Abstract
The adenovirus (Ad) fiber protein largely determines viral tropism through interaction with specific cell surface receptors. This molecule may also be involved in virion assembly or maturation, as some previously characterized fiber mutants were defective for processing of viral structural proteins. We previously described packaging cell lines that express Ad type 5 (Ad5) fiber and can complement the temperature-sensitive Ad fiber mutant H5ts142. We have now used these packaging cells to construct a new adenoviral vector (Ad5.βgal.ΔF) with E1, E3, and L5 (fiber) deleted and analyzed the fiber null phenotype. Ad5.βgal.ΔF growth was completely helper independent, and fiberless particles were produced by a single final round of growth in 293 cells. Cryoelectron microscopic studies and sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis showed that the structure and composition of these particles was nearly identical to those of first-generation Ad vectors. As expected, fiberless particles had reduced infectivity on epithelial cells, but they retained the ability to infect monocytic cells via an integrin-dependent pathway. These studies provide a novel approach to developing retargeted Ad gene therapy vectors.
Adenoviruses (Ads) are nonenveloped DNA viruses with icosahedral symmetry. There are at least 47 known Ad serotypes, many of which are associated with respiratory, gastrointestinal, or ocular disease (23). Ad has served as a model for the study of many biological processes and is in use as a vector for clinical gene therapy (26). Of the known serotypes, the closely related Ad type 2 (Ad2) and Ad5 have been most extensively studied. A multistage pathway by which Ad2 infects epithelial cells has been described elsewhere (21, 50).
The outer shell of the Ad capsid contains three major proteins, hexon, penton base, and fiber, along with several minor proteins. Cryoelectron microscopic (cryo-EM) structural studies have revealed the locations of most of these in the viral particle (42, 44) and in some cases have provided clues to their function (18). The majority of the capsid by mass is hexon, which forms the facets of the icosahedral particle (47). Each of the 12 vertices contains a complex of the penton base and fiber proteins (47). A ∼25-Å protrusion at the top of each penton base monomer contains an RGD sequence (43), which interacts with cellular αv integrins to mediate virus internalization and endosome disruption (49, 50). This interaction appears to be conserved across many Ad serotypes (31). In at least some cell types (for example, THP-1 monocytic cells), the penton base can also mediate virus attachment by binding to β2 integrins via its RGD motif (25).
The homotrimeric fiber protein forms a prominent spike that protrudes from each vertex of the capsid. Fiber is anchored to the penton base by its N terminus, while its C-terminal domain mediates attachment to cellular receptors (12, 30, 34). The fibers from Ad2 or Ad5 bind to a 46-kDa protein termed CAR (coxsackievirus and adenovirus receptor), expressed on the surface of many cells (4, 45), and the Ad2 fiber protein has also been reported to bind major histocompatibility class I antigens (22). The fiber from Ad3 binds to an as yet unidentified but more widely distributed receptor (14, 41). Since all of these virus serotypes are thought to be internalized via the integrin-penton interaction (31), the fiber-receptor interaction largely determines Ad cell tropism.
In addition to its role in targeting Ad infection, the fiber has been proposed to facilitate assembly or to stabilize the viral particles. A number of Ad proteins are synthesized as precursors which are then cleaved to their mature forms by the virally encoded L3 23-kDa protease (2, 3). Studies using fiber mutant viruses have suggested that a defect in the fiber protein might lead to defective proteolytic processing of proteins VI, VII, and VIII and therefore to accumulation of their uncleaved precursors (10, 15, 17). Other defects including abnormal sedimentation on CsCl gradients and incomplete packaging of viral DNA into the mutant particles were reported. These earlier studies led investigators to conclude that defective proteolysis due to lack of the fiber protein leads to a general block in virus maturation.
The inability to propagate viral mutants lacking the genes that encode structural proteins has hindered study of their roles in capsid assembly. The aforementioned studies were done with either temperature-sensitive (ts) fiber mutants (10, 15) or deletion mutants that were propagated in the presence of nondefective helper virus (17). Their interpretation may therefore be clouded by leaky expression of the ts protein or by residual helper virus in the preparations. A true null mutant which is completely helper independent would be useful in studying the fiber mutant phenotype.
We previously reported the generation of cell lines expressing a functional Ad5 fiber protein, which can complement fiber mutant Ads (48). In the studies reported here, we used these cell lines to generate a helper-independent gene transfer vector with E1, E3, and fiber deleted and have examined the structure and infectivity of Ad5 particles lacking the fiber protein. The fiberless virus, in combination with packaging cell systems that we have previously developed, should be useful in vector retargeting.
MATERIALS AND METHODS
Cell lines, virus propagation, and infectivity assays.
293 (Ad5-transformed human embryonic kidney [20]) and THP-1 monocytic cells were obtained from the American Type Culture Collection. 211B is a derivative of 293 which expresses the wild-type Ad5 fiber (48). Cells were grown at 37°C with 5% CO2 in Dulbecco modified Eagle medium (DMEM) or RPMI 1640 (for THP-1) supplemented with 10% fetal calf serum. For virus construction, cells were transfected with the indicated plasmids by using the Gibco calcium phosphate transfection system according to the manufacturer’s instructions and observed daily for evidence of cytopathic effect (CPE). For purified viral preparations, cells were infected with the indicated Ad and observed for completion of CPE. Cells were collected 2 to 5 days after infection, and virus was isolated by four rapid freeze-thaw cycles. Virus was then purified by centrifugation on preformed 15 to 40% CsCl gradients (111,000 × g for 3 h at 4°C). The bands were harvested, dialyzed into storage buffer (10 mM Tris [pH 8.1], 0.9% NaCl, 10% glycerol), aliquoted, and stored at −70°C.
Ad preparations were titered by plaque assay on 211B cells. Cells were plated on polylysine-coated six-well plates at 1.5 × 106 cells/well. Duplicate dilutions of virus stock were added to the plates in complete DMEM (1 ml/well). After a 5-h incubation at 37°C, virus was removed and the wells were overlaid with 2 ml of 0.6% low-melting-point agarose in medium 199 (Gibco). An additional 1 ml of overlay was added at 5-day intervals.
Infection of THP-1 cells was assayed by infecting 2 × 105 cells at the indicated multiplicity of infection in 0.5 ml of complete RPMI 1640. Forty-eight hours postinfection, the cells were fixed with glutaraldehyde and stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (24), and the percentage of stained cells was determined by light microscopy.
DNA constructs.
