Abstract
Recalcitrant infections pose a serious challenge by prolonging antibiotic therapies and contributing to the spread of antibiotic resistance, thereby threatening the successful treatment of bacterial infections. One potential contributing factor in persistent infections is antibiotic persistence, which involves the survival of transiently tolerant subpopulations of bacteria. This review summarizes the current understanding of antibiotic persistence, including its clinical significance and the environmental and evolutionary factors at play. Additionally, we discuss the emerging concept of persister regrowth and potential strategies to combat persister cells. Recent advances highlight the multifaceted nature of persistence, which is controlled by deterministic and stochastic elements and shaped by genetic and environmental factors. To translate in vitro findings to in vivo settings, it is crucial to include the heterogeneity and complexity of bacterial populations in natural environments. As researchers continue to gain a more holistic understanding of this phenomenon and develop effective treatments for persistent bacterial infections, the study of antibiotic persistence is likely to become increasingly complex.
Keywords: antibiotic persistence, evolution, persistent infections, persister recovery, tolerance
Subject Categories: Evolution & Ecology; Microbiology, Virology & Host Pathogen Interaction
This review discusses the current understanding of antibiotic persistence, including its clinical significance, the environmental and evolutionary factors at play, the emerging concept of persister regrowth, and potential strategies to combat persister cells.

Introduction
Bacterial species have evolved various survival strategies to deal with a myriad of stressors that impede their growth or survival (Hibbing et al, 2010). Among these strategies, antibiotic resistance has been well‐studied and allows bacteria to thrive in antibiotic‐rich environments. Despite widespread public awareness of antibiotic resistance, another critical, yet often overlooked survival strategy to antibiotics is persistence (Huemer et al, 2020). Antibiotic persistence is characterized by the ability of a bacterial subpopulation to tolerate a lethal antibiotic dose, while the majority of the isogenic population is rapidly killed, resulting in biphasic killing (Fig 1A). The surviving persister cells can give rise to a new population after antibiotic treatment is ceased (Balaban et al, 2019), provided that they have reverted to the normal, antibiotic‐sensitive state and have recovered from potential antibiotic‐inflicted damage (Wilmaerts et al, 2019b). From a clinical perspective, persistence could possibly lead to relapse of the infection, despite successful initial treatment of the patient (Fig 1B). Indeed, persistence has been linked to the chronic nature of various persistent infections (Fauvart et al, 2011). Moreover, the tolerance of persister cells promotes the emergence of resistance, further underpinning the importance of understanding bacterial persistence (Levin‐Reisman et al, 2017; Windels et al, 2019b; Bakkeren et al, 2020; Santi et al, 2021; Sulaiman & Lam, 2021).
Figure 1. Distinguishing between resistance, tolerance, and persistence and the possible clinical implication of persistence.
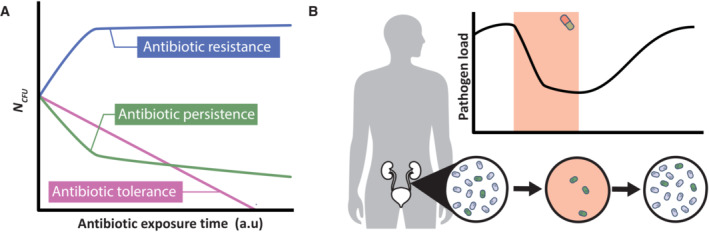
(A) Antibiotic‐resistant cells are characterized by their ability to grow during antibiotic treatment (blue). This is in contrast to antibiotic tolerance (pink) and persistence (green). In case of antibiotic tolerance, the decreased population‐wide sensitivity results in slower killing, which implies that prolonged antibiotic treatment is required to eradicate the population (pink). Antibiotic persistence is a survival strategy where only a small subpopulation is highly tolerant to the antibiotic. This results in characteristic biphasic killing, where the majority of sensitive cells are rapidly killed and the subpopulation of persister cells survives. However, note that killing of persister cells can still happen at a slow rate (green). (B) A patient suffering from, for example, a urinary tract infection receives antibiotic treatment. The pathogen load in the urinary tract rapidly decreases, resulting in a seemingly successful treatment. However, once the antibiotic treatment is ceased, surviving persister cells can again increase the pathogen load, resulting in a chronic infection.
In the past, most research efforts have focused on investigating stochastic and deterministic persister formation in vitro (Van den Bergh et al, 2017). However, recent studies have expanded to include the mechanisms underlying persister recovery and regrowth (Wilmaerts et al, 2019b, 2022; Semanjski et al, 2021), the clinical context of the host (Helaine et al, 2014; Stapels et al, 2018; Huemer et al, 2021; Wang & Jin, 2022), and ecological and evolutionary aspects of bacterial persistence (Bakkeren et al, 2020; Personnic et al, 2021; Verstraete et al, 2022b). Therefore, there is a need for a comprehensive understanding of these recent findings. In this review, we summarize recent advances on bacterial persistence and underscore its clinical relevance. We also provide a concise overview of the genetic mechanisms underlying persistence with a focus on recovery pathways. Finally, we examine ecological and evolutionary dynamics of persistence and discuss a range of potential strategies to combat persister cells, including the usefulness of combining antibiotics and potentiating compounds.
A persistent threat in the clinic
Persistent bacterial infections pose a significant burden to human health worldwide due to their chronic nature and their challenging, long‐lasting treatment (La Rosa et al, 2022). Although the term “persistent infection” suggests the presence of bacterial persister cells, it rather points toward infections that are not cleared by the host immune system or, by extension, by antibiotic treatment due to antibiotic survival strategies such as resistance, tolerance and persistence (Fig 1A; Balaban et al, 2019). Resistance is acquired through genetic changes and denotes the ability of the bacterial population to survive and reproduce in the presence of antibiotics and is characterized by an increase in the minimum inhibitory concentration (MIC) of the antibiotic. Tolerance, on the contrary, refers to a population's ability to survive longer exposure to antibiotics, typically due to restrictive growth either caused by genetic mutations or environmental conditions. Tolerance therefore prolongs the minimum duration for killing the population, while the MIC remains unchanged since tolerant cells do not grow in the presence of antibiotics. While resistance and tolerance apply to the entire population, persistence refers to a subpopulation that consists of phenotypic variants with increased levels of tolerance. The formation of this subpopulation proceeds either deterministically, stochastically or by a combination of both. Importantly, the survival of persister cells does not affect the population's MIC (Brauner et al, 2016; Balaban et al, 2019; Ronneau et al, 2021). Although a fully tolerant population is characterized by a high survival rate upon treatment and the persister phenotype merely applies to a small subpopulation, persistence can have a clear fitness advantage over population‐wide tolerance. While tolerance coincides with restricted growth in absence of antibiotics, a sensitive population containing a small fraction of persister cells retains colonization abilities of the host, while the persister cells ensure continuation of the infection after an antibiotic treatment (Michaux et al, 2022). For clarity, in the remainder of this work, the term “persistence” will be used in the context of bacterial persister cells.
The presence of antibiotic‐tolerant persisters in bacterial populations can have significant implications for the success of antibiotic therapy. These cells are able to survive antibiotic exposure and then recover and regrow once the antibiotic pressure is released, causing a relapse of infection (Fig 1B; Balaban et al, 2019). This threat is amplified when persister cells are shielded from the host immune response, mostly by residing within biofilms (Lewis, 2007; Ciofu et al, 2022). Pathogens like Pseudomonas aeruginosa, Staphylococcus aureus, uropathogenic Escherichia coli (UPEC), and some Salmonella species attach to host tissues or indwelling devices, where they have been shown to form biofilms (Steenackers et al, 2012; Mulcahy et al, 2014; Speziale et al, 2014; Narayanan et al, 2018; Ciofu & Tolker‐Nielsen, 2019). Apart from biofilm formation, UPEC has the ability to invade host bladder cells, thereby rendering the bacteria once again inaccessible for the host immune defense (Anderson et al, 2004). Mycobacterium tuberculosis and Salmonella spp., on the contrary, directly interact with the host immune system by reprogramming macrophages upon macrophage internalization. The specific intracellular conditions inside the macrophage subsequently trigger the bacteria to enter the persister state (Gengenbacher & Kaufmann, 2012; Helaine et al, 2014; Stapels et al, 2018). Research on pathogens isolated from patients with relapsing infections supports the notion that bacterial persistence plays a role in the chronic nature of these infections. For example, high‐persistence (Hip) mutants were identified in clinical isolates of P. aeruginosa, UPEC, and M. tuberculosis derived from patients that underwent repeated antibiotic treatment and experienced infection relapse (Mulcahy et al, 2010; Schumacher et al, 2015; Torrey et al, 2016; Bartell et al, 2020). In addition, S. aureus clinical isolates from persistent infections showed increased persister levels compared with laboratory strains (Huemer et al, 2021), and invasive nontyphoidal Salmonella clinical isolates that have undergone multiple rounds of treatment retain the characteristics of persistence rather than evolving toward tolerance (Hill et al, 2021). Lastly, another important threat that comes with the presence of persister cells is their ability to facilitate the emergence and spread of resistance (Levin‐Reisman et al, 2017; Windels et al, 2019c; Bakkeren et al, 2020; Santi et al, 2021; Sulaiman & Lam, 2021). To fully comprehend antibiotic persistence and its implications for clinical settings, it is essential to consider environmental and evolutionary factors.
Surviving environmental adversity
Persistence is a phenotype that is heavily influenced by environmental conditions. Bacteria can form persisters in response to various stressors, such as low oxygen levels, nutrient scarcity, heat, acidity, and exposure to toxic compounds, including antibiotics (Fig 2A; Boon & Dick, 2012; Gutierrez et al, 2017; Wang et al, 2017; Kubistova et al, 2018; Paranjape & Shashidhar, 2019; Van den Bergh et al, 2022). These so‐called induced or triggered persisters have been extensively studied in the laboratory and are predominantly present during stationary phase, when bacteria enter a low metabolic state to survive in a nutrient‐limited environment (Verstraeten et al, 2015; Brown, 2019). In contrast, persisters during exponential growth or following dilution in fresh medium are relatively few (Keren et al, 2004; Gutierrez et al, 2017; Salcedo‐Sora & Kell, 2020). This latter type of persisters is referred to as spontaneous persisters and are formed stochastically during steady‐state exponential growth when cells are most uniform (Fig 2A; Keren et al, 2004; Kussell et al, 2005; Balaban et al, 2019).
Figure 2. Formation and heterogeneity of antibiotic persisters.
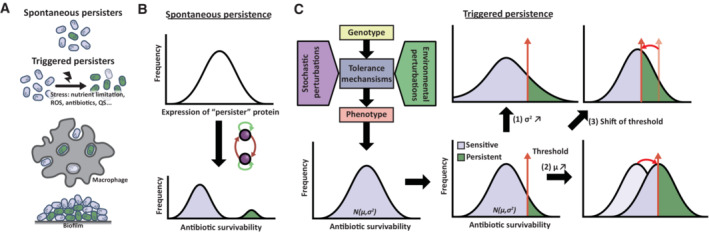
(A) Two categories of persister cells are depicted: spontaneous and triggered persistence. Spontaneous persisters arise stochastically due to cellular noise, whereas triggered persisters form in response to environmental stressors, such as abiotic stress, macrophage‐ or biofilm‐associated stresses. Abbreviations: ROS, reactive oxygen species; QS, Quorum sensing molecules. (B) and (C) present the models that explain how both types of persisters arise heterogeneously in a population. (B) Spontaneous persisters arise from random variation in persister protein expression, coupled with feedback loops, resulting in bistable phenotypic differentiation. (C) Triggered persisters, on the contrary, represent a subset of cells with the highest level of antibiotic tolerance within a population with varying levels of susceptibility arising from developmental noise in various tolerance mechanisms. The proportion of persisters depends on both the (1) mean (μ) and (2) variance (σ) of this distribution as well as the (3) specific antibiotic conditions and duration. Perturbing the mean, such as through environmental triggers or genetic mutations, can increase the fraction of cells surviving antibiotic exposure. Increasing the variance of this distribution, through environmental heterogeneity or genetic changes in buffering or potentiating genes, can also increase the fraction of survivors.
