To overcome some of the limitations of scRNA-seq, including modest cohort sizes, the authors use gene networks to define cell states that can deconvole bulk RNA-seq data, demonstrating convergence with scRNA-seq data, and characterizing immunotherapy response.
Abstract
Single-cell technologies have elucidated mechanisms responsible for immune checkpoint inhibitor (ICI) response, but are not amenable to a clinical diagnostic setting. In contrast, bulk RNA sequencing (RNA-seq) is now routine for research and clinical applications. Our workflow uses transcription factor (TF)–directed coexpression networks (regulons) inferred from single-cell RNA-seq data to deconvolute immune functional states from bulk RNA-seq data. Regulons preserve the phenotypic variation in CD45+ immune cells from metastatic melanoma samples (n = 19, discovery dataset) treated with ICIs, despite reducing dimensionality by >100-fold. Four cell states, termed exhausted T cells, monocyte lineage cells, memory T cells, and B cells were associated with therapy response, and were characterized by differentially active and cell state–specific regulons. Clustering of bulk RNA-seq melanoma samples from four independent studies (n = 209, validation dataset) according to regulon-inferred scores identified four groups with significantly different response outcomes (P < 0.001). An intercellular link was established between exhausted T cells and monocyte lineage cells, whereby their cell numbers were correlated, and exhausted T cells predicted prognosis as a function of monocyte lineage cell number. The ligand–receptor expression analysis suggested that monocyte lineage cells drive exhausted T cells into terminal exhaustion through programs that regulate antigen presentation, chronic inflammation, and negative costimulation. Together, our results demonstrate how regulon-based characterization of cell states provide robust and functionally informative markers that can deconvolve bulk RNA-seq data to identify ICI responders.
Introduction
Despite the great success of immune checkpoint inhibitors (ICI), only 12% of patients across all cancer indications are estimated to respond to therapy (1). The tumor microenvironment (TME) plays an important role in response outcomes to ICIs, exerting both immunostimulatory and immunosuppressive effects (2). ICI therapies mitigate the dysfunctional effects of the TME on T cells (3). Therefore, understanding which immune cell states are amenable to functional revival, which cell states are refractory, and how susceptible and resistant cell states dynamically interact, could provide new opportunities for disease management and therapeutic intervention.
Studies have analyzed the correlation between immune cell states and ICI response. For instance, effector and partially exhausted CD8+ T cells incite tumor control in response to ICIs (4). However, when differentiated into a terminally exhausted state, due to continued antigen stimulation, they fail to control tumor progression (5). Regulatory T cells (Treg) attenuate the activity of CD8+ T cells to maintain self-tolerance. Preclinical studies have demonstrated that CTLA-4–specific monoclonal antibodies may improve responses, at least in part, through modulation of Treg-mediated suppression of effector CD8+ T cells (6). Tumor-associated macrophages (TAM) represent a spectrum of phenotypes with M1 and M2 macrophages at either end. Inhibiting the activity of M2-like TAMs and redirecting their polarization towards the M1 phenotype can enhance response to ICIs (7). An outstanding question from such studies is whether the states of tumor-infiltrating immune cells, their transcriptional landscape, and cooccurrence patterns can effectively characterize patient response to ICIs.
Single-cell genomics at large-scale has emerged as a powerful technology for obtaining high-resolution transcriptomic data of tumor cellular ecosystems from primary tumor samples. However, constrained by prohibitive costs, single-cell RNA sequencing (scRNA-seq) cohorts are often modestly sized, making it difficult to validate biomarkers for predicting response to drug treatments.
Here, we leveraged the resolution offered by scRNA-seq to identify immune cell states associated with ICI response and the larger sample sizes of bulk RNA-seq datasets for validation. We demonstrated that a signature of transcription factor (TF)–directed gene networks, characterizing cell states associated with response, can identify patients likely to receive clinical benefit from ICI treatment. We explored dependencies between these cell states and established an intercellular link between monocyte-lineage cells (MLC) and exhausted T cells that impacts patient prognosis. Overall, these findings support the importance of considering the multifaceted nature of immune cell states present in the TME, and not limiting treatment strategies to T cells.
Materials and Methods
Patient data from previously published cohorts
Pretreatment samples with response information available in the form of RECIST criteria were selected from published studies (8–13). The overall survival and clinical response data, as originally reported, were used. A binary definition of response was implemented to standardize clinical response across studies: complete response (CR), partial response (PR), or stable disease (SD) for responders, and progressive disease (PD) for nonresponders.
scRNA-seq data preparation and regulon inference
For the discovery melanoma dataset, log-normalized transcripts per million (TPM) scRNA-seq data were accessed through NCBI's Gene Expression Omnibus (GEO) under accession number GSE120575 and processed as described in ref. 8. An additional filtering step removed genes expressed in <1% of cells or with <500 counts. A TF-directed coexpression network was inferred from the filtered gene expression matrix of pretreatment samples using GRNBoost2 in (https://github.com/tmoerman/arboreto) pySCENIC. RcisTarget (v1.6.0) was then used for cis-regulatory motif analyses on the coexpression network, removing indirect targets from TF modules to create regulons. The resulting regulon activity scores were calculated in the filtered gene expression matrix using AUCell with default parameters (v1.8.0; ref. 14).
The basal cell carcinoma dataset was accessed from GEO under the accession number GSE123814 (9). The raw unique molecular identifier counts (UMI) were log-normalized, and genes expressed in <1% of cells or with <500 counts were removed. Processed single-cell data for the external melanoma dataset was obtained from GEO under the accession number GSE72056, and no further processing steps were applied (15). The regulons identified in the discovery melanoma dataset were scored in both datasets using AUCell with default parameters.
Cell clustering
For the discovery melanoma dataset, clustering was performed independently on the gene expression and regulon activity feature space using the Seurat package (Version 4.0; ref. 16). The gene expression and regulon activity values were scaled using the “ScaleData” function (16). Clusters were identified using shared nearest neighbor (SNN)–based clustering, with the first 15 principal components, k = 20 (defines k for k-nearest neighbor algorithm) and resolution = 0.3 as inputs. The same principal components were used to generate uniform manifold approximation and projection (UMAP) projections. All remaining parameters were set to default values. The cell annotations were retained (8). Once isolated from the complete dataset, MLCs were clustered using the same protocol but with resolution = 0.055.
T-distributed stochastic neighbor embedding (T-SNE) was implemented using the Rtsne package (version 0.16) under default parameters on the regulon feature space of the external melanoma dataset. The cell phenotype annotations were retained (15).
Identifying and analyzing differentially abundant cell states
Differentially abundant (DA) cell states between responders and nonresponders were identified using the DAseq algorithm (17). The regulon UMAP projection generated from Seurat was provided as input to DAseq. DAseq uses a k-nearest neighbor algorithm to define the local neighborhood of a cell, from which its differential abundance score is calculated. The range of k values started at 50 and ended at 500, with a step of 50. A permutation test was performed with the “getDAcells” function in DAseq (as described in 17) to determine a threshold for identifying DA cells. All other DAseq parameters were set to default values.
