Abstract
Association of the cellular adhesive protein CD44 and the N-terminal (FERM) domain of cytoskeleton adaptors is critical for cell proliferation, migration, and signaling. Phosphorylation of the cytoplasmic domain (CTD) of CD44 acts as an important regulator of the protein association, but the structural transformation and dynamics mechanism remain enigmatic. In this study, extensive coarse-grained simulations were employed to explore the molecular details in the formation of CD44-FERM complex under S291 and S325 phosphorylation, a modification path known to exert reciprocal effects on the protein association. We find that phosphorylation of S291 inhibits complexation by causing the CTD of CD44 to adopt a more closed structure. In contrast, S325 phosphorylation liberates the CD44-CTD from the membrane surface and promotes the linkage with FERM. The phosphorylation-driven transformation is found to occur in a PIP2-dependent manner, with PIP2 effecting the relative stability of the closed and open conformation, and a replacement of PIP2 by POPS greatly abrogates this effect. The revealed interdependent regulation mechanism by phosphorylation and PIP2 in the association of CD44 and FERM further strengthens our understanding of the molecular basis of cellular signaling and migration.
Significance
It has been confirmed that two phosphorylated modifications alternatively occurring on the CTD of CD44 closely mediate its interaction with the FERM domain, but the details of structural transformation and molecular information remain elusive. Using high-throughput coarse-grained molecular simulations, we found that the discrepant effect of phosphorylation of S291 and S325 on the binding efficiency of CD44 and FERM is based on the conformational responsibility of CTD combined with PIP2 mediation. That is, the conformational changes of CTD caused by phosphorylation affect the binding efficiency of FERM, but the regulation of phosphorylation depends on the presence of PIP2. The molecular details provide new insights into the cell proliferation and migration involved in the actin cytoskeleton anchored in the CD44-FERM complex.
Introduction
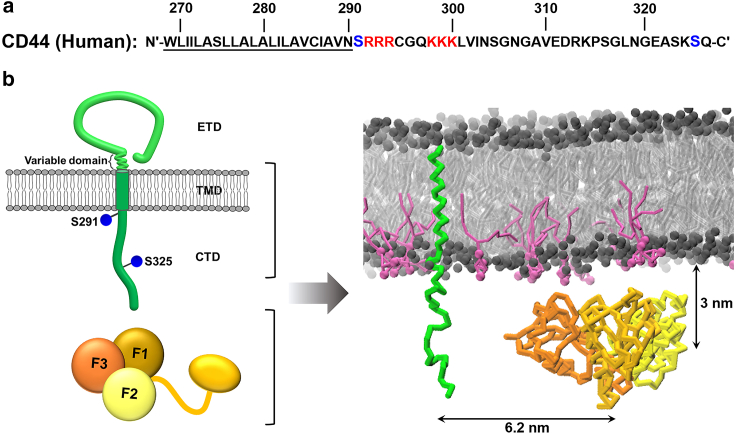
Cluster-of-differentiation-44 protein (CD44) is a single-pass cellular adhesion membrane protein with versatile cell functions (1,2). It is expressed on embryonic stem cells and other cell types or compartments like cell microvilli, playing an important role in cell proliferation and migration (3,4). An “inside-out” regulation pathway is proposed, stating that association between the CD44 cytoplasmic domain (CTD) and its cytoskeleton receptors is essential in cell signal transduction and binding of CD44 to the extracellular matrix via its extracellular domain (ETD) (5). The upregulated binding of ETD-hyaluronan is found to further promote tumor cell proliferation (6,7,8). Note that the transmembrane domain (TMD) and the CTD of CD44 are highly conserved (Fig. 1 a and b), which is critical for protein linkage and cell signaling (9,10). Through the CTD, CD44 interacts with the activated N-terminus (FERM) of Ezrin-Radixin-Moesin (ERM) protein (Fig. 1 b), one of important cytoskeleton junction receptors (11,12). The CD44-FERM complex anchors the CD44-related transmembrane receptor complex on the actin cytoskeleton, thereby coordinating the spatial and temporal localization and signal transduction of the receptor complex (10,13,14,15).
Figure 1.
System setup. (a) CD44 (human) sequence used in this study with the TMD highlighted by dark underline. The basic residues close to S291 are marked in red. (b) Left panel: schematic diagram of CD44 (upper) and FERM (lower) structure. The TMD and CTD are displayed in dark green, and the two conserved amino acids S291 and S325 are highlighted with blue spheres. The FERM is represented by three subdomains: F1 (gold), F2 (yellow), and F3 (orange). Right panel: snapshot of the starting configuration of the simulated system. The proteins are shown with backbone only. POPC and PIP2 lipids are presented in gray and mauve, respectively. Waters and ions are omitted for clarity. To see this figure in color, go online.
Phosphorylation acts as a ubiquitous posttranslational modification that regulates protein signaling and interaction (16,17,18). CD44-CTD contains two conserved phosphorylated sites, S291 and S325 (Fig. 1 b), and the phosphorylation/dephosphorylation switching between them is closely involved in association of CD44 and FERM (19,20). The protein association is prohibited by activation of protein kinase C (PKC) that phosphorylates S291 while dephosphorylating S325 (21). In addition to phosphorylation, the membrane environment plays a key role in protein activation (22). In particular, phosphatidylinositol 4,5-bisphosphate (PIP2) lipids are important in the formation of the CD44-FERM complex, as shown by small-angle neutron scattering experiments (23). Moreover, dynamic clustering and scattering of PIP2 are observed when proteins approach each other in the membrane cyto-proximity region (24). Given the multiple negative charges of PIP2 lipid headgroups, and the negative charge introduced by phosphorylation, it is not far-fetched to assume both factors are interdependent and together act as a regulatory mechanism in the association of FERM and CD44. To this end, revealing more information of protein affinity of FERM and CD44 regulated by a switching CTD phosphorylation in the juxta-membrane environment is crucial to understand cell proliferation and targeted tumor inhibition. As a complement to experimental approaches, molecular dynamics (MD) simulations show great power in unraveling protein-lipid and protein-protein interactions. The use of coarse-grained (CG) models, in particular the Martini force field, has proven valuable to efficiently explore the microsecond timescales required for protein complexation in a heterogeneous lipid environment. The Martini model relies on an average four-to-one mapping principle and experiment-matched thermodynamic data (25), and it has proven successfulin simulating a wide variety of protein-protein and protein-lipid interactions including the binding to PIP2 lipids and the effect of phosphorylation (26,27,28,29). A number of studies with Martini CG simulations show that PIP2 effects the membrane absorption of peripheral membrane proteins (30,31) and sorting of CD44 between phase-segregated membrane domains to recognize FERM (32). Simulations of L-selectin, a related single-span signaling protein, revealed that the membrane adhesion of its CTD is influenced by phosphorylated modifications (33). Our recent simulation work elaborated on the molecular mechanism in CD44 homodimerization influenced by palmitoylations and membrane microenvironments (34).
Here, we use MD simulations based on the Martini model to explore the structural basis of CD44-FERM binding regulated by the phosphorylation of S291 and S325 and in-depth elucidate the regulatory role of PIP2 in mediating the interaction. The molecular information obtained here will strengthen the regulation mechanism of CD44 associating with FERM affected by phosphorylation and PIP2 and further deepen our understanding for CD44-related cell proliferation and targeted inference in tumor prevention.
