Summary
Background
The integration of next-generation sequencing (NGS) comprehensive gene profiling (CGP) into clinical practice is playing an increasingly important role in oncology. Therefore, the HKU-HKSH Multi-disciplinary Molecular Tumour Board (MTB) was established to advance precision oncology in Hong Kong. A multicenter retrospective study investigated the feasibility of the HKU-HKSH MTB in determining genome-guided therapy for treatment-refractory solid cancers in Hong Kong.
Methods
Patients who were presented at the HKU-HKSH MTB between August 2018 and June 2022 were included in this study. The primary study endpoints were the proportion of patients who receive MTB-guided therapy based on genomic analysis and overall survival (OS). Secondary endpoints included the proportion of patients with actionable genomic alterations, objective response rate (ORR), and disease control rate (DCR). The Kaplan–Meier method was used in the survival analyses, and hazard ratios were calculated using univariate Cox regression.
Findings
122 patients were reviewed at the HKU-HKSH MTB, and 63% (n = 77) adopted treatment per the MTB recommendations. These patients achieved a significantly longer median OS than those who did not receive MTB-guided therapy (12.7 months vs. 5.2 months, P = 0.0073). Their ORR and DCR were 29% and 65%, respectively.
Interpretation
Our study demonstrated that among patients with heavily pre-treated advanced solid cancers, MTB-guided treatment could positively impact survival outcomes, thus illustrating the applicability of NGS CGPs in real-world clinical practice.
Funding
The study was supported by the Li Shu Pui Medical Foundation. Dr Aya El Helali was supported by the Li Shu Pui Medical Foundation Fellowship grant from the Li Shu Pui Medical Foundation. Funders had no role in study design, data collection, data analysis, interpretation, or writing of the report.
Keywords: Next-generation sequencing, Molecular tumour board, Sequence-directed therapy, Precision oncology
Research in context.
Evidence before this study
We searched PubMed for studies published, using the search terms “molecular tumor board” OR “molecular tumour board” AND “NGS” OR “next generation sequencing”. We found several studies that evaluated the clinical impact of MTBs. All published studies were performed in Europe or the United States, with the majority being single-institution studies. Of the previously published studies, 11–43% of patients received MTB-recommended targeted therapies. Furthermore, the ORR in the previously published studies ranged between 0 and 67%.
Added value of this study
We report the findings from the HKU-HKSH MTB cohort study of patients who underwent comprehensive genomic profiling. The primary study objective was to investigate the feasibility of the HKU-HKSH MTB to determine genome-guided therapy in tumour-agnostic advanced-stage disease. Results from our study demonstrated a high rate of patients harbouring actionable targets, and patients who adopted MTB-guided recommendations derived favourable ORR and DCR. To our knowledge, this is the first study reported in the region in a Chinese patient population.
Implications of all the available evidence
High-throughput NGS profiling is essential in delivering precision cancer medicine. However, the MTB bridges the gap and is an integral service for the safe and effective implementation of NGS CGP guided therapy into clinical practice. Prospective studies are needed to confirm the clinical importance of the MTB.
Introduction
High-throughput Next-generation sequencing (NGS) comprehensive gene profiling (CGP) is a vital component of the cancer care workflow of the 21st century. NGS CGPs have been incorporated into clinical practice to improve patient outcomes by identifying actionable drug targets.1 The widespread interrogation of the genomic landscape across cancer types may enable genome-guided therapy1 and drug discovery.2,3 NGS CGP has transformed the landscape of clinical trials in the era of precision cancer medicine by emphasising the importance of genomically stratified tumour-agnostic trials.4 Precision cancer medicine endorses a patient-centric approach, ensuring we adopt a tailor-made strategy unique to each cancer patient.5,6
Multi-disciplinary Molecular Tumour Boards (MTBs) have been established to aid frontline oncologists by integrating NGS CGPs to facilitate the implementation of precision cancer medicine into clinical practice.7, 8, 9 An integral role of the MTB is to enable the timely referral of patients to genomically stratified clinical trials.10,11 NGS testing is performed via commercial platforms, which differ significantly in their scope. For example, some studies have been limited to DNA-based testing of a selected group of known targetable cancer-related genes.11,12 Therefore, MTB harmonises treatment recommendations by comprehensively interrogating reported genomic alterations using an evidence-based approach.
