Abstract
Oxidative protein folding occurs in the endoplasmic reticulum (ER) to generate disulfide bonds, and the by‐product is hydrogen peroxide (H2O2). However, the relationship between oxidative protein folding and senescence remains uncharacterized. Here, we find that the protein disulfide isomerase (PDI), a key oxidoreductase that catalyzes oxidative protein folding, accumulated in aged human mesenchymal stem cells (hMSCs) and deletion of PDI alleviated hMSCs senescence. Mechanistically, knocking out PDI slows the rate of oxidative protein folding and decreases the leakage of ER‐derived H2O2 into the nucleus, thereby decreasing the expression of SERPINE1, which was identified as a key driver of cell senescence. Furthermore, we show that depletion of PDI alleviated senescence in various cell models of aging. Our findings reveal a previously unrecognized role of oxidative protein folding in promoting cell aging, providing a potential target for aging and aging‐related disease intervention.
Keywords: human mesenchymal stem cells (hMSCs), hydrogen peroxide (H2O2), oxidative protein folding, protein disulfide isomerase (PDI), senescence
Subject Categories: Molecular Biology of Disease, Translation & Protein Quality
Protein disulfide isomerase (PDI), a key oxidoreductase that catalyzes oxidative protein folding, accumulates in aged human mesenchymal stem cells. Depletion of PDI slows the rate of oxidative protein folding, decreases the leakage of ER‐derived H2O2 into the nucleus, and alleviates cell senescence.
Introduction
Stem cell exhaustion is considered one of the hallmarks of aging (Cai et al, 2022; Lopez‐Otin et al, 2023). Mesenchymal stem cells (MSCs) are adult stem cells that show self‐renewal capacity and can differentiate into different mesodermal lineages, such as chondrocytes, osteoblasts, and adipocytes (Ullah et al, 2015). Therefore, MSCs have been extensively applied to tissue engineering and regenerative medicine (Li et al, 2017; Han et al, 2019; Wang et al, 2022a). However, the functions of MSCs progressively decline during aging, a process involving lost maintenance of tissue homeostasis, which induces aging‐associated tissue degeneration. (Li et al, 2017; Deng et al, 2019; Fraile et al, 2022; Sun et al, 2022b). Therefore, understanding the molecular processes involved in MSCs senescence is crucial to identify novel therapeutic targets for the treatment of aging‐related degeneration.
Proteostasis enables healthy cell and organismal development and protects against disease (Balch et al, 2008). Impaired protein homeostasis is linked to aging and aging‐related diseases (Hipp et al, 2019). Studies have revealed that reducing protein synthesis (Tavernarakis, 2008; Papadopoli et al, 2019) and boosting the protein quality control system (Higuchi‐Sanabria et al, 2018; Ghosh et al, 2020; Zhang et al, 2022b) can ameliorate aging‐related diseases and extend the life span. However, little is known about the relationship between the protein folding rate and aging. A unique process of protein folding is oxidative protein folding, which is critical for the formation of disulfide bonds during the folding of nascent peptides into form native proteins. In eukaryotes, approximately one‐third of proteins are secretory or membrane proteins, which usually form disulfide bonds and fold in the endoplasmic reticulum (ER; Wang & Wang, 2023). Oxidative protein folding is mainly catalyzed by protein disulfide isomerase (PDI) family members, which play central roles in disulfide bond oxidation, reduction, and isomerization (Bulleid & Ellgaard, 2011; Wang & Wang, 2023). Protein disulfide isomerase introduces disulfides into substrate proteins by transferring electrons to an upstream oxidase, ER oxidoreductin 1 (Ero1). Ero1 is a sulfhydryl oxidase, which uses molecular oxygen as an electron acceptor and produces disulfide with an equimolar concentration of H2O2 (Bulleid & Ellgaard, 2011). It has been proposed that oxidative protein folding contributes approximately 25% of cellular reactive oxygen species (ROS) during protein synthesis (Tu & Weissman, 2004).
The excessive accumulation of ROS levels and decreased antioxidant systems can induce severe oxidative damage in lipids, DNA, and proteins, which promotes stem cell dysfunction with aging (Oh et al, 2014; Cai et al, 2022; Wang et al, 2022b). While previous studies of oxidative stress on cell senescence have mostly focused on mitochondria‐derived ROS (Li et al, 2013; Vasileiou et al, 2019; Kong et al, 2022), ROS generated in the ER have been neglected for a long time. In fact, little is known about the role of oxidative protein folding in cell senescence. Since PDI‐knockout mice are embryonic lethal (Xu et al, 2014; Parakh & Atkin, 2015), research into the role of PDI in aging has been hindered.
In this study, we first observed that PDI was accumulated in aged hMSCs. We generated PDI‐knockout (PDI −/− ) human embryonic stem cells (hESCs) and hMSCs, and found that PDI deficiency attenuated cell senescence in hMSCs. By using the ultrasensitive, genetically encoded H2O2 sensor HyPer7, we showed that PDI‐mediated oxidative protein folding triggered the release of H2O2 from the ER to the nucleus. PDI‐H2O2 axis induced the accumulation of SERPINE1, which induced cell senescence. Importantly, depletion of PDI alleviated senescence in various cell aging models, indicating that targeting oxidative protein folding is a promising strategy for aging and aging‐related disease intervention.
Results
PDI is upregulated in senescent hMSCs
To establish a link between cell senescence and PDI, we first detected the expression of PDI, the key enzyme catalyzing oxidative protein folding in the ER, in replicative senescent (RS) hMSCs, and Werner Syndrome (WS) hMSCs, which undergo premature senescence (Zhang et al, 2015). The results showed that PDI expression was upregulated in RS hMSCs and WS hMSCs (Fig 1A and B). In addition, PDI expression was also upregulated in the livers of aged mice (Fig 1C). Moreover, PDI was identified to be significantly upregulated with aging according to the published “senescence associated secretory phenotype (SASP) Atlas” (Basisty et al, 2020), and database of human plasma (Ritchie et al, 2021), while other members of the PDI family were not (Fig EV1A). These results suggest that PDI may be involved in cell senescence.
Figure 1. Generation and characterization of PDI −/− hESCs.
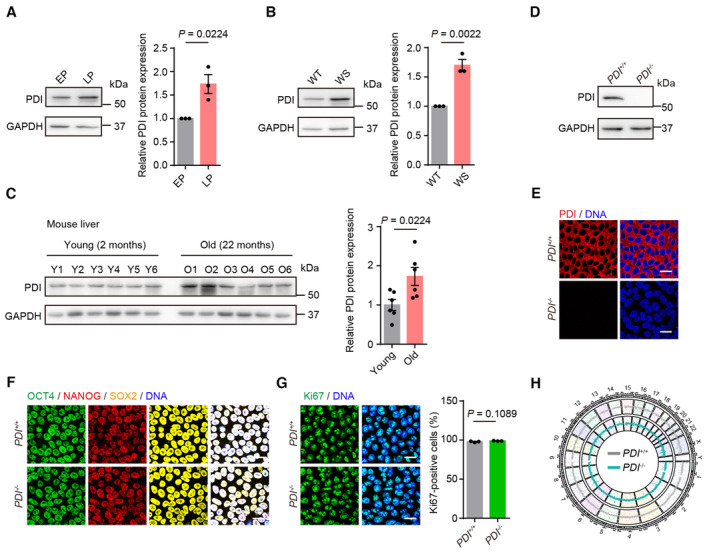
- Left, western blotting of PDI protein in replicative senescent hMSCs at early‐passage (EP) and late‐passage (LP). GAPDH was used as the loading control. Right, statistical analysis of relative PDI protein expression levels. n = 3 independent experiments.
- Left, western blotting of PDI protein in wild‐type (WT) and Werner syndrome (WS) hMSCs. GAPDH was used as the loading control. Right, statistical analysis of relative PDI protein expression levels. n = 3 independent experiments.
- Left, western blotting of PDI protein in the liver of mice at young (2 months) and old (22 months) ages. GAPDH was used as the loading control. Right, statistical analysis of relative PDI protein expression levels. n = 6 mice.
- Western blotting of PDI expression in PDI +/+ and PDI −/− hESCs. GAPDH was used as the loading control.
- Immunostaining of PDI in PDI +/+ and PDI −/− hESCs. Scale bar, 25 μm.
- Immunostaining of OCT4, NANOG and SOX2 in PDI +/+ and PDI −/− hESCs. Scale bar, 25 μm.
- Left, Ki67 staining of PDI +/+ and PDI −/− hESCs. Scale bar, 25 μm. Right, statistical analysis of Ki67‐positive cells. n = 3 independent experiments.
- Genome wide analysis of copy number variations (CNVs) of PDI +/+ and PDI −/− hESCs.
Data information: In (A–C, G), data are presented as mean ± SEM, two‐tailed Student's t‐test.
Source data are available online for this figure.
Figure EV1. Generation and characterization of PDI −/− hESCs.
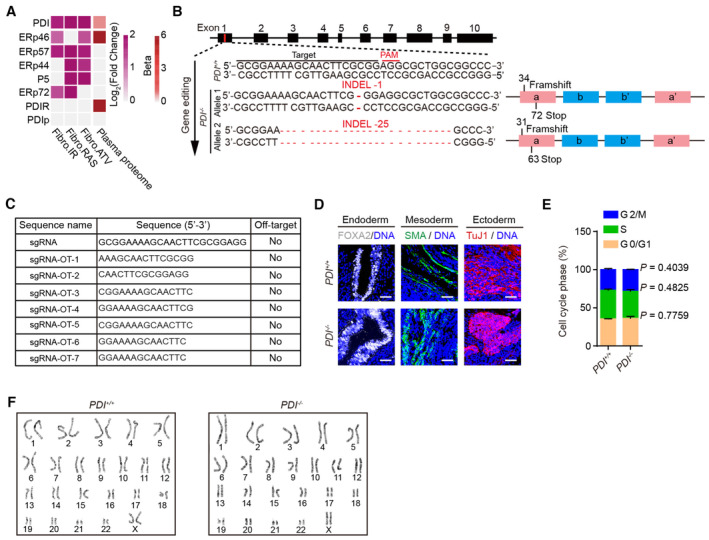
- Conjoint analysis showing the secretion levels of PDI and its family members in SASPs (www.saspatlas.com) from multiple senescence inducers and in plasma proteome from 3,087 healthy individuals (Basisty et al, 2020; Ritchie et al, 2021). ATV, atazanavir treatment; RAS, inducible RAS overexpression; IR, X‐irradiation. The color keys that change from white to dark purple represent the fold change of gene and protein, change from white to red represent the change in protein levels per unit increase in the covariate in plasma proteomic.
- Illustration of PDI gene editing (exon 1) via CRISPR/Cas9‐mediated non‐homologous end joining (NHEJ) in H9 hESCs. The sequences of the mutated alleles in PDI −/− hESCs depicting the deletions were shown. The INDELs caused a frameshift and leaded to an early termination.
- Analysis of the off‐target of PDI −/− hESCs using genomic DNA PCR and sequencing. Predicted off‐target sites against sgRNA and sgRNA sequences were presented in the table.
- Immunostaining of the FOXA2 (endoderm), SMA (mesoderm) and TuJ1 (ectoderm) in teratomas derived from PDI +/+ and PDI −/− hESCs. Scale bar, 50 μm.
- Cell cycle analysis of PDI +/+ and PDI −/− hESCs, Data are shown as mean ± SEM, n = 3 biological repeats, two‐tailed Student's t‐test.
- Karyotyping analysis of PDI +/+ and PDI −/− hESCs.
PDI‐deficient hESCs maintain pluripotency
To study the roles of PDI in human stem cell aging and homeostasis, we generated PDI‐deficient hESCs via CRISPR/Cas9‐mediated gene editing technology. We obtained PDI‐knockout (PDI −/− ) hESCs, which harbored frameshift mutations resulting in premature stop codons (Fig EV1B). The successful ablation of PDI protein was validated by western blotting and immunofluorescence staining (Fig 1D and E). No off‐target cleavage was detected in the seven sites predicted to be most likely affected (Fig EV1C). PDI −/− hESCs expressed pluripotency markers, including OCT4, NANOG, and SOX2 (Fig 1F). In addition, PDI −/− hESCs maintained in vivo differentiation potential toward three germ layer lineages (endoderm, mesoderm, and ectoderm), as shown by a teratoma assay (Fig EV1D). In addition, no remarkable difference in cell proliferation ability or cell cycle kinetics was detected between PDI −/− and PDI +/+ hESCs (Figs 1G and EV1E). Moreover, a genome‐wide copy number variation (CNV) analysis and karyotype analysis showed that PDI −/− hESCs maintained genomic stability and integrity (Figs 1H and EV1F). Taken together, these results indicate that the PDI −/− hESCs maintain normal self‐renewal capability and pluripotency.
PDI deficiency alleviates hMSCs senescence
To investigate the role of PDI in human adult stem cells, we differentiated PDI +/+ and PDI −/− hESCs into hMSCs, adult stem cells susceptible to senescence (Fig 2A). The PDI +/+ and PDI −/− hMSCs expressed typical hMSC markers, such as CD73, CD90, and CD105 (Fig EV2A), but not hMSC‐irrelevant markers, such as CD34, CD43, or CD45 (Fig EV2B). The differentiated hMSCs maintained genomic integrity, as revealed by a CNV analysis (Fig EV2C). Western blotting and immunofluorescence staining confirmed the loss of PDI protein in the PDI −/− hMSCs (Fig 2B and C). Since PDI is a structural subunit of collagen prolyl 4‐hydroxylases (P4H; Koivu et al, 1987; Hatahet & Ruddock, 2009), the results showing that the catalytic subunits of P4H (P4HA1 and P4HA2) were reduced in PDI −/− hMSCs validated the loss of PDI (Fig EV2D). However, we did not observe a significant change in the protein levels of other PDI family members, such as P5, ERp46, ERp72, ERp57, or ERp44, in the PDI −/− hMSCs (Fig 2D). The redundancy of PDI family proteins may account the proper oxidative protein folding and acquisition of PDI‐deficient hMSCs.
Figure 2. PDI deficiency alleviates hMSCs senescence.
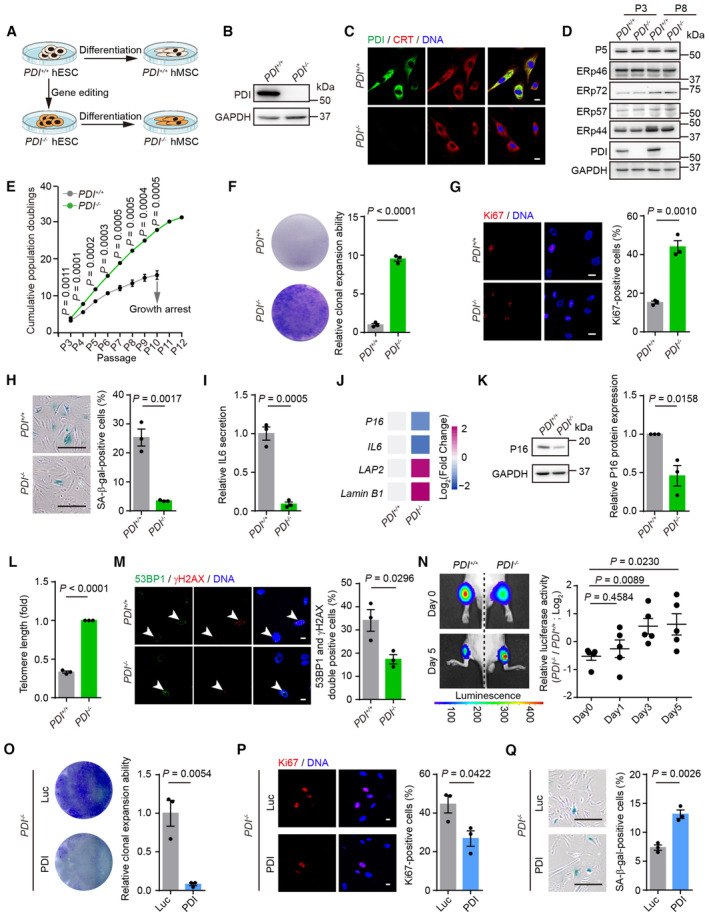
- Schematic representation of differentiation PDI +/+ and PDI −/− hESCs into hMSCs.
