells), followed by BL (140.37/106 cells), and BT (2.69/106 cells). Because of the high sensitivity and specificity of real-time PCR, our study strongly suggests the use of real-time PCR as a diagnostic tool for leprosy.
INTRODUCTION
The global prevalence of leprosy after introduction of multidrug therapy in 1982 has declined from 5 million cases in the 1980 s to less than 200,000 at the end of 2016.1 In India, the prevalence rate of 57.8 per 10,000 in 1983 was reduced to less than 1 per 10,000 by the end of 2005, when India was declared to have achieved the WHO target of elimination of the disease as a public health problem.1 In Tamil Nadu, the number of cases on record by the end of March 2015 was 2,888, which increased to 3,207 in 2017.2,3 According to the clinical spectrum proposed by Ridley and Jopling, leprosy is characterized by two polar forms, a paucibacillary (PB) tuberculoid and multibacillary (MB) lepromatous form, and there are also intermediate, borderline tuberculoid (BT), borderline (BB), and borderline lepromatous (BL) forms. Some patients exhibit rare form known as pure neural leprosy (PNL). Pure neural leprosy is difficult to diagnose because lesions are absent and acid-fast bacilli (AFB) are absent in slit smear.4 In PB cases, bacterial load is much lower, and thus early diagnosis is difficult.
In clinical practice, diagnosis is mainly based on clinical presentation along with bacteriological analysis by Ziehl–Neelsen (ZN) smear and histopathology (HP). However, acid-fast staining is less sensitive and requires at least 104 organisms per gram of tissue for reliable detection. A negative ZN smear indicates only that the bacillary load is < 10,000 bacilli/mL, and this does not mean a patient is not infected. This can be problematic for diagnosis of tuberculoid leprosy (TT) and PNL, in which bacterial burden is low or absent.5
In leprosy, early diagnosis is important to prevent transmission and to prevent the onset of disability. Recently, real-time polymerase chain reaction (PCR)-based methods have been developed for diagnosis with high sensitivity and specificity. This offers a culture-independent method that is sensitive enough for identification and confirmation of infection, especially in PB and PNL cases where bacillary load is low or absent. The purpose of this study was to determine usefulness of quantitative real-time PCR (qPCR) in clinically diagnosed cases of leprosy. This report describes the positivity of quantitative real-time PCR and bacterial DNA load in different types of leprosy.
MATERIALS AND METHODS
Study design and site.
The study was a cross-sectional descriptive study conducted in the Department of Microbiology at a tertiary care center in southern India. The study period was from December 2018 to January 2020.
Selection criteria.
Inclusion criteria were patients of all age groups who were clinically diagnosed with leprosy (including pure neuritic leprosy), attending the outpatient department of the Department of Dermatology.
Sample size.
Thirty-two patients during the study period were included. Ethical clearance was given from Institute Ethics Committee before the study commenced. Information concerning the study was given to participants in written format, and informed consent was obtained.
Patient categorization.
Leprosy patients were categorized based on histopathological and clinical findings according to the Ridley and Jopling classification into lepromatous leprosy (LL), BL, BT, and TT.
Samples included for study.
Slit skin smear (SSS) was collected for detection of AFB by acid-fast staining. Skin tissue biopsy from treated and untreated leprosy patients were collected at the time of enrollment into the study. If skin biopsy sample was sufficient, it was divided into two parts, one for histopathological examination including Fite staining and the other for qPCR for detection of Mycobacterium leprae DNA. If the skin biopsy sample was minimal, it was used for HP, and SSS was collected for qPCR.
Microbiological diagnosis.
DNA was extracted using commercially available DNA extraction kit for tissue samples. Real-time PCR (HELINI™ Mycobacterium leprae Real time PCR Kit Helini Biomolecules, Chennai, India) was performed using a commercial kit targeting M. leprae–specific insertion sequence element, according to the manufacturer’s instructions.
Quantitative PCR for M. leprae DNA.
Positive and negative controls were included along with a test run. Internal controls were added either to the sample, before extraction, or directly to the elute. The 25-µL PCR reaction consisted of Probe PCR master mix (10 µL), Leprae/IC PP mix (5 µL), and test DNA (10 µL).
Quantification of M. leprae DNA.