The ability of bacteria to persist is not only triggered by abiotic environmental challenges but also through communication with other bacteria (Vega et al, 2012; Personnic et al, 2023). Quorum sensing offers a means for bacteria to communicate with each other using signal molecules, enabling them to induce virulence factors and coordinate behavior, examples of which are swarming or biofilm formation. In Legionella pneumophila, quorum sensing is a major regulator of phenotypic heterogeneity and the Legionella quorum sensing system controls the ratio between growing and nongrowing—antibiotic‐tolerant—states (Personnic et al, 2021). Similarly, Streptococcus mutans and P. aeruginosa exhibit augmented antibiotic persistence attributed to the secretion of signaling molecules (Möker et al, 2010; Leung & Lévesque, 2012). Moreover, these signaling molecules may also trigger persistence in other species besides their own (Leung & Lévesque, 2012). These findings suggest that quorum sensing‐mediated persistence may represent a collective social behavior that enables dense bacterial populations to survive hostile environments. This behavior may play a role in the formation of persisters in biofilms, which are dense microbial communities that are notoriously difficult to treat with antibiotics, with persister levels up to 1,000‐fold higher than planktonic cells in exponential growth phase (Spoering & Lewis, 2001; Lewis, 2005). However, heterogenous environmental conditions, such as gradients in nutrient or oxygen availability leading to localized growth arrest, could also induce persistence within biofilms (Borriello et al, 2004; Nguyen et al, 2011; Flemming et al, 2016).
During bacterial infections, host immune cells employ various strategies to generate hostile conditions for pathogens. Examples include the production of reactive oxygen species (ROS), the sequestration of essential nutrients, or the release of inflammatory mediators and antimicrobial peptides (Foster, 1999; Fang, 2011; Becker & Skaar, 2014; Murdoch & Skaar, 2022). However, while these host immune‐mediated stress conditions are effective in controlling infections, they may unintentionally induce persister formation, which can negatively affect drug efficacy (Helaine et al, 2014; Liu et al, 2016; Beam et al, 2021). For example, macrophages induce persistence in phagocytosed S. aureus through the production of reactive oxygen and nitrogen species and by creating acid stress within the phagosome (Rowe et al, 2020; Beam et al, 2021; Huemer et al, 2021; Ronneau et al, 2023). In this case, persistence is likely induced through general stress responses (Peyrusson et al, 2020; Ranganathan et al, 2020). Other bacterial species, such as Salmonella enterica and M. tuberculosis, similarly display increased tolerance to antibiotics following internalization by macrophages (Helaine et al, 2014; Liu et al, 2016). Another mechanism by which the host immune system may induce bacterial persistence is through its use of antimicrobial host defense peptides, which represent an important antimicrobial component of the innate immune system (Mookherjee et al, 2020). However, sublethal doses of certain antimicrobial peptides prime bacterial cells and increase their tolerance and persistence (Rodríguez‐Rojas et al, 2021; Sandín et al, 2022). In conclusion, pathogens face fluctuating and unfavorable conditions during infection, partly due to the host's immune response, which can paradoxically promote pathogen persistence. To combat persistent infections, it is crucial to consider the interplay between pathogenic bacteria, the host immune response, and the heterogeneous infection environment.
Heterogeneity and bacterial persistence
It is important to note that, even in cases of triggered persistence, antibiotic persistence is subject to cell‐to‐cell differences since per definition only a subset of the population exhibits antibiotic tolerance. The question then arises as to how this heterogeneity in antibiotic survivability in genetically uniform populations emerges. In natural environments such as in the gut or the soil, or during an infection, microbial populations are exposed to significant environmental heterogeneity (Nguyen et al, 2021; Sokol et al, 2022). Such heterogeneity within the same spatial niche is likely to result in drastic differentiation of physiological states, potentially resulting in variations in antibiotic susceptibility that may explain recalcitrant infections in some situations. Although microfluctuations or subtle environmental gradients cannot be fully precluded, well‐mixed laboratory cultures generally display limited temporal and spatial variability (Junkins et al, 2022). However, even under these homogenous conditions, persister subpopulations can still emerge, highlighting the stochastic nature of antibiotic persistence.
In the absence of environmental or genetic variation, phenotypic heterogeneity can be attributed to stochastic cellular noise (Elowitz et al, 2002). The latter refers to random fluctuations in gene expression and biochemical processes within individual cells (Ackermann, 2015). This noise arises from various sources such as the inherent randomness of chemical reactions, stochastic partitioning during cell division, or cell cycle and age differences (Elowitz et al, 2002; Raser & O'Shea, 2005; Avery, 2006; Huh & Paulsson, 2011). Spontaneous persisters are believed to arise purely from stochastic fluctuations in gene expression via “persister” proteins that induce persister formation when expression stochastically reaches a specific threshold level (Fig 2B; Rotem et al, 2010; Dewachter et al, 2019). For instance, the expression of the toxin HipA can cause bistability of different states, allowing the bacterial cell to exist in either a susceptible or a persistent state (Balaban et al, 2004).
Like many biological phenomena, triggered persistence is the product of multiple parallel and interdependent processes, which are subject to their own mechanistic idiosyncrasies and triggers. Perturbing any of these processes (e.g., growth homeostasis, stress response, or membrane transport) can dramatically affect antibiotic susceptibility and persistence (Wilmaerts et al, 2019b). Moreover, it has been shown that these processes exhibit significant stochastic variation, leading to cell‐to‐cell heterogeneity (Raser & O'Shea, 2005; Ghosh et al, 2011; Kiviet et al, 2014; Amato & Brynildsen, 2015; Shan et al, 2017). To that end, it is likely that triggered persisters do not reflect a uniform subpopulation and may be formed through various parallel mechanisms and in response to different conditions (Fig 2C). Generally speaking, in case of antibiotic persistence, it is assumed that tolerance heterogeneity results in a distribution with two distinct phenotypic states consisting of a majority of susceptible cells and a minority of persister cells that arise via bistable switching. However, the probability distribution of individual cells' antibiotic susceptibility exhibits a monomodal Gaussian distribution (Scheler et al, 2020). Persistence may similarly represent a continuous quantitative trait rather than a binary persister–nonpersister state, which would be in line with it being a complex polygenic trait. This idea is related to the concept of “dormancy depth” (Pu et al, 2019; Bollen et al, 2021; Dewachter et al, 2021), which measures the extent of a cell's persistence through dormancy and is closely linked to its lag time. Dormancy refers to a reduced metabolic state that allows organisms to survive unfavorable conditions such as nutrient limitation, and although it can contribute to persistence, persister cells are not necessarily dormant and may use other mechanisms to survive antibiotics. Increasing dormancy depth within a population can be achieved by prolonging the stationary phase, leading to an increase in persisters and longer regrowth times (Pu et al, 2019), with viable but nonculturable cells representing the most extreme phenotype in that spectrum (Bollen et al, 2021; Dewachter et al, 2021). This concept could be extended beyond just persistence via dormancy toward triggered persistence and tolerance in general. In this manner, dormancy depth, along with other processes related to persistence including environmental factors, contribute to the overall phenotypic variation of a multifactorial phenotypic trait, for example, antibiotic survivability. Perturbations to this compound trait can arise from a variety of mechanisms including mutation, cellular noise, environmental fluctuations, and phenotypic plasticity, resulting in a shift in the mean or variance of this trait (Fig 2C). The persistence level of a population and the survival of individual cells then depends on the position of this distribution relative to a threshold determined by the experimental conditions in which the persistence level is measured. During antibiotic treatment, as the most susceptible cells are rapidly eliminated, the killing rate slows down, and only more tolerant cells remain. Depending on the experimental conditions and the threshold employed, this may result in a complete flattening of the killing curve. This perspective provides a useful unifying framework for understanding persistence and cell‐to‐cell tolerance heterogeneity and underscores the heterogeneous nature of persistence, not only between persisters and nonpersisters but also between individual persister cells.
Defense mechanisms of persister cells against antibiotics
The heterogenous nature of persisters is reflected in the variety of defense mechanisms that make them tolerant to antibiotics (Fig 3A). These mechanisms of antibiotic defense are commonly classified as either passive or active. Passive mechanisms primarily involve entering a dormant or quiescent state and thereby preventing the corrupting effects of antibiotics on vital cellular processes (Lewis, 2010), while active mechanisms involve other strategies such as decreasing intracellular antibiotic concentrations or actively preventing damage (Nguyen et al, 2011; Orman & Brynildsen, 2013; Pu et al, 2016). For a more in‐depth discussion of this topic, readers are referred to other reviews (Van den Bergh et al, 2017; Harms, 2019; Wilmaerts et al, 2019b).
Figure 3. Mechanisms of persistence defense, recovery, and regrowth.
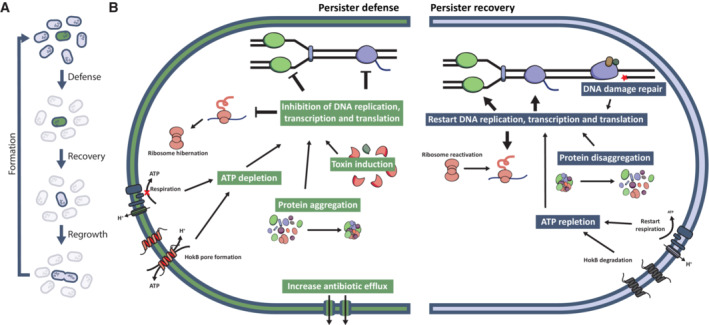
(A) During the course of antibiotic treatment, persister cells utilize various defense mechanisms to survive the effects of antibiotics. Once the treatment ceases, persister cells first recover from the inflicted damage before regrowing and forming a new population, where other cells can switch to the persister state. This recovery period involves repairing DNA damage and resuming critical cellular processes, such as DNA replication, transcription, and translation. (B) Persister cells can defend themselves from antibiotic damage by lowering their metabolism through inhibition of DNA replication, transcription, and translation. These pathways can be inhibited by expressing pathway‐specific toxins, by sequestering essential proteins of these pathways in aggregates or by depleting the ATP needed for their functioning. Additionally, persister cells might protect themselves from more antibiotic‐induced damage by increasing antibiotic efflux. For their recovery, persister cells need to repair the inflicted DNA damage. Moreover, they need to increase their metabolism to be able to regrow by restarting DNA replication, transcription and translation. To reactivate these processes, persister cells need to replete their ATP levels and remove the aggregates present in the cell.
Passive defense of persister cells against antibiotics
Most antibiotics need an active target to exert their function (Eng et al, 1991), which means that bacteria can become tolerant by reducing cellular activity (Hu & Coates, 2012; Balaban et al, 2019). Indeed, persister cells are often considered to be tolerant because they reside in a dormant state characterized by reduced metabolic activity and a reduction of global cellular processes and growth (Balaban et al, 2004; Amato et al, 2013). Notably, the different mechanisms involved in persistence often share genetic components, making it difficult to define a single cause of persistence by dormancy. One way to lower metabolic activity and increase persistence is to deplete ATP (Fig 3B), for example, by the addition of arsenate (Conlon et al, 2016), the reduction of the membrane potential (Kwan et al, 2013), or the induction of the toxins TisB (Dörr et al, 2010) or HokB (Wilmaerts et al, 2018). Furthermore, the activity of essential cellular processes can be reduced by directly targeting important proteins involved in DNA replication (Tripathi et al, 2012), transcription, or translation (Fig 3B; Kwan et al, 2013). Several toxins from type II toxin–antitoxin modules such as HipA and TacT have been shown to specifically inhibit these processes (Korch & Hill, 2006; Cheverton et al, 2016). However, it is important to note that a general role of type II toxin–antitoxin modules in persistence is controversial (Goormaghtigh et al, 2018; Kaldalu et al, 2020) and that overexpression of other toxic proteins that do not belong to toxin–antitoxin modules can also induce persistence (Vázquez‐Laslop et al, 2006). In addition to direct targeting of important proteins, persister cells have been associated with the presence of protein aggregates in which important proteins might be sequestered (Fig 3B; Pu et al, 2019; Yu et al, 2019; Dewachter et al, 2021; Goode et al, 2021). Indeed, various conditions, such as nutrient starvation, ATP depletion, heat shock and heterologous protein expression, not only increase persistence but also aggregation in the cells (Leszczynska et al, 2013; Mordukhova & Pan, 2014; Pu et al, 2019; Dewachter et al, 2021; Goode et al, 2021; Peyrusson et al, 2022). Moreover, adding osmolytes or buffering components to the growth medium decreases both persistence and protein aggregation (Leszczynska et al, 2013). Aggregates in persister cells contain many proteins that function in transcription, translation, and energy production (Pu et al, 2019; Dewachter et al, 2021; Huemer et al, 2021). It is therefore hypothesized that dormancy can be induced when sufficient essential proteins are contained in aggregates, thereby inhibiting the cell's overall functioning and preventing damage from antibiotics (Bollen et al, 2021; Dewachter et al, 2021).