Plasmids were constructed and propagated by standard methods (37). pDV44 was constructed in Escherichia coli DH10B (Gibco) to reduce problems associated with rearrangement of the large plasmid. DNA for transfections was isolated by using the Qiagen system.
pDV44 was constructed by removing the fiber gene and some of the residual E3 sequences from pBHG10 (Microbix Biosystems). To simplify manipulations, the 11.9-kb BamHI fragment including the rightmost part of the Ad5 genome was removed from pBHG10 and inserted into pBS/SK. The resulting plasmid is termed p11.3. A 3.4-kb DNA fragment corresponding to the E4 region and both inverted terminal repeats of Ad5 was amplified from pBHG10 by using the oligonucleotides 5′ CAC AAC GAG CTC AAT TAA TTA ATT GCC ACA TCC TC 3′ and 5′ TGT ACA CCG GAT CCG GCG CAC ACC 3′ and cloned into the vector pCR2.1 (Invitrogen) to create pDV42. pDV42 was digested with PacI, which cuts at a unique site (bold type) in one of the PCR primers, and with SalI, which cuts at a unique site in the pCR2.1 polylinker. This fragment was used to replace the corresponding PacI/XhoI fragment of p11.3 (the pBS polylinker adjacent to the Ad DNA fragment contains a unique XhoI site), creating pDV43. Finally, pDV44 was constructed by replacing the 11.9-kb BamHI fragment of pBHG10 by the analogous BamHI fragment of pDV43. pDV44 therefore differs from pBHG10 by the deletion of Ad5 nucleotides (nt) 30819 to 32743 (residual E3 sequences and all but the 3′-most 41 nt of the fiber open reading frame).
To create pΔE1Bβgal, an simian virus 40-driven β-galactosidase cassette was excised from pSVβgal (Promega) by digestion with VspI and BamHI and cloned into the EcoRV and BamHI sites in pΔE1sp1B (Microbix Biosystems).
Analysis of recombinant Ad particles.
Western blotting and immunofluorescent staining were performed as described elsewhere (48), using polyclonal rabbit antibodies against recombinant Ad2 fiber or penton base proteins. Viral DNA was isolated (52), and Southern blotting was performed by standard methods (37). DNA was transferred to nylon membranes (MSI), and the signal was detected with a Genius nonradioactive kit (Boehringer). Fiber and E4 sequences were detected by using labeled inserts from pCLF and pE4/Hygro, respectively (48).
Cryo-EM.
Purified viral preparations of wild-type (Ad5.βgal.wt) and fiberless (Ad5.βgal.ΔF) Ad5 were concentrated to ∼8 × 1011 particles/ml and cryofrozen on holey carbon grids (1). Briefly, a 4-μl droplet of sample was placed on a glow-discharged grid, blotted for 10 s with filter paper, and plunged immediately into ethane slush chilled by liquid nitrogen. The grid was then transferred to a prechilled Gatan 626 cryo-transfer holder and examined under a Philips CM120 transmission electron microscope equipped with cryo-EM accessories. Viral particle images were collected with a Gatan slow-scan charge-coupled device camera under low-dose conditions (<20 electrons/Å2) at a nominal magnification of ×45,000 and at two levels of defocus (−0.5 and −1.0 μm). The pixel sampling size was 4.1 Å as determined by calibration with a catalase crystal.
Individual particle images were extracted as 400- by 400-pixel fields by using the QVIEW software package (39). The IMAGIC package was used for all subsequent image processing and reconstruction steps (46). The technique of angular reconstitution was applied to calculate the Euler orientational angles within the icosahedral asymmetric triangle. The −0.5-μm and −1.0-μm defocus sets were then combined after two-dimensional correction for the contrast transfer function. The parameters for the modeled contrast transfer function equation (spherical aberration constant [Cs] = 2 mm, fraction of amplitude contrast = 0.1, kV = 120, decay constant = 20 nm2, Fermi filter resolution cutoff = 8.1 Å, filter width = 3 Å, defocus = −0.5 or −1.0 μm) were selected to minimize ringing effects in the particle images. Three-dimensional reconstruction was carried out by exact filtered back projection after two cycles of translational and nearest-neighbor orientational refinement (11). The resolution of the final structures was assessed with the Fourier shell correlation function using a 0.5 threshold cutoff (6, 13). The wild-type and fiberless Ad5 reconstructions were normalized based on the strong viral capsid density. The isosurface level was selected to correspond to the molecular edge where the total enclosed volume changed the least with a fixed change in contour level. Isosurface representations of the density maps were generated by using the AVS software package (Advanced Visualization Systems, Inc.).
RESULTS
Construction of the Ad5.βgal.ΔF virus.
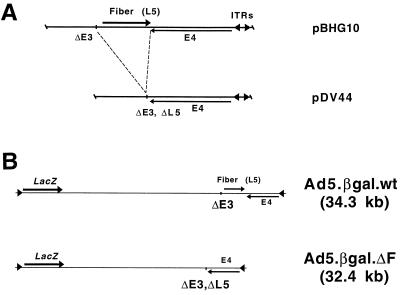
A fiberless Ad5 genomic plasmid (pDV44) was constructed by removing the fiber gene and some of the residual E3 sequences from pBHG10 (5) (Fig. 1A). pDV44 contains a wild-type E4 region, but only the last 41 nt of the fiber open reading frame (this sequence was retained to avoid affecting expression of the adjacent E4 transcription unit). Both pBHG10 and pDV44 contain unpackageable Ad5 genomes and must be rescued by cotransfection and subsequent homologous recombination with DNA carrying functional packaging signals (5). To generate vectors marked with a reporter gene, either pDV44 or pBHG10 was cotransfected with pΔE1Bβgal, which contains the left end of the Ad5 genome with a simian virus 40-driven β-galactosidase reporter gene inserted in place of the E1 region.
FIG. 1.
Fiber deletion in pDV44 and genomic structures of the Ad5.βgal.ΔF and Ad5.βgal.wt vectors. (A) pDV44 was constructed by removing the fiber gene and residual E3 sequences (nt 30819 to 32743 of Ad5) from pBHG10 (see Materials and Methods). ITRs, inverted terminal repeats. (B) Viruses constructed by cotransfection of either pBHG10 or pDV44 with pΔE1Bβgal. Both are E1/E3-lacking Ad5 vectors, and Ad5.βgal.ΔF has the additional fiber (L5) deletion as in pDV44.
