Abstract
Geminiviruses are plant viruses with circular single-stranded DNA (ssDNA) genomes encapsidated in double icosahedral particles. Tomato leaf curl geminivirus (ToLCV) requires coat protein (CP) for the accumulation of ssDNA in protoplasts and in plants but not for systemic infection and symptom development in plants. In the absence of CP, infected protoplasts accumulate reduced levels of ssDNA and increased amounts of double-stranded DNA (dsDNA), compared to accumulation in the presence of wild-type virus. To determine whether the gene 5 protein (g5p), a ssDNA binding protein from Escherichia coli phage M13, could restore the accumulation of ssDNA, ToLCV that lacked the CP gene was modified to express g5p or g5p fused to the N-terminal 66 amino acids of CP (CP66:6G:g5). The modified viruses led to the accumulation of wild-type levels of ssDNA and high levels of dsDNA. The accumulation of ssDNA was apparently due to stable binding of g5p to viral ssDNA. The high levels of dsDNA accumulation during infections with the modified viruses suggested a direct role for CP in viral DNA replication. ToLCV that produced the CP66:6G:g5 protein did not spread efficiently in Nicotiana benthamiana plants, and inoculated plants developed only very mild symptoms. In infected protoplasts, the CP66:6G:g5 protein was immunolocalized to nuclei. We propose that the fusion protein interferes with the function of the BV1 movement protein and thereby prevents spread of the infection.
Geminiviruses are plant pathogens that cause significant yield losses in crop plants in many countries (4, 14, 18, 35). Different members are transmitted by whiteflies or leafhoppers (9, 26). Most of the whitefly-transmitted geminiviruses have bipartite genomes, while all the leafhopper-transmitted geminiviruses and some of the whitefly-transmitted geminiviruses have monopartite genomes. The monopartite genomes (2,566 to 3,028 nucleotides [nt]) encode proteins required for replication, encapsidation, and movement, while in the bipartite viruses, movement functions are encoded by a second genome component of a similar size (9, 20, 50).
Geminiviruses replicate via a rolling-circle mechanism analogous to the replication of bacteriophages with single-stranded DNA (ssDNA) genomes (44, 46). The incoming geminivirus ssDNA is converted by host enzymes to double-stranded DNA (dsDNA), which in turn serves as a template for the transcription of early, replication-associated genes on the complementary-sense strand (13, 16, 17, 25, 48). Replication initiator protein (Rep or AC1) is the only viral protein required for replication (13, 16). In bipartite geminiviruses, a second protein (AC3) enhances replication (49). AC2, another early gene product, transactivates the expression of the coat protein (CP) gene on the virion-sense strand (47). While CP is not required for replication of the virus in protoplasts or plants, mutations in CP lead to dramatic decreases in the accumulation of ssDNA in protoplasts or plants without affecting the accumulation of dsDNA (5, 27, 52). On the other hand, tomato golden mosaic virus CP mutations have no effect on DNA accumulation in plants (6, 15) but reduce ssDNA accumulation and increase dsDNA accumulation in protoplasts (49). In plants, the lack of CP results in a complete loss of infectivity of monopartite viruses (3, 27, 38) but not bipartite viruses (6, 15, 32, 39).
CP may influence the ratios of ssDNA and dsDNA levels in a passive manner by depleting the ssDNA that is available for conversion to dsDNA through encapsidation, by modulating ssDNA synthesis, or both. No evidence is available for how CP influences ssDNA accumulation in geminiviruses. In tomato leaf curl virus from New Delhi (ToLCV-Nde, hereafter referred as ToLCV), a geminivirus with a bipartite genome, disrupting the synthesis of wild-type CP resulted in a drastic reduction in ssDNA accumulation and a three- to fivefold increase in dsDNA accumulation in infected protoplasts (33). Inoculated plants, however, developed severe symptoms and accumulated wild-type levels of dsDNA and low levels of ssDNA. To better understand the role of CP in replication, we determined whether a heterologous ssDNA binding protein could complement CP function in ssDNA accumulation. We show here that ToLCV modified to express the ssDNA binding gene 5 protein (g5p) from Escherichia coli phage M13 in place of CP accumulates ssDNA to wild-type levels in protoplasts but fails to move efficiently in plants.
MATERIALS AND METHODS
Plasmid constructs.
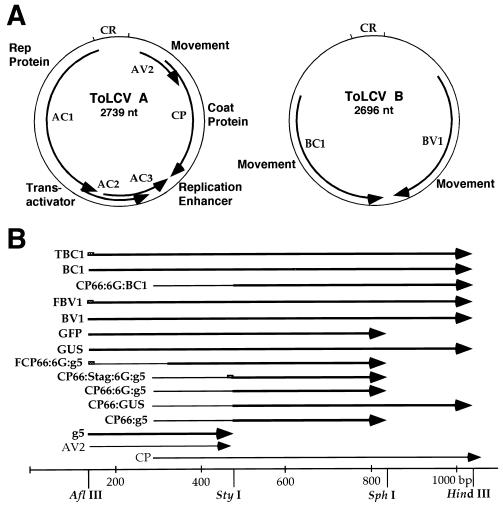
Infectious clones of the A and B components of ToLCV (32) were used to generate the virus constructs used in this study. The genome organization of ToLCV and a schematic representation of the virus constructs used in this study are shown in Fig. 1, and detailed descriptions and methods of construction of each of the plasmids are summarized in Table 1. Partial head-to-tail dimers made from these constructs were used to infect Nicotiana benthamiana plants and N. tabacum BY2 protoplasts.
FIG. 1.
Genome organization and schematic representation of constructs of ToLCV used in this study. (A) Genome organization of ToLCV showing the ORFs and their functions. CR, common region for both components. (B) Linear physical map of AV2 and CP regions of ToLCV with nucleotide positions and relevant restriction enzyme sites (bottom). The positions of different gene replacements are shown above the linear map. Note that the gene replacements shown are not to the scale. Descriptions of the constructs are given in Table 1.
TABLE 1.