Although tolerance of persisters to antibiotics is often attributed to their low metabolic state, dormancy alone is not sufficient to fully explain persistence. For example, there is not always a correlation between growth rate and cell survival during antibiotic treatment (Orman & Brynildsen, 2013; Wakamoto et al, 2013). Furthermore, some persister cells, such as intracellular persisters, still show metabolic activity and synthesize specific proteins that are important for their survival within the host cell (Orman & Brynildsen, 2013; Manina et al, 2015; Stapels et al, 2018; Wilmaerts et al, 2018; Peyrusson et al, 2020; Sulaiman & Lam, 2020; Semanjski et al, 2021; Mode et al, 2022; Ronneau et al, 2023). Additionally, while dormant persister cells are protected from some types of harm, they are not fully shielded from DNA damage (Völzing & Brynildsen, 2015; Wilmaerts et al, 2022). Moreover, it was shown that cells in stationary phase unavoidably acquire oxidative damage, the level of which is even higher in nongrowing cells (Nyström & Gustavsson, 1998; Desnues et al, 2003). Clearly, dormancy protects bacteria from damage to some extent, but active mechanisms are also at play.
Active defense of persister cells against antibiotics
Persister cells can actively protect themselves against antibiotics by either reducing the antibiotic concentration within the cell or by actively preventing antibiotic damage (Wilmaerts et al, 2019b). The former can be achieved by increasing efflux activity (Fig 3B; Pu et al, 2016) or preventing prodrug activation (Wakamoto et al, 2013). The latter can be accomplished by activating stress responses (Nguyen et al, 2011; Sulaiman & Lam, 2020; Semanjski et al, 2021; Van den Bergh et al, 2022). This increase in stress responses is often detected in intracellular persister cells as they need to survive exposure to a more complex environment of multiple small niches characterized by specific environmental conditions and stresses (Demarre et al, 2019; Peyrusson et al, 2020). In addition to activating stress responses, intracellular persisters have been observed to reduce the host's immune response by secreting specific effector molecules (Stapels et al, 2018). This indicates that intracellular persister cells use an array of mechanisms, at least part of which have already been observed in in vitro studies, and that they possibly depend on a combination of these mechanisms to survive in their challenging environment. This underscores the importance of verifying in vitro results in vivo (Box 1).
Box 1. In need of answers.
What is the timing and causal sequence of the processes involved in persister cell formation? To what extent are these processes independent of each other, or do they interact and influence each other in a coordinated manner?
What are the mechanisms of persistence in vivo and how do they relate to the persister mechanisms that have already been found in vitro?
In addition to repairing DNA damage, are there mechanisms in place for persister cells to repair any protein damage caused by antibiotics?
How do persister cells coordinate the switch from DNA repair to regrowth, and which effector proteins are involved in this process? What is the role of protein aggregates and protein synthesis in regulating this switch?
How do persister cells respond to environmental cues, and how does this affect their recovery and proliferation? What are the clinical implications of these interactions?
How do environmental factors, such as nutrient availability, host immune response, and interspecies interactions, influence the evolution of persistence?
How do the mechanisms of persister cell regrowth and proliferation differ from those of normal bacterial growth, and how can we exploit these differences for therapeutic purposes?
What are the specific mechanisms underlying the efficacy of proposed antipersister compounds, and how effective are they in vivo in targeting persister cells? Additionally, what are the potential side effects associated with their use and can these be mitigated?
Mechanisms of persister recovery and regrowth
Persisters cells need to start recovery and regrowth to be able to recolonize the environment once antibiotics are depleted (Fig 3A). Several environmental factors can stimulate regrowth of nongrowing bacteria, including persisters, such as fresh nutrients (Jõers et al, 2010), quorum sensing signals from growing cells (Nichols et al, 2008; D'Onofrio et al, 2010; Jõers et al, 2019) and the removal of certain host immune factors (Beatty et al, 1993; Mohan et al, 2001). However, the length of the lag phase between transfer of persisters to new medium and their regrowth depends on the concentration of the antibiotic and the duration of the treatment (Himeoka & Mitarai, 2021), as well as on the intensity and length of the persistence‐inducing condition (Kaplan et al, 2021; Cesar et al, 2022). Conditions of gradual stress were suggested to activate specific recovery pathways that lead to fast and homogeneous regrowth, while more acute or longer stresses induce random pathways that lead to slower and heterogeneous regrowth (Kaplan et al, 2021). Different mechanisms have been described to control the shift from persister to antibiotic‐sensitive state, such as reprogramming of cell metabolism and damage repair (Wilmaerts et al, 2019b). Although these two different mechanisms will be discussed separately, a combination of both is likely necessary for persister recovery and regrowth.
Persister recovery and regrowth requires reversion to a growth‐competent state
To recover from the action of the antibiotic and regrow, persisters first need to transition to a growth‐competent state by replenishing their energy level and reactivating crucial cellular pathways. Indeed, persister cells were shown to increase their ATP levels before reinitiating growth (Fig 3B; Huemer et al, 2021; Manuse et al, 2021). Accordingly, nutrient‐rich medium increases regrowth (Jõers et al, 2010; Yamasaki et al, 2020), and persister cells upregulate glycolysis to increase ATP levels (Semanjski et al, 2021). In the case of HokB‐induced persisters, which are ATP‐depleted as a result of membrane potential dissipation and direct ATP leaking through HokB pores (Wilmaerts et al, 2018), the HokB pores are degraded and the membrane is repolarized before regrowth (Wilmaerts et al, 2019a). Additionally, intracellular persister cells can be locked in the persister state with low respiration and ATP levels because of intoxication of the TCA cycle by reactive nitrogen species. These locked persister cells can resume growth when the reactive nitrogen species are reduced or inhibited (Ronneau et al, 2023).
In addition to ATP repletion, persister cells need to reactivate essential macromolecular pathways such as DNA replication and cell division before growth is initiated (Fig 3B). During persister recovery and regrowth, the cell division genes ftsW and amiA are upregulated (Belland et al, 2003), while genes encoding inhibitors of cell division and DNA replication, sulA and cspD, respectively, are degraded (Langklotz & Narberhaus, 2011; Mohiuddin et al, 2022). Furthermore, when shifting to the growing state, persister cells increase various anabolic pathways such as the biosynthesis of multiple amino acids, secondary metabolites, and proteins (Fig 3B; Semanjski et al, 2021).
The increase in protein biosynthesis during persister regrowth (Semanjski et al, 2021) can be achieved in multiple ways. First, persister cells increase their ribosomal content and translation by increasing the production of ribosomal proteins and rRNA (Kim et al, 2018a; Sulaiman & Lam, 2020; Semanjski et al, 2021). Second, various studies suggest that persister cells reactivate inactivated ribosomes during regrowth (Yamasaki et al, 2020; Song & Wood, 2020a; Semanjski et al, 2021). Persister cells are associated with ribosomes that are inactivated by binding with the ribosome inhibitor RaiA or by ribosome dimerization (McKay & Portnoy, 2015; Prossliner et al, 2018; Song & Wood, 2020a). The inactivation of these ribosomes might protect them from degradation such that they can be rapidly reactivated during regrowth (Cho et al, 2015). Moreover, RaiA‐induced ribosomes and ribosome dimers are associated with a fast and slow regrowth of stationary‐phase cells, respectively. This suggests that cells have a fast and a slower mechanism for ribosome recovery after inactivation (Lang et al, 2021). Third, persister cells harbor many translation‐related proteins in aggregates (Pu et al, 2019; Dewachter et al, 2021; Huemer et al, 2021). These aggregates are removed before persister regrowth with the help of chaperones DnaK and ClpB (Fig 3B; Pu et al, 2019; Cesar et al, 2022). This suggests that persister cells might use protein aggregates as temporary storage compartments from which proteins can be extracted and reused during regrowth, which is reminiscent of stress granules in quiescent eukaryotic cells (Narayanaswamy et al, 2009; Saad et al, 2017). Similarly in bacteria, FtsZ, a key cytoskeletal protein, is refolded, relocated, and reused when stationary‐phase cells restart growth upon transfer to fresh medium (Yu et al, 2019). Finally, persister cells can increase translation by reverting the detrimental effects of translation‐targeting toxins. Indeed, persister recovery and regrowth were observed following toxin inhibition by their cognate antitoxins (Pedersen et al, 2002; Korch & Hill, 2006; Cheverton et al, 2016; Rycroft et al, 2018) or following reversion of toxin‐induced effects (Christensen et al, 2003; Cheverton et al, 2016; Rycroft et al, 2018).
It is important to note that some of the mechanisms of recovery and regrowth discussed above are not necessarily unique to persister cells. Stationary‐phase cells also have a reduced metabolism, which requires reactivation upon transfer to fresh medium. Nevertheless, gaining more insight in these mechanisms is needed to understand how persister cells recover and regrow.
Persister recovery and regrowth depends on cellular damage repair
Besides reversion to a growth‐competent state, persister cells need to repair damage, inflicted either directly or indirectly by the antibiotic or by environmental stressors, to allow recovery and regrowth (Fig 3B; Wilmaerts et al, 2019b).
Fluoroquinolones offer the most evident example of an antibiotic class causing direct damage to persister cells. In E. coli, their primary target is DNA gyrase (Drlica & Zhao, 1997; Malik et al, 2006), which plays a critical role in the relaxation of DNA during replication and transcription. Several studies have indicated that, following fluoroquinolone treatment, persister cells suffer DNA damage and induce the SOS response, a DNA damage‐inducible DNA repair pathway, suggesting that DNA repair is essential for recovery and regrowth (Dörr et al, 2009; Völzing & Brynildsen, 2015; Barrett et al, 2019; Goormaghtigh & Van Melderen, 2019). Indeed, multiple SOS‐responsive genes implicated in homologous recombination repair, including recA, recB, ruvA, ruvB, and uvrD, have been found to be important for persister recovery (Theodore et al, 2013; Völzing & Brynildsen, 2015; Mok & Brynildsen, 2018; Lemma & Brynildsen, 2021; Wilmaerts et al, 2022). The dependence on homologous recombination repair is further supported by the finding that persister cells often originate from cells containing a second chromosome, which may serve as a homolog in recombinational repair (Murawski & Brynildsen, 2021). While SOS induction followed by expression of SOS‐responsive DNA repair genes is crucial for successful recovery from fluoroquinolone treatment, it is not considered a distinguishing factor for persistence (Mok & Brynildsen, 2018). Eventually, what distinguishes persister cells from nonpersister cells seems to be their ability to delay growth‐related processes until DNA repair has been completed (Mok & Brynildsen, 2018). Indeed, a few studies using cultures in exponential growth phase point toward a mechanism of SOS‐induced growth delay during fluoroquinolone treatment, either through increased production of the TisB toxin or the SulA cell division inhibitor (Dörr et al, 2010; Theodore et al, 2013; Edelmann & Berghoff, 2022).
Most bactericidal antibiotics also cause indirect damage to cells through the production of ROS (Kohanski et al, 2007; Foti et al, 2012; Dwyer et al, 2014; Van Acker & Coenye, 2017). The most important type of DNA damage caused by ROS is the incorporation of the mutagenic 8‐oxo‐guanine, formed by oxidation of the guanine nucleotide pool. While this lesion is not necessarily lethal, failed attempts to repair it prior to cell division may lead to cell death (Foti et al, 2012; Gruber & Walker, 2018). Successful repair of oxidative DNA damage may therefore be important for persister survival. When the level of oxidative damage is high, repair of the non‐helix‐distortive 8‐oxo‐guanine lesion is accomplished by nucleotide excision repair (NER; Gruber & Walker, 2018; Dhawale et al, 2021). In accordance with this, transcription‐coupled NER was shown to be important for persister survival following fluoroquinolone treatment, as knockout of NER genes results in impaired persister recovery and regrowth (Wilmaerts et al, 2022). Transcription‐coupled repair requires the removal of the RNA polymerase from the site of the lesion, either by UvrD‐mediated backtracking allowing rapid resumption of transcription following repair (Epshtein et al, 2014), or by Mfd‐mediated displacement, which terminates transcription (Park et al, 2002). Accordingly, UvrD‐mediated backtracking was shown to result in a short persister lag phase, whereas Mfd‐mediated displacement results in a longer persister lag phase following treatment. This difference could account for lag phase heterogeneity within the persister population and enable persister cells to optimize the timing of DNA damage repair versus growth resumption (Mok & Brynildsen, 2018; Wilmaerts et al, 2022). Although the study of Wilmaerts et al (2022) focused on fluoroquinolone antibiotics, NER may also be implicated in persister survival following treatment with other classes of bactericidal antibiotics. Indeed, transcriptomics, proteomics, and transposon insertion sequencing have revealed a role for the oxidative stress response and the SOS response, including NER, during β‐lactam treatment of high‐persistence hipA‐induced and cyaA‐mutant cells (Keren et al, 2004; Molina‐Quiroz et al, 2018; Sulaiman & Lam, 2020; Semanjski et al, 2021). However, a comprehensive study clearly linking NER of oxidative DNA damage by ROS in persister survival following treatment with β‐lactam antibiotics is currently lacking.