- Western blotting of PDI protein in PDI +/+ and PDI −/− hMSCs. GAPDH was used as the loading control.
- Immunostaining of PDI and calreticulin (CRT) in PDI +/+ and PDI −/− hMSCs, CRT was used as ER marker. Scale bar, 25 μm.
- Western blotting analysis of PDI family members expression in PDI +/+ and PDI −/− hMSCs at early‐passage (P3) and late‐passage (P8). GAPDH was used as the loading control.
- Growth curve showing the cumulative population doublings of PDI +/+ and PDI −/− hMSCs. n = 3 biological repeats.
- Single clonal formation of PDI +/+ and PDI −/− hMSCs at P8. n = 3 biological repeats.
- Left, Ki67 staining of PDI +/+ and PDI −/− hMSCs at P8. Scale bar, 20 μm. Right, statistical analysis of Ki67‐positive cells. n = 3 biological repeats.
- Left, SA‐β‐gal staining of PDI +/+ and PDI −/− hMSCs at P8. Scale bar, 200 μm. Right, statistical analysis of SA‐β‐gal‐positive cells. n = 3 biological repeats.
- ELISA analysis for IL6 secretion in PDI +/+ and PDI −/− hMSCs at P8. n = 3 biological repeats.
- Heatmap showing the mRNA levels of P16, IL6, LAP2 and Lamin B1 in PDI +/+ and PDI −/− hMSCs at P8. Colors from blue to red represent the levels of gene expression from low to high. n = 3 independent repeats.
- Left, western blotting showing the P16 expression in PDI +/+ and PDI −/− hMSCs at P8. GAPDH was used as the loading control. Right, statistical analysis of relative P16 protein expression levels. n = 3 independent experiments.
- qPCR analysis of the telomere length of PDI +/+ and PDI −/− hMSCs at P8. n = 3 independent experiments.
- Left, Immunostaining of 53BP1 and γH2AX in PDI +/+ and PDI −/− hMSCs at P8. White arrowheads indicate 53BP1 and γH2AX double‐positive cells. Right, statistical analysis of 53BP1 and γH2AX double‐positive cells. Scale bar, 10 μm. n = 3 biological repeats.
- Luciferase activity analysis in the tibialis anterior muscles of nude mice transplanted with PDI +/+ or PDI −/− hMSCs that expressed luciferase. Luciferase activity was detected at days 0, 1, 3, 5. Left, representative images showing the luciferase activity at day 0 and 5. Right, statistical analysis of the luciferase activity in each day. n = 5 mice.
- Single clonal formation analysis of the PDI −/− hMSCs transduced with lentiviruses expressing luciferase (Luc) or PDI. n = 3 biological repeats.
- Left, Ki67 staining of PDI −/− hMSCs transduced with lentiviruses expressing Luc or PDI. Scale bar, 10 μm. Right, statistical analysis of Ki67‐positive cells. n = 3 biological repeats.
- Left, SA‐β‐gal staining of PDI −/− hMSCs transduced with lentiviruses expressing Luc or PDI. Scale bar, 200 μm. Right, statistical analysis of SA‐β‐gal‐positive cells. n = 3 biological repeats.
Data information: In (E–I, K–Q), data are presented as mean ± SEM, two‐tailed Student's t‐test.
Source data are available online for this figure.
Figure EV2. Generation and characterization PDI +/+ and PDI −/− hMSCs.
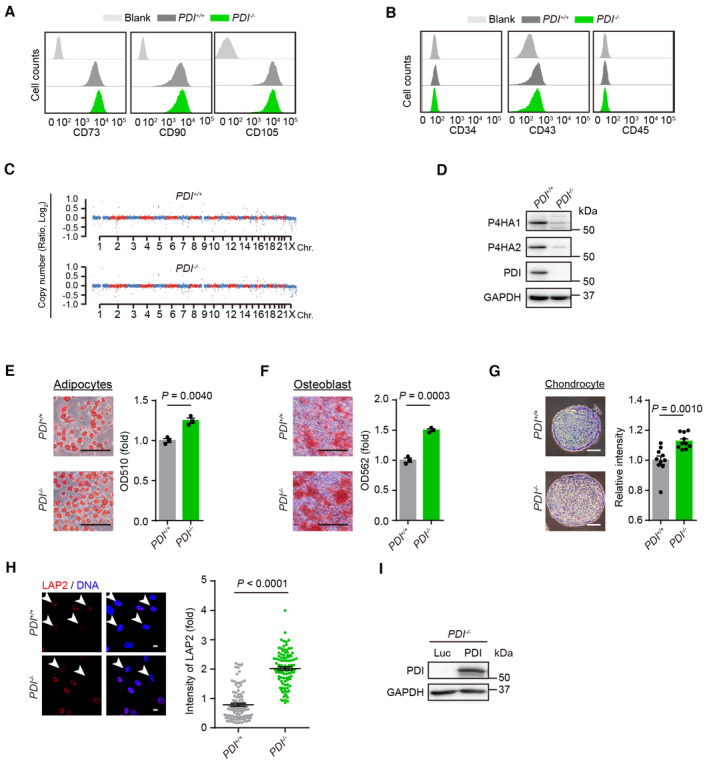
- FACS analysis for the PDI +/+ and PDI −/− hMSC positive markers including CD73, CD90 and CD105.
- FACS analysis for the PDI +/+ and PDI −/− hMSC negative markers including CD34, CD43 and CD45.
- Genome wide analysis of CNV of PDI +/+ and PDI −/− hMSCs.
- Western blotting of the P4HA1 and P4HA2 protein in PDI +/+ and PDI −/− hMSC. GAPDH was used as the loading control.
- Oil Red‐O staining was used to characterize adipogenesis of PDI +/+ and PDI −/− hMSC. The absorbance at 510 nm was measured. Scale bar, 200 μm. n = 3 biological repeats.
- Alizarin Red S staining was used to characterize osteogenesis of the PDI +/+ and PDI −/− hMSC. The absorbance at 562 nm was measured. Scale bar, 200 μm. n = 3 biological repeats.
- Toluidine blue staining was used to characterize chondrogenesis of PDI +/+ and PDI −/− hMSC. Scale bar, 100 μm. The intensity of the chondrocyte staining was measured. n = 10 biological repeats.
- Left, Immunostaining of the LAP2 expression in PDI +/+ and PDI −/− hMSCs at P8. White arrowheads indicate cells with decreased LAP2 expression. Scale bar, 20 μm. Right, statistical analysis of the fluorescence of LAP2. n = 100 cells.
- Western blotting of the PDI expression in PDI −/− hMSCs transduced with Luc or PDI. GAPDH was used as the loading control.
Data information: In (E–H), data are presented as mean ± SEM, two‐tailed Student's t‐test.
Next, we compared the multipotent differentiation potential of PDI +/+ and PDI −/− hMSCs and observed that the PDI −/− hMSCs exhibited enhanced differentiation ability toward adipocytes, osteoblasts and chondrocytes compared with that of the PDI +/+ hMSCs (Fig EV2E–G). Notably, we observed that PDI deficiency markedly increased the cumulative population‐doubling level of hMSCs during serial passaging. The growth arrest of the PDI +/+ hMSCs was evident in passage 10, but the PDI −/− hMSCs continued to grow until passage 12 (Fig 2E). Cell clonal expansion formation and Ki67 immunostaining confirmed the increased proliferation of the PDI −/− hMSCs (Fig 2F and G). Additionally, decreased senescence‐associated β‐gal (SA‐β‐gal) activity was detected in the PDI −/− hMSCs (Fig 2H). We also detected reduced secretion of interleukin‐6 (IL‐6; Fig 2I and J), upregulated expression of nuclear lamina proteins such as LAP2 and Lamin B1 (Figs 2J and EV2H), downregulated expression of the senescence‐associated marker P16 (Fig 2J and K), and increased telomere lengths (Fig 2L) in the PDI −/− hMSCs. In addition, the DNA damage response in the PDI −/− hMSCs was attenuated, as evidenced by a decreased percentage of 53BP1 and γH2AX double‐positive cells (Fig 2M). The tibialis anterior muscle of mice has a microenvironment suitable for the survival of mesoderm cells, and there is an accelerated attrition of senescent MSCs in vivo (Pan et al, 2016; Deng et al, 2019). Thus, we injected PDI +/+ and PDI −/− hMSCs transduced with a lentiviral vector expressing luciferase into the tibialis anterior muscles of immune‐deficient mice and observed that, compared with PDI +/+ hMSCs, PDI −/− hMSCs showed delayed in vivo attrition (Fig 2N). Reintroduction of PDI into PDI −/− hMSCs (Fig EV2I) restored senescent phenotypes in stem cells, as indicated by a decrease in clonal formation ability and the number of Ki67‐positive cells, as well as an increase in the number of SA‐β‐gal‐positive cells (Fig 2O–Q).
PDI deficiency slows the rate of oxidative protein folding and reduces the level of its byproduct, H2O2
Since PDI is a key oxidoreductase, we aimed to determine the effect of PDI on cellular redox homeostasis. ER‐, cytosol‐ and mitochondrion‐localized redox‐sensitive green fluorescent proteins (roGFPs) were used to monitor subcellular glutathione redox conditions in live cells. The ratio of fluorescence intensity at 390/465 nm (for roGFP‐ER) or 405/488 nm (for roGFP‐cytosol and roGFP‐mitochondria) excitation increases under oxidizing conditions and decreases under reducing conditions (Wu et al, 2019). The results showed that deletion of PDI slightly reduced the ER but exerted no effect on the glutathione redox state of the cytosol or mitochondria (Fig EV3A–C), probably due to the functional redundancy of PDI family proteins involved the maintenance of redox homeostasis. As Ero1α‐PDI constitutes the pivotal oxidative folding pathway in the ER (Bulleid & Ellgaard, 2011; Zhang et al, 2014), we wondered whether PDI deficiency affects the efficacy of oxidative protein folding. Similar to that of PDI, the expression of Ero1α was increased in RS hMSCs and was suppressed by PDI deletion (Fig EV3D). Furthermore, we used a dithiothreitol (DTT) pulse‐chase assay to monitor the oxidative folding process of the immunoglobulin J‐chain, which forms three intrachain disulfide bonds (Zhang et al, 2018). PDI deficiency slowed the rate at which the reduced J‐chain disappeared, and fewer oxidized dimers or high‐molecular‐weight species accumulated (Fig 3A and B). Similar to the folding of J‐chain, loss of PDI also slowed the oxidative folding rate of low‐density lipoprotein receptor (LDLR), a cysteine‐rich protein (Kadokura et al, 2020; Fig EV3E).
Figure EV3. ER‐to‐cytosol release of H2O2 is minimized by PDI deletion.
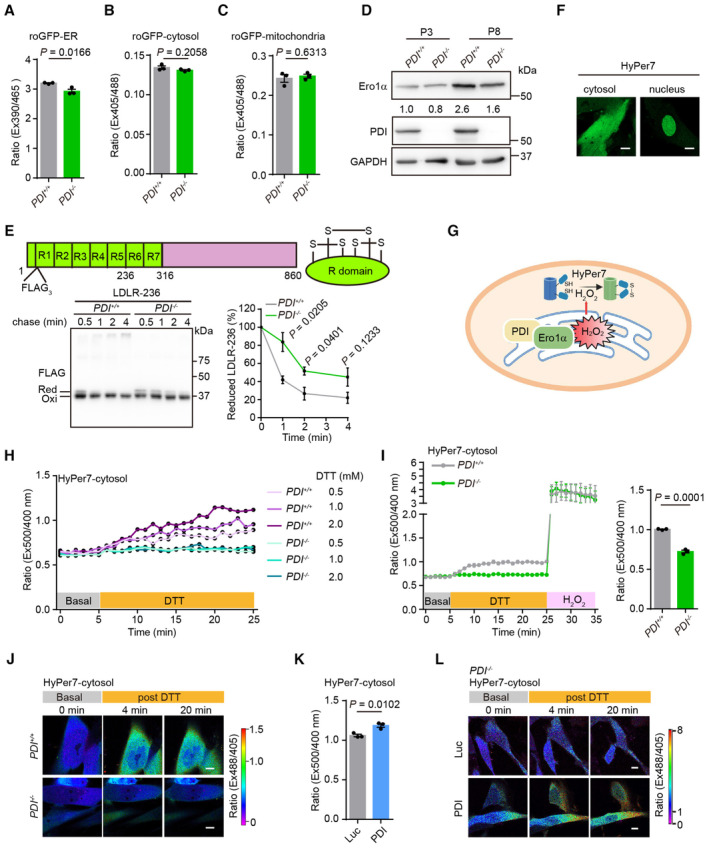
-
A–CGlutathione redox states in the ER (A), cytosol (B) and mitochondria (C) of PDI +/+ and PDI −/− hMSCs were determined by the fluorescence ratio of roGFP‐ER, roGFP‐cytosol or roGFP‐mitochondria, respectively. n = 3 biological repeats.
-
DWestern blotting analysis of Ero1α and PDI expression in PDI +/+ and PDI −/− hMSCs at early‐passage (P3) and late‐passage (P8). GAPDH was used as the loading control.
-
EUpper, domain organization of LDLR. LDLR contains N‐terminally located seven cysteine‐rich domains (R1‐R7). Lower, oxidative folding of a truncated form of LDLR including domains R1‐R5 (LDLR‐236). PDI +/+ and PDI −/− hMSCs were transfected with LDLR‐236 for 48 h, then were pulsed with 5 mM DTT and chased at indicated time after DTT removal. The lysates were analyzed using non‐reducing SDS‐10% PAGE and α‐FLAG western blotting. The mobilities of reduced LDLR‐236 monomers (Red), oxidized monomers (Oxi) are indicated. The fraction of reduced LDLR‐236 was quantified by densitometry. n = 3 independent experiments.
-
FConfirmation of HyPer7 localization to the cytosol and nucleus. Scale bar, 10 μm.
-
GSchematic diagram showing the detection of the cytosol H2O2 by HyPer7‐cytosol upon activation of the Ero1α‐PDI oxidative protein folding system in the ER.
-
HResponse of HyPer7‐cytosol probes in PDI +/+ and PDI −/− hMSCs. The fluorescence intensity ratio of HyPer7‐cytosol at 520 nm with excitation at 500/400 nm was firstly monitored at resting state for 5 min, followed by addition of different concentrations of DTT (yellow bar) for another 20 min.
-
ILeft, response of HyPer7‐cytosol probes in PDI +/+ and PDI −/− hMSCs upon addition of 2 mM DTT for 20 min, followed by addition of 100 μM H2O2. Right, statistical analysis of the fluorescence intensity ratio at 500/400 nm excitation of HyPer7‐cytosol probe at 20 min post DTT addition. n = 3 independent experiments.