A pDV44-derived virus is expected to be replication defective due to the fiber deletion, so that the cells in which it is grown must complement this defect. We previously reported production of 293-based cell lines which stably express a wild-type Ad5 fiber protein and can rescue a ts fiber mutant Ad as well complement an E1 deletion (48). One of these (211B) was used for rescue and propagation of the virus described here. pDV44 and pΔE1βgal were cotransfected into 211B cells, and the monolayers were observed for evidence of CPE. One of a total of 58 transfected dishes showed evidence of spreading cell death at day 15. A crude freeze-thaw lysate was prepared from these cells, and the resulting virus (termed Ad5.βgal.ΔF) was plaque purified twice and then expanded. As a control, the first-generation virus Ad5.βgal.wt, which is identical to Ad5.βgal.ΔF except for the fiber deletion, was constructed by cotransfection of pBHG10 and pΔE1Bβgal (Fig. 1B). In contrast to the low efficiency of recovery of the fiberless genome (1 of 58 dishes), all of the 9 dishes cotransfected with pΔE1Bβgal and pBHG10 produced virus.
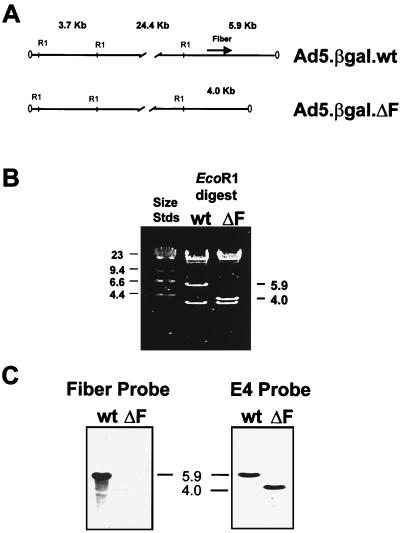
To confirm that the vector genomes had the expected structures and that the fiber gene was absent from the Ad5.βgal.ΔF chromosome, we analyzed DNA isolated from viral particles. Genomic DNA from both Ad5.βgal.wt and Ad5.βgal.ΔF produced the expected restriction patterns (Fig. 2A) following digestion with either EcoRI (Fig. 2B) or NdeI (data not shown). Southern blotting with labeled fiber DNA as a probe demonstrated the presence of fiber sequence in Ad5.βgal.wt but not in Ad5.βgal.ΔF DNA (Fig. 2C). As a positive control, the blot was stripped and reprobed with labeled E4 sequence. As expected, E4 signal was readily detectable in both genomes at equal intensities (Fig. 2C).
FIG. 2.
Analysis of the viral chromosomes. (A) Predicted EcoRI restriction maps of Ad5.βgal.wt and Ad5.βgal.ΔF. The 5.9-kb fragment at the right end of the Ad5.βgal.wt genome is reduced to 4.0 kb by the deletion of fiber sequences in Ad5.βgal.ΔF. (B) Ethidium bromide-stained gel of EcoRI-digested viral DNA. wt, wild-type virus; ΔF, fiberless virus. (C) Southern blot of the gel shown in panel B probed either with labeled fiber or E4 sequences. Positions of size standards (Stds) are indicated in kilobase pairs.
Characterization of the fiberless mutant.
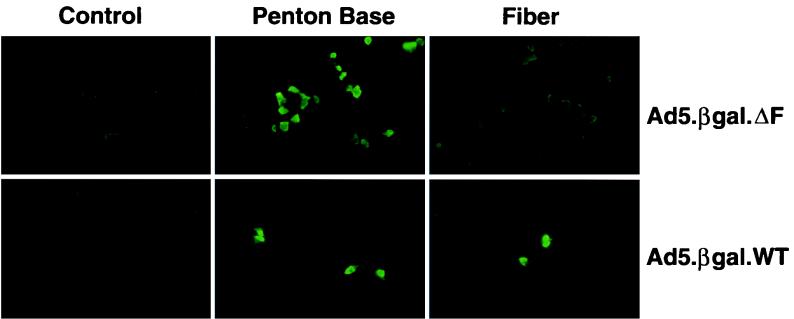
To verify that Ad5.βgal.ΔF was fiber defective, 293 cells (which are permissive for growth of E1-deleted Ad vectors but do not express fiber) were infected with Ad5.βgal.ΔF or Ad5.βgal.wt. Twenty-four hours postinfection, the cells were stained with polyclonal antibodies directed either against fiber or against the penton base protein (50). As shown in Fig. 3, cells infected with either virus were stained by the anti-penton base antibody, while only cells infected with the Ad5.βgal.wt control virus reacted with the antifiber antibody. This confirms that the fiberless Ad mutant does not direct the synthesis of fiber protein.
FIG. 3.
Penton base and fiber expression in Ad-infected cells. 293 cells were infected with Ad5.βgal.ΔF or Ad5.βgal.wt and stained with antibodies to the fiber or penton base proteins. As a control, infected 293 cells were stained without the incubation with primary antibody.
Growth of the fiber-deleted vector in complementing cells.
We found that Ad5.βgal.ΔF could readily be propagated in 211B cells. As assayed by protein concentration, CsCl-purified stocks of either Ad5.βgal.ΔF or Ad5.βgal.wt contained similar numbers of viral particles (Table 1), and the particles appeared to band normally on CsCl gradients. However, infectivity of the Ad5.βgal.ΔF particles was lower than that of the Ad5.βgal.wt control, as indicated by an increased particle/PFU ratio (Table 1). This is likely due to a reduced amount of fiber protein incorporated into mutant particles during growth in the 211B cells (see below and Discussion). We also found that Ad5.βgal.ΔF plaqued more slowly than the control virus. When plated on 211B cells, Ad5.βgal.wt plaques appeared within 5 to 7 days, while plaques of Ad5.βgal.ΔF continued to appear until as much as 15 to 18 days postinfection. Despite their slower formation, the morphology of Ad5.βgal.ΔF plaques was essentially normal.
TABLE 1.
Particle numbers and infectious titers of representative Ad preparations
| Virus | CsCl-purified prepn | Cell line | Particles/mla | PFU/mlb | Particle/PFU ratio | Fiber source |
|---|---|---|---|---|---|---|
| Ad5.βgal.wt | 1 | 211B | 7.4 × 1011 | 7.5 × 1010 | 10 | Ad chromosome |
| 2 | 211B | 3.0 × 1011 | 5.0 × 109 | 60 | Ad chromosome | |
| Ad5.βgal.ΔF | 3 | 211B | 7.7 × 1011 | 3.5 × 108 | 2,200 | Packaging cells |
| 4 | 211B | 1.9 × 1012 | 2.3 × 109 | 808 | Packaging cells | |
| 5 | 293 | 4.5 × 1011 | 9.5 × 106 | 47,400 | None | |
| 6 | 293 | 3.4 × 1011 | 3.5 × 107 | 9,700 | None |
Calculated from viral protein concentration (1 μg of protein = 4 × 109 particles).
