Abstract
The long non-coding RNAs (lncRNAs) constitute an important class of the human transcriptome. The discovery of lncRNAs provided one of many unexpected results of the post-genomic era and uncovered a huge number of previously ignored transcriptional events. In recent years, lncRNAs are known to be linked with human diseases, with particular focus on cancer. Growing evidence has indicated that dysregulation of lncRNAs in breast cancer (BC) is strongly associated with the occurrence, development, and progress. Increasing numbers of lncRNAs have been found to interact with cell cycle progression and tumorigenesis in BC. The lncRNAs can exert their effect as a tumor suppressor or oncogene and regulate tumor development through direct or indirect regulation of cancer-related modulators and signaling pathways. What is more, lncRNAs are excellent candidates for promising therapeutic targets in BC due to the features of high tissue and cell-type specific expression. However, the underlying mechanisms of lncRNAs in BC still remain largely undefined. Here, we concisely summarize and sort out the current understanding of research progress in relationships of the roles for lncRNA in regulating the cell cycle. We also summarize the evidence for aberrant lncRNA expression in BC, and the potential for lncRNA to improve BC therapy is also discussed. Together, lncRNAs can be considered as exciting therapeutic candidates whose expression can be altered to impede BC progression.
Keywords: lncRNAs, cell cycle, breast cancer
BACKGROUND
Long non-coding RNAs (lncRNAs) represent a large type of transcripts longer than 200 nucleotides in length that lack coding capacity. They are originally thought to be the noise of transcripts in the genome with no biological function.1 The lncRNAs were initially uncovered by large-scale sequencing of the mouse full-length cDNA libraries.2 The majority of them encompass RNA polymerase II transcribed RNAs that share transcription similarities with messenger RNAs (mRNAs) having a 5′-terminal 7-methylguanosine (m7 G) cap and a 3′ terminal poly(A) tail.3
The lncRNAs are well accepted that they do not code for proteins, but recent research has validated that part of the putative small open reading frame in lncRNAs can still be translated into a polypeptide.4 Emerging evidence has validated that lncRNAs are implicated in diverse biological processes in cancers, including breast cancer (BC).5 In particular, the functions of lncRNAs have attracted a lot of attention as a potentially new and vital layer of regulatory mechanisms in recent years, although only a few human known lncRNAs have been characterized to date.6 In the early 1990s, lncRNAs have been found to contribute to epigenetic regulation that belonged to some of the original findings of gene-specific regulatory roles of lncRNAs, such as H197 and X inactive-specific transcript (XIST).8,9
Currently, many researchers have reported that lncRNAs participate in the modulation of every layer of biological processes, including cell cycle progression, differentiation, proliferation, apoptosis, and chemoresistance of various cancers,5,10,11 which are closely related to the occurrence, development, and prognosis of various cancers, such as leukemia, colon, liver, lung, and BC.5,12–15
There are six types of lncRNAs according to their characteristics, gene loci, and relationship with their neighbor genes, such as intronic lncRNAs (within genes), intergenic lncRNAs (between genes), antisense lncRNAs, sense lncRNAs, enhancer LncRNAs, and bidirectional lncRNAs.16,17 Abnormally expressed lncRNAs have been determined in various cancer types. In BC, altered lncRNA expression was demonstrated.18 In this review, we talk about lncRNAs regulating cell cycle progress and acting as tumor suppressors and oncogenes in BC. Apart from this, the potential of lncRNAs for target therapy is also discussed.
lncRNAs REGULATE CELL CYCLE PROGRESSION
Nuclear paraspeckle assembly transcript 1
The nuclear paraspeckle assembly transcript 1 (NEAT1) is frequently up-regulated in a wide variety of human tumors.19 NEAT1 is encoded by the NEAT1 gene, which is located on chromosome 11q13.1.20 It is a highly abundant 4 kb lncRNA found in paraspeckles, nuclear domains that control sequestration of related proteins.21 The increased expression of NEAT1 is negatively associated with survival in cancer patients, including BC,22 which indicates that NEAT1 plays significant roles in tumor growth and cell proliferation.
It has been validated that NEAT1 knockdown can modulate E2F3 expression by competitively binding to miR-377 and lead to increased cell cycle arrest in the G1 phase in non-small cell lung cancer cell lines.23 It has also been observed that deletion of NEAT1 significantly induced cell cycle arrest and activated apoptosis to suppress cell proliferation in pancreatic cancer cells.24 In addition, NEAT1 knockdown resulted in cell cycle arrest in the G1 phase that correlated with the increased S-phase cell population by repressing cyclin E1 and D1 expression in triple-negative breast cancer (TNBC) cells.25
Taurine upregulated gene 1
lncRNA taurine upregulated gene 1 (TUG1) is also known as lncRNA00080, TI-227H, and LINC00080. TUG1 is a 7.5 kb nucleotide lncRNA sequence localized to chromosome 22q12.2 and was originally identified in a genomic screen of taurine-treated mouse retinal cells.26 TUG1 is actively involved in various physiological processes, including regulating genes at epigenetics, transcription, post-transcription, translation, and post-translation.27 In 2005, it was originally found as a highly expressed transcript in mouse retinal cells by taurine.28
Recently, more and more researchers have investigated that TUG1 is expressed at an abnormal level in many tumor formations. Downregulation of TUG1 may repress the invasion, migration, and proliferation of cancer cells and promote cell apoptosis according to the type of cancer.29 TUG1 silencing can also result in significantly accumulated cells at the G1 phase in small cell lung cancer (SCLC) cells that promote SCLC cell growth.30 What is more, TUG1 was observed to be expressed at a lower level in BC tissues and cells. TUG1 silencing enhances BC cell cycle progression through cyclin D1 and cyclin-dependent kinase 4 (CDK4) upregulation.31
LINC00628
Long intergenic non-protein coding RNA 628 (LINC00628) can be mapped to chromosome 1q32.1 and located in the second intron of the pleckstrin homology domain containing A6 (PLEKHA6). Recently, the maternally imprinted gene with a length of about 1.2 kb and that has a ploy A tail has become a research hotspot.32 LINC00628 is a relatively less characterized lncRNA. LINC00628 silencing can decrease cell proliferation, migration, and invasion as well as the drug resistance of lung adenocarcinoma cells to vincristine.33
LINC00628 has been validated to service as a tumor suppressor in gastric cancer (GC) that a remarkable increase of cells in G0/G1 phase was investigated after elevation of LINC00628 expression, and vice versa.32 A previous study has determined a decreased cell ratio of G0/G1 phase when the LINC00628 was knocked down via regulating p57 level, a cell cycle regulator protein, in colorectal cancer.34 In addition, highly expressed LINC00628 led to cell cycle arrest at the G0/G1 phase and induced cell apoptosis through modulating phosphatidylinositol 3-kinase (PI3K)/protein kinase B (AKT) signaling pathway in osteosarcoma.35 LINC00628 has also been determined to restrain the metastasis and growth of BC and trigger the apoptosis of BC cells in previous findings.36,37
Urothelial carcinoma associated 1
Cancer-resistant drug resistance gene of urothelial carcinoma associated 1 (UCA1) is mapped to human chromosome 19p13.12 with ∼2.3 kb in length. UCA1 was a novel lncRNA and originally recognized in human bladder cancer in 2006 and has been discovered among various types of human malignant cancers.38,39 UCA1 has been reported to interfere with cell cycle regulation at different levels.
It has been validated that UCA1 physically interacts with EZH2 and represses transcription through histone methylation (H3K27me3) on the promoter of cell cycle genes p21cip and p27Kip140–42 and stimulates cyclin D1 expression.42 UCA1 silencing has been found to arrest cells at the G0/G1 phase in CRC cells,43,44 GC,42 hepatocellular carcinoma (HCC),41 melanoma,45 bladder cancer,46 glioma,47 and medulloblastoma48 cells.
What is more, UCA1 has been found to modulate multiple key players in cell cycle progression. Cell cycle progression from the G1 to S phase is determined by activating E2F transcriptional regulation, which is mediated by the separation of E2F from the retinoblastoma (Rb)-complex after Rb was phosphorylated by CDK-cyclin. UCA1 promotes cyclin D1 expression because of this phosphorylation complex42,47 and maintains CDK phosphorylation stability by suppressing its inhibitors p21CIP and p27Kip.40,41,49–52
In addition, the cyclin D1 expression and the level of cyclin D1 and the CDK inhibitors complex were elevated during the G1 phase, which decreased these inhibitors binding to cyclin E/CDK2 complexes and promoted the cell cycle progression. This regulatory network is further accelerated by the findings that UCA1 can upregulate cMYC, either by interacting with cyclin D153 in the liver cancer cells, or by sequestering miR-135 in thyroid cancer cells,54 respectively.
In BC, UCA1 expression was found to be highly elevated in BC tissues and cells.55–57 Ectopic expression of UCA1 has been confirmed to increase the population of BC cells in the S phase, whereas it reduced cells in the G1 population through regulating miR-143, and vice versa.57 UCA1 has also been found to enhance trastuzumab resistance in BC cells by suppressing miR-18a repression of Yes-associated protein 1 (YAP1) to enhance cell apoptosis and restrain the cell cycle at G2/M phase.26
In addition, UCA1 can desensitize BC cells to tamoxifen in BC cells through a feedback regulatory loop of miR-18a-HIF1α,55 inhibition of Wnt/β-Catenin pathway,56 or modulation of the EZH2/p21 axis and the PI3K/AKT signaling pathway58 via enhancing cell apoptosis and arresting the cell cycle in the G2/M phase.
p53-inducible cancer-associated RNA transcript 1
LncRNA p53-inducible cancer-associated RNA transcript 1 (PICART1) is a novel lncRNA transcript encoded by a gene located at 17q21.33 with ∼2.5 kb in length that was confirmed by using transcriptome sequencing analysis.59 PICART1 is p53-inducible and functions as a tumor suppressor by modulating the AKT/GSK3β/β-catenin signaling pathway. P53 can upregulate PICART1 via binding to PICART at −1,808 to −1,783 bp.59
P53 is a well-accepted suppressor gene that can modulate the expression of multiple genes contributing to apoptosis, growth arrest, and inhibition of cell cycle progression. PICART1 was diminished in GC tissues and cells. PICART1 overexpressed GC cells were restrained in the G1/G0 phase in GC cells by modulating the MAPK/ERK and PI3K/AKT signaling pathways.60 PICART1 expression was validated to be downregulated in BC tissues and cells. Studies confirmed that cell proliferation repression and an arrest of cell cycle at the G2/M phase resulted from ectopic expression of PICART1. In contrast, PICART1 knockdown promoted cell proliferation via the AKT/GSK-3b/β-catenin signaling pathway.59
LINC00511
Long intergenic noncoding RNA 00511 (LINC00511, also known as onco-lncRNA-12) is a novel lncRNA that encoded on chromosome 17q24.3, which is overexpressed in multiple human malignancies and may function as a carcinogenic lncRNA.61 LINC00511 has been found to play a regulatory role in proliferation, apoptosis, invasion, and autophagy of trophoblast cells.62 Many findings have validated that LINC00511 can also affect and regulate cell cycle progression.
LINC00511 knockout hindered cell cycle progression in BC cells since LINC00511 silencing had an accumulation in G0/G1 and a reduction of the S phase to G2/M phase ratio in BC cells.63 Previous studies have also validated that LINC00511 can affect the cell cycle by modulating the cycle-related proteins to participate in chemotherapy resistance. Highly expressed LINC00511 was observed in cervical cancer (CC). Silencing LINC00511 expression led to more CC cells restrained at the G1 phase and suppressed cell resistance to paclitaxel.64
LINC00511 silencing has also been reported to enhance paclitaxel cytotoxicity in BC through modulating miR-29c/CDK6 axis.65 In addition, expression of CDK6, cell cycle-related protein, in TNBC cells was changed after down-regulation of LINC00511 and improved the chemosensitivity of TNBC in vitro.66
Abnormalities of the cell cycle are frequently observed. In cancer cells, one of the main properties is their ability to constantly upregulate proliferative activity.67 It is now well accepted that abnormalities in various positive and negative modulators of the cell cycle are frequent in different cancer types, including BC. Abnormalities such as upregulation of cyclins, as well as defective function of the Rb gene and CDK inhibitors (CKIs, such as p27), are often seen in BC.
There are two categories of cyclin-CDKs that modulate the transportation of mammalian cells from progression through G1, quiescence status, into the S phase of the cell cycle: activation of CDK4 or CDK6 by the D type cyclins and activation of CDK2 by cyclinE.68 In 1994, induction of cyclin D1 in BC cells was found to shorten G1 and was sufficient for cell cycle completion in cells previously arrested in G0.69 Cyclin D1 is one of the most important oncoproteins that is necessary for cancer cell proliferation and has roles in resistance to tamoxifen in BC.
The cyclin D1 also carries out roles in the modulation of the G1 to S phase progression in BC. Cyclin D1 forms active complexes when interacting with CDK4 and CDK6, forming cyclin D-CDK4/6 complex and Rb phosphorylation to promote cell cycle progression.70 Besides, cyclin E, encoded by the CCNE1 gene, is also extensively studied in BC. Downregulation of p27 (also known as KIP1) protein is necessarily associated with cyclin E-CDK2 catalytic activity. P27 overexpression negatively regulates cell proliferation through arresting the cell cycle.71
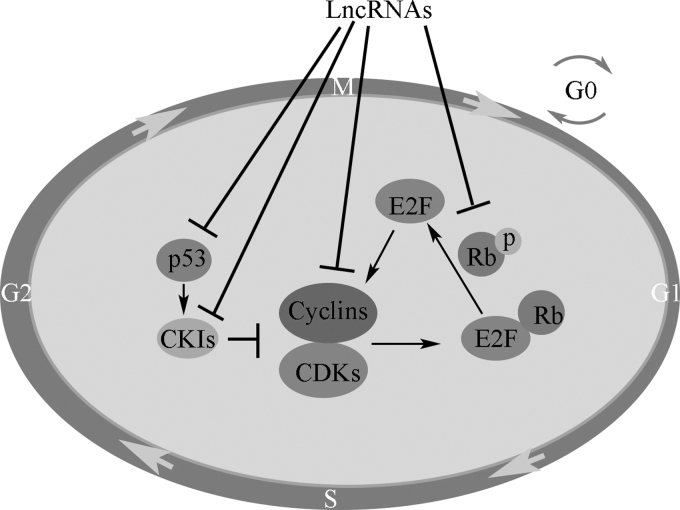
The deregulation of Cyclin E in conjunction with CDK2 results in cancer development.68 Overexpression of Cyclin E1 in TNBC has been observed to result in increased sensitivity to inhibit ATR and WEE1, which are cell cycle checkpoint kinases, and replication stress.72 All in all, lncRNAs interact with E2F, Rb, cyclins, CDKs, CKIs, and associated signaling pathways, providing the potential to regulate cell cycle progression (Fig. 1).73
Figure 1.
lncRNAs and cell cycle. lncRNAs interact with E2F, Rb, cyclins, CDKs, and CKIs, providing the potential to regulate cell cycle progression. CDK, cyclin-dependent kinase; CKIs, cyclin-dependent kinase inhibitor; lncRNAs, long non-coding RNAs; Rb, retinoblastoma.
The cell cycle regulation world of lncRNA is only one side of the coin. In recent years, significant progress and efforts have been made toward validating suppressor and oncogenic lncRNAs and their target therapy potential in BC. For instance, lncRNAs growth arrest-specific 5 (GAS5) in HCC,74 lncRNA named F630028O10Rik (abbreviated as F63) in lung cancer,75 lncRNA EPB41L4A-AS2 in BC,76 lncRNA colorectal neoplasia differentially expressed in HCC,77 etc., function as tumor suppressors. On the contrary, lncRNA estrogen inducible lncRNA (ERINA) in BC,78 lncRNA XIST in osteosarcoma,79 lncRNA metastasis-associated lung carcinoma transcript 1 (MALAT1) in GC,80 etc., function as tumor oncogenes.
HOX transcript antisense intergenic RNA
The lncRNA HOX transcript antisense intergenic RNA (HOTAIR) is located on the human chromosome 12q13.13. It spans a genomic region of ∼2.2 kb in length.81 It acts as a scaffold for chromatin-modifying complexes, such as polycomb repressive complex 2 (PRC2) and lysine-specific demethylase 1 (LSD1), leading to epigenetic modifications that affect gene expression patterns.82 lncRNA HOTAIR has been implicated in normal cellular processes. It participates in the regulation of embryonic development, X-chromosome inactivation, and neuronal differentiation.83 It can modulate gene expression and chromatin structure, influencing various non-cancer biological functions.
The HOTAIR has also been extensively studied for its role in regulating cell cycle progression in BC. HOTAIR has been reported to promote tumor growth and metastasis by influencing cell cycle progression in BC.84 The overexpression of HOTAIR has been associated with the downregulation of tumor suppressor genes, leading to the promotion of BC cell proliferation, invasion, and metastasis. In the MCF-7 cell line, downregulation of HOTAIR has been shown to elevate p53 expression while reducing the expression of AKT and JNK.
This resulted in induced apoptosis and limited capabilities for metastasis, invasion, and proliferation, potentially due to cell cycle arrest at the G1 phase.85,86 Further, HOTAIR has been implicated in the negative regulation of p53 and p21 expression in MCF-7 and MB-231 bCSCs, promoting cell cycle entry and proliferation. Conversely, downregulation of HOTAIR leads to p21 activation and cell cycle arrest at the G1 phase, likely through the inhibition of CDK1, CDK2, CDK4, and CDK6.85 The downregulation of HOTAIR has been shown to enhance the radiosensitivity of BC cells, which is accompanied by the induction of DNA damage, cell cycle arrest, and apoptosis by recruiting miR-218.87
Overall, the dysregulation of HOTAIR in BC can disrupt normal cell cycle control mechanisms, leading to aberrant cell proliferation and tumor progression. Targeting HOTAIR may hold therapeutic potential for the treatment of BC.
Mini-chromosome maintenance complex component 3 associated protein antisense RNA 1
The lncRNA mini-chromosome maintenance complex component 3 associated protein antisense RNA 1 (MCM3AP-AS1) is an RNA gene located on chr21:46,228,977–46,259,390 (GRCh38/hg38), plus strand. The corresponding cytogenetic band is 21q22.3. The lncRNA encoded by this gene has 15 splice variants, with sizes ranging from 572 bp (MCM3AP-AS1-209) to 3,213 bp (MCM3AP-AS1-213).88 The sense transcript derived from this genomic locus plays a dual role in the development of solid tumors and hematological malignancies.
In various studies, MCM3AP-AS1 has been found to act as a sponge for multiple microRNAs, such as miR-194-5p, miR-876-5p, and miR-126. MCM3AP-AS1 has also been shown to impact cancer-related signaling pathways such as PTEN/AKT, PI3K/AKT, and ERK1/2. Multiple lines of evidence from cell line studies, animal studies, and clinical studies consistently support the oncogenic role of MCM3AP-AS1 in various tissues, except for CC, where MCM3AP-AS1 acts as a tumor suppressor.88 It is also involved in RNA processing and cell cycle-related functions, and MCM3AP-AS1 is dysregulated in expression in various types of cancers.89
In fact, MCM3AP serves as a tumor suppressive protein in breast cancer.90 However, its functions in regulating the cell cycle in BC are not well characterized. Knockdown of MCM3AP-AS1 can induce cell cycle arrest at the G1 phase in MDA-MB-231 and ZR-75-30 cells via binding with ZFP36.91
LINC00993
LINC00993 maps on chromosome 10p11.21.92 A study has confirmed a significant correlation between the intergenic lncRNA LINC00993 and the expression of the ANKRD30A gene. Both genes are located adjacent to each other on chromosome 10 and exhibit breast-specific characteristics. The structural characteristics of ANKRD30A suggest that it could potentially interact with LINC00993 as a transcription factor, indicating that the expression of the ANKRD30A gene may be influenced by the epigenetic activity of LINC00993.93
Further, LINC00993 has been shown to upregulate the expression of p16INK4A and p14ARF, leading to G0/G1 cell cycle arrest through the E2F pathway in TNBC. This suggests that LINC00993 plays a role in regulating cell cycle progression and cell growth by modulating the activity of the E2F transcription factors and the expression of key cell cycle regulators such as p16INK4A and p14ARF in TNBC.92
LncRNAs in BC
The number of functional lncRNAs in BC still remains debated. Although there is still a lack of evidence to support the biological function for the majority of lncRNAs, recent studies suggest that a fast-growing number of lncRNAs are important for cellular functions in various cancer types, including BC.94 In BC, the regulatory roles of lncRNAs are considered important factors in various pathological and biological behavior, such as apoptosis, epithelial-mesenchymal transition (EMT), metastasis, invasion, and cell cycle progression.93,95–97
Abnormal expression of lncRNAs has been observed to cause biological dysfunction of some part or process in BC. Mitotically-associated lncRNA (MANCR; LINC00704) was found to be upregulated in BC and functionally linked with genomic stability, cell viability, and proliferation.93 lncRNA LINC00668 has been found to stimulate the progression of BC by suppressing apoptosis and accelerating cell cycle.98 lncRNA GAS5 expression was reduced in TNBC tissues and cells, whereas overexpressed GAS5 reduced the cell proliferation and the inhibitory concentration (IC50) value of TNBC, and accelerated their apoptosis, so as to restrain the progression of TNBC.99
The lncRNA can also alter the cell cycle by modulating the cycle-elated proteins in BC to participate in chemotherapy resistance, for instance, LINC00511, HIF1A-AS2, and AK124454.26,100,101
Suppressor lncRNAs in BC
The expression of suppressor lncRNAs was downregulated in BC tissue or cells, and these lncRNAs inversely correlated with multiple carcinogenesis-associated biological functions. lncRNA EPB41L4A-AS2 levels were observed to be decreased in BC tissues compared with corresponding normal tissues. Oppositely, high EPB41L4A-AS2 expression was related to better overall survival of BC patients, which is the same results as those observed in renal and lung cancer patients. In addition, overexpression of EPB41L4A-AS2 has been shown to restrain tumor cell proliferation in vitro.76 The mature of maternally expressed gene 3 (MEG3) partially shares DLK1-MEG3 locus and is mapped to human chromosome 14q32 with ∼1.6 kb in length.102
Growing evidence has confirmed that lncRNA MEG3 was downregulated many tumor types as a tumor suppressor related to malignancies, including BC.102 It has been shown that MEG3 upregulation repressed the tumorigenesis, EMT of BC by suppressing miR-21 through the PI3K/Akt pathway,102 sponging miR-421 targeting E-cadherin,103 up-regulating endoplasmic reticulum stress and inducing nuclear factor κB (NF-κB) and p53,104 and so on.
The lncRNAs heart and neural crest derivatives expressed 2 antisense RNA 1 (HAND2-AS1) was diminished in multiple tumor types and could be manipulated as an important tumor modulator to regulate tumor cells metastasis, invasion, proliferation, drug resistance, metabolism, and apoptosis.105 It has been reported that HAND2-AS1 expression was downregulated in BC and negatively correlated with clinical features and prognosis. Upregulation of HAND2-AS1 repressed invasion, migration, and cell viability in BC by interacting with miR-1275, altering SOX7 expression,106 and acting as ceRNA of miR-3118 via upregulating PHLPP2.107
Further, lncRNA HAND2-AS1 has been confirmed to restrain the growth of TNBC by decreasing the secretion of mesenchymal stem cells derived exosomal miR-106a-5p.108 TPT1-AS1 was largely downregulated in BC from publicly available datasets and collected BC tissue samples. TPT1-AS1 overexpression inhibited cell proliferation and sensitized BC cells to paclitaxel by modulating miR-3156-5p/caspase 2 axis in vitro.109
LINC00993 is a breast-specific lncRNA. It is especially largely downregulated in TNBC. Its higher expression is associated with better outcome. LINC00993 inhibits TNBC growth via regulating cell cycle-related genes both in vitro and in vivo.92 The earlier cited findings help with the understanding of BC development and may further help guide therapeutic strategy in the clinic.
Oncogenic lncRNAs in BC
Oncogenic lncRNAs were elevated in BC tissue and were negatively associated with survival outcomes. In TNBC, serum terminal differentiation-induced non-coding RNA (TINCR) has been found to be significantly overexpressed. High circulating lncRNA TINCR was negatively associated with clinical outcome and clinicopathological features of TNBC, which shows great promise as a robust biomarker to predict the prognosis of TNBC.110
TINCR was also reported to be overexpressed in BC tissue, which can accelerate metastasis and proliferation of BC cells, whereas downregulation of TINCR induces apoptosis and G1/G0 arrest by regulating OAS1.111 TINCR silencing greatly repressed invasion and migration in vitro as well as xenograft tumor growth in vivo.112 lncRNA PPP1R26 Antisense RNA 1 (PPP1R26-AS1) expression was also increased in BC tissues and cell lines, which was followed by worse overall survival in BC patients. Knockdown of lncRNA PPP1R26-AS1 led to a remarkable reduction in activities of migration, invasion, and proliferation by serving as ceRNA to interact with miR-1226-3p in BC.113
In addition, lncRNA down syndrome cell adhesion molecule-antisense RNA-1 (DSCAM-AS1) has also been identified to be overexpressed in BC tissue and cells. DSCAM-AS1 can promote proliferation and impair apoptosis of BC cells by decreasing miR-204-5p and increasing RRM2 expression. DSCAM-AS1/miR-204-5p/RRM2 could provide a promising therapeutic candidate for BC.114 Besides, MANCR has also been found to be expressed higher in BC tissues.
DSCAM-AS1 and MANCR lncRNAs can also be considered as a potential biomarker since the receiver operating characteristic curve analysis showed a satisfactory diagnostic efficacy.115 In addition, HOTAIR was found to be reversely associated with the radiosensitivity but positively associated with the malignancy of BC cells. lncRNA HOTAIR facilitates the expression of heat shock protein family A (Hsp70) member 1A (HSPA1A) by sequestering miR-449b-5p post-transcriptionally and thus endows BC with radiation resistance.116
Other than that, HOTAIR has been reported to indirectly inhibit miR-7 expression, which led to SETDB1 overexpression and EMT promotion in stem cells of BC.117 Notably, the progression and migration of BC is also mediated by the transforming growth factor (TGF)-β1/HOTAIR axis. Secretion of TGF-β1 by CAF activated the TGF-β1/Smad pathway in BC cells, elevating HOTAIR transcription and modifying histones in the CDK5 signaling pathway, which causes alternations in the tumor microenvironment.118
Moreover, lncRNA MCM3AP-AS1 has been validated as a cytoplasmic RNA and expressed at a high level in BC cells. MCM3AP-AS1/miR-28-5p/CENPF axis has been reported to facilitate BC progression.119 All in all, oncogenic lncRNAs could stimulate new strategies for the discovery of new prognostic biomarkers and improve therapeutic decision making for BC.
Target therapy potential in BC
Surgery, radiotherapy, endocrine therapy, and chemotherapy are the main therapeutic strategies of BC.120,121 Those main therapies combined with targeted therapy have strongly delayed tumor progression and improved the overall survival of BC.122,123 Recent studies have indicated that lncRNAs are especially attractive as diagnostic and prognostic indicators, as well as specific therapeutic targets because of its high specificity in tissue and cell type for certain cancers.
The lncRNA-based target therapy is similar to other targeted therapies, which includes transferring the tumor suppressor lncRNA into specifically targeted cells, blocking, silencing, or destroying carcinogenic lncRNA. lncRNAs possess multiple characteristics that make them a promising candidate in the treatment of diseases. lncRNAs have been suggested to function as an independent biomarker for diagnosis and prognosis in various cancer types, including pancreatic cancer,124 GC,125 HCC,126 and lung cancer.127
Researchers have validated that many lncRNAs are correlated with the occurrence and development of BC. However, the role and the underlying molecular mechanisms of many lncRNAs in the regulation of cellular processes still remain elusive in BC. With a deeper understanding of lncRNA in the cancer progression, its attempts in the application to treat BC will become more and more extensive.
Recent studies have observed that part of the putative small open reading frame in lncRNAs can be translated into a polypeptide. For example, the translation of micro-peptide CIP2A-BP has been found to be encoded by LINC00665, which was affected by TGF-β in BC cell lines. CIP2A-BP directly binds to the oncogene CIP2A to replace the B56γ subunit of PP2A, and it released PP2A activity to inhibit the PI3K/AKT/NF-κB pathway, resulting in a decrease in the expression levels of MMP2, MMP9, and Snail.
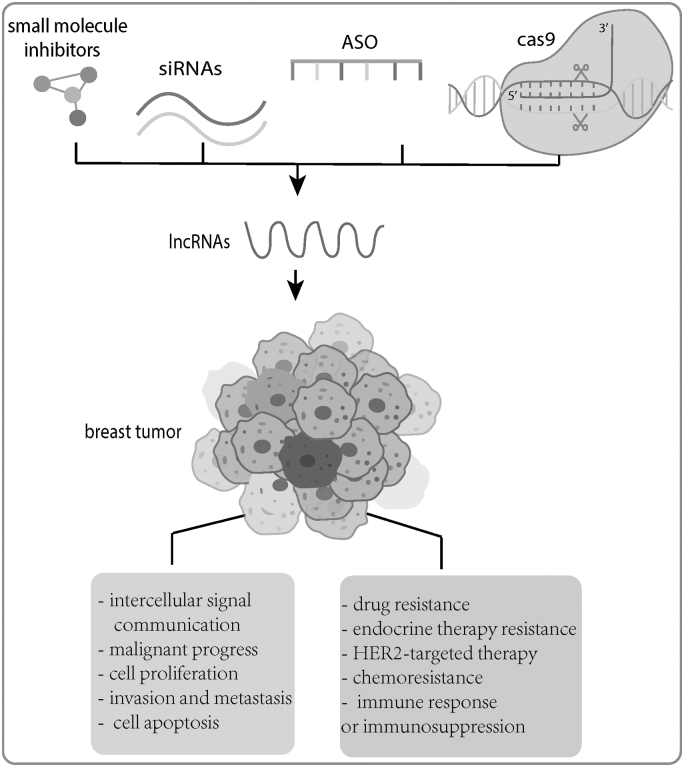
It indicated that lncRNAs that encode a polypeptide may become a potential target for cancer treatment.4 Besides, lncRNAs have also been validated to contribute to transcriptional128 and post-transcriptional level regulation that regulates mRNA splicing,129 mRNA stability,130 protein translation,131 and protein stability132 to control biological processes. In addition, lncRNAs have been reported to participate in epigenetic regulation,133,134 intercellular signal communication,135 malignant progress,136 cell proliferation,137 invasion and metastasis,138 cell apoptosis,139 drug resistance,140 endocrine therapy resistance,141 HER2-targeted therapy,131 chemoresistance,142 immune response or immunosuppression,142 and so on (Fig. 2).
Figure 2.
Potential target therapy in BC. BC, breast cancer.
In recent years, since the increased success in exploring lncRNA function and its structural information, the potential of lncRNA-based cancer therapy has been widely recognized. Targeting lncRNAs for the treatment of cancer represents a novel and promising therapeutic approach, leveraging their combined therapeutic effectiveness, high specificity, and minimal side effects.143 Several molecular drugs against lncRNAs, along with nanoparticle vectors for efficient delivery, have been explored as potential strategies in this field.
Various delivery modalities are being explored for developing lncRNAs as therapeutic and diagnostic markers. Some common approaches include viral vectors, liposomes, nanoparticles, exosomes, and chemically modified oligonucleotides. Viral vectors were among the first engineered nanoscale delivery systems utilized for transporting RNA products to specific tissues.
They have been recognized for their ability to efficiently transfer genetic material into the interior of cells and evade immune detection by infected cells. The use of viral vectors as delivery vehicles allows for precise and effective delivery of RNA payloads, enabling targeted modulation of gene expression. These versatile tools have played a crucial role in advancing RNA-based therapeutics and have contributed to significant progress in the field of gene therapy.144
Viral vectors, such as lentiviruses, can efficiently deliver lncRNAs to target cells.145 Non-viral vectors offer advantages such as lower immunogenicity and safety, with lipid-based vectors, particularly cationic liposomes, demonstrating high transfection efficiency. Liposome modulation, including PEG-lipid conjugation and ligand/antibody addition, further enhances delivery specificity for targeted therapies.146–152
Exosomes serve as practical biological vectors in cancer treatment, offering stability, immune compatibility, and specific targeting capabilities. As a biological vector, exosomes are less likely to activate the immune response and are highly stable in vivo. The construction process involves selecting suitable parental cells, transfecting them with desired ligands, isolating and packaging exosomes, and injecting them into the body for targeted delivery. Successful applications include delivering small interfering RNA (siRNA) for Alzheimer's disease and BC treatment.
Targeting lncRNA DANCR for therapy shows promise, with RNAi suitable for cytoplasmic DANCR and non-viral nanoparticles delivering siDANCR demonstrating inhibitory effects on tumor cells.145,153–159 Chemically modified oligonucleotides, including antisense oligonucleotides (ASOs),160,161 provide a means to specifically target lncRNAs for therapeutic or diagnostic purposes (Fig. 2). The ASOs are synthetic strands that bind to target RNAs, recruiting ribonuclease H (Rnase H) for degradation.
Chemical modifications, such as phosphorylation and 2′-sugar modifications, enhance stability and cellular uptake. Locked nucleic acid (LNA) improves RNA affinity, whereas GAPMER combines DNA and LNA for efficient RNA targeting. Nusinersen is an approved ASO therapy for spinal muscular atrophy. The ASOs are unstable in vivo combined with their relatively high charge and hydrophilicity.162–169 In addition, advances in CRISPR/Cas9 technology enable precise genome editing of lncRNA sequences.170
The siRNAs target lncRNAs and induce gene silencing. Patisiran, an siRNA-based drug, treats hereditary transthyretin amyloidosis. The siRNAs can be chemically synthesized or derived from DNA-based hairpin structures (shRNA) for prolonged silencing. Chemical modifications enhance stability, and synthetic carriers aid cellular uptake.171–178 Moreover, some small-molecule inhibitors179 and indirect modulators of lncRNAs also advance solid tumor treatment in the future (Fig. 2).
The small molecule combined with drug in vitro and in orthotopic BC180 or as a single agent in xenograft model of TNBC181 can target lncRNA to realize much better killing. The choice of delivery modality depends on factors such as the specific lncRNA target, desired delivery site, and the intended therapeutic or diagnostic application. Continued research and development in this field aims at enhancing the efficiency, specificity, and safety of lncRNA delivery modalities for clinical applications.
CONCLUDING REMARKS
Gene expression is regulated by lncRNAs at multiple levels. Since lncRNAs are involved in BC from the onset of the malignant state through the progression to metastasis, lncRNAs and their loci may be ideally targeted for the design and synthesis of new drug therapies to treat BC patients. To date, the functional mechanisms of a few lncRNAs have been studied clearly in preliminary research. However, the underlying functional mechanisms regarding how most of lncRNAs regulate BC still remain elusive and that warrants deeper investigation.
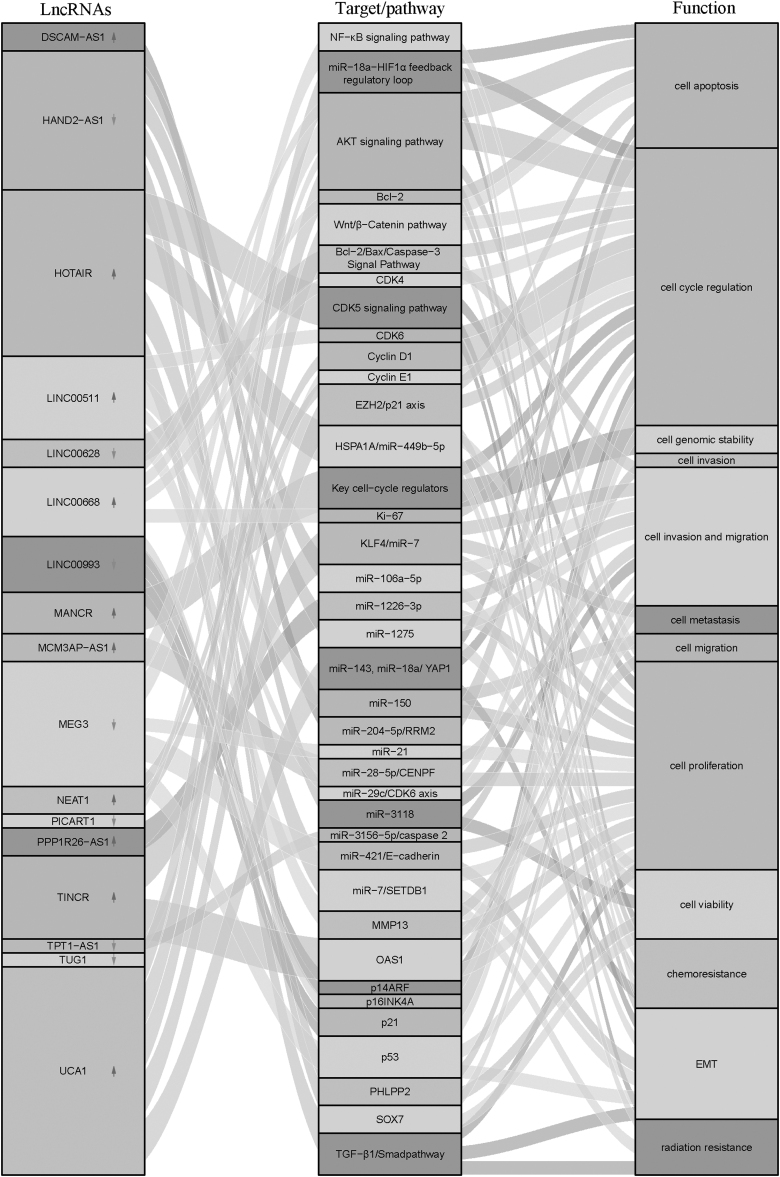
Aberrant expression of lncRNAs has been found in BC that are correlated with cell cycle progression (Figs. 1 and 3 and Table 1), although the upregulation or downregulation of an lncRNA expression in cancers depends on the individual lncRNA itself and tumor type. Practically, lncRNAs can function as inhibitors or promoters in BC cell proliferation by regulating cell cycle modulators.
Figure 3.
Sankey diagram reveals the mechanism of lncRNAs in BC progression.
Table 1.
Long non-coding RNAs in breast cancer in this review
| lncRNA | Expression in BC | Target/Pathway | Function | Reference |
|---|---|---|---|---|
| NEAT1 | Upregulation | Cyclin E1 and D1 | Cell cycle regulation | 25 |
| TUG1 | Downregulation | Cyclin D1 | Cell cycle regulation | 31 |
| LINC00628 | Downregulation | Bcl-2/Bax/Caspase-3 Signal Pathway | Cell cycle regulation, invasion, and migration | 36 |
| UCA1 | Upregulation | miR-143, miR-18a/YAP1, miR-18a-HIF1α feedback regulatory loop, Wnt/β-Catenin pathway, EZH2/p21 axis and the AKT signaling pathway | Cell cycle regulation, apoptosis, and chemoresistance | 26,55–58 |
| PICART1 | Downregulation | AKT signaling pathway | Cell cycle regulation | 59 |
| LINC00511 | Upregulation | miR-29c/CDK6 axis, CDK6 | Cell cycle regulation | 26,66 |
| MANCR | Upregulation | Key cell-cycle regulators | Cell cycle regulation, viability, and genomic stability | 93 |
| LINC00668 | Upregulation | Ki-67, CDK4 and Bcl-2, p21, AKT signaling pathway | Cell cycle regulation, apoptosis | 98 |
| GAS5 | Downregulation | N/A | Cell proliferation, chemoresistance, and apoptosis | 99 |
| LINC00511 | Upregulation | miR-150 and MMP13 | Cell proliferation, migration | 101 |
| HIF1A-AS2 | Upregulation | N/A | Metabolism, cell division | 100 |
| AK124454 | Upregulation | N/A | Metabolism, cell division | 100 |
| EPB41L4A-AS2 | Downregulation | N/A | Cell proliferation | 76 |
| MEG3 | Downregulation | miR-21, Akt signaling pathway, miR-421/E-cadherin, NF-κB and p53 | Cell proliferation, EMT | 102–104 |
| HAND2-AS1 | Downregulation | miR-1275, SOX7, miR-3118, PHLPP2, miR-106a-5p | Cell viability, migration, and invasion | 106–108 |
| TPT1-AS1 | Downregulation | miR-3156-5p/caspase 2 | Cell proliferation, chemoresistance | 109 |
| LINC00993 | Downregulation | p16INK4A, p14ARF, p53, and p21 | Cell cycle regulation | 92 |
| TINCR | Upregulation | OAS1, KLF4/miR-7 | Cell proliferation, metastasis, migration, and invasion | 110–112 |
| PPP1R26-AS1 | Upregulation | miR-1226-3p | Cell proliferation, migration, and invasion | 113 |
| DSCAM-AS1 | Upregulation | miR-204-5p/RRM2 | Cell proliferation, apoptosis | 114 |
| MANCR | Upregulation | N/A | N/A | 115 |
| DSCAM-AS1 | Upregulation | N/A | N/A | 115 |
| HOTAIR | Upregulation | P53, p21, HSPA1A/miR-449b-5p, miR-7/SETDB1, TGF-β1/Smad pathway, CDK5 signaling pathway | Cell cycle regulation, radiation resistance, EMT, and cell proliferation | 85–87,116–118 |
| MCM3AP-AS1 | Upregulation | ZFP36, miR-28-5p/CENPF | Cell cycle regulation, migration, and invasion | 91,119 |
BC, breast cancer; CDK, cyclin-dependent kinase; DSCAM-AS1, down syndrome cell adhesion molecule-antisense RNA-1; EMT, epithelial-mesenchymal transition; GAS5, growth arrest-specific 5; HOTAIR, HOX transcript antisense intergenic RNA; HSPA1A, heat shock protein family A (Hsp70) member 1A; lncRNA, long non-coding RNA; MANCR, mitotically-associated lncRNA; MCM3AP-AS1, mini-chromosome maintenance complex component 3 associated protein antisense RNA 1; MEG3, maternally expressed gene 3; N/A, not applicable; NEAT1, nuclear paraspeckle assembly transcript 1; NF-κB, nuclear factor κB; PICART1, p53-inducible cancer-associated RNA transcript 1; TGF-β1, transforming growth factor-beta 1; TINCR, terminal differentiation-induced non-coding RNA; TUG1, taurine upregulated gene 1; UCA1, urothelial carcinoma associated 1; YAP1, Yes-associated protein 1.
In addition, lncRNAs are capable of acting as an oncogene or a tumor suppressor (Fig. 3 and Table 1) by interacting with the promoter or enhancer region of a gene, the regulatory modulators, or associated signaling pathways to modulate the gene expression. What is more, lncRNAs can also regulate a protein's function. Studies conducted to decode the functions of lncRNA in BC will increase our understanding of molecular mechanisms of BC, and they will offer a rationale for targeting lncRNAs in BC.
The specific expression of lncRNAs also provides the possibility of killing cancer cells selectively. As a result, lncRNAs with high specificity and accuracy have the outstanding opportunity to be considered as new important therapeutic targets to combat various aspects of BC progression.
However, the secondary structure of lncRNAs is more complex than that of mRNA, and the regulation of lncRNA expression is more stringent. Moreover, some lncRNAs have dual functions as coding polypeptide products. Thus, the mechanisms of functional lncRNAs are apparently very complicated. This requires further research depth and width. The lncRNAs are more stable than mRNA since the actual number of lncRNA exceeds that of protein-coding genes.
Because of the recent development and improvement of high-throughput sequencing technologies, detection of lncRNAs is faster and more convenient. Although companies such as the Curna Inc., MiNA Therapeutics Ltd. and RaNA Therapeutics Inc. are taking steps for the development of lncRNA-based strategies that may provide a viable alternative to overcome the limitations of targeting those protein,182 it still needs to resolve the difficulty about designing small-molecule drugs against lncRNAs, which limits the application of lncRNAs as a therapy target in cancer, including BC. Therefore, more research is necessary before lncRNAs can be placed in front of cancer targets.
AUTHORs' CONTRIBUTIONS
Q.Y. collated the data and wrote drafts of the manuscript. X.Z. and Y.M. conceptualized the study, contributed to the data collection, and edited the manuscript. H.M. and G.B. edited the table and figures. All authors read and approved the final manuscript.
AUTHOR DISCLOSURE
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
FUNDING INFORMATION
This study was financially supported by Fundamental Research Funds for the Henan University of Science and Technology (Q.Y.) and A-type Doctoral Talent Project of the Henan University of Science and Technology (X.Z.).
REFERENCES
- 1. Deniz E, Erman B. Long noncoding RNA (lincRNA), a new paradigm in gene expression control. Funct Integr Genomics 2017;17:135–143. [DOI] [PubMed] [Google Scholar]
- 2. Okazaki Y, Furuno M, Kasukawa Tet al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 2002;420:563–573. [DOI] [PubMed] [Google Scholar]
- 3. Marchese FP, Raimondi I, Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol 2017;18:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guo B, Wu S, Zhu X, et al. Micropeptide CIP2A-BP encoded by LINC00665 inhibits triple-negative breast cancer progression. EMBO J 2020;39:e102190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sideris N, Dama P, Bayraktar S, et al. LncRNAs in breast cancer: A link to future approaches. Cancer Gene Ther 2022;29:1866–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Borkiewicz L, Kalafut J, Dudziak K, et al. Decoding LncRNAs. Cancers (Basel) 2021;13(11):2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brannan CI, Dees EC, Ingram RS, et al. The product of the H19 gene may function as an RNA. Mol Cell Biol 1990;10:28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brockdorff N, Ashworth A, Kay GF, et al. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 1992;71:515–526. [DOI] [PubMed] [Google Scholar]
- 9. Brown CJ, Hendrich BD, Rupert JL, et al. The human XIST gene: Analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 1992;71:527–542. [DOI] [PubMed] [Google Scholar]
- 10. Wilusz JE. Long noncoding RNAs: Re-writing dogmas of RNA processing and stability. Biochim Biophys Acta 2016;1859:128–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xia H, Hui KM. Mechanism of cancer drug resistance and the involvement of noncoding RNAs. Curr Med Chem 2014;21:3029–3041. [DOI] [PubMed] [Google Scholar]
- 12. Dong S, Qu X, Li W, et al. The long non-coding RNA, GAS5, enhances gefitinib-induced cell death in innate EGFR tyrosine kinase inhibitor-resistant lung adenocarcinoma cells with wide-type EGFR via downregulation of the IGF-1R expression. J Hematol Oncol 2015;8:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huang MD, Chen WM, Qi FZ, et al. Long non-coding RNA ANRIL is upregulated in hepatocellular carcinoma and regulates cell apoptosis by epigenetic silencing of KLF2. J Hematol Oncol 2015;8:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang X, Xiao R, Pan S, et al. Uncovering the roles of long non-coding RNAs in cancer stem cells. J Hematol Oncol 2017;10:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zeng C, Yu X, Lai J, et al. Overexpression of the long non-coding RNA PVT1 is correlated with leukemic cell proliferation in acute promyelocytic leukemia. J Hematol Oncol 2015;8:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: A new paradigm. Cancer Res 2017;77:3965–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell 2011;43:904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Berteaux N, Lottin S, Monte D, et al. H19 mRNA-like noncoding RNA promotes breast cancer cell proliferation through positive control by E2F1. J Biol Chem 2005;280:29625–29636. [DOI] [PubMed] [Google Scholar]
- 19. Dong P, Xiong Y, Yue J, et al. Long non-coding RNA NEAT1: A novel target for diagnosis and therapy in human tumors. Front Genet 2018;9:471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ma P, Pan Y, Yang F, et al. KLF5-modulated lncRNA NEAT1 contributes to tumorigenesis by acting as a scaffold for BRG1 to silence GADD45A in gastric cancer. Mol Ther Nucleic Acids 2020;22:382–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Romero-Barrios N, Legascue MF, Benhamed M, et al. Splicing regulation by long noncoding RNAs. Nucleic Acids Res 2018;46:2169–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Choudhry H, Albukhari A, Morotti M, et al. Tumor hypoxia induces nuclear paraspeckle formation through HIF-2alpha dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene 2015;34:4482–4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang J, Li Y, Dong M, et al. Long non-coding RNA NEAT1 regulates E2F3 expression by competitively binding to miR-377 in non-small cell lung cancer. Oncol Lett 2017;14:4983–4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang B, Liu C, Wu Q, et al. Long non-coding RNA NEAT1 facilitates pancreatic cancer progression through negative modulation of miR-506-3p. Biochem Biophys Res Commun 2017;482:828–834. [DOI] [PubMed] [Google Scholar]
- 25. Shin VY, Chen J, Cheuk IW, et al. Long non-coding RNA NEAT1 confers oncogenic role in triple-negative breast cancer through modulating chemoresistance and cancer stemness. Cell Death Dis 2019;10:270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhu HY, Bai WD, Ye XM, et al. Long non-coding RNA UCA1 desensitizes breast cancer cells to trastuzumab by impeding miR-18a repression of Yes-associated protein 1. Biochem Biophys Res Commun 2018;496:1308–1313. [DOI] [PubMed] [Google Scholar]
- 27. Guo C, Qi Y, Qu J, et al. Pathophysiological functions of the lncRNA TUG1. Curr Pharm Des 2020;26:688–700. [DOI] [PubMed] [Google Scholar]
- 28. Young TL, Matsuda T, Cepko CL. The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr Biol 2005;15:501–512. [DOI] [PubMed] [Google Scholar]
- 29. Zhang Q, Geng PL, Yin P, et al. Down-regulation of long non-coding RNA TUG1 inhibits osteosarcoma cell proliferation and promotes apoptosis. Asian Pac J Cancer Prev 2013;14:2311–2315. [DOI] [PubMed] [Google Scholar]
- 30. Niu Y, Ma F, Huang W, et al. Long non-coding RNA TUG1 is involved in cell growth and chemoresistance of small cell lung cancer by regulating LIMK2b via EZH2. Mol Cancer 2017;16:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fan S, Yang Z, Ke Z, et al. Downregulation of the long non-coding RNA TUG1 is associated with cell proliferation, migration, and invasion in breast cancer. Biomed Pharmacother 2017;95:1636–1643. [DOI] [PubMed] [Google Scholar]
- 32. Zhang ZZ, Zhao G, Zhuang C, et al. Long non-coding RNA LINC00628 functions as a gastric cancer suppressor via long-range modulating the expression of cell cycle related genes. Sci Rep 2016;6:27435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xu SF, Zheng Y, Zhang L, et al. Long non-coding RNA LINC00628 interacts epigenetically with the LAMA3 promoter and contributes to lung adenocarcinoma. Mol Ther Nucleic Acids 2019;18:166–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang MC, Zhang L, Zhang MQ, et al. Downregulated LINC00628 aggravates the progression of colorectal cancer via inhibiting p57 level. Eur Rev Med Pharmacol Sci 2020;24:1763–1770. [DOI] [PubMed] [Google Scholar]
- 35. He R, Wu JX, Zhang Y, et al. LncRNA LINC00628 overexpression inhibits the growth and invasion through regulating PI3K/Akt signaling pathway in osteosarcoma. Eur Rev Med Pharmacol Sci 2018;22:5857–5866. [DOI] [PubMed] [Google Scholar]
- 36. Chen DQ, Zheng XD, Cao Y, et al. Long non-coding RNA LINC00628 suppresses the growth and metastasis and promotes cell apoptosis in breast cancer. Eur Rev Med Pharmacol Sci 2017;21:275–283. [PubMed] [Google Scholar]
- 37. Ghafouri-Fard S, Abak A, Tondro Anamag F, et al. The emerging role of non-coding RNAs in the regulation of PI3K/AKT pathway in the carcinogenesis process. Biomed Pharmacother 2021;137:111279. [DOI] [PubMed] [Google Scholar]
- 38. Wang XS, Zhang Z, Wang HC, et al. Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clin Cancer Res 2006;12:4851–4858. [DOI] [PubMed] [Google Scholar]
- 39. Xue M, Chen W, Li X. Urothelial cancer associated 1: A long noncoding RNA with a crucial role in cancer. J Cancer Res Clin Oncol 2016;142:1407–1419. [DOI] [PubMed] [Google Scholar]
- 40. Cai Q, Jin L, Wang S, et al. Long non-coding RNA UCA1 promotes gallbladder cancer progression by epigenetically repressing p21 and E-cadherin expression. Oncotarget 2017;8:47957–47968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang ZQ, Cai Q, Hu L, et al. Long noncoding RNA UCA1 induced by SP1 promotes cell proliferation via recruiting EZH2 and activating AKT pathway in gastric cancer. Cell Death Dis 2017;8:e2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Han Y, Yang YN, Yuan HH, et al. UCA1, a long non-coding RNA up-regulated in colorectal cancer influences cell proliferation, apoptosis and cell cycle distribution. Pathology 2014;46:396–401. [DOI] [PubMed] [Google Scholar]
- 43. Ni B, Yu X, Guo X, et al. Increased urothelial cancer associated 1 is associated with tumor proliferation and metastasis and predicts poor prognosis in colorectal cancer. Int J Oncol 2015;47:1329–1338. [DOI] [PubMed] [Google Scholar]
- 44. Hu JJ, Song W, Zhang SD, et al. HBx-upregulated lncRNA UCA1 promotes cell growth and tumorigenesis by recruiting EZH2 and repressing p27Kip1/CDK2 signaling. Sci Rep 2016;6:23521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wei Y, Sun Q, Zhao L, et al. lncRNA UCA1-miR-507-FOXM1 axis is involved in cell proliferation, invasion and G0/G1 cell cycle arrest in melanoma. Med Oncol 2016;33:88. [DOI] [PubMed] [Google Scholar]
- 46. Zhen S, Hua L, Liu YH, et al. Inhibition of long non-coding RNA UCA1 by CRISPR/Cas9 attenuated malignant phenotypes of bladder cancer. Oncotarget 2017;8:9634–9646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhao W, Sun C, Cui Z. A long noncoding RNA UCA1 promotes proliferation and predicts poor prognosis in glioma. Clin Transl Oncol 2017;19:735–741. [DOI] [PubMed] [Google Scholar]
- 48. Zhengyuan X, Hu X, Qiang W, et al. Silencing of urothelial carcinoma associated 1 inhibits the proliferation and migration of medulloblastoma cells. Med Sci Monit 2017;23:4454–4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Huang J, Zhou N, Watabe K, et al. Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1). Cell Death Dis 2014;5:e1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jahangiri B, Khalaj-Kondori M, Asadollahi E, et al. Cancer-associated fibroblasts enhance cell proliferation and metastasis of colorectal cancer SW480 cells by provoking long noncoding RNA UCA1. J Cell Commun Signal 2019;13:53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pan J, Li X, Wu W, et al. Long non-coding RNA UCA1 promotes cisplatin/gemcitabine resistance through CREB modulating miR-196a-5p in bladder cancer cells. Cancer Lett 2016;382:64–76. [DOI] [PubMed] [Google Scholar]
- 52. Wang X, Gong Y, Jin B, et al. Long non-coding RNA urothelial carcinoma associated 1 induces cell replication by inhibiting BRG1 in 5637 cells. Oncol Rep 2014;32:1281–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pu H, Zheng Q, Li H, et al. CUDR promotes liver cancer stem cell growth through upregulating TERT and C-Myc. Oncotarget 2015;6:40775–40798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang Y, Hou Z, Li D. Long noncoding RNA UCA1 promotes anaplastic thyroid cancer cell proliferation via miR-135a-mediated c-myc activation. Mol Med Rep 2018;18:3068–3076. [DOI] [PubMed] [Google Scholar]
- 55. Li X, Wu Y, Liu A, et al. Long non-coding RNA UCA1 enhances tamoxifen resistance in breast cancer cells through a miR-18a-HIF1alpha feedback regulatory loop. Tumour Biol 2016;37:14733–14743. [DOI] [PubMed] [Google Scholar]
- 56. Liu H, Wang G, Yang L, et al. Knockdown of long non-coding RNA UCA1 increases the tamoxifen sensitivity of breast cancer cells through inhibition of Wnt/beta-catenin pathway. PLoS One 2016;11:e0168406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tuo YL, Li XM, Luo J. Long noncoding RNA UCA1 modulates breast cancer cell growth and apoptosis through decreasing tumor suppressive miR-143. Eur Rev Med Pharmacol Sci 2015;19:3403–3411. [PubMed] [Google Scholar]
- 58. Ma C, Zu X, Liu K, et al. Knockdown of pyruvate kinase M inhibits cell growth and migration by reducing NF-kB activity in triple-negative breast cancer cells. Mol Cells 2019;42:628–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cao Y, Lin M, Bu Y, et al. p53-inducible long non-coding RNA PICART1 mediates cancer cell proliferation and migration. Int J Oncol 2017;50:1671–1682. [DOI] [PubMed] [Google Scholar]
- 60. Li JF, Li WH, Xue LL, et al. Long non-coding RNA PICART1 inhibits cell proliferation by regulating the PI3K/AKT and MAPK/ERK signaling pathways in gastric cancer. Eur Rev Med Pharmacol Sci 2019;23:588–597. [DOI] [PubMed] [Google Scholar]
- 61. Yu CL, Xu XL, Yuan F. LINC00511 is associated with the malignant status and promotes cell proliferation and motility in cervical cancer. Biosci Rep 2019;39(9):BSR20190903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Dong X, Zhang Y, Chen X, et al. Long noncoding RNA LINC00511 regulates the proliferation, apoptosis, invasion and autophagy of trophoblast cells to mediate pre-eclampsia progression through modulating the miR-31-5p/homeobox protein A7 axis. J Obstet Gynaecol Res 2020;46:1298–1309. [DOI] [PubMed] [Google Scholar]
- 63. Azadbakht N, Doosti A, Jami MS. CRISPR/Cas9-mediated LINC00511 knockout strategies, increased apoptosis of breast cancer cells via suppressing antiapoptotic genes. Biol Proced Online 2022;24:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mao BD, Xu P, Xu P, et al. LINC00511 knockdown prevents cervical cancer cell proliferation and reduces resistance to paclitaxel. J Biosci 2019;44(2):44. [PubMed] [Google Scholar]
- 65. Zhang H, Zhao B, Wang X, et al. LINC00511 knockdown enhances paclitaxel cytotoxicity in breast cancer via regulating miR-29c/CDK6 axis. Life Sci 2019;228:135–144. [DOI] [PubMed] [Google Scholar]
- 66. Yuan Y, Li E, Zhao J, et al. Highly penetrating nanobubble polymer enhances LINC00511-siRNA delivery for improving the chemosensitivity of triple-negative breast cancer. Anticancer Drugs 2021;32:178–188. [DOI] [PubMed] [Google Scholar]
- 67. Gutschner T, Diederichs S. The hallmarks of cancer: A long non-coding RNA point of view. RNA Biol 2012;9:703–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hwang HC, Clurman BE. Cyclin E in normal and neoplastic cell cycles. Oncogene 2005;24:2776–2786. [DOI] [PubMed] [Google Scholar]
- 69. Musgrove EA, Lee CS, Buckley MF, et al. Cyclin D1 induction in breast cancer cells shortens G1 and is sufficient for cells arrested in G1 to complete the cell cycle. Proc Natl Acad Sci U S A 1994;91:8022–8026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Alao JP. The regulation of cyclin D1 degradation: Roles in cancer development and the potential for therapeutic invention. Mol Cancer 2007;6:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sheaff RJ, Groudine M, Gordon M, et al. Cyclin E-CDK2 is a regulator of p27Kip1. Genes Dev 1997;11:1464–1478. [DOI] [PubMed] [Google Scholar]
- 72. Guerrero Llobet S, van der Vegt B, Jongeneel E, et al. Cyclin E expression is associated with high levels of replication stress in triple-negative breast cancer. NPJ Breast Cancer 2020;6:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Subramanian M, Jones MF, Lal A. Long Non-coding RNAs embedded in the Rb and p53 pathways. Cancers (Basel) 2013;5:1655–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wang C, Ke S, Li M, et al. Downregulation of LncRNA GAS5 promotes liver cancer proliferation and drug resistance by decreasing PTEN expression. Mol Genet Genomics 2020;295:251–260. [DOI] [PubMed] [Google Scholar]
- 75. Qin L, Zhong M, Adah D, et al. A novel tumour suppressor lncRNA F630028O10Rik inhibits lung cancer angiogenesis by regulating miR-223-3p. J Cell Mol Med 2020;24:3549–3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Xu S, Wang P, You Z, et al. The long non-coding RNA EPB41L4A-AS2 inhibits tumor proliferation and is associated with favorable prognoses in breast cancer and other solid tumors. Oncotarget 2016;7:20704–20717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Xie SC, Zhang JQ, Jiang XL, et al. LncRNA CRNDE facilitates epigenetic suppression of CELF2 and LATS2 to promote proliferation, migration and chemoresistance in hepatocellular carcinoma. Cell Death Dis 2020;11:676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Fang Z, Wang Y, Wang Z, et al. ERINA is an estrogen-responsive LncRNA that drives breast cancer through the E2F1/RB1 pathway. Cancer Res 2020;80:4399–4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Li H, Cui J, Xu B, et al. Long non-coding RNA XIST serves an oncogenic role in osteosarcoma by sponging miR-137. Exp Ther Med 2019;17:730–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Xu W, Ding M, Wang B, et al. Molecular mechanism of the canonical oncogenic lncRNA MALAT1 in gastric cancer. Curr Med Chem 2021;28:8800–8809. [DOI] [PubMed] [Google Scholar]
- 81. Hajjari M, Salavaty A. HOTAIR: An oncogenic long non-coding RNA in different cancers. Cancer Biol Med 2015;12:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Tsai MC, Manor O, Wan Y, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010;329:689–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Pauli A, Rinn JL, Schier AF. Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet 2011;12:136–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Shi Y, Huang Q, Kong X, et al. Current knowledge of long non-coding RNA HOTAIR in breast cancer progression and its application. Life (Basel) 2021;11(6):483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Deng J, Yang M, Jiang R, et al. Long non-coding RNA HOTAIR regulates the proliferation, self-renewal capacity, tumor formation and migration of the cancer stem-like cell (CSC) subpopulation enriched from breast cancer cells. PLoS One 2017;12:e0170860. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 86. Yu Y, Lv F, Liang D, et al. HOTAIR may regulate proliferation, apoptosis, migration and invasion of MCF-7 cells through regulating the P53/Akt/JNK signaling pathway. Biomed Pharmacother 2017;90:555–561. [DOI] [PubMed] [Google Scholar]
- 87. Hu X, Ding D, Zhang J, et al. Knockdown of lncRNA HOTAIR sensitizes breast cancer cells to ionizing radiation through activating miR-218. Biosci Rep 2019;39(4):BSR20181038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ghafouri-Fard S, Khoshbakht T, Hussen BM, et al. A review on the role of MCM3AP-AS1 in the carcinogenesis and tumor progression. Cancer Cell Int 2022;22:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ma T, Wu FH, Wu HX, et al. Long non-coding RNA MCM3AP-AS1: A crucial role in human malignancies. Pathol Oncol Res 2022;28:1610194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kuwahara K, Yamamoto-Ibusuki M, Zhang Z, et al. GANP protein encoded on human chromosome 21/mouse chromosome 10 is associated with resistance to mammary tumor development. Cancer Sci 2016;107:469–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Tang TP, Qin CX, Yu H. MCM3AP-AS1 regulates proliferation, apoptosis, migration, and invasion of breast cancer cells via binding with ZFP36. Transl Cancer Res 2021;10:4478–4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Guo S, Jian L, Tao K, et al. Novel breast-specific long non-coding RNA LINC00993 acts as a tumor suppressor in triple-negative breast cancer. Front Oncol 2019;9:1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Tracy KM, Tye CE, Ghule PN, et al. Mitotically-associated lncRNA (MANCR) affects genomic stability and cell division in aggressive breast cancer. Mol Cancer Res 2018;16:587–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Jin H, Du W, Huang W, et al. lncRNA and breast cancer: Progress from identifying mechanisms to challenges and opportunities of clinical treatment. Mol Ther Nucleic Acids 2021;25:613–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Gooding AJ, Zhang B, Jahanbani FK, et al. The lncRNA BORG drives breast cancer metastasis and disease recurrence. Sci Rep 2017;7:12698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Zhang M, Wu WB, Wang ZW, et al. lncRNA NEAT1 is closely related with progression of breast cancer via promoting proliferation and EMT. Eur Rev Med Pharmacol Sci 2017;21:1020–1026. [PubMed] [Google Scholar]
- 97. Zheng S, Li M, Miao K, et al. lncRNA GAS5-promoted apoptosis in triple-negative breast cancer by targeting miR-378a-5p/SUFU signaling. J Cell Biochem 2020;121:2225–2235. [DOI] [PubMed] [Google Scholar]
- 98. Qiu X, Dong J, Zhao Z, et al. LncRNA LINC00668 promotes the progression of breast cancer by inhibiting apoptosis and accelerating cell cycle. Onco Targets Ther 2019;12:5615–5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Li J, Li L, Yuan H, et al. Up-regulated lncRNA GAS5 promotes chemosensitivity and apoptosis of triple-negative breast cancer cells. Cell Cycle 2019;18:1965–1975. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 100. Jiang YZ, Liu YR, Xu XE, et al. Transcriptome analysis of triple-negative breast cancer reveals an integrated mRNA-lncRNA signature with predictive and prognostic value. Cancer Res 2016;76:2105–2114. [DOI] [PubMed] [Google Scholar]
- 101. Shi G, Cheng Y, Zhang Y, et al. Long non-coding RNA LINC00511/miR-150/MMP13 axis promotes breast cancer proliferation, migration and invasion. Biochim Biophys Acta Mol Basis Dis 2021;1867:165957. [DOI] [PubMed] [Google Scholar]
- 102. Zhu M, Wang X, Gu Y, et al. MEG3 overexpression inhibits the tumorigenesis of breast cancer by downregulating miR-21 through the PI3K/Akt pathway. Arch Biochem Biophys 2019;661:22–30. [DOI] [PubMed] [Google Scholar]
- 103. Zhang W, Shi S, Jiang J, et al. LncRNA MEG3 inhibits cell epithelial-mesenchymal transition by sponging miR-421 targeting E-cadherin in breast cancer. Biomed Pharmacother 2017;91:312–319. [DOI] [PubMed] [Google Scholar]
- 104. Zhang Y, Wu J, Jing H, et al. Long noncoding RNA MEG3 inhibits breast cancer growth via upregulating endoplasmic reticulum stress and activating NF-kappaB and p53. J Cell Biochem 2019;120:6789–6797. [DOI] [PubMed] [Google Scholar]
- 105. Huang Z, Wang Z, Xia H, et al. Long noncoding RNA HAND2-AS1: A crucial regulator of malignancy. Clin Chim Acta 2023;539:162–169. [DOI] [PubMed] [Google Scholar]
- 106. Wang Y, Cai X. Long noncoding RNA HAND2-AS1 restrains proliferation and metastasis of breast cancer cells through sponging miR-1275 and promoting SOX7. Cancer Biomark 2020;27:85–94. [DOI] [PubMed] [Google Scholar]
- 107. Dong G, Wang X, Jia Y, et al. HAND2-AS1 Works as a ceRNA of miR-3118 to suppress proliferation and migration in breast cancer by upregulating PHLPP2. Biomed Res Int 2020;2020:8124570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Xing L, Tang X, Wu K, et al. LncRNA HAND2-AS1 suppressed the growth of triple negative breast cancer via reducing secretion of MSCs derived exosomal miR-106a-5p. Aging (Albany NY) 2020;13:424–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Huang Y, Zheng Y, Shao X, et al. Long non-coding RNA TPT1-AS1 sensitizes breast cancer cell to paclitaxel and inhibits cell proliferation by miR-3156-5p/caspase 2 axis. Hum Cell 2021;34:1244–1254. [DOI] [PubMed] [Google Scholar]
- 110. Wang X, Li S, Xiao H, et al. Serum lncRNA TINCR serve as a novel biomarker for predicting the prognosis in triple-negative breast cancer. Technol Cancer Res Treat 2020;19:1533033820965574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Lu D, Di S, Zhuo S, et al. The long noncoding RNA TINCR promotes breast cancer cell proliferation and migration by regulating OAS1. Cell Death Discov 2021;7:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Liu Y, Du Y, Hu X, et al. Up-regulation of ceRNA TINCR by SP1 contributes to tumorigenesis in breast cancer. BMC Cancer 2018;18:367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Zhou S, Zhang S, Zhang H, et al. Clinical potential of lncRNA PPP1R26-AS1 in breast cancer and its contribution to cancer progression. Mol Biotechnol 2022;64:660–669. [DOI] [PubMed] [Google Scholar]
- 114. Liang WH, Li N, Yuan ZQ, et al. DSCAM-AS1 promotes tumor growth of breast cancer by reducing miR-204-5p and up-regulating RRM2. Mol Carcinog 2019;58:461–473. [DOI] [PubMed] [Google Scholar]
- 115. Tahmouresi F, Razmara E, Pakravan K, et al. Upregulation of the long noncoding RNAs DSCAM-AS1 and MANCR is a potential diagnostic marker for breast carcinoma. Biotechnol Appl Biochem 2021;68:1250–1256. [DOI] [PubMed] [Google Scholar]
- 116. Zhang S, Wang B, Xiao H, et al. LncRNA HOTAIR enhances breast cancer radioresistance through facilitating HSPA1A expression via sequestering miR-449b-5p. Thorac Cancer 2020;11:1801–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Zhang H, Cai K, Wang J, et al. MiR-7, inhibited indirectly by lincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of breast cancer stem cells by downregulating the STAT3 pathway. Stem Cells 2014;32:2858–2868. [DOI] [PubMed] [Google Scholar]
- 118. Ren Y, