Abstract
BACKGROUND:
Multiple sclerosis (MS), a central nervous system disease in which nerve signals are disrupted by scarring and demyelination, is classified into phenotypes depending on the patterns of cognitive or physical impairment progression: relapsing-remitting MS (RRMS), primary-progressive MS (PPMS), secondary-progressive MS (SPMS), or progressive-relapsing MS (PRMS). The phenotype is important in managing the disease and determining appropriate treatment. The ICD-9-CM code 340.0 is uninformative about MS phenotype, which increases the difficulty of studying the effects of phenotype on disease.
OBJECTIVE:
To identify MS phenotype using natural language processing (NLP) techniques on progress notes and other clinical text in the electronic medical record (EMR).
METHODS:
Patients with at least 2 ICD-9-CM codes for MS (340.0) from 1999 through 2010 were identified from nationwide EMR data in the Department of Veterans Affairs. Clinical experts were interviewed for possible keywords and phrases denoting MS phenotype in order to develop a data dictionary for NLP. For each patient, NLP was used to search EMR clinical notes, since the first MS diagnosis date for these keywords and phrases. Presence of phenotype-related keywords and phrases were analyzed in context to remove mentions that were negated (e.g., “not relapsing-remitting”) or unrelated to MS (e.g., “RR” meaning “respiratory rate”). One thousand mentions of MS phenotype were validated, and all records of 150 patients were reviewed for missed mentions.
RESULTS:
There were 7,756 MS patients identified by ICD-9-CM code 340.0. MS phenotype was identified for 2,854 (36.8%) patients, with 1,836 (64.3%) of those having just 1 phenotype mentioned in their EMR clinical notes: 1,118 (39.2%) RRMS, 325 (11.4%) PPMS, 374 (13.1%) SPMS, and 19 (0.7%) PRMS. A total of 747 patients (26.2%) had 2 phenotypes, the most common being 459 patients (16.1%) with RRMS and SPMS. A total of 213 patients (7.5%) had 3 phenotypes, and 58 patients (2.0%) had 4 phenotypes mentioned in their EMR clinical notes. Positive predictive value of phenotype identification was 93.8% with sensitivity of 94.0%.
CONCLUSIONS:
Phenotype was documented for slightly more than one third of MS patients, an important but disappointing finding that sets a limit on studying the effects of phenotype on MS in general. However, for cases where the phenotype was documented, NLP accurately identified the phenotypes. Having multiple phenotypes documented is consistent with disease progression. The most common misidentification was because of ambiguity while clinicians were trying to determine phenotype. This study brings attention to the need for care providers to document MS phenotype more consistently and provides a solution for capturing phenotype from clinical text.
What is already known about this subject
Multiple sclerosis (MS) is an autoimmune disease of the central nervous system that is classified into phenotypes depending on the patterns of inflammation, demyelination of the central nervous system, and/or disability progression.
Observational, epidemiological research studies using electronic medical record (EMR) data have primarily made use of information in structured format, such as ICD-9-CM codes.
Because no ICD-9-CM codes exist to identify MS phenotype, existing large-scale studies of MS patients have lacked the ability to make any inferences at the phenotype level.
What this study adds
This study uses natural language processing technology on progress notes and other clinical text in an EMR to identify MS phenotype to determine phenotype prevalence in a large population database.
Of 7,756 MS patients, 2,854 (36.8%) patients had at least 1 phenotype in their clinical notes.
Among the 2,854 MS patients with at least 1 identified phenotype, the most common phenotype pattern was relapsing-remitting MS only (39.2%), followed by the progression to secondary-progressive MS (16.1%), secondary-progressive MS only (13.1%), and primary-progressive MS only (11.4%).