To bridge the gap between scientific development and clinical application, a monthly multi-disciplinary MTB was established in 2018 through a collaboration between the University of Hong Kong (HKU) and the Hong Kong Sanatorium & Hospital (HKSH). The HKU-HKSH MTB aims to implement an evidence-based precision cancer medicine program to facilitate treatment selection for patients with advanced cancer in Hong Kong. The HKU-HKSH MTB is a territory-wide pan-cancer service for interrogating tumour genomic profiling in Hong Kong. The MTB is supported by specialists in oncology, surgery, pathology, genetics, bioinformatics, data science, and pharmacology. The mission of the HKU-HKSH MTB is to provide a platform for discussing and interpreting complex genomic findings with scientific experts and recommendations on optimal drug selection by clinical experts based on the latest evidence and information from ongoing clinical trials to integrate precision cancer medicine into clinical practice in Hong Kong. The vision of the HKU-HKSH MTB is to offer a service of excellence by providing personalised and cutting-edge cancer treatment to our patients.
This study conducted a retrospective clinical outcome analysis of patients with advanced solid cancers presented at the HKU-HKSH-MTB for review and treatment recommendations. The primary objective of our study was to demonstrate the survival impact of a comprehensively curated real-world retrospective MTB dataset to facilitate the management of patients with cancer in the era of precision oncology.
Methods
Study description
The Protocol was approved by the institutional ethics review boards of the University of Hong Kong/Hong Kong West Cluster (UW 22-514), Hong Kong Sanatorium (REC-2022-14), and the Hong Kong Children's Hospital (HKCH-REC-2020-068). This multicentre retrospective study was performed in accordance with the International Conference on Harmonization Guidelines for Good Clinical Practice and the Declaration of Helsinki. The primary study objective was to investigate the feasibility of the HKU-HKSH MTB to determine genome-guided therapy in tumour-agnostic treatment-refractory advanced-stage disease. Patients with NGS CGP reports evaluated at the HKU-HKSH MTB between August 2018 and June 2022 were deemed eligible for inclusion in this study. There were no prespecified exclusion criteria.
Study endpoints and data collection
The primary study endpoints were the proportion of patients who received MTB-guided therapy and overall survival (OS). OS was defined as the date of MTB discussion to the date of death or last follow-up. The data cut-off point was the 31st of October, 2022. Patients lost to follow-up or alive within the study duration were considered right censored. Secondary endpoints included the following: the proportion of patients with actionable genomic alterations [defined as a gene which may confer sensitivity to a clinically available therapy], objective response rate (ORR) [defined as partial response (PR) plus complete response (CR)], and disease control rate (DCR) [defined as stable disease (SD) ≥6 months plus PR plus CR]. Treatment responders were defined as patients who derived a DCR of at least SD ≥6 months, while non-responders were defined as patients with a best response of SD <6 months or progress disease (PD). We defined rapid progression as PD or death within 6 weeks from discussion at the MTB.
Electronic medical records were reviewed, and data were collected on patient demographics, oncology-related treatment history, tumour characteristics (e.g., tumour type, biopsy site, and tumour content), and reported NGS genomic variants. In addition, data from the HKU-HKSH MTB report were reviewed, and MTB recommendations were extracted. Response data based on MTB recommendations were also documented. NGS panel reports implemented in clinical practice in Hong Kong and evaluated at the MTB were issued by one of the following NGS platforms: Foundation One (http://www.foundationmedicine.com), ACT Genomics (http://www.actgenomics.com), Lucence (http://www.lucence.com), or Gaurdant 360 (http://www.guardanthealth.com). Notably, the scope of the HKU-HKSH MTB was not limited to the above NGS platforms. Across the NGS CGPs described above, profiling of tumour tissue (324–572 genes) or blood-derived circulating tumour DNA (ctDNA) (73–311 genes) was conducted by hybrid-capture or amplicon-based NGS with a 0.1–0.4% limit of detection of variant allele frequency and mean sequencing depth of ≥500X.
HKU-HKSKH molecular tumour board
A monthly multi-disciplinary MTB reviewed, interpreted, and discussed the results of the patients with genomically complex NGS reports. The HKU-HKSH MTB consists of multi-disciplinary and interdisciplinary expert panel members: oncologists, physicians with cancer subspecialties, molecular pathologists, cancer biologists, pharmacists, clinical geneticists, bioinformaticians, biostatisticians, and data scientists. The expert panel comprehensively discussed the genomic variants that were the likely tumour drivers, assessed the potential for matched therapies, and differentiated between missense mutations of unknown significance and putative drivers as follows: patient registration [structured MTB referral form and signed patient consent form are submitted to the HKU-HKSH MTB], discussion at the HKU-HKSH MTB, development of a personalised sequence specific recommendation [which included a referral to clinical trials, alignment to available target therapies], and longitudinal follow-up every 6 months to capture treatment response and survival data. Recommendations were summarised in a structured report format for all patients reviewed within 48 h of the case discussion. The considerations for targeted therapy included in the final MTB reports were informed using variant annotation databases.