Materials and methods
Model setup
In this study, all systems were simulated by CG-MD based on the Martini 2.2 force field (35,36). Experiments showed that two phosphorylation sites (S291 and S325) existed in the intracellular CTD of CD44 (19). The serine phosphorylation parameters of Martini originate from Pluhackova et al. (37). All phosphates in the phosphorylation sites were doubly negatively charged. The phosphorylated serine parameters referenced are summarized Table S1. Three models of CD44 (wild-type (WT), S291p, S325p) were constructed using Pymol software and subsequently converted to CG models using the martinize.py (http://www.cgmartini.nl) script. The three types of CD44 models studied here are composed of the TMD and a shortened CTD of 36 amino acid residues, which covers the FERM-interaction domain and two conserved phosphorylated sites. The amino acid sequence of the truncated CD44 is shown in Fig. 1 a. Based on the available experimental data, the TMD was modeled as α-helix, whereas the CTD remained flexible and was defined as random coil (38). The atomic structure of Radixin-FERM was extracted from the protein database (PDB: 2ZPY) and transformed into CG model using the martinize.py tool. A default elastic network was applied to the FERM to maintain the protein structural stability (39).
Two types of lipid bilayers were constructed using the insane.py script (40). The outer leaflet was composed of 100% POPC, and the inner leaflet was 95% POPC/5% PIP2 (–4e) and 80% POPC/20% POPS (–1e), respectively (41). CD44 was inserted into the bilayer parallel with the membrane normal, and the CTD was exposed to the cytoplasmic solution. In the system simulating the interaction between CD44 and FERM, the box size is set as 15 × 15 × 15 nm3, and each leaflet of the lipid bilayer contains 361 lipid molecules. At the beginning of the simulation, the FERM was located below the plasma membrane with a distance of 3.0 nm from its center of mass to the average position of the lipid phosphate groups of the lower leaflet. CD44 and FERM were placed along the diagonal of the simulation box with the distance between the centroids of the two proteins equal to 6.2 nm (Fig. 1 b), in order to avoid unnecessary interaction caused by the system settings. In the simulation system with sole CD44, it is placed in the center of the plasma membrane, with a small size of 12 × 12 × 15 nm3 to save the computation cost, and each leaflet contains 225 lipid molecules. The systems containing FERM only utilize the same size (15 × 15 × 15 nm3) as the systems containing both CD44 and FERM. All systems were solvated with standard CG water beads (W) and neutralized with sodium (Ion) counterions. The detailed molecular information of the three simulation systems can be seen in Table 1.
Table 1.
An overview of the components of the simulation systems used in the study
| Protein | WT-CD44/FERM S291p-CD44/FERM S325p-CD44/FERM |
WT-CD44 S291p-CD44 S325p-CD44 |
FERM | |||
|---|---|---|---|---|---|---|
| Lipids | ||||||
| Upper leaflet | 100% POPC (361) | 100% POPC (225) | 100% POPC (361) | |||
| Lower leaflet | 95% POPC /5% PIP2 (342/18) |
80% POPC /20% POPS (288/72) |
95% POPC /5% PIP2 (213/11) |
80% POPC /20% POPS (180/44) |
95% POPC /5% PIP2 (342/18) |
80% POPC /20% POPS (288/72) |
| W | 19,674 | 19,788 | 12,648 | 12,709 | 19,668 | 19,782 |
| Ion | 62 (WT) 64 (S291p, S325p) | 38 (WT) 40 (S291p, S325p) | 68 | |||
| Box | 15 × 15 × 15 nm3 | 12 × 12 × 15 nm3 | 15 × 15 × 15 nm3 | |||
| Time | 3 μs × 10 (each simulation system) | |||||
| 420 μs (total) | ||||||
Simulation details
All simulations were performed using the Gromacs-5.1.2 software package (42). The systems first underwent an energy minimization for 5000 steps using the steepest-descent method, followed by an equilibration of 50 ns using the Berendsen method for coupling of temperature and pressure (43). The warm bath was conducted with a reference temperature (ref_t) of 320 K and a time constant (tau_t) of 1.0 ps, whereas the pressure bath was conducted in a semiisotropic coupling type with a reference pressure (ref_p) of 1.0 bar, a compressibility constant of 4.5 × 10−5 bar−1, and a time constant (tau_p) of 5.0 ps. During the equilibration process, the position of the backbone of the protein was restricted by a harmonic constraint with a force constant of 1000 kJ mol−1·nm−2. Finally, after the position limitation was removed, a random initial velocity was set, and a production simulation was performed for 3.0 μs. To ensure stability of simulation, the coupling methods for temperature and pressure were transferred to the Nosé-Hoover (44,45) (the same ref_t and tau_t as the equilibration above) and Parrinello-Rahman (46) with a ref_p of 1.0 bar, a compressibility constant of 3.0 × 10−4 bar−1, and a tau_p of 12.0 ps. In all cases, periodic boundary conditions were applied, the neighbor list was updated every 10 steps, and the integration time step was set to 0.02 ps. A shift function was used for the nonbonded electrostatic and Lennard-Jones interactions. The former decreased to zero from the distance of 0–1.2 nm, and the latter were shifted to zero from 0.9 to 1.2 nm, following standard procedures for the Martini force field (47). A total of 14 systems were used in this study, where each system was simulated for 3 μs and with 10 replicas for a total of 420 μs.
Analysis
The binding free energy measures the strength of the interaction between proteins (48). In this paper, the umbrella sampling method is used to characterize the free energy of interaction between CD44/FERM by calculating the potential of mean force (PMF) as a function of the center-of-mass distance between the two proteins. The root mean-square deviation of the CD44/FERM complex was calculated, and the conformation with continuous and stable binding between the two proteins at about 2 μs was selected as the initial conformation (Fig. S1). To create the starting positions for the individual umbrella windows, a nonequilibrium pulling simulation was performed. The debinding process of proteins can be viewed in the Supporting material (Video S1). Given the strong binding of FERM to the CD44-CTD, only the cytoplasmic tail of CD44 is pulled away from the FERM during the umbrella sampling protocol, reducing the sampling error as the CD44/FERM interface remains intact. During the pulling process, a force of 1000 kJ mol−1·nm−2 was applied to the FERM for position control, and a force of 1000 kJ mol−1·nm−2 was set for CD44 in the opposite direction. The centers of mass of the two proteins were pulled apart during 1 ns, at a rate of 0.01 nm/ps. Conformations were extracted at intervals of approximately 0.15 nm, generating between 31 and 33 separate windows (system dependent) covering an overall separation distance of 5.0 nm. Each umbrella window was then equilibrated for 20 ns, using a harmonic potential with a force constant of 1000 kJ mol−1·nm−2, followed by a 3.0-μs production simulation. The weighted histogram analysis method in Gromacs was used to calculate the resulting binding free energy, and the bootstrap analysis method was used to calculate the statistical error (49).
The two-dimensional density map was calculated by the densmap tool of Gromacs. The contact intensity for estimating interaction between proteins was determined with the gmx mindist tool, using a cutoff distance of 0.6 nm from the referring protein to the targeted protein. All simulation snapshots in this study were made using VMD software package (50). The heat map of the interaction between CD44-CTD residues and FERM was drawn using the Python plotting library matplotlib (27). All analysis results were calculated by averaging over 10 replicas, and statistical error bars were calculated from the standard deviation of the 10 values.