Evolution of antibiotic persistence
All bacterial species investigated to date exhibit some level of persistence, illustrative of its universal nature. Even fungal pathogens have been observed to show persistence in response to antimicrobials (LaFleur et al, 2006; Bojsen et al, 2017). The adaptive advantage of persistence in the context of antibiotic treatment for infections is readily apparent. By investing in a diversity of phenotypes that increase their chances of long‐term survival in an unpredictably changing environment, bacteria are able to hedge their bets against the possibility of encountering lethal concentrations of antibiotics (Kussell & Leibler, 2005; Grimbergen et al, 2015). However, the evolutionary benefits of persistence in natural environments are less straightforward as bacteria typically do not encounter such high concentrations of antibiotics in their natural ecosystems (Stepanyan et al, 2015). The level of persistence is determined by both genetic and environmental factors (Bakkeren et al, 2020; Verstraete et al, 2022b). Adaptive laboratory evolution has shown that antibiotic persistence is a highly evolvable trait and populations can rapidly evolve an increased persister fraction in response to periodic exposure to high antibiotic concentrations (Van den Bergh et al, 2016; Levin‐Reisman et al, 2019; Windels et al, 2021). Given that persistence results in a reproductive advantage upon antibiotic exposure and is evolvable, it is tempting to look for an adaptive—evolutionary—explanation of antibiotic persistence. However, the mere existence of utility or function does not imply adaptation (Gould & Lewontin, 1979). As such, persistence is not necessarily an adaptation to the current environment of bacteria, but could also represent a byproduct of other adaptive—or nonadaptive—pressures that occurred in the past, that is, induction of dormancy in response to hostile environments, or a byproduct of cell‐to‐cell variability resulting from inherent errors in biological processes (Johnson & Levin, 2013; Levin et al, 2014). This nonadaptive view on persistence does not preclude it from being co‐opted to increase survival in a different context (Gould & Vrba, 1982). For instance, selection may act directly on increasing persistence during adaptive laboratory evolution of persistence or within a host undergoing antibiotic treatment. However, also here, increased antibiotic persistence may have been the result of indirect or bystander selection for survival to hostile environments, provided by the immune system, rather than direct selection toward increased antibiotic tolerance (Bakkeren et al, 2020).
Despite the speculative nature of the evolutionary origin of persistence, the ability of persisters to survive antibiotics makes them a potential reservoir of genetic diversity that can be utilized when the environment changes, enabling the population to adapt quickly to new conditions (Windels et al, 2019b, 2020). In particular, the evolution of increased persistence or tolerance enables populations to evolve resistance more rapidly (Levin‐Reisman et al, 2017). This is especially significant in conditions with high antibiotic concentrations, where enhanced tolerance allows the survival of resistance‐conferring mutants beyond the mutation prevention concentration. This is facilitated through multiple mechanisms, including the increase in effective population size during antibiotic exposure, which enhances the mutation supply and increases the likelihood of acquiring resistance‐conferring mutations (Levin & Rozen, 2006; Levin‐Reisman et al, 2017), or through synergistic epistasis between tolerance and resistance mutations (Levin‐Reisman et al, 2019). In addition, it has been found that persisters exhibit increased mutation rates, which further boost the mutational supply, enabling faster adaptation (Windels et al, 2019c). Persisters could also serve as a refuge for costly resistance‐conferring plasmids, which can be transferred to other bacteria through horizontal gene transfer if conditions permit (Bakkeren et al, 2019). For a more comprehensive discussion on the evolution of persistence and its link to resistance, we encourage the reader to consult other reviews (Bakkeren et al, 2020; Verstraete et al, 2022b). Understanding how persistence evolves and affects the evolution of antibiotic resistance may ultimate allow us to develop new strategies to combat antibiotic persistence and limit resistance. Evolutionary research can aid in the development of such strategies by identifying the mechanisms and factors that promote their evolution and the interaction between persistence and resistance evolution.
Approaches for combating antibiotic persistence
As persister cells can complicate treatment and current antibiotics have proven insufficient to completely eradicate them, there is an ongoing effort to develop new antipersister strategies. While combinatorial antibiotic therapy is a promising approach (Fig 4; Keren et al, 2004; Aedo et al, 2019; Windels et al, 2019a), its efficacy likely varies depending on the species or strain of bacteria (Brochado et al, 2018), highlighting the importance of reliable identification and sensitivity profiling. However, even with optimal treatment, complete eradication of all persister cells may not be achievable due to their multiantibiotic tolerance. Therefore, there is a need for new antimicrobial compounds that can target cellular processes that are not protected in persisters (Fig 4). For example, compounds that affect cell viability by crosslinking DNA (Kwan et al, 2015), degrading proteins nonspecifically (Conlon et al, 2013) or disrupting the cell membrane (Hurdle et al, 2011; Defraine et al, 2018; Kim et al, 2018b; Hamad et al, 2022) or cell wall (Briers et al, 2014) can be considered. However, such antibiotics are often nonspecific and may therefore pose a risk of cytotoxic side effects. This highlights the need for further research and discovery of antimicrobial compounds that specifically target persisters and not host cells.
Figure 4. Strategies to combat persistence.
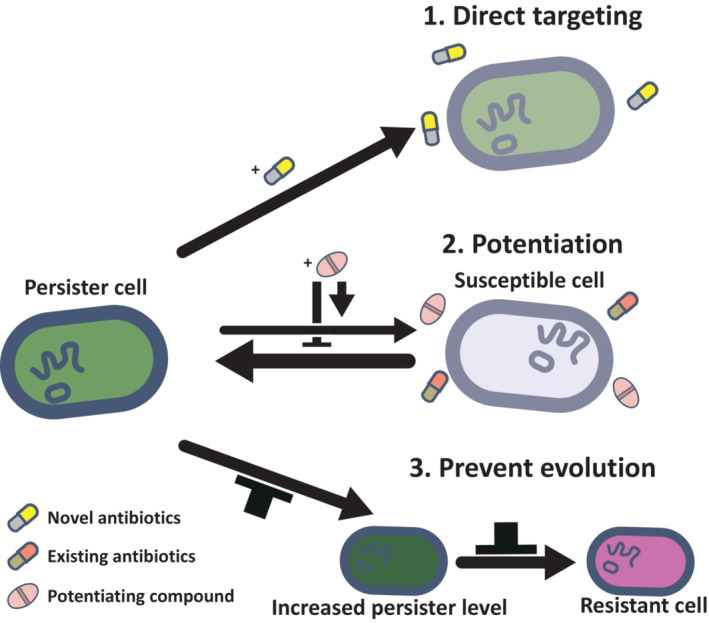
(1) Direct targeting of persister cells using single‐drug therapy with novel antibiotics or combinatorial therapy with existing antibiotics. (2) Combining existing antibiotics with a potentiating compound to enhance the efficacy of the antibiotic against the whole population, including both persister and nonpersister cells. Potentiating compounds can inhibit persister formation, trigger persister recovery and regrowth, and increase antibiotic uptake or decrease antibiotic efflux. (3) Preventing evolution of increased levels of persisters, which may also limit the evolution and spread of resistance via persistence.
Another option to decrease the number of persisters is the use of nonantibiotic potentiating compounds as adjuvants to existing antibiotics, which can improve the efficacy of the antibiotics against a population of both sensitive and persister cells (Fig 4). One possibility to increase the killing of both sensitive and persister cells is to increase the intracellular antibiotic concentration. Antibody–antibiotic conjugates, where an antipathogen antibody is linked to an antibiotic, increase the specificity of their delivery, while minimizing potential toxic side effects (Lehar et al, 2015; Zhou et al, 2016; Mariathasan & Tan, 2017). Moreover, an increased intracellular antibiotic concentration can be achieved by increasing the uptake of the antibiotic with compounds that cause direct membrane permeabilization such as colistin (Cui et al, 2016; Chung & Ko, 2019) or substances that activate mechanosensitive ion channels (Jiafeng et al, 2015; Chen et al, 2019; Lv et al, 2022). Additionally, efflux pump inhibitors can also increase intracellular antibiotic concentrations (Adams et al, 2011; Pu et al, 2016). The effectiveness of an antibiotic therapy on a population of bacteria may also be improved by decreasing the number of antibiotic‐tolerant persister cells. One strategy to lower persister levels is by preventing their formation with quorum sensing inhibitors (Starkey et al, 2014; Allegretta et al, 2017), antioxidants (Rowe et al, 2020) or by inhibiting toxin–antitoxin systems using toxin inhibitors (Li et al, 2016). However, a notable downside of using antioxidants is that this could result in concomitantly lower beneficial ROS‐mediated mechanisms of the immune system (Dumas & Knaus, 2021), leading to increased infections and longer infection durations. Preventing active persistence mechanisms by decreasing active stress responses such as the SOS response using RecA inhibitors (Alam et al, 2016; Yamamoto et al, 2022), the general stress response using mesalamine (Dahl et al, 2017) or the stringent response by limiting ppGpp production (Nguyen et al, 2011; Dutta et al, 2019) has also been associated with lower persister levels and an improved treatment in animal infection models. Another strategy to decrease the number of persisters in a population is by resensitizing them to antibiotics by inducing their regrowth. Persister regrowth can be stimulated by administering signaling molecules like indoles (Song & Wood, 2020b), although the effects are strain and antibiotic specific (Sun et al, 2020). Moreover, many carbon sources are known to restart the metabolism and thereby either increase the PMF and the uptake of the antibiotic in the cell (Allison et al, 2011; Kitzenberg et al, 2022) or induce persister regrowth (Allison et al, 2011; Vilchèze et al, 2017).
Another strategy, parallel to the ones mentioned earlier, could be to restrict or reverse the evolution of antibiotic persistence (Fig 4). Combination therapy or antibiotic cycling has been suggested as potential approaches to prevent the evolution of resistance against commonly used antibiotics (Michel et al, 2008; Baym et al, 2016). These strategies rely on negative evolutionary interactions between antibiotics, known as collateral resistance, where a mutation that confers resistance to one drug concomitantly decreases resistance to another. Conversely, resistance to one drug leading to resistance to another is referred to as cross‐resistance. However, it is typically observed that the evolution of increased persistence to a specific drug is accompanied by increased persistence to other drugs, including those of other classes, thus limiting the potential use of mixing or cycling antibiotic (Michiels et al, 2016; Van den Bergh et al, 2016; Lázár et al, 2022). Nonetheless, the patterns of cross‐persistence and collateral persistence interactions remain poorly understood and further research may identify promising antibiotic combinations. Additional methods, such as blocking evolvability factors, may also be promising to prevent persistence evolution. For instance, targeting mutagenesis‐promoting factors, such as Mfd, the mycobacterial mutasome or SOS response regulators, could be a potential approach to prevent the evolution of persistence (Ragheb et al, 2019; Merrikh & Kohli, 2020). By reducing the number of persisters or limiting high‐persistence evolution, the proposed strategies can not only combat antibiotic persistence and their role in persistent infections but may also slow down the evolution of antibiotic resistance. This could prolong the effectiveness of existing antibiotics and help to preserve the usefulness of these critical drugs in the future, which is particularly important given the decreasing rate of discovery of novel antibiotics.