Differentially expressed genes (DEG) were identified [Bonferroni-corrected q-value ≤0.05 and log2(fold-change) > 0] for cell phenotypes predominating a cell state based on their inclusion or exclusion in that cell state. Gene set enrichment analysis (GSEA) was performed using hallmark gene sets retrieved from gsea-msigdb.org (18). The cell phenotypes included monocytes/macrophages, B cells, memory T cells, exhausted CD8 T cells, exhausted heat shock CD8 T cells, and lymphocytes exhausted/cell cycle (16).
Characterizing cell states of response and regulon pruning
Differentially active regulons between DA cell states were calculated using the Wilcoxon test through Seurat (16). To quantify the cell-state specificity of a regulon, an entropy-based strategy (Jensen–Shannon divergence) was adapted, as described previously (19). Regulons with the highest cell state–specific and differential activity scores were selected as essential regulators.
Regulon modules were identified on the basis of the connection specificity index (CSI; ref. 20), which is a context-dependent measure for identifying specific associating partners. The CSI calculation was used as described previously (19).
To reduce regulon complexity whilst maintaining a cell type–specific effect, regulon target genes were pruned if not upregulated in the predominant immune cell phenotype of its associated cell state. Marker genes for immune cell phenotypes were determined according to Sade–Feldman and colleagues (8).
Validation datasets: bulk RNA-seq
Bulk RNA-seq data from melanoma biopsies were obtained from the following studies: (i) a skin cutaneous melanoma cohort from The Cancer Genome Atlas (TCGA; ref. 21); (ii) Van Allen and colleagues, an advanced melanoma anti–CTLA-4 treated cohort (10); (iii) Hugo and colleagues, an advanced melanoma anti–PD-1 treated cohort (12); (iv) Riaz and colleagues, an advanced melanoma anti–PD-1 treated cohort (11); and (v) Gide and colleagues, an advanced melanoma anti–PD-1 or combined ICI-treated cohort (13). The combination of ICI studies was chosen because the distribution of treatments received modeled the scRNA-seq discovery cohort (Supplementary Tables S1 and S2).
For Hugo and colleagues and Riaz and colleagues, raw FASTQ files were obtained from the Sequence Read Archive (SRP067938, SRP094781; refs. 11, 12). The RASflow pipeline was used for trimming, alignment, and feature counting, as described previously (22). For TCGA cohort, Gide and colleagues and Van Allen and colleagues, raw FASTQ files were preprocessed as described previously (23), and the counts were accessed at (https://zenodo.org/record/4661265, https://www.cbioportal.org/study?id=skcm_tcga). Gene counts were normalized using the standard DEseq2 protocol (24).
Regulon scoring, batch effect correction, and patient clustering
The pruned regulon gene sets were scored independently in each validation dataset using Single Sample GSEA (ssGSEA; ref.25). Median centering and scaling were performed on each dataset, which were then combined. Hierarchical clustering of patients based on their regulon scores was performed using the Pheatmap R package. Average linkage clustering was applied with a correlation distance metric. Associations between cluster assignment and patient response or survival outcome (deceased/living) were assessed using the chi-squared test. For continuous variables, associations with cluster assignment were determined using the Kruskal–Wallis test or ANOVA.
Modelling intercellular communication
The R package nichenetr (NicheNet; v1.0.0) was used to infer cell communication between the MLC and exhausted T-cell states (26). The MLCs were defined as “sender” cells, and exhausted T cells were considered as the “receiver” population. Genes with mean expression >2 log2(TPM) in either cell type were retained, and those expressed in exhausted T cells were used as “background genes.” The “gene set of interest” required by nichenetr was defined as the RUNX3 and PRDM1 regulons. The top five ligands were selected according to their regulatory potential score.
Trajectory inference
Cytotoxic lymphocytes, exhausted CD8 T cells, exhausted heat shock CD8 T cells, lymphocytes exhausted/cell cycle, and memory T cells were included in trajectory inference. The first and second principal components of the gene expression matrix containing these cell phenotypes were used in a Gaussian mixture model to identify pseudo-time–dependent cell clusters (Mclust package; ref. 27).
Slingshot was used to fit a minimum spanning tree (MST) to these clusters and determine the approximate trajectory (28). Memory T cells were set as the starting cluster. This piecewise linear trajectory was smoothed using simultaneous principal curves to obtain the final trajectories and pseudo-time values. Associations between pseudo-time and individual gene expression levels were described by fitting negative binomial general additive models via the tradeSeq package (29).
Defining MLC subclusters and doublet detection
scRNA-seq data of genes elevated in blood and immune cells compared with other cell types was downloaded from The Human Protein Atlas (http://www.proteinatlas.org/; ref. 30). The “FindAllMarkers” function in Seurat identified DEGs [P value ≤0.01 and log2(fold-change) > 0.25] between the MLC subclusters. Immune cells from The Human Protein Atlas were ordered according to their mean expression of the DEGs. Doublet detection was implemented on the full melanoma discovery dataset (n = 16,291) using the Chord algorithm (31). The doublet rate was set to 0.08, overkill was enabled, and the remaining parameters were set to default.
Statistical analysis
Survival curves
Kaplan–Meier curves were generated using the Survival R package (Version 3.2–13) and compared using the log-rank test.
Cox regression model
The regulon activities of RUNX3, PRDM1, and MAFB were scored in the Van Allen dataset using ssGSEA (10). Patients were split into high- and low-MLC cohorts according to the mean (+1 SD) MAFB regulon score. A Cox regression model was used to estimate the HRs of deaths associated with a patient's mean activity of RUNX3 and PRDM1 while accounting for their membership in either the high or low MLC cohorts.
Regression models
The top 5 regulatory potential scoring MLC ligands according to nichenetr, and a CIBERSORT “Absolute Score,” were used as explanatory variables in a multiple linear regression modelling the mean activity of RUNX3 and PRDM1 in the Van Allen dataset (10). The MLC ligands were scored using ssGSEA (25), and the “Absolute Score” was determined at CIBERSORTx (stanford.edu), using the LM22 Source GEP (32). For the anti–CTLA-4 treatment group, a multiple logistic regression with ssGSEA inferred regulon scores as the predictors and treatment response as the dependent variable was used.
Data availability
All data relevant to the study are included in the article and its supplementary data files, or upon request from the corresponding author. The public datasets used and/or analyzed during this study are available in the GEO or Zenodo databases (https://www.ncbi.nlm.nih.gov/geo/; https://zenodo.org/record/4661265).
Results
Regulon inference and cell states of response
The presence or absence of certain immune cells in the TME influences response to ICIs (2). Identifying and characterizing these immune cells can provide avenues for understanding and predicting response to ICIs. We analyzed scRNA-seq data derived from tumor-resident immune cells of metastatic melanoma patient samples (n = 19) collected prior to ICI treatment (8).
scRNA-seq data are inherently noisy due to sparse and shallow sequencing with missing values, which can compromise the clear, robust, and reproducible distinction of different cell types. To circumvent this problem, we inferred TF-directed gene networks (hereafter referred to as regulons) using the SCENIC workflow (14). Regulons act as a buffer against noisy expression fluctuations at the individual gene level, remaining stable despite numerous resampling and multiple sample batches (33). A regulon is denoted by the upstream TF, contains its direct downstream gene targets, and reflects the activity of the TF in a cell.