Results
Binding of CD44 and FERM depends on the interplay of phosphorylation and membrane compositions
In order to reveal the effect of phosphorylation of two serine residues (S291 and S325) at the CTD of CD44 on the formation of the CD44-FERM complex, we performed a series of CG-MD simulations. The setup consisted of either the CD44 WT, the phosphorylated S291 (S291p), or the phosphorylated S325 (S325p) protein embedded in a lipid bilayer patch (15 × 15 × 15 nm3) composed of 95% POPC/5% PIP2, with PIP2 being present in the cytosolic leaflet only (Fig. 1 b). The FERM domain was placed on the cytosolic side, 3.0 nm below the average position of the lipid phosphate groups of the lower leaflet. The CD44 studied here consists of the TMD and a truncated CTD of 36 residues that covers the two conserved phosphorylated serine sites. The initial separation between CD44 and FERM was set to 6.2 nm, and the lipid components were distributed randomly in the beginning, which allowed for an unbiased observation of PIP2-mediated protein complexation (Fig. 1 b). 10 replicas simulations (each of 3 μs) were performed for each of the two phosphorylated systems as well as the WT for reference.
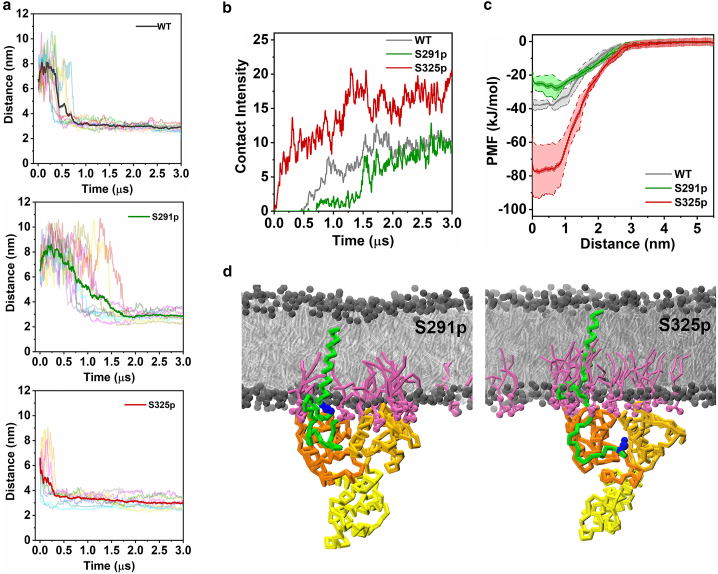
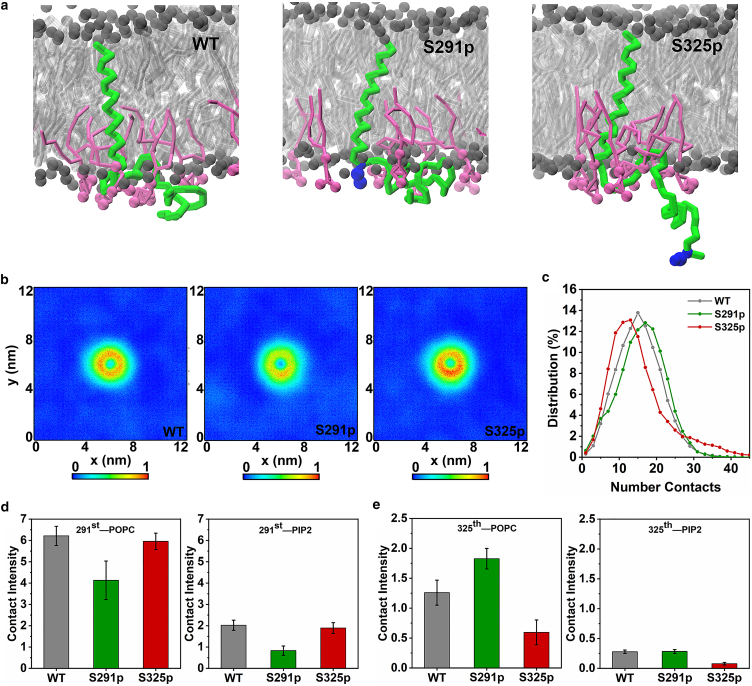
In all three conditions, the FERM domain first attached to the PIP2-containing membrane leaflet and then complexed with CD44, however, with different association rates depending on the phosphorylation state. According to the distance evolution of CD44 and the FERM under different phosphorylated states (Fig. 2 a), S291p slows down the formation rate of the CD44-FERM complex, with an average binding time of 1.3 μs, compared with 0.6 μs for the CD44-WT. In contrast, S325p significantly accelerates the protein interaction, with complexation observed within 0.2 μs for all 10 replicas. Correspondingly, contact intensity analysis shows that the interaction between CD44-S325p and FERM is remarkably stronger than the cases of WT and S291p, whereas the S291p presents the weakest (Figs. 2 b and S3). In order to further verify the influence of different phosphorylation sites on the protein association efficiency, the binding free energy was extracted by calculating the PMF between the centers of mass of each protein. The energy required for separating the protein complex of CD44-WT/FERM was 38 (±1.0) kJ/mol, and it decreases to 27 (±1.9) kJ/mol for S291p. In contrast, the binding energy increases to 78 (±2.1) kJ/mol in the case of S325p (Figs. 2 c and S2). The differences in binding energy cost for the three systems thus confirms the different roles of phosphorylation in the strength of CD44 packing with the cytoskeleton receptor: S291 phosphorylation is unfavorable for the formation of the CD44-FERM complex, whereas S325 phosphorylation exhibits a promoting factor, which is consistent with the existing conclusions based on experimental data (19,51).
Figure 2.
Association of phosphorylated CD44 and FERM. (a) Evolution of protein distance between the centers of mass of CD44-CTD and FERM. The protein complex was considered to be established when the distance is about 4 nm. 10 replicas were analyzed and averaged by a bold line for the three systems. (b) Contact intensity of CD44 of different phosphorylated states with FERM, averaged over 10 replicas (Fig. S3). The contact intensity was calculated by using a cutoff distance of 0.6 nm. (c) PMF profiles as a function of distance between CD44-CTD and FERM. An average of PMFs from two randomly bound states was presented here (more details can be seen in Fig. S2). The peripheral error regions represent the standard deviation of two umbrella sampling simulations, whereas the interior error regions represent the statistical error within an umbrella sampling calculated by bootstrap analysis. (d) Representative snapshots of FERM binding CD44 for S291p (left) and S325p (right) in the 95% POPC/5% PIP2 membrane. Phosphorylation sites S291 and S325 are labeled by blue spheres. To see this figure in color, go online.