Concluding remarks
Bacterial pathogens are notorious for their remarkable adaptability in hostile environments, including their ability to persist in the presence of otherwise lethal doses of antibiotics. In this review, we have outlined recent advances in our understanding of antibiotic persistence. Current research focused on persister formation, recovery, and regrowth has reinforced the notion that persistence is a complex multifaceted and adaptive phenomenon that is significantly influenced by environmental cues. This highlights the importance of considering the ecological context in which bacteria exist, including intra‐ and interspecies communication and interactions with the immune system. Acknowledging this wider ecological framework is essential for a better understanding of antibiotic persistence as an emergent property of the microbial community in its natural environment, rather than solely determined by the characteristics of individual bacterial cells. This will require researchers moving beyond conventional methods that rely on homogeneous isogenic cultures and instead develop techniques that can capture the heterogeneity and complexity of bacterial populations in vivo. However, there exists a relative paucity of antibiotic persistence research beyond well‐mixed laboratory conditions. Therefore, it is critical to contextualize basic research findings in in vivo mammalian models or clinical settings (Box 1). Recent research conducted with murine models appears to hold great potential for translating in vitro findings (Newson et al, 2022; preprint: Verstraete et al, 2022a). Additionally, several promising strategies have been proposed to combat antibiotic persistence, but their efficacy is yet to be validated. As a first step toward this validation, further development and utilization of murine models may prove to be valuable. In conclusion, further research is necessary to obtain a more holistic understanding of bacterial persistence and its implications in clinical settings, potentially leading to the development of effective therapies for persistent bacterial infections.
Author contributions
Celien Bollen: Conceptualization; writing – original draft. Elen Louwagie: Conceptualization; writing – original draft. Natalie Verstraeten: Conceptualization; funding acquisition; writing – review and editing. Jan Michiels: Conceptualization; funding acquisition; writing – review and editing. Philip Ruelens: Conceptualization; writing – original draft; writing – review and editing.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Acknowledgements
The work was supported by grants from Fonds Wetenschappelijk Onderzoek (G0B0420N, G0C4322N, G0I1522N), KU Leuven (C16/17/006) and Vlaams Instituut voor Biotechnology (VIB). CB and EL are recipients of a PhD fellowship from Fonds Wetenschappelijk Onderzoek (1S88319N, 1126120N).
EMBO reports (2023) 24: e57309
Contributor Information
Jan Michiels, Email: jan.michiels@kuleuven.be.
Philip Ruelens, Email: philip.ruelens@kuleuven.be.
References
- Ackermann M (2015) A functional perspective on phenotypic heterogeneity in microorganisms. Nat Rev Microbiol 13: 497–508 [DOI] [PubMed] [Google Scholar]
- Adams KN, Takaki K, Connolly LE, Wiedenhoft H, Winglee K, Humbert O, Edelstein PH, Cosma CL, Ramakrishnan L (2011) Drug tolerance in replicating Mycobacteria mediated by a macrophage‐induced efflux mechanism. Cell 145: 39–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aedo SJ, Orman MA, Brynildsen MP (2019) Stationary phase persister formation in Escherichia coli can be suppressed by piperacillin and PBP3 inhibition. BMC Microbiol 19: 140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam MK, Alhhazmi A, DeCoteau JF, Luo Y, Geyer CR (2016) RecA inhibitors potentiate antibiotic activity and block evolution of antibiotic resistance. Cell Chem Biol 23: 381–391 [DOI] [PubMed] [Google Scholar]
- Allegretta G, Maurer CK, Eberhard J, Maura D, Hartmann RW, Rahme L, Empting M (2017) In‐depth profiling of MvfR‐regulated small molecules in Pseudomonas aeruginosa after quorum sensing inhibitor treatment. Front Microbiol 8: 924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison KR, Brynildsen MP, Collins JJ (2011) Metabolite‐enabled eradication of bacterial persisters by aminoglycosides. Nature 473: 216–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato SM, Brynildsen MP (2015) Persister heterogeneity arising from a single metabolic stress. Curr Biol 25: 2090–2098 [DOI] [PubMed] [Google Scholar]
- Amato SM, Orman MA, Brynildsen MP (2013) Metabolic control of persister formation in Escherichia coli . Mol Cell 50: 475–487 [DOI] [PubMed] [Google Scholar]
- Anderson GG, Martin SM, Hultgren SJ (2004) Host subversion by formation of intracellular bacterial communities in the urinary tract. Microbes Infect 6: 1094–1101 [DOI] [PubMed] [Google Scholar]
- Avery SV (2006) Microbial cell individuality and the underlying sources of heterogeneity. Nat Rev Microbiol 4: 577–587 [DOI] [PubMed] [Google Scholar]
- Bakkeren E, Huisman JS, Fattinger SA, Hausmann A, Furter M, Egli A, Slack E, Sellin ME, Bonhoeffer S, Regoes RR et al (2019) Salmonella persisters promote the spread of antibiotic resistance plasmids in the gut. Nature 573: 276–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkeren E, Diard M, Hardt W‐D (2020) Evolutionary causes and consequences of bacterial antibiotic persistence. Nat Rev Microbiol 18: 479–490 [DOI] [PubMed] [Google Scholar]
- Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S (2004) Bacterial persistence as a phenotypic switch. Science 305: 1622–1625 [DOI] [PubMed] [Google Scholar]
- Balaban NQ, Helaine S, Lewis K, Ackermann M, Aldridge B, Andersson DI, Brynildsen MP, Bumann D, Camilli A, Collins JJ et al (2019) Definitions and guidelines for research on antibiotic persistence. Nat Rev Microbiol 17: 441–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett TC, Mok WWK, Murawski AM, Brynildsen MP (2019) Enhanced antibiotic resistance development from fluoroquinolone persisters after a single exposure to antibiotic. Nat Commun 10: 1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartell JA, Cameron DR, Mojsoska B, Haagensen JAJ, Pressler T, Sommer LM, Lewis K, Molin S, Johansen HK (2020) Bacterial persisters in long‐term infection: emergence and fitness in a complex host environment. PLoS Pathog 16: e1009112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baym M, Stone LK, Kishony R (2016) Multidrug evolutionary strategies to reverse antibiotic resistance. Science 351: aad3292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beam JE, Wagner NJ, Shook JC, Bahnson ESM, Fowler VG, Rowe SE, Conlon BP (2021) Macrophage‐produced peroxynitrite induces antibiotic tolerance and supersedes intrinsic mechanisms of persister formation. Infect Immun 89: e00286‐21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty WL, Byrne GI, Morrison RP (1993) Morphologic and antigenic characterization of interferon gamma‐mediated persistent chlamydia trachomatis infection in vitro . Proc Natl Acad Sci USA 90: 3998–4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker KW, Skaar EP (2014) Metal limitation and toxicity at the interface between host and pathogen. FEMS Microbiol Rev 38: 1235–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belland RJ, Nelson DE, Virok D, Crane DD, Hogan D, Sturdevant D, Beatty WL, Caldwell HD (2003) Transcriptome analysis of chlamydial growth during IFN‐gamma‐mediated persistence and reactivation. Proc Natl Acad Sci USA 100: 15971–15976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojsen R, Regenberg B, Folkesson A (2017) Persistence and drug tolerance in pathogenic yeast. Curr Genet 63: 19–22 [DOI] [PubMed] [Google Scholar]
- Bollen C, Dewachter L, Michiels J (2021) Protein aggregation as a bacterial strategy to survive antibiotic treatment. Front Mol Biosci 8: 669664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon C, Dick T (2012) How Mycobacterium tuberculosis goes to sleep: the dormancy survival regulator DosR a decade later. Future Microbiol 7: 513–518 [DOI] [PubMed] [Google Scholar]
- Borriello G, Werner E, Roe F, Kim AM, Ehrlich GD, Stewart PS (2004) Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrob Agents Chemother 48: 2659–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauner A, Fridman O, Gefen O, Balaban NQ (2016) Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat Rev Microbiol 14: 320–330 [DOI] [PubMed] [Google Scholar]
- Briers Y, Walmagh M, Grymonprez B, Biebl M, Pirnay J‐P, Defraine V, Michiels J, Cenens W, Aertsen A, Miller S et al (2014) Art‐175 is a highly efficient antibacterial against multidrug‐resistant strains and persisters of Pseudomonas aeruginosa . Antimicrob Agents Chemother 58: 3774–3784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochado AR, Telzerow A, Bobonis J, Banzhaf M, Mateus A, Selkrig J, Huth E, Bassler S, Zamarreño Beas J, Zietek M et al (2018) Species‐specific activity of antibacterial drug combinations. Nature 559: 259–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DR (2019) Nitrogen starvation induces persister cell formation in Escherichia coli . J Bacteriol 201: e00622‐18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesar S, Willis L, Huang KC (2022) Bacterial respiration during stationary phase induces intracellular damage that leads to delayed regrowth. iScience 25: 103765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Gao Y, Lv B, Sun F, Yao W, Wang Y, Fu X (2019) Hypoionic shock facilitates aminoglycoside killing of both nutrient shift‐ and starvation‐induced bacterial persister cells by rapidly enhancing aminoglycoside uptake. Front Microbiol 10: 2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverton AM, Gollan B, Przydacz M, Wong CT, Mylona A, Hare SA, Helaine S (2016) A Salmonella toxin promotes persister formation through acetylation of tRNA. Mol Cell 63: 86–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J, Rogers J, Kearns M, Leslie M, Hartson SD, Wilson KS (2015) Escherichia coli persister cells suppress translation by selectively disassembling and degrading their ribosomes. Mol Microbiol 95: 352–364 [DOI] [PubMed] [Google Scholar]
- Christensen SK, Pedersen K, Hansen FG, Gerdes K (2003) Toxin–antitoxin loci as stress‐response‐elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J Mol Biol 332: 809–819 [DOI] [PubMed] [Google Scholar]
- Chung ES, Ko KS (2019) Eradication of persister cells of Acinetobacter baumannii through combination of colistin and amikacin antibiotics. J Antimicrob Chemother 74: 1277–1283 [DOI] [PubMed] [Google Scholar]
- Ciofu O, Tolker‐Nielsen T (2019) Tolerance and resistance of Pseudomonas aeruginosa biofilms to antimicrobial agents‐how P. aeruginosa can escape antibiotics. Front Microbiol 10: 913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofu O, Moser C, Jensen PØ, Høiby N (2022) Tolerance and resistance of microbial biofilms. Nat Rev Microbiol 20: 621–635 [DOI] [PubMed] [Google Scholar]
- Conlon BP, Nakayasu ES, Fleck LE, LaFleur MD, Isabella VM, Coleman K, Leonard SN, Smith RD, Adkins JN, Lewis K (2013) Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature 503: 365–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon BP, Rowe SE, Gandt AB, Nuxoll AS, Donegan NP, Zalis EA, Clair G, Adkins JN, Cheung AL, Lewis K (2016) Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat Microbiol 1: 16051 [DOI] [PubMed] [Google Scholar]
- Cui P, Niu H, Shi W, Zhang S, Zhang H, Margolick J, Zhang W, Zhang Y (2016) Disruption of membrane by colistin kills uropathogenic Escherichia coli persisters and enhances killing of other antibiotics. Antimicrob Agents Chemother 60: 6867–6871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl J‐U, Gray MJ, Bazopoulou D, Beaufay F, Lempart J, Koenigsknecht MJ, Wang Y, Baker JR, Hasler WL, Young VB et al (2017) The anti‐inflammatory drug mesalamine targets bacterial polyphosphate accumulation. Nat Microbiol 2: 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defraine V, Liebens V, Loos E, Swings T, Weytjens B, Fierro C, Marchal K, Sharkey L, O'Neill AJ, Corbau R et al (2018) 1‐((2,4‐dichlorophenethyl)amino)‐3‐phenoxypropan‐2‐ol kills Pseudomonas aeruginosa through extensive membrane damage. Front Microbiol 9: 129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarre G, Prudent V, Schenk H, Rousseau E, Bringer M‐A, Barnich N, Tran Van Nhieu G, Rimsky S, De Monte S, Espéli O (2019) The Crohn's disease‐associated Escherichia coli strain LF82 relies on SOS and stringent responses to survive, multiply and tolerate antibiotics within macrophages. PLoS Pathog 15: e1008123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnues B, Cuny C, Grégori G, Dukan S, Aguilaniu H, Nyström T (2003) Differential oxidative damage and expression of stress defence regulons in culturable and non‐culturable Escherichia coli cells. EMBO Rep 4: 400–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewachter L, Fauvart M, Michiels J (2019) Bacterial heterogeneity and antibiotic survival: understanding and combatting persistence and heteroresistance. Mol Cell 76: 255–267 [DOI] [PubMed] [Google Scholar]