-
JConfocal imaging of PDI +/+ and PDI −/− hMSCs expressing Hyper7‐cytosol at steady state and upon addition of 2 mM DTT. The fluorescence intensity ratio at 488/405 nm obtained from individual cells was calculated at indicated times with a representative false color image. Scale bar, 10 μm.
-
KH2O2 levels in PDI −/− hMSCs transduced with lentiviruses expressing Luc or PDI were determined by the fluorescence ratio of HyPer7‐cytosol at 500/400 nm excitation after the addition of 2 mM DTT for 20 min. n = 3 independent experiments.
-
LConfocal imaging of PDI −/− hMSCs transduced with lentiviruses expressing Luc or PDI and Hyper7‐cytosol at steady state and upon addition of 2 mM DTT. The fluorescence intensity ratio at 488/405 nm obtained from individual cells was calculated at indicated times with a representative false color image. Scale bar, 10 μm.
Data information: In (A–C, E, I, K), data are presented as mean ± SEM, two‐tailed Student's t‐test.
Figure 3. Deficiency of PDI slows the rate of oxidative protein folding and reduces its byproduct H2O2 .
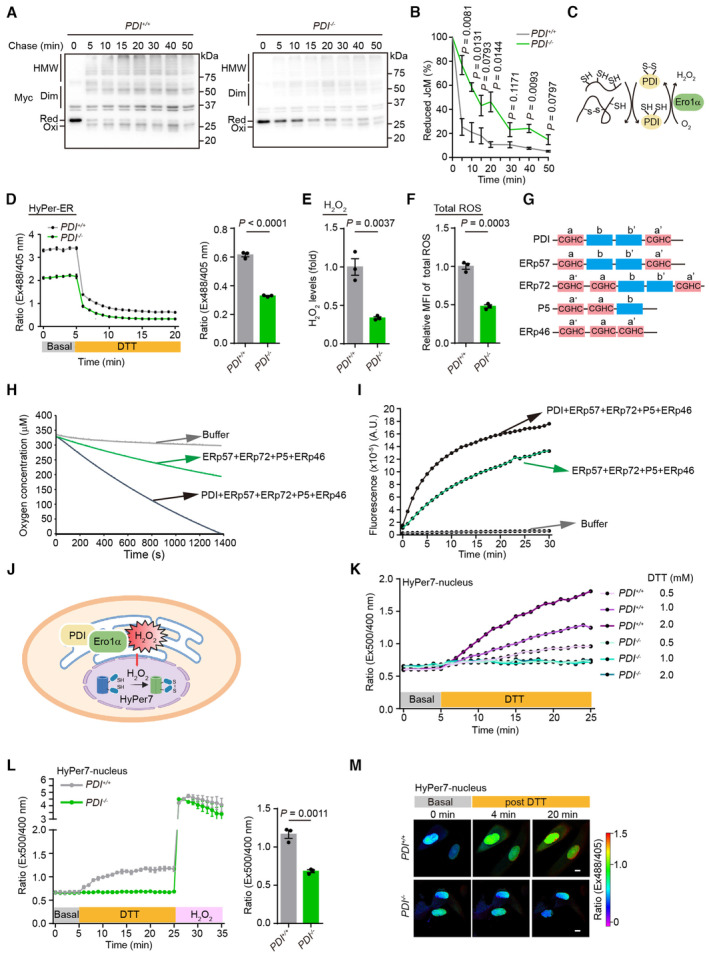
- Oxidative folding of Myc‐tagged J‐Chain (JcM). PDI +/+ and PDI −/− hMSCs were transfected with JcM for 48 h, then were pulsed with DTT and chased at indicated time after DTT removal. The lysates were analyzed using non‐reducing SDS‐15% PAGE and α‐Myc western blotting. The mobilities of reduced JcM monomers (Red), oxidized monomers (Oxi), homodimers (Dim), and high‐molecular‐weight (HMW) species are indicated.
- The fraction of reduced JcM (Red/[Red + Oxi]) in (A) was quantified by densitometry. n = 3 independent experiments.
- Model of the Ero1α‐PDI oxidative protein folding pathway.
- Time‐dependent changes of H2O2 levels in the ER of PDI +/+ and PDI −/− hMSCs at P8. Left, the fluorescence intensity ratio of HyPer‐ER at 535 nm with excitation at 488 and 405 nm was first monitored at resting state for 5 min, followed by addition of 0.5 mM DTT (yellow bar) for another 15 min. Right, statistical analysis of the fluorescence intensity ratio at 488/405 nm excitation of HyPer‐ER probe at 15 min post DTT addition. n = 3 independent experiments.
- Analysis of the H2O2 levels in PDI +/+ and PDI −/− hMSCs at P8 using Amplex Red assays. n = 3 biological repeats.
- Analysis of the cellular total ROS levels in PDI +/+ and PDI −/− hMSCs at P8 using H2DCFDA probe. n = 3 biological repeats.
- Schematic representation of the PDI family members. The ‐CGHC‐ active sites in the catalytic domains are indicated.
- Oxygen consumption by 1 μM purified Ero1α protein was monitored by using an oxygen electrode in the presence of PDI proteins (each 10 μM) and 10 mM GSH.
- H2O2 produced by Ero1α‐PDIs system were detected using Amplex Red assays.
- Schematic diagram showing the detection of the nuclear H2O2 by HyPer7‐nucleus upon activation of the Ero1α‐PDI oxidative protein folding system in the ER.
- Response of HyPer7‐nucleus probes in PDI +/+ and PDI −/− hMSCs. The fluorescence intensity ratio of HyPer7‐nucleus at 520 nm with excitation at 500/400 nm was firstly monitored at resting state for 5 min, followed by addition of different concentrations of DTT (yellow bar) for another 20 min.
- Left, response of HyPer7‐nucleus probes in PDI +/+ and PDI −/− hMSCs upon addition of 2 mM DTT for 20 min, followed by addition of 100 μM H2O2. Right, statistical analysis of the fluorescence intensity ratio at 500/400 nm excitation of HyPer‐nucleus probe at 20 min post‐DTT addition. n = 3 independent experiments.
- Confocal imaging of PDI +/+ and PDI −/− hMSCs expressing Hyper7‐nucleus at steady state and upon addition of 2 mM DTT. The fluorescence intensity ratio at 488/405 nm obtained from individual cells was calculated at indicated times with a representative false color image. Scale bar, 10 μm.
Data information: In (B–F, L), data are presented as mean ± SEM, two‐tailed Student's t‐test.
Source data are available online for this figure.
The by‐product of Ero1α‐PDI‐mediated oxidative protein folding is H2O2 (Fig 3C). Since the accumulation of excess H2O2 has been reported to cause oxidative stress and induce hMSCs senescence (Burova et al, 2013), we reasoned that the slow oxidative protein folding in the absence of PDI helps to attenuate the excessive accumulation of H2O2 and thus delay cell senescence. Therefore, the ER‐localized H2O2 probe HyPer‐ER was used to measure the levels of H2O2 in the ER. The fluorescence intensity ratio at 488/405 nm excitation largely decreased in PDI −/− hMSCs, indicating relatively a low oxidation rate of the HyPer‐ER probe (Fig 3D). Because the HyPer‐ER probe can be oxidized by either H2O2 or by disulfides in the ER (Ramming et al, 2014), a low dose of DTT (0.5 mM) was added to exclude the oxidative effect exerted by disulfides (Zhang et al, 2019b). Then, we observed that the levels of H2O2 in the ER lumen of the PDI −/− hMSCs were much lower than those in the PDI +/+ cells (Fig 3D). An Amplex Red assay confirmed the decreased overall H2O2 levels in the lysates of PDI −/− hMSCs (Fig 3E). Moreover, the total ROS levels were reduced in the PDI −/− hMSCs, as determined by H2DCFDA staining (Fig 3F).
To validate the finding showing that the decrease in H2O2 levels in the PDI −/− hMSCs was mediated by PDI, we reconstituted the oxidative protein folding system in vitro with purified proteins. In addition to PDI, four other constitutively expressed PDI family members, P5, ERp46, ERp72, and ERp57, each of which harbors at least two ‐CGHC‐ active sites critical for catalyzing the thiol‐disulfide interchange reaction (Wang et al, 2021), were analyzed (Fig 3G). An oxygen electrode was used to monitor the oxygen consumption during the oxidation of the PDIs mediated by Ero1α in the presence of GSH, the electron donor. Ero1α oxidized the other PDIs slowly, and the addition of PDI markedly increased the oxygen consumption rate (Fig 3H). Moreover, the presence of PDI increased the production of H2O2, as monitored by Amplex Red assay (Fig 3I). Altogether, our data indicate that PDI deficiency slows the rate of oxidative protein folding and reduces the rate of H2O2 production in the ER.
The ER‐to‐nucleus release of H2O2 is attenuated by PDI deletion
Whether ER‐derived H2O2 is released and affects surrounding organelles, including the nucleus to regulate the gene expression profile during aging, remained unclear. As H2O2 can be rapidly eliminated by peroxidases and peroxiredoxins in the cytosol and nucleus, we employed a recently developed ultrasensitive H2O2 sensor, HyPer7 (Pak et al, 2020; Hoehne et al, 2022), to detect the diffusion of ER‐derived H2O2. HyPer7 was targeted to the nucleus or cytosol (Fig EV3F), and the fluorescence intensity ratio at 500/400 nm excitation was monitored (Figs 3J and EV3G). DTT treatment mimics the burst of free thiols during protein synthesis and profoundly activates H2O2 generation by the Ero1α‐PDI oxidative protein folding system (Ramming et al, 2014). At the resting state, the HyPer7‐nucleus probe was mainly in its reduced state in both PDI +/+ and PDI −/− hMSCs, indicating that the nucleus was harboring a low level of H2O2. Notably, HyPer7‐nucleus in the PDI +/+ hMSCs responded to DTT challenge in a dose‐dependent manner, whereas the probe in the PDI −/− hMSCs was not oxidized at all (Fig 3K). HyPer7‐nucleus in both cell lines was fully oxidized when exogenous H2O2 was added, suggesting that the probe was active (Fig 3L). Live imaging with confocal microscopy confirmed the abovementioned results at the single‐cell level, which clearly showed that the nucleus was flooded with H2O2 upon DTT challenge, while PDI deletion abrogated this effect (Fig 3M). Similar results were observed when HyPer7 was targeted to the cytosol, although the probe was oxidized to a lesser extent, probably due to the powerful antioxidant system in the cytosol (Fig EV3H–J). These results suggest that the H2O2 generated by oxidative protein folding can be efficiently released into the nucleus and cytosol and that knocking out PDI diminishes the release of ER‐derived H2O2.
PDI deficiency attenuates cell senescence through the downregulation of SERPINE1 expression
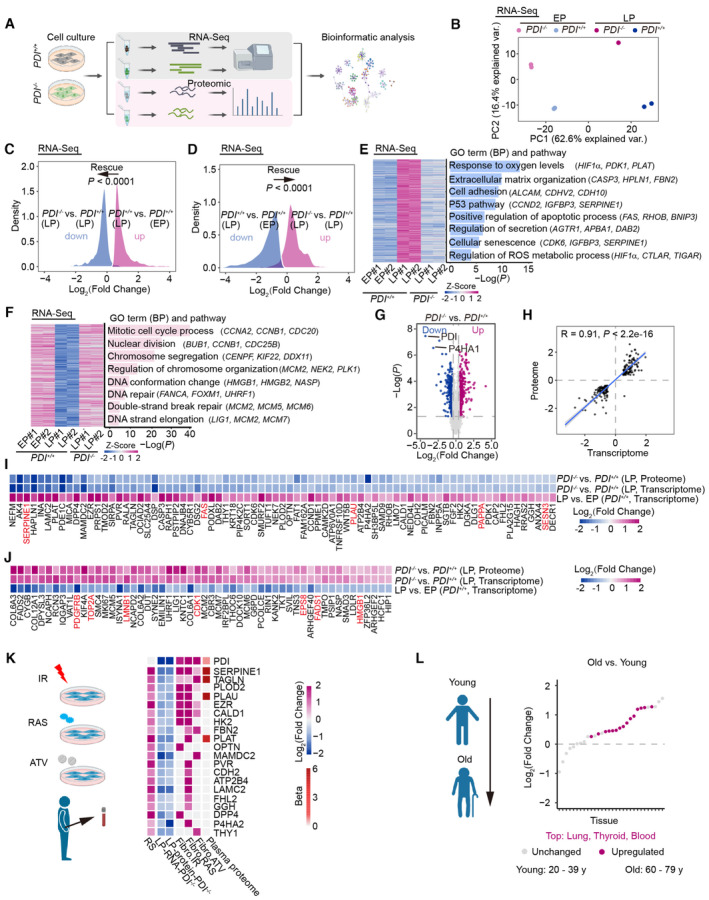
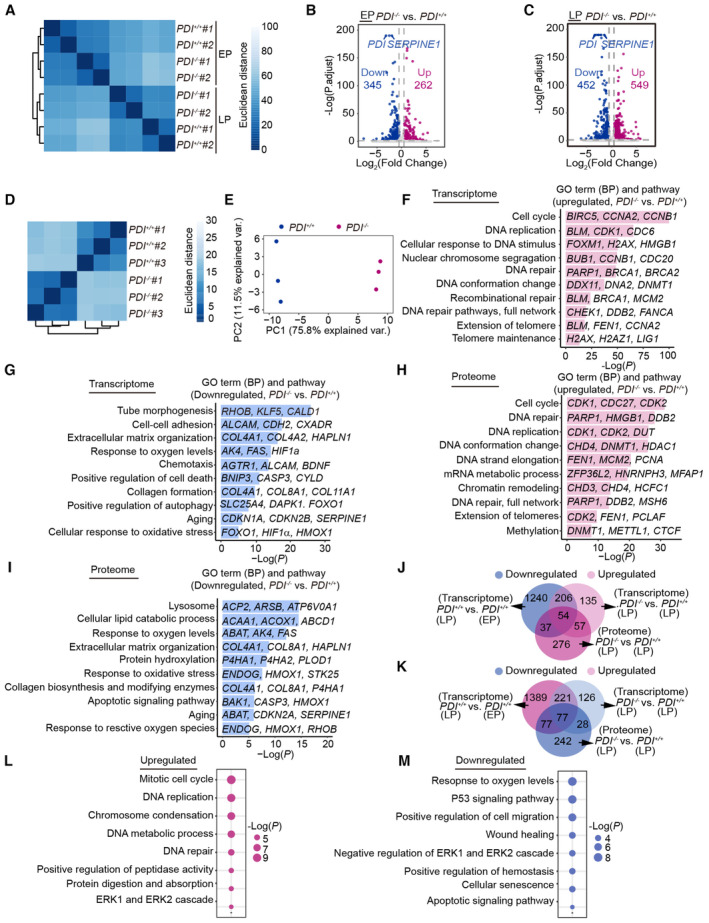
To determine changes in gene expression during PDI‐regulated cell senescence, we performed genome‐wide RNA sequencing (RNA‐Seq; Fig 4A). High reproducibility of transcriptome expression profiles between replicates was confirmed by principal component analysis (PCA) and measurements of Euclidean distance (Figs 4B and EV4A). By performing differential expression analysis, we identified 607 and 1,001 differentially expressed genes (DEGs) in early‐passage (EP) and late‐passage (LP) hMSCs, respectively (Fig EV4B and C). The RNA‐Seq results also revealed that PDI deficiency partially restored the expression of DEGs in LP hMSCs to that in EP hMSCs (Fig 4C and D). A biology pathway analysis showed that DEGs enriched in “response to oxygen levels,” “P53 pathways,” “cellular senescence,” and “regulation of ROS metabolic process” were upregulated during cell senescence and downregulated by PDI depletion. In contrast, DEGs enriched in “mitotic cell cycle process” and “DNA repair” that had been downregulated during cell senescence were reactivated by PDI deletion (Fig 4E and F).