Additionally, we used local databases to identify genomically matched clinical trials. MTB treatment recommendations followed the ESMO Scale for Clinical Actionability of Molecular Targets (ESCAT),13 providing clinical evidence-based criteria to prioritise sequence-directed therapy, clinical trial referral, or standard of care treatment based on physician choice and additional testing, including recommendations for germline testing. Notably, HKU-HKSH did not endorse any advice based on the preclinical evidence.
Data analysis
The details of our study are reported according to the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) Reporting Guidelines. Patient demographics, baseline characteristics, target treatment implemented, and tumour characteristics were presented as descriptive statistics and summarised as median (range) or count (percentage). Overall survival was calculated from the date of the MTB discussion to the last follow-up date. The data cut-off point was the 31st of October, 2022. Genetic alterations with frequency >5% are listed, and actionable targets detected are displayed as an oncoplot. For the co-occurrence of gene mutations, the magnitude of the co-mutation was expressed as a log odds ratio, and chi-square tests were performed; multiple testing adjustment was performed using the Benjamini-Hochberg method.14 The Kaplan–Meier method was used in the survival analyses. In addition, the proportional hazards assumption for Cox regression was tested, and hazard ratios were calculated using univariate Cox regression. Statistical analyses were performed using R (version 4.2.2) and survival analysis and proportional hazards assumption test using the Grambsch-Therneau score test implemented in R package survival (version 3.4.0). Oncoplot was illustrated using the R package ComplexHeatmap (version 2.15.1).
Funding source
The study was supported by the Li Shu Pui Medical Foundation. Aya El Helali was supported by the Li Shu Pui Fellowship grant from the Li Shu Pui Medical Foundation. Funders had no role in study design, data collection, data analysis, interpretation, or writing of the report.
Results
Patient demographics and baseline molecular characteristics
One hundred twenty-two clinically complex cases were discussed at the HKU-HKSH MTB between August 2018 and June 2022 (Table 1). 64 patients (52.5%) were male, and 58 (47.5%) were female (Table 1). The median patient age was 60 (Range 11–95) (Table 1).
Table 1.
Demonstrates the patient demographics in the overall population.
| Patient demographics and baseline charactericstics | |
| Patients | 122 |
| Age | 60 (11–95) |
| Line of treatment | 3 (2–4) |
| Gender | |
| Female | 58 (47.5) |
| Male | 64 (52.5) |
| Tumour type | |
| Breast cancer | 9 (7.4) |
| Endocrine cancer | 2 (1.6) |
| Gastrointestinal cancer | 7 (5.7) |
| Genitourinary cancer | 7 (5.7) |
| Gynecologic cancer | 9 (7.4) |
| Head & neck cancer | 4 (3.3) |
| Hepato-biliary pancreatic cancer | 22 (18.0) |
| Pediatric malignancy | 3 (2.5) |
| Primary CNS tumour | 23 (18.9) |
| Sarcoma | 8 (6.6) |
| Thoracic cancer | 20 (16.4) |
| Carcinoma of unknown origin | 8 (6.6) |
| MS-status | |
| MSI-L | 2 (1.6) |
| MSS | 99 (81.1) |
| MSI-H | 3 (2.5) |
| Not reported | 18 (14.8) |
| TMB | |
| TMB-L | 91 (74.6) |
| TMB-H | 17 (13.9) |
| Not reported | 14 (11.5) |
| Germline mutation | |
| Detected | 6 (4.9) |
| Not detected | 84 (68.9) |
| Testing not performed | 32 (26.2) |
MS-status: Microsatelite status, MSS: Microsatelite status, MSI-L: Microsatelite instable low, MSI-H: Microsatelite instable high, TMB: Tumour mutational burden, TMB-L: Low tumour mutational burden, TMB-H: High tumour mutational burden.
Among the patient population, the most common tumour types were primary central nervous system (CNS) (n = 23, 18.9%), hepato-biliary-pancreatic (HBP) (n = 22, 18.0%), and thoracic cancers (n = 20, 16.4%) (Table 1). Microsatellite instability was detected in 2.5% (n = 3) (Table 1). A high tumour mutation burden (TMB-H) was detected in 13.9% of patients (n = 17) (Table 1). Furthermore, germline mutation status was confirmed in 6 patients (4.9%) (Table 1). Of these, germline mutations in mismatch repair (MMR) (n = 4), BRCA2 (n = 1), and ATM (n = 1) were validated in the tissues and blood, respectively.