- Dewachter L, Bollen C, Wilmaerts D, Louwagie E, Herpels P, Matthay P, Khodaparast L, Khodaparast L, Rousseau F, Schymkowitz J et al (2021) The dynamic transition of persistence toward the viable but nonculturable state during stationary phase is driven by protein aggregation. MBio 12: e00703‐21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawale A, Bindal G, Rath D, Rath A (2021) DNA repair pathways important for the survival of Escherichia coli to hydrogen peroxide mediated killing. Gene 768: 145297 [DOI] [PubMed] [Google Scholar]
- D'Onofrio A, Crawford JM, Stewart EJ, Witt K, Gavrish E, Epstein S, Clardy J, Lewis K (2010) Siderophores from neighboring organisms promote the growth of uncultured bacteria. Chem Biol 17: 254–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörr T, Lewis K, Vulić M (2009) SOS response induces persistence to fluoroquinolones in Escherichia coli . PLoS Genet 5: e1000760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörr T, Vulić M, Lewis K (2010) Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli . PLoS Biol 8: e1000317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drlica K, Zhao X (1997) DNA gyrase, topoisomerase IV, and the 4‐quinolones. Microbiol Mol Biol Rev 61: 377–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas A, Knaus UG (2021) Raising the ‘good’ oxidants for immune protection. Front Immunol 12: 698042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta NK, Klinkenberg LG, Vazquez M‐J, Segura‐Carro D, Colmenarejo G, Ramon F, Rodriguez‐Miquel B, Mata‐Cantero L, Porras‐De Francisco E, Chuang Y‐M et al (2019) Inhibiting the stringent response blocks Mycobacterium tuberculosis entry into quiescence and reduces persistence. Sci Adv 5: eaav2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer DJ, Belenky PA, Yang JH, MacDonald IC, Martell JD, Takahashi N, Chan CTY, Lobritz MA, Braff D, Schwarz EG et al (2014) Antibiotics induce redox‐related physiological alterations as part of their lethality. Proc Natl Acad Sci USA 111: E2100–E2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann D, Berghoff BA (2022) A shift in perspective: a role for the type I toxin TisB as persistence‐stabilizing factor. Front Microbiol 13: 871699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz MB, Levine AJ, Siggia ED, Swain PS (2002) Stochastic gene expression in a single cell. Science 297: 1183–1186 [DOI] [PubMed] [Google Scholar]
- Eng RH, Padberg FT, Smith SM, Tan EN, Cherubin CE (1991) Bactericidal effects of antibiotics on slowly growing and nongrowing bacteria. Antimicrob Agents Chemother 35: 1824–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epshtein V, Kamarthapu V, McGary K, Svetlov V, Ueberheide B, Proshkin S, Mironov A, Nudler E (2014) UvrD facilitates DNA repair by pulling RNA polymerase backwards. Nature 505: 372–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang FC (2011) Antimicrobial actions of reactive oxygen species. MBio 2: e00141‐11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauvart M, de Groote VN, Michiels J (2011) Role of persister cells in chronic infections: clinical relevance and perspectives on anti‐persister therapies. J Med Microbiol 60: 699–709 [DOI] [PubMed] [Google Scholar]
- Flemming H‐C, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S (2016) Biofilms: an emergent form of bacterial life. Nat Rev Microbiol 14: 563–575 [DOI] [PubMed] [Google Scholar]
- Foster JW (1999) When protons attack: microbial strategies of acid adaptation. Curr Opin Microbiol 2: 170–174 [DOI] [PubMed] [Google Scholar]
- Foti JJ, Devadoss B, Winkler JA, Collins JJ, Walker GC (2012) Oxidation of the guanine nucleotide pool underlies cell death by bactericidal antibiotics. Science 336: 315–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gengenbacher M, Kaufmann SHE (2012) Mycobacterium tuberculosis: success through dormancy. FEMS Microbiol Rev 36: 514–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Sureka K, Ghosh B, Bose I, Basu J, Kundu M (2011) Phenotypic heterogeneity in mycobacterial stringent response. BMC Syst Biol 5: 18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode O, Smith A, Łapińska U, Bamford R, Kahveci Z, Glover G, Attrill E, Carr A, Metz J, Pagliara S (2021) Heterologous protein expression favors the formation of protein aggregates in persister and viable but nonculturable bacteria. ACS Infect Dis 7: 1848–1858 [DOI] [PubMed] [Google Scholar]
- Goormaghtigh F, Van Melderen L (2019) Single‐cell imaging and characterization of Escherichia coli persister cells to ofloxacin in exponential cultures. Sci Adv 5: eaav9462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goormaghtigh F, Fraikin N, Putrinš M, Hallaert T, Hauryliuk V, Garcia‐Pino A, Sjödin A, Kasvandik S, Udekwu K, Tenson T et al (2018) Reassessing the role of type II toxin‐antitoxin systems in formation of Escherichia coli type II persister cells. MBio 9: e00640‐18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, Lewontin RC (1979) The spandrels of san Marco and the panglossian paradigm: a critique of the adaptationist programme. Proc R Soc Lond B Biol Sci 205: 581–598 [DOI] [PubMed] [Google Scholar]
- Gould SJ, Vrba ES (1982) Exaptation – a missing term in the science of form. Paleobiology 8: 4–15 [Google Scholar]
- Grimbergen AJ, Siebring J, Solopova A, Kuipers OP (2015) Microbial bet‐hedging: the power of being different. Curr Opin Microbiol 25: 67–72 [DOI] [PubMed] [Google Scholar]
- Gruber CC, Walker GC (2018) Incomplete base excision repair contributes to cell death from antibiotics and other stresses. DNA Repair (Amst) 71: 108–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez A, Jain S, Bhargava P, Hamblin M, Lobritz MA, Collins JJ (2017) Understanding and sensitizing density‐dependent persistence to quinolone antibiotics. Mol Cell 68: 1147–1154 [DOI] [PubMed] [Google Scholar]
- Hamad M, Al‐Marzooq F, Srinivasulu V, Omar HA, Sulaiman A, Zaher DM, Orive G, Al‐Tel TH (2022) Antibacterial activity of small molecules which eradicate methicillin‐resistant Staphylococcus aureus persisters. Front Microbiol 13: 823394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms A (2019) The biology of persister cells in Escherichia coli . In Persister Cells and Infectious Disease, Lewis K (ed), pp 39–57. Cham: Springer International Publishing; [Google Scholar]
- Helaine S, Cheverton AM, Watson KG, Faure LM, Matthews SA, Holden DW (2014) Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science 343: 204–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbing ME, Fuqua C, Parsek MR, Peterson SB (2010) Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol 8: 15–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill PWS, Moldoveanu AL, Sargen M, Ronneau S, Glegola‐Madejska I, Beetham C, Fisher RA, Helaine S (2021) The vulnerable versatility of Salmonella antibiotic persisters during infection. Cell Host Microbe 29: 1757–1773 [DOI] [PubMed] [Google Scholar]
- Himeoka Y, Mitarai N (2021) When to wake up? The optimal waking‐up strategies for starvation‐induced persistence. PLoS Comput Biol 17: e1008655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Coates A (2012) Nonmultiplying bacteria are profoundly tolerant to antibiotics. In Antibiotic Resistance, Coates ARM (ed), pp 99–119. Berlin, Heidelberg: Springer; [DOI] [PubMed] [Google Scholar]
- Huemer M, Mairpady Shambat S, Brugger SD, Zinkernagel AS (2020) Antibiotic resistance and persistence‐implications for human health and treatment perspectives. EMBO Rep 21: e51034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huemer M, Mairpady Shambat S, Bergada‐Pijuan J, Söderholm S, Boumasmoud M, Vulin C, Gómez‐Mejia A, Antelo Varela M, Tripathi V, Götschi S et al (2021) Molecular reprogramming and phenotype switching in Staphylococcus aureus lead to high antibiotic persistence and affect therapy success. Proc Natl Acad Sci USA 118: e2014920118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh D, Paulsson J (2011) Non‐genetic heterogeneity from stochastic partitioning at cell division. Nat Genet 43: 95–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurdle JG, O'Neill AJ, Chopra I, Lee RE (2011) Targeting bacterial membrane function: an underexploited mechanism for treating persistent infections. Nat Rev Microbiol 9: 62–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiafeng L, Fu X, Chang Z (2015) Hypoionic shock treatment enables aminoglycosides antibiotics to eradicate bacterial persisters. Sci Rep 5: 14247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jõers A, Kaldalu N, Tenson T (2010) The frequency of persisters in Escherichia coli reflects the kinetics of awakening from dormancy. J Bacteriol 192: 3379–3384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jõers A, Vind K, Hernández SB, Maruste R, Pereira M, Brauer A, Remm M, Cava F, Tenson T (2019) Muropeptides stimulate growth resumption from stationary phase in Escherichia coli . Sci Rep 9: 18043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson PJT, Levin BR (2013) Pharmacodynamics, population dynamics, and the evolution of persistence in Staphylococcus aureus . PLoS Genet 9: e1003123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junkins EN, McWhirter JB, McCall L‐I, Stevenson BS (2022) Environmental structure impacts microbial composition and secondary metabolism. ISME Commun 2: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaldalu N, Hauryliuk V, Turnbull KJ, La Mensa A, Putrinš M, Tenson T (2020) In vitro studies of persister cells. Microbiol Mol Biol Rev 84: e00070‐20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan Y, Reich S, Oster E, Maoz S, Levin‐Reisman I, Ronin I, Gefen O, Agam O, Balaban NQ (2021) Observation of universal ageing dynamics in antibiotic persistence. Nature 600: 290–294 [DOI] [PubMed] [Google Scholar]
- Keren I, Shah D, Spoering A, Kaldalu N, Lewis K (2004) Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli . J Bacteriol 186: 8172–8180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J‐S, Yamasaki R, Song S, Zhang W, Wood TK (2018a) Single cell observations show persister cells wake based on ribosome content. Environ Microbiol 20: 2085–2098 [DOI] [PubMed] [Google Scholar]
- Kim W, Zhu W, Hendricks GL, Van Tyne D, Steele AD, Keohane CE, Fricke N, Conery AL, Shen S, Pan W et al (2018b) A new class of synthetic retinoid antibiotics effective against bacterial persisters. Nature 556: 103–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitzenberg DA, Lee JS, Mills KB, Kim J‐S, Liu L, Vázquez‐Torres A, Colgan SP, Kao DJ (2022) Adenosine awakens metabolism to enhance growth‐independent killing of tolerant and persister bacteria across multiple classes of antibiotics. MBio 13: e00480‐22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiviet DJ, Nghe P, Walker N, Boulineau S, Sunderlikova V, Tans SJ (2014) Stochasticity of metabolism and growth at the single‐cell level. Nature 514: 376–379 [DOI] [PubMed] [Google Scholar]
- Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ (2007) A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130: 797–810 [DOI] [PubMed] [Google Scholar]
- Korch SB, Hill TM (2006) Ectopic overexpression of wild‐type and mutant hipA genes in Escherichia coli: effects on macromolecular synthesis and persister formation. J Bacteriol 188: 3826–3836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubistova L, Dvoracek L, Tkadlec J, Melter O, Licha I (2018) Environmental stress affects the formation of Staphylococcus aureus persisters tolerant to antibiotics. Microb Drug Resist 24: 547–555 [DOI] [PubMed] [Google Scholar]
- Kussell E, Leibler S (2005) Phenotypic diversity, population growth, and information in fluctuating environments. Science 309: 2075–2078 [DOI] [PubMed] [Google Scholar]
- Kussell E, Kishony R, Balaban NQ, Leibler S (2005) Bacterial persistence: a model of survival in changing environments. Genetics 169: 1807–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan BW, Valenta JA, Benedik MJ, Wood TK (2013) Arrested protein synthesis increases persister‐like cell formation. Antimicrob Agents Chemother 57: 1468–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan BW, Chowdhury N, Wood TK (2015) Combatting bacterial infections by killing persister cells with mitomycin C. Environ Microbiol 17: 4406–4414 [DOI] [PubMed] [Google Scholar]