Assayed by plaquing on 211B cells (see Materials and Methods).
Production of fiberless Ad particles.
Fiber mutant Ads have been reported to be defective both for proteolytic processing of viral proteins and for particle maturation (10, 15, 17). As Ad5.βgal.ΔF represents a true fiber null mutation and its stocks are free of helper virus, it provided an opportunity to reevaluate the fiber mutant phenotype. A single round of growth in cells (such as 293) which do not produce fiber should generate a homogeneous preparation of fiberless Ad, thereby allowing us to determine whether such particles would be stable and/or infectious. Either Ad5.βgal.wt or Ad5.βgal.ΔF was grown in 293 or 211B cells, and the resulting particles were purified on CsCl gradients. Ad5.βgal.ΔF particles could readily be produced in 293 cells at approximately the same level as the control virus and behaved similarly on the gradients, suggesting that there was not a gross defect in morphogenesis of fiberless capsids (Table 1).
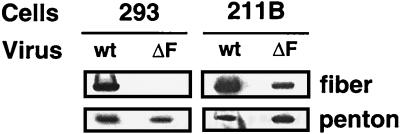
As shown in Fig. 4, particles of either virus contained similar amounts of penton base regardless of the cell type in which they were grown. This demonstrated that fiber is not required for assembly of the penton base complex into virions. However, as predicted, the Ad5.βgal.ΔF particles produced in 293 cells did not contain fiber protein. 211B-grown Ad5.βgal.ΔF also contained less fiber than the Ad5.βgal.wt control virus (Fig. 4). Importantly, the infectivities of the different viral preparations on epithelial cells (Table 1) correlated with the amount of fiber protein present. The fiberless Ad particles were several thousand-fold less infectious than the first-generation vector control on a per-particle basis, while infectivity of 211B-grown Ad5.βgal.ΔF was only 50- to 100-fold less than that of Ad5.βgal.wt. These studies confirmed fiber’s crucial role in infection of epithelial cells via CAR binding.
FIG. 4.
Analysis of vertex proteins in the viral particles. 293 (non-fiber-expressing) or 211B (fiber-expressing) cells were infected with Ad5.βgal.wt (wt) or Ad5.βgal.ΔF (ΔF), and the resulting viral particles were purified on CsCl gradients; 10 μg of purified virions was then electrophoresed on 5 to 16% gradient gels and Western blotted. Proteins were detected with polyclonal antifiber or anti-penton base antibodies.
Composition and structure of the fiberless particles.
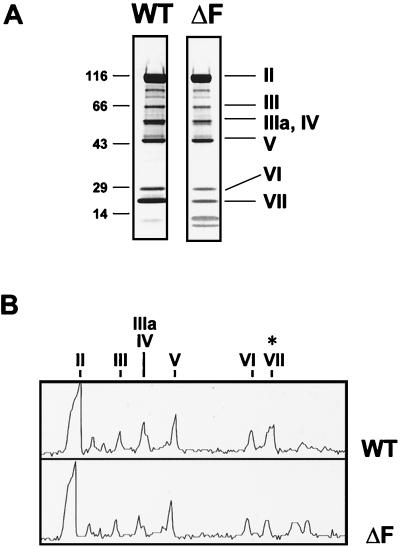
We compared the proteins contained in particles of 293-grown Ad5.βgal.ΔF to those in Ad5.βgal.wt to determine whether proteolysis or particle assembly was defective in this fiber null mutant (Fig. 5). The overall pattern of proteins in the fiberless particles was observed to be quite similar to that of a first-generation vector, with the exception of reduced intensity of the composite band resulting from both proteins IIIa and IV (fiber) (Fig. 5B). The fiberless particles also had a reduced level of protein VII, in agreement with previous reports (10, 15, 17). Although we did not see the substantial amounts of uncleaved precursors to proteins VI, VII, and VIII which some workers have observed in fiberless particles (17), it is possible that the low-molecular-weight bands migrating ahead of protein VII (Fig. 5) represent either aberrantly cleaved viral proteins or their breakdown products.
FIG. 5.
Protein composition of fiberless Ad particles. (A) Five micrograms of Ad5.βgal.wt (WT; a standard E1-lacking Ad vector) or of Ad5.βgal.ΔF (ΔF; grown in the absence of fiber) was electrophoresed on a 5 to 16% gradient gel and stained with Coomassie blue. Positions of molecular weight markers and viral proteins II (hexon), III (penton base), IIIa/IV (fiber is not resolved from protein IIIa in this system), V, VI, and VII are indicated. (B) Densitometer trace of the gel shown in panel A. Protein VII is indicated by ∗.
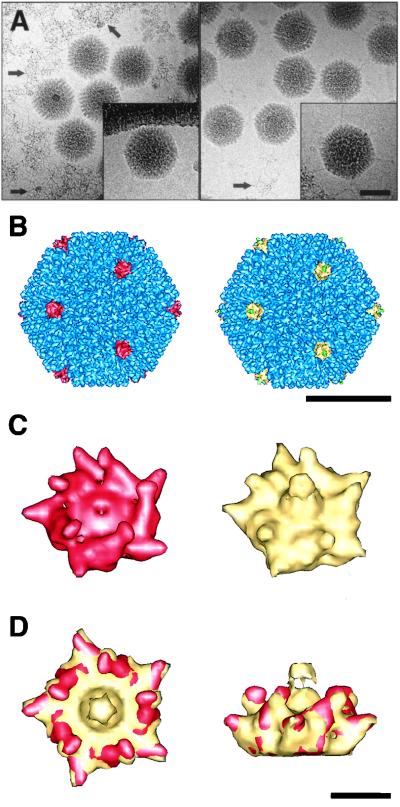
To more closely examine the structures of the 293-grown Ad5.βgal.ΔF (fiberless particles) and Ad5.βgal.wt (wild-type particles), we used cryo-EM. The fiber, which consists of an extended stalk with a knob at the end, was faintly visible in favorable orientations of wild-type Ad5 particles but not in images of the fiberless particles (Fig. 6A, inset). Filamentous material likely corresponding to free viral DNA was seen in micrographs of fiberless particles. This material was also present in micrographs of the first-generation control virus, albeit at much lower levels.