The cells were clustered using either gene expression or regulon activity. A comparison of both approaches revealed that information was not sacrificed for the dimensionality reduction introduced by regulons: expression variability remained between immune cell phenotypes (Fig. 1A).
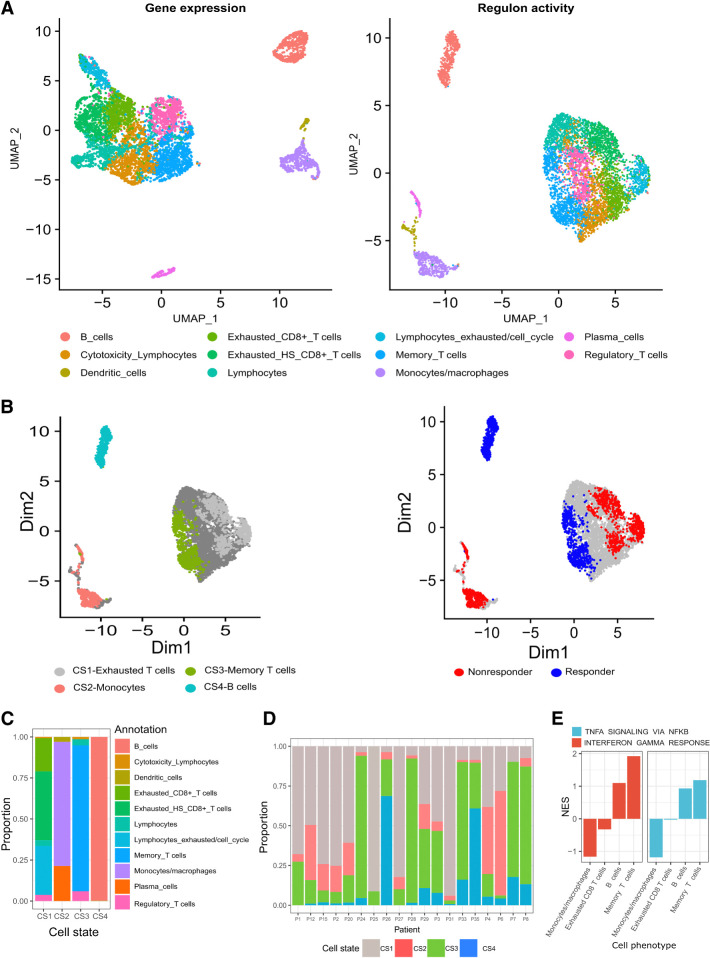
Figure 1.
Analysis of scRNA-seq data from samples taken from 19 patients with melanoma before ICI therapy (GSE120575; ref. 8). A, Dimensionality reduction, clustering and UMAP projection of tumor-resident immune cells. Left, Clustering according to gene expression profiles. Right, Clustering according to regulon activity profiles. B, UMAP embeddings of pretreatment cells, annotated on the basis of DAseq identified cell states (left), and their association with response (right). C, Distribution of immune cell phenotypes in the four DAseq identified cell states. D, The proportion of the DAseq identified cell states in each patient sample. E, The normalized enrichment scores (NES) of IFNG and TNFA pathways according to DEGs between cells from a cell phenotype (x-axis labels) that are either included or excluded from their associated DAseq cell state. Scores were calculated using GSEA.
Leveraging the DAseq method (17), four distinct pretreatment cell states were identified as DA between responders and nonresponders (Fig. 1B). RECIST criteria were used to define response, with SD, CR, and PR representing responders, PD representing nonresponders. Each cell state was annotated according to its predominant immune cell phenotype (Fig. 1C). Cell state 1 (CS1), termed exhausted T cells, was associated with nonresponders and contained three distinct exhaustion phenotypes: exhausted CD8 T cells, exhausted heat shock CD8 T cells, and lymphocytes exhausted/cell cycle. Annotated as MLCs, cell state 2 (CS2) contained predominately monocytes/macrophages and was associated with nonresponders. Cell state 3 (CS3) was abundant in responders and consisted of memory T cells. B cells made up cell state 4 (CS4), which was also associated with responders. The proportion of cell states in patient samples was analyzed to ensure multiple patients contributed to its association with response. Each cell state contained cells from the majority of patients (Fig. 1D).
Certain cell states are primarily constituted by one cell phenotype, but no cell phenotype is fully present in any given cell state (Supplementary Table S3). Hence, cells within a phenotype differ depending on their inclusion in a cell state. Cells included in cell states of response (B cells and memory T cells) had increased IFN-gamma (IFNG) and TNF alpha (TNFA) pathway activity compared with their excluded counterparts. If the cell state was linked to nonresponse (monocytes and exhausted T cells), included cells downregulated these pathways (Fig. 1E). IFNG is pivotal to ICI response and marks a point of divergence between cell states and the phenotypes from which they were derived.
Regulons defining cell states of response
During immune cell differentiation from multipotent hematopoietic stem cells, lineage-restricted TFs are induced, which in turn establish the identity of a specific cell type (34). We posit that regulons can characterize the transcriptional state of immune cells, providing a robust and functionally informative marker for deconvolving the TME. To identify regulons capable of characterizing the DAseq-identified cell states, we employed two metrics: differential regulon activity using a Wilcoxon test; and regulon specificity according to the Jensen–Shannon divergence (35). A differentially active regulon for a particular cell state has a statistically significant increase in median activity compared with all other cell states. Regulon specificity measures how distinct the distribution of regulon scores is for a specific cell state. Regulons scoring highly for both metrics in a cell state were selected.
The PRDM1 and RUNX3 regulons were significantly upregulated and specific to the exhausted T-cell state (Fig. 2A and B; Supplementary Fig. S1). Both TFs modulate terminal differentiation of effector CD8+ T cells, with RUNX3 being required for accessibility to PRDM1 TF motifs (36, 37). The LEF1 and FOXP1 TFs demonstrated enhanced expression in memory T-cell phenotypes (38). Their associated regulons were highly specific and differentially active in the memory T-cell state (Fig. 2A and B; Supplementary Fig. S1). MAFB and PAX5 scored highly for both metrics in the MLCs and B-cell states, respectively (Fig. 2A and B; Supplementary Fig. S1). MAFB is an inducer of monocyte differentiation (39), while PAX5 is a master regulator of B-cell development (40). A proportion of the exhausted and memory T-cell states (CS1 and CS3 clusters) identified by DAseq contained Tregs (Fig. 1C). Tregs represent an important cell type in ICI response. To quantify this cell type in future deconvolution steps, SMAD1 was identified as being specific to and differentially active in Tregs (Fig. 2A and B). TGFβ-dependent recruitment of SMAD has been identified as a key component in the generation of Tregs by participating in the “on-and-off” switch of FOXP3, and manipulation of SMAD activity is sufficient to alter the Treg differentiation program (41).
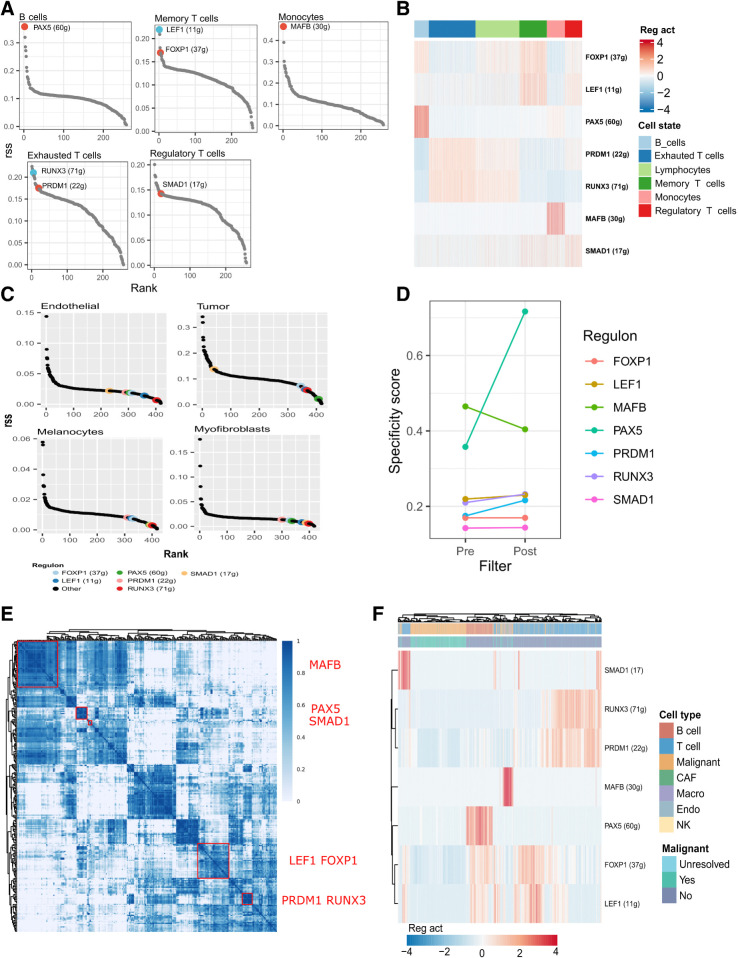
Figure 2.
The regulons that define cell states of response. A, The rank of regulons in their associated cell state based on regulon specificity score (RSS). The numbers in parenthesis denote the number of genes in the regulon before pruning. B, Relative activity level of each regulon across each of the DAseq-identified cell states. C, The rank of regulons in nonimmune cells from an external basal cell carcinoma ICI dataset based on rss (GSE123814; ref. 9). D, RSS for each regulon pre- and postfiltering of target genes according to gene markers of their associated cell type. E, Identified regulon modules based on regulon CSI matrix, along with the representative TFs. The color scale bar denotes the CSI score. F, Relative activity (scaled) of the pruned regulons in single cell expression profiles of malignant and nonmalignant cell types from an external metastatic melanoma dataset (GSE72056; ref. 15). CAF, cancer-associated fibroblasts; macro, macrophages; endo, endothelial cells; NK, natural killer.
To distinguish immune cells and their functional states, regulons must be cell state–specific and not coexpressed in unrelated cells residing in the TME. An independent scRNA-seq dataset of basal cell carcinoma samples confirmed low regulon specificity scores in tumor cells, melanocytes, endothelial cells, and myofibroblasts, suggesting that the presence of nonimmune cells does not impact regulon specificity (Fig. 2C; ref. 9).
Regulons inferred using regression are context dependent and at risk of overfitting the data. Complex regulons containing many genes are more susceptible to this phenomenon. To counteract this and improve generalizability, regulon sizes were reduced by pruning genes not significantly upregulated [Bonferroni-corrected q-value ≤0.05 and log2(fold-change) > 0.5] in the immune cell phenotypes making up the regulons associated cell state. To illustrate, PAX5 target genes were filtered according to genes upregulated in B cells. The upregulated genes were determined independently of the cell state and regulon inference, thereby offering an orthogonal metric to mitigate spurious connections present in a regulon. This approach reduced the complexity of each regulon and increased the overall regulon specificity (Fig. 2D; Supplementary Table S4).
Finally, we tested if the pruned regulons quantified an independent signal and remained functional in an external dataset. A pairwise similarity network of regulon activity scores was generated on the basis of the CSI in the discovery melanoma dataset (see Materials and Methods). The regulons clustered into distinct network modules, suggesting no redundancy between them (Fig. 2E). Unsupervised clustering of an independent scRNA-seq metastatic melanoma dataset using the pruned regulons separated immune from malignant cells and delineated immune cell subsets (Fig. 2F). A t-SNE of these cells, colored according to regulon activity, revealed differential regulon activity scores in their equivalent cell states compared with malignant cells (Supplementary Fig. S1). Both findings demonstrate the capacity of regulons to characterize the TME despite changing cellular ecosystems.
Regulon validation in bulk RNA-seq datasets
To determine if these regulons characterize response to ICIs in melanoma, we collated four datasets with pretreatment transcriptomic data and therapy response information (n = 209; refs. 10–13). The distribution of patients who received mono- or combination therapy in the validation dataset (80% anti–PD-1 or anti–CTLA-4, 20% anti–CTLA-4+anti–PD-1) matched the discovery scRNA-seq dataset (78% anti–PD-1 or anti–CTLA-4, 22% anti–CTLA-4+anti–PD-1; Supplementary Tables S1 and S2).
The relative activity of each regulon was scored in each dataset independently using ssGSEA (25). Median centering and scaling of regulon scores were performed to reduce batch effects between datasets (Fig. 3A), which were then combined for hierarchical clustering.
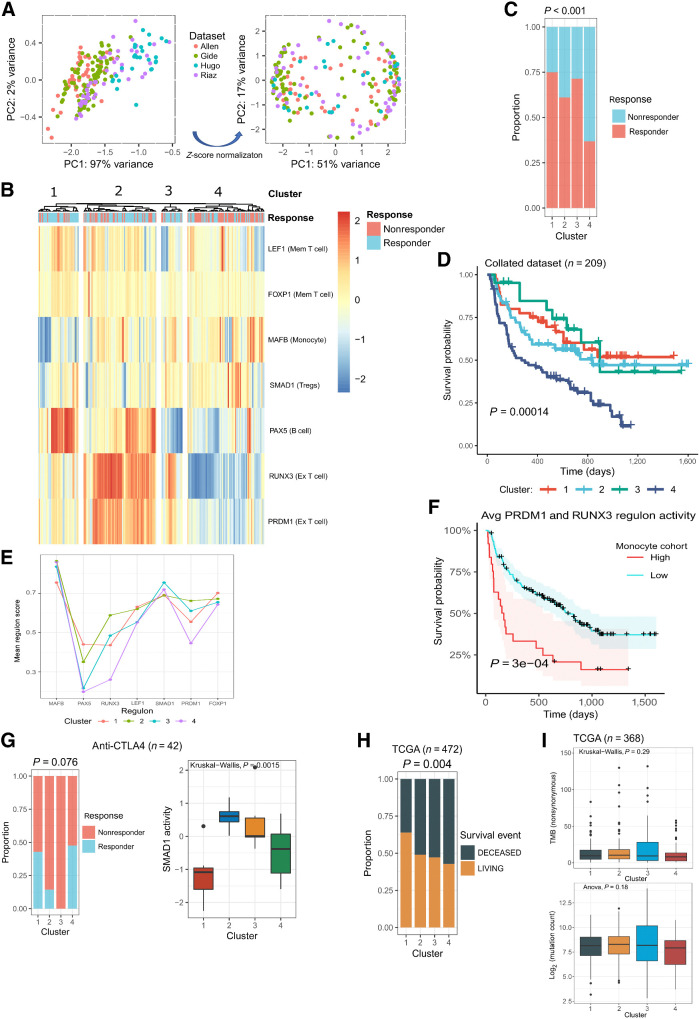
Figure 3.
The regulons correlate with response in four independent bulk RNA-seq datasets (10–13). A, PCA and clustering of patients from four datasets according to their regulon scores using ssGSEA, before (left image) and after (right image) z-score normalization. B, Hierarchical clustering of patients according to their ssGSEA inferred regulon scores, which are represented by the color scale bar. Pearson correlation was used as the distance metric. C, The proportion of responders and nonresponders in each cluster. The P value was calculated with a chi-squared test. D, Kaplan–Meier survival curves for each cluster, compared using a log-rank test. E, Mean regulon scores for each cluster identified using hierarchical clustering. F, Cox regression analysis comparing the association of PRDM1 and RUNX3 mean activity and survival in patients with high and low activity scores for MLCs. Statistical significance was inferred using the log rank test. G, Patients treated with anti-CTLA4 monotherapy. Left, The proportion of responders and nonresponders in each cluster following hierarchical clustering with regulon scores. Right, The relative activity of the SMAD1 regulon in each identified cluster (Kruskal–Wallis test). H, The proportion of TCGA samples that experienced a survival event (living/deceased) in each cluster identified using regulon scores (chi-squared test). I, Tumor mutational burden by nonsynonymous mutations (top, Kruskal–Wallis) and mutation count in exome sequencing data (bottom, ANOVA) in patients from TCGA across the identified clusters. For each box plot, the central black lines represent the median, whereas boxes extend from the first to the third quartiles. The whiskers extend to 1.5 times the interquartile range beyond the box.
Four clusters with significantly different response outcomes (chi-squared test, P < 0.001) were identified (Fig. 3B and C). The majority of patients in clusters 1, 2, and 3 were responders (75%, 61%, and 71%, respectively), whereas cluster 4 contained primarily nonresponders (63%; Fig. 3B and C). Overall survival (OS) following ICI treatment was significantly improved in clusters containing primarily responders (Fig. 3D).
Regulon scores were low in cluster 4, suggesting an immune desert phenotype in which all compartments of the tumor are sparse for immune cells (Fig. 3B and E; ref. 42). Cluster 1 contained high levels of productive memory T- and B-cell states, with concomitant low values for immunosuppressive cell states (MLCs and Tregs), likely representing the infiltrated-inflamed phenotype (Fig. 3B and E; ref. 42). Clusters 2 and 3 differed from cluster 1 in the number of MLCs and exhausted T cells.
We explored the relationship between these two cell states and found by Cox-regression analysis that the exhausted T-cell state predicted prognosis as a function of MLCs: higher numbers of MLCs were associated with worse survival (Fig. 3F). This may explain why cluster 3, and to a greater extent cluster 2, exhibited lower proportions of responders compared with cluster 1 (Fig. 3C). It could also clarify the shorter durability of responses in these clusters (Fig. 3D). As such, clusters 2 and 3 likely represent the altered-immunosuppressed phenotype, with high infiltration of MLCs coinciding with T-cell exhaustion, resulting in worse patient outcomes (42).
The standard of care in the first-line setting is both anti–PD-1 monotherapy or combined anti–CTLA-4 and anti–PD-1 regimens (43). However, exploration of anti–CTLA-4 agents remains warranted given their durable dose responsiveness and potential ability to deplete Tregs from cancers (43). We explored regulon characterization at each treatment level, stratifying patients who received anti–PD-1, anti–CTLA-4, or combination therapy. Clusters were enriched for responders in the anti–PD-1 and combination therapy groups (P = 0.033 and P = 0.025, respectively; Supplementary Fig. S2), but for nonresponders in the anti–CTLA-4 treatment group (P = 0.76; Fig. 3G). A strong dependency existed between the Treg-cell state, defined by SMAD1, and clusters 2 and 3, which primarily contained nonresponders to anti–CTLA-4 (P = 0.0015; Fig. 3G). Indeed, a logistic regression model determined that SMAD1 was the most significant predictor of anti–CTLA-4 response (coefficient = −1.32; P = 0.03), suggesting that certain cell states are more receptive to specific ICI regimens.
Tumor-infiltrating immune cells mediate tumor progression and, in some cases, elicit spontaneous regression (44). Therefore, regulons characterizing ICI response may depend on identifying patients with an underlying prognostic TME. We tested this by clustering skin cutaneous melanoma samples (n = 472) from TCGA according to their regulon activities (21). Four clusters with a significant difference in survival outcomes (chi-squared test, P = 0.004) were identified (Fig. 3H; Supplementary Fig. S2). Statistical significance was less than previously reported for response outcomes, suggesting the regulon-defined cell states influence ICI response more than prognosis. No association existed between cluster assignment and mutation count or TMB from synonymous mutations (Fig. 3I, n = 368). This is not surprising given the complex interaction between the developing tumor and the host immune system.
MLCs and exhausted T cells: relationship dynamics
We investigated why PRDM1 and RUNX3 were associated with worse prognosis when MLCs were abundant. Patients from the scRNA-seq dataset were split into high and low cohorts according to the proportion (median cut-off point) of cells in the exhausted T-cell state. The high cohort had significantly more MLCs (Fig. 4A), suggesting that the exhausted T-cell state may be linked to the activity of MLCs.
Figure 4.
The relationship between MLCs and exhausted T cells. A, The percentage of MLCs in patients with either high or low proportions of the exhausted T-cell state, compared using a Wilcoxon test. B, NicheNet inferred interactions between genes expressed in MLCs and the PRDM1 and RUNX3 regulons in exhausted T cells. C, The average expression pattern of MLC target genes identified by NicheNet across a pseudo-time trajectory in memory (Mem), cytotoxic (Cyto), and exhausted (Ex) T cells. The line represents the genes average expression with respect to pseudo-time, as inferred by a generative additive model. The cells are annotated as Memory, Exhausted, or Cyto T cells, and whether they were identified by DAseq. D, Pearson correlation between the top five MLC ligands from NicheNet, scored in the Van Allen dataset using ssGSEA, and the activity of RUNX3 and PRDM1. E, UMAP plot of MLC state after subclustering. The cells are colored by identified clusters. F, The expression level (log TPM) of the top MLC ligands from NicheNet in the MLC subclusters (two-sided Wilcoxon). G, The percentage of cells from the MLC subclusters in patients with either high or low proportions of the exhausted T-cell state (two-sided Wilcoxon). The central black line represents the median; boxes extend from the first to the third quartiles; whiskers extend to 1.5 times the interquartile range beyond the box. H, The proportion of total cells from MLC subclusters in nonresponder patients. I, The expression levels of DEGs [Bonferroni-corrected q-value ≤0.05 and log2 (fold-change) > 0.25] between MLC subclusters in cell phenotypes from the protein atlas. The cell types are in increasing order based on the mean expression values of the DEGs.