Molecular profile of the MTB cohort
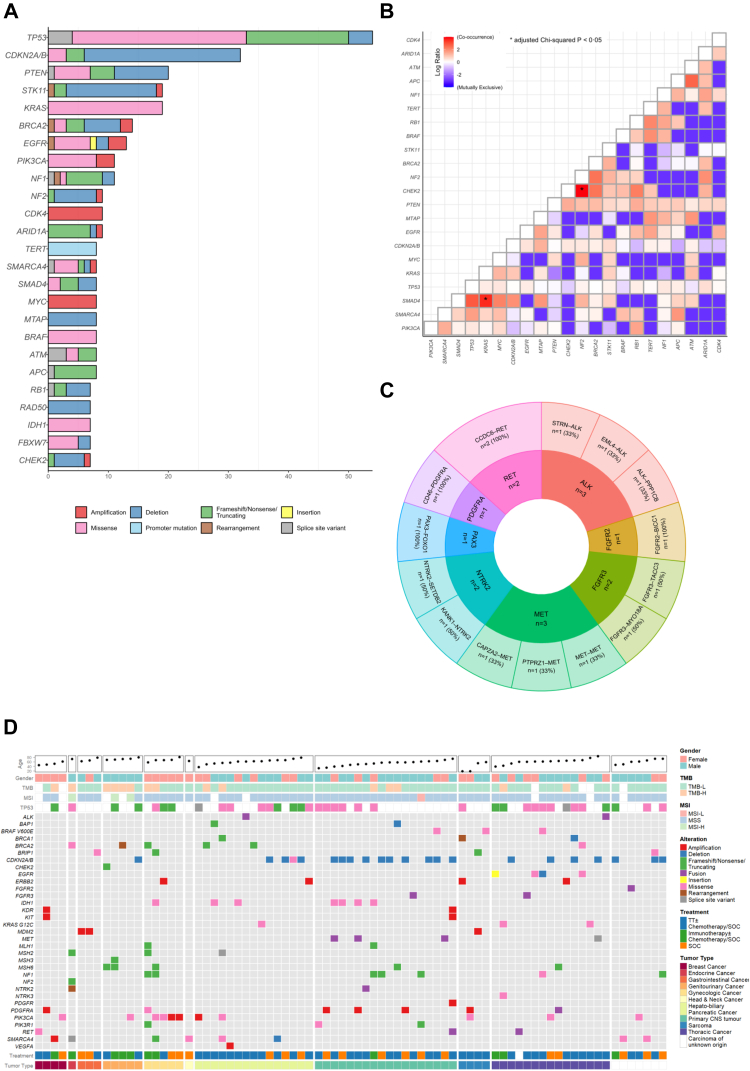
Genes reported in the MTB cohort with a frequency of >5% are shown in Fig. 1A. Missense mutations and deletions are the most frequent alterations reported in the patient population. The most frequently mutated gene was TP53 (44.3%), more than half of which were missense mutations. This was followed by CDKN2A/B (26.2%), with the majority being deletions, PTEN (16.4%), KRAS (15.6%), and STK11 (15.6%), each with variable alterations. Missense mutations in TP53 were mainly observed in mutational hotspots R248 and R273. CDKN2A/B deletions were most commonly detected in 52.2% of primary CNS tumours, while PTEN deletions were found in 25% of sarcomas and 21.7% of primary CNS tumours. Co-occurring mutations were most frequently observed between KRAS:SMAD4 (adjusted P = 0.049) and NF2:CHEK2 (adjusted P = 0.020) (Fig. 1B). The most frequently reported gene fusions were MET (n = 3) and ALK (n = 3) (Fig. 1C). Patients with primary CNS cancers had the highest frequency of actionable fusion genes (MET n = 3; RET, n = 1; FGFR3 n = 1; NTRK2 n = 1), followed by those with non-small cell lung cancer (RET n = 1; PDGFRA n = 1; ALK n = 1; FGFR3 n = 1) (Supplemental Table S1).
Fig. 1.
(A) demonstrates the genes reported in the overall patient population with a frequency >5%, (B) demonstrates the co-occurring genes in the overall patient population, (C) demonstrates the gene fusions reported in the overall patient population, (D) the oncoplot, clustered based on tumor type, illustrates the actionable mutations in the patient. TMB-L: Tumour mutational burden low, TMB-H: Tumour mutational burden high, MSI-L: Microsatellite instability low, MSS: Microsatellite stable, MSI-H: Microsatellite instability high, TT: Target therapy, SOC: Standard of care, ∗ represents adjusted chi-squared P < 0.05.
The patient population consisted of 12 tumour types (Table 1), and over 500 alterations were detected; 33 (6.4%) were actionable targets (Fig. 1D). In the overall patient population, 78 (63.9%) patients harboured one or more actionable targets; the median number of targets in each patient was one (IQR:1–2). Nine actionable targets (BRCA1, BRCA2, BRIP1, CHEK2, MLH1, MSH2, MSH3, MSH6, and BAP1) were associated with DNA Damage Response (DDR). In addition, the actionable targets frequently detected in our patient cohort were CDKN2A/B (28.2%), PIK3CA (14.1%), and SMARCA4 (10.3%). The tumour types with a high incidence of actionable targets were primary CNS (n = 18, 14.8%), thoracic (n = 15, 12.3%), and HPB (n = 15, 12.3%) cancers.
