Abstract
The sequences involved in the replication and packaging of transmissible gastroenteritis virus (TGEV) RNA have been studied. The structure of a TGEV defective interfering RNA of 9.7 kb (DI-C) was described previously (A. Mendez, C. Smerdou, A. Izeta, F. Gebauer, and L. Enjuanes, Virology 217: 495–507, 1996), and a cDNA with the information to encode DI-C RNA was cloned under the control of the T7 promoter. The molecularly cloned DI-C RNA was replicated in trans upon transfection of helper virus-infected cells and inhibited 20-fold the replication of the parental genome. A collection of 14 DI-C RNA deletion mutants (TGEV minigenomes) was synthetically generated and tested for their ability to be replicated and packaged. The smallest minigenome (M33) that was replicated by the helper virus and efficiently packaged was 3.3 kb. A minigenome of 2.1 kb (M21) was also replicated, but it was packaged with much lower efficiency than the M33 minigenome, suggesting that it had lost either the sequences containing the main packaging signal or the required secondary structure in the packaging signal due to alteration of the flanking sequences. The low packaging efficiency of the M21 minigenome was not due to minimum size restrictions. The sequences essential for minigenome replication by the helper virus were reduced to 1,348 nt and 492 nt at the 5′ and 3′ ends, respectively. The TGEV-derived RNA minigenomes were successfully expressed following a two-step amplification system that couples pol II-driven transcription in the nucleus to replication supported by helper virus in the cytoplasm, without any obvious splicing. This system and the use of the reporter gene β-glucuronidase (GUS) allowed minigenome detection at passage zero, making it possible to distinguish replication efficiency from packaging capability. The synthetic minigenomes have been used to design a helper-dependent expression system that produces around 1.0 μg/106 cells of GUS.
Transmissible gastroenteritis virus (TGEV) is a member of the Coronaviridae family (19, 37) with a plus-stranded, polyadenylated RNA genome of 28.5 kb (18). There is limited information on the mechanism of TGEV replication and transcription. The construction of a cDNA encoding an infectious RNA would be very useful to understand gene expression in TGEV. Unfortunately, this cDNA has not been assembled for technical reasons. To overcome this limitation, it would be useful to obtain defective minigenomes from which a cDNA could be produced.
Mouse hepatitis virus (MHV) defective RNAs have been very useful to identify the sequences required for MHV RNA replication, to study viral gene function, and to express heterologous genes (8, 37). In addition, defective RNAs derived from infectious bronchitis virus (IBV) and bovine coronavirus (BCV) have been characterized and used as templates for the construction of cDNA clones (12, 54, 55). Although there are some differences in the sequences required for replication and packaging, a basic consensus is that the replication of the MHV minigenomes requires the 5′ end 467 to 1,100 nucleotides (nt) and the 3′-terminal 447 nt, respectively (66).
To engineer cDNAs encoding TGEV-defective RNAs, three deletion mutants of 22, 10.6, and 9.7 kb (DI-A, DI-B, and DI-C, respectively) maintaining the cis signals required for replication and packaging by helper virus were isolated (47). DI-C RNA was the most abundant and was selected to generate a cDNA that could be used to study the replication and transcription of the TGEV genome. In addition, these synthetic minigenomes can be used for the tissue-specific expression of antigens or molecules interfering with virus replication. In fact, MHV has been previously used to express high amounts of heterologous antigens (38), with the limitation of unstable expression with virus passage.
TGEV-derived constructs may have several advantages as vectors to induce mucosal immunity over expression systems that do not replicate within mucosal tissues, since (i) TGEV infects enteric and respiratory mucosal areas (19), (ii) its tropism may be controlled by modifying the spike (S) gene (3), (iii) nonpathogenic strains are available to develop a helper virus-dependent expression system (59), and (iv) coronaviruses are RNA cytoplasmic viruses that replicate without a DNA intermediate (37), making their integration into the cellular chromosomes unlikely. Vector systems for the expression of heterologous genes have been developed from full-length cDNA clones of positive-strand RNA viruses such as alphaviruses, including Sindbis virus, Semliki Forest virus (SFV), and Venezuelan equine encephalitis virus (VEE) (22, 39, 56). These systems have been very useful to elicit humoral and cellular immune responses.
In this study, we report the construction of a full-length cDNA clone encoding a synthetic TGEV DI-C RNA, which is replicated and packaged upon transfection into helper virus-infected cells. The molecularly cloned DI-C minigenome interfered with TGEV replication. The sequences required for DI-C RNA replication and packaging have been determined by differentiating these two processes. With the TGEV-derived minigenomes, an expression system producing around 1.0 μg/106 cells of the heterologous protein has been developed. The availability of the TGEV-derived synthetic minigenomes reported in this study will be very helpful for the study of replication and gene expression in porcine coronaviruses.
MATERIALS AND METHODS
Cells and viruses.
Viruses were grown in swine testis (ST) cells (46). TGEV PUR46-MAD strain (60) was grown and titrated as described previously (30).
Construction of cDNAs encoding RNA minigenomes.
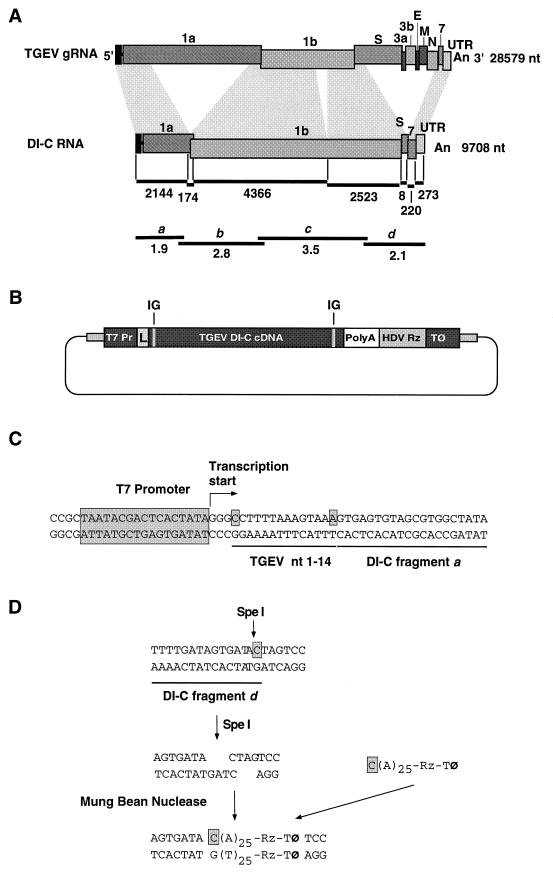
A cDNA encoding DI-C RNA was assembled into plasmid pDI-C. A 9.7-kb band corresponding to DI-C RNA was purified from a gel similar to the one shown in Fig. 1 (47). The four DI-C reverse transcription (RT)-PCR-derived overlapping fragments a, b, c, and d (Fig. 2A) (47) were corrected for point mutations introduced by the RT-PCR procedure and were assembled into plasmid pSL1190 (Pharmacia). The T7 promoter and TGEV nt 1 to 14 were inserted by PCR upstream of DI-C fragment a by using a 69-mer synthetic oligonucleotide (5′-GTGGCGCGCGGCCGCTAATACGACTCACTATAGGGCCTTTTAAAGTAAAGTAGTGAGTGTAGCGTGGCTATA-3′). This oligonucleotide includes BssHII and NotI restriction endonuclease sites (underlined) at the 5′ end to facilitate the cloning of the amplified fragment and the DI-C fragment a first 20 nt at the 3′ end. Three extra bases of nonviral origin (GGG) were included between the promoter and DI-C cDNA, to increase T7 RNA polymerase transcription levels (44). The first TGEV nucleotide assembled was a C (Fig. 2C). To construct the 3′ end, DI-C fragment d was digested with SpeI and blunt ended with mung bean nuclease (Boehringer Mannheim) to remove all plasmid sequences (Fig. 2D). To restore the last viral nucleotide (C) and to include a synthetic poly(A) tract, a 48-mer oligonucleotide (5′-CA25GGGTCGGCATGGCATCTCCACC-3′) was synthesized. This oligonucleotide included the last TGEV nucleotide (C) plus a synthetic poly(A25) tract and the 5′ end of the hepatitis delta virus antigenomic ribozyme (HDV Rz) sequences (63). With this oligonucleotide, the HDV Rz and the T7 terminator sequences (17) were cloned by PCR from transcription vector 2.0 (51), kindly provided by A. Ball, University of Alabama, and incorporated at the DI-C 3′ end. The reverse oligonucleotide was 20-mer (5′-CAAGCTTGCATGCCTGCAGG-3′). The resulting plasmid, pDI-C (13,273 nt), contains the full-length cDNA clone of DI-C RNA (9,708 nt) under the T7 promoter.
FIG. 1.
Northern blot analysis of TGEV defective minigenomes. The analysis of the TGEV PUR46-MAD strain RNAs alone (lane a) or after 40 passages at a high multiplicity of infection (50 to 100 PFU/cell), that resulted in the selection of three defective RNAs (DI-A, DI-B, and DI-C) (lane b), was performed by Northern blotting with a probe specific for the 3′ UTR. Numbers and letters on the left indicate the position of the genomic (gRNA) and viral mRNAs. S, Spike; 3a, gene 3a; E, envelope; M, membrane; N, nucleoprotein; 7, gene 7. Arrows and numbers on the right indicate the position and approximate size of the defective RNAs.
FIG. 2.
Assembly of a cDNA encoding DI-C RNA. (A) The four overlapping cDNA fragments: a, b, c, and d (thin bars) cloned in order to assemble the cDNA encoding DI-C RNA are indicated. These fragments are depicted at the approximate location of the RNA used as template (DI-C RNA) and in relationship to the genomic RNA from which they are derived (TGEV gRNA), indicated by shadowed rectangles. The numbers and letters above the rectangles indicate the viral genes. The numbers below the rectangles indicate the length in nucleotides of the RNA fragments that constitute the DI-C minigenome. The numbers to the right of the TGEV gRNA and the DI-C RNA rectangles indicate the length in nucleotides of the TGEV genome and of DI-C RNA. S, spike gene; 3a and 3b, nonstructural proteins 3a and 3b genes; E, envelope protein gene; M, membrane protein gene; N, nucleoprotein gene; 7, gene 7; UTR, untranslated regions; An, poly(A). (B) Schematic structure of the plasmid containing the cDNA encoding DI-C RNA. A cDNA complementary to the DI-C RNA was assembled downstream of the T7 promoter (T7 Pr). L, leader. At the DI-C 3′ end, a synthetic poly(A) tract with 25 residues was added (PolyA). HDV Rz, hepatitis delta virus ribozyme; T⊘, T7 transcription terminator. IG, intergenic sequence. (C) Sequence of DI-C cDNA 5′ end. The T7 promoter (large shadowed box) and the 5′ TGEV nucleotides 1 to 14 (between the two shadowed nucleotides) were placed upstream of DI-C fragment a, which started at TGEV nt 15 (47). TGEV nt 1 to 14 and DI-C fragment a first 20 nt are underlined. The position of the first nucleotide transcribed is indicated by the arrow (2). (D) Scheme of the construction of the 3′ end of the cDNA encoding DI-C RNA. DI-C fragment d was digested with SpeI restriction endonuclease and blunt ended with mung bean nuclease, and the resulting fragment ligated to the indicated PCR fragment containing a C (shadowed box), the poly(A) tail (A)25, the HDV Rz (Rz) and the T7 transcription terminator sequences (T⊘).
Fourteen DI-C RNA internal deletion mutants were generated by removing the sequences between one or two restriction endonuclease sites, as summarized in Table 1, by standard procedures (57).
TABLE 1.
Restriction sites used to generate pDI-C-derived deletion mutants
| Minigenomea | Deleted nucleotidesb | Enzyme(s) | Procedure |
|---|---|---|---|
| M94 | 8896–9198 | HpaI | Partial digestion and religation |
| M90 | 5829–6539 | StuI | Religated |
| M88 | 8013–8984 | NsiI | Religated |
| M80 | 3281–5050 | PstI | Religated |
| M67 | 2880–5894 | XcmI | Religated |
| M64 | 2880–5894 | XcmI, HpaI | M67 digested with HpaI and religated |
| 8896–9198 | |||
| M62 | 1363–4937 | BsmI | Filled in with Klenow and religated |
| M54 | 3809–8112 | NdeI | Religated (two sites within the deletion) |
| M50 | 2403–7102 | AflII | Partial digestion and religation |
| M42 | 2709–8271 | EcoRI | Partial digestion and religation |
| M39 | 2709–8271 | EcoRI, HpaI | M42 digested with HpaI and religated |
| 8896–9198 | |||
| M33 | 2713–9198 | EcoRI + HpaI | EcoRI site filled in with T4 DNA pol and ligated with HpaI site |
| M22 | 1659–9198 | BglII + HpaI | See CMV-M22-GUS in Materials and Methods |
| M21 | 1349–8986 | SphI | Religated (two sites within the deletion) |
The number indicates the approximate size of the minigenome in hundreds of nucleotides.
Numbering according to DI-C (47).
To increase minigenome RNA expression levels, the cDNAs were next cloned after the cytomegalovirus (CMV) promoter (16, 52). The CMV promoter was amplified from pcDNA3.1 (Invitrogen) and joined directly to the 5′ end of DI-C cDNA (52). The first TGEV nucleotide was replaced in this case by an A, the first nucleotide of the Purdue strain of TGEV (18). The T7 terminator was replaced by a cDNA coding for the bovine growth hormone (BGH) terminator sequences (52). Plasmids pCMV-DI-C, pCMV-M54, pCMV-M39, pCMV-M33, and pCMV-M21 were constructed with the high-copy number plasmid pSL1190.
To evaluate the expression levels using the minigenomes, E. coli K12 β-glucuronidase (GUS) was used as a reporter gene (29, 62). The GUS gene was cloned after TGEV N gene transcription regulatory sequences (TRS). These sequences, composed of the N gene intergenic (IG) sequence CUAAAC plus the 5′ upstream 88 nt flanking the consensus sequence, were amplified by PCR from a plasmid containing the M and N genes of strain PUR46-MAD of TGEV (58). To this end, two oligonucleotides that included four restriction endonuclease sites (the MluI, BlnI, SwaI, and Bsu36I sites are underlined) at the 5′ end of the PCR fragment (5′-AACGCGTCCTAGGATTTAAATCCTAAGGCTATGTAAAATCTAAAGCTGG-3′) and four restriction endonuclease sites (the NotI, SgrAI, StuI, and SalI sites are underlined) at the 3′ end (5′-GACGTCGACAGGCCTCGCCGGCGCGGCCGCGTTTAGTTATACCATATG-3′) were used. The GUS gene was amplified by PCR from plasmid pGUS1 (Plant Genetic Systems) with a forward 40-mer oligonucleotide (5′-GCGGCCGCAGGCCTGTCGACGACCATGGTCCGTCCTGTAG-3′) which included the NotI, StuI, and SalI restriction endonuclease sites (underlined nucleotides). The GUS initiation codon is shown in bold. Nucleotides shown in italics were included to fit the consensus motif of the ribosome scanning model (36). The reverse primer was 41 nt long (5′-GGTACCGCGCGCCTGGGCTAGCGCGATCATAGGCGTCTCGC-3′) and included NheI, BssHII, and KpnI restriction sites. The GUS gene was cloned into M39, M33, and M21 minigenomes within the most 3′ deletion introduced during the generation of each minigenome. The resulting plasmids were named pCMV-M39-GUS, pCMV-M33-GUS, and pCMV-M21-GUS. The GUS gene was also cloned in two cases in the reverse orientation, leading to plasmids pCMV-M33-SUG and pCMV-M21-SUG (Fig. 5B).
FIG. 5.
Replication and packaging of DI-C RNA deletion mutants. (A) The schematic comparison of the structure of three DI-C deletion mutants, one (M22) rescued with very low efficiency, and two others (M62 and M33) rescued with high efficiency is shown. F1 and F2 indicate the large fragments that differentiate M33 and M62, respectively, from M22. An, poly(A); HDV Rz, hepatitis delta virus ribozyme. Δ, position of the deletions introduced during the selection of DI-C RNA. Rescue efficiencies are reported as indicated in Fig. 4. RE, rescue efficiency. (B) GUS activity per 106 cells at passage P0 expressed by a helper-dependent expression system. The cDNAs encoding the minigenomes were cloned after the CMV promoter and were flanked at the 3′ end by a poly(A) tail (An), the hepatitis delta virus ribozyme, and the bovine growth hormone termination and polyadenylation sequences (BGH). The GUS gene was cloned within the deletion introduced to create the minigenomes. Two plasmids identical to M33 and M21 but with a deletion of 171 nt starting at nt 192 from the 3′ end were also constructed. Control plasmids were used with the GUS sequences in the reverse orientation (SUG). The relative GUS activity shown is the average of three independent experiments with the standard deviation. The background (RLUs in the presence of the buffer but in the absence of cell extracts) has been subtracted. GUS values below the discontinuous line are nonsignificant. Mock, untransfected cultures.  , genes S, 7 and 3′ UTR sequences. (C) GUS activity per 106 cells in the presence of helper virus shown by CMV-M39-GUS (▴), CMV-M33-GUS (•), CMV-M22-GUS (♦), and CMV-M21-GUS (■) at different passages. The background (RLUs in the presence of the buffer but in the absence of cell extracts) has been subtracted. No significant activity was observed in the absence of the helper virus. The data shown correspond to one of three representative experiments with similar results.
, genes S, 7 and 3′ UTR sequences. (C) GUS activity per 106 cells in the presence of helper virus shown by CMV-M39-GUS (▴), CMV-M33-GUS (•), CMV-M22-GUS (♦), and CMV-M21-GUS (■) at different passages. The background (RLUs in the presence of the buffer but in the absence of cell extracts) has been subtracted. No significant activity was observed in the absence of the helper virus. The data shown correspond to one of three representative experiments with similar results.
Plasmid pCMV-M22-GUS was derived from plasmid pCMV-M33-GUS by digestion with BglII and Bsu36I to remove a 1,063-nt fragment, blunt ended with T4 DNA pol, and religated. Plasmid pCMV-M22-GUS included DI-C nt 1 to 1659, which were cloned upstream of the N gene intergenic sequence, plus the GUS gene and the same sequences at the 3′ end of the M33 minigenome (Table 1).
To generate minigenomes that could not be replicated in trans by the helper virus, a 171-nt deletion was introduced into the 3′ untranslated region (UTR) starting at 192 nt from the 3′ end of pCMV-M33-GUS and pCMV-M21-GUS minigenomes. This deletion removes sequences that include motifs essential for MHV replication and likely for the replication of the other coronaviruses as well (27, 28, 40, 41). The deletion was introduced between two restriction endonuclease sites, BbrPI (TGEV nt 28388) and Asp700 (nt 28559) (53). The resulting plasmids were named pCMV-M33Δ171-GUS and pCMV-M21Δ171-GUS, respectively (Fig. 5B). All plasmid constructs were sequenced at the cloning junctions by using an Applied Biosystems 373 DNA sequencer and the appropriate oligonucleotides to ensure that no nucleotide changes had been introduced.
In vitro transcription.
In vitro transcription of linearized DNA templates was performed with T7 RNA polymerase (Promega), according to the manufacturer’s instructions. All plasmids were linearized with XhoI restriction endonuclease, downstream of the T7 terminator. The length of the in vitro-transcribed RNAs was estimated in 1% agarose–Tris-borate-EDTA (TBE)–0.1% sodium dodecyl sulfate (SDS) gels (57).
Rescue of T7-driven transcripts by electroporation of helper virus-infected ST cells.
ST cells were grown to confluence and infected with TGEV PUR46-MAD at a multiplicity of infection (MOI) of 10. At 4 to 6 h postinfection, cells were trypsinized and resuspended in ice-cold phosphate-buffered saline, pH 7.2 (PBS). The cells were electroporated (200 V, 500 μF, single pulse) with in vitro-transcribed RNA (5 μg/106 cells) by using a Gene Pulser apparatus (BioRad). The electroporated cells were resuspended in Dulbecco modified Eagle medium (DMEM) supplemented with 2% fetal calf serum and incubated at 37°C for 18 h. Supernatants from these cultures were passed with fresh ST cells at least six times in order to amplify the virions containing the minigenomes. Within each passage, the virus was grown for 22 to 24 h. After the last passage, the RNA was extracted as described previously (47).
Rescue of RNA pol II-driven transcripts.
ST cells grown to 50% confluence in 35-mm-diameter dishes were transfected with 5 to 10 μg of plasmid DNA encoding CMV-driven minigenomes and 15 μl of Lipofectin reagent in Optimem medium (Gibco-BRL), according to the manufacturer’s instructions. The cells were infected with TGEV PUR46-MAD (MOI, 5) at 4 to 6 h posttransfection. Supernatants from these cultures were used to infect fresh ST cell monolayers at 22 to 24 h postinfection, and several passages were performed to amplify the RNA.
RNA analysis by Northern hybridization and Northern blotting.
Cytoplasmic RNA was extracted from helper virus-infected and RNA-transfected ST cells at different passages, as described previously (47). Northern hybridization was performed directly on partially dried gels with a leader-specific oligonucleotide complementary to nt 66 to 91 of the TGEV genome as described previously (55). Northern blot analysis was performed after the RNAs were blotted onto nylon membranes (Duralon-UV; Stratagene) with a [α-32P]dATP-labeled 3′ UTR-specific single-stranded (ss)DNA probe, complementary to nt 28300 to 28544 of the TGEV PUR46-MAD strain genome (53), following standard procedures (57). In both cases, RNAs were separated in denaturing 1% agarose-2.2 M formaldehyde gels.
In vitro activity of HDV ribozyme.
M50 RNA was transcribed in vitro with T7 RNA polymerase for 1 h at 37°C. The transcription mixture was analyzed by Northern blotting with the probe complementary to the leader.
Chemiluminescent detection of β-glucuronidase in cell extracts.
Expression of β-glucuronidase in cell extracts was detected by a chemiluminescent assay (GUS-Light kit; Tropix), according to the manufacturer’s instructions. GUS-encoding minigenome-transfected or mock-transfected cells were infected with helper virus (MOI, 5). At 22 to 24 h postinfection, cells grown in 35-mm-diameter dishes were washed with PBS and resuspended in 200 μl of lysis buffer (100 mM sodium phosphate [pH 7.8], 0.2% Triton X-100, 1 mM dithiothreitol [DTT]). Undiluted or serially diluted cell extracts (6.7 μl) were incubated for 1 h at room temperature with 60 μl of Glucuron chemiluminescent substrate (9–11), 1:100 diluted in GUS reaction buffer (100 mM sodium phosphate [pH 7.0], 10 mM EDTA) by using the appropriate luminometer tubes (Sarstedt no. 55.476). Tubes containing the reaction mixture were placed in the luminometer chamber, and the reaction was enhanced by the injection of 100 μl of Light Emission Accelerator (11). The light emission over the first 10 s of reaction enhancement was recorded with a Lumat LB 9501 luminometer (Berthold Systems). A background of 2,000 to 4,500 relative luminometric units (RLU) was obtained under our assay conditions and was subtracted from each reading. The amount of protein expressed was estimated by using standard calibration curves generated with purified β-glucuronidase provided by Sigma (106 RLU/0.35 ng of β-glucuronidase).
Nucleotide sequence accession number.
The sequence of DI-C RNA has been submitted to the EMBL database under accession no. AJ011482.
RESULTS
Construction of a synthetic DI-C RNA derived from TGEV.
Three subgenomic RNA species were generated after passage of the TGEV PUR46-MAD strain in ST cells at a high MOI; these were named DI-A, DI-B, and DI-C and were 22, 10.6, and 9.7 kb, respectively (Fig. 1) (47). DI-C RNA was purified from the gel (Fig. 1), and a cDNA complementary to DI-C RNA was assembled. Four overlapping fragments comprising full-length DI-C RNA were cloned (Fig. 2A), and their sequence was corrected to generate cDNA copies with the consensus sequence of the PUR46-MAD strain of TGEV (53). These fragments were assembled into a cDNA encoding DI-C RNA and were cloned downstream of T7 promoter with three extra bases (GGG) of nonviral origin between the T7 promoter and the 5′ end of DI-C cDNA (Fig. 2B and C). Then, a C was introduced in the first position of DI-C cDNA instead of the A present in the wt genome (18, 47). The 3′ end of the DI-C cDNA clone was flanked by a poly(A25) tail followed by a cDNA encoding the HDV Rz to generate the correct 3′ end, and by the T7 transcription terminator (Fig. 2D).
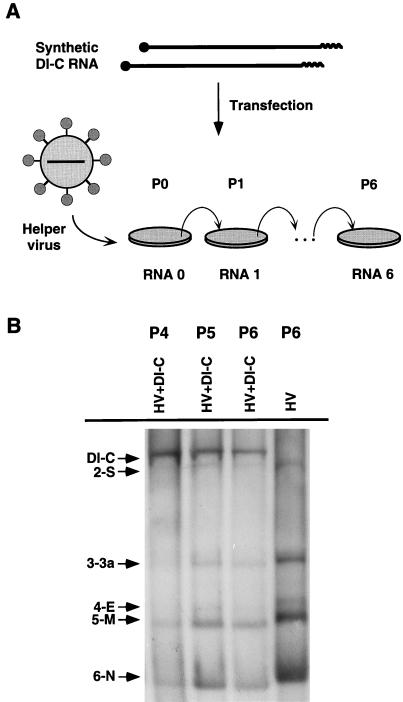
To rescue synthetic DI-C RNA, TGEV PUR46-MAD-infected ST cells were electroporated with in vitro-transcribed DI-C RNA (Fig. 3A). Supernatants of these cultures were passed onto fresh ST cell monolayers, and cytoplasmic RNA was extracted. A synthetic DI-C RNA with the expected size (9.7 kb), not present in the helper virus-infected, untransfected cultures, was detected at passage P2 by Northern analysis (data not shown). This band was clearly seen at passage P4 by using a probe complementary to the virus leader or to the 3′ end. This RNA remained stable for at least 20 passages (Fig. 3B and data not shown). These data indicated that a synthetic, replicating TGEV-derived RNA was engineered.
FIG. 3.
Rescue of synthetic DI-C RNA. With the plasmid described in Fig. 2, T7 transcripts with the information for DI-C RNA were obtained in vitro (A). The RNAs were transfected into ST cells that were previously infected with the helper virus (TGEV PUR46-MAD strain). Supernatants of the infected cultures were passaged six times (from P0 to P6), the RNAs of passages P4, P5, and P6 were extracted and analyzed by Northern hybridization with a probe complementary to the leader sequence (B). In lanes P4, P5, and P6, a band with the expected size of DI-C RNA was identified. The RNA from passage P6 of ST cells infected with the helper virus (HV) but untransfected was also analyzed. The numbers and letters on the left of the gel indicate number of the RNA and the protein that is encoded by this mRNA. S, spike; 3a, non-structural protein 3a; E, envelope protein; M, membrane protein; N, nucleoprotein.
DI-C RNA accumulated to higher levels than the viral RNAs. In fact, an interference with the expression of the helper virus RNAs was clearly observed in comparisons of the relative production of the viral RNAs in the absence or the presence of DI-C (Fig. 3B) (47). The reduction in the amount of helper virus RNAs persisted for at least 20 passages (data not shown). The titer of the helper virus was reduced 20-fold in the presence of DI-C RNA. The inhibition was approximately the same for all helper virus mRNA as shown in Fig. 3 and as previously described (47). These data indicated that the molecularly cloned DI-C RNA interfered with helper virus replication and most likely was the result of competition over repeated passages of the helper virus plus the minigenome.
The HDV Rz cloned at the 3′ end of DI-C RNA was active in vitro as determined by analyzing the processing of minigenome M50 RNA, a deletion mutant of DI-C RNA (data not shown). The RNA corresponding to the M50 minigenome was in vitro transcribed by using T7 polymerase. The RNAs were studied by Northern blotting with a probe complementary to both the unprocessed and the processed RNA transcripts. Two bands with the size expected for both RNAs (5.3 and 5.0 kb) were detected in the same proportion, indicating that around 50% of the RNA molecules were self-cleaved to generate a minigenome RNA with the desired 3′ end.
Delimitation of the sequences required for the replication and packaging of DI-C.
A collection of 14 TGEV minigenomes was generated, and their ability to be replicated and packaged (i.e., rescued) was tested with PUR46-MAD as helper virus (Fig. 4 and minigenome M22 shown in Fig. 5). Minigenomes are named by a number that indicates their size in hundreds of nucleotides. All minigenomes but the two smallest ones of 2.1 kb (M21) and 2.2 kb (M22) were efficiently rescued (Fig. 4 and 5).
FIG. 4.
Rescue efficiency of DI-C deletion mutants. A collection of 13 deletion mutants was constructed with the restriction enzymes indicated in Table 1. The location of the deletions, referred to the parental DI-C structure, is indicated by empty boxes (A). The letters above the boxes indicate the genes from which DI-C was derived. The numbers below these boxes indicate the nucleotides of each gene that were included in DI-C RNA. The rescue efficiency of each mutant was estimated by Northern hybridizations at passage P8, as the molar ratio to mRNA E and represents the average of three different experiments. To estimate the relative amount of M39 RNA, which overlaps with mRNA 3a, the amount of M39 RNA was calculated by subtracting from the density of this band that of gene 3a from lane M42. The amount subtracted represented less than 10% of the total. A probe complementary to nucleotides 66 to 91 of the TGEV genome was used. Grey boxes, sequence of the minigenomes. Empty boxes, deleted fragments. Minigenomes detected in or over a 10-fold molar excess were assigned (+++), 5-fold molar excess (++) and less than 1 molar ratio (+). Rescue efficiency of M21 was assigned as (+/−) because, although it was not detected by this technique, it was detected by RT-PCR at passage 3. IG, intergenic sequence. (B) Northern blot of the minigenome RNAs rescued by the strain PUR46-MAD of TGEV and performed at passage 8. A set of the minigenomes rescued in the same experiment are presented as an example. The positions of the bands corresponding to the minigenomes are indicated by arrowheads. Lanes 1 to 6 correspond to minigenomes DI-C, M54, M42, M39, M33, and M21; lane 7, the TGEV PUR46-MAD strain without a minigenome; lane 8, uninfected ST cells. The position of the helper virus mRNAs is indicated to the left by their numbers and by letters indicating the protein encoded by these RNAs.
The smallest minigenome rescued with high efficiency was M33, with 3.3 kb, comprising (i) the 5′-end 2,144 nt and the 3′ end 174 nt of ORF1a; (ii) the 5′-end 394 nt and the 3′-end 11 nt of ORF1b; (iii) the 5′-end 8 nt of S gene; and (iv) the 492 3′-most-end nucleotides of the virus, including part of gene 7 and the complete 3′ UTR. This indicates that minigenome replication requires only the 3′ end 492 nt or less from the 3′ end of the infectious genome. In addition, only the first 1,348 nt of ORF1a are essential for minigenome replication, since these are the only ORF1a nt present within minigenome M21, which replicates efficiently (see below).
Some of the minigenomes were rescued with higher efficiency than the original DI-C RNA (Fig. 4). All minigenomes but two (M21 and M22) were detected by Northern hybridization and Northern blot analysis for more than 15 passages. Minigenome M21 was detected by RT-PCR analysis at passage P1 but only sometimes at P3 (data not shown). The best evidence for minigenome M21 replication was obtained by using GUS as a reporter gene (see below).
DI-C RNA contains two consensus IG sequences followed by the corresponding open reading frames (ORFs), located either at the 5′ or the 3′ end of DI-C RNA (Fig. 5A). A sequence fragment of 303 nt containing the IG sequence located at the 3′ end, starting at nt 813 from the 3′ end of minigenome DI-C, preceding the S gene in the parental virus, was removed from three minigenomes (DI-C of 9.7 kb, M67, and M42) to generate minigenomes M94, M64, and M39, respectively. T7 RNA transcripts of the three pairs of minigenomes were transfected into ST cells and rescued by using TGEV helper virus, and the extent of their replication after eight passages in cell culture was determined by Northern hybridization analysis. In all cases the rescue of the minigenomes with one IG sequence was increased between three- and sixfold in relationship to the minigenomes with two IG sequences (results not shown).
M21 RNA was rescued with a very low efficiency compared with the other minigenomes (Fig. 4). In order to determine whether the lower rescue was due either to a lower replication or packaging efficiency, it was convenient to evaluate the extent of replication of these minigenomes at passage zero (P0) either by metabolically labeling (32P) the viral RNAs or by Northern blot analysis. While the helper virus genome and the mRNAs were synthesized at levels that permitted their detection by both procedures at P0, the sensitivity of the assays did not reveal the extent of minigenome replication (data not shown). To overcome this limitation, the M21, M33, and M39 cDNAs were cloned behind the CMV promoter (52), instead of the T7 promoter, in order to amplify the minigenome RNA in two steps, one in the nucleus with the CMV promoter and the cellular RNA polymerase II and another in the cytoplasm with the viral polymerase (Fig. 5B). In addition, the transformation efficiency with a DNA plasmid, in contrast to an in vitro-transcribed RNA, was increased 40- to 50-fold. To further increase the sensitivity of the assay, the GUS gene was inserted in the minigenomes as a reporter gene. The GUS gene, under the control of N gene consensus IG sequence (CUAAAC) preceded by the 88 nt upstream of the IG sequence, was cloned into M21, M33, and M39 minigenomes, to generate plasmids CMV-M21-GUS, CMV-M33-GUS, and CMV-M39-GUS (Fig. 5B). Minigenome cDNAs were flanked at the 3′ end by HDV Rz and the transcription termination and polyadenylation signals from the BGH. The two-step amplification system allowed minigenome detection at passage P0.
M21 minigenome lacks 14 nt at the ORF 1a (nt 1350 to 1363) in relationship to minigenome M62 and to all the other minigenomes that have been rescued. To study whether this sequence was responsible for the difference in packaging efficiency between minigenomes M21 and M62, the minigenome CMV-M22-GUS was constructed. This minigenome was derived from the M33 and has the most 5′ end 1659 nt of ORF 1a sequence (i.e., it includes the 14 nt missing in the M21 in relationship to the M62 minigenome). The replication level of minigenomes M21, M22, M33, and M62 at P0 was estimated by studying the expression of GUS with the constructs CMV-M21-GUS, CMV-M22-GUS, CMV-M33-GUS, and CMV-M62-GUS. All these minigenomes expressed similar levels of GUS at P0 (Fig. 5B and C), indicating that they were replicated to similar levels.
Two control plasmids with the sequence encoding GUS but in the reverse sense (CMV-M21-SUG and CMV-M33-SUG) were also generated. As expected, no GUS activity was expressed with these minigenomes. In addition, two other minigenomes with a deletion of 171 nt starting at nt 192 from the 3′ end of both minigenomes were constructed. This deletion should abrogate minigenome replication dependent on the viral polymerase (27), but not the nuclear amplification mediated by the cellular RNA polymerase II. The deletion drastically reduced the GUS expression by M33 and M21 RNAs (Fig. 5B). The presence of the helper virus was essential for the replication and transcription of the minigenomes (Fig. 5B and C).
Interestingly, the rescue of minigenomes CMV-M21-GUS and CMV-M22-GUS was reduced by about 100- to 1,000-fold in the third passage, suggesting that both minigenomes were efficiently replicated but not packaged (Fig. 5C). These results indicate that the deletion of 14 nt in the M21 minigenome in relationship to the M62 is not responsible for the lack of efficient packaging, since the presence of the 14 nt in minigenome M22 did not improve the packaging efficiency of this minigenome.
The minigenomes M33 and M62 that were efficiently replicated and packaged differed from M22 in either fragment 1 (F1) of 1.0 kb or fragment 2 (F2) of 4.1 kb which do not overlap or have any obvious sequence homology, indicating that at least one of these two ORF1 fragments was required for efficient packaging, although neither of them was essential.
Expression of heterologous genes by using a helper virus-dependent expression system based on TGEV-derived minigenomes.
In order to develop an expression system, the GUS gene was cloned into the M39 minigenome. This defective genome was selected because it showed one of the highest rescue levels among the 13 minigenomes as determined by Northern hybridization analysis (Fig. 4).
To facilitate the cloning of foreign genes, a small polylinker containing the restriction endonuclease sites NruI, MluI, Acc65I, and KpnI with the sequence TCGCGACGCGTGGTACC was introduced at position 510 from the 3′ end of the M39 minigenome to generate M39-L defective RNA. The rescue of this RNA by the PUR46-MAD strain of TGEV virus was determined by Northern hybridization analysis. The introduction of the polylinker did not affect the replication level of the minigenome (data not shown).
The expression of GUS activity downstream of the N gene intergenic sequences was evaluated by using the CMV-M39-GUS vector. This IG sequence included the consensus CUAAAC sequence and the 88 nt flanking the 5′ end of the consensus sequence. High levels of GUS activity were detected following three passages posttransfection (Fig. 5C). The GUS activity detected corresponds to protein expression levels around 1.0 μg/106 cells. These expression levels have been confirmed by studying the GUS protein expressed by using a Western blot assay (data not shown).
DISCUSSION
In this study the rescue of synthetic minigenomes derived from defective TGEV RNAs is described. The sequences of the RNA minigenome required for replication and packaging were reduced to 3.3 kb. For efficient minigenome replication, but with deficient packaging, these sequences were reduced to 2.1 kb. The synthetic minigenomes have been used to develop an efficient helper-dependent expression system. In addition, it has been shown that a molecularly cloned TGEV-derived defective minigenome interferes with TGEV replication.
The synthetic DI-C derived RNAs were constructed by adding three nucleotides (GGG) not present in the sequence of the wild-type TGEV to its 5′ end (47). The presence of three G’s downstream of T7 promoter facilitates the transcription of the sequences thereafter (1, 2). The first nucleotide introduced in the synthetic RNAs was either an A after the CMV promoter, as in the Purdue virus strain of TGEV (18), or a C, which has been previously described in another TGEV isolate (50) and was introduced into the minigenomes expressed downstream of the T7 promoter. Nevertheless, both types of synthetic RNAs replicated very efficiently, in fact, most of them to a higher level than the DI-C minigenome. This is not surprising, since minor modifications of the RNA 5′ end in the construction of viral cDNAs have frequently resulted in infectious clones (6).
The 3′ end of DI-C RNA was flanked by the HDV Rz to remove any sequence after the poly(A), particularly in the minigenome expressed after CMV promoter. We have shown that around 50% of the molecules were self-cleaved in vitro by the ribozyme. It is not known whether this cleavage efficiency is maintained in cells.
The 3′ end sequences required for the replication of TGEV-derived RNA minigenomes have been reduced to 492 nt, although it is not excluded that it could be further slightly reduced by deletion mutagenesis. The size of the 3′ end sequence required for TGEV RNA replication is similar to the 3′ end replication signal of MHV defective RNAs which comprises the 3′ terminal 447 nt. In MHV the most 3′ end 55 nt of the positive-strand RNA plus the poly(A) tail comprises the cis-acting signal for the initiation of minus-strand synthesis (40). These data suggest that part of the 3′ end terminal 447 nt, that was identified as the 3′ replication signal are required for positive strand synthesis. In addition, in MHV the sequences within nt 270 to 305 from the 3′-end abrogate gene expression, suggesting a rigid sequence requirement for transcription in this region (41).
The 5′ end sequences required for minigenome RNA replication by TGEV included the first 1,348 nt. Other gene 1 sequences present within minigenome M62 were considered as non-essential, since they were not present in the M21 nor M33 minigenomes which also replicated efficiently. Probably, this 5′ end sequence could be further reduced by deletion mutagenesis. The 5′ sequences in BCV have been reduced to 498 nt (25, 45) and in MHV to 467 nt (32, 45, 66). In addition, the requirement for an internal sequence that could act as an enhancer has also been described for the MHV-JHM strain (33).
The essential sequences required to rescue defective IBV RNAs have been determined by deletion mutagenesis (14, 54, 55). By this technique on a cloned IBV defective RNA (CD-91), the smallest defective RNA rescued to date has been 3.9 kb. Within this RNA, a 1.4-kb region, derived from the 3′ end of ORF1b, may contain an IBV packaging signal or a cis-acting sequence essential for replication.
A collection of 13 DI-C RNA deletion mutants was constructed, and their relative replication efficiencies in relation to the synthesis of a viral mRNA of similar size were determined at passage 8, when the minigenomes have reached maximum amplification. The replication of the defective RNAs diverged by a factor of 10, and no correlation between rescue efficiency and minigenome ORF length was observed. Minigenomes containing more information were not necessarily rescued with higher efficiency, suggesting that the secondary structure of minigenome RNAs may have a significant influence on their rescue.
The removal of a sequence fragment of 303 nt containing the IG sequences located at the 3′ end, starting at nt 813 from the 3′ end of minigenome DI-C, increased the rescue efficiency of three minigenomes of different length between 3- to 6-fold in relationship to the minigenomes with two IG sequences. One possible explanation is that the increase in the rescue was due to the removal of the IG sequences, although the deletion of an unknown motif present within the sequences flanking the IG site is also possible. The increase of DI-C genome synthesis could be the result of a decrease in the polymerase pausing when it encounters a functional downstream IG sequence during the negative strand synthesis. This interpretation would support that the IG sequence acts as a terminator rather than as a transcription initiator (35, 37, 61). It is known for MHV that removal of 3′ terminal IG sequences increases synthesis of the next larger mRNAs and genome RNA synthesis (31, 67).
In order to increase the RNA levels, the minigenomes were cloned after the CMV promoter, leading to a two-step amplification system that uses the cellular RNA polymerase II and the viral polymerase (52). Although the transport of RNAs of cytoplasmic viruses from the nucleus to the cytoplasm involves the risk of RNA modification by splicing, no apparent alteration of the TGEV defective RNAs was observed, as determined by evaluating the minigenome size in gels. In addition, the system with two amplification steps was successfully used to express GUS activity. Similar results were previously obtained in expression systems based on other cytoplasmic viruses as Sindbis and Semliki forest viruses (4, 16). These changes made possible minigenome detection at passage P0, when it is of practical interest to study coronavirus replication.
Minigenomes M21, M22, M33, and M39 were replicated to a similar extent, as determined by the expression of GUS at passage P0. In contrast, M21 and M22 minigenomes differed markedly from minigenomes M33 and M39 in their efficiency to be passaged in cell cultures, suggesting that the packaging efficiency of these minigenomes was much higher than that of M21 and M22, although it is not possible to rule out whether the lower packaging observed is due to a lower stability of the minigenomes M21 and M22. This possibility is being investigated.
Efficient packaging of minigenome M33 required a fragment of about 1.0 kb from the 5′ end of TGEV RNA (fragment F1, Fig. 5A), not present in the M22 minigenome. Alternatively, minigenome M62 required a fragment (F2) of about 4.1 kb located at the 3′ end, present in minigenome M62 but not in M21 or M22 (Fig. 5A). These results suggest that either both F1 and F2 fragments contain a packaging signal or that their presence is required for the proper folding of a packaging signal present within the 2.1- and 2.2-kb RNAs. Currently available data in coronaviruses and other viral systems are compatible with both the presence of one or more packaging signals, and with the need of proper folding of the packaging signals to maintain their function (5, 23, 48, 68).
In MHV a defined packaging signal of 61 nt has been described (21). This sequence has been sufficient to pack a nonviral RNA encoding the chloramphenicol acetyl transferase (CAT) gene into virions (68). Nevertheless, the efficiency of this packaging signal was not determined, and it is not known whether a combination of multiple genomic regions is needed. Similarly, it was shown that a subgenomic MHV RNA containing the packaging signal was encapsidated, but the efficiency was lower than the encapsidation efficiency of the corresponding DI RNA (5), suggesting that additional sequences were probably required for optimum packaging. In fact, several nucleoprotein binding sites have been identified in coronavirus RNAs. These binding sites map within the leader sequence (49), the 3′ terminus of IBV (69) and MHV (26), and the packaging signal described in MHV (48). In human immunodeficiency virus type 1 (HIV-1), a defined sequence of 120 nt of the viral RNA containing four stem-loop structures is important for efficient encapsidation (13). One of these loops is sufficient to direct the recognition and packaging of heterologous RNAs into virus-like particles (24). Nevertheless, the deletion of this loop from the native genome does not fully abrogate packaging. Deletion of different combinations of the stem loops reduces packaging up to 5% of that found in the wild-type virus. Thus, it is likely that in vivo packaging involves more than one interaction (13).
In general, packaging signals have frequently been associated with secondary structures and not to a defined primary nucleotide sequence (15). This suggests that sequences flanking the packaging signals are probably important to provide sufficient flexibility to allow the packaging signal to form its specific structure (48). The introduction of deletions or the insertion of sequences in a minigenome may affect its packaging efficiency by interfering with the proper folding of the packaging signal.
The inefficient packaging of minigenomes M21 and M22 was not due to a size restriction, since the same low packaging capacity of these RNAs was observed after inserting the GUS gene into M21 RNA, leading to an RNA of 4.1 kb, larger than that of minigenomes M33 and M39 and similar to M42 RNA, which were efficiently encapsidated. In MHV, BCV, and IBV, minigenomes with similar small sizes are also efficiently packed (7, 14, 25, 32, 42, 43, 45, 64). Packaging of subgenomic RNAs of about 0.8 and 1.5 kb into MHV virions has been reported, although with low efficiency (5, 68).
The synthetic minigenomes constructed, in collaboration with a helper virus, were useful to express significant amounts (around 1.0 μg/106 cells) of a heterologous protein (GUS). The amount of GUS protein expressed was confirmed by Western Blot (unpublished results). In addition, improvement of this expression system using specific transcription regulatory sequences has led to a 5- to 10-fold increase in the expression levels. The transcription of a minigenome derived mRNA with the information for GUS was studied by Northern blot analysis using two probes, one complementary to the 3′ end that hybridizes with the helper genome, the helper derived mRNAs, and the minigenome, and a second probe specific for GUS gene, only complementary to the minigenome and the mRNA derived from it. The GUS mRNA was detected only in passages of maximum GUS expression and using optimized transcription regulatory sequences (unpublished data). PCR analysis confirmed that the band detected by Northern blot analysis corresponded to GUS mRNA.
Coronavirus-derived minigenomes have been previously used to express heterologous genes. MHV defective RNA was used to express hemagglutinin-esterase (HE) protein, although the vector was only stable for 3 passages, in contrast to the TGEV derived minigenomes that expressed the heterologous gene for up to 10 passages (data not shown). IBV-derived defective RNAs have also been used as expression vectors (20), although the expression levels have not been reported. Coronaviruses might be the base for the development of safe expression vectors with large cloning capacity (>20 kb). This is particularly attractive with TGEV-derived expression systems due to the high TGEV titers (>1010 PFU/ml) in tissue culture. The availability of TGEV-derived synthetic RNAs will be very useful to modify the genome of TGEV by site-directed recombination (34, 65) in order to correlate genome structure with biological function and to develop tissue-specific expression systems to interfere with virus replication at mucosal surfaces.
ACKNOWLEDGMENTS
We thank I. Sola and V. Buckwold for critically reading the manuscript.
This work has been supported by grants from the Comisión Interministerial de Ciencia y Tecnología (CICYT), La Consejería de Educación y Cultura de la Comunidad de Madrid, and Fort Dodge Veterinaria from Spain, and the European Communities (Biotechnology and FAIR projects). A.I. and S.A. received fellowships from the Department of Education, University and Research of the Gobierno Vasco.
REFERENCES
- 1.Ball L A. Cellular expression of a functional nodavirus RNA replicon from vaccinia virus vectors. J Virol. 1992;66:2335–2345. doi: 10.1128/jvi.66.4.2335-2345.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ball L A. Requirements for the self-directed replication of flock house virus RNA 1. J Virol. 1995;69:720–727. doi: 10.1128/jvi.69.2.720-727.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballesteros M L, Sanchez C M, Enjuanes L. Two amino acid changes at the N-terminus of transmissible gastroenteritis coronavirus spike protein result in the loss of enteric tropism. Virology. 1997;227:378–388. doi: 10.1006/viro.1996.8344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berglund P, Smerdou C, Fleeton M N, Tubulekas I, Liljestrom P. Enhancing immune responses using suicidal DNA vaccines. Nat Biotechnol. 1998;16:562–565. doi: 10.1038/nbt0698-562. [DOI] [PubMed] [Google Scholar]
- 5.Bos E C W, Dobbe J C, Luytjes W, Spaan W J M. A subgenomic mRNA transcript of the coronavirus mouse hepatitis virus strain A59 defective interfering (DI) RNA is packaged when it contains the DI packaging signal. J Virol. 1997;71:5684–5687. doi: 10.1128/jvi.71.7.5684-5687.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyer J C, Haenni A L. Infectious transcripts and cDNA clones of RNA viruses. Virology. 1994;198:415–426. doi: 10.1006/viro.1994.1053. [DOI] [PubMed] [Google Scholar]
- 7.Brian D A, Chang R Y, Hofmann M A, Sethna P B. Role of subgenomic minus strand RNA in coronavirus replication. Arch Virol. 1994;9:173–180. doi: 10.1007/978-3-7091-9326-6_17. [DOI] [PubMed] [Google Scholar]
- 8.Brian D A, Spaan W J M. Recombination and coronavirus defective interfering RNAs. Semin Virol. 1997;8:101–111. doi: 10.1006/smvy.1997.0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bronstein I, Fortin J, Stanley P E, Stewart G S A B, Kricka L J. Chemiluminescent and bioluminescent reporter gene assays. Anal Biochem. 1994;219:169–181. doi: 10.1006/abio.1994.1254. [DOI] [PubMed] [Google Scholar]
- 10.Bronstein I, Fortin J J, Voyta J C, Juo R-R, Edwards B, Olenses C E M, Lijam N, Kricka L J. Chemiluminescent reporter gene assays: sensitive detection of the GUS and SEAP gene products. BioTechniques. 1994;17:172–177. [PubMed] [Google Scholar]
- 11.Bronstein I, Martin C S, Fortin J J, Olesen C E M, Voyta J C. Chemiluminescence: sensitive detection technology for reporter gene assays. Clin Chem. 1996;42:1542–1546. [PubMed] [Google Scholar]
- 12.Chang R Y, Hofmann M A, Sethna P B, Brian D A. A cis-acting function for the coronavirus leader in defective interfering RNA replication. J Virol. 1994;68:8223–8231. doi: 10.1128/jvi.68.12.8223-8231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clever J L, Parslow T G. Mutant human immunodeficiency virus type I genomes with defects in RNA dimerization or encapsidation. J Virol. 1997;71:3407–3414. doi: 10.1128/jvi.71.5.3407-3414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dalton K, Penzes Z, Wroe C, Stirrups K, Evans S, Shaw K, Brown T D K, Britton P, Cavanagh D. Sequence elements involved in the rescue of IBV defective RNA CD-91. Adv Exp Med Biol. 1998;440:253–258. doi: 10.1007/978-1-4615-5331-1_32. [DOI] [PubMed] [Google Scholar]
- 15.De Guzman R N, Wu Z-R, Stalling C C, Pappalardo L, Borer P N, Summers M F. Structure of the HIV-1 nucleocapsid protein bound to the SL3 Ψ-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 16.Dubensky J, W. T, Driver D A, Polo J M, Belli B A, Latham E M, Ibanez C E, Chada S, Brumm D, Banks T A, Mento S J, Jolly D J, Chang S M W. Sindbis virus DNA-based expression vectors: utility for in vitro and in vivo gene transfer. J Virol. 1996;70:508–519. doi: 10.1128/jvi.70.1.508-519.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunn J J, Studier F W. Complete nucleotide sequence of bacteriophage T7 DNA and locations of T7 genetic elements. J Mol Biol. 1983;166:477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- 18.Eleouet J F, Rasschaert D, Lambert P, Levy L, Vende P, Laude H. Complete sequence (20 kilobases) of the polyprotein-encoding gene 1 of transmissible gastroenteritis virus. Virology. 1995;206:817–822. doi: 10.1006/viro.1995.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Enjuanes L, Van der Zeijst B A M. Molecular basis of transmissible gastroenteritis coronavirus epidemiology. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 337–376. [Google Scholar]
- 20.Evans S, Stirrups K, Dalton K, Shaw K, Cavanagh D, Britton P. Utilising a defective IBV RNA for heterologous gene expression with potential prophylactic application. Adv Exp Med Biol. 1998;440:687–692. doi: 10.1007/978-1-4615-5331-1_88. [DOI] [PubMed] [Google Scholar]
- 21.Fosmire J A, Hwang K, Makino S. Identification and characterization of a coronavirus packaging signal. J Virol. 1992;66:3522–3530. doi: 10.1128/jvi.66.6.3522-3530.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frolov I, Hoffman T A, Prágai B M, Dryga S A, Huang H V, Schlesinger S, Rice C M. Alphavirus-based expression vectors: Strategies and applications. Proc Natl Acad Sci USA. 1996;93:11371–11377. doi: 10.1073/pnas.93.21.11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frolova E, Frolov I, Schlesinger S. Packaging signals in alphaviruses. J Virol. 1997;71:248–258. doi: 10.1128/jvi.71.1.248-258.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayashi T, Shioda T, Iwakura Y, Shibuta H. RNA packaging signal of human immunodeficiency virus type 1. Virology. 1992;188:590–599. doi: 10.1016/0042-6822(92)90513-o. [DOI] [PubMed] [Google Scholar]
- 25.Hofmann M A, Sethna P B, Brian D A. Bovine coronavirus mRNA replication continues throughout persistent infection in cell culture. J Virol. 1990;64:4108–4114. doi: 10.1128/jvi.64.9.4108-4114.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hogue B G, Cologna R. Fifth International Symposium on Positive Strand RNA Viruses, St. Petersburg, Fla. 1998. Nucleocapsid protein binding sites within coronavirus defective genomes. [Google Scholar]
- 27.Hsue B, Masters P S. A bulged stem-loop structure in the 3′ untranslated region of the genome of the coronavirus mouse hepatitis virus is essential for replication. J Virol. 1997;71:7567–7578. doi: 10.1128/jvi.71.10.7567-7578.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsue B, Masters P S. An essential secondary structure in the 3′ untranslated region of the mouse hepatitis virus genome. Adv Exp Med Biol. 1998;440:297–302. doi: 10.1007/978-1-4615-5331-1_39. [DOI] [PubMed] [Google Scholar]
- 29.Jefferson R A, Burgess S M, Hirsh D. β-glucuronidase from Escherichia coli as a gene-fusion marker. Proc Natl Acad Sci USA. 1986;83:8447–8451. doi: 10.1073/pnas.83.22.8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiménez G, Correa I, Melgosa M P, Bullido M J, Enjuanes L. Critical epitopes in transmissible gastroenteritis virus neutralization. J Virol. 1986;60:131–139. doi: 10.1128/jvi.60.1.131-139.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joo M, Makino S. The effect of two closely inserted transcription consensus sequences on coronavirus transcription. J Virol. 1995;69:272–280. doi: 10.1128/jvi.69.1.272-280.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim Y, Jeong Y, Makino S. Analysis of cis-acting sequences essential for coronavirus defective interfering RNA replication. Virology. 1993;197:53–63. doi: 10.1006/viro.1993.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim Y-N, Makino S. Characterization of a murine coronavirus defective interfering RNA internal cis-acting replication signal. J Virol. 1995;69:4963–4971. doi: 10.1128/jvi.69.8.4963-4971.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koetzner C A, Parker M M, Ricard C S, Sturman L S, Masters P S. Repair and mutagenesis of the genome of a deletion mutant of the coronavirus mouse hepatitis virus by targeted RNA recombination. J Virol. 1992;66:1841–1848. doi: 10.1128/jvi.66.4.1841-1848.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Konings D A M, Bredenbeek P J, Noten J F H, Hogeweg P, Spaan W J M. Differential premature termination of transcription as a proposed mechanism for the regulation of coronavirus gene expression. Nucleic Acids Res. 1988;16:10849–10860. doi: 10.1093/nar/16.22.10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lai M M C, Cavanagh D. The molecular biology of coronaviruses. Adv Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liao C L, Zhang X, Lai M M C. Coronavirus defective-interfering RNA as an expression vector: the generation of a pseudorecombinant mouse hepatitis virus expressing hemagglutinin-esterase. Virology. 1995;208:319–327. doi: 10.1006/viro.1995.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liljeström P. Alphavirus expression systems. Curr Opin Biotechnol. 1994;5:495–500. doi: 10.1016/0958-1669(94)90063-9. [DOI] [PubMed] [Google Scholar]
- 40.Lin Y-J, Liao C L, Lai M M C. Identification of the cis-acting signal for minus-strand RNA synthesis of a murine coronavirus: implications for the role of minus-strand RNA in RNA replication and transcription. J Virol. 1994;68:8131–8140. doi: 10.1128/jvi.68.12.8131-8140.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin Y-J, Zhang X, Wu R-C, Lai M M C. The 3′ untranslated region of coronavirus RNA is required for subgenomic mRNA transcription from a defective interfering RNA. J Virol. 1996;70:7236–7240. doi: 10.1128/jvi.70.10.7236-7240.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin Y J, Lai M M C. Deletion mapping of a mouse hepatitis virus defective interfering RNA reveals the requirement of an internal and discontiguous sequence for replication. J Virol. 1993;67:6110–6118. doi: 10.1128/jvi.67.10.6110-6118.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Makino S, Yokomori K, Lai M M C. Analysis of efficiently packaged defective interfering RNAs of murine coronavirus: localization of a possible RNA-packaging signal. J Virol. 1990;64:6045–6053. doi: 10.1128/jvi.64.12.6045-6053.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin C T, Coleman J E. Kinetic analysis of T7 RNA polymerase-promoter interactions with small synthetic promoters. Biochemistry. 1987;26:2690–2696. doi: 10.1021/bi00384a006. [DOI] [PubMed] [Google Scholar]
- 45.Masters P S, Koetzner C A, Kerr C A, Heo Y. Optimization of targeted RNA recombination and mapping of a novel nucleocapsid gene mutation in the coronavirus mouse hepatitis virus. J Virol. 1994;68:328–337. doi: 10.1128/jvi.68.1.328-337.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McClurkin A W, Norman J O. Studies on transmissible gastroenteritis of swine. II. Selected characteristics of a cytopathogenic virus common to five isolates from transmissible gastroenteritis. Can J Comp Vet Sci. 1966;30:190–198. [PMC free article] [PubMed] [Google Scholar]
- 47.Mendez A, Smerdou C, Izeta A, Gebauer F, Enjuanes L. Molecular characterization of transmissible gastroenteritis coronavirus defective interfering genomes: packaging and heterogeneity. Virology. 1996;217:495–507. doi: 10.1006/viro.1996.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Molenkamp R, Spaan W J M. Identification of a specific interaction between the coronavirus mouse hepatitis virus A59 nucleocapsid protein and packaging signal. Virology. 1997;239:78–86. doi: 10.1006/viro.1997.8867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nelson G W, Stohlman S A. Localization of the RNA-binding domain of mouse hepatitis virus nucleocapsid protein. J Gen Virol. 1993;74:1975–1979. doi: 10.1099/0022-1317-74-9-1975. [DOI] [PubMed] [Google Scholar]
- 50.Page K W, Britton P, Boursnell M E G. Sequence analysis of the leader RNA of two porcine coronaviruses: transmissible gastroenteritis coronavirus and porcine respiratory coronavirus. Virus Genes. 1990;4:289–301. doi: 10.1007/BF00570024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pattnaik A K, Ball L A, LeGrone A W, Wertz G W. Infectious defective interfering particles of VSV from transcripts of a cDNA clone. Cell. 1992;69:1011–1020. doi: 10.1016/0092-8674(92)90619-n. [DOI] [PubMed] [Google Scholar]
- 52.Penzes Z, Gonzalez J M, Izeta A, Muntion M, Enjuanes L. Progress towards the construction of a transmissible gastroenteritis coronavirus self-replicating RNA using a two-layer expression system. Adv Exp Med Biol. 1998;440:319–327. doi: 10.1007/978-1-4615-5331-1_42. [DOI] [PubMed] [Google Scholar]
- 53.Penzes, Z., A. Izeta, C. Smerdou, A. Mendez, M. L. Ballesteros, and L. Enjuanes. Complete nucleotide sequence of transmissible gastroenteritis coronavirus strain PUR46-MAD. Submitted for publication.
- 54.Penzes Z, Tibbles K, Shaw K, Britton P, Brown T D K, Cavanagh D. Characterization of a replicating and packaged defective RNA of avian coronavirus infectious bronchitis virus. Virology. 1994;203:286–293. doi: 10.1006/viro.1994.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Penzes Z, Wroe C, Brown T D K, Britton P, Cavanagh D. Replication and packaging of coronavirus infectious bronchitis virus defective RNAs lacking a long open reading frame. J Virol. 1996;70:8660–8668. doi: 10.1128/jvi.70.12.8660-8668.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pushko P, Parker M, Ludwing G V, Davis N L, Johnston R E, Smith J F. Replication-helper systems from attenuated Venezuelan equine encephalitis virus: expression of heterologous genes in vitro and immunization against heterologous pathogens in vivo. Virology. 1997;239:389–401. doi: 10.1006/viro.1997.8878. [DOI] [PubMed] [Google Scholar]
- 57.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 58.Sánchez, C. M., and L. Enjuanes. 1998. Unpublished results.
- 59.Sánchez C M, Gebauer F, Suñé C, Méndez A, Dopazo J, Enjuanes L. Genetic evolution and tropism of transmissible gastroenteritis coronaviruses. Virology. 1992;190:92–105. doi: 10.1016/0042-6822(92)91195-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sánchez C M, Jiménez G, Laviada M D, Correa I, Suñé C, Bullido M J, Gebauer F, Smerdou C, Callebaut P, Escribano J M, Enjuanes L. Antigenic homology among coronaviruses related to transmissible gastroenteritis virus. Virology. 1990;174:410–417. doi: 10.1016/0042-6822(90)90094-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sawicki S G, Sawicki D L. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J Virol. 1990;64:1050–1056. doi: 10.1128/jvi.64.3.1050-1056.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schlaman H R M, Risseeuw E, Franke-van Dijk M E I, Hooykaas P J J. Nucleotide sequence corrections of the uidA open reading frame encoding β-glucuronidase. Gene. 1994;138:259–260. doi: 10.1016/0378-1119(94)90820-6. [DOI] [PubMed] [Google Scholar]
- 63.Sharmeen L, Kuo M Y P, Dinter-Gottlieb G, Taylor J. Antigenomic RNA of human hepatitis delta virus can undergo self-cleavage. J Virol. 1988;62:2674–2679. doi: 10.1128/jvi.62.8.2674-2679.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Van der Most R G, Bredenbeek P J, Spaan W J M. A domain at the 3′ end of the polymerase gene is essential for encapsidation of coronavirus defective interfering RNAs. J Virol. 1991;65:3219–3226. doi: 10.1128/jvi.65.6.3219-3226.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Van der Most R G, Heijnen L, Spaan W J M, Degroot R J. Homologous RNA recombination allows efficient introduction of site-specific mutations into the genome of coronavirus MHV-A59 via synthetic co-replicating RNAs. Nucleic Acids Res. 1992;20:3375–3381. doi: 10.1093/nar/20.13.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Van der Most R G, Spaan W J M. Coronavirus replication, transcription, and RNA recombination. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 11–31. [Google Scholar]
- 67.Van Marle G, Luytjes W, Van der Most R G, van der Straaten T, Spaan W J M. Regulation of Coronavirus mRNA transcription. J Virol. 1995;69:7851–7856. doi: 10.1128/jvi.69.12.7851-7856.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Woo K, Joo M, Narayanan K, Kim K H, Makino S. Murine coronavirus packaging signal confers packaging to nonviral RNA. J Virol. 1997;71:824–827. doi: 10.1128/jvi.71.1.824-827.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou M, Collisson E W. Fifth International Symposium on Positive Strand RNA Viruses, St. Petersburg, Fla. 1998. Determination of RNA binding domains in the IBV nucleocapsid protein. [Google Scholar]