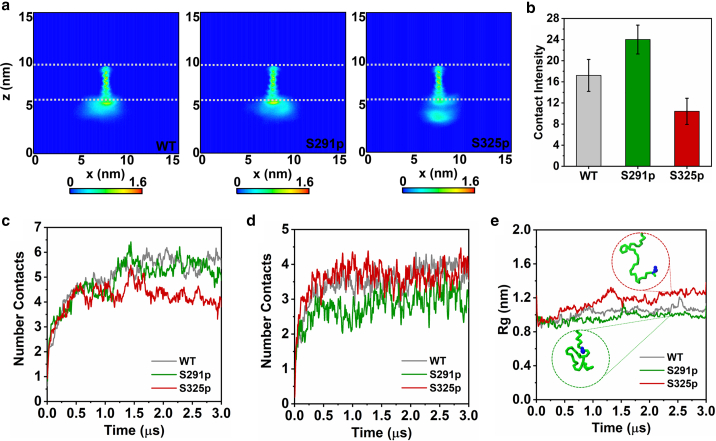
In order to explain the reason why the phosphorylation at different positions leads to distinct binding modes of CD44-FERM, we picked out the representative CD44-FERM binding modes under different phosphorylation conditions (Fig. 2 d). It can be observed that the CTDs of S291p and S325p exhibit different conformation modes. By analyzing the two-dimensional density map of the CTDs position relative to bilayer headgroups (Figs. 3 a and S4 a), it can be seen that S291p-CTD tends to be close to the PIP2-containing membrane leaflet, whereas S325p-CTD is more detached from the membrane. This phenomenon is supported by the contact intensity of CTD with the inner leaflet (Fig. 3 b). S325p can attenuate the binding of CTD to the membrane, which facilitates for a further binding to FERM. Additionally, enrichment of PIP2 around CD44-CTD is observed to be affected by different phosphorylation conditions (Fig. 2 d). Compared with S325p-CTD, the number of contacts between S291p-CTD and PIP2 is significantly increased (Figs. 3 c, S4 b, and S5), which is consistent with the above result that S291p-CTD attaches more tightly to the inner leaflet. In contrast, S291p reduces the PIP2 molecules in vicinity of CD44-TMD, whereas the interaction of S325p-TMD with PIP2 is significantly increased (Figs. 3 d, S4 c, and S6). This indicates that S291 phosphorylation repels PIP2 molecules, resulting in an opposite trend in the contact distribution of PIP2 around the CTD and TMD.
Figure 3.
Conformational characteristics of CTD in different phosphorylation states. (a) The lateral density maps of the CD44-WT (left), CD44-S291p (middle), and CD44-S325p (right) relative to the lipid headgroups. The gray dashed line shows the average position of the lipid phosphate headgroups. (b) Intensity of contact between CTD and phosphates of POPC lipid headgroups. Number of contacts between (c) the CTD and (d) the TMD with the lipids of PIP2, averaged over 10 replicas (individual traces shown in Figs. S5 and S6). (e) Radius of gyration of the CTD under different phosphorylation conditions, averaged over 10 replicas (individual traces in Fig. S7a). To see this figure in color, go online.
In addition to interactions with lipid molecules, conformational changes also play a key role in the signaling pathways of transmembrane proteins. It can be observed that, in contrast with the coiled structure of the S291p, the S325p-CTD adopts a more extended form (Fig. 2 d). It is found that the radius of gyration of S291p-CTD is significantly smaller than that of S325p-CTD in the process of protein binding (Figs. 3 e and S7 a). S325p avoids unnecessary intramolecular interaction among the CTD residues and binds FERM in an extended shape (Fig. S7 b), which allows the protein complex to be more stable.
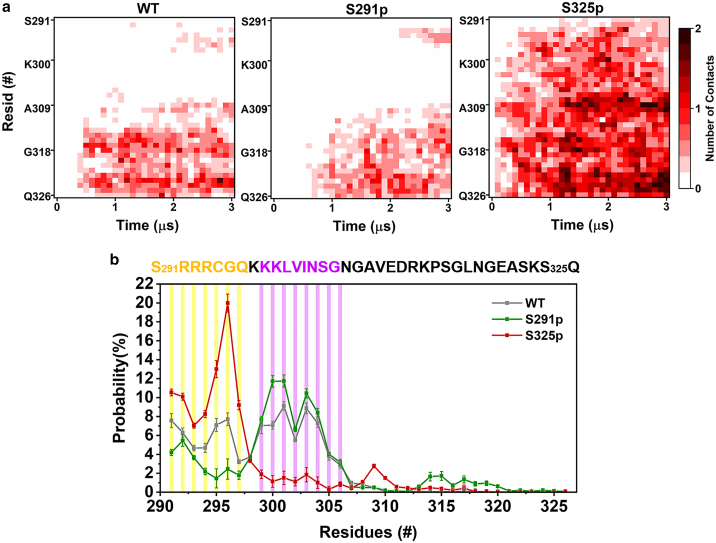
To gain more detailed information how the structural transformation of CD44-CTD induced by phosphorylations affects the complex formation, we calculated the residue interaction heatmap between CD44-CTD and FERM along all 10 simulation trajectories (Figs. 4 a and S8). It reveals that a short juxta-membrane region (R293–K298) and a membrane-remote CTD tail (N307–Q326) of CD44-WT contribute to FERM binding. Phosphorylation of S291 basically abrogates the juxta-membrane FERM-binding region, whereas a high-contact-density map is observed for S325p, implying its entire CTD domain is capable of interacting with FERM throughout the simulations. Interestingly, the residue fragment (K299–G306) in S291p-CTD that cannot access FERM exhibits an enhanced interaction with lipid molecules (Fig. 4 b). It indicates that the conformation leads to a slower binding rate and reduced interaction strength between S291p-CTD and FERM. Collectively, our data show that the phosphorylation of CTD at specific sites can determine protein recognition and binding intensity via the modified interactive motifs and conformational modes.
Figure 4.
Residue analysis of CD44 revealing contacts to FERM and the membrane. (a) Contact heatmap plots between the residues of CD44-CTD and FERM during the entire 3-μs simulation for the WT (left), S291p (middle), and S325p (right) CD44 variants. An average of 10 replicas is shown here (individual replicas are shown in Fig. S8). (b) The probability distribution of the CTD residues in different phosphorylation states to be in contact with lipid phosphates. To see this figure in color, go online.
PIP2 plays a crucial role in priming CD44 for binding to FERM
The distinct conformation of CD44-CTD described above reflects the importance of membrane regulation as CD44-S291p shows an unfavorable FERM package state with a membrane concealing of the residue fragment (K299–G306). It is speculated that the negative-charged phosphorylation of S291 regulates the distribution of anionic PIP2 lipids, thereby enhancing the interaction of K299–G306 with the membrane. In order to explore the conformational changes of CD44-CTD affected by PIP2 before CD44 meets FERM, we constructed simulation systems (12 × 12 × 15 nm3) merely containing CD44 in the 95% POPC/5% PIP2 lipid bilayer (Fig. 5 a). In line with our previous simulation results (32), we observe a strong enrichment of PIP2 lipids around CD44 (Fig. 5 a). Compared with S325p that shows a remarkable enrichment of PIP2 near the TMD region, PIP2 enrichment around S291p is less evident (Figs. 5 b and S9 a). Nevertheless, the contact intensity of PIP2 on the entire CTD for the S291p variant is higher than S325p (Fig. S9 b and c), implying the phosphorylation of S291 mostly repels PIP2 to a more remote CTD region that keeps a considerable capacity in binding PIP2. This in turn leads to an enhanced attachment of the S291p-CTD to the membrane, adopting a closed conformation supported by the graphical image (Fig. 5 a), the 2D density maps of CD44 near the membrane (Fig. S9 d), and the protein-lipid contact intensity (Fig. 5 c). In contrast, the contact frequency between the S325p-CTD and PIP2 decreases (Fig. S9 c), as the S325p-CTD tends to separate away from the inner leaflet (Figs. 5 a, c, and S9 d). The difference in PIP2 aggregation can be attributed to the position of the negative charge upon phosphorylation. In case of S291, phosphorylation takes place close to a stretch of basic residues, which is mainly responsible for PIP2 binding (Fig. 1 a). The negative charge of phosphorylation forces the contact of the 291st site with PIP2 to decrease significantly (Fig. 5 d). For the S325p case, the electrostatic repulsion with the PIP2 lipids of S325 phosphorylation can be relieved by the CTD moving away from the inner membrane surface (Fig. 5 e), leading to a strong PIP2 binding of the basic stretch (Fig. 5 a). The results thus indicate that, before CD44 binds to FERM, the phosphorylation modification and presence of PIP2 coordinate to present an alternative CTD structure that controls the FERM-interaction potency.