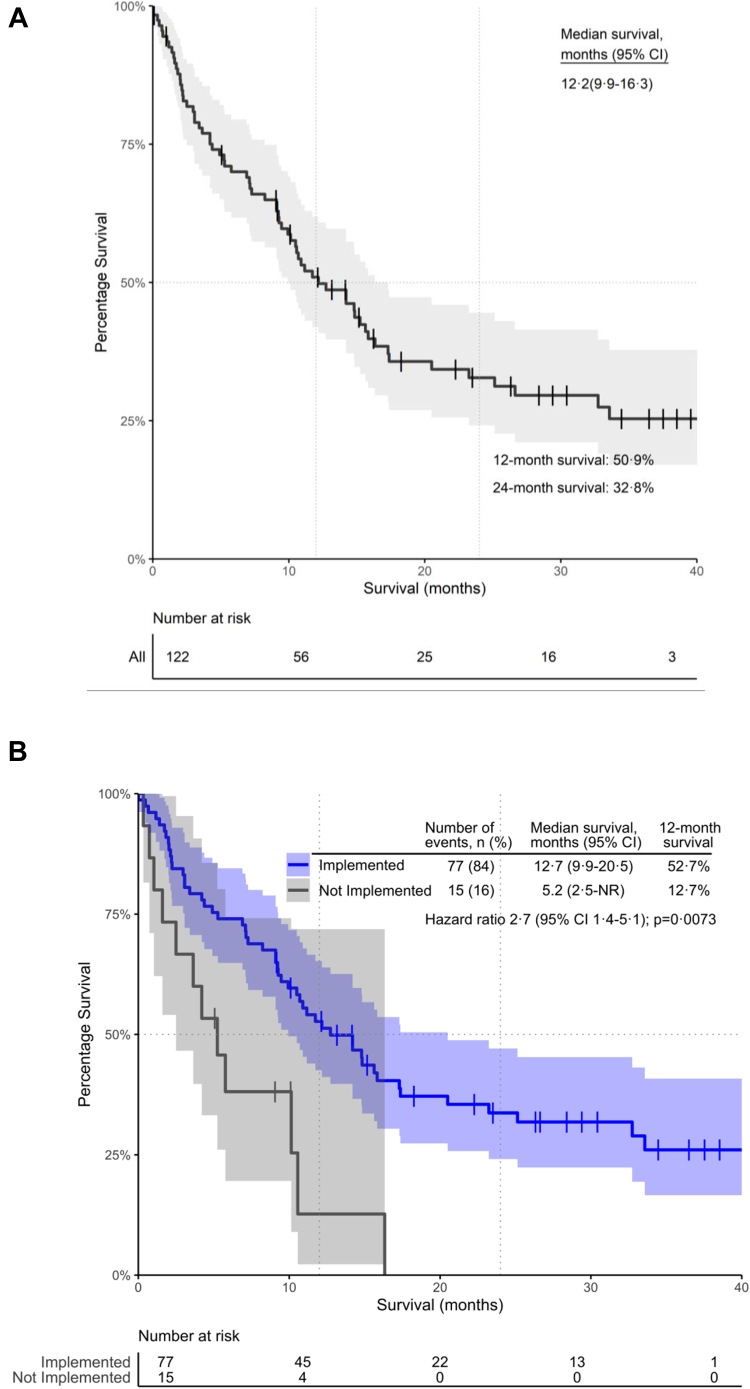
Impact of the MTB on survival
The median OS of the retrospective MTB patient population was 12.2 months (95% CI: 9.9–16.3). The 12- and 24-month overall survival rates were 50.9% and 32.8%, respectively (Fig. 2A). We evaluated survival outcomes based on whether MTB recommendations were implemented in the patient's cancer care pathway (Fig. 2B). Notably, 23 (18.9%) patients were lost to follow-up, 7 (5.7%) patients had not yet exhausted standard of care (SOC) options and therefore had not adopted the MTB recommendations at the time of data cut-off, and an additional 15 (12.3%) patients did not proceed with treatment recommendations as per the MTB report. The leading justification for not integrating MTB recommendations into clinical practice is either patient choice (n = 4) or the financial burden of the treatment strategy (n = 7). Among the patients where MTB recommendations were implemented, the median survival was 12.7 months, compared to only 5.2 months in patients who did not proceed with the MTB treatment strategy (P = 0.0073, HR = 2.7, 95% CI: 1.4–5.1; proportional hazard assumption test P = 0.61) (Fig. 2B).
Fig. 2.
