ABSTRACT.
Staphylococcus aureus causes a wide range of illnesses, from skin infections and persistent bone infections to life-threatening septicemia and endocarditis. Methicillin-resistant S. aureus (MRSA) is one of the most common bacteria that cause nosocomial and community-acquired infections. Clindamycin is one of the most effective treatments for several bacterial infections. Despite this, these infections may develop inducible clindamycin resistance during treatment, leading to treatment failure. This study determined the incidence of inducible clindamycin resistance among S. aureus clinical isolates. A total of 800 S. aureus strains were identified from clinical samples collected from several university hospitals in Egypt. All isolates were examined for the presence of MRSA using cefoxitin (30 μg) and the Kirby Bauer disk diffusion technique. The induction phenotypes of all 800 S. aureus strains were evaluated using the disk approximation test (D test), as recommended by the Clinical and Laboratory Standard Institute. Of the 800 strains of S. aureus, 540 (67.5%) were identified as MRSA and 260 (32.5%) were classified as methicillin-sensitive S. aureus (MSSA). In MRSA infections, clindamycin constitutive and inducible resistance was more frequent than in MSSA infections (27.8% versus 11.5% and 38.9% versus 15.4%, respectively). Clindamycin-sensitive strains were more prevalent in MSSA (53.8%) than in MRSA (20.4%) infections. In conclusion, the frequency of constitutive and inducible clindamycin resistance in MRSA isolates emphasizes the need to use the D test in routine antimicrobial susceptibility testing to evaluate clindamycin susceptibility, as the inducible resistance phenotype can inhibit the action of clindamycin and thus affect treatment efficacy.
INTRODUCTION
Staphylococcus aureus is a major cause of nosocomial and community-acquired infections, from simple skin and soft tissue infections to life-threatening systemic infections. The incidence of methicillin-resistant S. aureus (MRSA) is on the rise. As antimicrobial resistance patterns increase, so do treatment options for infections, necessitating that physicians modify the antimicrobials they prescribe, including the use of macrolide–lincosamide–streptomycin group B (MLSB) antibiotics.1 Because of its better pharmacokinetics and pharmacodynamics, the MLSB antibiotic clindamycin is recommended for the treatment of MRSA infections.2,3 Erythromycin induces clindamycin resistance via boosting erythromycin ribosome methylase synthesis (erm).4 Using the disk diffusion approach, five isolates with inducible clindamycin resistance exhibit erythromycin resistance, but a false sensitivity to clindamycin.5 Clinically, inducible clindamycin resistance is a significant problem because of the difficulty in recognizing the infections caused by these isolates using standard laboratory procedures.6 Inability to identify inducible clindamycin-resistant S. aureus may result in clindamycin overdose and therapy failure.6,7
The Clinical and Laboratory Standards Institute (CLSI) recommends the erythromycin–clindamycin disk approximation test (D test) for the identification of the clindamycin-resistant inducible phenotype in S. aureus isolates.8
We also wondered whether this phenotype was more common in patients with diabetes mellitus than patients who do not have diabetes mellitus. In 2010, type II diabetes accounted for 90% of the world’s 285 million patients with diabetes mellitus. The number of these patients is expected to increase by 2030.9
The trend of clindamycin resistance among S. aureus clinical isolates was not detected by our literature research. Our purpose was to determine the prevalence of constitutive and inducible clindamycin resistance in clinical isolates of S. aureus from many locations, including Al-Hussein University Hospital. This finding might prevent doctors from misusing clindamycin and may enhance patient outcomes clinically.
MATERIALS AND METHODS
This prospective study was conducted on 800 patients at highly specialized intensive care unit (ICU) centers concerned with infectious disease management, including the Microbiology Unit, Clinical Pathology Department, Al-Hussein University Hospital; National Liver Institute; and hospitals of Menoufia University, Al-Azhar University, Helwan University, Ain Shams University, Tanta University, Alexandria University, Suez University, and Shebin El-Kom Teaching Hospital for 2 years (October 2019–October 2021). Diabetes mellitus was diagnosed by laboratory testing in accordance with the criteria set by the WHO and the American Diabetes Association—namely, a fasting blood glucose level of ≥ 126 mg/dL or a 2-hour postprandial blood glucose level of ≥ 200 mg/dL.9
The clinical samples (including pus, urine, blood, sputum, endotracheal aspirates, and others) were received during the study period from various inpatient units, such as ICUs, neonatal ICUs, and hospital wards, were processed according to standard bacteriological techniques for the isolation and identification of bacterial pathogens. Pus sources were skin infections, surgical wound infections, orthopedic surgical wounds, urological infections, or burns.10
Early identification of S. aureus was based on colony characteristics of organisms studied on Mannitol salt agar, gram reactions, coagulase detection on slides or in test tubes, and catalase detection on slides.