Multiple sclerosis (MS) is an autoimmune disease of the central nervous system that causes a range of symptoms, such as numbness and weakness in the limbs or muscle spasms, as nerve signals are disrupted by scarring and demyelination.1,2 MS is classified into phenotypes depending on the patterns of inflammation, demyelination of the central nervous system, and/or disability progression. MS phenotype designation, which dictates disease management strategies and appropriate treatment recommendations, is important in the real-world care of MS patients.3-5 Relapsing-remitting MS (RRMS) is characterized by unpredictable relapses followed by periods of remission lasting as long as several years with no new signs of disease activity and describes the initial course of 80% of individuals with MS.2 Roughly 65% of patients initially diagnosed with RRMS will advance to secondary-progressive MS (SPMS). These patients begin to have progressive neurologic decline between acute attacks without any definite periods of remission. The median time between disease onset and conversion from RRMS to SPMS is 19 years.1,6 Primary-progressive MS (PPMS) patients are those who never have remission after their initial MS symptoms. This characterizes approximately 10%-15% of MS patients.7 Progressive-relapsing MS (PRMS) characterizes patients who, from onset, have a steady neurologic decline but also suffer clear superimposed attacks. This is the least common MS phenotype.1
The widespread adoption of electronic medical records (EMRs) has enabled researchers to gain a wealth of valuable information on patient conditions and diagnoses. Observational, epidemiological research studies using EMR data have primarily made use of information in structured format, such as International Classification of Diseases, Ninth Revision, Clinical Modification (ICD-9-CM) codes. However, MS is wholly contained as a single ICD-9-CM code, 340.0, without distinction of MS phenotype. As a consequence, existing large-scale studies of MS patients have lacked the ability to make any inferences at the phenotype level. This coarse granularity was continued to the next version of ICD, ICD-10-CM, so even the mandated switch to ICD-10-CM that occurred in October 2015 does not enable stratification of MS phenotype. Unstructured data in free text, such as progress notes and other clinical text, represent a potential source of MS phenotype information that can be used to supplement structured data. The rationale is that information not used for coding or billing, but that is still clinically important, is documented by the care providers. While extracting information from unstructured formats using manual chart review can be prohibitively costly and time consuming, natural language processing (NLP), a set of methods to identify and interpret meaning from written text, can efficiently analyze and extract information from very large datasets.8
The objective of this study was to use NLP techniques on progress notes and other clinical text in an EMR to identify MS phenotype. To our knowledge, this is the first study to make use of NLP technology to determine phenotype prevalence in a large patient population.
Methods
Patient Selection
This study used nationwide EMR data from the Veterans Health Administration (VHA) to identify patients with at least 2 ICD-9-CM codes for MS (340.0) who sought care in the VHA system from 1999 through 2010. The date of the first MS diagnosis was assigned as the index date. Patients who did not have at least 180 days of observation in the VHA before the index date or who had the ICD-9-CM code for other demyelinating disease (341) were excluded from this analysis.
NLP Development
The purpose of the NLP development in this study was to identify documented mentions of MS phenotypes in clinical notes. The system was built using the Unstructured Information Management Architecture Asynchronous Scaleout as a document-processing pipeline.9 The system used a set of libraries that enabled programmatic configuration of the pipeline and easy manipulation of text in context.10 Each module in the pipeline identified keywords and phrases associated with a particular MS phenotype and output structured annotations that were overlaid on the text.
Three clinical experts (a registered nurse, a physical therapist, and a licensed family counselor) contributed possible keywords and phrases denoting MS phenotype. This list was added to through multiple iterations in which human reviewers examined clinical notes for visits where MS was discussed and reviewed the context around where known MS phenotype keywords were found. A nonexhaustive version of this list is found in Table 1. Because this list of keywords and phrases includes terms that could be used to identify non-MS phenotype-related concepts (e.g., the acronym “RR” in a clinical note could mean “relapsing-remitting” but could also mean “respiratory rate”), these terms were only accepted when in proximity to an MS reference. Cases where a phenotype was mentioned, but explicitly stated as not present for the patient, were also excluded (e.g., “not relapsing-remitting”).
TABLE 1.
Words and Phrases used in NLP
| Term |
| RR |
| RRMS |
| Relapsing remitting |
| PP |
| PPMS |
| Primary progressive |
| SP |
| SPMS |
| Secondary progressive |
| PR |
| PRMS |
| Progressive relapsing |
MS = multiple sclerosis; NLP = natural language processing; PPMS = primary-progressive MS; PRMS = progressive-relapsing MS; RRMS = relapsing-remitting MS; SPMS = secondary-progressive MS.