Description and method of construction of viral DNAs used in this study
| Construct | Description and method of construction |
|---|---|
| AV2−CP− | A double mutant of AV2 and CP in which the Met1 codon of AV2 was changed to a termination codon and the Arg66 codon of CP was frameshifted. The mutant was described earlier as M1te/R66fr (33). |
| g5AV2−CP− | A 264-bp sequence coding for g5p from the bacteriophage M13mp18 vector was amplified by PCR (10 cycles) and cloned between the AflIII (nt 125) and StyI (nt 479) sites, resulting in the replacement of the AV2 ORF and overlapping 5′ CP ORF sequences with the g5p gene. |
| g5−AV2−CP− | A negative control for the g5AV2−CP− construct in which the Met1 codon of g5p was mutated to a termination codon. |
| CP− | A mutant of CP made by end filling and religation at the unique StyI site (nt 479), causing a frameshift at the Arg66 codon and termination after amino acid (aa) 69. The mutant was described earlier as R66fr (33). |
| CP66:g5 | A 264-bp sequence coding for g5p from the M13mp18 vector was amplified by PCR (10 cycles) and cloned between the StyI (nt 479) and SphI (nt 836) sites, resulting in the fusion of the g5p sequence to the Arg66 codon of CP. |
| CP66:6G:g5 | Similar to CP66:g5, except that an oligonucleotide coding for 6 glycines was inserted between the codons for Arg66 of CP and Met1 of g5p. |
| CP66:g5− | A negative control in which the Arg66 codon of CP66:g5 was frameshifted. |
| CP66:Stag:6G:g5 | Similar to CP66:6G:g5, except that a sequence coding for the 15-aa Stag peptide epitope (KETAAAKFERQHMDS [23]) was inserted after the Arg66 codon of CP. The Stag epitope was inserted to immunolocalize the CP66:6G:g5 protein in protoplasts by use of S protein coupled to FITC. |
| FCP66:6G:g5 | A sequence coding for the 9-aa Flag peptide epitope (MDYKDDDDK [19]) was added before the Met1 codon of CP66:6G:g5 and cloned between AflIII (nt 125) and SphI (nt 836). The AV2 ORF was deleted. The Flag epitope was added to immunoprecipitate the CP66:6G:g5 protein from protoplasts by use of anti-Flag antibody. |
| CP66:GUS | A 1,806-bp DNA fragment coding for GUS (21) was PCR amplified (10 cycles) and cloned between the StyI (nt 479) and HindIII (nt 1041) sites of the A component. The HindIII site was created at the codon for Tyr251 of CP (15 bp before the termination codon [33]), facilitating the replacement of the CP sequence with other sequences. |
| GUSAV2−CP− | A 1,869-bp NcoI-EcoRI DNA fragment coding for GUS was cloned between the AflIII (nt 125) and HindIII (nt 1041) sites of the A component after blunt ending the EcoRI site on the GUS gene and the HindIII site on the A-component DNA. |
| GFPAV2−CP− | A 717-bp NcoI-BamHI DNA fragment coding for GFP (S65C, M153T, V163A [37]) was cloned between the AflIII (nt 125) and SphI (nt 836) sites of the A component after blunt ending the BamHI site on the GFP gene and the SphI site on the A-component DNA. |
| BV1AV2−CP− | An 849-bp sequence coding for BV1 from the B component of ToLCV was amplified by PCR (10 cycles) and cloned between the AflIII (nt 125) and HindIII (nt 1041) sites of the A component. |
| FBV1AV2−CP− | Similar to BV1AV2−CP−, except that the sequence coding for the 9-aa Flag peptide was added before the Met1 codon of BV1. The Flag epitope was added to immunolocalize the BV1 protein in protoplasts by use of anti-Flag antibody. |
| BC1AV2−CP− | An 882-bp sequence coding for BC1 from the B component of ToLCV was amplified by PCR (10 cycles) and cloned between the AflIII (nt 125) and HindIII (nt 1041) sites of the A component. |
| TBC1AV2−CP− | Similar to BC1AV2−CP−, except that the sequence coding for the 11-aa T7 epitope (MASMTGGQQMG [24]) was added before the Met1 codon of BC1. The T7 tag epitope was added to immunolocalize the BC1 protein in protoplasts by use of anti-T7 tag antibody. |
| CP66:6G:BC1 | A 900-bp sequence coding for 6 glycines and BC1 from the B component of ToLCV was amplified by PCR (10 cycles) and cloned between the StyI (nt 479) and HindIII (nt 1041) sites. |
| BC1− | B-component DNA in which a frameshift mutation of BC1 was created by deletion of the 3′ overhang and religation at the PstI site (nt 2075). The mutant was described earlier as BC1M (33). |
Protoplast and plant inoculations.
N. benthamiana plants (2-week-old seedlings grown in Magenta boxes) and protoplasts isolated from suspensions of BY2 cells were infected with viral DNAs as described earlier (32, 33). Protoplasts were collected from cultures 48 h postinoculation for DNA isolation, immunoprecipitation reactions, and Western blot analysis. Plants were scored for symptoms, and the newly formed upper leaves were collected for Southern blot analysis 22 to 25 days following inoculation. To study the local and systemic movements of the virus expressing green fluorescent protein (GFP) (8), bottom leaves of 4-week old seedlings (10 plants per construct) were inoculated. Inoculated leaves and noninoculated upper leaves were observed at 3-day intervals for 15 days under a fluorescence microscope for the detection of fluorescence emitted by GFP. In all experiments that involved plants, wild-type B-component DNA, which is essential for systemic spread and symptom development, was included.
Southern blotting.
Total DNA was isolated from protoplasts (28) and plants (11), electrophoresed in 1% agarose gels (without ethidium bromide), and transferred to Hybond nylon membranes (Amersham, Arlington Heights, Ill.) by standard protocols (41). Hybridization reactions were performed with a randomly primed 32P-labeled A-component-specific probe (the 900-bp AflII-PstI fragment containing open reading frames [ORFs] for AC1, AC2, and AC3). The amounts of viral ssDNA and dsDNA (supercoiled, linear, open circular, and dimeric forms) were quantitated by exposing the Southern blots to storage phosphor screen plates and determining counts on a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.). The ssDNA form was confirmed by its susceptibility to S1 and mung bean nucleases (33). In the absence of ethidium bromide, the supercoiled viral DNA form migrates ahead of the ssDNA form.
Immunoprecipitation and Western blotting.
For immunoprecipitation reactions, protoplasts infected with the virus A component expressing the CP66:6G:g5 protein tagged with the Flag epitope (FCP66:6G:g5) (Table 1) were lysed by use of a hand-held Polytron with Nonidet P-40 (NP-40) buffer (50 mM Tris-HCl [pH 7.5], 1% NP-40, 0.15, 0.25, 0.50, 0.75, or 1.0 M NaCl) or radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate) containing a cocktail of protease inhibitors (Boehringer Mannheim Biochemicals, Indianapolis, Ind.). Cell debris was removed by centrifugation at 4°C for 10 min at 15,000 × g. Lysates were immunoprecipitated with anti-Flag monoclonal antibody M2 covalently linked to agarose (Sigma, St. Louis, Mo.). Immune complexes were washed four times with NP-40 or RIPA buffer and once with Tris-buffered saline (50 mM Tris-HCl [pH 7.5], 150 mM NaCl). Half of each sample was heated in Laemmli sample buffer, fractionated by SDS-polyacrylamide gel electrophoresis (13% acrylamide), and transferred to a polyvinylidene difluoride membrane (Schleicher & Schuell, Inc., Keene, N.H.). Immunoprecipitated protein was visualized with anti-Flag antibody M2 by use of enhanced chemiluminescence-Western blot reagents (Pierce, Rockford, Ill.). The remaining half of each immune complex collected by this procedure was used for isolating viral DNA. Whole-cell protein extracts for direct Western blotting were prepared by boiling the protoplast pellets with an equal volume of 2× Laemmli sample buffer.
Immunofluorescence.
