ABSTRACT.
Histidine-rich protein 2– (HRP2-) based rapid diagnostic tests (RDTs) are widely used to detect Plasmodium falciparum in sub-Saharan Africa. Reports of parasites with pfhrp2 and/or pfhrp3 (pfhrp2/3) gene deletions in Africa raise concerns about the long-term viability of HRP2-based RDTs. We evaluated changes in pfhrp2/3 deletion prevalence over time using a 2018–2021 longitudinal study of 1,635 enrolled individuals in Kinshasa Province, Democratic Republic of the Congo (DRC). Samples collected during biannual household visits with ≥ 100 parasites/µL by quantitative real-time polymerase chain reaction were genotyped using a multiplex real-time PCR assay. Among 2,726 P. falciparum PCR-positive samples collected from 993 participants during the study period, 1,267 (46.5%) were genotyped. No pfhrp2/3 deletions or mixed pfhrp2/3-intact and -deleted infections were identified in our study. Pfhrp2/3-deleted parasites were not detected in Kinshasa Province; ongoing use of HRP2-based RDTs is appropriate.
INTRODUCTION
Progress toward malaria control and elimination in Africa requires prompt diagnosis and treatment with effective antimalarial drugs. Rapid diagnostic tests (RDTs) are widely used to identify individuals infected with Plasmodium; their deployment has enabled significant improvements in malaria diagnostic testing across the continent over the past decade.1,2 The majority of RDTs used to diagnose falciparum malaria in sub-Saharan Africa detect Plasmodium falciparum histidine-rich protein 2 (HRP2) and its paralog HRP3, encoded by the pfhrp2 and pfhrp3 genes, respectively.1 Histidine-rich protein 2–based RDTs are generally more sensitive and heat stable than RDTs detecting other antigens.3 However, test-and-treat strategies that rely upon HRP2-based RDTs are threatened by the emergence of P. falciparum strains that escape detection owing to deletion of the pfhrp2 and/or pfhrp3 (pfhrp2/3) genes.1,4 High prevalence of pfhrp2/3-deleted parasites in Eritrea,5 Ethiopia,6,7 Djibouti,8,9 and surrounding countries recently prompted changes in malaria diagnostic testing policies. Reports from sub-Saharan countries outside of the Horn of Africa, however, indicate lower prevalence.10
The Democratic Republic of the Congo (DRC) has one of the highest malaria burdens in the world, accounting for 12% of global malaria cases and deaths.1 In the DRC, we previously reported 6.4% pfhrp2 deletion prevalence in samples from a 2013–2014 nationally representative cross-sectional survey of asymptomatic children under 5 years of age.11 However, no pfhrp2/3 deletions were observed in our 2017 cross-sectional study of symptomatic children and adults across three DRC provinces.12 These studies were both cross-sectional and did not provide information about how the prevalence of deletions may be changing over time. This study aimed to estimate pfhrp2/3 deletion prevalence and changes over time in Kinshasa Province, the most populous province in the DRC.