Figure 4. Deficiency of PDI alleviates cell senescence through downregulation of SERPINE1 expression.
-
AWork flow for the RNA‐Seq and proteomic analysis.
-
BPrincipal component analysis (PCA) of transcriptome of early‐passage (EP) and late‐passage (LP) PDI +/+ and PDI −/− hMSCs.
-
C, DDensity plots showing that upregulated (C) or downregulated (D) genes in the LP hMSCs (PDI +/+ , LP vs. PDI +/+ , EP) that were restored after PDI deletion (PDI −/− , LP vs. PDI +/+ , LP). Comparisons were performed with two‐sided Wilcoxon signed‐rank test.
-
E, FHeatmap showing upregulated (E) or downregulated (F) genes in the LP hMSCs (PDI +/+ , LP vs. PDI +/+ , EP) that were restored after PDI deletion (PDI −/− , LP vs. PDI +/+ , LP). Representative GO terms, pathways and genes are shown on the right.
-
GVolcano plots showing differentially expressed proteins (DEPs) between PDI −/− and PDI +/+ hMSCs at LP.
-
HCorrelation analysis of the expression changes at the proteomic and transcriptional levels.
-
I, JHeatmaps showing the upregulated (I) or downregulated (J) genes in the LP hMSCs (PDI +/+ , LP vs. PDI +/+ , EP) that were restored after PDI deletion at both the proteomic and transcriptional levels (PDI −/− , LP vs. PDI +/+ , LP). Genes marked red represent the aging‐related genes from “Aging Atlas” database (https://ngdc.cncb.ac.cn/aging). The list of rescued genes was ranked based on Log2 (fold change) in corresponding comparison.
-
KConjoint analysis showing the secretion levels of PDI and those in (I) and various SASPs from multiple senescence inducers and in plasma proteome from 3,087 healthy individuals (Basisty et al, 2020; Ritchie et al, 2021). RS, replicative senescence (PDI +/+, LP vs. PDI +/+, EP). ATV, atazanavir treatment; RAS, inducible RAS overexpression; IR, X‐irradiation. The color keys that change from dark blue to dark purple represent the fold change of gene and protein, change from white to red represent the change in protein levels per unit increase in the covariate in plasma proteomic.
-
LDot plots showing the upregulated expression of SERPINE1 in different human tissues during aging. These tissue‐specific gene expression data were obtained from Genotype‐Tissue Expression Project (GTEx). Red dots represent significantly upregulated expression in corresponding age groups (P < 0.05). Comparisons were performed with two‐sided Wilcoxon rank‐sum test.
Figure EV4. Deficiency of PDI alleviates cell senescence through downregulation of SERPINE1 expression.
-
AHeatmap showing the Euclidian distance of transcriptome between replicates of PDI +/+ and PDI −/− hMSCs at EP and LP.
-
B, CVolcano plots showing the DEGs between the PDI −/− and PDI +/+ hMSCs at EP (B) or LP (C).
-
DHeatmap showing the Euclidian distance of proteome between replicates of PDI +/+ and PDI −/− hMSCs at LP.
-
EPCA of PDI +/+ and PDI −/− hMSCs proteome at LP.
-
F, GRepresentative GO terms and pathways enrichment analysis based on upregulated (F) or downregulated (G) DEGs between PDI −/− and PDI +/+ hMSCs at LP.
-
H, IRepresentative GO terms and pathways enrichment analysis based on upregulated (H) or downregulated (I) DEPs between PDI −/− and PDI +/+ hMSCs at LP.
-
JVenn diagrams showing downregulated DEGs (PDI +/+ , LP vs PDI +/+ EP), upregulated DEGs (PDI −/− LP, vs PDI +/+ ) and upregulated DEPs (PDI −/− , LP vs PDI +/+ , LP).
-
KVenn diagrams showing upregulated DEGs (PDI +/+ , LP vs PDI +/+ EP), downregulated DEGs (PDI −/− LP, vs. PDI +/+ ) and downregulated DEPs (PDI −/− , LP vs. PDI +/+ , LP).
-
L, MRepresentative GO terms and pathways enrichment analysis based on upregulated (L) or downregulated (M) DEGs and DEPs in the LP hMSCs (PDI +/+ , LP vs. PDI +/+ , EP) that were restored after PDI deletion at both the proteomic and transcriptional levels (PDI −/− , LP vs. PDI +/+ , LP).
We next performed quantitative proteomic analysis of LP PDI +/+ and PDI −/− hMSCs to identify changes in protein levels mediated by PDI knockout (Fig 4A and G). PCA and Euclidean distance measurements showed high reproducibility between replicates (Fig EV4D and E). In line with the results of RNA‐Seq, we found that differentially expressed proteins (DEPs) and DEGs enriched in pathways such as “cell cycle,” “DNA replication,” and “DNA repair” were upregulated in the PDI −/− hMSCs. In contrast, DEPs and DEGs enriched in pathways such as “response to oxygen levels” and “response to oxidative stress” were downregulated in the PDI −/− hMSCs, consistent with results showing that deletion of PDI reduced H2O2 levels. In addition, “aging”‐related DEGs and DEPs were downregulated, consistent with the phenotypes observed in the PDI −/− hMSCs (Fig EV4F–I).
To better explore the molecular mechanisms of PDI‐regulated cell senescence, we performed an integrated analysis with transcriptomic and proteomic data. A correlation analysis of DEGs and DEPs showed high congruence (R = 0.91, Fig 4H) between the transcriptomic and proteomic data. We further identified and ranked genes with expression that changed at both the transcriptional and proteomic levels. The analysis revealed 54 upregulated and 77 downregulated DEPs/DEGs in RS hMSCs, and the expression of these genes was reversed by PDI deletion (Fig EV4J and K). Moreover, after PDI deletion, the restored DEGs and DEPs were mainly enriched in “mitotic cell cycle,” “DNA repair,” “response to oxygen levels,” and “cellular senescence” terms (Fig EV4L and M). Next, we performed an integrative comparative analysis between the restored genes and annotated hotspot aging‐related genes obtained from an Aging Atlas gene set (Aging Atlas, 2021) and found that 12 of the restored genes were also hotspot aging‐related genes (Fig 4I and J). Among these 12 genes, SERPINE1 was found to be a key candidate with the greatest degree of recovery. In addition, in line with the expression of PDI, SERPINE1 is a component in the SASP and a prominent plasma biomarker of aging (Fig 4K), underlining the potential correlation between dysregulation of SERPINE1 and PDI expression during aging. Moreover, by analyzing the transcriptome profile of human tissues in different age groups, we found that the expression of SERPINE1 was upregulated in most aged human tissues, such as the lung, thyroid, and blood (Fig 4L), implying that SERPINE1 is closely related with physiological aging. Notably, SERPINE1 is also a redox‐regulated gene (Swiatkowska et al, 2002; Oszajca et al, 2008). Therefore, we reasoned that PDI‐H2O2‐SERPINE1 may constitute a key axis that regulates stem cell aging.
SERPINE1 is a key driver of cell senescence
The SERPINE1 gene encodes plasminogen activator inhibitor‐1 (PAI‐1), which negatively regulates the activities of two plasminogen activators (tissue‐type PA, tPA, and urokinase‐type PA, uPA) and therefore inhibits fibrinolysis (Rahman & Krause, 2020). In addition, PAI‐1 plays an important role in regulating cell death, cell senescence and inflammation (Rahman & Krause, 2020). The mRNA and protein levels of SERPINE1 were increased in senescent hMSCs, and this increase was not evident in PDI −/− hMSCs (Fig 5A and B). To determine whether the geroprotective effect conferred by PDI deletion was due to the downregulation of SERPINE1 expression, we induced the expression of SERPINE1 in PDI −/− hMSCs using a CRISPR/dCas9 transcriptional activation system (Joung et al, 2017; Fig 5C). The endogenous activation efficiency of two guide RNAs (gRNAs) targeting the promoter region of the SERPINE1 gene was validated by western blotting (Fig 5D). Activation of endogenous SERPINE1 expression in PDI −/− hMSCs accelerated cell senescence, as manifested by decreased single clonal formation ability, fewer Ki67‐positive cells and increased SA‐β‐gal‐positive cells (Fig 5E–G). Moreover, the activation of SERPINE1 expression in PDI +/+ hMSCs accelerated cell senescence (Fig EV5A–C), suggesting that SERPINE1 is a driving force that mediates cell aging.
Figure 5. PDI‐H2O2‐SERPINE1 axis plays a key role in cell senescence.
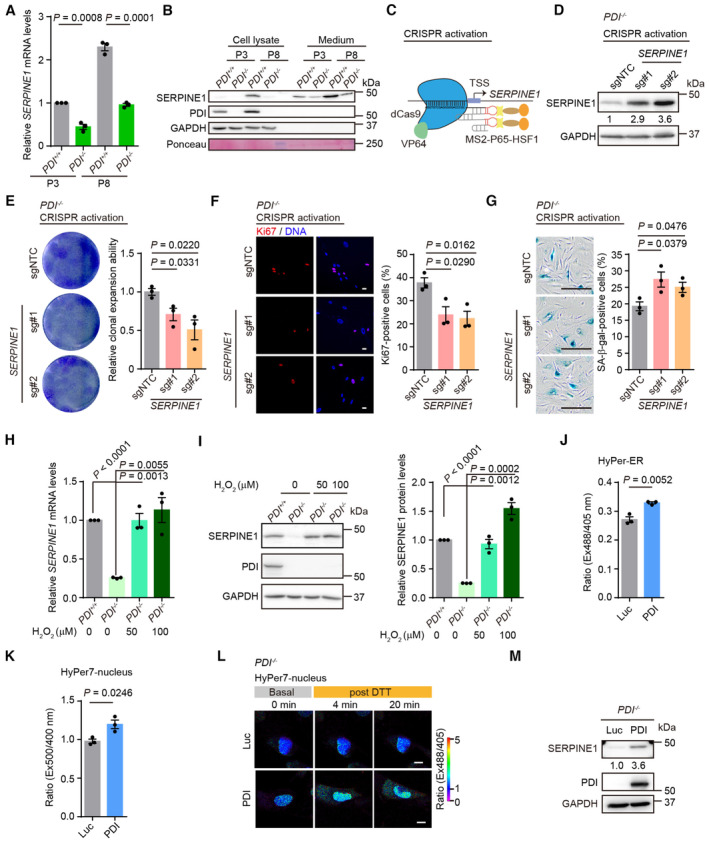
- qPCR analysis of SERPINE1 expression in PDI +/+ and PDI −/− hMSCs at EP and LP. n = 3 independent experiments.
- Western blotting analysis of SERPINE1 protein in PDI +/+ and PDI −/− hMSCs at EP and LP. GAPDH was used as the loading control of cell lysates. Ponceau was used as the loading control of conditional medium.
- Schematic of CRISPR/dCas9 mediated transcriptional activation systems.
- Western blotting of SERPINE1 protein in PDI −/− hMSCs transduced with two SERPINE1‐targeting activation sgRNAs or sgNTC. GAPDH was used as the loading control.
- Single clonal formation assay in PDI −/− hMSCs transduced with sgNTC or SERPINE1‐targeting activation sgRNAs. n = 3 biological repeats.
- Left, Ki67 staining in PDI −/− hMSCs transduced with sgNTC or SERPINE1‐targeting activation sgRNAs. Scale bar, 20 μm. Right, statistical analysis of Ki67‐positive cells. n = 3 independent experiments.
- Left, SA‐β‐gal staining in PDI −/− hMSCs transduced with sgNTC or SERPINE1‐targeting activation sgRNAs. Scale bar, 200 μm. Right, statistical analysis of SA‐β‐gal‐positive cells. n = 3 biological repeats.
- qPCR analysis of the SERPINE1 mRNA expression in PDI +/+ and PDI −/− hMSCs treated with the indicated concentration H2O2 for 4 days. n = 3 independent experiments.
- Left, western blotting analysis of the SERPINE1 protein expression in PDI +/+ and PDI −/− hMSCs treated with the indicated concentration H2O2 for 4 days. GAPDH was used as the loading control. Right, statistical analysis of relative SERPINE1 protein levels. n = 3 independent experiments.
- H2O2 levels in PDI −/− hMSCs transduced with lentiviruses expressing Luc or PDI were determined by the fluorescence ratio of HyPer‐ER at 488/405 nm excitation after the addition of 0.5 mM DTT for 15 min. n = 3 biological repeats.
- H2O2 levels in PDI −/− hMSCs transduced with lentiviruses expressing Luc or PDI were determined by the fluorescence ratio of HyPer7‐nucleus at 500/400 nm excitation after the addition of 2 mM DTT for 20 min. n = 3 independent experiments.
- Confocal imaging of PDI −/− hMSCs transduced with lentiviruses expressing Luc or PDI and Hyper7‐nucleus at steady state and upon addition of 2 mM DTT. The fluorescence intensity ratio at 488/405 nm obtained from individual cells was calculated at indicated times with a representative false color image. Scale bar, 10 μm.
- Western blotting of SERPINE1 expression in PDI −/− hMSCs transduced with lentiviruses expressing Luc or PDI. GAPDH was used as the loading control.
Data information: In (A, E–K), data are presented as mean ± SEM, two‐tailed Student's t‐test.
Source data are available online for this figure.
Figure EV5. Upregulation of SERPINE1 induces cell senescence.
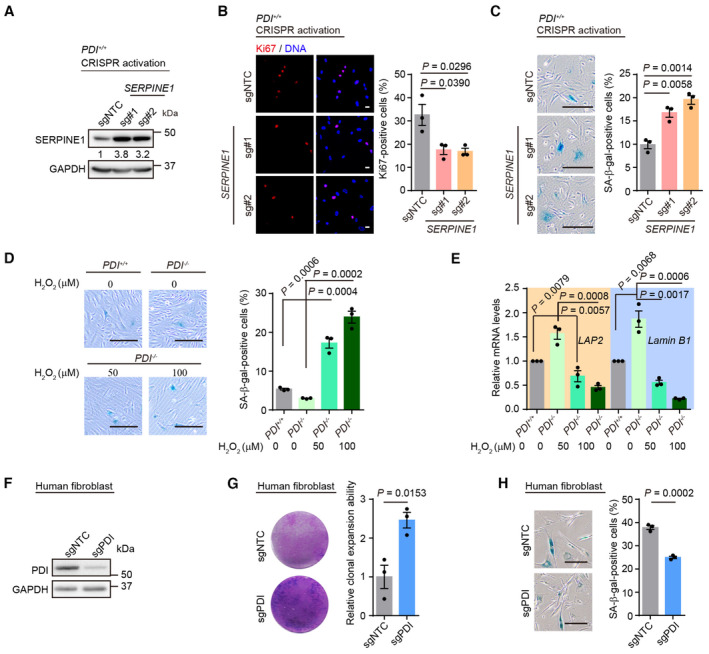
- Western blotting of SERPINE1 protein in PDI +/+ hMSCs transduced with two SERPINE1‐targeting activation sgRNAs or sgNTC. GAPDH was used as the loading control.
- Left, Ki67 staining in PDI +/+ hMSCs transduced with sgNTC or SERPINE1‐targeting activation sgRNAs. Scale bar, 20 μm. Right, statistical analysis of Ki67‐positive cells. n = 3 biological repeats.
- Left, SA‐β‐gal staining in PDI +/+ hMSCs transduced with sgNTC or SERPINE1‐targeting activation sgRNAs. Scale bar, 200 μm. Right, statistical analysis of SA‐β‐gal‐positive cells. n = 3 biological repeats.
- Left, SA‐β‐gal staining of PDI +/+ and PDI −/− hMSCs treated with the indicated concentration H2O2 for 4 days. Scale bar, 200 μm. Right, statistical analysis of SA‐β‐gal‐positive cells. n = 3 biological repeats.
- qPCR analysis of aging‐related genes expression in PDI +/+ and PDI −/− hMSCs treated with the indicated concentration H2O2 for 4 days. Data are shown as mean ± SEM, n = 3 independent experiments.
- Western blotting of PDI expression in human fibroblast transduced with sgNTC or PDI‐targeting sgRNA. GAPDH was used as the loading control.
- Single clonal formation assay in human fibroblasts transduced with sgNTC or PDI‐targeting sgRNA. n = 3 biological repeats.
- SA‐β‐gal staining in human fibroblasts transduced with sgNTC or PDI‐targeting sgRNA. Scale bar, 200 μm. n = 3 biological repeat.
Data information: In (B–E, G, H), data are presented as mean ± SEM, two‐tailed Student's t‐test.
The PDI‐H2O2 axis regulates SERPINE1 expression
Next, we wondered whether the expression of SERPINE1 was mediated by H2O2 levels. The addition of H2O2 increased both the transcriptional and protein levels of SERPINE1 in PDI −/− hMSCs to an extent similar to those in PDI +/+ hMSCs (Fig 5H and I). Importantly, the same concentration of H2O2 induced the senescence of PDI −/− hMSCs (Fig EV5D and E). Furthermore, reintroduction of PDI into PDI −/− hMSCs increased the H2O2 levels in the ER (Fig 5J) and enhanced the release of H2O2 into the nucleus (Fig 5K and L) and cytosol (Fig EV3K and L). Replenishment of PDI also led to the upregulated expression of SERPINE1 (Fig 5M). Therefore, we conclude that PDI regulates the expression of SERPINE1 via H2O2 and that PDI‐H2O2‐SERPINE1 axis constitutes an important pathway that contributes to cell senescence.
Targeting PDI alleviates senescence in diverse cell types
To explore the geroprotective role of PDI depletion in a wider range of cell types, RS hMSCs, premature‐senescent hMSCs (Hutchinson‐Gilford Progeria Syndrome, HGPS; and WS hMSCs), and primary hMSCs from an elderly individual were edited via CRISPR/Cas9 technology to knockdown PDI expression (Fig 6A). In all the hMSCs models, PDI knockdown alleviated cell senescence, as manifested by increased clonal expansion ability and a decreased percentage of SA‐β‐gal‐positive cells (Fig 6B and C). Moreover, PDI knockdown mitigated wild‐type hMSCs senescence induced by different cellular stressors, including ultraviolet radiation (UV) and oncogene overexpression (Fig 6D–G). Additionally, PDI knockdown inhibited the senescence of human fibroblasts (Fig EV5F–H). Altogether, these results indicate that PDI is a potential target for alleviating cell senescence in different types of hMSCs and human fibroblasts.
Figure 6. Targeting PDI alleviates senescence in diverse cell types.
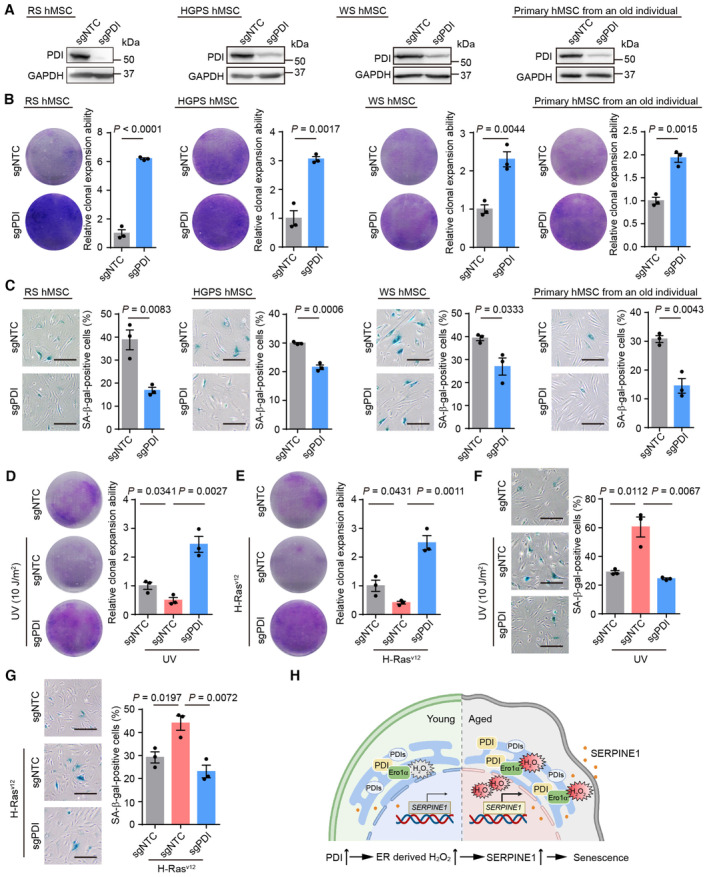
-
AWestern blotting of PDI expression in replicative senescent (RS) hMSCs, Hutchinson‐Gilford Progeria Syndrome (HGPS) hMSCs, WS hMSCs and human primary hMSCs from an old individual transduced with sgNTC or PDI‐targeting sgRNA. GAPDH was used as the loading control.
-
BSingle clonal formation assay in RS hMSCs, HGPS hMSCs, WS hMSCs and human primary hMSCs from an old individual transduced with sgNTC or PDI‐targeting sgRNA. n = 3 biological repeats.
-
CSA‐β‐gal staining in RS hMSCs, HGPS hMSCs, WS hMSCs and human primary hMSCs from an old individual transduced with sgNTC or PDI‐targeting sgRNA. n = 3 biological repeats. Scale bar, 200 μm.
-
D–G(D, F) Single clonal formation assay (D) and SA‐β‐gal staining assay (F) in RS hMSCs transduced with sgNTC or PDI‐targeting sgRNA after treatment with UV irradiation (10 J/m2) and then cultured for another 2 days. n = 3 biological repeats. (E, G) Single clonal formation assay (E) and SA‐β‐gal staining assay (G) in RS hMSCs transduced with sgNTC or PDI‐targeting sgRNA after overexpressing of H‐RasV12. n = 3 biological repeats. Scale bar, 200 μm.
-
HA model showing PDI and its upstream oxidase Ero1α are upregulated during aging, and Ero1α‐PDI mediated oxidative protein folding produces excess H2O2 in the ER as well as in the nucleus, upregulates the expression of SASP including SERPINE1, disrupts hMSCs homeostasis and promotes senescence.
Data information: In (B–G), data are presented as mean ± SEM, two‐tailed Student's t‐test.
Source data are available online for this figure.
Discussion
In this study, we established a novel link between oxidative protein folding and cell aging. We observed that the expression of PDI, a key enzyme catalyzing disulfide bonds formation in the ER, was upregulated during replicative and premature hMSCs senescence. Increased PDI accelerated oxidative protein folding, resulting in the accumulation of the byproduct H2O2 in both the ER and nucleus. ER‐derived oxidative stress induced the expression of SERPINE1, which promoted cell senescence furtherly (Fig 6H).
Since PDI is widely considered to be important for disulfide bonds formation and PDI‐knockout mice are embryonic lethal (Xu et al, 2014; Parakh & Atkin, 2015), the viability of PDI‐knockout hMSCs is surprising. Moreover, they were healthier and younger than wild‐type cells. The high redundancy of PDI family proteins in the ER may explain this outcome. We showed that the recombination of four PDI homologs, namely ERp57, P5, ERp72, and ERp46, led to disulfide formation in cooperation with Ero1α oxidase, but the oxygen consumption and H2O2 production rates were slower than with PDI. In addition, the rate of oxidative folding of the J‐chain in PDI −/− hMSCs was slowed, and fewer high‐molecular‐weight species due to indiscriminate oxidation by H2O2 accumulated (Wang et al, 2014). Moreover, PDI knockout attenuated the upregulated expression of Ero1α during aging, further lowering the risk of oxidative stress. Thus, PDI deletion resulted in slower but safer oxidative protein folding in hMSCs. However, this outcome does not mean that PDI is dispensable in all developmental stage and all types of cells. For example, the PDI knockout mice have lethal phenotypes occurring at E9.5 (https://www.mousephenotype.org/data/genes/MGI:97464), which is later than the blastocyst‐stage where the hESCs derived, and PDI −/− hESCs cannot be differentiated into human vascular endothelial cells (hVECs) efficiently (our unpublished observation), suggesting that PDI may play an important role in angiogenesis. Besides, in pancreas β‐cells, PDI is required for the correct disulfide formation of proinsulin (Jang et al, 2019). In addition, PDI associates with Drp1 to protect endothelial cells against mitochondrial fragmentation and endothelial dysfunction (Kim et al, 2018).
According to the free radical theory of aging, accumulating ROS induce oxidative stress and promote cell senescence (Harman, 1956; Kudryavtseva et al, 2016). Yet, previous studies of oxidative stress on cell senescence have mostly focused on mitochondrion‐derived ROS (Li et al, 2013; Vasileiou et al, 2019), the role of ER‐generated ROS has been less extensively studied. In fact, the ER may be the most significant ROS producer in some cell types, and among purified organelles, ER‐derived ROS accounts for approximately 60% of all the ROS produced (Colton & Gilbert, 2007). Previous results have shown that upregulation of Ero1 resulted in excessive ROS production, while knockdown of Ero1 reduced the peroxide levels in worms, extending their lifespan by 32% (Curran & Ruvkun, 2007). Ero1‐generated H2O2 can be eliminated by ER‐resident glutathione peroxidase 7 (GPx7; Wang et al, 2014). Our previous study showed that GPx7 knockdown promoted cell senescence (Fang et al, 2018), further emphasizing the role played by ER‐derived H2O2 in aging. However, whether H2O2 can readily pass through the ER membrane and affect other organelles has been contentiously debated (Konno et al, 2015; Appenzeller‐Herzog et al, 2016). In this study, by using the ultrasensitive H2O2 probe HyPer7, we clearly showed that the H2O2 generated by oxidative protein folding was released into the nucleus and cytosol. As a mobile signaling molecule, H2O2 can participate in the regulation of gene expression during stem cell aging (Barandalla et al, 2017). How the H2O2 generated in the ER is released into the nucleus or cytosol remains unclear. One candidate is ER membrane‐localized aquaporin 11 (AQP11), as indicated by downregulation of AQP11 severely disrupting the flux of H2O2 through the ER (Appenzeller‐Herzog et al, 2016; Basisty et al, 2020).
The lack of an animal or a cell model has largely limited the investigation of PDI and oxidative protein folding in the regulation of aging. In this study, we established PDI‐knockout hESCs and showed that PDI −/− hESCs maintained pluripotency and self‐renewal capacity. In addition, PDI −/− hESCs were differentiated into hMSCs. Thus, our study provides valuable cell models for exploring the biological function of PDI in various human stem cells and a potential strategy for generating genetic enhancement human stem cells for cell therapies. The geroprotection conferred by PDI depletion was validated in various types of hMSCs and human fibroblasts. The unexpected function of PDI in regulating cell senescence highlights PDI as a newly discovered potential target for alleviating aging. Recently, PDI has emerged as a potential target for cancer and thrombosis treatment (Xu et al, 2014; Bekendam & Flaumenhaft, 2016; Wang et al, 2021). It will be interesting to investigate whether anticancer and antithrombotic PDI inhibitors can be used for aging intervention.
Furthermore, we revealed that the redox and aging‐related gene SERPINE1 is regulated by the PDI‐H2O2 axis and functions as a key driver of cell senescence. SERPINE1 is regarded as a vital component of the senescence‐related secretome (Khan et al, 2017). In line with previous reports indicating that SERPINE1 expression is regulated via H2O2 (Swiatkowska et al, 2002; Oszajca et al, 2008), our study indicated that exogenous H2O2 or ER‐derived H2O2 promoted the expression of SERPINE1, which further accelerated cell senescence. Therefore, it is likely that the leaked H2O2 from the ER during oxidative protein folding activates oxidative stress‐related transcription factors such as NF‐κB and HIF‐1 in the nucleus, and upregulates SERPINE1 gene expression (Oszajca et al, 2008; Rahman & Krause, 2020). Since it is a secretory protein, SERPINE1 can be used as a reliable and convenient biomarker of aging. Moreover, as both PDI (Kim et al, 2013; Wang et al, 2022c) and SERPINE1 (Westrick & Eitzman, 2007; Hisada et al, 2021) have been well‐documented to be associated with thrombosis, targeting the PDI‐H2O2‐SERPINE1 axis may be a more effective strategy for the prevention and treatment of vascular disease.
Materials and Methods
Cell culture
PDI +/+ hESCs (Line H9, WA09, WiCell Research Institute, Inc.) and PDI −/− hESCs were cultured on Mitomycin C (MMC; Selleck)‐inactivated mouse embryonic fibroblast (MEF) feeder cells in the hESCs culture medium supplemented with DMEM/F12 medium (Gibco), 20% KnockOut Serum Replacement (Gibco), 2 mM GlutaMAX (Gibco), 0.1 mM nonessential amino acids (NEAA, Gibco), 100 μg/ml streptomycin and 100 U/ml penicillin (Thermo Fisher Scientific), 55 μM β‐mercaptoethanol (Thermo Fisher Scientific), and 10 ng/ml FGF2 (Joint Protein Central, JPC) or on Matrigel (Corning) in mTeSR medium (STEMCELL Technologies). hMSCs were cultured in 0.1% Gelatin‐coated plates in hMSC medium containing α‐MEM basal medium (Gibco), 10% fetal bovine serum (FBS, Gibco), 0.1 mM NEAA (Gibco), 100 μg/ml streptomycin and 100 U/ml penicillin (Thermo Fisher Scientific), and 1 ng/ml FGF2 (JPC). Human fibroblasts (BJ‐1, ATCC) and HEK293T cells were cultured in high‐glucose DMEM (Thermo Fisher Scientific) supplemented with 10% FBS and 100 μg/ml streptomycin and 100 U/ml penicillin. There was no mycoplasma contamination observed during cell culture.
Generation of PDI −/− hESCs
The PDI gene knockout was performed with the CRISPR/Cas9 genome editing system (Hu et al, 2020; Zhang et al, 2022a). In brief, the guide sequence targeting the exon 2 of the PDI gene was cloned into the pCAG‐mCherry‐gRNA vector (Addgene #87110). H9 hESCs were cultured in Matrigel‐coated plates in mTeSR medium and were pretreated with the ROCK inhibitor Y‐27632 (Selleck) for 24 h before plasmid electroporation. The H9 hESCs were dissociated with TrypLE (Thermo Fisher Scientific), and 5 × 106 H9 ESCs were electroporated with 14 μg pCAG‐1BPNLS‐Cas9‐1BPNLS‐2AGFP (Addgene #87109) vectors and 7 μg pCAGmCherry‐PDI‐gRNA vectors using a 4D Nucleofector (Lonza). After 48 h culture in mTeSR medium, GFP and mCherry double‐positive cells were sorted by FACS (BD, Aria II) and then cultured on MMC‐inactivated MEF cells in hESC culture medium. After 2 weeks in culture, emerging colonies were picked into a 24‐well culture plate and expanded for genomic DNA extraction and sequencing identification.