Using NicheNet (26), we conducted intercellular communication analysis and identified genes whose expression in MLCs was associated with the expression of PRDM1 and RUNX3 gene targets in the exhausted T-cell state. HLA-A, HMGB1/2, CD86, and TNF were the top scoring genes in MLCs and interacted with cytotoxicity-related genes, such as PRF1, GZMA, and CCL5 (Fig. 4B). We performed trajectory inference on memory, cytotoxic, and exhausted T cells and modeled the expression dynamics of cytotoxicity-related genes along a dynamic biological timeline. The expression levels of GZMA, PRDM1, CCL5, and VCAM1 were low at early time points, increased to a maximum, and then declined. Exhausted T cells were the most prominent cell type beyond the maximum expression point—when gene expression is decreasing (Fig. 4C). These suggest that MLCs are a causative agent in an “activation-dependent exhaustion program” such as those reported previously (15).
The Van Allen dataset confirmed the relationship of HLA-A, HMGB2, CD86, HMGB1, and TNF with PRDM1 and RUNX3 regulon activity: an ssGSEA inferred score for the MLC genes was strongly correlated with PRDM1 and RUNX3 activity (R = 0.63, R = 0.69, respectively; Fig. 4D and E). Immune-related genes are expected to be correlated if they quantify immune cells in the TME. We accounted for this by fitting a multiple linear regression: the relationship of RUNX3 and PRDM1 activity with the MLC score was adjusted for a CIBERSORT inferred “Absolute Score,” which measures the total immune component present in a tumor. The MLC score remained a significant predictor of RUNX3 and PRDM1 activities (P < 0.001), suggesting the relationship is not confounded by the number of immune cells in the TME.
Resolving monocyte heterogeneity is essential for understanding their role in ICI response. We clustered the MLC state at a finer granularity, identifying two distinct subclusters (denoted as 0 and 1; Fig. 4E). The top-scoring MLC effector genes identified previously by NicheNet were upregulated in subcluster 0, suggesting greater interactions with the exhausted T-cell state (Fig. 4F). Indeed, patients in the high exhausted T-cell cohort had significantly higher proportions of subcluster 0 compared to subcluster 1 (Fig. 4G). Both subclusters retained the association between MLCs and nonresponders (Fig. 4H). However, the DEGs between subclusters (Supplementary Fig. S2) demonstrated elevated expression in markedly different cell types from the human protein atlas (http://www.proteinatlas.org/; ref. 30). Genes upregulated in subcluster 0 were highly expressed in prototypical monocyte populations, whereas upregulated genes in subcluster 1 were highly expressed in B-cell populations (Fig. 4I). B-cell marker expression in subcluster 1 was not an artifact of doublet formation between monocytes and B cells (Supplementary Fig. S2). In fact, CD79a activation on myeloid derived suppressor cells (MDSC) has demonstrated immunosuppressive properties in metastatic melanoma (45), and may explain the relationship between subcluster 1 and nonresponders.
Discussion
Building on prior single-cell transcriptomic efforts to understand the relationship between the TME and ICI response (2, 8), we comprehensively analyzed the immune compartment in patients with metastatic melanoma prior to ICI treatment. Unlike previous approaches, and in response to the inherent noise of scRNA-seq data, we used regulons for the characterization of cellular states. Our results showed that regulons constitute robust guides of cellular identity and have comparable performance with gene expression data in delineating immune cell phenotypes. Importantly, as the scoring of regulons can overcome batch and technical effects (14), our approach is more effective for validating findings from single-cell analyses in a bulk RNA-seq context.
Four distinct immune cell states were enriched in either responder or nonresponder tumor samples. As these cell states were not constrained to any predefined clusters by DAseq, they were not previously identified by Sade-Feldman and colleagues (8). The exhausted CD8+ T-cell state was enriched in nonresponders and was defined by RUNX3 and PRDM1, both of which demonstrated increased chromatin accessibility and expression in terminally exhausted cells (37). Terminally exhausted cells are the progeny of polyfunctional “progenitor exhausted” cells and are considered unresponsive to ICIs (4, 5).
MDSCs present a major obstacle to ICI efficacy, but their underlying heterogeneity remains only partially understood (42). Our study identified an MLC state that induces terminal exhaustion in CD8+ T cells through two distinct subpopulations. One subpopulation, representing monocytes, upregulates ligands involved in antigen presentation (HLA-A), chronic inflammation (TNF), and negative costimulation (HMGB1, which is a TIM3 ligand), whereas the second subpopulation has elevated expression of B-cell markers (CD79a). Stimulation of MDSCs through CD79a enhances their suppressive effect on T-cell proliferation, stimulates their migration, and induces protumorigenic cytokine secretion (45). This explains their association with T-cell exhaustion and ICI resistance and prompts their exploration in additional datasets. The decline in the expression of cytotoxic (GZMA and VCAM1) and activation (CCL5) genes that interact with MLC ligands in the exhausted CD8+ T-cell state is preceded by a maximum expression value, suggesting that these cells may be stimulated until they become desensitized to the costimulatory pathway signal. In the context of ICI response, exhausted CD8+ T cells were associated with worse outcomes if high MLC levels were present, suggesting that exhausted T cells and ICI resistance is inextricably linked to MLCs.
Two cell states were associated with response to ICIs: B cells and memory T cells, the latter of which were defined by the LEF1 and FOXP1 regulons; LEF1 and FOXP1 are TFs linked to early differentiated memory T cells. By mapping the organizational structure of CD8+ memory T cells, a memory-cell state characterized by LEF1 and FOXP1 was identified (46). This cell state potentially facilitates an immunotherapy response through its self-renewal capabilities and propensity for activation and effector differentiation (46). Although the memory-cell state identified here may not be transcriptionally identical, which is a potential consequence of different cellular ecosystems used for identifying the cell state (normal physiology vs. melanoma), previous studies have reported the importance of early differentiated, stem cell–like memory cells for response to ICIs (4).
Reports on the impact of B cells on ICI response were initially inconsistent. It is now apparent that B cells can either bolster or negate ICIs, depending on their phenotype (47). In response to autologous melanoma secretomes, B cells differentiate into a plasmablast-like B-cell population with upregulation of PAX5. In concordance with our PAX5-defined B-cell state, the frequency of plasmablast-like B cells predicted response and survival to ICI via increasing PD-1+ T-cell activation (48). We hypothesize that PAX5 regulates a differentiation program in B cells exposed to melanoma stimuli that drives tumor clearance in response to an ICI.
Studies have reported greater reproducibility and insights into cellular heterogeneity from regulatory networks compared with gene expression–based approaches (14, 49). Networks also add functional context from gene interactions, facilitating the biological interpretation of findings. We did not benchmark our approach against gene expression–based methods; however, future work should systematically compare gene markers with network approaches for characterizing cellular states, deconvoluting bulk RNA-seq, and translating findings between omics platforms. Our study focused on pretreatment metastatic melanoma, and it remains to be seen whether the regulons function similarly in other cancers. Given the complexity of the TME, we propose implementing this method in a context-dependent manner.
Identifying highly resolved cellular states has numerous potential clinical benefits: administering ICI therapy based on the distribution of responsive cell states in the TME; and reprogramming resistant cells into productive cellular states. The development of cell modification strategies is an active research area, particularly in macrophages (50), and it requires a deeper understanding of the complex interplay between cellular states within the tumor-immune ecosystem. Our approach offers a valuable tool to accelerate therapy development in this field.
Supplementary Material
Supplementary Figures S1 and S2
Supplementary Tables S1 and S2
Acknowledgments
This publication has emanated from research conducted with the financial support of Precision Oncology Ireland, a Consortium of five Irish Universities, six Irish Charities, and seven Industry Partners, which was funded by the Science Foundation Ireland Strategic Partnership Programme, under Grant number [18/SPP/3522]. This project has received cofunding from the Astra Zeneca.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Immunology Research Online (http://cancerimmunolres.aacrjournals.org/).
Authors' Disclosures
D. Egan reports grants from Science Foundation Ireland and AstraZeneca during the conduct of the study. M. Kreileder reports grants from Science Foundation Ireland and AstraZeneca during the conduct of the study. M. Nabhan reports grants from Science Foundation Ireland and AstraZeneca during the conduct of the study. L.F. Iglesias-Martinez reports grants from Science Foundation Ireland and AstraZeneca during the conduct of the study. S.J. Dovedi reports other support from AstraZeneca during the conduct of the study; other support from AstraZeneca outside the submitted work; and reports employment with AstraZeneca and ownership of AstraZeneca stock. V. Valge-Archer reports other support from AstraZeneca outside the submitted work; and employment with AstraZeneca and ownership of AstraZeneca stock. T. Slidel employment with AstraZeneca and ownership of AstraZeneca stock AstraZeneca Plc. C. Bendtsen reports grants from SFI during the conduct of the study; personal fees from AstraZeneca AB outside the submitted work. I.P. Barrett reports employment with AstraZeneca. D.J. Brennan reports grants from Astra Zeneca during the conduct of the study; grants and personal fees from GSK and personal fees from MSD outside the submitted work. W. Kolch reports grants from Science Foundation Ireland and AstraZeneca during the conduct of the study. V. Zhernovkov reports grants from Science Foundation Ireland and AstraZeneca during the conduct of the study. No disclosures were reported by the other authors.
Authors' Contributions
D. Egan: Conceptualization, resources, data curation, software, formal analysis, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. M. Kreileder: Conceptualization, resources, supervision, funding acquisition, methodology, project administration, writing–review and editing. M. Nabhan: Conceptualization, resources, supervision, project administration, writing–review and editing. L.F. Iglesias-Martinez: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, investigation, methodology, project administration, writing–review and editing. S.J. Dovedi: Conceptualization, writing–review and editing. V. Valge-Archer: Conceptualization, writing–review and editing. A. Grover: Writing–review and editing. R.W. Wilkinson: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, investigation, methodology, project administration, writing–review and editing. T. Slidel: Writing–review and editing. C. Bendtsen: Conceptualization, resources, supervision, funding acquisition, methodology, project administration, writing–review and editing. I.P. Barrett: Conceptualization, resources, supervision, funding acquisition, project administration, writing–review and editing. D.J. Brennan: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, investigation, methodology, project administration, writing–review and editing. W. Kolch: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, investigation, methodology, project administration, writing–review and editing. V. Zhernovkov: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, investigation, methodology, project administration, writing–review and editing.
References
- 1. Haslam A, Prasad V. Estimation of the percentage of US patients with cancer who are eligible for and respond to checkpoint inhibitor immunotherapy drugs. JAMA Network Open 2019;2:e192535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bagaev A, Kotlov N, Nomie K, Svekolkin V, Gafurov A, Isaeva O, et al. Conserved pan-cancer microenvironment subtypes predict response to immunotherapy. Cancer Cell 2021;39:845–65. [DOI] [PubMed] [Google Scholar]
- 3. Speiser DE, Ho P-C, Verdeil G. Regulatory circuits of T cell function in cancer. Nat Rev Immunol 2016;16:599–611. [DOI] [PubMed] [Google Scholar]
- 4. Siddiqui I, Schaeuble K, Chennupati V, Fuertes Marraco SA, Calderon-Copete S, Pais Ferreira D, et al. Intratumoral Tcf1+PD-1+CD8+ T cells with stem-like properties promote tumor control in response to vaccination and checkpoint blockade immunotherapy. Immunity 2019;50:195–211. [DOI] [PubMed] [Google Scholar]
- 5. Miller BC, Sen DR, Al Abosy R, Bi K, Virkud YV, LaFleur MW, et al. Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol 2019;20:326–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wing K, Onishi Y, Prieto-Martin P, Yamaguchi T, Miyara M, Fehervari Z, et al. CTLA-4 control over Foxp3+ regulatory T cell function. Science 2008;322:271–5. [DOI] [PubMed] [Google Scholar]
- 7. Rodell CB, Arlauckas SP, Cuccarese MF, Garris CS, Li R, Ahmed MS, et al. TLR7/8-agonist-loaded nanoparticles promote the polarization of tumour-associated macrophages to enhance cancer immunotherapy. Nat Biomed Eng 2018;2:578–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sade-Feldman M, Yizhak K, Bjorgaard SL, Ray JP, de Boer CG, Jenkins RW, et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell. 2018;175:998–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yost KE, Satpathy AT, Wells DK, Qi Y, Wang C, Kageyama R, et al. Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat Med 2019;25:1251–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 2015;350:207–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Riaz N, Havel JJ, Makarov V, Desrichard A, Urba WJ, Sims JS, et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell 2017;171:934–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell 2016;165:35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gide TN, Quek C, Menzies AM, Tasker AT, Shang P, Holst J, et al. Distinct immune cell populations define response to anti-PD-1 monotherapy and anti-PD-1/anti-CTLA-4 combined therapy. Cancer Cell 2019;35:238–55. [DOI] [PubMed] [Google Scholar]
- 14. Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods 2017;14:1083–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tirosh I, Izar B, Prakadan SM, Wadsworth MH, Treacy D, Trombetta JJ, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016;352:189–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018;36:411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhao J, Jaffe A, Li H, Lindenbaum O, Sefik E, Jackson R, et al. Detection of differentially abundant cell subpopulations in scRNA-seq data. Proc Natl Acad Sci U S A 2021;118:e2100293118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Suo S, Zhu Q, Saadatpour A, Fei L, Guo G, Yuan G-C. Revealing the critical regulators of cell identity in the mouse cell atlas. Cell Rep 2018;25:1436–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bass JIF, Diallo A, Nelson J, Soto JM, Myers CL, Walhout AJM. Using networks to measure similarity between genes: association index selection. Nat Methods 2013;10:1169–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Akbani R, Akdemir KC, Aksoy BA, Albert M, Ally A, Amin SB, et al. Genomic classification of cutaneous melanoma. Cell 2015;161:1681–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang X, Jonassen I. RASflow: an RNA-seq analysis workflow with snakemake. BMC Bioinf 2020;21:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee JS, Nair NU, Dinstag G, Chapman L, Chung Y, Wang K, et al. Synthetic lethality-mediated precision oncology via the tumor transcriptome. Cell 2021;184:2487–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinf 2013;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Browaeys R, Saelens W, Saeys Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat Methods 2020;17:159–62. [DOI] [PubMed] [Google Scholar]