- La Rosa R, Johansen HK, Molin S (2022) Persistent bacterial infections, antibiotic treatment failure, and microbial adaptive evolution. Antibiotics 11: 419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFleur MD, Kumamoto CA, Lewis K (2006) Candida albicans biofilms produce antifungal‐tolerant persister cells. Antimicrob Agents Chemother 50: 3839–3846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang M, Krin E, Korlowski C, Sismeiro O, Varet H, Coppée J‐Y, Mazel D, Baharoglu Z (2021) Sleeping ribosomes: bacterial signaling triggers RaiA mediated persistence to aminoglycosides. iScience 24: 103128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langklotz S, Narberhaus F (2011) The Escherichia coli replication inhibitor CspD is subject to growth‐regulated degradation by the Lon protease. Mol Microbiol 80: 1313–1325 [DOI] [PubMed] [Google Scholar]
- Lázár V, Snitser O, Barkan D, Kishony R (2022) Antibiotic combinations reduce Staphylococcus aureus clearance. Nature 610: 540–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehar SM, Pillow T, Xu M, Staben L, Kajihara KK, Vandlen R, DePalatis L, Raab H, Hazenbos WL, Hiroshi Morisaki J et al (2015) Novel antibody–antibiotic conjugate eliminates intracellular S. aureus . Nature 527: 323–328 [DOI] [PubMed] [Google Scholar]
- Lemma AS, Brynildsen MP (2021) Toxin induction or inhibition of transcription or translation posttreatment increases persistence to fluoroquinolones. MBio 12: e01983‐21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leszczynska D, Matuszewska E, Kuczynska‐Wisnik D, Furmanek‐Blaszk B, Laskowska E (2013) The formation of persister cells in stationary‐phase cultures of Escherichia coli is associated with the aggregation of endogenous proteins. PLoS ONE 8: e54737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung V, Lévesque CM (2012) A stress‐inducible quorum‐sensing peptide mediates the formation of persister cells with noninherited multidrug tolerance. J Bacteriol 194: 2265–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin BR, Rozen DE (2006) Non‐inherited antibiotic resistance. Nat Rev Microbiol 4: 556–562 [DOI] [PubMed] [Google Scholar]
- Levin BR, Concepción‐Acevedo J, Udekwu KI (2014) Persistence: a copacetic and parsimonious hypothesis for the existence of non‐inherited resistance to antibiotics. Curr Opin Microbiol 21: 18–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin‐Reisman I, Ronin I, Gefen O, Braniss I, Shoresh N, Balaban NQ (2017) Antibiotic tolerance facilitates the evolution of resistance. Science 355: 826–830 [DOI] [PubMed] [Google Scholar]
- Levin‐Reisman I, Brauner A, Ronin I, Balaban NQ (2019) Epistasis between antibiotic tolerance, persistence, and resistance mutations. Proc Natl Acad Sci USA 116: 14734–14739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K (2005) Persister cells and the riddle of biofilm survival. Biochemistry 70: 267–274 [DOI] [PubMed] [Google Scholar]
- Lewis K (2007) Persister cells, dormancy and infectious disease. Nat Rev Microbiol 5: 48–56 [DOI] [PubMed] [Google Scholar]
- Lewis K (2010) Persister cells. Annu Rev Microbiol 64: 357–372 [DOI] [PubMed] [Google Scholar]
- Li T, Yin N, Liu H, Pei J, Lai L (2016) Novel inhibitors of toxin HipA reduce multidrug tolerant persisters. ACS Med Chem Lett 7: 449–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Tan S, Huang L, Abramovitch RB, Rohde KH, Zimmerman MD, Chen C, Dartois V, VanderVen BC, Russell DG (2016) Immune activation of the host cell induces drug tolerance in Mycobacterium tuberculosis both in vitro and in vivo . J Exp Med 213: 809–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv B, Zeng Y, Zhang H, Li Z, Xu Z, Wang Y, Gao Y, Chen Y, Fu X (2022) Mechanosensitive channels mediate hypoionic shock‐induced aminoglycoside potentiation against bacterial persisters by enhancing antibiotic uptake. Antimicrob Agents Chemother 66: e01125‐21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik M, Zhao X, Drlica K (2006) Lethal fragmentation of bacterial chromosomes mediated by DNA gyrase and quinolones. Mol Microbiol 61: 810–825 [DOI] [PubMed] [Google Scholar]
- Manina G, Dhar N, Mckinney JD (2015) Stress and host immunity amplify Mycobacterium tuberculosis phenotypic heterogeneity and induce nongrowing metabolically active forms. Cell Host Microbe 17: 32–46 [DOI] [PubMed] [Google Scholar]
- Manuse S, Shan Y, Canas‐Duarte SJ, Bakshi S, Sun W‐S, Mori H, Paulsson J, Lewis K (2021) Bacterial persisters are a stochastically formed subpopulation of low‐energy cells. PLoS Biol 19: e3001194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S, Tan M‐W (2017) Antibody–antibiotic conjugates: a novel therapeutic platform against bacterial infections. Trends Mol Med 23: 135–149 [DOI] [PubMed] [Google Scholar]
- McKay SL, Portnoy DA (2015) Ribosome hibernation facilitates tolerance of stationary‐phase bacteria to aminoglycosides. Antimicrob Agents Chemother 59: 6992–6999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrikh H, Kohli RM (2020) Targeting evolution to inhibit antibiotic resistance. FEBS J 287: 4341–4353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaux C, Ronneau S, Giorgio RT, Helaine S (2022) Antibiotic tolerance and persistence have distinct fitness trade‐offs. PLoS Pathog 18: e1010963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel J‐B, Yeh PJ, Chait R, Moellering RC, Kishony R (2008) Drug interactions modulate the potential for evolution of resistance. Proc Natl Acad Sci USA 105: 14918–14923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michiels J, Van den Bergh B, Verstraeten N, Fauvart M, Michiels J (2016) In vitro emergence of high persistence upon periodic aminoglycoside challenge in the ESKAPE pathogens. Antimicrob Agents Chemother 60: 4630–4637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mode S, Ketterer M, Québatte M, Dehio C (2022) Antibiotic persistence of intracellular Brucella abortus . PLoS Negl Trop Dis 16: e0010635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan VP, Scanga CA, Yu K, Scott HM, Tanaka KE, Tsang E, Tsai MC, Flynn JL, Chan J (2001) Effects of tumor necrosis factor alpha on host immune response in chronic persistent tuberculosis: possible role for limiting pathology. Infect Immun 69: 1847–1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohiuddin SG, Massahi A, Orman MA (2022) lon deletion impairs persister cell resuscitation in Escherichia coli . MBio 13: e02187‐21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok WWK, Brynildsen MP (2018) Timing of DNA damage responses impacts persistence to fluoroquinolones. Proc Natl Acad Sci USA 115: E6301–E6309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möker N, Dean CR, Tao J (2010) Pseudomonas aeruginosa increases formation of multidrug‐tolerant persister cells in response to quorum‐sensing signaling molecules. J Bacteriol 192: 1946–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina‐Quiroz RC, Silva‐Valenzuela C, Brewster J, Castro‐Nallar E, Levy SB, Camilli A (2018) Cyclic AMP regulates bacterial persistence through repression of the oxidative stress response and SOS‐dependent DNA repair in uropathogenic Escherichia coli . MBio 9: e02144‐17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mookherjee N, Anderson MA, Haagsman HP, Davidson DJ (2020) Antimicrobial host defence peptides: functions and clinical potential. Nat Rev Drug Discov 19: 311–332 [DOI] [PubMed] [Google Scholar]
- Mordukhova EA, Pan JG (2014) Stabilization of homoserine‐o‐succinyltransferase (MetA) decreases the frequency of persisters in Escherichia coli under stressful conditions. PLoS ONE 9: e110504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulcahy LR, Burns JL, Lory S, Lewis K (2010) Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J Bacteriol 192: 6191–6199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulcahy LR, Isabella VM, Lewis K (2014) Pseudomonas aeruginosa biofilms in disease. Microb Ecol 68: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murawski AM, Brynildsen MP (2021) Ploidy is an important determinant of fluoroquinolone persister survival. Curr Biol 31: 2039–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch CC, Skaar EP (2022) Nutritional immunity: the battle for nutrient metals at the host–pathogen interface. Nat Rev Microbiol 20: 657–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan A, Nair MS, Muyyarikkandy MS, Amalaradjou MA (2018) Inhibition and inactivation of uropathogenic Escherichia coli biofilms on urinary catheters by sodium selenite. Int J Mol Sci 19: 1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanaswamy R, Levy M, Tsechansky M, Stovall GM, O'Connell JD, Mirrielees J, Ellington AD, Marcotte EM (2009) Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc Natl Acad Sci USA 106: 10147–10152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newson JP, Gaissmaier MS, McHugh SC, Hardt W‐D (2022) Studying antibiotic persistence in vivo using the model organism Salmonella typhimurium. Curr Opin Microbiol 70: 102224 [DOI] [PubMed] [Google Scholar]
- Nguyen D, Joshi‐Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y et al (2011) Active starvation responses mediate antibiotic tolerance in biofilms and nutrient‐limited bacteria. Science 334: 982–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen J, Lara‐Gutiérrez J, Stocker R (2021) Environmental fluctuations and their effects on microbial communities, populations and individuals. FEMS Microbiol Rev 45: fuaa068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols D, Lewis K, Orjala J, Mo S, Ortenberg R, O'Connor P, Zhao C, Vouros P, Kaeberlein T, Epstein SS (2008) Short peptide induces an “uncultivable” microorganism to grow in vitro . Appl Environ Microbiol 74: 4889–4897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyström T, Gustavsson N (1998) Maintenance energy requirement: what is required for stasis survival of Escherichia coli? Biochim Biophys Acta 1365: 225–231 [DOI] [PubMed] [Google Scholar]
- Orman MA, Brynildsen MP (2013) Dormancy is not necessary or sufficient for bacterial persistence. Antimicrob Agents Chemother 57: 3230–3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paranjape SS, Shashidhar R (2019) Comparison of starvation‐induced persister cells with antibiotic‐induced persister cells. Curr Microbiol 76: 1495–1502 [DOI] [PubMed] [Google Scholar]
- Park J‐S, Marr MT, Roberts JW (2002) E. coli transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Cell 109: 757–767 [DOI] [PubMed] [Google Scholar]
- Pedersen K, Christensen SK, Gerdes K (2002) Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol Microbiol 45: 501–510 [DOI] [PubMed] [Google Scholar]
- Personnic N, Striednig B, Hilbi H (2021) Quorum sensing controls persistence, resuscitation, and virulence of legionella subpopulations in biofilms. ISME J 15: 196–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Personnic N, Doublet P, Jarraud S (2023) Intracellular persister: a stealth agent recalcitrant to antibiotics. Front Cell Infect Microbiol 13: 1141868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyrusson F, Varet H, Nguyen TK, Legendre R, Sismeiro O, Coppée J‐Y, Wolz C, Tenson T, Van Bambeke F (2020) Intracellular Staphylococcus aureus persisters upon antibiotic exposure. Nat Commun 11: 2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyrusson F, Nguyen TK, Najdovski T, Van Bambeke F (2022) Host cell oxidative stress induces dormant Staphylococcus aureus persisters. Microbiol Spectr 10: e02313‐21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prossliner T, Skovbo Winther K, Sørensen MA, Gerdes K (2018) Ribosome hibernation. Annu Rev Genet 52: 321–348 [DOI] [PubMed] [Google Scholar]
- Pu Y, Zhao Z, Li Y, Zou J, Ma Q, Zhao Y, Ke Y, Zhu Y, Chen H, Baker MAB et al (2016) Enhanced efflux activity facilitates drug tolerance in dormant bacterial cells. Mol Cell 62: 284–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu Y, Li Y, Jin X, Tian T, Ma Q, Zhao Z, Lin S, Chen Z, Li B, Yao G et al (2019) ATP‐dependent dynamic protein aggregation regulates bacterial dormancy depth critical for antibiotic tolerance. Mol Cell 73: 143–156 [DOI] [PubMed] [Google Scholar]
- Ragheb MN, Thomason MK, Hsu C, Nugent P, Gage J, Samadpour AN, Kariisa A, Merrikh CN, Miller SI, Sherman DR et al (2019) Inhibiting the evolution of antibiotic resistance. Mol Cell 73: 157–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganathan N, Johnson R, Edwards AM (2020) The general stress response of Staphylococcus aureus promotes tolerance of antibiotics and survival in whole human blood. Microbiology 166: 1088–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raser JM, O'Shea EK (2005) Noise in gene expression: origins, consequences, and control. Science 309: 2010–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez‐Rojas A, Baeder DY, Johnston P, Regoes RR, Rolff J (2021) Bacteria primed by antimicrobial peptides develop tolerance and persist. PLoS Pathog 17: e1009443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronneau S, Hill PW, Helaine S (2021) Antibiotic persistence and tolerance: not just one and the same. Curr Opin Microbiol 64: 76–81 [DOI] [PubMed] [Google Scholar]