The isolates of identified S. aureus organisms were subjected to the following tests. First, using the disk diffusion method, MRSA was identified phenotypically based on its resistance to cefoxitin (30 μg) (Oxoid, Basingstoke, UK) on Muller-Hinton agar (Oxoid). On the basis of the CLSI’s 2019 guidelines,8 the zone of inhibition was determined, and the S. aureus bacteria were categorized as methicillin sensitive (diameter, ≥ 22 mm) or methicillin resistant (diameter, ≤ 21 mm). Second, inducible clindamycin resistance was found using a D test in accordance with CLSI standards.8 A test for disk approximation was conducted. The plates were incubated after a 2-μg clindamycin disk was put 21 mm from the edge of a 15-μg erythromycin disk. After an overnight incubation at 37°C, three distinct phenotypes were observed. The D test was positive for the inducible MLSB (iMLSB) phenotype. The iMLSB S. aureus isolates were resistant to erythromycin (zone size ≤ 13 mm) but sensitive to clindamycin (zone size ≥ 21 mm) and exhibited a D-shaped zone of inhibition surrounding the clindamycin disk that flattened toward the erythromycin disk (i.e., D test positive; Figure 1). Staphylococcus aureus isolates exhibited resistance to erythromycin (zone size ≤ 13 mm) and clindamycin (zone size ≤ 14 mm), with a circular zone of inhibition around the clindamycin (Figure 2). For the methicillin-sensitive (MS) phenotype, S. aureus isolates exhibited a resistance to erythromycin (zone size ≤ 13 mm) but a sensitivity to clindamycin (zone size ≥ 21 mm) and a circular zone of inhibition surrounding the clindamycin; they were negative for the D test (Figures 3 and 4).
Figure 1.
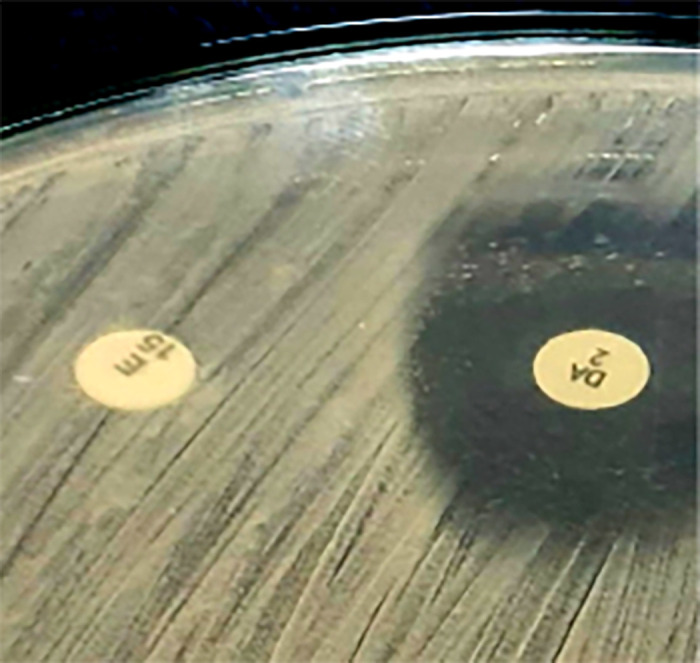
Disk approximation test positive for the inducible macrolide-lincosamide-streptogramin B phenotype. Staphylococcus aureus isolates showed resistance to erythromycin.
Figure 2.
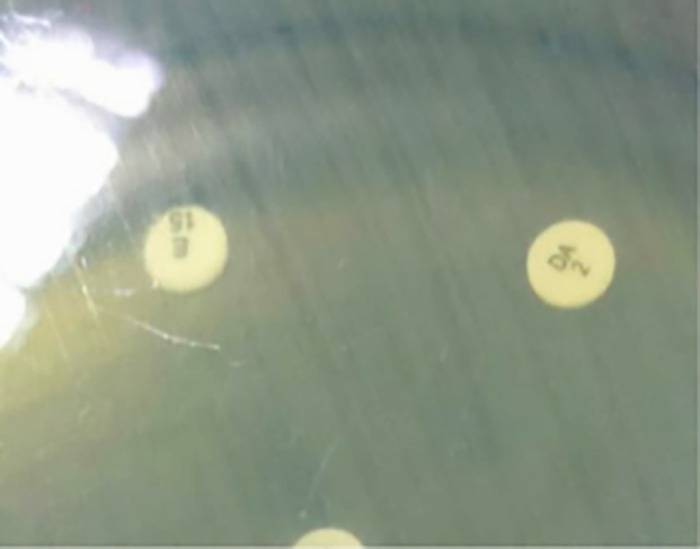
Constitutive macrolide-lincosamide-streptogramin B phenotype. Staphylococcus aureus isolates show resistance to both erythromycin and clindamycin.
Figure 3.

Methicillin-sensitive phenotype. Staphylococcus aureus isolates exhibit resistance to erythromycin but sensitivity to clindamycin.
Figure 4.

Methicillin-sensitive phenotype. Staphylococcus aureus isolates exhibit resistance to both clindamycin and erythromycin.
Statistical methods.
The descriptive statistics for quantitative variables included the mean and SD, whereas those for categorical variables included frequencies and simple percentages. The variables that were categorical were examined using the χ2 test. The Kolmogorov–Smirnov test demonstrated that normality exists. There was no variable found to have a normal distribution. Consequently, the Kruskal–Wallis test was used to assess the group effect. The Dwass–Steel–Critchlow–Fligner multiple comparison approach was used for pairwise two-sample comparisons among the three groups. Spearman’s rank correlation coefficients were determined. For data analysis, version 9.4 of the SAS software was used (SAS Institute, Cary, NC).
RESULTS
We enrolled 800 patients in our study. The patients were 12 to 70 years old; 460 were male (57.5%) and 340 were female (42.5%). From diverse clinical specimens, 800 strains of S. aureus were isolated then identified. Table 1 shows the different clinical samples from which S. aureus was isolated. Staphylococcus aureus strains were isolated most commonly from pus (40%), blood (27.5%), and tracheal aspirates (15%). All S. aureus strains were tested for cefoxitin (30 μg) sensitivity. Table 2 shows the sensitivity pattern of S. aureus for cefoxitin. Baseline comorbidities were also recorded.
Table 1.
Sample distribution in relation to hospital wards (N = 800 patients)
| Type of specimen | Ward type, n (%) | n | % | |||||
|---|---|---|---|---|---|---|---|---|
| Intensive care unit | Surgical | Medical | ||||||
| Females (n = 135) | Males (n = 245) | Females (n = 170) | Males (n = 160) | Females (n = 40) | Males (n = 50) | |||
| Pus | 15 (11.1) | 25 (10.2) | 134 (78.8) | 146 (91.3) | – | – | 320 | 40 |
| Blood | 43 (31.9) | 97 (39.6) | 15 (8.8) | 5 (3.1) | 25 (62.5) | 35 (70.0) | 220 | 27.5 |
| Tracheal aspirate | 40 (39.6) | 80 (32.7) | – | – | – | – | 120 | 15 |
| Sputum | 12 (8.9) | 38 (15.5) | 5 (2.9) | 5 (3.1) | 8 (20.0) | 12 (24.0) | 80 | 10 |
| Urine | 25 (18.5) | 5 (2.0) | 16 (9.4) | 4 (2.5) | 7 (17.5) | 3 (6.0) | 60 | 7.5 |
Table 2.
Cefoxitin susceptibility pattern in patients with and without diabetes mellitus
| Susceptibility | No. of isolates | % | Interpretation |
|---|---|---|---|
| Resistant (≤ 21-mm diameter) | 540 | 67.5 | Methicillin–resistant Staphylococcus aureus |
| Diabetic | 405 | 74 | – |
| Nondiabetic | 145 | 26 | – |
| Sensitive (≥ 22-mm diameter) | 260 | 32.5 | Methicillin-sensitive S. aureus |
| Diabetic | 95 | 36.5 | – |
| Nondiabetic | 165 | 63.5 | – |
Of the 800 S. aureus strains tested for resistance to erythromycin (15 μg) and clindamycin (2 μg) in combination (i.e., they were tested with the D test), 180 strains (22.5%) were positive on the D test (i.e., they were the inducible clindamycin-resistant iMLSB phenotype; see Figure 1), 130 strains (16.25%) were negative on the D test (i.e., they were the MS phenotype; see Figure 3), 220 strains (27.5%) were resistant to both erythromycin and clindamycin (i.e., they were the constitutive MLSB [cMLSB] phenotype; see Figure 2), and 270 strains (33.75%) were sensitive to both clindamycin and erythromycin (i.e., they were the S phenotype; Figure 4). There was no isolate that showed sensitivity to erythromycin and resistance to clindamycin (Table 3).
Table 3.
Different phenotypes by induction test in patients with and without diabetes Mellitus
| Resistant phenotype | Induction test phenotype | ERY susceptibility | CLI susceptibility | No. of patients with diabetes | Total no. of isolates | % |
|---|---|---|---|---|---|---|
| iMLSB | D test positive | R | S | 118 | 180 | 22.5 |
| MS | D test negative | R | S | 82 | 130 | 16.25 |
| cMLSB | R | R | R | 176 | 220 | 27.5 |
| No resistance | S | S | S | 124 | 270 | 33.75 |
CLI = clindamycin; cMLSB = constitutive macrolide–lincosamide–streptomycin group B; D test = disk approximation test; ERY = erythromycin; iMLSB = inducible macrolide–lincosamide–streptomycin group B; MS = methicillin sensitive; R = resistant; S = sensitive.
Table 4 showed the resistance phenotypes observed among MRSA isolates. Of the 540 MRSA isolates, 150 (27.8%) had iMLSB phenotypes and 210 (38.9%) had cMLSB phenotypes, 110 (20.4%) showed no resistance, and 70 (12.9%) showed the MS phenotype. The resistance traits of the methicillin-sensitive S. aureus (MSSA) isolates are shown in Table 5. Of the 260 MSSA isolates, 30 (11.5%) had an iMLSB phenotype, 50 (19.3%) had the MS phenotype, 40 (15.4%) had the cMLSB phenotype, and 140 (53.8%) showed no resistance. Table 6 showed the sample distribution among patients with diabetes mellitus (62.5%) and patients without diabetes mellitus (37.5%). In people with diabetes mellitus, MRSA infections are prevalent and more common.
Table 4.
Resistant phenotypes in methicillin-resistant Staphylococcus aureus
| Resistant phenotype | Induction test phenotype | ERY susceptibility | CLI susceptibility | No. of isolates | % |
|---|---|---|---|---|---|
| iMLSB | D test positive | R | S | 150 | 27.8 |
| MS | D test Negative | R | S | 70 | 12.9 |
| cMLSB | R | R | R | 210 | 38.9 |
| No resistance | S | S | S | 110 | 20.4 |
CLI = clindamycin; cMLSB = constitutive macrolide–lincosamide–streptomycin group B; D test = disk approximation test; ERY = erythromycin; iMLSB = inducible macrolide–lincosamide–streptomycin group B; MS = methicillin sensitive; R = resistant; S = sensitive.
Table 5.
Resistant phenotypes in methicillin-sensitive Staphylococcus aureus
| Resistant phenotype | Induction test phenotype | ERY susceptibility | CLI susceptibility | No. of isolates | % |
|---|---|---|---|---|---|
| iMLSB | D test positive | R | S | 30 | 11.5 |
| MS | D test negative | R | S | 50 | 19.3 |
| cMLSB | R | R | R | 40 | 15.4 |
| No resistance | S | S | S | 140 | 53.8 |
CLI = clindamycin; cMLSB = constitutive macrolide–lincosamide–streptomycin group B; D test = disk approximation test; ERY = erythromycin; iMLSB = inducible macrolide–lincosamide–streptomycin group B; MS = methicillin sensitive; R = resistant; S = sensitive.
Table 6.
Sample distribution in patients with and without diabetes mellitus (N = 800)
| Type of specimen | Group | |||
|---|---|---|---|---|
| Diabetes mellitus (n = 500) | No diabetes mellitus (n = 300) | |||
| N | % | n | % | |
| Pus | 195 | 39 | 125 | 41.7 |
| Blood | 130 | 26 | 90 | 30 |
| Tracheal aspirate | 70 | 14 | 50 | 16.7 |
| Sputum | 55 | 11 | 25 | 8.3 |
| Urine | 50 | 10 | 10 | 3.3 |
DISCUSSION
Staphylococcus aureus is one of the most prevalent organisms that causes pneumonia, skin and soft tissue infections, abscesses, osteomyelitis, and endocarditis in hospitals and in the community.11,12
Throughout the duration of the study, a total of 800 S. aureus bacteria were isolated and identified from various clinical samples. Staphylococcus aureus was isolated most often from pus (320 isolates, or 40%), followed by blood (220 instances, or 27.5%), tracheal aspirates (120 instances, or 15%), and urine isolates (60 instances, or 7.5%). Similar to the findings of Deepak et al.,13 Adhikari et al.,14 and Lyall et al.,15 pus had the greatest rate of S. aureus isolation, followed by blood, with a little percentage difference.
Multiple antimicrobial-resistant S. aureus is problematic and restricts therapy choices. Clindamycin is considered a viable option for the treatment of staphylococcal infections because of due to its oral and parenteral availability, 90% oral bioavailability, low cost, and excellent tissue penetration.16 It accumulates in deep abscesses and suppresses the manufacture of S. aureus toxins.17 The S. aureus resistance pathways include 23S ribosomal RNA target tissue alteration mediated by erm genes (A, B, C, and E), resistance mutation, and efflux pump expression.18,19 In our analysis of 800 strains of S. aureus, 540 (67.5%) were classified as MRSA and 260 (32.5%) as MSSA. Various geographic locations have reported varying rates of MRSA prevalence, as shown by many studies.
Toleti et al.20 reported a prevalence rate of 64.70%, whereas Kishk et al.17 reported a prevalence rate of 61.3%. In their research done in Jordan, Jarajreh et al.21 also observed a greater prevalence rate of 77.5%. In contrast, Singh et al.22 and Adhikari et al.14 observed substantially lower rates in their studies: 37.8% and 25.1%, respectively.
Methicillin-resistant S. aureus may have developed as a result of inappropriate infection prevention and control methods, the misuse of antibiotics, and deficient procedures in hospital ICUs and critical care units regarding the use of urinary catheters, mechanical breathing devices, and intravascular catheters. Regional and national discrepancies in MRSA frequency may be explained sporadically by differences in the adoption of infection prevention and control methods, monitoring systems, and antibiotic legislation among nations.
In our analysis, erythromycin and clindamycin resistance were detected in 66.25% and 27.5% of Isolates, respectively. A total of 54.5% of isolates were resistant to erythromycin, whereas 38.6% were resistant to clindamycin, according to Kishk et al.17 Deepak et al.13 found that erythromycin resistance was widespread in 61.4% of isolates.
A trial done in India revealed that 39.14% of isolates exhibited erythromycin resistance.23 In Iran, Mansouri and Sadeghi24 observed a high frequency of erythromycin resistance. These disparities in erythromycin resistance may be attributable to regional differences in antibiotic policy, and macrolide and lincosamide use rates.
Clindamycin resistance and erythromycin resistance are often demonstrated simultaneously. When isolates seem sensitive to clindamycin yet resistant to erythromycin, it may be challenging to detect the iMLSB phenotype using standard laboratory techniques. If administered to patients, this erroneous sensitivity of iMLSB isolates to clindamycin might result in clindamycin treatment failure. Twenty-five percent to 40% of erythromycin-resistant S. aureus isolates exhibited the iMLSB phenotype, 41.5% displayed the cMLSB phenotype, and 24.5% expressed the MS phenotype.
In the many studies carried out to evaluate clindamycin resistance in S. aureus resistant to erythromycin, Kishk et al.17 reported greater rates of cMLSB than iMLSB (38.6% and 13.6%, respectively). Bansal et al.25 reported isolates with the phenotypes iMLSB, cMLSB, and MS at 65%, 22.5%, and 20%, respectively. Deepak et al.13 demonstrated cMLSB resistance in 31.67% of isolates, iMLSB resistance in 21.1% of isolates, and MS resistance in 47.20% of isolates. Regha et al.26 found iMLSB, cMLSB, and MS phenotypes of 12.7%, 8.1%, and 41.8%, respectively. Steward et al.27 reported that the iMLSB phenotype was most frequent, at 16.4%, followed by the cMLSB phenotype at 12.5% and the MS phenotype at 7.8%.