Kaplan–Meier Curve illustrating (A) the Overall Survival In the overall MTB patient population, (B) the Overall Survival stratified based on the implementation of the MTB recommendations in the overall population. 95% CI: 95% confidence interval. The shaded area represents the confidence interval bands.
63% (n = 77) of the MTB patient population adopted the treatment strategies. 36 (46.8%) patients received targeted therapy with or without SOC, 28 (36.3%) received SOC, and 13 (16.9%) received immunotherapy with or without SOC, as per the MTB report. Sequence-directed tyrosine kinase inhibitors targeting actionable BRAF V600E (n = 1), CDKN2A/B (n = 3), FGFR (n = 2), ERBB2 (n = 2), KIT (n = 1), KRAS (n = 1), MEK (n = 3), MET (n = 2), PIK3CA (n = 1), SMARCB (n = 1), and RTK (n = 7) were implemented in our cohort (Supplemental Table S2). Additionally, PARP inhibitors with or without SOC were administered to 11 patients (Supplemental Table S2). Three patients with primary CNS cancer received crizotinib and derived survival benefit (median survival 13 months, range 10.7–15.6 months) (Supplemental Table S1). Interestingly, one patient with MSI, TMB-H papillary thyroid cancer harbouring an NTRK 2 fusion, received pembrolizumab with an OS of 42.6 months at the time of data cut-off (Supplemental Table S1).
Furthermore, we evaluated survival outcomes based on treatment responses according to the MTB recommendations (Table 2). The ORR was 28.6% (median OS 25.1 month, 95% CI: 15.6—NR) (Table 2). Furthermore, the disease control rate (DCR) was 65% (median OS 26.6 months, 95% CI: 17.3—NR) (Table 2).
Table 2.
Demonstrates the tumour reponse data, illustrating the tumour response data, ORR and DCR.
| Tumour response data | Number of events n (%) | Median survival months (95% CI) | 12-month survival |
|---|---|---|---|
| CR | 6 (7.8) | ||
| PR | 16 (20.8) | ||
| SD | 28 (36.3) | ||
| PD | 27 (35.1) | ||
| ORR | 22 (28.6) | 25.1 (15.6-NR) | 81.1% |
| DCR | 50 (65.0) | 32.7 (15.8-NR) | 75.4% |
CR: Complete response, PR: Partial response, SD: Stable disease ≥6 months, PD: Progressive disease, ORR: Objective response rate (CR + PR), DCR: Disease control rate (CR + PR + SD ≥6 months), CI: Confidence interval, NR: Not reached, n: Number of events.
Discussion
This study advocates for a multi-disciplinary evidence-based MTB program. The HKU-HKSH MTB is a first-in-kind multicentre pan-cancer precision oncology service delivered to a predominantly Chinese population. The HKU-HKSH MTB has set a gold standard clinical service of excellence and cancer management model for other institutions to follow within the region. The data presented in this study exhibits the pivotal role of multi-disciplinary MTBs in facilitating the delivery of precision oncology and guiding the management of cancer patients.
Firstly, the results of our study demonstrated the clinical benefit of integrating high-throughput NGS CGPs into real-world clinical practice. Notably, a large proportion of potentially actionable genetic alterations, including mutations and gene fusions, have been identified by NGS, leading to the recommendation of sequence-directed therapy in 63% (n = 77) of the MTB patient population. This illustrates that genomic profiling of patients with advanced treatment refractory cancer is feasible and effective. Furthermore, patients who received an MTB-guided treatment strategy had statistically significant survival benefits, with a considerable disease control rate (65.0%), thus demonstrating the success of integrative genomic profiling in patients with advanced solid cancer in improving their clinical outcomes. Our study findings align with the published literature demonstrating the feasibility of establishing and implementing a molecular tumour board, increasing the appropriate prescription of MTB-guided therapy, and establishing a unique N-of-1 genome-matched strategy.15 Of the previously published studies, 11–43%15 of patients received MTB-recommended therapies. In contrast, 63% of our population received and adopted the MTB-guided strategies. Furthermore, the ORR and DCR in the previously published studies ranged between 0–67% and 42–100%, respectively,15 while our study demonstrates an ORR of 28.6% and a DCR of 65.0%. In addition, our study demonstrates that 78 patients (63.9%) harboured actionable targets, consistent with the current published data, where 36–100% of patients discussed at the MTB harboured actionable mutations.15 Furthermore, the practical implementation of precision oncology in our study and others could be attributed to the presence of the MTB. Throughout or study period, genomic profiles were individually interpreted and reviewed at monthly MTB meetings to facilitate the selection of sequence-directed therapy. This corresponds with other findings that the MTB could improve treatment decisions and patient management.16, 17, 18, 19, 20, 21, 22, 23, 24, 25
Importantly, our work highlights an important public health issue, specifically the health disparity and social inequality amplified in the era of precision-guided treatment. Seven patients were unable to proceed with the MTB recommendations due to the financial impact of sequence-matched target therapy. These public health issues will be further explored in our ongoing prospective study.