Molecular grade water (450 µL) was added into three 1.5-mL microcentrifuge tubes, labeled as QS2, QS3, and QS4. Fifty microliters of QS1 was added into QS2 and mixed. Serial dilution was carried out by transferring 50 µL of QS2 into QS3, and this was repeated for QS4. Ten microliters of QS1 (200,000 copies/µL), QS2 (20,000 copies/µL), QS3 (2,000 copies/µL) and QS4 (200 copies/µL) (in duplicates) were used for each run.
Cycle threshold (Ct) value for all four standards were noted (mean value of duplicates was taken), and standard curve was put up against the DNA copy number in each standard. DNA copy number was converted to log number and used for plotting standard curve against Ct value. Similarly, Ct value of M. leprae DNA in all test samples were noted and extrapolated against the standards using standard curve. Any of the preparations (QS1–QS4) was used as positive control.
Normalization of DNA copy number with a reference gene (Beta2 microglobulin).
Beta2 microglobulin (B2M) is a reference gene (housekeeping gene) frequently used to normalize DNA or RNA levels between samples and is present in all nucleated cells. Thus, normal human blood (white blood cells [WBCs]) was used for preparation of standards. Five milliliters of venous blood was collected in an EDTA anticoagulated tube and total WBC count was determined using Sysmex XT (Sysmex Corporation, Kobe, Hyogo, Japan) 2000i. Total WBC count was 6,000 cells/µL of blood. For DNA extraction (QIAamp DNA minikit, Qiagen India Ltd., New Delhi, India), 200 µL of blood was used. The final elution volume was 200 µL, which will have 1,200,000 WBCs. Ten microliters of elute was used as QS1 (60,000 cells),4 which was serially diluted (1 in 10 dilution) to obtain three dilutions (QS2, QS3, QS4) with a cell number of 6,000, 600, and 60, respectively. Real-time PCR was conducted for these four standards, and Ct values were determined.
The 25-µL PCR reaction consisted of master mix (12 µL), forward primer (1 µL), reverse primer (1 µL), probe (0.4 µL), DNA (10 µL), and nuclease free water (0.1 µL). Final DNA copy numbers were expressed in terms of DNA copy number/number of cells. The Ct values of all these standards against corresponding cell number (logarithmic value) were used to plot standard curve. Expression of B2M in all clinical samples were determined by real-time PCR, and Ct values were extrapolated in the standard curve to obtain final cell number.
Data analysis.
Patient data, including demographic details, clinical type, WHO classification and HP report, was noted at time of enrolment into study. Categorical variables such as gender, type of leprosy, HP findings, and quantitative real time PCR positivity were expressed as frequency and percentages. Continuous variables such as age (years) and bacterial DNA load (copies/mL) were expressed as mean ± SD and median. For comparison of positivity of quantitative real time PCR in different types of leprosy, χ2 test or Fisher Exact test were used. For comparison of bacterial DNA load in different types of leprosy, independent t test was used.
RESULTS
A total of 32 clinically suspected leprosy patients were included in this study. The mean age of patients was 44.84 ± 12.26 (range, 26–65), Figure 1. Of these, 25 (78.1%) were male and 7 (21.9%) were female. These patients were clinically categorized according to Ridley Jopling classification. Of the 32 patients, 9 (28.1%) were classified as BT, 6 (18.8%) as BL, 14 (43.8%) as LL and 3 (9.4%) as PNL. According to WHO classification, 14 (43.7%) had smear positive MB leprosy, 15 (46.9%) had smear negative MB leprosy and 3 (9.4%) had PNL.
Figure 1.
Mean age of patients with leprosy.
The mean age and gender of different clinical types of leprosy are shown in Tables 1 and 2, respectively.
Table 1.
Mean age of different clinical types of leprosy
| Type of leprosy | Mean age | P value |
|---|---|---|
| Borderline tuberculoid (n = 9) | 47.0 ± 10.49 | 0.717 |
| Borderline lepromatous (n = 6) | 39.67 ± 13.68 | |
| Lepromatous leprosy (n = 14) | 45.79 ± 12.13 | |
| Pure neuritic leprosy (n = 3) | 44.33 ± 18.88 |
Table 2.