The sequence for the PDI guide RNA (gRNA) was 5′‐GCGGAAAAGCAACUUCGCGG‐3′. The primers for clone identification were 5′‐CAGGATTTATAAAGGCGAGGC‐3′ (forward) and 5′‐CTCACAGAACTCCACCAGCA‐3′ (reverse).
Generation and characterization of PDI +/+ and PDI −/− hMSCs
Differentiation of hESCs into hMSCs was performed as described previously (Cheng et al, 2019; Shan et al, 2022). Briefly, embryoid bodies (EBs) were derived from hESCs and plated onto Matrigel‐coated plates in hMSC differentiation medium containing 90% α‐MEM, 10% FBS, 0.1 mM NEAA, 100 μg/ml streptomycin and 100 U/ml penicillin, 10 ng/ml FGF2 (JPC) and 5 ng/ml TGFβ (HumanZyme). After about 10 days, the fibroblast‐like cells appeared. The cells were then transferred into gelatin‐coated plates in hMSC culture medium. Then, CD73, CD90, and CD105 tri‐positive cells were sorted as hMSCs by FACS (BD, Aria II). Identification of hMSC‐specific markers with antibodies including anti‐human CD73‐PE (BD, 550257), anti‐human CD90‐FITC (BD, 555595), and anti‐human CD105‐APC (BD, 323208). CD43, CD34, and CD45 were used as negative markers for verifying hMSCs by FACS (BD FACS Calibur). Negative hMSC markers with antibodies including anti‐human CD34‐FITC (BD, 555821), anti‐human CD43‐APC (BD, 560198), and anti‐human CD45‐FITC (BD, 555482). To evaluate the multiple‐lineage differentiation abilities, differentiation of hMSCs toward chondrocytes, osteoblasts, and adipocytes and characterized by Toluidine blue (Sigma, 89,640, chondrogenesis), Alizarin Red S staining (Sigma, A5533, osteogenesis), and Oil red O (Sigma, O1391, adipogenesis) staining.
Isolation and culture of primary hMSCs
The primary hMSCs were isolated from a 76‐year‐old healthy male gingiva tissues according to previous report (Hu et al, 2020; He et al, 2022). Briefly, the gingiva tissues were cut into granule and digested in TrypLE (Thermo Fisher Scientific) and Dispase IV (Thermo Fisher Scientific) at 37°C for 30 min. Then, the digested tissues were filtered through a 70‐μm cell strainer (Falcon) and centrifuged at 500 g for 10 min. The pellets were cultured in the hMSCs culture medium for about 14 days in gelatin‐coated plates. Primary hMSCs were isolated from the dental pulp tissues of different individuals with the approval from the Ethics Committee of the 306 Hospital of PLA in Beijing (Ren et al, 2019).
Copy number variation (CNV) identification
The genomic DNA from hESCs or hMSCs was extracted using a DNeasy Blood & Tissue Kit (TIANGEN). To obtain DNA fragments of approximately 300 base pairs, the extracted genomic DNA was subjected to ultrasonication by Covaris. Sequencing libraries were constructed with the Next DNA Library Prep Reagent Set for Illumina (NEB). WGS data analysis was performed as previously described (Liu et al, 2022b). In brief, we trimmed raw reads with TrimGalore. Then, the clean reads were mapped to the human hg19 genome. The mapped reads were further filtered using samtools and Picard software. Remained reads were counted for each 500‐kb window using read counter function in HMMcopy_utils (https://github.com/shahcompbio/hmmcopy_utils). The R/Bioconductor package HMMcopy (v1.26.0) was used to correct copy number, GC content and mappability.
Clonal expansion assay
Clonal expansion assay was performed as described previously (Liang et al, 2021; Lei et al, 2022; Zhao et al, 2022). In brief, about 2,000 cells were seeded on a 12‐well plate coated with gelatin. Approximately 10 days later, the cells were washed with PBS, fixed with 4% paraformaldehyde (PFA) at room temperature for 30 min, and stained with 0.1% crystal violet solution (Sigma) at room temperature for 30 min. After wash with water, images were captured by Epson Perfection V370 Photo and relative cell density was quantified using ImageJ.
Senescence‐associated β‐galactosidase (SA‐β‐gal) staining
SA‐β‐gal staining assay was carried out as described previously (Ren et al, 2019; Hu et al, 2020; Liu et al, 2023). In brief, hMSCs on six‐well plates were washed with PBS twice, and then fixed in fixation buffer containing 2% (w/v) formaldehyde and 0.2% (w/v) glutaraldehyde for 5 min at room temperature. After that, cells were washed with PBS twice and stained with SA‐β‐gal staining buffer containing 5 mM K4[Fe(CN)6], 5 mM K3[Fe(CN)6], 150 mM NaCl, 2 mM MgCl2, 40 mM citric acid/Na phosphate buffer, 1 mg/ml X‐gal (AMRESCO) at 37°C overnight. Stained cells were then captured with a digital microscope camera (Olympus) and the number of SA‐β‐gal‐positive cells was quantified by ImageJ.
Animal experiments
All animal experiments conducted in this research were approved by the Institutional Biomedical Research Ethics Committee of the Institute of Biophysics and Institute of Zoology, CAS. The mice were raised and housed at 25°C in the Laboratory of Immunodeficiency Animals of Institute of Biophysics and Institute of Zoology, CAS, on a 12‐h light–dark cycle. For the western blotting assay, C57BL/6J male mice (SiPeiFu) were euthanized and tissues were collected. For the teratoma assay, hESCs were harvested in a Matrigel/mTeSR (1:4) solution and the mixtures were injected into the inguinal region of NOD/SCID mice (male, 4–6 weeks, GemPharmatech). After about 2 months, the teratomas were collected and analyzed using immunofluorescence staining. The hMSC transplantation assay was carried out as previously described (Bi et al, 2020). Briefly, about 1 × 106 hMSCs expressing luciferase were injected into the tibialis anterior (TA) muscle of nude mice (male, 6–8 weeks, GemPharmatech). The luciferase activity of mice was imaged using IVIS spectrum imaging system (XENOGEN, Caliper) every 2 days after injection.
Immunofluorescence microscopy
Immunostaining was conducted as described previously (Shan et al, 2022; Wang et al, 2022d). Briefly, cells on the coverslips were washed twice with PBS and fixed with 4% PFA at room temperature for 15 min, washed with PBS twice, and permeabilized with 0.4% Triton X‐100 in PBS at room temperature for another 5 min and then blocked with 10% donkey serum (Jackson ImmunoResearch Labs) in PBS at room temperature for 1 h. The cells were then incubated with primary antibodies in blocking solution at 4°C overnight and then incubated the corresponding fluorescent secondary antibodies in dark at room temperature for 1 h. Images were captured using a confocal microscope (Leica SP5 and Nikon). The primary antibodies used were as follows (company, catalog number): anti‐OCT4 (Santa Cruz, sc‐5279), anti‐SOX2 (Santa Cruz, sc‐17320), anti‐NANOG (Abcam, ab21624), anti‐Ki67 (ZSGB‐Bio, ZM‐0166), anti‐γH2AX (Millipore, 05–636), anti‐53BP1 (Bethyl Laboratories, A300‐273A), anti‐β‐tubulin III (TuJ1, Sigma, T2200), anti‐SMA (Abcam, ab32575), anti‐FOXA2 (Cell Signaling Technology, 8186S), anti‐PDI (Abcam, ab2792), anti‐calreticulin (Abcam, ab2907), and anti‐LAP2 (BD, 611000).
Cell cycle analysis
Cell cycle analysis was performed as previously described (Zhang et al, 2019b). Cells were harvested and washed twice with PBS, and then were fixed in 70% ethanol at −20°C overnight. The fixed cells were washed with PBS and stained with 200 μL propidium iodide (Invitrogen) for 30 min in dark at room temperature. Samples were then analyzed by BD flow cytometry, and cell‐cycle phase distributions were analyzed by the ModFit software.
Determination of cytosol, mitochondria, and ER redox states
PDI +/+ and PDI −/− hMSCs were infected with PLE4‐roGFP‐cytosol, PLE4‐roGFP‐mitochondria or PLE4‐roGFP‐ER lentiviruses for 72 h, harvested and washed with Hanks' Balanced Salt Solution (HBSS, Gibco), and then transferred into a flat‐bottom 96‐well plates. For the roGFP‐cytosol and roGFP‐mitochondrial, the fluorescence intensity was measured at 525 nm with excitation at 405 and 488 nm using an EnSpire Multimode Plate Reader (Perkin Elmer), and the ratio of 405/488 nm was calculated. For the roGFP‐ER, the fluorescence intensity was measured at 525 nm with excitation at 390 and 465 nm, and the ratio of 390/465 nm was calculated.
In vivo oxidative protein folding assays
The in vivo myc‐tagged Ig‐J chains (JcM) folding assay was performed as described (Zhang et al, 2018). In brief, PDI +/+ and PDI −/− hMSCs transfected with pcDNA3.1‐JcM for 48 h were harvested and incubated in MSC culture medium containing 5 mM DTT for 5 min at 37°C. The cells were washed twice with ice‐cold PBS immediately, and then aliquoted and resuspended with fresh culture medium at 20°C. Aliquots were taken at different time points, quenched by 20 mM NEM on ice, and lysed in RIPA buffer containing protease inhibitor cocktail and 20 mM NEM for 30 min on ice. The supernatants were resolved by nonreducing SDS‐15% PAGE and analyzed by western blotting. The in vivo Flag‐tagged LDLR (1–236) folding assay was similar to the JcM folding assay.
Determination of H2O2 levels in the ER
The H2O2 levels in the ER were detected using HyPer‐ER probe according to previous reports (Zhang et al, 2019b). In brief, hMSCs infected with PLE4‐HyPer‐ER for 72 h were washed twice with HBSS, and then transferred into a flat‐bottom 96‐well plates. The fluorescence intensity of the cells was measured at 530 nm with excitation at 405 and 488 nm for 5 min, and then for another 15 min in the presence of 0.5 mM DTT using an EnSpire Multimode Plate Reader. The ratio of 488/405 nm was calculated.
Determination of the release of H2O2 to the nucleus or cytosol
PDI +/+ and PDI −/− hMSCs were infected with the viruses of PLE4‐HyPer7‐nucleus or PLE4‐HyPer7‐cytosol. 72 h later, the cells were harvested and resuspended with HBSS, and then transferred into a flat‐bottom 96‐well plates. The fluorescence intensities were measured at 520 nm with excitation at 400 and 500 nm using EnSpire Multimode Plate Reader for 5 min as steady state. To investigate the release of H2O2 in the ER, 0.5, 1 or 2 mM DTT was added into different wells and the fluorescence intensity was measured for about 30 min. At last, 100 μM H2O2 was added into the wells, and then the ratio of 500/400 nm was calculated.
For imaging experiments, PDI +/+ and PDI −/− hMSCs were infected with the viruses of PLE4‐HyPer7‐nucleus or PLE4‐HyPer7‐cytosol and then were seeded into 35‐mm glass bottom dishes. Cell imaging was performed using a Nikon confocal microscopy. Fluorescence of HyPer7 probes was excited sequentially via 405 and 488 nm and with 500–550 nm emission. After 2–4 images were acquired, 2 mM DTT was added. The ratio images were calculated using the confocal software according to fluorescence intensity.
Lentivirus production
Lentivirus production was performed as previously described (Wu et al, 2020; Jing et al, 2022). Briefly, HEK293T cells were co‐transfected with corresponding lentiviral vectors, psPAX2 (Addgene, #12260) and pMD2G (Addgene, #12259) using LipofectamineTM 3000 Transfection Reagent (Thermo Fisher Scientific). Viral particles were collected by ultracentrifugation at 19,400 g for 2.5 h at 4°C.
Lentiviral CRISPR/Cas9‐mediated gene knockout or CRISPR/dCas9‐mediated gene activation
Lentiviral CRISPR/Cas9‐mediated gene knockout and CRISPR/dCas9‐mediated gene activation were performed as previously described (Wu et al, 2020; Sun et al, 2022a; Wang et al, 2022d). To construct CRISPR/Cas9‐mediated gene knockout lentiviral vectors, the sgRNA targeting PDI was inserted in the lentiCRISPRv2 (Addgene, #52961). For knockout of the expression of PDI, hMSCs were transduced with Lenti‐CRISPRv2‐PDI. 48 h later, the cells were selected with puromycin (Invitrogen, ant‐pr‐1) for 7 days. CRISPRv2‐NTC (nontargeting control) was used as negative control. The sequence of PDI sgRNA was 5′‐AGGCAGAAGGUUCCGAGAUC‐3′. To induce endogenous SERPINE1 expression, sgRNA targeting the transcriptional start site (TSS) of SERPINE1 was inserted into lentiSAM v2 vector (Addgene, #75112). LentiSAM v2‐NTC (non‐targeting control) was used as negative control. For the induction of endogenous expression of SERPINE1, hMSCs were co‐transduced with the LentiSAM v2 and LentiMPH v2 (Addgene, #89308) to transcriptionally activate the expression of SERPINE1. 48 h later, the cells were selected with blasticidin (Sigma, 15205) and hygromycin (Invitrogen, ant‐hg‐1) for 7 days. The sequences of SERPINE1 sgRNA were used as follows:
SERPINE1‐sgRNA#1: 5′‐UCCCUCUCUGGGACUUGCUG‐3′,
SERPINE1‐sgRNA#2: 5′‐AGGUGUCGAGGGGGACCCGC‐3′.
Western blotting
Western blotting was performed as previously described (Liu et al, 2022a). Briefly, cells were lysed in lysis buffer (Millipore) containing phosphatase and protease inhibitor cocktail (Roche) for 30 min on ice. Then, cell lysates were centrifuged at 17,000 g for 30 min at 4°C, and protein concentration was determined by a BCA protein quantification kit. 20 μg sample was loaded and separated by SDS–PAGE and then transferred to PVDF membranes (Millipore). The membranes were then blocked in TBST buffer containing 5% (w/v) nonfat powdered milk for 1 h. Subsequently, the membranes were incubated with primary antibodies overnight at 4°C followed by HRP‐conjugated secondary antibodies at room temperature for 1 h. The imaging was performed using a ChemiScope Mini imaging system (Clinx Science) with enhanced chemiluminescence (Thermo Fisher). The primary antibodies used for western blotting in this study were anti‐PDI (Abcam, ab2792), anti‐SERPINE1 (Santa Cruz, sc‐5297), anti‐GAPDH‐HRP (Sigma‐Aldrich, G9295), anti‐Myc (Sigma, M4439), anti‐ERp44 (Cell Signaling Technology, #3798), anti‐ERp46 (Animal Facility, Institute of Genetics and Developmental Biology, CAS, rabbit serum), anti‐ERp57 (Cell Signaling Technology, #2881), anti‐ERp72 (OriGene, TA503904), P5 (PTM BIO, PTM‐5512), anti‐P4HA1 (PTM BIO, PTM‐5859), anti‐P4HA2 (PTM BIO, PTM‐6033), anti‐FLAG (Sigma, 220,616), P16 (BD, 550834), Ero1α (Sigma‐Aldrich, MAB T376).