Despite the survival impact illustrated in this study, some limitations must be addressed. First, the study was retrospective with small sample size, and thus, may be subject to various biases and confounding factors. Diverse patient demographics, cancer types, and genomic characteristics make it difficult to include an appropriately matched control group. Moreover, this retrospective cohort study did not evaluate specific patient characteristics, including performance status and comorbidities. Data on MS status and TMB were unavailable in some NGS CGP reports at the time of MTB discussion; these two characteristics were only presented descriptively and not included in the time-to-event analyses. Thus, the potential problems of biased estimates and loss of power to detect associations caused by missing data in covariates are minimised. Rapid disease progression is a potential confounding factor in the survival analysis in a subset of patients, n = 3 patients in the “not implemented” cohort and n = 5 in the “implemented” cohort. In collaboration with the University of Hong Kong Shenzhen hospital and the HKU-HKSH MTB, we are conducting a prospective study to validate our MTB findings and address the potential social and geographical disparities.
In conclusion, our study suggests that integrating NGS CGPS into the cancer care pathway by implementing a multi-disciplinary MTB can positively impact survival. However, further optimisation of genomic profiling and the MTB platform is necessary to advance the delivery of precision oncology.
Contributors
Conceptualization: AEH, TCL, EYLK, DJHS, JWHW, LWTC, APYL, VHFL, WTN, AWML, ESKM.
Methodology: AEH, TCL, EYLK, DJHS, JWHW, LWTC, APYL, VHFL, WTN, AWML, ESKM, CKC, HHL, STSL, GCFC, CHLW.
Formal analysis: AEH, EYLK, DJHS, JWHW, LWTC, CHLW.
Investigation: AEH, TCL, EYLK, DJHS, JWHW, LWTC, APYL, VHFL, WTN, AWML, ESKM.
Resources: AEH, TCL, EYLK.
Data Curation: AEH, TCL, EYLK, DJHS, JWHW, LWTC, APYL.
Writing–Original Draft: AEH, EYLK, CKC, WTN, AWML, ESKM, CHLW.
Writing–Review & Editing: AEH, TCL, EYLK, DJHS, CKC, JWHW, LWTC, JKSL, APYL, HHL, STAL, GCFC, VHFL, WTN, AWML, ESKM, CHLW.
Visualization: AEH, TCL, EYLK, DJHS, CKC, JWHW, LWTC, JKSL, APYL, HHL, STAL, GCFC, VHFL, WTN, AWML, ESKM, CHLW.
Supervision: AEH, DJHS, JWHW, LWTC.
Project administration: AEH.
Funding acquisition: AEH.
Data sharing statement
The data analysed during the current study are available from the corresponding author on reasonable request.
Declaration of interests
All authors declare no competing interests.
Acknowledgements
We want to acknowledge the administrative support by Ms. Ryon Heung.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.lanwpc.2023.100775.
Contributor Information
Aya El Helali, Email: ahelali@hku.hk.
Anne W.M. Lee, Email: awmlee@hku.hk.
Edmond S.K. Ma, Email: eskma@hksh.com.
Appendix A. Supplementary data
References
- 1.Mosele F., Remon J., Mateo J., et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: a report from the ESMO Precision Medicine Working Group. Ann Oncol. 2020;31(11):1491–1505. doi: 10.1016/j.annonc.2020.07.014. [DOI] [PubMed] [Google Scholar]
- 2.Szustakowski J.D., Balasubramanian S., Kvikstad E., et al. Advancing human genetics research and drug discovery through exome sequencing of the UK Biobank. Nat Genet. 2021;53(7):942–948. doi: 10.1038/s41588-021-00885-0. [DOI] [PubMed] [Google Scholar]
- 3.Mateo J., Steuten L., Aftimos P., et al. Delivering precision oncology to patients with cancer. Nat Med. 2022;28(4):658–665. doi: 10.1038/s41591-022-01717-2. [DOI] [PubMed] [Google Scholar]
- 4.Subbiah V., Cassier P.A., Siena S., et al. Pan-cancer efficacy of pralsetinib in patients with RET fusion–positive solid tumors from the phase 1/2 ARROW trial. Nat Med. 2022;28(8):1640–1645. doi: 10.1038/s41591-022-01931-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malone E.R., Oliva M., Sabatini P.J.B., Stockley T.L., Siu L.L. Molecular profiling for precision cancer therapies. Genome Med. 2020;12(1):8. doi: 10.1186/s13073-019-0703-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fountzilas E., Tsimberidou A.M., Vo H.H., Kurzrock R. Clinical trial design in the era of precision medicine. Genome Med. 2022;14(1):101. doi: 10.1186/s13073-022-01102-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartoletti M., Bergamini A., Giannone G., et al. A fully virtual and nationwide molecular tumor board for gynecologic cancer patients: the virtual experience of the MITO cooperative group. Int J Gynecol Cancer. 2022 doi: 10.1136/ijgc-2022-003425. [DOI] [PubMed] [Google Scholar]