- 27. Scrucca L, Fop M, Murphy TB, Raftery AE. mclust 5: clustering, classification and density estimation using gaussian finite mixture models. R J 2016;8:289–317. [PMC free article] [PubMed] [Google Scholar]
- 28. Street K, Risso D, Fletcher RB, Das D, Ngai J, Yosef N, et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. Bmc Genomics 2018;19:477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Van den Berge K, Roux de Bézieux H, Street K, Saelens W, Cannoodt R, Saeys Y, et al. Trajectory-based differential expression analysis for single-cell sequencing data. Nat Commun 2020;11:1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Uhlen M, Karlsson MJ, Zhong W, Tebani A, Pou C, Mikes J, et al. A genome-wide transcriptomic analysis of protein-coding genes in human blood cells. Science 2019;366:eaax9198. [DOI] [PubMed] [Google Scholar]
- 31. Xiong K-X, Zhou H-L, Lin C, Yin J-H, Kristiansen K, Yang H-M, et al. Chord: an ensemble machine learning algorithm to identify doublets in single-cell RNA sequencing data. Commun Biol 2022;5:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Newman AM, Steen CB, Liu CL, Gentles AJ, Chaudhuri AA, Scherer F, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol 2019;37:773–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lim K, Wong L. Finding consistent disease subnetworks using PFSNet. Bioinformatics 2014;30:189–96. [DOI] [PubMed] [Google Scholar]
- 34. Hosokawa H, Rothenberg EV. How transcription factors drive choice of the T cell fate. Nat Rev Immunol 2021;21:162–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 2011;25:1915–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Behr FM, Chuwonpad A, Stark R, van Gisbergen KPJM. Armed and ready: transcriptional regulation of tissue-resident memory CD8 T cells. Front Immunol 2018;9:1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang D, Diao H, Getzler AJ, Rogal W, Frederick MA, Milner J, et al. The transcription factor runx3 establishes chromatin accessibility of cis-regulatory landscapes that drive memory cytotoxic T lymphocyte formation. Immunity 2018;48:659–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lu H, Wang H, Yan L, Shao H, Zhang W, Shen H, et al. Overexpression of early T-cell differentiation-specific transcription factors transforms the terminally differentiated effector T cells into less differentiated state. Cell Immunol 2020;353:104118. [DOI] [PubMed] [Google Scholar]
- 39. Kelly LM, Englmeier U, Lafon I, Sieweke MH, Graf T. MafB is an inducer of monocytic differentiation. EMBO J 2000;19:1987–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Medvedovic J, Ebert A, Tagoh H, Busslinger M. Chapter 5 - Pax5: a master regulator of B cell development and leukemogenesis. Adv Immunol 2011;111:179–206. [DOI] [PubMed] [Google Scholar]
- 41. Ruan Q, Kameswaran V, Tone Y, Li L, Liou H-C, Greene MI, et al. Development of foxp3+ regulatory T cells is driven by the c-rel enhanceosome. Immunity 2009;31:932–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ugel S, Canè S, De Sanctis F, Bronte V. Monocytes in the tumor microenvironment. Annu Rev Pathol 2021;16:93–122. [DOI] [PubMed] [Google Scholar]
- 43. Switzer B, Puzanov I, Skitzki JJ, Hamad L, Ernstoff MS. Managing metastatic melanoma in 2022: a clinical review. JCO Oncol Pract 2022;18:335–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jochems C, Schlom J. Tumor-infiltrating immune cells and prognosis: the potential link between conventional cancer therapy and immunity. Exp Biol Med (Maywood) 2011;236:567–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Luger D, Yang Y, Raviv A, Weinberg D, Banerjee S, Lee M-J, et al. Expression of the B-Cell receptor component CD79a on immature myeloid cells contributes to their tumor promoting effects. PLoS One 2013;8:e76115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Galletti G, De Simone G, Mazza EMC, Puccio S, Mezzanotte C, Bi TM, et al. Two subsets of stem-like CD8+ memory T cell progenitors with distinct fate commitments in humans. Nat Immunol 2020;21:1552–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sharonov GV, Serebrovskaya EO, Yuzhakova DV, Britanova OV, Chudakov DM. B cells, plasma cells and antibody repertoires in the tumour microenvironment. Nat Rev Immunol 2020;20:294–307. [DOI] [PubMed] [Google Scholar]
- 48. Griss J, Bauer W, Wagner C, Simon M, Chen M, Grabmeier-Pfistershammer K, et al. B cells sustain inflammation and predict response to immune checkpoint blockade in human melanoma. Nat Commun 2019;10:4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Obradovic A, Graves D, Korrer M, Wang Y, Roy S, Naveed A, et al. Immunostimulatory cancer-associated fibroblast subpopulations can predict immunotherapy response in head and neck cancer. Clin Cancer Res 2022;28:2094–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Poltavets AS, Vishnyakova PA, Elchaninov AV, Sukhikh GT, F TKh. Macrophage modification strategies for efficient cell therapy. Cells. 2020;9:1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1 and S2
Supplementary Tables S1 and S2
Data Availability Statement
All data relevant to the study are included in the article and its supplementary data files, or upon request from the corresponding author. The public datasets used and/or analyzed during this study are available in the GEO or Zenodo databases (https://www.ncbi.nlm.nih.gov/geo/; https://zenodo.org/record/4661265).



![Figure 4. The relationship between MLCs and exhausted T cells. A, The percentage of MLCs in patients with either high or low proportions of the exhausted T-cell state, compared using a Wilcoxon test. B, NicheNet inferred interactions between genes expressed in MLCs and the PRDM1 and RUNX3 regulons in exhausted T cells. C, The average expression pattern of MLC target genes identified by NicheNet across a pseudo-time trajectory in memory (Mem), cytotoxic (Cyto), and exhausted (Ex) T cells. The line represents the genes average expression with respect to pseudo-time, as inferred by a generative additive model. The cells are annotated as Memory, Exhausted, or Cyto T cells, and whether they were identified by DAseq. D, Pearson correlation between the top five MLC ligands from NicheNet, scored in the Van Allen dataset using ssGSEA, and the activity of RUNX3 and PRDM1. E, UMAP plot of MLC state after subclustering. The cells are colored by identified clusters. F, The expression level (log TPM) of the top MLC ligands from NicheNet in the MLC subclusters (two-sided Wilcoxon). G, The percentage of cells from the MLC subclusters in patients with either high or low proportions of the exhausted T-cell state (two-sided Wilcoxon). The central black line represents the median; boxes extend from the first to the third quartiles; whiskers extend to 1.5 times the interquartile range beyond the box. H, The proportion of total cells from MLC subclusters in nonresponder patients. I, The expression levels of DEGs [Bonferroni-corrected q-value ≤0.05 and log2 (fold-change) > 0.25] between MLC subclusters in cell phenotypes from the protein atlas. The cell types are in increasing order based on the mean expression values of the DEGs.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/3fd5/10398358/0f8331eaa50a/1125fig4.jpg)