MATERIALS AND METHODS
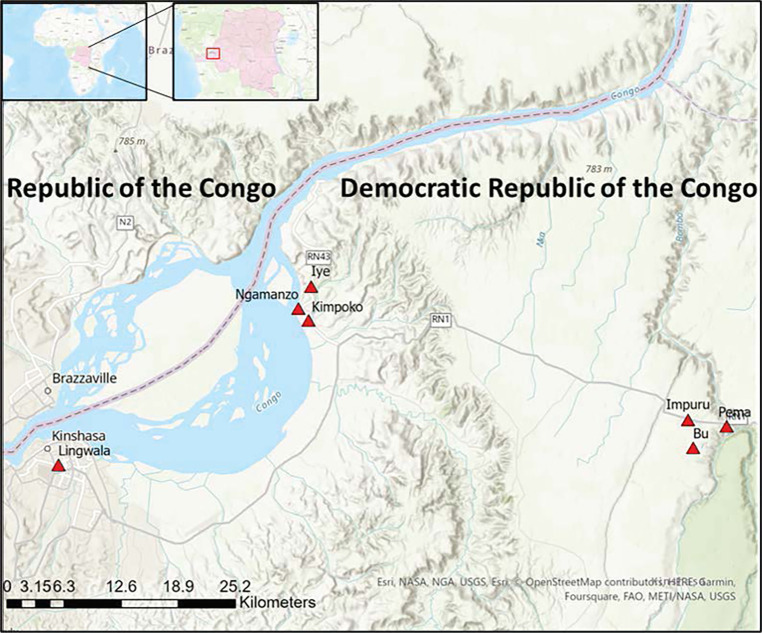
This study includes samples collected as part of a 2018–2021 longitudinal study of malaria conducted at seven sites across Kinshasa Province, DRC. A total of 1,635 participants were enrolled in one urban neighborhood, three peri-urban villages, and three rural villages (Figure 1). These study sites experience varying malaria endemicities, reflective of the heterogeneity in malaria transmission observed in the DRC and other sub-Saharan African countries. Study visits were conducted as part of twice-yearly household surveys in the dry and rainy seasons (active surveillance) and as part of routine care at local health centers (passive surveillance) as previously described.13,14 At each visit, a comprehensive questionnaire on malaria symptoms and treatment and bed net usage was administered. A finger- or heel-prick sample was obtained at each visit for RDT (SD Bioline Ag P.f./Pan RDT [05FK60], Alere, Gyeonggi-do, Republic of Korea, or CareStart Malaria Pf HRP2 Ag [02571], Access Bio, Somerset, NJ) and dried blood spot (DBS) preparation for future molecular investigation. Rapid diagnostic test–positive patients were treated according to national guidelines.
Figure 1.
Study sites in Kinshasa Province, Democratic Republic of the Congo.
DNA was extracted from DBS using Chelex-100 (Sigma-Aldrich, St Louis, MO) and saponin or Tween (Bio-Rad Laboratories, Hercules, CA) as previously described.15,16 Quantitative real-time polymerase chain reaction (qPCR) targeting the P. falciparum lactate dehydrogenase (pfldh) gene was used to estimate P. falciparum parasitemia, using serial dilutions of DNA extracted from a mock DBS made with cultured P. falciparum 3D7 or FCR3 strain parasites at known parasite density. Samples with ≥ 100 parasites/µL were selected for pfhrp2/3 deletion identification using a multiplex real-time PCR assay that detects pfldh, pfhrp2, pfhrp3, and human beta-tubulin (HumTuBB).17 We used this parasite density threshold to reduce the risk of unintentional misclassification of deletions in the setting of low DNA concentrations.18,19 Positive calls required cycle threshold (Ct) values < 35. Samples positive for HumTuBB and pfldh but negative for pfhrp2 or pfhrp3 were subjected to a confirmatory real-time PCR targeting the P. falciparum beta-tubulin (PfBtubulin) gene. Deletion calls were limited to samples positive for HumTuBB and both single-copy P. falciparum genes (pfldh and pftubulin), but negative for pfhrp2 and/or pfhrp3. Mixed infections of pfhrp2/3-intact and -deleted strains were defined conservatively as samples in which (pfhrp2 Ct − pfldh Ct) > 3 or (pfhrp3 Ct − pfldh Ct) > 3.17 All assays included P. falciparum 3D7, DD2, and HB3 strain DNA as wildtype, pfhrp2-deleted, and pfhrp3-deleted controls, respectively. All participants provided informed assent/consent. Ethical approval for this study was granted by the Institutional Review Boards of the University of North Carolina-Chapel Hill and the Kinshasa School of Public Health.
RESULTS
A total of 1,267 samples collected from 649 individuals in 179 households between 2018 and 2021 were included in this study (Figure 2). Among these, the median number of P. falciparum infections with ≥ 100 parasites/µL detected per participant was 2.0 (interquartile range [IQR]: 1–3). The median age at enrollment in 2018 was 9 years (IQR: 5–15 years of age); 48.8% reported female gender. At the time of enrollment, 48.7% reported malaria in the preceding 6 months, and 42.7% of those reported more than one episode in that 6-month period. The median household size was 8 (IQR: 6–10), with high bed net coverage across the study population (90.2%). The baseline characteristics of the study population are summarized in Table 1.
Figure 2.
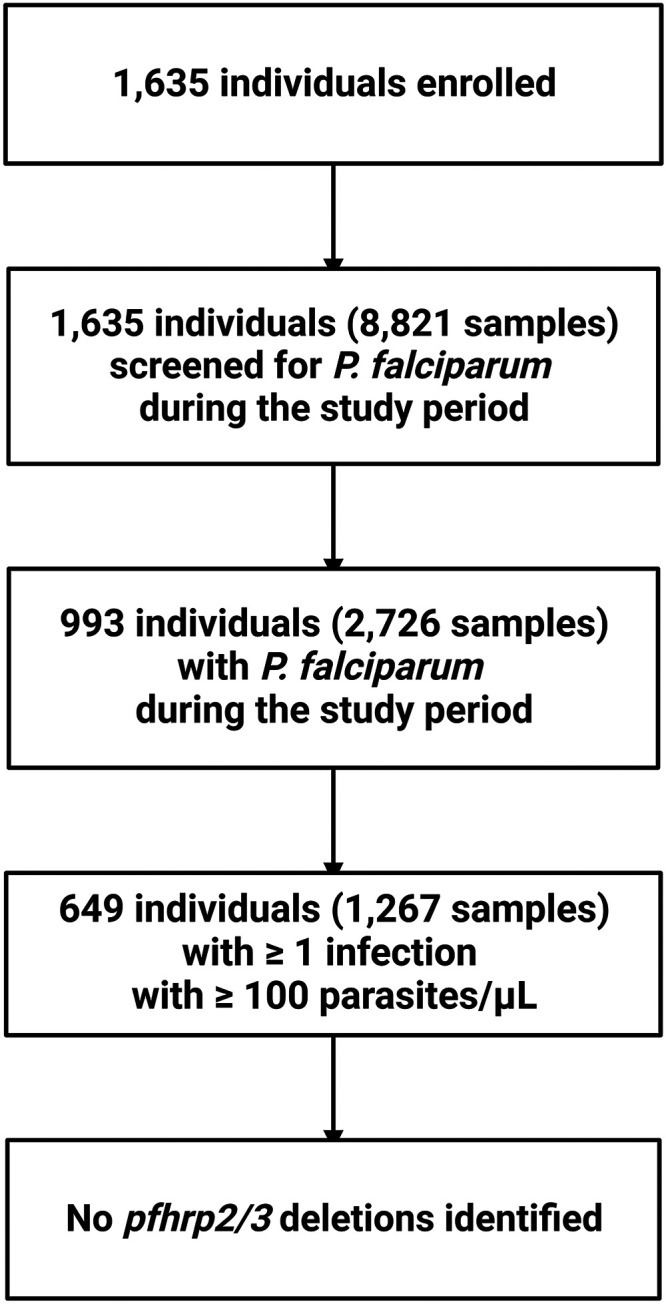
Flow diagram for the detection of pfhrp2/3 deletion in 1,267 samples from 649 participants during household visits of the longitudinal study (2018–2021) in Kinshasa Province, Democratic Republic of the Congo.
Table 1.
Baseline characteristics of the study population included in this analysis
| Participants (N = 649) | |
|---|---|
| Age in years, median (IQR) | 9 (5–15) |
| Age strata, n (%) | |
| < 1 year | 15 (2.3) |
| 1–5 years | 166 (25.6) |
| 6–10 years | 191 (29.5) |
| 11–15 years | 119 (18.4) |
| 16–25 years | 76 (11.7) |
| > 25 years | 81 (12.5) |
| Sex, n (%) | |
| Female | 317 (48.8) |
| Male | 332 (51.2) |
| Malaria in previous 6 months, n (%) | |
| No | 311 (51.3) |
| Yes–once | 169 (27.9) |
| Yes–many | 126 (20.8) |
| Slept under bed net previous night, n (%) | |
| No | 47 (9.8) |
| Yes | 432 (90.2) |
| Household size, median (IQR) | 8 (6–10) |
| Site of residence, n (%) | |
| Bu (rural) | 87 (13.4) |
| Impuru (rural) | 123 (19.0) |
| Pema (rural) | 121 (18.6) |
| Ngamanzo (peri-urban) | 108 (16.6) |
| Iye (peri-urban) | 60 (9.2) |
| Kimpoko (peri-urban) | 122 (18.8) |
| Lingwala (urban) | 28 (4.3) |
| Number of samples per participant, median (IQR) | 2 (1–3) |
IQR = interquartile range.