Protoplasts transfected with viral constructs were cultured on chamber slides (Nalge Nunc, Rochester, N.Y.) for 48 h, fixed with 3% paraformaldehyde in PBSEM (50 mM phosphate [pH 6.95], 150 mM NaCl, 5 mM EGTA, 5 mM MgSO4) for 30 min, and permeabilized with 100% methanol at −20°C for 10 min. The cells were washed two times with PBSEM containing 0.5% Tween 20 for 30 min each time. CP66:6G:g5 protein tagged with the Stag epitope (CP66:Stag:6G:g5) (Table 1) was detected with the S protein coupled to fluorescein isothiocyanate (FITC) (Novagen, Madison, Wis.). The 15-amino-acid-long Stag peptide was inserted after Arg66 of CP to construct the CP66:Stag:6G:g5 protein. Flag epitope-tagged BV1, T7 epitope-tagged BC1, CP, and β-glucuronidase (GUS) (Table 1) were detected with anti-Flag antibody M2 (Sigma), anti-T7 tag antibody (Novagen), anti-CP antisera (33), and anti-GUS antisera (5′-3′, Boulder, Colo.) diluted 1:100 in phosphate-buffered saline, respectively. After incubation with the primary antibody for 1 h at 30°C, the cells were washed as before and incubated with FITC- or rhodamine-conjugated immunoglobulin G (Pierce) at a dilution of 1:100. The cells were mounted in Fluoromount G (Electron Microscopy Sciences, Fort Washington, Pa.) and viewed with a Nikon fluorescence microscope or an Olympus confocal microscope (for detecting T7 epitope-tagged BC1 protein).
RESULTS
ToLCV expressing g5p or CP66:6G:g5 protein accumulates ssDNA to wild-type levels in protoplasts.
Our earlier work with ToLCV showed that viral CP and AV2 are not required for virus replication in protoplasts, whereas AV2 is required for efficient movement in plants (33). CP is not essential for systemic movement and symptom development in ToLCV. However, mutations in the CP sequence caused a marked decrease in ssDNA accumulation in N. bentamiana and tomato plants and in BY2 protoplasts while increasing dsDNA accumulation in protoplasts. Virus that contained mutations in AV2 plus CP behaved like AV2 mutant virus in plants (i.e., poor virus movement and very mild symptoms) and like CP mutant virus in protoplasts (i.e., decrease in ssDNA and increase in dsDNA accumulation).
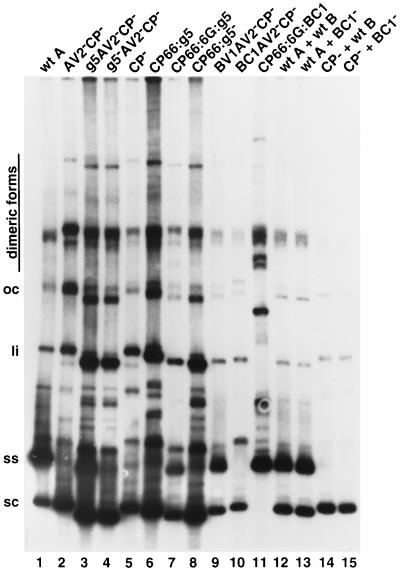
Here we investigated the effects of g5p from E. coli phage M13 (40) on the replication of ToLCV. Each of the mutations is described in Table 1 and Fig. 1. The AV2 ORF and the overlapping 5′ portion of the CP ORF were replaced with g5p, and its effect on virus replication in protoplasts was assayed. In these experiments, protoplasts were inoculated with the wild type or mutants as described below. Surprisingly, the modified A component, designated g5AV2−CP−, led to the accumulation of ssDNA to the same levels as did the wild-type A component (Table 2 and Fig. 2, lanes 1 and 3). However, dsDNA accumulation was high (three- to sixfold higher than wild-type levels) and similar to the accumulation in the presence of mutations in CP (Table 2 and Fig. 2, lanes 2 to 4). Infection by virus in which the g5p gene was mutated to prevent its translation (g5−AV2− CP−) (Table 1) behaved like virus infections with A-component mutants AV2−CP− and CP− (Table 2 and Fig. 2, lane 4).
TABLE 2.
Effect of g5p on the replication and movement of ToLCV in N. tabacum protoplasts and N. benthamiana plants
| Virus | Protoplast inoculations
|
Plant inoculations
|
||||
|---|---|---|---|---|---|---|
| ssDNAa | dsDNAa | No. of plants inoculated | Symptom type | ssDNAb | dsDNAb | |
| Wild type | 100 | 100 | 20 | Severe | 100 | 100 |
| AV2−CP− | <1 (0–0.03) | 506 (427–584) | 10 | Very mildc | 0.3 (0.05–0.5) | 11 (9.6–17) |
| g5AV2−CP− | 102 (79–133) | 409 (349–573) | 20 | Very mildc | 0.6 (0.1–2.7) | 15.2 (6.2–49.2) |
| g5−AV2−CP− | 7 (5–12) | 384 (210–779) | 20 | Very mildc | 0.1 (0.0–0.2) | 5.7 (0.0–11.4) |
| CP− | 5 (2–7) | 241 (148–369) | 20 | Severed | 4.3 (2.6–6.5) | 102 (65–139) |
| CP66:g5 | 17 (8–27) | 442 (345–576) | 20 | Mild | 2.2 (0.8–4.2) | 30.6 (15.3–55.1) |
| CP66:6G:g5 | 118 (34–234) | 517 (133–784) | 30 | Very mildc | 0.9 (0.4–1.7) | 10.9 (5.5–14.7) |
| CP66:g5− | 9 (3–14) | 424 (179–789) | 20 | Severed | 4.0 (1.8–6.1) | 139.7 (56.0–197.7) |
The values represent the average amount (range) of A-component DNA in five independent protoplast transfections per mutant. Protoplasts (∼106) were transfected with 2 μg of A-component DNA and 40 μg of herring sperm DNA. Viral DNA was quantitated on Southern blots with a PhosphorImager (Molecular Dynamics). Values are relative to those for the wild type, which was assigned a value of 100.
The values represent the average amount (range) of viral DNA in 12 inoculated plants per virus construct, except for AV2−CP−, for which the values represent the average in four plants. Each plant was inoculated with 0.5 μg of A-component DNA and 0.5 μg of wild-type B-component DNA, which is essential for viral movement and symptom development. Values are relative to those for the wild type, which was assigned a value of 100.
Many plants did not show symptoms.
Severe symptoms like those in plants inoculated with the wild-type virus but without intense chlorosis.
FIG. 2.
Replication of ToLCV constructs in infected BY2 protoplasts. Southern blot analysis was performed as described in Materials and Methods. The viral constructs used for infecting protoplasts are shown above the lanes. Protoplasts were inoculated with A-component DNA alone (lanes 1 to 11) or coinoculated with A- and B-component DNAs (lanes 12 to 15). Each lane contained 4 μg of DNA prepared from protoplasts (single transfection). Viral DNA was detected with a radioactively labeled probe from A-component DNA. The positions of supercoiled (sc), single-stranded (ss), linear (li), and open circular (op) viral DNA forms are indicated. Note that the positions of supercoiled and other viral DNA forms in lane 11 are shifted upward due to the larger size of the CP66:6G:BC1 construct. wt, wild type.
Since AV2 is required for efficient virus movement in plants, we made another construct in which g5p was fused to CP at Arg66 without affecting the AV2 ORF (CP66:g5) (Table 1). The CP66:g5 virus A component also led to the accumulation of ssDNA, but to lower levels than did g5AV2−CP− (Table 2 and Fig. 2, lane 6). To address the possibility that the N-terminal 66 amino acids (aa) of CP interfered with the ability of g5p to bind DNA, a linker of six glycine residues was introduced between Arg66 of CP and g5p to separate the CP domain from the g5p domain (CP66:6G:g5). The addition of the linker restored the ability of the CP66:6G:g5 virus A component to accumulate ssDNA to levels comparable to those of g5AV2−CP− (Table 2 and Fig. 2, lane 7). A control construct in which the g5p portion of the fusion protein was not translated (CP66:g5−) failed to accumulate ssDNA (Table 2 and Fig. 2, lane 8). That the ability of the virus A component expressing the CP66:6G:g5 protein to accumulate ssDNA was not due to the N-terminal 66 aa of CP was suggested by the facts that the virus A component expressing g5p alone accumulated ssDNA and the virus A component expressing CP66:6G:BC1 (see below) or CP66:6G:AV2 (data not shown) failed to accumulate ssDNA.
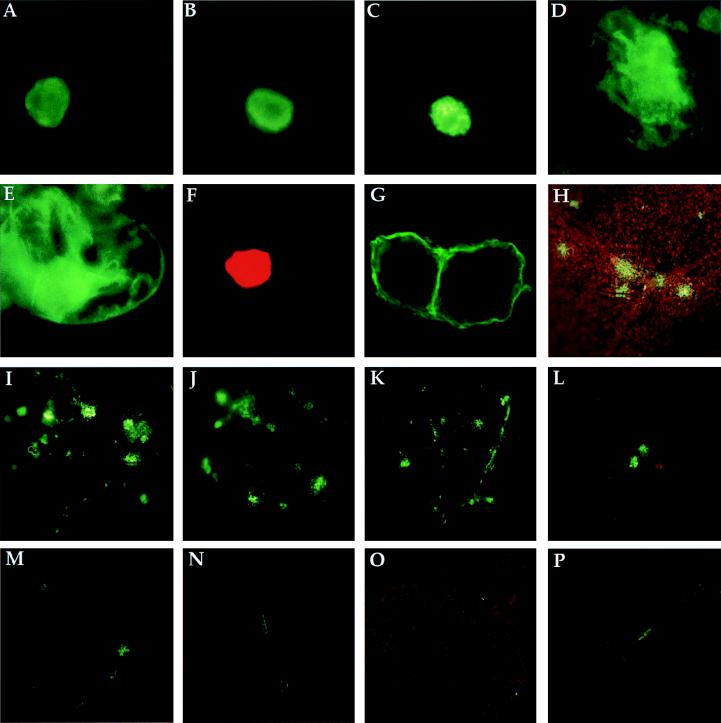
Geminiviruses replicate in the nucleus (1, 29), so it is likely that in order to cause the accumulation of ssDNA, the CP66:6G:g5 and g5 proteins must be present in the nucleus. To immunolocalize the CP66:6G:g5 fusion protein in protoplasts, we inserted the Stag epitope between Arg66 of CP and the glycine linker (CP66:Stag:6G:g5) (Table 1). At 48 h after infection, protoplasts were fixed and subjected to reactions with S protein coupled to FITC. The CP66:Stag:6G:g5 protein as well as wild-type CP (detected with anti-CP antisera) were localized to the nucleus (Fig. 3A and B). When GUS was produced as a fusion protein with the N-terminal 66 aa of CP (CP66:GUS), GUS (detected with anti-GUS antisera) was also localized to the nucleus (Fig. 3C). This result indicated that the N-terminal 66 aa of CP contains a nuclear localization signal. We also determined if g5p contains a nuclear localization signal by fusing the g5p coding sequence to the GUS coding sequence at the N terminus. The g5:GUS fusion protein (expressed in the g5:GUSAV2−CP− virus A component) (Table 1) and the unfused GUS protein (expressed in the GUSAV2−CP− virus A component) (Table 1) remained in the cytoplasm (Fig. 3D and E), suggesting that g5p has no nuclear localization signal. It is possible that g5p may have entered the nucleus in a passive manner, as its size (9.7 kDa) is smaller than the permeability barrier of the nuclear envelope (12).
FIG. 3.
Indirect immunofluorescence of proteins expressed in protoplasts (A to G) and fluorescence of GFP expressed in plants (H to P). Protoplasts were transfected, and antigens were visualized with different primary antibodies and FITC- or rhodamine-conjugated secondary antibodies. GFP fluorescence in plants was monitored every 3 days for 15 days, and the area shown in each panel corresponds to a leaf area measuring 2.5 by 2.5 mm. (A) Protoplast infected with CP66:Stag:6G:g5 virus and stained with S protein coupled to FITC. (B) Protoplast infected with wild-type virus and stained with anti-CP antisera. (C) Protoplast infected with CP66:GUS virus and stained with anti-GUS antisera. (D) Protoplast infected with g5:GUSAV2−CP− virus and stained with anti-GUS antisera. (E) Protoplast infected with GUSAV2−CP− virus and stained with anti-GUS antisera. (F) Protoplast infected with FBV1AV2−CP− virus and stained with anti-Flag antibody. (G) Protoplasts infected with TBC1AV2−CP− virus and stained with anti-T7 tag antibody. Note that two cells are shown in this micrograph. (H and I) Inoculated leaf (H) and systemically infected leaf (I) of a plant infected with GFPAV2−CP− and CP66:g5− viruses 6 days postinoculation (dpi). (J and K) Inoculated leaf (J) and systemically infected leaf (K) of a plant infected with GFPAV2−CP− and CP66:g5− viruses 15 dpi. (L and M) Inoculated leaf (L) and systemically infected leaf (M) of a plant infected with GFPAV2−CP− and CP66:6G:g5 viruses 6 dpi. (N to P) Inoculated leaf (N) and systemically infected leaves (O and P) of a plant infected with GFPAV2−CP− and CP66:6G:g5 viruses 15 dpi.
Movement of ToLCV expressing CP66:6G:g5 protein is impaired in plants.
N. benthamiana plants were inoculated with selected virus constructs to determine the effect of g5p on virus spread. In these studies, the B component was coinoculated with the A component onto N. benthamiana seedlings. As expected, plants inoculated with A-component mutant AV2−CP−, g5AV2−CP−, or g5−AV2−CP− plus the B component showed very mild or no symptoms, and all inoculated plants accumulated low levels of viral DNA (Table 2). A previously reported ToLCV mutant (33) that did not produce CP but produced AV2 (CP−) resulted in severe disease symptoms and wild-type levels of dsDNA in systemic infections (Table 2). Surprisingly, plants inoculated with the virus expressing the CP66:6G:g5 protein showed very mild or no symptoms, even though the virus contained an intact AV2 gene (Table 2). These plants accumulated low levels of viral DNA, similar to plants inoculated with the AV2−CP− virus (Table 2). Plants inoculated with the virus expressing the CP66:g5 protein (which accumulated ssDNA to a lower level than CP66:6G:g5 virus in protoplasts) showed mild symptoms and accumulated moderate levels of dsDNA. We also considered the possibility that the impaired movement of the virus expressing g5p was due to possible toxic effects of g5p. We did not detect any differences in protoplast viability or in the appearance of plant leaves inoculated with wild-type virus or virus expressing g5p that might suggest toxicity of g5p.