Figure 5.
Behavior of CD44-CTD before FERM binding. (a) Representative conformations of CD44-WT, CD44-S291p, and CD44-S325p in the 95% POPC/5% PIP2 membrane. (b) Density maps of the distribution of PIP2 relative to the TMD. Results were obtained from averaging the last 2.5 μs of all 10 replicas. The CD44 is prefixed in the box center for analysis. (c) Protein-lipid contact distribution of the CTD with the lipid phosphates under different phosphorylation states. Contact intensity of (d) the 291st residue and (e) the 325th residue with lipid phosphates of POPC (left) and PIP2 (right) in the three phosphorylation states. To see this figure in color, go online.
To further test the specificity of PIP2 in the conformational switching of the CD44-CTD under different phosphorylation modifications, we replaced the 5% PIP2 component by 20% POPS to maintain the same bilayer charge concentration (Fig. S10 a). It is found that POPS molecules cluster only weakly in vicinity of CD44 under either of the two phosphorylation states (Figs. S10 b and S11 a). Moreover, the interaction of the CTD with the membrane surface is remarkably reduced compared with the cases of PIP2, and little difference is found between the two phosphorylation conditions (Figs. S10 c and S11 b–d). Therefore, PIP2 appears to be a specific mediator in maintaining the distinct structure of CD44-CTD under different phosphorylation modifications.
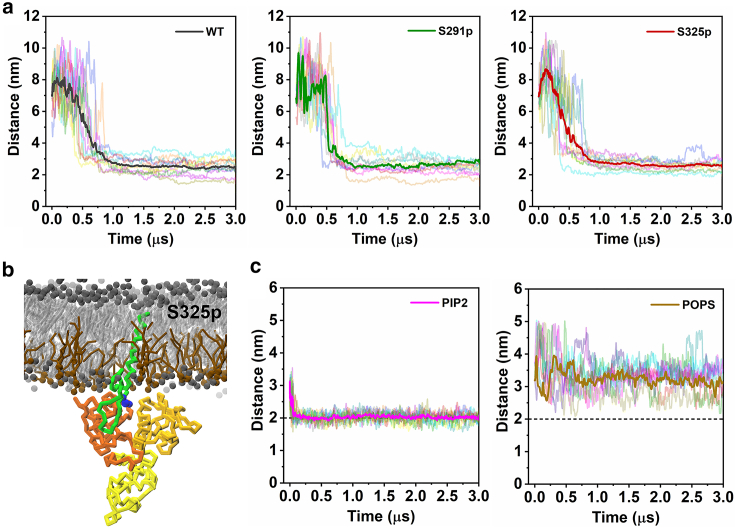
We subsequently recruited FERM in the system of 80% POPC/20% POPS membrane environment. Binding still occurs, as is evident from the temporal distance plots between FERM and the CD44-CTD (Fig. 6 a) and from the snapshot of the bound state (Fig. 6 b); however, the position of the phosphorylation site no longer plays a role. It is noteworthy that, in the POPS environment, membrane adhesion of FERM is rarely observed before it binds to CD44 (Figs. 6 c and S12), consistent with our previous work on complexation of FERM and L-selectin (24). The replacement of PIP2 by POPS also results in a larger equilibrium binding distance (Fig. 6 a) and presumably a weaker binding free energy as previously quantified for the FERM/L-selectin complex. In absence of PIP2, association of the two proteins just occurs after a random mobility of FERM in the cytoplasmic space, and successful binding is more dependent on the structure accessibility of the CD44-CTD, which is no longer affected by phosphorylation (Fig. S13). In contrast, in the PIP2 membrane environment, FERM quickly binds to the membrane, diffuses laterally, and recognizes the binding site of CTD in a phosphorylation-dependent manner. Therefore, the membrane environment containing PIP2 plays an important role in the rapid and efficient formation of the CD44-FERM complex.
Figure 6.
PIP2 effects CD44 structure in a phosphorylation-dependent manner. (a) Distance evolution of the phosphorylated CD44-CTDs with FERM in the bilayer of 80% POPC/20% POPS. (b) The representative binding mode of CD44-S325p and FERM in the 80% POPC/20% POPS membrane. (c) Distance evolution between the centers of mass of FERM and the lower lipid headgroups along the membrane normal in the membranes composed of 95% POPC/5% PIP2 (left) and 80% POPC/20% POPS (right). 10 replicas are analyzed and averaged by a bold line for each system. To see this figure in color, go online.
Taken together, our data demonstrate that the phosphorylation of S291 and S325 affects the conformational state of the CD44-CTD, which further regulates the recognition and binding of CD44 to the cytoplasmic protein acceptor. The comparison of POPS and PIP2 membrane environments reveals that PIP2 and the phosphorylated residues cooperate to determine the structure of CD44-CTD, indicating that the regulation of phosphorylation functions in a PIP2-dependent manner. The molecular details of conformational changes and protein complexation under the coregulation of phosphorylation and PIP2 are significant for understanding the cell proliferation activity involving CD44 and provide a theoretical basis for the design of inhibitory drugs for the potential mechanism of tumor cell migration.
Discussion
Phosphorylation modifications play an important role in an adjustable protein association of CD44 and the cytoskeletal receptor protein. In this study, we have unraveled the structure differences of CD44-CTD upon phosphorylation and how they contribute to the altered binding behavior in CD44 and FERM. The S291-phosphorylated CTD variant attaches on the membrane surface, adopting a curled conformation that disturbs further recognition and association with FERM. In contrast, the extended form of the S325-phosphorylated CTD variant exposes the FERM-binding fragment and promotes the formation of CD44/FERM complexes (Figs. 3 and 5). CD44-FERM binding is known to drive downstream signaling and increase cell proliferation events (52,53). A recent study has demonstrated that blocking the binding of CD44 and moesin in breast cancer cells prevented the migration and invasion of cancer cells (54). The binding of CD44-FERM can regulate the disease environment of abnormal proliferation of cancer cells, so understanding the regulatory mechanism of CD44 phosphorylation warrants further investigation. More and more studies have begun to focus on the potential application value of CD44 and cytoskeletal protein abnormal binding in the early diagnosis of tumor, metastasis potential assessment, treatment, and prognosis (55). CD44 is expected to be a potentially valuable biomarker and therapeutic target, especially in CD44 overexpressed tumor cells (56).