H2O2 measurement in PDI +/+ and PDI −/− hMSCs via Amplex red
The cellular H2O2 levels were quantified by Amplex Red Hydrogen Peroxide/Peroxidase Assay Kit (Invitrogen, A22188) according to the manufacturer's instructions. PDI +/+ and PDI −/− hMSCs were harvested and lysed in lysis buffer on ice for 30 min. Then the cell lysates were centrifuged at 17,000 g for 10 min at 4°C, and the protein concentration was quantified by BCA Kit. Then, the same amount of protein of supernatants was transferred to a black 96‐well plate. 50 μM Amplex Red reagent and 0.1 U/ml horseradish peroxidase (HRP) in HBSS were added into each well and incubated at 37°C for 30 min. Fluorescence intensity was measured at 590 nm with excitation at 545 nm.
H2O2 measurement in vitro
To measure the production of H2O2 under different conditions, DTT‐reduced PDI, ERp57, P5, ERp72, and ERp46 (10 μM each) were mixed in 50 mM sodium phosphate buffer, pH 7.4 in a 96‐well microtiter plate. Ero1α (1 μM) together with horseradish peroxidase (0.1 U/ml) and Amplex Red (5 μM) were added to the PDI mixture to initiate H2O2 production. The production of H2O2 was monitored kinetically at 550 nm excitation and 590 nm emission for 30 min.
Oxygen consumption assay
Oxygen consumption was monitored using an Oxygraph Clark‐type oxygen electrode (Hansatech Instruments) as previously described (Wang et al, 2011). Reactions were initiated by adding Ero1α protein to a final concentration of 1 μM into buffer B (100 mM Tris‐HAc, pH 8.0, 50 mM NaCl, 2 mM EDTA) containing PDI, ERp57, P5, ERp72, and ERp46 proteins (10 μM each) and 10 mM glutathione (GSH).
Measurement of cellular reactive oxygen species (ROS)
Cellular total ROS levels were measured using 2.5 μM H2DCFDA (Invitrogen, C6827), as previously reported (Diao et al, 2021). Cells were stained with H2DCFDA for about 15 min at 37°C in the dark, and then analyzed by flow cytometry (BD).
ELISA analysis of the secretion of IL6
ELISA analysis of the secretion of IL6 was performed as previously described (Hu et al, 2020; Liu et al, 2023). Briefly, the supernatants from PDI +/+ and PDI −/− hMSCs were collected and centrifuged at 500 g for 5 min. Then, the supernatants were incubated with IL6 antibody‐coated ELISA plates according to the manufacturer's manual (BioLegend). Finally, the absorbance at 450 nm was detected using a Multimode Plate Reader and the data were normalized to cell numbers.
Telomere length analysis
Analysis of the telomere length by qPCR was performed as previously described (Zhang et al, 2019a). Briefly, the genomic DNA of PDI +/+ and PDI −/− hMSCs was extracted using a DNeasy Blood & Tissue Kit (TIANGEN), and qPCR was performed to analyze the telomere length using the following primers. Telomere‐36B4u was used as the internal reference.
Telomere‐tel‐F: 5′‐GGTTTTTGAGGGTGAGGGTGAGGGTGAGGGTGAGGGT‐3′,
Telomere‐tel‐R: 5′‐TCCCGACTATCCCTATCCCTATCCCTATCCCTATCCCTA‐3′,
Telomere‐36B4u‐F: 5′‐CAGCAAGTGGGAAGGTGTAATCC‐3′,
Telomere‐36B4u‐R: 5′‐CCCATTCTATCATCAACGGGTACAA‐3′.
RNA extraction and RT‐qPCR
RNA extraction and RT‐qPCR were performed as previously described (Bi et al, 2020). In brief, total RNA was extracted by TRIzol reagent (Invitrogen). 2 μg of RNA was used for cDNA synthesis using the GoScript Reverse Transcription System (Promega). qPCR was performed using SYBR Select Master Mix (Thermo Fisher Scientific) on a QuantStudio 7 Flex machine (Applied Biosystems). The relative expression of genes was normalized to the GAPDH transcript. The qPCR primers used are as follows:
GAPDH forward primer, 5′‐GGAGCGAGATCCCTCCAAAAT‐3′,
GAPDH reverse primer, 5′‐GGCTGTTGTCATACTTCTCATGG‐3′,
SERPINE1 forward primer, 5′‐ACCGCAACGTGGTTTTCTCA‐3′,
SERPINE1 reverse primer, 5′‐TTGAATCCCATAGCTGCTTGAAT‐3′,
LAP2 forward primer, 5′‐CCCCTCGGTCCTGACAAAAG‐3′,
LAP2 reverse primer, 5′‐CGCTCTTCGTCACTGGAGAA‐3′,
Lamin B1 forward primer, 5′‐GAAAAAGACAACTCTCGTCGCA‐3′,
Lamin B1 reverse primer, 5′‐GTAAGCACTGATTTCCATGTCCA‐3′,
P16 forward primer, 5′‐ATGGAGCCTTCGGCTGACT‐3′,
P16 reverse primer, 5′‐GTAACTATTCGGTGCGTTGGG‐3′,
IL6 forward primer, 5′‐ACTCACCTCTTCAGAACGAATTG‐3′,
IL6 reverse primer, 5′‐CCATCTTTGGAAGGTTCAGGTTG‐3′.
RNA‐seq library construction and sequencing
Briefly, total RNA was extracted using TRIzol reagent, and genomic DNA was removed using a DNA‐free Kit. Library preparation was conducted using a NEBNext® UltraTM Directional RNA Library Prep Kit for Illumina (New England Biolabs). Quality control and sequencing on Illumina HiSeq X Ten platforms were performed by Novogene Bioinformatics Technology Co., Ltd.
RNA‐seq data processing
RNA‐seq data processing was performed as previously described (Liu et al, 2022b). In brief, low‐quality reads and adaptors were first trimmed using TrimGalore (v0.4.4_dev). The remaining clean reads were mapped to the UCSC human hg19 genome using HISAT2 software (v2.2.1). Reads on each annotated gene were counted using featureCounts (v2.0.1). Differentially expressed genes (DEGs) were calculated using DESeq2, if their adjusted P‐value < 0.05 and absolute Log2(Fold Change) > 0.5. Gene Ontology (GO) term and pathway enrichment analysis were performed using Metascape (https://metascape.org/) with P‐value less than 0.05. The principal component analysis (PCA) and Euclidian distance were performed using R based on Log2 (FPKM+1). The list of aging‐related genes was obtained from the Aging Atlas (Aging Atlas, 2021). These DEGs are listed in Datasets [Link], [Link].
4D label‐free proteomics
Protein extraction
hMSCs were harvested and lysed with 4 volumes of lysis buffer (8 M urea, 1% protease inhibitor). The cell lysates were centrifuge at 12,000 g for 10 min at 4°C, and protein quantification was measured with a BCA kit.
Trypsin digestion
Equal amounts of protein of each sample was used for enzymatic digestion. Then, 20% trichloroacetic acid (TCA) was added slowly, mixed by vortex, and precipitated at 4°C for 2 h. Protein pellets were obtained by centrifugation at 4500 g for 5 min and wash 2–3 times with precooled acetone. After drying the precipitate, added Triethylamonium bicarbonate (TEAB) to a final concentration of 200 mM. The samples were digested with trypsin at a ratio of 1:50 (protease:protein, m/m) overnight, and then reduced by DTT (5 mM) at 56°C for 30 min. Then, 11 mM iodoacetamide (IAA) was added, and incubated at room temperature for 15 min in the dark to remove excess DTT.
Liquid chromatography‐mass spectrometry analysis
The peptides were dissolved in phase A of liquid chromatography mobile phase and separated using NanoElute ultra‐high performance liquid phase system. Mobile phase A was supplemented with 0.1% formic acid and 2% acetonitrile; Mobile phase B was supplemented with 0.1% formic acid and 100% acetonitrile. The liquid set was: 0–70 min, 6~24% B; 70–84 min, 24~35% B; 84–87 min, 35~80% B; 87–90 min, 80% B. The flow rate was maintained at 450 nL/min. The peptides were separated by ultra‐high performance liquid phase system and injected into Capillary ion source for ionization and then analyzed by timsTOF Pro mass spectrometer. The voltage of ion source was set at 1.75 kV, and the peptide precursor ions and their secondary fragments were detected and analyzed using high‐resolution TOF. The scan range of MS was set to 100–1,700. The data acquisition mode was Parallel Accumulation Serial Fragmentation (PASEF) mode. After a first‐order mass spectrometer was acquired, PASEF mode was performed for 10 times to collect the second‐order spectrum with the charge number of the precursor ion in the range of 0–5, and the dynamic exclusion time of the tandem mass spectrometry scan was set to 30 s to avoid repeated scanning of the precursor ion.
Data analysis
Differentially expressed proteins (DEPs) between PDI −/− vs. PDI +/+ were calculated by statistical analysis using unpaired Student's t‐test, with the cut‐off for P‐value < 0.05 and absolute Log2(Fold Change) > 0.3. Pathway enrichment analysis was performed using Metascape. The DEPs are listed in Dataset EV4.
Statistical analysis
Data were presented as mean ± SEM. Statistical analyses were performed using GraphPad Prism software. Comparisons were performed with two‐tailed Student's t‐test or two‐sided Wilcoxon test.
Author contributions
Fang Cheng: Formal analysis; investigation; visualization; methodology; writing – original draft. Qianzhao Ji: Data curation; formal analysis; visualization. Lu Wang: Investigation; methodology. Chih‐chen Wang: Conceptualization; supervision. Guang‐Hui Liu: Conceptualization; supervision; funding acquisition; writing – review and editing. Lei Wang: Conceptualization; supervision; funding acquisition; writing – original draft; writing – review and editing.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Dataset EV3
Dataset EV4
PDF+
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 5
Source Data for Figure 6
Acknowledgments
We thank Xi'e Wang for administrative assistance, Xi Wang and Ping Liu for helpful discussion, Junying Jia and Shu Meng (Institute of Biophysics, CAS) for their help in the flow cytometry experiments, Lijuan Huang (Institute of Zoology, CAS) for her help in optical in vivo imaging. This work was supported by the National Key R&D Program of China (2022YFA1303000, 2021YFA1300800, 2020YFA0804000, 2020YFA0112200); the National Natural Science Foundation of China (32271204, 92254305, 32022033, 81921006, 92149301, 92168201); the Strategic Priority Research Program of CAS (XDB37020303, XDA16010000); the Project for Young Scientists in Basic Research of CAS (YSBR‐075, YSBR‐076); Tencent Foundation (2021‐1045); and the Youth Innovation Promotion Association, CAS, to L.W. Cartoons in Figs 2A, 3J, 4A, 4K, 4L, 6H and EV3G were created with BioRender.
EMBO reports (2023) 24: e56439
Contributor Information
Guang‐Hui Liu, Email: ghliu@ioz.ac.cn.
Lei Wang, Email: wanglei@ibp.ac.cn.
Data availability
The high‐throughput sequencing data including RNA‐seq and whole‐genome sequencing (WGS) generated in this study have been deposited in the Genome Sequence Archive (GSA) in the National Genomics Data Center, Beijing Institute of Genomics (China National Center for Bioinformation) of the Chinese Academy of Sciences under the accession number HRA002973 (https://ngdc.cncb.ac.cn/gsa‐human/browse/HRA002973). The proteomic data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD037244 (http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD037244).