- Ronneau S, Michaux C, Helaine S (2023) Decline in nitrosative stress drives antibiotic persister regrowth during infection. Cell Host Microbe 31: 1–14 [DOI] [PubMed] [Google Scholar]
- Rotem E, Loinger A, Ronin I, Levin‐Reisman I, Gabay C, Shoresh N, Biham O, Balaban NQ (2010) Regulation of phenotypic variability by a threshold‐based mechanism underlies bacterial persistence. Proc Natl Acad Sci USA 107: 12541–12546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe SE, Wagner NJ, Li L, Beam JE, Wilkinson AD, Radlinski LC, Zhang Q, Miao EA, Conlon BP (2020) Reactive oxygen species induce antibiotic tolerance during systemic Staphylococcus aureus infection. Nat Microbiol 5: 282–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rycroft JA, Gollan B, Grabe GJ, Hall A, Cheverton AM, Larrouy‐Maumus G, Hare SA, Helaine S (2018) Activity of acetyltransferase toxins involved in Salmonella persister formation during macrophage infection. Nat Commun 9: 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad S, Cereghetti G, Feng Y, Picotti P, Peter M, Dechant R (2017) Reversible protein aggregation is a protective mechanism to ensure cell cycle restart after stress. Nat Cell Biol 19: 1202–1213 [DOI] [PubMed] [Google Scholar]
- Salcedo‐Sora JE, Kell DB (2020) A quantitative survey of bacterial persistence in the presence of antibiotics: towards antipersister antimicrobial discovery. Antibiotics 9: 508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandín D, Valle J, Morata J, Andreu D, Torrent M (2022) Antimicrobial peptides can generate tolerance by lag and interfere with antimicrobial therapy. Pharmaceutics 14: 2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi I, Manfredi P, Maffei E, Egli A, Jenal U (2021) Evolution of antibiotic tolerance shapes resistance development in chronic Pseudomonas aeruginosa infections. MBio 12: e03482‐20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheler O, Makuch K, Debski PR, Horka M, Ruszczak A, Pacocha N, Sozański K, Smolander O‐P, Postek W, Garstecki P (2020) Droplet‐based digital antibiotic susceptibility screen reveals single‐cell clonal heteroresistance in an isogenic bacterial population. Sci Rep 10: 3282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher MA, Balani P, Min J, Chinnam NB, Hansen S, Vulić M, Lewis K, Brennan RG (2015) HipBA‐promoter structures reveal the basis of heritable multidrug tolerance. Nature 524: 59–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semanjski M, Gratani FL, Englert T, Nashier P, Beke V, Nalpas N, Germain E, George S, Wolz C, Gerdes K et al (2021) Proteome dynamics during antibiotic persistence and resuscitation. mSystems 6: e0054921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Y, Gandt AB, Rowe SE, Deisinger JP, Conlon BP, Lewis K (2017) ATP‐dependent persister formation in Escherichia coli . MBio 8: e02267‐16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol NW, Slessarev E, Marschmann GL, Nicolas A, Blazewicz SJ, Brodie EL, Firestone MK, Foley MM, Hestrin R, Hungate BA et al (2022) Life and death in the soil microbiome: how ecological processes influence biogeochemistry. Nat Rev Microbiol 20: 415–430 [DOI] [PubMed] [Google Scholar]
- Song S, Wood TK (2020a) ppGpp ribosome dimerization model for bacterial persister formation and resuscitation. Biochem Biophys Res Commun 523: 281–286 [DOI] [PubMed] [Google Scholar]
- Song S, Wood TK (2020b) Combatting persister cells with substituted indoles. Front Microbiol 11: 1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speziale P, Pietrocola G, Foster TJ, Geoghegan JA (2014) Protein‐based biofilm matrices in Staphylococci . Front Cell Infect Microbiol 4: 171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoering AL, Lewis K (2001) Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J Bacteriol 183: 6746–6751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapels DAC, Hill PWS, Westermann AJ, Fisher RA, Thurston TL, Saliba AE, Blommestein I, Vogel J, Helaine S (2018) Salmonella persisters undermine host immune defenses during antibiotic treatment. Science 362: 1156–1160 [DOI] [PubMed] [Google Scholar]
- Starkey M, Lepine F, Maura D, Bandyopadhaya A, Lesic B, He J, Kitao T, Righi V, Milot S, Tzika A et al (2014) Identification of anti‐virulence compounds that disrupt quorum‐sensing regulated acute and persistent pathogenicity. PLoS Pathog 10: e1004321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenackers H, Hermans K, Vanderleyden J, De Keersmaecker SCJ (2012) Salmonella biofilms: an overview on occurrence, structure, regulation and eradication. Food Res Int 45: 502–531 [Google Scholar]
- Stepanyan K, Wenseleers T, Duéñez‐Guzmán EA, Muratori F, Van Den Bergh B, Verstraeten N, De Meester L, Verstrepen KJ, Fauvart M, Michiels J (2015) Fitness trade‐offs explain low levels of persister cells in the opportunistic pathogen Pseudomonas aeruginosa . Mol Ecol 24: 1572–1583 [DOI] [PubMed] [Google Scholar]
- Sulaiman JE, Lam H (2020) Proteomic study of the survival and resuscitation mechanisms of filamentous persisters in an evolved Escherichia coli population from cyclic ampicillin treatment. mSystems 5: e00462‐20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulaiman JE, Lam H (2021) Evolution of bacterial tolerance under antibiotic treatment and its implications on the development of resistance. Front Microbiol 12: 617412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Bian M, Li Z, Lv B, Gao Y, Wang Y, Fu X (2020) 5‐methylindole potentiates aminoglycoside against gram‐positive bacteria including Staphylococcus aureus persisters under hypoionic conditions. Front Cell Infect Microbiol 10: 84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodore A, Lewis K, Vulić M (2013) Tolerance of Escherichia coli to fluoroquinolone antibiotics depends on specific components of the SOS response pathway. Genetics 195: 1265–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrey HL, Keren I, Via LE, Lee JS, Lewis K (2016) High persister mutants in Mycobacterium tuberculosis . PLoS ONE 11: e0155127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi A, Dewan PC, Barua B, Varadarajan R (2012) Additional role for the ccd operon of F‐plasmid as a transmissible persistence factor. Proc Natl Acad Sci USA 109: 12497–12502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Acker H, Coenye T (2017) The role of reactive oxygen species in antibiotic‐mediated killing of bacteria. Trends Microbiol 25: 456–466 [DOI] [PubMed] [Google Scholar]
- Van den Bergh B, Michiels J, Wenseleers T, Windels EM, Boer PV, Kestemont D, Kestemont D, De Meester L, Verstrepen KJ, Verstraeten N et al (2016) Frequency of antibiotic application drives rapid evolutionary adaptation of Escherichia coli persistence. Nat Microbiol 1: 16020 [DOI] [PubMed] [Google Scholar]
- Van den Bergh B, Fauvart M, Michiels J (2017) Formation, physiology, ecology, evolution and clinical importance of bacterial persisters. FEMS Microbiol Rev 41: 219–251 [DOI] [PubMed] [Google Scholar]
- Van den Bergh B, Schramke H, Michiels JE, Kimkes TEP, Radzikowski JL, Schimpf J, Vedelaar SR, Burschel S, Dewachter L, Lončar N et al (2022) Mutations in respiratory complex I promote antibiotic persistence through alterations in intracellular acidity and protein synthesis. Nat Commun 13: 546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez‐Laslop N, Lee H, Neyfakh AA (2006) Increased persistence in Escherichia coli caused by controlled expression of toxins or other unrelated proteins. J Bacteriol 188: 3494–3497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega NM, Allison KR, Khalil AS, Collins JJ (2012) Signaling‐mediated bacterial persister formation. Nat Chem Biol 8: 431–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstraete L, Aizawa J, Govaerts M, Vooght LD, Michiels J, den Bergh BV, Cos P (2022a) In vitro persistence level reflects in vivo antibiotic survival of natural Pseudomonas aeruginosa isolates in a murine lung infection model. bioRxiv 10.1101/2022.10.21.513228 [PREPRINT] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstraete L, Van den Bergh B, Verstraeten N, Michiels J (2022b) Ecology and evolution of antibiotic persistence. Trends Microbiol 30: 466–479 [DOI] [PubMed] [Google Scholar]
- Verstraeten N, Knapen WJ, Kint CI, Liebens V, Van den Bergh B, Dewachter L, Michiels JE, Fu Q, David CC, Fierro AC et al (2015) Obg and membrane depolarization are part of a microbial bet‐hedging strategy that leads to antibiotic tolerance. Mol Cell 59: 9–21 [DOI] [PubMed] [Google Scholar]
- Vilchèze C, Hartman T, Weinrick B, Jain P, Weisbrod TR, Leung LW, Freundlich JS, Jacobs WR (2017) Enhanced respiration prevents drug tolerance and drug resistance in Mycobacterium tuberculosis . Proc Natl Acad Sci USA 114: 4495–4500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Völzing KG, Brynildsen MP (2015) Stationary‐phase persisters to ofloxacin sustain DNA damage and require repair systems only during recovery. MBio 6: e00731‐15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakamoto Y, Dhar N, Chait R, Schneider K, Signorino‐Gelo F, Leibler S, McKinney JD (2013) Dynamic persistence of antibiotic‐stressed Mycobacteria . Science 339: 91–95 [DOI] [PubMed] [Google Scholar]
- Wang C, Jin L (2022) Microbial persisters and host: recent advances and future perspectives. Crit Rev Microbiol 1–13 10.1080/1040841X.2022.2125286 [DOI] [PubMed] [Google Scholar]
- Wang T, El Meouche I, Dunlop MJ (2017) Bacterial persistence induced by salicylate via reactive oxygen species. Sci Rep 7: 43839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmaerts D, Bayoumi M, Dewachter L, Knapen W, Mika JT, Hofkens J, Dedecker P, Maglia G, Verstraeten N, Michiels J (2018) The persistence‐inducing toxin hokB forms dynamic pores that cause ATP leakage. MBio 9: e00744‐18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmaerts D, Dewachter L, De Loose P‐J, Bollen C, Verstraeten N, Michiels J (2019a) HokB monomerization and membrane repolarization control persister awakening. Mol Cell 75: 1031–1042 [DOI] [PubMed] [Google Scholar]
- Wilmaerts D, Windels EM, Verstraeten N, Michiels J (2019b) General mechanisms leading to persister formation and awakening. Trends Genet 35: 401–411 [DOI] [PubMed] [Google Scholar]
- Wilmaerts D, Focant C, Matthay P, Michiels J (2022) Transcription‐coupled DNA repair underlies variation in persister awakening and the emergence of resistance. Cell Rep 38: 110427 [DOI] [PubMed] [Google Scholar]
- Windels EM, Ben Meriem Z, Zahir T, Verstrepen KJ, Hersen P, Van den Bergh B, Michiels J (2019a) Enrichment of persisters enabled by a ß‐lactam‐induced filamentation method reveals their stochastic single‐cell awakening. Commun Biol 2: 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windels EM, Michiels JE, Bergh BVD, Fauvart M (2019b) Antibiotics: combatting tolerance to stop resistance. MBio 10: e02095‐19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windels EM, Michiels JE, Fauvart M, Wenseleers T, Van Den Bergh B, Michiels J (2019c) Bacterial persistence promotes the evolution of antibiotic resistance by increasing survival and mutation rates. ISME J 13: 1239–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windels EM, den Bergh BV, Michiels J (2020) Bacteria under antibiotic attack: different strategies for evolutionary adaptation. PLoS Pathog 16: e1008431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windels EM, Fox R, Yerramsetty K, Krouse K, Wenseleers T, Swinnen J, Matthay P, Verstraete L, Wilmaerts D, Van den Bergh B et al (2021) Population bottlenecks strongly affect the evolutionary dynamics of antibiotic persistence. Mol Biol Evol 38: 3345–3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto N, Ohno Y, Tsuneda S (2022) ldhA‐induced persister in Escherichia coli is formed through accidental SOS response via intracellular metabolic perturbation. Microbiol Immunol 66: 225–233 [DOI] [PubMed] [Google Scholar]
- Yamasaki R, Song S, Benedik MJ, Wood TK (2020) Persister cells resuscitate using membrane sensors that activate chemotaxis, lower cAMP levels, and revive ribosomes. iScience 23: 100792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Liu Y, Yin H, Chang Z (2019) Regrowth‐delay body as a bacterial subcellular structure marking multidrug‐tolerant persisters. Cell Discov 5: 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Lehar S, Gutierrez J, Rosenberger CM, Ljumanovic N, Dinoso J, Koppada N, Hong K, Baruch A, Carrasco‐Triguero M et al (2016) Pharmacokinetics and pharmacodynamics of DSTA4637A: a novel THIOMAB™ antibody antibiotic conjugate against Staphylococcus aureus in mice. MAbs 8: 1612–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]