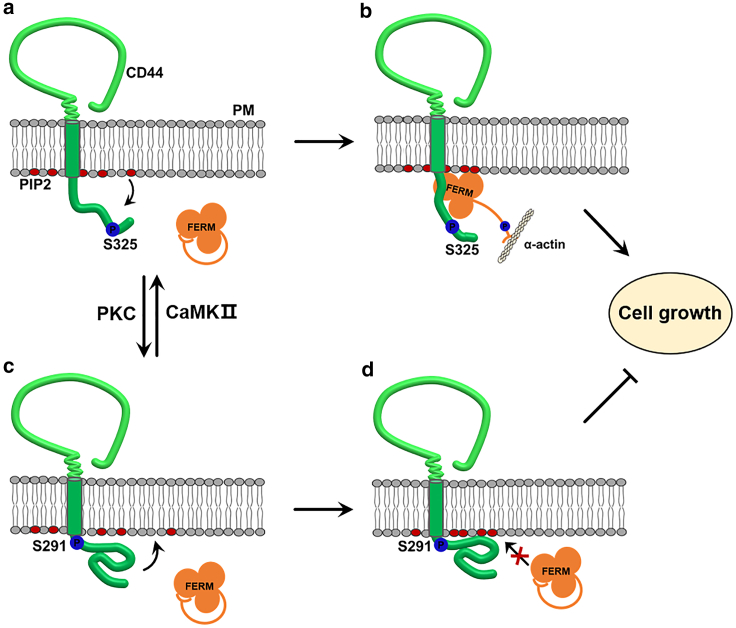
CD44-CTD changes its structure in different ways during phosphorylation modification, primarily driven by electrostatic interaction that modulates the conformational change to a large extent. It was reported that loss of ezrin binding was dependent on the presence of a negative charge at the 291st site as no impairment was observed in mutant with a neutral amino acid (21). Similarly, our study shows that a negative charge introduced via S291 phosphorylation repels PIP2, resulting in its preferential interaction with K299-G304 region instead (Figs. 5 a and S10 c), causing structural folding of CTD and the membrane-proximal region to adhere closely on the membrane surface, so that the binding of FERM to CD44 and related cell proliferation are inhibited (Fig. 7 c and d). In contrast, a repulsive effect between PIP2 and S325 phosphorylation can coordinate allosterically to promote the extension of CTD (Fig. 7 a) and release of CTD from the membrane, which together promote the binding of CD44 and FERM, which is known to be essential for cell proliferation activity (57) (Fig. 7 b). Based on dynamic simulations, Newe et al. also reported that L-selectin, a protein similar to CD44, is desorbed from the plasma membrane due to the repulsive force provided by the phosphorylation of S367, which is the basis for ERM binding and then signaling processes (33).
Figure 7.
Schematic presentation of phosphorylation alternative on S291 or S325 modulates CD44/ERM interaction modulated by CD44-CTD conformations and PIP2. (a) CaMK II is known to phosphorylate the tail of CD44. Phosphorylation of S325, but not S291, drives repulsion of the CD44 tail from the inner leaflet of the plasma membrane. CD44-CTD is presented in an extended shape. (b) Phosphorylation of S325 encourages binding of CD44-ERM, and PIP2 acts as a promoter. ERM binds to the CD44-CTD at the proximity of inner membrane, whereas the C-terminal domain of ERM is able to connect with the α-actinin cytoskeleton protein, which promotes cell growth. (c) As S291 is phosphorylated by PKC, the first half of CD44-CTD adheres to the inner membrane due to strong electrostatic interactions with PIP2, whereas the structure of the second half of CD44-CTD is folding. (d) The membrane concealing conformation and rolled structure of CD44-CTD will block the further binding to ERM and inhibit cell proliferation. To see this figure in color, go online.
Given the increasing reports reflecting the positive role of PIP2 in association of CD44 and FERM, we aim here for elucidation of the unknown function of PIP2, together with phosphorylation, in the conformational change of CD44. We find that the regulatory effect of phosphorylation of S291 and S325 occurs only in the presence of PIP2 (Figs. 6, S10, and S11). Compared with the case of PIP2, phosphorylation does not regulate the binding efficiency of CTD and FERM when the inner membrane contains POPS lipids at the same charge concentration (Fig. 6 a and b), and CTD does not undergo the related conformational changes (Fig. S11). PIP2, however, is able to sense the different phosphorylation states, inducing conformational changes in CTD, which is an important primer for the protein interaction between CD44 and FERM (Figs. 3, 5, and 7). In addition, a previous study shows that there are specific PIP2 binding sites between the F1 and F3 subunits of FERM, which are necessary for significant membrane attachment (58). Rey-Gallardo et al. found that blocking moesin-PIP2 binding significantly reduces T558 phosphorylation and activation of moesin in monocytes (59). Our simulation results show that FERM hardly binds to the inner membrane containing POPS (Figs. 6 b and S12), implying FERM cannot recognize and bind to CD44 through a membrane translocation pathway (i.e., 2D diffusion on the membrane) in the absence of PIP2. It therefore provides another reason for the low efficiency of binding between the CTD and FERM under POPS condition. PIP2 not only regulates CD44, but the binding of PIP2 to ERM is necessary for ERM activation (60). Pluhackova et al. used MD simulations to investigate how phosphorylation and acidic lipids affect the structure and dynamics of the transmembrane protein β2AR. Phosphorylation of residues S246, S261, and S262 in the intracellular loop (ICL3) drives ICL3 release into the cytoplasm to interact with arrestin, but the absence of acidic lipids reverses the phosphorylation regulation (37). It implies that PIP2 is critical for protein regulation by phosphorylation in more than one way.
In summary, we provide in-depth molecular investigations revealing that specific phosphorylation of S291 or S325 regulates the conformational changes of CD44-CTD, which affects the further recognition and combination of CD44-like proteins and cytoplasmic protein acceptors such as FERM. The comparison of POPS and PIP2 membrane environments shows that PIP2 and phosphorylated residues cooperate to maintain the regulatory structure, indicating that the regulation of phosphorylation requires the participation of PIP2 molecules. In addition, PIP2 drives the binding of FERM to the membrane, also crucial for subsequent formation of the CD44-FERM complex. The molecular details of conformational changes and protein translocation under the coregulation of phosphorylation and PIP2 are significant for understanding the cell proliferation activity of CD44, and they provide a theoretical basis for the design of inhibitory drugs for the potential mechanism of tumor cell migration.
Author contributions
F.S., S.J.M., and H.A. designed the research. F.S. and M.R. performed the simulations; F.S. and M.R. contributed the analytic methods. M.R. and L.Z. analyzed the simulation results. M.R., F.S., S.J.M., and H.A. wrote the article.
Acknowledgments
This work was supported by the Natural Science Foundation of China (No. 32201022), the Science and Technology Project of Hebei Education Department (No. ZD2020140), the Natural Science Foundation of Hebei Province (No. C2019202244 and No. H2020202005), and the Natural Science Foundation of Tianjin City (No. 19JCYBJC28300).
Declaration of interests
The authors declare no competing interests.
Editor: Alan Grossfield.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2023.05.021.
Contributor Information
Hailong An, Email: hailong_an@hebut.edu.cn.
Siewert Jan Marrink, Email: s.j.marrink@rug.nl.