Gender of different clinical types of leprosy
| Type of leprosy | Male | Female |
|---|---|---|
| Pure neuritic leprosy (n = 3) | 3 (100%) | 0 |
| Borderline tuberculoid (n = 9) | 4 (44.4%) | 5 (55.6%) |
| Borderline lepromatous (n = 6) | 5 (83.3%) | 1 (16.7%) |
| Lepromatous leprosy (n = 14) | 13 (92.9%) | 1 (7.1%) |
Slit skin smear.
The SSS was positive in 14 (43.8%) of these leprosy patients: two (22.2%) BT patients, five (83.3%) BL patients, and seven (50%) LL patients.
Fite stain.
Skin biopsy was collected from all patients for Fite stain and HP. Of the 32 patients, 17 (53.1%) were positive for AFB by Fite stain. Fite stain was positive in 1 (11.1%) BT patient, four (66.7%) BL patients, 11 (78.6%) LL, and 1 (33.3%) PNL patient. Both SSS and Fite stain positivity were statistically significant (P = 0.029; Table 3).
Table 3.
Association between SSS positivity and Fite stain positivity
| Fite stain | Total | |||
|---|---|---|---|---|
| Positive | Negative | |||
| SSS | Positive | 11 | 3 | 14 |
| Negative | 6 | 12 | 18 | |
| Total | 17 | 15 | 32 | |
SSS = slit skin smear.
Histopathology.
Of the 32 clinically suspected cases of leprosy, 29 (90.6%) had characteristic features suggestive of leprosy, and three (9.4%) lacked findings suggestive of leprosy. One BL leprosy patient who has completed 12 months of treatment showed evidence of only small-vessel vasculitis, and two pure neuritic leprosy were negative histopathologically.
Quantitative real-time PCR.
Quantitative real-time PCR was performed on samples collected from all 32 patients: skin biopsy from 24 (75.0%) patients, SSS from six (18.8%) patients, and nerve biopsy from two (6.3%) patients. We studied the cell count of various samples. The median cell counts of skin biopsy, nerve biopsy, and SSS were 28,125, 4,546, and 114, respectively. The positivity of qPCR was 89.7% (26/29) in MB leprosy and 33.3% (1/3) in PNL. The positivity of qPCR in BT, BL, LL, and PNL were 77.8% (7/9), 83.3% (5/6), 100% (14/14), and 33.3% (1/3), respectively.
Sensitivity and specificity of qPCR.
The sensitivity and specificity of qPCR using HP as the gold standard was 93.1% and 100%, respectively. The qPCR result was negative in two histopathologically confirmed cases of leprosy (Table 4). Both these cases were reported as BT type based on presence of epitheloid granuloma in one and granuloma around nerve bundles and periadnexal structures along with occasional multinucleated giant cell in the other. Both were SSS and Fite-stain negative.
Table 4.
Comparison of histopathology and real-time PCR report
| Histopathology report | Total | |||
|---|---|---|---|---|
| Positive | Negative | |||
| Realtime PCR report | DNA detected | 27 | 0 | 27 |
| DNA not detected | 2 | 3 | 5 | |
| Total | 29 | 3 | 32 | |
PCR = polymerase chain reaction.
Bacterial DNA load.
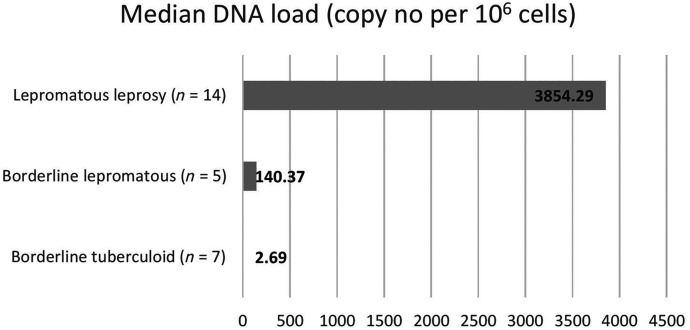
The median M. leprae DNA load and mean Ct value of different types of leprosy are summarized in Table 5. Among the three pure neuritic leprosy cases, only one was positive by qPCR with the DNA load of 2,717.92 copy no per 106 cells and Ct value of 30.9.
Table 5.
Mycobacterium leprae DNA load and mean Ct value of different types of leprosy
| Type of leprosy | Median DNA load* | Mean Ct value |
|---|---|---|
| Borderline tuberculoid (n = 7) | 2.69 | 34.79 ± 6.62 |
| Borderline lepromatous (n = 5) | 140.37 | 32.02 ± 4.74 |
| Lepromatous leprosy (n = 14) | 3,854.29 | 31.59 ± 5.69 |
Ct = cycle threshold.