We next examined the cell-to-cell and long-distance movement of ToLCV expressing the CP66:6G:g5 protein by using green fluorescent protein (GFP) as a visible marker for virus movement. Plants were inoculated with A-component DNA expressing GFP in place of AV2 and CP (GFPAV2−CP−) alone or coinoculated with A-component DNA of the wild-type, CP66:6G:g5, or CP66:g5− construct. GFPAV2−CP− virus was expected to move inefficiently in plants, as it does not carry AV2; it was expected to move efficiently when complemented by another virus carrying AV2. GFP could not be detected in plants by day 3 postinoculation, but it was present on inoculated and upper leaves by day 6 in the majority of the plants inoculated with GFPAV2−CP− plus wild-type A-component viruses or GFPAV2−CP− plus CP66:g5− viruses (Fig. 3H and I; only data on plants inoculated with GFPAV2−CP− plus CP66:g5− viruses are shown). The virus expressing GFP continued to spread to upper and newly emerging leaves in these plants (Fig. 3J and K). GFP was observed in veins, the mesophyll, and epidermal cells and was present in large areas of the leaves in plants inoculated with GFPAV2−CP− plus CP66:g5− viruses. In contrast, GFP was restricted to small spots on the inoculated leaves of most of the plants inoculated with GFPAV2−CP− or GFPAV2−CP− plus CP66:6G:g5 viruses (Fig. 3L and M; only data on plants inoculated with GFPAV2−CP− plus CP66:6G:g5 viruses are shown). These plants also showed GFP staining in some adjacent and newly emerging leaves, mostly restricted to veins (Fig. 3N, O, and P). These results indicated that the expression of g5p in place of CP decreased the efficiency of virus systemic movement.
In vivo binding of CP66:6G:g5 protein to viral DNA.
The accumulation of viral ssDNA in protoplasts inoculated with the virus A component expressing g5p or CP66:6G:g5 protein suggested that g5p binds to ssDNA. To test this possibility, we inoculated protoplasts with the virus A component expressing the Flag epitope-tagged CP66:6G:g5 protein (FCP66:6G:g5) (Table 1), immunoprecipitated the Flag epitope-tagged CP66:6G:g5 protein with anti-Flag antibody, and characterized the viral DNA that coimmunoprecipitated with the CP66:6G:g5 protein by Southern blotting. The immunoprecipitations were performed under different salt (1% NP-40 buffer with 0.15 to 1.0 M NaCl) conditions and in the presence of 0.1% sodium dodecyl sulfate, 0.5% deoxycholate, and 1% NP-40 detergents (RIPA buffer) to assay the affinity of binding. The Flag epitope-tagged CP66:6G:g5 protein was immunoprecipitated under all of the buffer conditions tested; the amount of protein immunoprecipitated increased with increasing salt concentration (Fig. 4A). The amount of coimmunoprecipitated ssDNA increased up to a 0.5 M salt concentration and decreased at higher concentrations (Fig. 4B), indicating that the g5p-ssDNA complex was destabilized in buffer that contained 1 M salt. Immunoprecipitation in RIPA buffer also resulted in a reduced amount of precipitated ssDNA (Fig. 4B). These results showed that g5p bound to viral ssDNA and that 1 M salt (in NP-40 buffer) dissociated g5p from the viral ssDNA.
FIG. 4.
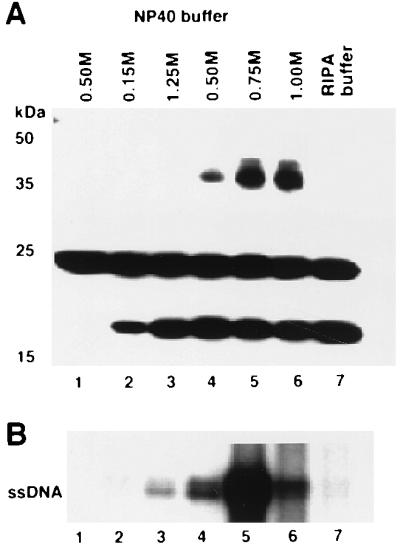
In vivo binding of g5p to ToLCV DNA. (A) Flag epitope-tagged CP66:6G:g5 protein expressed in protoplasts was immunoprecipitated with anti-Flag antibody coupled to agarose after lysis of protoplasts in NP-40 buffer containing different concentrations of NaCl (shown above lanes 1 to 6) or RIPA buffer (lane 7), and the immunoprecipitated protein was detected on a Western blot with anti-Flag antibody (lanes 2 to 7). Lane 1 contained protein immunoprecipitated from protoplasts transfected with wild-type virus as a control. The protein band present in all lanes at ∼24 kDa is the light chain of anti-Flag antibody used for immunoprecipitations. The immunoprecipitated CP66:6G:g5 protein was detected at two different molecular masses corresponding to monomeric and dimeric forms. Positions of molecular mass markers are indicated in kilodaltons on the left. (B) Viral ssDNA that coimmunoprecipitated with the Flag epitope-tagged CP66:6G:g5 protein was detected on a Southern blot with 32P-labeled A-component DNA as a probe. Lanes 1 to 7 were given the same treatments as in panel A.
Role of BV1 and BC1 movement proteins in the spread of ToLCV.
The above results indicate that the CP66:6G:g5 protein is localized to the nucleus and binds stably to ToLCV virus DNA in vivo and that ToLCV expressing CP66:6G:g5 does not move efficiently in plants. The inefficient movement of ToLCV expressing the CP66:6G:g5 protein may have been due to an interference of g5p with the function of the BV1 or BC1 movement protein of ToLCV. In squash leaf curl virus (SLCV), BV1 (referred to as BR1 in SLCV) but not BC1 (referred to as BL1 in SLCV) binds to ssDNA in vitro (34). BR1 and BL1 of SLCV interact with each other in a cooperative manner; in protoplasts, BR1 localizes to the nucleus in the absence of BL1 but localizes to the cell periphery in the presence of BL1 (42, 43). Both BV1 and BC1 are required for the systemic spread and symptom development of ToLCV (33). To determine if BV1 and BC1 of ToLCV have functions similar to those of BR1 and BL1 of SLCV, we immunolocalized BV1 and BC1 of ToLCV and examined their ability to complement the viral ssDNA accumulation of CP mutants. For these experiments, the BV1 and BC1 genes were fused to sequences coding for the Flag epitope tag and the T7 epitope tag, respectively, and inserted in place of AV2 and CP in the A component (FBV1AV2−CP− and TBC1AV2−CP−) (Table 1). In protoplasts inoculated with the FBV1AV2−CP− construct, the BV1 protein accumulated in the nucleus (detected with anti-Flag antibody) (Fig. 3F), while in protoplasts inoculated with TBC1AV2−CP−, the BC1 protein was localized to the cell periphery (detected with anti-T7 tag antibody) (Fig. 3G). Expression of the BV1 protein in place of the AV2 and CP proteins (BV1AV2−CP−) also led to the accumulation of ssDNA by the A-component virus (Table 3 and Fig. 2, lane 9). The binding affinity of the BV1 protein tagged with the Flag epitope for viral DNA in protoplasts inoculated with FBV1AV2−CP− DNA was determined by immunoprecipitation reactions similar to those shown in Fig. 4. The binding affinity of the BV1 protein for viral ssDNA was similar to the binding affinity of the CP66:6G:g5 protein for viral ssDNA (data not shown). In contrast to results obtained with virus A component expressing BV1, A-component virus expressing BC1 in place of AV2 and CP (BC1AV2−CP−) did not accumulate ssDNA (Table 3 and Fig. 2, lane 10). Since the BC1 protein was localized to the cell periphery, we fused BC1 to the N-terminal 66 aa of CP (CP66:6G:BC1) to direct it to the nucleus. Virus A component expressing the CP66:6G:BC1 protein also did not accumulate ssDNA (Table 3 and Fig. 2, lane 11), showing that the BC1 movement protein may not bind to viral ssDNA or that the binding affinity may not be strong enough to result in the accumulation of ssDNA. These results show that BV1 is localized to the nucleus in the absence of BC1 and that BV1 binds to viral ssDNA in vivo.