Fude Sun, Email: sunfd@hebut.edu.cn.
Supporting material
References
- 1.Chen C., Zhao S., et al. Freeman J.W. The biology and role of CD44 in cancer progression: therapeutic implications. J. Hematol. Oncol. 2018;11:64. doi: 10.1186/s13045-018-0605-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weng X., Maxwell-Warburton S., et al. Kang L. The membrane receptor CD44: novel insights into metabolism. Trends Endocrinol. Metab. 2022;33:318–332. doi: 10.1016/j.tem.2022.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Domev H., Amit M., et al. Itskovitz-Eldor J. Efficient engineering of vascularized ectopic bone from human embryonic stem cell-derived mesenchymal stem cells. Tissue Eng. Part A. 2012;18:2290–2302. doi: 10.1089/ten.TEA.2011.0371. [DOI] [PubMed] [Google Scholar]
- 4.Thapa R., Wilson G.D. The importance of CD44 as a stem cell biomarker and therapeutic target in cancer. Stem Cells Int. 2016;2016 doi: 10.1155/2016/2087204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mellor L., Knudson C.B., et al. Knudson W. Intracellular domain fragment of CD44 alters CD44 function in chondrocytes. J. Biol. Chem. 2013;288:25838–25850. doi: 10.1074/jbc.M113.494872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yusupov M., Privat-Maldonado A., et al. Bogaerts A. Oxidative damage to hyaluronan-CD44 interactions as an underlying mechanism of action of oxidative stress-inducing cancer therapy. Redox Biol. 2021;43 doi: 10.1016/j.redox.2021.101968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim S.I., Koo J.S. Expression of cancer stem cell markers in breast phyllodes tumor. Cancer Biomark. 2020;29:235–243. doi: 10.3233/CBM-191276. [DOI] [PubMed] [Google Scholar]
- 8.Misra S., Hascall V.C., et al. Ghatak S. Interactions between hyaluronan and its receptors (CD44, rhamm) regulate the activities of inflammation and cancer. Front. Immunol. 2015;6:201. doi: 10.3389/fimmu.2015.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ponta H., Sherman L., Herrlich P.A. CD44: from adhesion molecules to signalling regulators. Nat. Rev. Mol. Cell Biol. 2003;4:33–45. doi: 10.1038/nrm1004. [DOI] [PubMed] [Google Scholar]
- 10.Li L.L., Bao J., et al. Guo X. Biomechanics and functional regulations of CD44-ligand interactions. Chin. J. Theor. Appl. Mech. 2021;56:539–548. [Google Scholar]
- 11.Mori T., Kitano K., et al. Hakoshima T. Structural basis for CD44 recognition by ERM proteins. J. Biol. Chem. 2008;283:29602–29612. doi: 10.1074/jbc.M803606200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.García-Ortiz A., Serrador J.M. ERM proteins at the crossroad of leukocyte polarization, migration and intercellular adhesion. Int. J. Mol. Sci. 2020;21:1502. doi: 10.3390/ijms21041502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shao Y., Li L., et al. Sun B. CD44/ERM/f-actin complex mediates targeted nuclear degranulation and excessive neutrophil extracellular trap formation during sepsis. J. Cell Mol. Med. 2022;26:2089–2103. doi: 10.1111/jcmm.17231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yaghobi Z., Movassaghpour A., et al. Baradaran B. The role of CD44 in cancer chemoresistance: a concise review. Eur. J. Pharmacol. 2021;903 doi: 10.1016/j.ejphar.2021.174147. [DOI] [PubMed] [Google Scholar]
- 15.Spadea A., Rios de la Rosa J.M., et al. Mehibel M. Evaluating the efficiency of hyaluronic acid for tumor targeting via CD44. Mol. Pharm. 2019;16:2481–2493. doi: 10.1021/acs.molpharmaceut.9b00083. [DOI] [PubMed] [Google Scholar]
- 16.Wang C., Okita Y., et al. Kato M. Glycoprotein non-metastatic melanoma protein b functions with growth factor signaling to induce tumorigenesis through its serine phosphorylation. Cancer Sci. 2021;112:4187–4197. doi: 10.1111/cas.15090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vitrac H., Mallampalli V.K.P.S., Dowhan W. Importance of phosphorylation/dephosphorylation cycles on lipid-dependent modulation of membrane protein topology by posttranslational phosphorylation. J. Biol. Chem. 2019;294:18853–18862. doi: 10.1074/jbc.RA119.010785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bahl S., Seto E. Regulation of histone deacetylase activities and functions by phosphorylation and its physiological relevance. Cell. Mol. Life Sci. 2021;78:427–445. doi: 10.1007/s00018-020-03599-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thorne R.F., Legg J.W., Isacke C.M. The role of the CD44 transmembrane and cytoplasmic domains in co-ordinating adhesive and aignalling events. J. Cell Sci. 2004;117:373–380. doi: 10.1242/jcs.00954. [DOI] [PubMed] [Google Scholar]
- 20.Lewis C.A., Townsend P.A., Isacke C.M. Ca(2+)/calmodulin-dependent protein kinase mediates the phosphorylation of CD44 required for cell migration on hyaluronan. Biochem. J. 2001;357:843–850. doi: 10.1042/0264-6021:3570843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Legg J.W., Lewis C.A., et al. Isacke C.M. A novel pkc-regulated mechanism controls CD44 ezrin association and directional cell motility. Nat. Cell Biol. 2002;4:399–407. doi: 10.1038/ncb797. [DOI] [PubMed] [Google Scholar]
- 22.Renard K., Byrne B. Insights into the role of membrane lipids in the structure, function and regulation of integral membrane proteins. Int. J. Mol. Sci. 2021;22:9026. doi: 10.3390/ijms22169026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen X., Khajeh J.A., et al. Bu Z. Phosphatidylinositol 4,5-bisphosphate clusters the cell adhesion molecule CD44 and assembles a specific CD44-ezrin heterocomplex, as revealed by small angle neutron scattering. J. Biol. Chem. 2015;290:6639–6652. doi: 10.1074/jbc.M114.589523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun F., Schroer C.F.E., et al. Luo S.Z. Molecular dynamics of the association of L-selectin and FERM regulated by PIP2. Biophys. J. 2018;114:1858–1868. doi: 10.1016/j.bpj.2018.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marrink S.J., Monticelli L., et al. Souza P.C. Two decades of martini: better beads, broader scope. WIREs Comput. Mol. Sci. 2022;13 [Google Scholar]
- 26.Corradi V., Mendez-Villuendas E., et al. Tieleman D.P. Lipid-protein interactions are unique fingerprints for membrane proteins. ACS Cent. Sci. 2018;4:709–717. doi: 10.1021/acscentsci.8b00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chavent M., Karia D., et al. Sansom M.S.P. Interactions of the EphA2 kinase domain with pips in membranes: implications for receptor function. Structure. 2018;26:1025–1034.e2. doi: 10.1016/j.str.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kirsch S.A., Kugemann A., et al. Dietrich P. Phosphatidylinositol-3,5-bisphosphate lipid-binding-induced activation of the human two-pore channel 2. Cell. Mol. Life Sci. 2018;75:3803–3815. doi: 10.1007/s00018-018-2829-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ingólfsson H.I., Neale C., et al. Streitz F.H. Machine learning-driven multiscale modeling reveals lipid-dependent dynamics of ras signaling proteins. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2113297119. e2113297119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thallmair V., Schultz L., et al. Thallmair S. Two cooperative binding sites sensitize PI(4,5)P2 recognition by the tubby domain. Sci. Adv. 2022;8:9471–9482. doi: 10.1126/sciadv.abp9471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herzog F.A., Braun L., et al. Vogel V. Structural insights how PIP2 imposes preferred binding orientations of FAK at lipid membranes. J. Phys. Chem. B. 2017;121:3523–3535. doi: 10.1021/acs.jpcb.6b09349. [DOI] [PubMed] [Google Scholar]