FIG. 6.
Cryo-EM of fiberless and wild-type Ad particles. The fiberless particles (293-grown Ad5.βgal.ΔF) are shown on the left, the wild-type particles (Ad5.βgal.wt) are shown on the right, in panels A to C. (A) Cryo-electron micrographs of Ad particles suspended in vitreous ice. Arrows mark free viral DNA. Insets show single representative particles with density corresponding to fiber protein present in Ad5.βgal.wt but absent in Ad5.βgal.ΔF. (B) Reconstructed Ad capsids viewed along an icosahedral threefold axis. The fiberless penton base protein (red), the wild-type penton base (yellow), the reconstructed portion of the flexible fiber (green), and the remaining capsid density (blue) are indicated. (C) Enlarged views of the penton base. Note that the fiber is shown in yellow, as its boundaries are not well defined. (D) Top and side views of the overlapping fiberless (red) and wild-type (yellow) penton base. The scale bars are 500 Å (A and B) and 50 Å (D).
Three-dimensional image reconstructions of fiberless and wild-type particles at ∼12-Å resolution showed similar sizes and overall features, with the exception that fiberless particles lacked density corresponding to the fiber protein (Fig. 6B and C). The densities corresponding to other capsid proteins, including penton base and proteins IIIa, VI, and IX, were comparable in the two structures. This finding confirms that absence of fiber does not prevent assembly of these components into virions. The fiber is truncated in the wild-type structure, as only the lower portion of its flexible shaft follows icosahedral symmetry. Note that the RGD protrusions on the fiberless penton base are angled slightly inward relative to those of the wild-type structure (Fig. 6D). Another difference between the two penton base proteins is that there is a ∼30-Å-diameter depression in the fiberless penton base around the fivefold axis where the fiber would normally sit. Similar observations were made for the Ad3 penton base dodecahedron both with and without the fiber (38). The Ad5 reconstructions confirm that capsid assembly, including addition of penton base to the vertices, is able to proceed in the complete absence of fiber.
Integrin-dependent infectivity of fiberless Ad particles.
While attachment via the viral fiber protein is a critical step in the infection of epithelial cells, an alternative pathway for infection of certain hematopoietic cells has been described. In this case, penton base mediates both binding to the cells (via β2 integrins) and internalization (through interaction with αv integrins) (25). Particles lacking fiber might therefore be expected to be competent for infection of these cells, even though on a per-particle basis they are several thousand-fold less infectious than normal Ad vectors on epithelial cells.
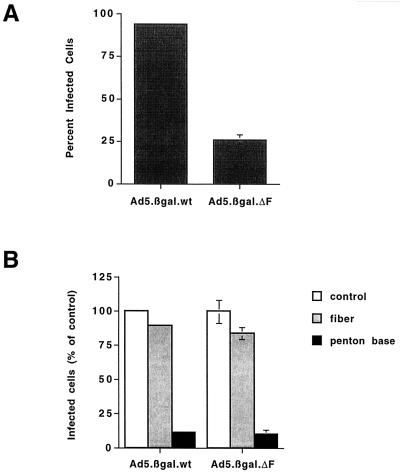
To investigate this issue, we infected THP-1 monocytic cells with Ad5.βgal.wt or with Ad5.βgal.ΔF grown in the absence of fiber. The fiberless particles were only a fewfold less infectious than first-generation Ad on THP-1 cells (Fig. 7A). In contrast, very large differences were seen in plaquing efficiency on epithelial (211B) cells (Table 1). Infection of THP-1 cells by either Ad5.βgal.ΔF or Ad5.βgal.wt was not blocked by an excess of soluble recombinant fiber protein but could be inhibited by the addition of recombinant penton base (Fig. 7B). These results indicate that the fiberless Ad particles use a fiber-independent pathway to infect these cells. Furthermore, the lack of fiber protein did not prevent Ad5.βgal.ΔF from internalizing into the cells and delivering its genome to the nucleus, demonstrating that fiberless particles are properly assembled and are capable of uncoating.
FIG. 7.
Infectivity of Ad particles on THP-1 monocytic cells. (A) THP-1 cells were infected with Ad5.βgal.wt or with fiberless Ad5.βgal.ΔF at 100,000 particles/cell. Forty-eight hours after infection, cells were fixed and stained with X-Gal, and the fraction of infected cells was determined by light microscopy. (B) Cells were infected with 1,000 particles of Ad5.βgal.wt or 100,000 particles of Ad5.βgal.ΔF per cell. As indicated, cells were pretreated with 100 μg of recombinant penton base or 20 μg of recombinant Ad2 fiber per ml.
DISCUSSION
Using the fiber-expressing 211B cell line, we were able for the first time to construct and propagate an Ad5-based gene delivery vector lacking the E1, E3, and L5 (fiber) genes. By growing Ad5.βgal.ΔF in 293 cells (which do not express fiber), fiberless Ad particles could also be produced for evaluation of their phenotype. As expected, particles that lacked fiber were greatly reduced in the ability to infect epithelial cells via the CAR-dependent pathway.
Although infectious particles of Ad5.βgal.ΔF were produced by growth in 211B cells, their ability to form plaques on epithelial cells was 50- to 100-fold reduced relative to wild-type virions (the first-generation vector Ad5.βgal.wt). We attribute this to the reduced amount of fiber protein incorporated into the virus particles. There is cell-to-cell variation in fiber expression by the 211B line (48), and particles produced by individual cells expressing a low level of fiber might be essentially noninfectious. Alternatively, a fewfold reduction in the number of fibers per virion might substantially reduce the overall avidity of the virus for its cell surface receptors. The findings of this study are consistent with our previous result that 211B cells partially complemented a ts fiber mutant even though the level of fiber that they produce is within three- to fivefold of that seen in an infected cell (48). Another group found that an E4 mutation which coincidentally reduced the level of fiber protein synthesis also drastically reduced the infectious yield of the vector produced (7). Together, these studies suggest that the level of fiber production is critical for production of fully infectious virus.
While fiberless particles were several thousand-fold less infectious than wild-type Ad in terms of plaquing on epithelial cells, their ability to infect monocytic cells by the fiber-independent pathway was reduced only about fivefold. Expression of the β-galactosidase reporter gene in Ad5.βgal.ΔF after infection of THP-1 cells confirms that fiberless viral particles are competent not only to bind to the cells but also to enter the cell, escape the endosome, and deliver the viral chromosome to the nucleus. Consistent with previous findings by Huang et al. (25), infection of these cells likely occurs via an integrin-penton base interaction.
Structural studies of wild-type viral particles have been useful in assigning roles for some viral proteins (for example, the role of protein IX as a cement which holds together the group-of-nine hexons [18]). However, while the cryo-EM reconstruction of the wild-type particles showed density corresponding to the basal part of the fiber, it did not provide information about how fiber might contribute to particle assembly or stability (44). Our ability to produce fiberless particles allowed us to directly address this question. Using cryo-EM and image reconstruction, we were able to analyze the structures of both wild-type and fiberless particles at ∼12-Å resolution. The overall structures of the viral capsids are quite similar, and there are no obvious differences in locations of the major (hexon and penton) or minor (such as proteins IIIa, VI, and IX) capsid proteins. A possible role for fiber in particle assembly might be to act as a scaffolding factor for penton base, as the 100-kDa protein does for hexon assembly (9). However, the structure of fiberless particles clearly demonstrates that the presence of fiber protein is not required for assembly or incorporation of penton base at the icosahedral vertices. Cryo-EM structures of the Ad3 penton dodecahedron showed that there is no central hole in the penton base where the fiber would normally attach, as was previously assumed on the basis of negative-stain EM images (36, 38). Our structure of Ad5.βgal.ΔF similarly showed no large hole at the fivefold axis. The orientations of the five RGD protrusions of the Ad5 fiberless virus particle appear to be shifted toward the fivefold axis compared to the wild-type particles. Our study of THP-1 infection further demonstrates that the penton base of fiberless particles is competent to interact with cellular integrins. The shift in the orientation of the RGD protrusions might account in part for the modest decrease in infectivity observed for fiberless particles relative to wild-type particles.
Falgout and Ketner found that in particles of fiberless Ad, uncleaved precursors to proteins VI, VII, and VIII were readily detectable and that the levels of the mature proteins were correspondingly reduced (17). We did not see a significantly reduced level of protein VI in fiberless particles, but the level of protein VII was much lower than in wild-type particles. However, their uncleaved precursor proteins were not present at dramatically elevated levels. This might be due to a lack of a proteolysis defect or perhaps to leakage of the uncleaved precursor from the fiberless particles. The fiberless Ad preparations differed from wild type by the presence of elevated amounts of what appears to be extraviral DNA on the EM grids. This may reflect a slightly reduced particle stability, and at the present time we do not know whether DNA leakage occurs during sample preparation or during virus purification. Since protein VII is associated with the viral chromosome (16, 33), its reduced level in the particles may be related to leakage of DNA rather than to any decreased synthesis or incorporation during viral assembly. Chee-Sheung and Ginsberg reported that the left end of the genome was preferentially represented in particles of the ts142 fiber mutant, which they interpreted as incomplete packaging of the viral DNA (10). Our results suggest that this might instead be due to loss of the genome via particle instability. The fiber protein may therefore play a role in sealing the vertex region following DNA packaging.
In general, the Ad5.βgal.ΔF phenotype that we observed was less severe than that previously reported for fiber mutants. The 293 cells which we used may somehow be more permissive than the KB or Vero cells used in the earlier experiments. We previously (48) found that the difference in infectious yield of the ts fiber mutant virus H5ts142 between the permissive and restrictive temperatures was considerably smaller in 293 cells than was reported by Chee-Sheung and Ginsberg, who used KB cells (10). Propagation of Ad5.βgal.ΔF in other non-fiber-expressing cell lines should help to resolve this discrepancy. Our fiber mutant genome differs from those previously used by the deletion of E1 and E3 regions, and another possibility would be that these changes somehow affect the fiber mutant phenotype.
The multiply deleted vector described here will be useful in improving Ad-based gene therapy strategies. Ad5.βgal.ΔF has an increased capacity to accept foreign DNA (deletion of the fiber gene should allow insertion of an additional 1.8 kb of sequence without exceeding the Ad packaging limit) relative to the first-generation vectors now widely used. Restoring a functional E3 region in Ad vectors may be beneficial in terms of reducing immunogenicity and prolonging transgene expression (8, 29, 35), and deletion of the fiber gene would allow E3 to be retained without compromising vector capacity. Ad5.βgal.ΔF is also more replication defective than first-generation vectors, and the presence of the second (fiber) deletion decreases the chance of generating fully replication-competent Ad via recombination in the packaging cells. As a replication-competent Ad generated by recombination at the E1 region would still be fiberless, it would be unable to spread efficiently via CAR-dependent mechanisms. However, it might be able to infect cells such as monocytes by the fiber-independent pathway.
Since any fiber in an Ad5.βgal.ΔF preparation is produced in trans by the packaging cells, this system should simplify the use of fiber modifications in vector retargeting. Several different strategies for manipulating the fiber protein have been explored. Vectors containing fibers of a different Ad serotype, or a chimeric fiber containing the receptor-binding domain of another serotype, have been reported (19, 28, 40). Additions of short protein epitopes to surface loops or at the C terminus of fiber, some of which have conferred altered binding properties, have also been constructed (27, 32, 51).
Such retargeted or pseudotyped vectors have so far been produced by incorporating a different or modified fiber gene into the vector chromosome. A system such as we describe here would allow the use of a single fiberless vector with a number of packaging lines expressing different fibers. For example, an Ad vector carrying a therapeutic transgene might be retargeted to different tumor types by growth in cells expressing fiber genes relevant for targeting neurons, hepatocytes, or lung epithelium. Since the fiber protein in a given preparation of vector is determined by the final round of growth, this system will also allow the production of Ad vectors with fibers that do not bind the packaging cells and therefore could not be grown in the usual manner.
Finally, infection of THP-1 cells by the fiberless particles suggests that a vector preparation which does not bind to the fiber receptor (CAR) might be useful in selectively targeting hematopoietic cells. Our results, along with those previously published (10, 15, 17), indicate that a complete lack of fiber protein may lead to particle instability and perhaps to loss of the genome. However, it should be possible to circumvent this problem using packaging lines which express a detargeted fiber protein which does not bind to its cognate receptor.
ACKNOWLEDGMENTS
We thank Joan Gausepohl for assistance with the manuscript and members of the Nemerow laboratory for helpful discussions.
This work was supported by grants from the National Institutes of Health (HL54352-04 to Glen R. Nemerow and AI42929 to Phoebe L. Stewart) and Genetic Therapy Inc./Novartis grant SFP1089. Charles Y. Chiu was supported by a fellowship from the Life and Health Insurance Medical Research fund, an NIH-MSTP training grant (GM08042), and the Aesculapians fund of the UCLA School of Medicine.
REFERENCES
- 1.Adrian M, Dubochet J, Lepault J, McDowall A W. Cryo-electron microscopy of viruses. Nature. 1984;308:32–36. doi: 10.1038/308032a0. [DOI] [PubMed] [Google Scholar]
- 2.Anderson C W. The proteinase polypeptide of adenovirus serotype 2 virions. Virology. 1990;177:259–272. doi: 10.1016/0042-6822(90)90479-b. [DOI] [PubMed] [Google Scholar]
- 3.Anderson C W, Baum P R, Gesteland R F. Processing of adenovirus 2-induced proteins. J Virol. 1973;12:241–252. doi: 10.1128/jvi.12.2.241-252.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergelson J M, Cunningham J A, Droguett G, Kurt-Jones E A, Krithivas A, Hong J S, Horwitz M S, Crowell R L, Finberg R W. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 5.Bett A J, Haddara W, Prevec L, Graham F L. An efficient and flexible system for construction of adenovirus vectors with insertions or deletions in early regions 1 and 3. Proc Natl Acad Sci USA. 1994;91:8802–8806. doi: 10.1073/pnas.91.19.8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Böttcher B, Wynne S A, Crowther R A. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature. 1997;386:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- 7.Brough D E, Lizonova A, Hsu C, Kulesa V A, Kovesdi I. A gene transfer vector-cell line system for complete functional complementation of adenovirus early regions E1 and E4. J Virol. 1996;70:6497–6501. doi: 10.1128/jvi.70.9.6497-6501.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruder J T, Jie T, McVey D L, Kovesdi I. Expression of gp19K increases the persistence of transgene expression from an adenovirus vector in the mouse lung and liver. J Virol. 1997;71:7623–7628. doi: 10.1128/jvi.71.10.7623-7628.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cepko C L, Sharp P A. Assembly of adenovirus major capsid protein is mediated by a nonvirion protein. Cell. 1982;31:407–415. doi: 10.1016/0092-8674(82)90134-9. [DOI] [PubMed] [Google Scholar]
- 10.Chee-Sheung C C, Ginsberg H S. Characterization of a temperature-sensitive fiber mutant of type 5 adenovirus and effect of the mutations on virion assembly. J Virol. 1982;42:932–950. doi: 10.1128/jvi.42.3.932-950.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiu, C. Y., P. Mathias, G. R. Nemerow, and P. L. Stewart. Binding and assembly of an integrin complex on human adenovirus. Submitted for publication.
- 12.Chroboczek J, Ruigrok R W, Cusack S. Adenovirus fiber. Curr Top Microbiol Immunol. 1995;199:164–200. doi: 10.1007/978-3-642-79496-4_10. [DOI] [PubMed] [Google Scholar]
- 13.Conway J F, Cheng N, Zlotnick A, Wingfield P T, Stahl S J, Steven A C. Visualization of a 4-helix bundle in the hepatitis B virus capsid by cryo-electron microscopy. Nature. 1997;386:91–94. doi: 10.1038/386091a0. [DOI] [PubMed] [Google Scholar]
- 14.Defer C, Belin M-T, Caillet-Boudin M-L, Boulanger P. Human adenovirus-host cell interactions: comparative study with members of subgroups B and C. J Virol. 1990;64:3661–3673. doi: 10.1128/jvi.64.8.3661-3673.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D’Halluin J-C, Milleville M, Martin G R, Boulanger P. Morphogenesis of human adenovirus type 2 studied with fiber- and fiber and penton base-defective temperature-sensitive mutants. J Virol. 1980;33:88–99. doi: 10.1128/jvi.33.1.88-99.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Everitt E, Lutter L, Philipson L. Structural proteins of adenoviruses. XII. Location and neighbor relationship among proteins of adenovirion type 2 as revealed by enzymatic iodination, immunoprecipitation, and chemical cross-linking. Virology. 1975;67:197–208. doi: 10.1016/0042-6822(75)90417-1. [DOI] [PubMed] [Google Scholar]
- 17.Falgout B, Ketner G. Characterization of adenovirus particles made by deletion mutants lacking the fiber gene. J Virol. 1988;62:622–625. doi: 10.1128/jvi.62.2.622-625.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furcinitti P S, van Oostrum J, Burnett R M. Adenovirus polypeptide IX revealed as capsid cement by difference images from electron microscopy and crystallography. EMBO J. 1989;8:3563–3570. doi: 10.1002/j.1460-2075.1989.tb08528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gall J, Kass-Eisler A, Leinwand L, Falck-Pedersen E. Adenovirus type 5 and 7 capsid chimera: fiber replacement alters receptor tropism without affecting primary immune neutralization epitopes. J Virol. 1996;70:2116–2123. doi: 10.1128/jvi.70.4.2116-2123.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graham F L, Smiley J, Russell W C, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–72. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 21.Greber U F, Willetts M, Webster P, Helenius A. Stepwise dismantling of adenovirus 2 during entry into cells. Cell. 1993;75:477–486. doi: 10.1016/0092-8674(93)90382-z. [DOI] [PubMed] [Google Scholar]
- 22.Hong S S, Karayan L, Tournier J, Curiel D T, Boulanger P A. Adenovirus type 5 fiber knob binds to MHC class I α2 domain at the surface of human epithelial and B lymphoblastoid cells. EMBO J. 1998;16:2294–2306. doi: 10.1093/emboj/16.9.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horwitz M S. Adenoviruses. In: Fields B N, Knipe D M, Howley P M, editors. Virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2149–2170. [Google Scholar]
- 24.Huang S, Endo R I, Nemerow G R. Upregulation of integrins αvβ3 and αvβ5 on human monocytes and T lymphocytes facilitates adenovirus-mediated gene delivery. J Virol. 1995;69:2257–2263. doi: 10.1128/jvi.69.4.2257-2263.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang S, Kamata T, Takada Y, Ruggeri Z M, Nemerow G R. Adenovirus interaction with distinct integrins mediates separate events in cell entry and gene delivery to hematopoietic cells. J Virol. 1996;70:4502–4508. doi: 10.1128/jvi.70.7.4502-4508.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozarsky K F, Wilson J M. Gene therapy: adenovirus vectors. Curr Opin Genet Dev. 1993;3:499–503. doi: 10.1016/0959-437x(93)90126-a. [DOI] [PubMed] [Google Scholar]