Copy no. per 106 cells.
DISCUSSION
Leprosy is a chronic granulomatous disease caused by a slow growing, obligately intracellular organism, M. leprae. Diagnosis continues to be challenging because the organism is noncultivable, and conventional methods have low sensitivity. In clinical practice, diagnosis is mainly based on clinical presentation along with bacteriological analysis by ZN smear and HP. However, low sensitivity of acid-fast staining further compromises diagnosis, especially in cases of tuberculoid leprosy and pure neural leprosy where AFB are rare or absent.5
Thirty-two patients were included in the study; of these, 25 (78.1%) were male and seven (21.9%) were female, with a mean age of 44.84 ± 12.26 (range: 26–65). This is in concordance with other studies that document a male preponderance of leprosy, with a 2:1 ratio. For instance, Liu et al. reported a prevalence of 71.6% (2,075/2,900) in males and 28.4% (825/2,900) in females.6 In an earlier study conducted by Kumar et al., male patients accounted for 45.3% with a mean age of 34.2 years.7 Similarly, in another study by Bang et al. involving 69 patients, 47 were men and 22 were women, with an average age group of 32.5 and 33.4 years in males and females, respectively.8 In a country like India, sociocultural variables such as illiteracy and low status accorded to females may contribute to decreased reporting of cases in women. With regard to bacillary load, studies report a higher percentage of multibacillary cases in males.9 Similar to this, we have noted overall prevalence of LL, characterized by high bacillary load, of 92.9% (13/14) in males and 7.1% (1/14) in females (Table 2).
Slit skin smear is the primary method for leprosy diagnosis in underdeveloped nations, where a majority of cases are detected. It is affordable and less invasive but has a low sensitivity.10 Thus, a selective approach in use of SSS diagnostics in leprosy patients is preferred, for example, in decision-making about changing patient treatment. For instance, a positive SSS in PB patients may indicate MB status, hence changing treatment duration; however, disease cannot be ruled out in an MB patient with negative SSS due to its low sensitivity.11 In our study, 15 (46.9%) were SSS positive by modified ZN staining. Other studies have demonstrated an SSS positivity of 33.3% (20/60), 53.6% (37/69), and 54% (36/66).8,12,13 A higher microscopic positivity (60.78%, 80.15%) has been reported by few researchers; this may be due to the use of a biopsy specimen rather than SSS.14,15 Also, a study on revisiting the role of SSS with critical analysis of the applicability of PCR in diagnosis determined acid-fast positivity of 100% in LL, whereas it was negative in tuberculoid, pure neuritic, and indeterminate cases.16 In another study by Goulart et al., SSS was positive for M. leprae in LL (95.23%) and BL (91.6%) patients.17 Our study reported the highest SSS positivity (83.3%) in BL, followed by LL (50%) and BT (22.2%).
We also noted a 53.1% (17/32) positivity for AFB by Fite stain. It is lower compared with a study by Bang et al., who found positivity of 81.6% (40/69).8 Reja et al. also demonstrated 60% (99/165) positivity for Fite staining.18 In the present study, Fite stain positivity was 78.6% for LL, 66.7% for BL patients, 11% for BT patients, and 33.3% for PNL patients. There was significant association between SSS positivity and Fite stain positivity (P = 0.029) in our analysis (Table 3).
In our study, qPCR was performed in all 32 samples, and positivity was 89.7% (26/29) for MB and 33.3% (1/3) for PNL. As we did not have any PB cases, we could not analyze positivity in this population. Various studies also demonstrate a substantially higher positivity rate in MB patients8,12,13,16,19 compared with PB, but development of real-time PCR methods has still made the rapid detection of M. leprae possible, even in samples with microscopically undetectable AFB. Routine use in these cases would ensure a proper diagnosis, particularly in distinguishing leprosy from other illness with similar symptoms.20 Martinez et al. published findings on skin biopsy samples in which highest positivity was obtained in MB (LL, BL, BB) patients (100%), followed by PB (BT; 62.5%) and PNL (50%) patients.4 Similarly, we determined that positivity rate of real-time PCR in LL was 100%, followed by BL (83.3%), BT (77.8%), and PNL (33.3%) in our cases.
We have analyzed sensitivity and specificity of real-time PCR using HP as the gold standard and found it to have sensitivity of 93.1% and specificity of 100%. In a study by Goulart et al., real-time PCR had a sensitivity of 91.3%.17 Similarly Bang et al. reported PCR specificity of 100%, which correlates with our findings8; the authors compared PCR results with HP examination and found PCR to be positive in all HP-positive cases; in addition, 45% (13/29) of HP-negative cases were positive by PCR. In our study, qPCR was negative in two HP confirmed cases of leprosy (Table 4); both these case were reported as BT based on microscopical examination of tissue section. Both were also negative for SSS and Fites stain. In contrast to conventional methods such as SSS, sensitivity of PCR is undoubtedly much higher, particularly in negative SSS cases. Kramme et al., on analysis of sensitivity compared with SSS, found PCR sensitivity as 88.9% in patients with positive SSS and 33.3% in patients with negative SSS.19 In another study on rapid detection and quantitation of M. leprae in skin specimens, real-time PCR was found to have 100% sensitivity in MB and 50% sensitivity in PB patients.13 Hence, our work highlights the significance and usefulness of PCR in early case detection of leprosy and limiting deformities. We also quantified DNA load in different types of leprosy and found the highest load in LL (3,854.29/106 cells), followed by BL (140.37/106 cells) and BT (2.69/106 cells), as shown in Figure 2. In a study by Lini et al., the maximum DNA copy number was obtained in LL followed by BL and BT, which is in agreement with our study.21
Figure 2.
Mycobacterium leprae DNA load of different types of leprosy.
In conclusion, we found a significant association between SSS positivity and Fite stain positivity; therefore, SSS can be used as an earlier detection tool for the diagnosis of leprosy. In our study, real-time PCR positivity for LL is 100%; hence, LL can be easily detected by PCR compared with other clinical diagnostics. In our study, 77.8% of BL cases were positive by real-time PCR, whereas Fite stain was less sensitive. Because of the high sensitivity and specificity of real-time PCR, our study strongly suggests the use of real-time PCR as a diagnostic tool for Leprosy.
ACKNOWLEDGMENTS
We thank the Jawaharlal Institute of Postgraduate Medical Education and Research (JIPMER) for supporting this project with the intramural research grant. The American Society of Tropical Medicine and Hygiene (ASTMH) assisted with publication expenses.
REFERENCES
- 1. Rao Pn, Suneetha S, 2018. Current situation of leprosy in India and its future implications. Indian Dermatol Online J 9: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Central Leprosy Division , 2016. National Leprosy Eradication Programme Annual Report 2015–2016. New Delhi, India: CLD. [Google Scholar]
- 3. Central Leprosy Division , 2017. National Leprosy Eradication Programme Annual Report 2016–2017. New Delhi, India: CLD. [Google Scholar]
- 4. Martinez AN, Britto CFPC, Nery JAC, Sampaio EP, Jardim MR, Sarno EN, Moraes MO, 2006. Evaluation of real-time and conventional PCR targeting complex 85 genes for detection of Mycobacterium leprae DNA in skin biopsy samples from patients diagnosed with leprosy. J Clin Microbiol 44: 3154–3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Truman RW, Andrews PK, Robbins NY, Adams LB, Krahenbuhl JL, Gillis TP, 2008. Enumeration of Mycobacterium leprae using real-time PCR. PLoS Negl Trop Dis 2: e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu Y-Y, Yu M-W, Ning Y, Wang H, 2018. A study on gender differences in newly detected leprosy cases in Sichuan, China, 2000–2015. Int J Dermatol 57: 1492–1499. [DOI] [PubMed] [Google Scholar]