In 34.9% of MRSA strains, the cMLSB phenotype predominated, whereas in MSSA bacteria, the sensitive phenotype predominated (53.8%). Kishk et al.17 found that cMLSB is prevalent in MRSA (56%). Azap et al.28 discovered that MRSA isolates responded better to inducible resistance than MSSA isolates (5.7% and 3.7%, respectively). Bottega et al.29 discovered that MRSA had a greater prevalence of constitutive and inducible resistance than MSSA (68.9% versus 4.5% and 10.3% versus 7.2%, respectively). This disagreement across research about the kind of resistance shown by induction D testing might be attributable to regional variations in the susceptibility of bacteria. In addition, the use of various antimicrobials for empirical treatment may affect the trajectory of resistance.
There is a close relationship between hyperglycemia and infection. Increased blood glucose levels of 11.1 mmol/L or higher (≥ 200 mg/dL) have been associated closely with a decrease in neutrophil activity.30,31 Antibiotic resistance rates in patients with diabetes mellitus have been proved to be greater than in persons without diabetes mellitus in a number of studies. In our study, the susceptibility of MRSA to cefoxitin was 74% among individuals with diabetes mellitus and 26% among patients without diabetes mellitus. Seventy percent of antibiotic-resistant bacteria in India were isolated from patients with diabetes with chronic wounds. Escherichia coli, Streptococcus pyogenes, S. aureus, Klebsiella pneumoniae, Proteus mirabilis, Enterococcus faecalis, and Pseudomonas aeruginosa caused asymptomatic bacteriuria in 32 patients with diabetes mellitus in Cameroon.32 Fifty percent of the staphylococcal strains isolated from patients with diabetes mellitus in India were resistant to methicillin, and 33% were resistant to MLSB.33
Methicillin-resistant S. aureus infections are common among staphylococcal infections and are on the rise in patients with diabetes mellitus. According to French researchers, S. aureus, the most common cause of diabetic foot infections, was detected in 36.5% of the isolates, of which 37.4% were MRSA.25
Standard susceptibility testing procedures cannot detect MLSB-induced resistance. The bacteria are resistant to erythromycin, but sensitive to clindamycin, according to routine susceptibility tests. In such instances, clindamycin treatment in vivo resulted in therapeutic failure.25 The CLSI recommended that all staphylococcal isolates be tested regularly with the D test for iMLSB.9
For the MRSA isolates in our study, the incidence of iMLSB was 27.8% (i.e., 150 of 530 MRSA isolates). Consequently, if the D test is not included in routine testing for the identification of the iMLSB phenotype, these isolates will be reported as clindamycin susceptible, resulting in MLSB antibiotic treatment failure.
The D test is usually done on erythromycin-resistant and clindamycin-sensitive isolates. It takes another day or 18 hours for the result of the D test to become available. Thus, there can be a delay in reporting to the clinician.25
Normal susceptibility testing methods are incapable of revealing the iMLSB resistance mechanism, and its occurrence varies across hospitals and different locations. During regular antibiotic susceptibility testing, all S. aureus clinical isolates may be submitted to the straightforward and cost-effective D test.
CONCLUSION
Methicillin-resistant S. aureus has become a serious global health concern, particularly in hospitals. Because of the antibiotic resistance of several bacteria, hospitals must apply proper control measures. Whether cMLSB or iMLSB, clindamycin resistance inhibits MRSA treatment options.
The D test is a straightforward, reliable, economical, and easy method for determining iMLSB resistance. To advise clinicians on the proper use of clindamycin, we propose including the D test in regular antimicrobial susceptibility testing to detect clindamycin susceptibility.
Our study has some limitations. Options other than the D test for detecting iMLSB were not studied, and the research had a relatively small sample size.
ACKNOWLEDGMENT
The American Society of Tropical Medicine and Hygiene (ASTMH) assisted with publication expenses.