A total of 1,267 samples with ≥ 100 P. falciparum parasites/µL from 649 participants during enrollment in the Kinshasa longitudinal study.
All 1,267 samples had detectable human DNA, as indicated by amplification of HumTuBB with Ct < 35. Multiplex PCR confirmed P. falciparum parasitemia in all but two samples that failed to amplify pfldh. Both pfhrp2 and pfhrp3 were negative in one sample (Ct > 35 for both gene targets). However, pfldh was negative in the multiplex assay and Pftubulin was negative in follow-up testing, indicating that the original pfldh qPCR result was a false-positive. Thus, this sample did not meet criteria to be considered a pfhrp2/3-deleted parasite. No mixed infections of pfhrp2/3-intact and -deleted strains were identified in our study population.
DISCUSSION
Overall, we did not find evidence of pfhrp2/3 deletion in parasites sampled during our longitudinal cohort study in Kinshasa Province. Our conservative approach to calling pfhrp2/3 deletions prevented us from detecting low-density infections and thus could underestimate the true prevalence of these strains. These study results differ from our previous DRC pfhrp2/3 deletion prevalence estimates; this discrepancy could be attributed to spatial heterogeneity in prevalence or incorrect deletion calls in low-density samples during our previous study. The multiplex real-time PCR assay and conservative calling approach used here reduced the risk of incorrect deletion calls. Our finding of no deletions is in line with our more recent study of symptomatic individuals and a similar study of asymptomatic and symptomatic school-aged children that showed no to little evidence of pfhrp2/3 deletion in the DRC.12,20 Together, these results support the continued use of HRP2-based RDTs for the diagnosis of malaria in the DRC.
ACKNOWLEDGMENTS
We thank the study participants and field teams who conducted study visits. We also express our gratitude to the late Prof. Steven Meshnick for mentorship and his role in the longitudinal study upon which this analysis was based. The following reagents were obtained through BEI Resources, National Institute of Allergy and Infectious Diseases, NIH: Genomic DNA from P. falciparum strain 3D7, MRA-102G, contributed by Daniel J. Carucci; P. falciparum strain HB3, MRA-155G, contributed by Thomas E. Wellems; and P. falciparum strain Dd2, MRA-150G, contributed by David Walliker.
REFERENCES
- 1.Geneva: World Health Organization, 2021. World malaria report 2021. Angew Chem Int Ed 6: 951–952. [Google Scholar]
- 2. Aidoo M, Incardona S, 2022. Ten years of universal testing: how the rapid diagnostic test became a game changer for malaria case management and improved disease reporting. Am J Trop Med Hyg 106: 29–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chiodini PL. et al. , 2007. The heat stability of Plasmodium lactate dehydrogenase-based and histidine-rich protein 2-based malaria rapid diagnostic tests. Trans R Soc Trop Med Hyg 101: 331–337. [DOI] [PubMed] [Google Scholar]
- 4. World Health Organization , 2021. Statement by the Malaria Policy Advisory Group on the Urgent Need to Address the High Prevalence of pfhrp2/3 Gene Deletions in the Horn of Africa and Beyond. Available from: https://www.who.int/news/item/28-05-2021-statement-by-the-malaria-policy-advisory-group-on-the-urgent-need-to-address-the-high-prevalence-of-pfhrp2-3-gene-deletions-in-the-horn-of-africa-and-beyond. Accessed June 2, 2022.
- 5. Berhane A. et al. , 2018. Major threat to malaria control programs by Plasmodium falciparum lacking histidine-rich protein 2, Eritrea. Emerg Infect Dis 24: 462–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Feleke SM. et al. , 2021. Plasmodium falciparum is evolving to escape malaria rapid diagnostic tests in Ethiopia. Nat Microbiol 6: 1289–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alemayehu GS, Blackburn K, Lopez K, Dieng CC, Lo E, Janies D, Golassa L, 2021. Detection of high prevalence of Plasmodium falciparum histidine-rich protein 2/3 gene deletions in Assosa zone, Ethiopia: implication for malaria diagnosis. Malar J 20: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Iriart X, Menard S, Chauvin P, Mohamed HS, Charpentier E, Mohamed MA, Berry A, Aboubaker MH, 2020. Misdiagnosis of imported falciparum malaria from African areas due to an increased prevalence of pfhrp2/pfhrp3 gene deletion: the Djibouti case. Emerg Microbes Infect 9: 1984–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rogier E. et al. , 2022. Plasmodium falciparum pfhrp2 and pfhrp3 gene deletions and relatedness to other global isolates, Djibouti, 2019–2020. Emerg Infect Dis 28: 2043–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Thomson R, Parr JB, Cheng Q, Chenet S, Perkins M, Cunningham J, 2020. Prevalence of Plasmodium falciparum lacking histidine-rich proteins 2 and 3: a systematic review. Bull World Health Organ 98: 558–568F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Parr JB. et al. , 2017. Pfhrp2-deleted Plasmodium falciparum parasites in the Democratic Republic of the Congo: a national cross-sectional survey. J Infect Dis 216: 36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Parr JB. et al. , 2021. Analysis of false-negative rapid diagnostic tests for symptomatic malaria in the Democratic Republic of the Congo. Sci Rep 11: 6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mwandagalirwa MK, Levitz L, Thwai KL, Parr JB, Goel V, Janko M, Tshefu A, Emch M, Meshnick SR, Carrel M, 2017. Individual and household characteristics of persons with Plasmodium falciparum malaria in sites with varying endemicities in Kinshasa Province, Democratic Republic of the Congo. Malar J 16: 456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carrel M. et al. , 2021. Individual, household and neighborhood risk factors for malaria in the Democratic Republic of the Congo support new approaches to programmatic intervention. Health Place 70: 102581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Plowe CV, Djimde A, Bouare M, Doumbo O, Wellems TE, 1995. Pyrimethamine and proguanil resistance-conferring mutations in Plasmodium falciparum dihydrofolate reductase: polymerase chain reaction methods for surveillance in Africa. Am J Trop Med Hyg 52: 565–568. [DOI] [PubMed] [Google Scholar]
- 16. Teyssier NB, Chen A, Duarte EM, Sit R, Greenhouse B, Tessema SK, 2021. Optimization of whole-genome sequencing of Plasmodium falciparum from low-density dried blood spot samples. Malar J 20: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Grignard L. et al. , 2020. A novel multiplex qPCR assay for detection of Plasmodium falciparum with histidine-rich protein 2 and 3 (pfhrp2 and pfhrp3) deletions in polyclonal infections. EBioMedicine 55: 102757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Parr JB, Anderson O, Juliano JJ, Meshnick SR, 2018. Streamlined, PCR-based testing for pfhrp2- and pfhrp3-negative Plasmodium falciparum. Malar J 17: 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Beshir KB, Parr JB, Cunningham J, Cheng Q, Rogier E, 2022. Screening strategies and laboratory assays to support Plasmodium falciparum histidine-rich protein deletion surveillance: where we are and what is needed. Malar J 21: 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nundu SS. et al. , 2022. Low prevalence of Plasmodium falciparum parasites lacking pfhrp2/3 genes among asymptomatic and symptomatic school-age children in Kinshasa, Democratic Republic of Congo. Malar J 21: 126. [DOI] [PMC free article] [PubMed] [Google Scholar]