- 7. Kumar A, Girdhar A, Girdhar BK, 2012. Six months fixed duration multidrug therapy in paucibacillary leprosy: risk of relapse and disability in Agra PB cohort study. BMJ Open 2: e001403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bang PD, Suzuki K, Phuong LT, Chu TM, Ishii N, Khang TH, 2009. Evaluation of polymerase chain reaction-based detection of Mycobacterium leprae for the diagnosis of leprosy. J Dermatol 36: 269–276. [DOI] [PubMed] [Google Scholar]
- 9. Sarkar R, Pradhan S, 2016. Leprosy and women. Int J Womens Dermatol 2: 117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Siwakoti S, Rai K, Bhattarai NR, Agarwal S, Khanal B, 2016. Evaluation of polymerase chain reaction (PCR) with slit skin smear examination (SSS) to confirm clinical diagnosis of leprosy in eastern Nepal. PLoS Negl Trop Dis 10: e0005220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Demsiss W, Van Henten S, Takarinda KC, Kamau EM, Abdela SG, 2022. Slit-skin smear for the classification of leprosy; are we wasting time and resource? J Infect Dev Ctries 16: 3S–7S. [DOI] [PubMed] [Google Scholar]
- 12. Torres P, Camarena JJ, Gomez JR, Nogueira JM, Gimeno V, Navarro JC, Olmos A, 2003. Comparison of PCR mediated amplification of DNA and the classical methods for detection of Mycobacterium leprae in different types of clinical samples in leprosy patients and contacts. Leprosy 74: 18–30. [PubMed] [Google Scholar]
- 13. Rudeeaneksin J, Srisungngam S, Sawanpanyalert P, Sittiwakin T, Likanonsakul S, Pasadorn S, Palittapongarnpim P, Brennan PJ, Phetsuksiri B, 2008. LightCyclerTM real-time PCR for rapid detection and quantitation of Mycobacterium leprae in skin specimens. FEMS Immunol Med Microbiol 54: 263–270. [DOI] [PubMed] [Google Scholar]
- 14. Yoon KH, Cho SN, Lee MK, Abalos RM, Cellona RV, Fajardo TT, Jr., Guido LS, Dela Cruz EC, Walsh GP, Kim JD, 1993. Evaluation of polymerase chain reaction amplification of Mycobacterium leprae-specific repetitive sequence in biopsy specimens from leprosy patients. J Clin Microbiol 31: 895–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Azevedo MCS, Ramuno NM, Fachin LR, Tassa M, Rosa PS, Belone AF, Diório SM, Soares CT, Garlet GP, Trombone AP, 2017. qPCR detection of Mycobacterium leprae in biopsies and slit skin smear of different leprosy clinical forms. Braz J Infect Dis 21: 71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Banerjee S, Biswas N, Kanti Das N, Sil A, Ghosh P, Hasanoor Raja AH, Dasgupta S, Kanti Datta P, Bhattacharya B, 2011. Diagnosing leprosy: revisiting the role of the slit-skin smear with critical analysis of the applicability of polymerase chain reaction in diagnosis: PCR versus slit-skin smear in diagnosis of leprosy. Int J Dermatol 50: 1522–1527. [DOI] [PubMed] [Google Scholar]
- 17. Goulart IMB, Cardoso AM, Santos MS, Gonçalves MA, Pereira JE, Goulart LR, 2007. Detection of Mycobacterium leprae DNA in skin lesions of leprosy patients by PCR may be affected by amplicon size. Arch Dermatol Res 299: 267–271. [DOI] [PubMed] [Google Scholar]
- 18. Reja AH, Biswas N, Biswas S, Dasgupta S, Chowdhury IH, Banerjee S, Chakraborty T, Dutta PK, Bhattacharya B, 2013. Fite–Faraco staining in combination with multiplex polymerase chain reaction: a new approach to leprosy diagnosis. Indian J Dermatol Venereol Leprol 79: 693–700. [DOI] [PubMed] [Google Scholar]
- 19. Kramme S, Bretzel G, Panning M, Kawuma J, Drosten C, 2004. Detection and quantification of Mycobacterium leprae in tissue samples by real-time PCR. Med Microbiol Immunol (Berl) 193: 189–193. [DOI] [PubMed] [Google Scholar]
- 20. Yan W, Xing Y, Yuan LC, Tan FY, De Yang R, Zhang Y, Li H-Y, 2014. Application of RLEP real-time PCR for detection of M. leprae DNA in paraffin-embedded skin biopsy specimens for diagnosis of paucibacillary leprosy. Am J Trop Med Hyg 90: 524–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lini N, Shankernarayan NP, Dharmalingam K, 2009. Quantitative real-time PCR analysis of Mycobacterium leprae DNA and mRNA in human biopsy material from leprosy and reactional cases. J Med Microbiol 58: 753–759. [DOI] [PubMed] [Google Scholar]