- 27.Krasnykh V, Dmitriev I, Mikheeva G, Miller C R, Belousova N, Curiel D T. Characterization of an adenovirus vector containing a heterologous peptide epitope in the HI loop of the fiber knob. J Virol. 1998;72:1844–1852. doi: 10.1128/jvi.72.3.1844-1852.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krasnykh V N, Mikheeva G V, Douglas J T, Curiel D T. Generation of recombinant adenovirus vectors with modified fibers for altering viral tropism. J Virol. 1996;70:6839–6846. doi: 10.1128/jvi.70.10.6839-6846.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee M G, Abina M A, Haddada H, Perricaudet M. The constitutive expression of the immunomodulatory gp 19K protein in E1-, E3-adenoviral vectors strongly reduces the host cytotoxic T cell response against the vector. Gene Ther. 1995;2:256–262. [PubMed] [Google Scholar]
- 30.Louis N, Fender P, Barge A, Kitts P, Chroboczek J. Cell-binding domain of adenovirus serotype 2 fiber. J Virol. 1994;68:4104–4106. doi: 10.1128/jvi.68.6.4104-4106.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mathias P, Wickham T J, Moore M, Nemerow G. Multiple adenovirus serotypes use αv integrins for infection. J Virol. 1994;68:6811–6814. doi: 10.1128/jvi.68.10.6811-6814.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michael S I, Hong J S, Curiel D T, Engler J A. Addition of a short peptide ligand to the adenovirus fiber protein. Gene Ther. 1995;2:660–668. [PubMed] [Google Scholar]
- 33.Mirza M A, Weber J. Structure of adenovirus chromatin. Biochim Biophys Acta. 1982;696:76–86. doi: 10.1016/0167-4781(82)90012-4. [DOI] [PubMed] [Google Scholar]
- 34.Philipson L, Lonberg-Holm K, Pettersson U. Virus-receptor interaction in an adenovirus system. J Virol. 1968;2:1064–1075. doi: 10.1128/jvi.2.10.1064-1075.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poller W, Schneider-Rasp S, Liebert U, Merklein F, Thalheimer P, Haack A, Schwaab R, Schmitt C, Brackmann H-H. Stabilization of transgene expression by incorporation of E3 region genes into an adenoviral factor IX vector and by transient anti-CD4 treatment of the host. Gene Ther. 1996;3:521–530. [PubMed] [Google Scholar]
- 36.Ruigrok R W H, Barge A, Albiges-Rizo C, Dayan S. Structure of adenovirus fibre II. Morphology of single fibres. J Mol Biol. 1990;215:589–596. doi: 10.1016/S0022-2836(05)80170-6. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Schoehn G, Fender P, Chroboczek J, Hewat E A. Adenovirus 3 penton dodecahedron exhibits structural changes of the base on fibre binding. EMBO J. 1996;15:6841–6846. [PMC free article] [PubMed] [Google Scholar]
- 39.Shah A K, Stewart P L. QVIEW: software for rapid selection of particles from digital electron micrographs. J Struct Biol. 1998;123:17–21. doi: 10.1006/jsbi.1998.4011. [DOI] [PubMed] [Google Scholar]
- 40.Stevenson S C, Rollence M, Marshall-Neff J, McClelland A. Selective targeting of human cells by a chimeric adenovirus vector containing a modified fiber protein. J Virol. 1997;71:4782–4790. doi: 10.1128/jvi.71.6.4782-4790.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stevenson S C, Rollence M, White B, Weaver L, McClelland A. Human adenovirus serotypes 3 and 5 bind to two different cellular receptors via the fiber head domain. J Virol. 1995;69:2850–2857. doi: 10.1128/jvi.69.5.2850-2857.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stewart P L, Burnett R M, Cyrklaff M, Fuller S D. Image reconstruction reveals the complex molecular organization of adenovirus. Cell. 1991;67:145–154. doi: 10.1016/0092-8674(91)90578-m. [DOI] [PubMed] [Google Scholar]
- 43.Stewart P L, Chiu C Y, Huang S, Muir T, Zhao Y, Chait B, Mathias P, Nemerow G R. Cryo-EM visualization of an exposed RGD epitope on adenovirus that escapes antibody neutralization. EMBO J. 1997;16:1189–1198. doi: 10.1093/emboj/16.6.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stewart P L, Fuller S D, Burnett R M. Difference imaging of adenovirus: bridging the resolution gap between X-ray crystallography and electron microscopy. EMBO J. 1993;12:2589–2599. doi: 10.1002/j.1460-2075.1993.tb05919.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomko R P, Xu R, Philipson L. HCAR and MAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc Natl Acad Sci USA. 1997;94:3352–3356. doi: 10.1073/pnas.94.7.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Heel M, Harauz G, Orlova E V, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 47.van Oostrum J, Burnett R M. Molecular composition of the adenovirus type 2 virion. J Virol. 1985;56:439–448. doi: 10.1128/jvi.56.2.439-448.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Von Seggern D J, Kehler J, Endo R, Nemerow G R. Complementation of a fiber mutant adenovirus by packaging cell lines stably expressing the Ad5 fiber protein. J Gen Virol. 1998;79:1461–1468. doi: 10.1099/0022-1317-79-6-1461. [DOI] [PubMed] [Google Scholar]
- 49.Wickham T J, Filardo E J, Cheresh D A, Nemerow G R. Integrin αvβ5 selectively promotes adenovirus mediated cell membrane permeabilization. J Cell Biol. 1994;127:257–264. doi: 10.1083/jcb.127.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wickham T J, Mathias P, Cheresh D A, Nemerow G R. Integrins αvβ3 and αvβ5 promote adenovirus internalization but not virus attachment. Cell. 1993;73:309–319. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]
- 51.Wickham T J, Tzeng E, Shears II L L, Roelvink P W, Li Y, Lee G M, Brough D E, Lizonova A, Kovesdi I. Increased in vitro and in vivo gene transfer by adenovirus vectors containing chimeric fiber proteins. J Virol. 1997;71:8221–8229. doi: 10.1128/jvi.71.11.8221-8229.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang W, Fang X, Branch C D, Mazur W, French B A, Roth J A. Generation and identification of recombinant adenovirus by liposome-mediated transfection and PCR analysis. BioTechniques. 1993;15:868–872. [PubMed] [Google Scholar]