References
- Aging Atlas C (2021) Aging Atlas: a multi‐omics database for aging biology. Nucleic Acids Res 49: D825–D830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appenzeller‐Herzog C, Banhegyi G, Bogeski I, Davies KJ, Delaunay‐Moisan A, Forman HJ, Gorlach A, Kietzmann T, Laurindo F, Margittai E et al (2016) Transit of H2O2 across the endoplasmic reticulum membrane is not sluggish. Free Radic Biol Med 94: 157–160 [DOI] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, Kelly JW (2008) Adapting proteostasis for disease intervention. Science 319: 916–919 [DOI] [PubMed] [Google Scholar]
- Barandalla M, Shi H, Xiao H, Colleoni S, Galli C, Lio P, Trotter M, Lazzari G (2017) Global gene expression profiling and senescence biomarker analysis of hESC exposed to H2O2 induced non‐cytotoxic oxidative stress. Stem Cell Res Ther 8: 160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basisty N, Kale A, Jeon OH, Kuehnemann C, Payne T, Rao C, Holtz A, Shah S, Sharma V, Ferrucci L et al (2020) A proteomic atlas of senescence‐associated secretomes for aging biomarker development. PLoS Biol 18: e3000599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekendam RH, Flaumenhaft R (2016) Inhibition of protein disulfide isomerase in thrombosis. Basic Clin Pharmacol Toxicol 119: 42–48 [DOI] [PubMed] [Google Scholar]
- Bi S, Liu Z, Wu Z, Wang Z, Liu X, Wang S, Ren J, Yao Y, Zhang W, Song M et al (2020) SIRT7 antagonizes human stem cell aging as a heterochromatin stabilizer. Protein Cell 11: 483–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulleid NJ, Ellgaard L (2011) Multiple ways to make disulfides. Trends Biochem Sci 36: 485–492 [DOI] [PubMed] [Google Scholar]
- Burova E, Borodkina A, Shatrova A, Nikolsky N (2013) Sublethal oxidative stress induces the premature senescence of human mesenchymal stem cells derived from endometrium. Oxid Med Cell Longev 2013: 474931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Song W, Li J, Jing Y, Liang C, Zhang L, Zhang X, Zhang W, Liu B, An Y et al (2022) The landscape of aging. Sci China Life Sci 65: 2354–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, Wang S, Song M, Liu Z, Liu P, Wang L, Wang Y, Zhao Q, Yan K, Chan P et al (2019) DJ‐1 is dispensable for human stem cell homeostasis. Protein Cell 10: 846–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colton C, Gilbert D (2007) Reactive oxygen species in biological systems: an interdisciplinary approach. New York, NY: Springer Science & Business Media; [Google Scholar]
- Curran SP, Ruvkun G (2007) Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet 3: e56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Ren R, Liu Z, Song M, Li J, Wu Z, Ren X, Fu L, Li W, Zhang W et al (2019) Stabilizing heterochromatin by DGCR8 alleviates senescence and osteoarthritis. Nat Commun 10: 3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao Z, Ji Q, Wu Z, Zhang W, Cai Y, Wang Z, Hu J, Liu Z, Wang Q, Bi S et al (2021) SIRT3 consolidates heterochromatin and counteracts senescence. Nucleic Acids Res 49: 4203–4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Yang J, Wu X, Zhang G, Li T, Wang X, Zhang H, Wang CC, Liu GH, Wang L (2018) Metformin alleviates human cellular aging by upregulating the endoplasmic reticulum glutathione peroxidase 7. Aging Cell 17: e12765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraile M, Eiro N, Costa LA, Martin A, Vizoso FJ (2022) Aging and mesenchymal stem cells: basic concepts, challenges and strategies. Biology (Basel) 11: 1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh R, Vinod V, Symons JD, Boudina S (2020) Protein and mitochondria quality control mechanisms and cardiac aging. Cell 9: 933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Li X, Zhang Y, Han Y, Chang F, Ding J (2019) Mesenchymal stem cells for regenerative medicine. Cell 8: 886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harman D (1956) Aging: a theory based on free radical and radiation chemistry. J Gerontol 11: 298–300 [DOI] [PubMed] [Google Scholar]
- Hatahet F, Ruddock LW (2009) Protein disulfide isomerase: a critical evaluation of its function in disulfide bond formation. Antioxid Redox Signal 11: 2807–2850 [DOI] [PubMed] [Google Scholar]
- He Y, Ji Q, Wu Z, Cai Y, Yin J, Zhang Y, Zhang S, Liu X, Zhang W, Liu GH et al (2022) Counteracts human mesenchymal stem cell senescence via maintaining mitochondrial homeostasis. Protein Cell 14: 202–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi‐Sanabria R, Frankino PA, Paul JW 3rd, Tronnes SU, Dillin A (2018) A futile Battle? Protein quality control and the stress of aging. Dev Cell 44: 139–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipp MS, Kasturi P, Hartl FU (2019) The proteostasis network and its decline in ageing. Nat Rev Mol Cell Biol 20: 421–435 [DOI] [PubMed] [Google Scholar]
- Hisada Y, Garratt KB, Maqsood A, Grover SP, Kawano T, Cooley BC, Erlich J, Moik F, Flick MJ, Pabinger I et al (2021) Plasminogen activator inhibitor 1 and venous thrombosis in pancreatic cancer. Blood Adv 5: 487–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoehne MN, Jacobs L, Lapacz KJ, Calabrese G, Murschall LM, Marker T, Kaul H, Trifunovic A, Morgan B, Fricker M et al (2022) Spatial and temporal control of mitochondrial H2O2 release in intact human cells. EMBO J 41: e109169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Ji Q, Song M, Ren J, Liu Z, Wang Z, Liu X, Yan K, Hu J, Jing Y et al (2020) ZKSCAN3 counteracts cellular senescence by stabilizing heterochromatin. Nucleic Acids Res 48: 6001–6018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang I, Pottekat A, Poothong J, Yong J, Lagunas‐Acosta J, Charbono A, Chen Z, Scheuner DL, Liu M, Itkin‐Ansari P et al (2019) PDIA1/P4HB is required for efficient proinsulin maturation and ss cell health in response to diet induced obesity. Elife 8: e44528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing Y, Zuo Y, Yu Y, Sun L, Yu Z, Ma S, Zhao Q, Sun G, Hu H, Li J et al (2022) Single‐nucleus profiling unveils a geroprotective role of the FOXO3 in primate skeletal muscle aging. Protein Cell 22: pwac061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung J, Konermann S, Gootenberg JS, Abudayyeh OO, Platt RJ, Brigham MD, Sanjana NE, Zhang F (2017) Genome‐scale CRISPR‐Cas9 knockout and transcriptional activation screening. Nat Protoc 12: 828–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadokura H, Dazai Y, Fukuda Y, Hirai N, Nakamura O, Inaba K (2020) Observing the nonvectorial yet cotranslational folding of a multidomain protein, LDL receptor, in the ER of mammalian cells. Proc Natl Acad Sci U S A 117: 16401–16408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan SS, Shah SJ, Klyachko E, Baldridge AS, Eren M, Place AT, Aviv A, Puterman E, Lloyd‐Jones DM, Heiman M et al (2017) A null mutation in SERPINE1 protects against biological aging in humans. Sci Adv 3: eaao1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Hahm E, Li J, Holbrook LM, Sasikumar P, Stanley RG, Ushio‐Fukai M, Gibbins JM, Cho J (2013) Platelet protein disulfide isomerase is required for thrombus formation but not for hemostasis in mice. Blood 122: 1052–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YM, Youn SW, Sudhahar V, Das A, Chandhri R, Cuervo Grajal H, Kweon J, Leanhart S, He L, Toth PT et al (2018) Redox regulation of mitochondrial fission protein Drp1 by protein disulfide isomerase limits endothelial senescence. Cell Rep 23: 3565–3578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivu J, Myllyla R, Helaakoski T, Pihlajaniemi T, Tasanen K, Kivirikko KI (1987) A single polypeptide acts both as the beta subunit of prolyl 4‐hydroxylase and as a protein disulfide‐isomerase. J Biol Chem 262: 6447–6449 [PubMed] [Google Scholar]
- Kong M, Guo L, Xu W, He C, Jia X, Zhao Z, Gu Z (2022) Aging‐associated accumulation of mitochondrial DNA mutations in tumor origin. Life Med 1: 149–167 [Google Scholar]
- Konno T, Pinho Melo E, Lopes C, Mehmeti I, Lenzen S, Ron D, Avezov E (2015) ERO1‐independent production of H2O2 within the endoplasmic reticulum fuels Prdx4‐mediated oxidative protein folding. J Cell Biol 211: 253–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudryavtseva AV, Krasnov GS, Dmitriev AA, Alekseev BY, Kardymon OL, Sadritdinova AF, Fedorova MS, Pokrovsky AV, Melnikova NV, Kaprin AD et al (2016) Mitochondrial dysfunction and oxidative stress in aging and cancer. Oncotarget 7: 44879–44905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J, Jiang X, Li W, Ren J, Wang D, Ji Z, Wu Z, Cheng F, Cai Y, Yu ZR et al (2022) Exosomes from antler stem cells alleviate mesenchymal stem cell senescence and osteoarthritis. Protein Cell 13: 220–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Fang P, Mai J, Choi ET, Wang H, Yang XF (2013) Targeting mitochondrial reactive oxygen species as novel therapy for inflammatory diseases and cancers. J Hematol Oncol 6: 19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wu Q, Wang Y, Li L, Bu H, Bao J (2017) Senescence of mesenchymal stem cells (review). Int J Mol Med 39: 775–782 [DOI] [PubMed] [Google Scholar]
- Liang C, Liu Z, Song M, Li W, Wu Z, Wang Z, Wang Q, Wang S, Yan K, Sun L et al (2021) Stabilization of heterochromatin by CLOCK promotes stem cell rejuvenation and cartilage regeneration. Cell Res 31: 187–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Wang X, Sun Y, Zhao H, Cheng F, Wang J, Yang F, Hu J, Zhang H, Wang CC et al (2022a) SARS‐CoV‐2 ORF8 reshapes the ER through forming mixed disulfides with ER oxidoreductases. Redox Biol 54: 102388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Ji Q, Ren J, Yan P, Wu Z, Wang S, Sun L, Wang Z, Li J, Sun G et al (2022b) Large‐scale chromatin reorganization reactivates placenta‐specific genes that drive cellular aging. Dev Cell 57: 1347–1368 [DOI] [PubMed] [Google Scholar]
- Liu X, Liu Z, Wu Z, Ren J, Fan Y, Sun L, Cao G, Niu Y, Zhang B, Ji Q et al (2023) Resurrection of endogenous retroviruses during aging reinforces senescence. Cell 186: 287–304 [DOI] [PubMed] [Google Scholar]
- Lopez‐Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G (2023) Hallmarks of aging: an expanding universe. Cell 186: 243–278 [DOI] [PubMed] [Google Scholar]
- Oh J, Lee YD, Wagers AJ (2014) Stem cell aging: mechanisms, regulators and therapeutic opportunities. Nat Med 20: 870–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oszajca K, Bieniasz M, Brown G, Swiatkowska M, Bartkowiak J, Szemraj J (2008) Effect of oxidative stress on the expression of t‐PA, u‐PA, u‐PAR, and PAI‐1 in endothelial cells. Biochem Cell Biol 86: 477–486 [DOI] [PubMed] [Google Scholar]
- Pak VV, Ezerina D, Lyublinskaya OG, Pedre B, Tyurin‐Kuzmin PA, Mishina NM, Thauvin M, Young D, Wahni K, Martinez Gache SA et al (2020) Ultrasensitive genetically encoded indicator for hydrogen peroxide identifies roles for the oxidant in cell migration and mitochondrial function. Cell Metab 31: 642–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H, Guan D, Liu X, Li J, Wang L, Wu J, Zhou J, Zhang W, Ren R, Zhang W et al (2016) SIRT6 safeguards human mesenchymal stem cells from oxidative stress by coactivating NRF2. Cell Res 26: 190–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoli D, Boulay K, Kazak L, Pollak M, Mallette F, Topisirovic I, Hulea L (2019) mTOR as a central regulator of lifespan and aging. F1000Res 8: F1000 Faculty Rev‐998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parakh S, Atkin JD (2015) Novel roles for protein disulphide isomerase in disease states: a double edged sword? Front Cell Dev Biol 3: 30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman FA, Krause MP (2020) PAI‐1, the plasminogen system, and skeletal muscle. Int J Mol Sci 21: 7066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramming T, Hansen HG, Nagata K, Ellgaard L, Appenzeller‐Herzog C (2014) GPx8 peroxidase prevents leakage of H2O2 from the endoplasmic reticulum. Free Radic Biol Med 70: 106–116 [DOI] [PubMed] [Google Scholar]
- Ren X, Hu B, Song M, Ding Z, Dang Y, Liu Z, Zhang W, Ji Q, Ren R, Ding J et al (2019) Maintenance of nucleolar homeostasis by CBX4 alleviates senescence and osteoarthritis. Cell Rep 26: 3643–3656 [DOI] [PubMed] [Google Scholar]
- Ritchie SC, Lambert SA, Arnold M, Teo SM, Lim S, Scepanovic P, Marten J, Zahid S, Chaffin M, Liu Y et al (2021) Integrative analysis of the plasma proteome and polygenic risk of cardiometabolic diseases. Nat Metab 3: 1476–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan H, Geng L, Jiang X, Song M, Wang J, Liu Z, Zhuo X, Wu Z, Hu J, Ji Z et al (2022) Large‐scale chemical screen identifies Gallic acid as a geroprotector for human stem cells. Protein Cell 13: 532–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Zheng Y, Fu X, Zhang W, Ren J, Ma S, Sun S, He X, Wang Q, Ji Z et al (2022a) Single‐cell transcriptomic atlas of mouse cochlear aging. Protein Cell 14: 180–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Li Q, Kirkland JL (2022b) Targeting senescent cells for a healthier longevity: the roadmap for an era of global aging. Life Med 1: 103–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiatkowska M, Szemraj J, Al‐Nedawi KN, Pawlowska Z (2002) Reactive oxygen species upregulate expression of PAI‐1 in endothelial cells. Cell Mol Biol Lett 7: 1065–1071 [PubMed] [Google Scholar]
- Tavernarakis N (2008) Ageing and the regulation of protein synthesis: a balancing act? Trends Cell Biol 18: 228–235 [DOI] [PubMed] [Google Scholar]
- Tu BP, Weissman JS (2004) Oxidative protein folding in eukaryotes: mechanisms and consequences. J Cell Biol 164: 341–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah I, Subbarao RB, Rho GJ (2015) Human mesenchymal stem cells – current trends and future prospective. Biosci Rep 35: e00191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasileiou PVS, Evangelou K, Vlasis K, Fildisis G, Panayiotidis MI, Chronopoulos E, Passias PG, Kouloukoussa M, Gorgoulis VG, Havaki S (2019) Mitochondrial homeostasis and cellular senescence. Cell 8: 686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhu L, Wang CC (2011) The endoplasmic reticulum sulfhydryl oxidase Ero1beta drives efficient oxidative protein folding with loose regulation. Biochem J 434: 113–121 [DOI] [PubMed] [Google Scholar]
- Wang L, Zhang L, Niu Y, Sitia R, Wang CC (2014) Glutathione peroxidase 7 utilizes hydrogen peroxide generated by Ero1alpha to promote oxidative protein folding. Antioxid Redox Signal 20: 545–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Yu J, Wang CC (2021) Protein disulfide isomerase is regulated in multiple ways: consequences for conformation, activities, and pathophysiological functions. Bioessays 43: e2000147 [DOI] [PubMed] [Google Scholar]
- Wang H, Jiang C, Cai J, Lu Q, Qiu Y, Wang Y, Huang Y, Xiao Y, Wang B, Wei X et al (2022a) Nestin prevents mesenchymal stromal cells from apoptosis in LPS‐induced lung injury via inhibition of unfolded protein response sensor IRE1α. Life Med 1: 359–371 [Google Scholar]
- Wang J, Zhou X‐F, Wang Y‐J (2022b) Continuous antioxidant drug exposure: a bridge from ideal world to real world of therapy for amyotrophic lateral sclerosis. Life Med 2: lnac042 [Google Scholar]
- Wang L, Wang X, Lv X, Jin Q, Shang H, Wang CC, Wang L (2022c) The extracellular Ero1alpha/PDI electron transport system regulates platelet function by increasing glutathione reduction potential. Redox Biol 50: 102244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Cheng F, Ji Q, Song M, Wu Z, Zhang Y, Ji Z, Feng H, Belmonte JCI, Zhou Q et al (2022d) Hyperthermia differentially affects specific human stem cells and their differentiated derivatives. Protein Cell 13: 615–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang C (2023) Oxidative protein folding fidelity and redoxtasis in the endoplasmic reticulum. Trends in Biochemical Sciences 48: 40–52 [DOI] [PubMed] [Google Scholar]
- Westrick RJ, Eitzman DT (2007) Plasminogen activator inhibitor‐1 in vascular thrombosis. Curr Drug Targets 8: 966–1002 [DOI] [PubMed] [Google Scholar]
- Wu X, Zhang L, Miao Y, Yang J, Wang X, Wang CC, Feng J, Wang L (2019) Homocysteine causes vascular endothelial dysfunction by disrupting endoplasmic reticulum redox homeostasis. Redox Biol 20: 46–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Shi Y, Lu M, Song M, Yu Z, Wang J, Wang S, Ren J, Yang YG, Liu GH et al (2020) METTL3 counteracts premature aging via m6A‐dependent stabilization of MIS12 mRNA. Nucleic Acids Res 48: 11083–11096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Sankar S, Neamati N (2014) Protein disulfide isomerase: a promising target for cancer therapy. Drug Discov Today 19: 222–240 [DOI] [PubMed] [Google Scholar]
- Zhang L, Niu Y, Zhu L, Fang J, Wang X, Wang L, Wang CC (2014) Different interaction modes for protein‐disulfide isomerase (PDI) as an efficient regulator and a specific substrate of endoplasmic reticulum oxidoreductin‐1alpha (Ero1alpha). J Biol Chem 289: 31188–31199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Li J, Suzuki K, Qu J, Wang P, Zhou J, Liu X, Ren R, Xu X, Ocampo A et al (2015) Aging stem cells. A Werner syndrome stem cell model unveils heterochromatin alterations as a driver of human aging. Science 348: 1160–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhu Q, Wang X, Yu J, Chen X, Wang J, Wang X, Xiao J, Wang CC, Wang L (2018) Secretory kinase Fam20C tunes endoplasmic reticulum redox state via phosphorylation of Ero1alpha. EMBO J 37: e98699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Liu Z, Liu X, Wang S, Zhang Y, He X, Sun S, Ma S, Shyh‐Chang N, Liu F et al (2019a) Telomere‐dependent and telomere‐independent roles of RAP1 in regulating human stem cell homeostasis. Protein Cell 10: 649–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li T, Zhang L, Shangguan F, Shi G, Wu X, Cui Y, Wang X, Wang X, Liu Y et al (2019b) Targeting the functional interplay between endoplasmic reticulum oxidoreductin‐1alpha and protein disulfide isomerase suppresses the progression of cervical cancer. EBioMedicine 41: 408–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Wu Z, Shi Y, Wang S, Ren J, Yu Z, Huang D, Yan K, He Y, Liu X et al (2022a) FTO stabilizes MIS12 and counteracts senescence. Protein Cell 13: 954–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu X, Klionsky DJ, Lu B, Zhong Q (2022b) Manipulating autophagic degradation in human diseases: from mechanisms to interventions. Life Med 1: 120–148 [Google Scholar]
- Zhao H, Ji Q, Wu Z, Wang S, Ren J, Yan K, Wang Z, Hu J, Chu Q, Hu H et al (2022) Destabilizing heterochromatin by APOE mediates senescence. Nat Aging 2: 303–316 [DOI] [PubMed] [Google Scholar]