REFERENCES
- 1. Vivek JS, Rajesh GN, Mukesh S, Manpreet K, Misra RN, Matnani GB, Ujagare MT, Saikat B, Ajay K, 2011. The prevalence of inducible clindamycin resistance among community and hospital-associated Staphylococcus aureus isolates in a tertiary care hospital in India. Biomed Res 22: 465–469. [Google Scholar]
- 2. Deotale V, Mendiratta DK, Raut U, Narang P, 2010. Inducible clindamycin resistance in Staphylococcus aureus isolated from clinical samples. Indian J Med Microbiol 28: 124–126. [DOI] [PubMed] [Google Scholar]
- 3. Fiebelkorn KR, Crawford SA, Mcelmeel ML, Jorgensen JH, 2003. Practical disk diffusion method for detection of inducible clindamycin resistance in Staphylococcus aureus and coagulase-negative staphylococci. J Clin Microbiol 41: 4740–4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kader AA, Kumar A, Krishna A, 2005. Induction of clindamycin resistance in erythromycin-resistant, clindamycin susceptible and methicillin-resistant clinical staphylococcal isolates. Saudi Med J 26: 1914–1917. [PubMed] [Google Scholar]
- 5. Prabhu K, Rao S, Rao V, 2011. Inducible clindamycin resistance in Staphylococcus aureus isolated from clinical samples. J Lab Physicians 3: 25–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vandana KE, Singh J, Chiranjay M, Bairy I, 2009. Inducible clindamycin resistance in Staphylococcus aureus: reason for treatment failure. J Glob Infect Dis 1: 76–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Patel M, Waites KB, Moser SA, Cloud GA, Hoesley CJ, 2006. Prevalence of inducible clindamycin resistance among community- and hospital-associated Staphylococcus aureus isolates. J Clin Microbiol 44: 2481–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. CLSI , 2019. CLSI Performance Standards for Antimicrobial Susceptibility Testing, 29th edition. Wayne, PA: Clinical and Laboratory Standard Institute. [Google Scholar]
- 9. American Diabetes Association , 2019. Standard of medical care in diabetes 2019. Diabetes Care 42 (Suppl 1): S4–S6. [DOI] [PubMed] [Google Scholar]
- 10. Collee JG, Mackie TJ, McCartney JE, 1996. Mackie and McCartney Practical Medical Microbiology, 14th edition. New York, NY: Churchill Livingstone. [Google Scholar]
- 11. Winn W, Allen S, Janda W, Koneman E, Procop G, Schreckenberger P, Woods G, 2006. Koneman’s Color Atlas and Textbook of Diagnostic Microbiology, 6th Edition. New York, NY: Lippincott Williams and Wilkins. [Google Scholar]
- 12. Ryan KJ, Ryan KJ, Ray CG. Sherris Medical Microbiology, 4th edition. New York, NY: McGraw Hill, 21–71. [Google Scholar]
- 13. Deepak KG, Anita P, Bhaskar Th, Kalpana C, Sonat J, 2019. Occurrence of inducible clindamycin resistance in clinical isolates of Staphylococcus aureus in a tertiary care hospital. Int J Health Sci Res 9: 71–77. [Google Scholar]
- 14. Adhikari RP, Shrestha S, Barakoti A, Amatya R, 2017. Inducible clindamycin and methicillin resistant Staphylococcus aureus in a tertiary care hospital, Kathmandu, Nepal. BMC Infect Dis 17: 483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lyall KS, Gupta V, Chhina D, 2013. Inducible clindamycin resistance among clinical isolates of Staphylococcus aureus. J Mahatma Gandhi Inst Med Sci 18: 112–115. [Google Scholar]
- 16. Bradely JR, 2005. Newer anti staphylococcal agents. Curr Opin Pediatr 17: 71–77. [DOI] [PubMed] [Google Scholar]
- 17. Kishk RM, Anani MM, Nemr NA, Soliman NM, Fouad MM. 2020. Inducible clindamycin resistance in clinical isolates of Staphylococcus aureus in Suez Canal University Hospital, Ismailia, Egypt. J Infect Dev Ctries 14: 1281–1287. [DOI] [PubMed] [Google Scholar]
- 18. Majhi S, Dash M, Mohapatra D, Mohapatra A, Chayani N, 2016. Nirupama Chayani: Detection of inducible and constitutive clindamycin resistance among Staphylococcus aureus isolates in a tertiary care hospital, Eastern India. Avicenna J Med 6: 75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mazloumi MJ, Akbari R, Yousefi SJM, Letters B, 2021. Detection of inducible clindamycin resistance genes (ermA, ermB, and ermC) in Staphylococcus aureus and Staphylococcus epidermidis . Microbiol Biotechnol Lett 49: 449–457. [Google Scholar]
- 20. Toleti S, Bobbillapati JR, Kollipaka SR, Myneni RB, 2015. Detection of inducible clindamycin resistance and susceptibilities to other antimicrobial agents in clinical isolates of Staphylococcus aureus. Int J Res Med Sci 3: 612–616. [Google Scholar]
- 21. Jarajreh D, Aqel A, Alzoubi H, Al-Zereini W, 2017. Prevalence of inducible clindamycin resistance in methicillin-resistant Staphylococcus aureus: the first study in Jordan. J Infect Dev Ctries 11: 350–354. [DOI] [PubMed] [Google Scholar]
- 22. Singh T, Deshmukh AB, Chitnis V, Bajpai T, 2016. Inducible clindamycin resistance among the clinical isolates of Staphylococcus aureus in a tertiary care hospital. Int J Health Allied Sci 5: 111–114. [Google Scholar]
- 23. Mokta K, Verma S, Chauhan D, Ganju SA, Singh D, Kanga A, Kumari A, Mehta V, 2015. Inducible clindamycin resistance among clinical isolates of Staphylococcus aureus from sub Himalayan region of India. J Clin Diagn Res 9: DC20–DC23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mansouri S, Sadeghi J, 2014. Inducible clindamycin resistance in methicillin-resistant and -susceptible Staphylococcus aureus isolated from south east of Iran. Jundishapur J Microbiol 7: e11868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bansal VP, Gadekar KS, Mulay MV, Mishra JK, 2019. Incidence of inducible clindamycin resistance in clinical isolates of staphylococci and their antibiotic sensitivity pattern. Int J Adv Res (Indore) 7: 1066–1073. [Google Scholar]
- 26. Regha IR, Harichandran D, Sulekha B, 2016. Inducible clindamycin resistance among clinical isolates of Staphylococcus aureus in a tertiary care centre, Kerala, India. Curr Microbiol App Sci 5: 929–934. [Google Scholar]
- 27. Steward CD, Raney PM, Morrell AK, Williams PP, McDougal LK, Jevitt L, McGowan JE, Jr., Tenover FC, 2005. Testing for induction of clindamycin resistance in erythromycin-resistant isolates of Staphylococcus aureus. J Clin Microbiol 43: 1716–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Azap OK, Arslan H, Timurkaynak F, Yapar G, Oruc E, Gagir U, 2005. Incidence of inducible clindamycin resistance in erythromycin-resistant isolates of Staphylococcus aureus. J Clin Microbiol 43: 1716–1721.15814990 [Google Scholar]
- 29. Bottega A, Rodrigues Md A, Carvalho FA, Wagner TF, Leal IA, Santos SO, Rampelotto RF, Hörner R, 2014. Evaluation of constitutive and inducible resistance to clindamycin in clinical samples of Staphylococcus aureus from a tertiary hospital. Rev Soc Bras Med Trop 47: 589–592. [DOI] [PubMed] [Google Scholar]
- 30. Kawahito S, Kitahata H, Oshita S, 2009. Problems associated with glucose toxicity: role of hyperglycemia-induced oxidative stress. World J Gastroenterol 15: 4137–4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Basu S, Ramchuran Panray T, Bali Singh T, Gulati AK, Shukla VK, 2009. A prospective, descriptive study to identify the microbiological profile of chronic wounds in outpatients. Ostomy Wound Manage 55: 14–20. [PubMed] [Google Scholar]
- 32. Njunda AL, Assob NJC, Nsagha SD, Nde FP, Kamga FHL, Nkume AF, Kwenti TE, 2012. Uropathogens from diabetic patients with asymptomatic bacteriuria and urinary tract infections. Sci J Microbiol 1: 141–146. [Google Scholar]
- 33. Rawat V, Singhai M, Kumar A, Jha PK, Goyal R, 2012. Bacteriological and resistance profile in isolates from patients with diabetes. N Am J Med Sci 4: 563–568. [DOI] [PMC free article] [PubMed] [Google Scholar]