- 32.Sun F., Schroer C.F.E., et al. Marrink S.J. Molecular mechanism for bidirectional regulation of CD44 for lipid raft affiliation by palmitoylations and PIP2. PLoS Comput. Biol. 2020;16:e1007777. doi: 10.1371/journal.pcbi.1007777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newe A., Rzeniewicz K., et al. Ivetic A. Serine phosphorylation of L-selectin regulates ERM binding, clustering, and monocyte protrusion in transendothelial migration. Front. Immunol. 2019;10:2227. doi: 10.3389/fimmu.2019.02227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma Z., Shi S., et al. Sun F. Molecular mechanism of CD44 homodimerization modulated by palmitoylation and membrane environments. Biophys. J. 2022;121:2671–2683. doi: 10.1016/j.bpj.2022.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Jong D.H., Singh G., et al. Marrink S.J. Improved parameters for the martini coarse-grained protein force field. J. Chem. Theory Comput. 2013;9:687–697. doi: 10.1021/ct300646g. [DOI] [PubMed] [Google Scholar]
- 36.Marrink S.J., Risselada H.J., et al. de Vries A.H. The martini force field: coarse grained model for biomolecular simulations. J. Phys. Chem. B. 2007;111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 37.Pluhackova K., Wilhelm F.M., Müller D.J. Lipids and phosphorylation conjointly modulate complex formation of β2-adrenergic receptor and β-arrestin2. Front. Cell Dev. Biol. 2021;9:3610. doi: 10.3389/fcell.2021.807913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mori T., Kitano K., et al. Hakoshima T. Crystallographic characterization of the radixin FERM domain bound to the cytoplasmic tail of adhesion molecule CD44. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2007;63:844–847. doi: 10.1107/S1744309107041152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Periole X., Cavalli M., et al. Ceruso M.A. Combining an elastic network with a coarse-grained molecular force field: structure, dynamics, and intermolecular recognition. J. Chem. Theory Comput. 2009;5:2531–2543. doi: 10.1021/ct9002114. [DOI] [PubMed] [Google Scholar]
- 40.Wassenaar T.A., Ingólfsson H.I., et al. Marrink S.J. Computational lipidomics with insane: a versatile tool for generating custom membranes for molecular simulations. J. Chem. Theory Comput. 2015;11:2144–2155. doi: 10.1021/acs.jctc.5b00209. [DOI] [PubMed] [Google Scholar]
- 41.James D.J., Khodthong C., et al. Martin T.F.J. Phosphatidylinositol 4,5-bisphosphate regulates snare-dependent membrane fusion. J. Cell Biol. 2008;182:355–366. doi: 10.1083/jcb.200801056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van der Spoel D., Lindahl E., et al. Berendsen H.J.C. Gromacs: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 43.Berendsen H.J.C., Postma J.P.M., et al. Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 44.Hoover W.G. Canonical dynamics: equilibrium phase-space distributions. Phys. Rev. A. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 45.Nosé S. A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 1984;81:511–519. [Google Scholar]
- 46.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 47.de Jong D.H., Baoukina S., et al. Marrink S.J. Martini straight: boosting performance using a shorter cutoff and gpus. Comput. Phys. Commun. 2016;199:1–7. [Google Scholar]
- 48.Corey R.A., Stansfeld P.J., Sansom M.S.P. The energetics of protein-lipid interactions as viewed by molecular simulations. Biochem. Soc. Trans. 2020;48:25–37. doi: 10.1042/BST20190149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hub J.S., de Groot B.L., van der Spoel D. G_wham-a free weighted histogram analysis implementation including robust error and autocorrelation estimates. J. Chem. Theory Comput. 2010;6:3713–3720. [Google Scholar]
- 50.Humphrey W., Dalke A., Schulten K. Vmd: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 51.Jokela T., Oikari S., et al. Tammi R. Interleukin-1β-induced reduction of CD44 ser-325 phosphorylation in human epidermal keratinocytes promotes CD44 homomeric complexes, binding to ezrin, and extended, monocyte-adhesive hyaluronan coats. J. Biol. Chem. 2015;290:12379–12393. doi: 10.1074/jbc.M114.620864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Terawaki S.I., Kitano K., et al. Hakoshima T. Mt1-mmp recognition by ERM proteins and its implication in CD44 shedding. Gene Cell. 2015;20:847–859. doi: 10.1111/gtc.12276. [DOI] [PubMed] [Google Scholar]
- 53.Orian-Rousseau V., Morrison H., et al. Ponta H. Hepatocyte growth factor-induced Ras activation requires ERM proteins linked to both cd44v6 and f-actin. Mol. Biol. Cell. 2007;18:76–83. doi: 10.1091/mbc.E06-08-0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu S., Shi X., et al. Gao F. CD44 cross-linking increases malignancy of breast cancer via upregulation of p-moesin. Cancer Cell Int. 2020;20:563. doi: 10.1186/s12935-020-01663-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Muntimadugu E., Kumar R., et al. Khan W. CD44 targeted chemotherapy for co-eradication of breast cancer stem cells and cancer cells using polymeric nanoparticles of salinomycin and paclitaxel. Colloids Surf. B Biointerfaces. 2016;143:532–546. doi: 10.1016/j.colsurfb.2016.03.075. [DOI] [PubMed] [Google Scholar]
- 56.Morath I., Hartmann T.N., Orian-Rousseau V. CD44: more than a mere stem cell marker. Int. J. Biochem. Cell Biol. 2016;81:166–173. doi: 10.1016/j.biocel.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 57.Chen L., Fu C., et al. Wei Q. The role of CD44 in pathological angiogenesis. FASEB J. 2020;34:13125–13139. doi: 10.1096/fj.202000380RR. [DOI] [PubMed] [Google Scholar]
- 58.Michie K.A., Bermeister A., et al. Curmi P.M.G. Two sides of the coin: ezrin/radixin/moesin and merlin control membrane structure and contact inhibition. Int. J. Mol. Sci. 2019;20:1996. doi: 10.3390/ijms20081996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rey-Gallardo A., Tomlins H., et al. Ivetic A. Sequential binding of ezrin and moesin to L-selectin regulates monocyte protrusive behaviour during transendothelial migration. J. Cell Sci. 2018;131 doi: 10.1242/jcs.215541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ben-Aissa K., Patino-Lopez G., et al. Shaw S. Activation of moesin, a protein that links actin cytoskeleton to the plasma membrane, occurs by phosphatidylinositol 4,5-bisphosphate (PIP2) binding sequentially to two sites and releasing an autoinhibitory linker. J. Biol. Chem. 2012;287:16311–16323. doi: 10.1074/jbc.M111.304881. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.