NLP Validation
To validate the NLP system at the patient level, this study calculated the sensitivity, specificity, negative predictive value (NPV), and positive predictive value (PPV) in 145 randomly selected veterans who had been identified as having MS in the structured data. The reference standard for these calculations was human annotation of every document of these 145 veterans. This human annotation consisted of one of our team members searching all documents for each of these 145 veterans for any mention of any of the 4 MS phenotypes. Each instance of identified phenotype was then verified by a second annotator. Sensitivity, specificity, NPV, and PPV were calculated separately for RRMS and any other phenotype. For these calculations, an MS phenotype mention identified by the NLP system was considered a true positive if the same mention was also identified by the human annotation. A false positive was an instance in which the NLP system identified a particular phenotype that was not captured through human annotation. Likewise, a true negative was a case in which neither the NLP system nor the human annotator identified a particular phenotype, while a false negative occurred when the NLP system failed to identify a specific phenotype that was captured by the human annotator. In addition, human annotators reviewed 1,000 NLP-extracted MS phenotype mentions to determine the PPV at the mention-level.
All relevant ethical safeguards have been met in relation to patient or subject protection. Institutional review board (IRB) approval for this study was obtained through the University of Utah’s IRB and the Department of Veteran Affair’s Office of Research and Development; therefore, this study was performed in accordance with the ethical standards contained in the 1964 Declaration of Helsinki and its later amendments.
Results
A total of 38,234 patients had at least 1 diagnosis for MS between January 1, 1999, and December 31, 2010 (Figure 1). Of those, 12,376 (32.4%) patients did not have a second MS diagnosis during the same time period and were excluded. An additional 17,026 patients (44.5%) were excluded because they did not have at least 6 months observation within the VHA system before the index diagnosis, and an additional 1,076 (2.8%) patients had an ICD-9-CM code for other demyelinating disease between January 1, 1999, and December 31, 2010.
FIGURE 1.
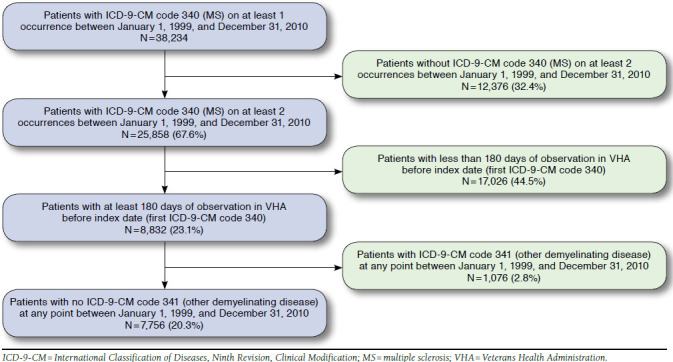
Attrition Summary
Of the 7,756 MS patients in this study, 2,854 (36.8%) patients had at least 1 identifiable phenotype in their clinical notes. The mean age of the MS patients across the entire cohort was 53.8 years, and mean age was 49.6 years for those with at least 1 phenotype. Also, of the 7,756 MS patients, 80.8% were male, while 78.9% of those with at least 1 phenotype were male. A total of 1,836 patients (64.3% of patients with phenotypes in clinical notes) had only 1 phenotype mentioned, while 747 (26.2%), 213 (7.5%), and 58 (2.0%) had 2, 3, and 4 phenotypes mentioned, respectively (Table 2). Mentions of multiple phenotypes in the same patient’s clinical notes may happen for several reasons. For example, there may be uncertainty surrounding the patient’s true diagnosis; there could be speculation as to what the diagnosis could be; or there may have simply been errors in previous clinical notes that were corrected in future notes. Among MS patients with at least 1 phenotype identified, the most common phenotype pattern was RRMS only (1,118, 39.2%; Table 2). The second most common phenotype pattern was the presence of RRMS and SPMS (459, 16.1%), which corresponds to actual disease progression. SPMS only (374, 13.1%) and PPMS only (325, 11.4%) were the next most common phenotype patterns found.
TABLE 2.