- 8.Tamborero D., Dienstmann R., Rachid M.H., et al. The Molecular Tumor Board Portal supports clinical decisions and automated reporting for precision oncology. Nat Cancer. 2022;3(2):251–261. doi: 10.1038/s43018-022-00332-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kohno T., Kato M., Kohsaka S., et al. C-CAT: the national datacenter for cancer genomic medicine in Japan. Cancer Discov. 2022;12(11):2509–2515. doi: 10.1158/2159-8290.CD-22-0417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klein H., Mazor T., Siegel E., et al. MatchMiner: an open-source platform for cancer precision medicine. NPJ Precis Oncol. 2022;6(1):69. doi: 10.1038/s41698-022-00312-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keller R.B., Mazor T., Sholl L., et al. Programmatic precision oncology decision support for patients with gastrointestinal cancer. JCO Precis Oncol. 2023;7 doi: 10.1200/PO.22.00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller R.W., Hutchcraft M.L., Weiss H.L., et al. Molecular tumor board–assisted care in an advanced cancer population: results of a phase II clinical trial. JCO Precis Oncol. 2022;6 doi: 10.1200/PO.21.00524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mateo J., Chakravarty D., Dienstmann R., et al. A framework to rank genomic alterations as targets for cancer precision medicine: the ESMO Scale for Clinical Actionability of molecular Targets (ESCAT) Ann Oncol. 2018;29(9):1895–1902. doi: 10.1093/annonc/mdy263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodol) 1995;57(1):289–300. [Google Scholar]
- 15.Larson K.L., Huang B., Weiss H.L., et al. Clinical outcomes of molecular tumor boards: a systematic review. JCO Precis Oncol. 2021;5:1122–1132. doi: 10.1200/PO.20.00495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwaederle M., Parker B.A., Schwab R.B., et al. Molecular tumor board: the university of California-san diego moores cancer center experience. Oncologist. 2014;19(6):631–636. doi: 10.1634/theoncologist.2013-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tafe L.J., Gorlov I.P., de Abreu F.B., et al. Implementation of a molecular tumor board: the impact on treatment decisions for 35 patients evaluated at dartmouth-hitchcock medical center. Oncologist. 2015;20(9):1011–1018. doi: 10.1634/theoncologist.2015-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dalton W.B., Forde P.M., Kang H., et al. Personalised medicine in the oncology clinic: implementation and outcomes of the johns hopkins molecular tumor board. JCO Precis Oncol. 2017;1:1–19. doi: 10.1200/PO.16.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knepper T.C., Bell G.C., Hicks J.K., et al. Key lessons learned from moffitt's molecular tumor board: the clinical genomics action committee experience. Oncologist. 2017;22(2):144–151. doi: 10.1634/theoncologist.2016-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bourien H., Lespagnol A., Campillo-Gimenez B., et al. Implementation of a molecular tumor board at a regional level to improve access to targeted therapy. Int J Clin Oncol. 2020;25(7):1234–1241. doi: 10.1007/s10147-020-01661-6. [DOI] [PubMed] [Google Scholar]
- 21.Kato S., Kim K.H., Lim H.J., et al. Real-world data from a molecular tumor board demonstrates improved outcomes with a precision N-of-One strategy. Nat Commun. 2020;11(1):4965. doi: 10.1038/s41467-020-18613-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heinrich K., Miller-Phillips L., Ziemann F., et al. Lessons learned: the first consecutive 1000 patients of the CCCMunichLMU Molecular Tumor Board. J Cancer Res Clin Oncol. 2022;149(5):1905–1915. doi: 10.1007/s00432-022-04165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charo L.M., Eskander R.N., Sicklick J., et al. Real-world data from a molecular tumor board: improved outcomes in breast and gynecologic cancers patients with precision medicine. JCO Precis Oncol. 2022;6 doi: 10.1200/PO.20.00508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Louie B.H., Kato S., Kim K.H., et al. Precision medicine-based therapies in advanced colorectal cancer: the university of California san diego molecular tumor board experience. Mol Oncol. 2022;16(13):2575–2584. doi: 10.1002/1878-0261.13202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dorman K., Zhang D., Heinrich K., et al. Precision oncology in pancreatic cancer: experiences and challenges of the CCCMunichLMU molecular tumor board. Target Oncol. 2023;18(2):257–267. doi: 10.1007/s11523-023-00950-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.