TABLE 3.
Complementation by BV1 and BC1 movement proteins of the accumulation of ToLCV ssDNA in protoplastsa
| A component | B component | ssDNA | dsDNA |
|---|---|---|---|
| Wild type | None | 100 | 100 |
| BV1AV2−CP− | None | 86 (50–121) | 230 (119–195) |
| BC1AV2−CP− | None | 2 (1–3) | 224 (162–288) |
| CP66:6G:BC1 | None | 5 (1–10) | 214 (180–267) |
| Wild type | Wild type | 100 | 100 |
| Wild type | BC1− | 84 (66–100) | 82 (66–98) |
| CP− | Wild type | 4 (3–6) | 164 (128–198) |
| CP− | BC1− | 5 (3–6) | 173 (160–185) |
Protoplasts were transfected with 2 μg of A-component DNA with or without 10 μg of B-component DNA. Viral DNA was quantitated on Southern blots with a PhosphorImager. The values represent the average amount (range) of viral DNA in two to five independent transfections, relative to a value of 100 assigned to the wild type.
In plants inoculated with the ToLCV A component containing CP66:6G:g5 plus the wild-type B component, the expression of the CP66:6G:g5 protein is controlled by the relatively strong CP promoter. The CP66:6G:g5 protein produced from the A component may outcompete the BV1 protein (expressed from the B component) for DNA binding if the amount of BV1 made under the control of its own promoter is relatively low. We conducted an experiment to determine if BV1, expressed under the control of its own promoter on the B component, can lead to the accumulation of ssDNA. Note that BV1 led to the accumulation of ssDNA when expressed in place of CP on the A component (Table 3). However, very little viral ssDNA accumulated in protoplasts coinoculated with A-component DNA with a mutation in CP (CP−) plus wild-type B-component DNA (i.e., expressing both BV1 and BC1) or B-component DNA with a mutation in BC1 (BC1−) (i.e., expressing only BV1) (Table 3 and Fig. 2, lanes 12 to 15). The failure of BV1 to cause the accumulation of ssDNA when expressed from the B component appeared to be due to low levels of BV1 protein being made; no BV1 protein was detected in protoplasts coinoculated with A-component DNA and B-component DNA expressing Flag epitope-tagged BV1 by immunolocalization and Western blotting procedures (data not shown). These results show that the B-component promoter driving the expression of BV1 is not as strong as when the gene is expressed from the CP promoter on the A component.
DISCUSSION
Previous work done by our group showed that in the absence of CP, ToLCV failed to accumulate ssDNA but produced levels of dsDNA severalfold higher than wild-type levels in protoplasts (33). Reduced levels of ssDNA have been observed for other geminiviruses when CP is not produced (5, 27, 49, 52). This observation raised the question as to whether the accumulation of ssDNA is due solely to encapsidation by CP or whether CP has some additional role in viral replication. We tested these possibilities by expressing a nonspecific ssDNA binding protein in place of CP and monitoring the accumulation of ssDNA to determine if it could serve as a substitute for CP in this putative function. g5p from E. coli phage M13 was chosen because of its small size (9.7 kDa) and lack of any enzymatic function in DNA replication. The role of g5p in the replication of M13 and other filamentous phages has been extensively studied (36), and its structure has been determined (45). g5p binds newly formed viral ssDNA tightly, cooperatively, and in a sequence-independent manner and protects it from degradation by E. coli nucleases (7, 31, 40).
In this report, we demonstrated that g5p can bind to ToLCV ssDNA in plant cells and that ToLCV expressing g5p or g5p fused to the N-terminal 66 aa of CP can accumulate ssDNA to wild-type levels. The binding of g5p to viral ssDNA in vivo was similar to the binding of g5p to M13 ssDNA in vitro (2). Although g5p compensated for the lack of CP by causing an increase in the accumulation of ToLCV ssDNA, it did not reduce the amount of dsDNA to wild-type levels. BV1 movement protein (when expressed in place of CP) also behaved like g5p in that it did not down-regulate dsDNA to wild-type levels. If CP regulates the levels of ssDNA and dsDNA by depleting the ssDNA available for conversion to dsDNA, the expression of g5p or BV1 could be expected to result in normal amounts of dsDNA. The fact that it did not suggests that CP may have a direct role in regulating viral replication, possibly by inhibiting minus-strand synthesis or by regulating gene expression. The CP of alfalfa mosaic virus, a virus with a plus-strand ssRNA genome, has been shown to play a direct role in the regulation of plus- and minus-strand RNA syntheses (10). The alfalfa mosaic virus CP was found in tight association with the viral RNA polymerase and inhibited minus-strand synthesis while stimulating plus-strand synthesis. Recent results obtained with SLCV suggest that CP acts to signal the switch from viral dsDNA replication to ssDNA replication or to sequester virion ssDNA from replication pools without fully encapsidating it (25a). Purification of geminivirus replication complexes is needed to directly assess the role of CP in replication.
Why do plants infected with a virus encoding the CP66:6G:g5 protein show very mild symptoms and accumulate low levels of viral DNA when infected protoplasts accumulate high levels of viral DNA? One likely possibility is that by binding to viral ssDNA, g5p affects virus movement by interfering with the function of the BV1 movement protein. BV1 of ToLCV was localized to the nucleus in infected protoplasts and bound to viral ssDNA in vivo; BC1 was localized to the cell periphery and did not complement viral ssDNA accumulation, even when it was directed to the nucleus as a fusion to the nuclear localization signal of CP. Recent studies on the roles of BR1 and BL1 in SLCV movement have shown that BR1 is localized to the nucleus, binds to ssDNA in vitro, and functions as a nuclear shuttle protein (34, 42). BL1 of SLCV is localized to the cell periphery in protoplasts and is associated with endoplasmic reticulum-derived tubules in developing phloem cells of systemically infected pumpkin seedlings (20, 43, 51). Based on these results, a model for SLCV was proposed in which BL1-containing tubules serve as a conduit for the transport of BR1 and its associated viral ssDNA from one cell to another (51). Studies with tomato golden mosaic virus have shown that BR1 interacts with viral ssDNA in vivo and that BR1 and BL1 have distinct and essential roles in cell-to-cell movement as well as systemic movement (22). It is likely that ToLCV uses a similar strategy in moving from cell to cell. The poor movement of ToLCV that produces the CP66:6G:g5 protein may be due to reduced binding of BV1 to viral ssDNA. It should be noted that BV1 did not lead to the accumulation of ssDNA of the A component that lacked CP when BV1 was expressed under the control of its own promoter from the B component. In plants coinoculated with the A component producing CP66:6G:g5 plus the A component producing GFP, GFP staining was mostly restricted to small areas on both inoculated and systemically infected leaves, showing an overall reduction in the efficiency of viral movement rather than specific interference with cell-to-cell spread or long-distance movement.
In contrast to the model presented for the movement of SLCV, a different model was proposed for bean dwarf mosaic virus in which BC1 binds to dsDNA and moves it through plasmadesmata by increasing their size exclusion limit (30). Interference with ToLCV movement due to binding of g5p to viral ssDNA suggests that in this virus, ssDNA moves from cell to cell. Our results also suggest that the expression of g5p in transgenic plants may afford a novel way of controlling geminiviruses and that such resistance may be effective against all geminiviruses.
ACKNOWLEDGMENTS
We thank Sondra Lazarowitz and Hal Padget for critically reading the manuscript.
This work was supported by financial assistance from the U.S. Agency for International Development (grant DAN-4197-A-00-1126-00); Maharashtra Hybrid Seeds Company, Jalna, India (grant 5-98378); and Institut Français de Recherche Scientifique pour le Développement en Coopération (ORSTOM), Paris, France.
REFERENCES
- 1.Accotto G P, Mullineaux P M, Brown S C, Marie D. Digitaria streak geminivirus replicative forms are abundant in S-phase nuclei of infected cells. Virology. 1993;195:257–259. doi: 10.1006/viro.1993.1369. [DOI] [PubMed] [Google Scholar]
- 2.Anderson R A, Nakashima Y, Coleman J E. Chemical modifications of functional residues of fd gene 5 DNA-binding protein. Biochemistry. 1975;14:907–917. doi: 10.1021/bi00676a006. [DOI] [PubMed] [Google Scholar]
- 3.Boulton M I, Steinkellner J, Donson J, Markham P G, King D I, Davies J W. Mutational analysis of virion-sense genes of maize streak virus. J Gen Virol. 1989;70:2309–2323. doi: 10.1099/0022-1317-70-9-2309. [DOI] [PubMed] [Google Scholar]
- 4.Briddon R W, Markham P G. Geminiviridae. In: Murphy F A, editor. Virus taxonomy. Sixth report of the International Committee on Taxonomy of Viruses. Vienna, Austria: Springer-Verlag; 1995. pp. 158–165. [Google Scholar]
- 5.Briddon R W, Watts J, Markham P G, Stanley J. The coat protein of beet curly top virus is essential for infectivity. Virology. 1989;172:628–633. doi: 10.1016/0042-6822(89)90205-5. [DOI] [PubMed] [Google Scholar]
- 6.Brough C L, Hayes R J, Morgan A J, Coutts R H A, Buck K W. Effects of mutagenesis in vitro on the ability of cloned tomato golden mosaic virus DNA to infect Nicotiana benthamiana plants. J Gen Virol. 1988;69:503–514. [Google Scholar]
- 7.Cavalieri S, Neet K, Goldthwait D. Gene 5 protein of bacteriophage fd: a dimer which interacts co-operatively with DNA. J Mol Biol. 1976;102:697–711. doi: 10.1016/0022-2836(76)90286-2. [DOI] [PubMed] [Google Scholar]
- 8.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 9.Davies J W, Stanley J. Geminivirus genes and vectors. Trends Genet. 1989;5:77–81. doi: 10.1016/0168-9525(89)90030-9. [DOI] [PubMed] [Google Scholar]
- 10.De Graaff M, Man in’t Veld M R, Jaspars E M. In vitro evidence that the coat protein of alfalfa mosaic virus plays a direct role in the regulation of plus and minus RNA synthesis: implications for the life cycle of alfalfa mosaic virus. Virology. 1995;208:583–589. doi: 10.1006/viro.1995.1189. [DOI] [PubMed] [Google Scholar]
- 11.Dellaporta S L, Wood J, Hicks J B. A plant DNA minipreparation: version II. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- 12.Dingwall C, Laskey R A. Protein import into the cell nucleus. Annu Rev Cell Biol. 1986;2:367–390. doi: 10.1146/annurev.cb.02.110186.002055. [DOI] [PubMed] [Google Scholar]
- 13.Elmer J S, Brand L, Sunter G, Gardiner W, Bisaro D M, Rogers S G. Genetic analysis of the tomato golden mosaic virus. II. The product of the AL1 coding sequence is required for replication. Nucleic Acids Res. 1988;16:7043–7060. doi: 10.1093/nar/16.14.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frischmuth T, Stanley J. Strategies for the control of geminivirus diseases. Semin Virol. 1993;4:329–337. [Google Scholar]
- 15.Gardiner W E, Sunter G, Brand L, Elmer L S, Rogers S G, Bisaro D M. Genetic analysis of tomato golden mosaic virus: the coat protein is not required for systemic spread or symptom development. EMBO J. 1988;7:899–904. doi: 10.1002/j.1460-2075.1988.tb02894.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanley-Bowdoin L, Elmer J S, Rogers S G. Expression of functional replication protein from tomato golden mosaic virus in transgenic tobacco plants. Proc Natl Acad Sci USA. 1990;87:1446–1450. doi: 10.1073/pnas.87.4.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanley-Bowdoin L, Elmer J S, Rogers S G. Functional expression of the leftward open reading frames of the A component of tomato golden mosaic virus in transgenic tobacco plants. Plant Cell. 1989;1:1057–1067. doi: 10.1105/tpc.1.11.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrison B D. Advances in geminivirus research. Annu Rev Phytopathol. 1985;23:55–82. [Google Scholar]
- 19.Hopp T P. Protein surface analysis: methods for identifying antigenic determinants and other interaction sites. J Immunol Methods. 1986;88:1–18. doi: 10.1016/0022-1759(86)90045-1. [DOI] [PubMed] [Google Scholar]
- 20.Ingham D J, Pascal E, Lazarowitz S G. Both bipartite geminivirus movement proteins define viral host range, but only BL1 determines viral pathogenicity. Virology. 1995;207:191–204. doi: 10.1006/viro.1995.1066. [DOI] [PubMed] [Google Scholar]
- 21.Jefferson R A. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep. 1987;5:387–405. [Google Scholar]
- 22.Jeffrey J L, Pooma W, Petty I T D. Genetic requirements for local and systemic movement of tomato golden mosaic virus in infected plants. Virology. 1996;223:208–218. doi: 10.1006/viro.1996.0469. [DOI] [PubMed] [Google Scholar]
- 23.Kim J S, Raines R T. Ribonuclease S-peptide as a carrier in fusion proteins. Protein Sci. 1993;2:348–356. doi: 10.1002/pro.5560020307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krek W, Ewen M E, Shorodka S, Arany Z, Kaelin W G, Livingston D M. Negative regulation of the growth-promoting transcription factor E2F-1 by a stably bound cyclin A-dependent protein kinase. Cell. 1994;78:161–172. doi: 10.1016/0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 25.Laufs J, Traut W, Heyraud F, Matzeit V, Rogers S G, Schell J, Gronenborn B. In vitro cleavage and joining at the viral origin of replication by the replicator initiator protein of tomato yellow leaf curl virus. Proc Natl Acad Sci USA. 1995;92:3879–3883. doi: 10.1073/pnas.92.9.3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25a.Lazarowitz, S. Personal communication.
- 26.Lazarowitz S G. Geminiviruses: genome structure and gene function. Crit Rev Plant Sci. 1992;11:327–349. [Google Scholar]
- 27.Lazarowitz S G, Pinder A J, Damsteegt V D, Rogers S G. Maize streak virus genes essential for systemic spread and symptom development. EMBO J. 1989;8:1023–1032. doi: 10.1002/j.1460-2075.1989.tb03469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mettler I J. A simple and rapid method for minipreparation of DNA from tissue cultured plant cells. Plant Mol Biol Rep. 1987;5:346–349. [Google Scholar]
- 29.Nagar S, Pedersen T J, Carrick K M, Hanley-Bowdoin L, Robertson D. A geminivirus induces expression of a host DNA synthesis protein in terminally differentiated plant cells. Plant Cell. 1995;7:705–719. doi: 10.1105/tpc.7.6.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noueiry A O, Lucas W J, Gilbertson R L. Two proteins of a plant DNA virus coordinate nuclear and plasmodesmal transport. Cell. 1994;76:925–932. doi: 10.1016/0092-8674(94)90366-2. [DOI] [PubMed] [Google Scholar]
- 31.Oey J, Knippers R. Properties of the isolated gene 5 protein of bacteriophage fd. J Mol Biol. 1972;68:125–138. doi: 10.1016/0022-2836(72)90268-9. [DOI] [PubMed] [Google Scholar]
- 32.Padidam M, Beachy R N, Fauquet C M. Tomato leaf curl geminivirus from India has a bipartite genome and coat protein is not essential for infectivity. J Gen Virol. 1995;76:25–35. doi: 10.1099/0022-1317-76-1-25. [DOI] [PubMed] [Google Scholar]
- 33.Padidam M, Beachy R N, Fauquet C M. The role of AV2 (“precoat”) and coat protein in viral replication and movement in tomato leaf curl geminivirus. Virology. 1996;224:390–404. doi: 10.1006/viro.1996.0546. [DOI] [PubMed] [Google Scholar]
- 34.Pascal E, Sanderfoot A A, Ward B M, Medville R, Turgeon R, Lazarowitz S G. The geminivirus BR1 movement protein binds single-stranded DNA and localizes to the cell nucleus. Plant Cell. 1994;6:995–1006. doi: 10.1105/tpc.6.7.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Polston J E, Anderson P K. The emergence of whitefly-transmitted geminiviruses in tomato in the western hemisphere. Plant Dis. 1997;81:1358–1369. doi: 10.1094/PDIS.1997.81.12.1358. [DOI] [PubMed] [Google Scholar]
- 36.Rasched I, Oberer E. Ff coliphages: structural and functional relationships. Microbiol Rev. 1986;50:401–427. doi: 10.1128/mr.50.4.401-427.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reichel C, Mathur J, Eckes P, Langenkemper K, Koncz C, Schell J, Reiss B, Maas C. Enhanced green fluorescence by the expression of an Aequorea victoria green fluorescent protein mutant in mono- and dicotyledonous plant cells. Proc Natl Acad Sci USA. 1996;93:5888–5893. doi: 10.1073/pnas.93.12.5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rigden J E, Dry I B, Mullineaux P M, Rezaian M A. Mutagenesis of the virion-sense open reading frames of tomato leaf curl geminivirus. Virology. 1993;193:1001–1005. doi: 10.1006/viro.1993.1215. [DOI] [PubMed] [Google Scholar]
- 39.Rochester D E, Fauquet C M, Depaulo J J, Beachy R N. Complete nucleotide sequence of the geminivirus tomato yellow leaf curl virus, Thailand isolate. J Gen Virol. 1994;75:477–485. doi: 10.1099/0022-1317-75-3-477. [DOI] [PubMed] [Google Scholar]
- 40.Salstrom J, Pratt D. Role of coliphage M13 gene 5 in single-stranded DNA production. J Mol Biol. 1971;61:489–501. doi: 10.1016/0022-2836(71)90061-1. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 42.Sanderfoot A A, Ingham D J, Lazarowitz S G. A viral movement protein as a nuclear shuttle. Plant Physiol. 1996;110:23–33. doi: 10.1104/pp.110.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanderfoot A A, Lazarowitz S G. Cooperation in viral movement: the geminivirus BL1 movement protein interacts with BR1 and redirects it from the nucleus to the cell periphery. Plant Cell. 1995;7:1185–1194. doi: 10.1105/tpc.7.8.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saunders K, Lucy A, Stanley J. DNA forms of the geminivirus African cassava mosaic virus consistent with a rolling circle mechanism of replication. Nucleic Acids Res. 1991;19:2325–2330. doi: 10.1093/nar/19.9.2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skinner M M, Zhang H, Leschnitzer D H, Guan Y, Bellamy H, Sweet R M, Gray C W, Konings R N H, Wang A H J, Terwillinger T C. Structure of the gene V protein of bacteriophage f1 determined by multiwavelength x-ray diffraction on selenomethionyl protein. Proc Natl Acad Sci USA. 1994;91:2071–2075. doi: 10.1073/pnas.91.6.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stenger D C, Revington G N, Stevenson M C, Bisaro D M. Replicational release of geminivirus genomes from tandemly repeated copies: evidence for rolling circle replication of a plant viral DNA. Proc Natl Acad Sci USA. 1991;88:8029–8033. doi: 10.1073/pnas.88.18.8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sunter G, Bisaro D M. Transactivation of geminivirus AR1 and BR1 gene expression by the viral AL2 gene product occurs at the level of transcription. Plant Cell. 1992;4:1321–1331. doi: 10.1105/tpc.4.10.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sunter G, Gardiner W E, Bisaro D M. Identification of tomato golden mosaic virus-specific RNAs in infected plants. Virology. 1989;170:243–250. doi: 10.1016/0042-6822(89)90372-3. [DOI] [PubMed] [Google Scholar]
- 49.Sunter G, Hartitz M D, Hormudzi S G, Brough C L, Bisaro D M. Genetic analysis of tomato golden mosaic virus: ORF AL2 is required for coat protein accumulation while ORF AL3 is necessary for efficient DNA replication. Virology. 1990;179:69–77. doi: 10.1016/0042-6822(90)90275-v. [DOI] [PubMed] [Google Scholar]
- 50.Timmermans M C P, Das O P, Messing J. Geminiviruses and their uses as extrachromosomal replicons. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:79–112. [Google Scholar]
- 51.Ward B M, Medville R, Lazarowitz S G, Turgeon R. The geminivirus BL1 movement protein is associated with endoplasmic reticulum-derived tubules in developing phloem cells. J Virol. 1997;71:3726–3733. doi: 10.1128/jvi.71.5.3726-3733.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woolston C J, Reynolds H V, Stacey N J, Mullineaux P M. Replication of wheat dwarf virus-DNA in protoplasts and analysis of coat protein mutants in protoplasts and plants. Nucleic Acids Res. 1989;17:6029–6041. doi: 10.1093/nar/17.15.6029. [DOI] [PMC free article] [PubMed] [Google Scholar]