Summary of Patients by NLP-Identified Phenotype Classification
| Phenotype | Number | % of Total | % of Total with Phenotype Identified | |
|---|---|---|---|---|
| Total MS patients | – | 7,756 | – | – |
| Total MS patients with any phenotype identified | – | 2,854 | 36.8 | – |
| 1 phenotype identified | RRMS | 1,118 | 14.4 | 39.2 |
| PPMS | 325 | 4.2 | 11.4 | |
| SPMS | 374 | 4.8 | 13.1 | |
| PRMS | 19 | 0.2 | 0.7 | |
| 2 phenotypes identified | RRMS and PPMS | 141 | 1.8 | 4.9 |
| RRMS and SPMS | 459 | 5.9 | 16.1 | |
| PPMS and SPMS | 18 | 0.2 | 0.6 | |
| PPMS and PRMS | 112 | 1.4 | 3.9 | |
| PPMS and PRMS | 7 | 0.1 | 0.2 | |
| SPMS and PRMS | 10 | 0.1 | 0.4 | |
| 3 phenotypes identified | RRMS, PPMS, and SPMS | 158 | 2.0 | 5.5 |
| RRMS, PPMS, and PRMS | 9 | 0.1 | 0.3 | |
| RRMS, SPMS, and PRMS | 35 | 0.5 | 1.2 | |
| PPMS, SPMS, and PRMS | 11 | 0.1 | 0.4 | |
| 4 phenotypes identified | RRMS, PPMS, SPMS, and PRMS | 58 | 0.7 | 2.0 |
MS = multiple sclerosis; NLP = natural language processing; PPMS = primary-progressive MS; PRMS = progressive-relapsing MS; RRMS = relapsing-remitting MS; SPMS = secondary-progressive MS..
The PPV of the NLP system at the patient level was 86.5% in identifying any MS phenotype and 84.0% in identifying RRMS phenotype. Specificity of the system was 94.7% for any MS phenotype and 96.5% for RRMS (Table 3).
TABLE 3.
NLP Test Characteristics
| Test Characteristic | Any MS Phenotype | RRMS Phenotype |
|---|---|---|
| Sensitivity | 0.627 | 0.656 |
| Specificity | 0.947 | 0.965 |
| PPV | 0.865 | 0.840 |
| NPV | 0.824 | 0.908 |
MS = multiple sclerosis; NLP = natural language processing; NPV = negative predictive value; PPV = positive predictive value; RRMS = relapsing-remitting MS.
At the mention level, the PPV of the system was 93.8% across the 1,000 NLP-extracted mentions reviewed. Of the 62 phenotype mentions extracted by NLP that were found to be incorrect, the largest class of error occurred when the phenotype was documented as a differential diagnosis, or the physician was unclear of the current state of the disease (e.g., “likely a primary or secondary progressive course” or “MS, relapsing-remitting; now secondary progressive?”), 25 instances (40.3%). Other classes of errors included use of uncommon or nontraditional phenotype names (e.g., “RR Multiple Sclerosis with secondary progression”), 10 instances (16.1%); use of templated text or formatting issues (e.g., “MS Type: Relapsing Remitting Primary Progressive Secondary Progressive Yes”), 9 instances (14.5%); a misinterpreted acronym for the Short Portable Mental Status Questionnaire (e.g., “monitor for worsening MS; consider SPMSQ q 6 months”), 12 instances (19.4%); and mention of a phenotype that the patient does not have or referring to a family member (e.g., “patient does not appear to have converted to SPMS” or “patient’s sibling had RRMS”), 6 instances (9.7%).
Discussion
MS phenotype is an important consideration in clinical care for MS patients. Advances in treatments may continue to improve quality of life and may slow disability progression of MS,11 but results have been shown to vary by phenotype.4 In this context, the methods demonstrated here could be used in several important areas of future research. First, outcomes studies could be performed to assess the effectiveness of MS treatment in real-world settings, with phenotype as an important patient characteristic. Second, epidemiological studies could be conducted to assess the progression of MS phenotype over time. Third, studies could identify the patient characteristics and disease severity measures that are predictive of progression from RRMS to SPMS phenotypes.
While this study is the first to use NLP to extract MS phenotype information from MS patient EMRs, other studies have prospectively identified this information from a cohort of MS patients. For example, Bergamaschi et al. (2012) reviewed charts at 3 medical centers across Italy to determine the incidence and prevalence of the progression to SPMS (defined as “continuing deterioration [for at least one year] severe enough to lead to an increase of at least one point of the Expanded Disability Status Scale (EDSS), without substantial remission or exacerbation … assessed retrospectively, at least one year after the onset of the gradual worsening”11), as well as its risk factors, in a cohort of 1,078 treated and untreated relapsing-remitting patients who had had an MS diagnosis for at least 10 years.11 At the 10-year mark, 87.9% of the patients were determined to be RRMS and 12.1% SPMS; by the end of the study period, 68.1% were RRMS and 31.9% were SPMS. Our results support this finding. A cross-sectional survey study of depression in 451 U.S. veterans with MS documented phenotype with or without a progressive component; of these, 59.4% had MS with a progressive component.12 This is consistent with our finding of 60.8% of patients with a progressive component.
The accuracy of the NLP system is consistent with or exceeds the performance of systems built to identify other similar clinical information.13-15 The 5 error types identified in the false positive and false negative instances can be addressed with the addition of keywords and phrases to represent the missed mentions and creation of rules to more thoroughly interpret the context in which the keywords and phrases are used.
One of the surprising findings in this study was that nearly two thirds of MS patients had no documentation of phenotype in their medical records. This makes sense for patients that receive specialist care outside the VHA, since it is likely that MS neurologists would record phenotype more frequently than primary care physicians and other care providers. It is also likely that MS patients with relatively stable symptomology may be managed only by a primary care provider. This finding may suggest that in addition to using NLP to find explicit mentions of phenotype in the notes, a patient’s medication regimen (e.g., dimethyltryptamine as a proxy for RRMS) and pattern of health care utilization tied to MS could be used to infer phenotype. While these are not perfect surrogates for phenotype, they may be used in addition to NLP in order to take advantage of all evidence in the medical record and may be the only option when phenotype is not documented in clinical notes.
Limitations
This study was conducted exclusively within the VHA system which, while advantageous for a number of reasons (e.g., access to data for a large number of MS patients or availability of clinical notes on which to perform NLP to identify MS phenotype), also brings with it the limitation that the veteran MS population likely has different characteristics than the general MS population. First, in our MS population, 80.8% were male; however, 2-3 times as many women as men in the general population are diagnosed with MS.16 In working with a primarily male MS population, our study may have inadvertently selected for sex-related differences in MS that are not present in a more gender-balanced population (such as the higher rate of PPMS in men compared with women), or it may have inadvertently captured gender-based differences in care and treatment for MS. O’Donovan et al. (2015) observed that MS occurs at a higher rate in the veteran population than in the general population (0.10% vs. 0.01%, respectively),17 but the reasons remain obscure. Although Wallin et al. (2014) have observed that military service itself is not a risk factor for MS,18 Vollmer et al. (2002) have noted that veterans with MS are more likely to be older, unemployed, more disabled, less financially stable, and less educated than nonveterans with MS.19 Veterans aged under 50 years with MS also are more likely to suffer from chronic comorbidities such as diabetes, heart disease, and stroke.20 Additionally, there may be systematic or administrative differences between patients with mentions of MS phenotype in their notes and those without phenotype mentions. For example, patients without mentions of MS phenotype in their notes may receive most of their specialty care outside the VHA.
Conclusions
This study demonstrated the feasibility of extracting MS phenotype from clinical text using NLP methods from a nationally representative sample of veterans with MS. It highlights the low frequency with which phenotype is documented in the patient record and calls for care providers to more consistently record this important piece of clinical information in their clinical notes. Because MS phenotype is often missing from clinical notes, and despite our efforts to find it when using NLP, this study indicates that it is difficult to conduct prevalence studies of MS phenotype within an EMR database. Future studies should use and expand on these methods in order to conduct outcomes research studies that incorporate phenotype information on MS patients. Tools may be developed in the future so as to require the capture of phenotype within EMR data.
Acknowledgments
This study is the result of work supported with resources and the use of facilities at the George E. Wahlen Department of Veterans Affairs Medical Center, Salt Lake City, Utah; the IDEAS 2.0 Center (I50HX001240); the VA Informatics and Computing Infrastructure (VA HSR RES 13-457); and the Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development, Health Services Research and Development (CDA 11-210).
References
- 1. Lublin FD, Reingold SC. Defining the clinical course of multiple sclerosis: results of an international survey. National Multiple Sclerosis Society (USA) Advisory Committee on Clinical Trials of New Agents in Multiple Sclerosis. Neurology. 1996;46(4):907-11. [DOI] [PubMed] [Google Scholar]
- 2. Compston A, Coles A. Multiple sclerosis. Lancet. 2008;372(9648):1502-17. [DOI] [PubMed] [Google Scholar]
- 3. Goodin DS, Frohman EM, Garmany GP Jr, et al. . Disease modifying therapies in multiple sclerosis: report of the Therapeutics and Technology Assessment Subcommittee of the American Academy of Neurology and the MS Council for Clinical Practice Guidelines. Neurology. 2002;58(2):169-78. [DOI] [PubMed] [Google Scholar]
- 4. Bitsch A, Bruck W. Differentiation of multiple sclerosis subtypes: implications for treatment. CNS Drugs. 2002;16(6):405-18. [DOI] [PubMed] [Google Scholar]
- 5. De Jager PL. Identifying patient subtypes in multiple sclerosis and tailoring immunotherapy: challenges for the future. Ther Adv Neurol Disord. 2009;2(6):8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rovaris M, Confavreux C, Furlan R, Kappos L, Comi G, Filippi M. Secondary progressive multiple sclerosis: current knowledge and future challenges. Lancet Neurol. 2006;5(4):343-54. [DOI] [PubMed] [Google Scholar]
- 7. Miller DH, Leary SM. Primary-progressive multiple sclerosis. Lancet Neurol. 2007;6(10):903-12. [DOI] [PubMed] [Google Scholar]
- 8. Hou JK, Imler TD, Imperiale TF. Current and future applications of natural language processing in the field of digestive diseases. Clin Gastroenterol Hepatol. 2014;12(8):1257-61. [DOI] [PubMed] [Google Scholar]
- 9. Ferrucci D, Lally A. UIMA: an architectural approach to unstructured information processing in the corporate research environment. Nat Lang Eng. 2004;10(3-4):327-48. [Google Scholar]
- 10. DuVall SL, Patterson O, Ginter T, Cornia R, Nebeker JR. FLAP: flying towards real-time decision support. Paper presented at: NIH Workshop on Natural Language Processing: State of the Art, Future Directions and Applications for Enhancing Clinical Decision-Making; April 23-24, 2012; Bethesda, MD.
- 11. Bergamaschi R, Quaglini S, Tavazzi E, et al. . Immunomodulatory therapies delay disease progression in multiple sclerosis. Mult Scler. Published online May 31, 2012. [DOI] [PubMed] [Google Scholar]
- 12. Williams RM, Turner AP, Hatzakis M Jr, Bowen JD, Rodriquez AA, Haselkorn JK. Prevalence and correlates of depression among veterans with multiple sclerosis. Neurology. 2005;64(1):75-80. [DOI] [PubMed] [Google Scholar]
- 13. Carrell DS, Halgrim S, Tran DT, et al. . Using natural language processing to improve efficiency of manual chart abstraction in research: the case of breast cancer recurrence. Am J Epidemiol. 2014;179 (6):749-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wu ST, Sohn S, Ravikumar KE, et al. . Automated chart review for asthma cohort identification using natural language processing: an exploratory study. Ann Allergy Asthma Immunol. 2013;111(5):364-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xia Z, Secor E, Chibnik LB, et al. . Modeling disease severity in multiple sclerosis using electronic health records. PLoS One. 2013;8(11):e78927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.National Multiple Sclerosis Society. Initiative on gender differences in MS. 2015. Available at: http://www.nationalmssociety.org/Research/Research-We-Fund/Past-Research-Efforts-section-1. Accessed October 24, 2016.
- 17. O’Donovan A, Cohen BE, Seal KH, et al. . Elevated risk for autoimmune disorders in Iraq and Afghanistan veterans with posttraumatic stress disorder. Biol Psychiatry. 2015;77(4): 365-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wallin MT, Kurtzke JF, Culpepper WJ, et al. . Multiple sclerosis in gulf war era veterans. 2. Military deployment and risk of multiple sclerosis in the First Gulf War. Neuroepidemiology. 2014;42(4):226-34. [DOI] [PubMed] [Google Scholar]
- 19. Vollmer TL, Hadjimichael O, Preiningerova J, Ni W, Buenconsejo J. Disability and treatment patterns of multiple sclerosis patients in United States: a comparison of veterans and nonveterans. J Rehabil Res Dev. 2002;39(2):163-74. [PubMed] [Google Scholar]
- 20. Lavela SL, Prohaska TR, Furner S, Weaver FM. Chronic diseases in male veterans with multiple sclerosis. Prev Chronic Dis. Published online February 9, 2012. [PMC free article] [PubMed] [Google Scholar]


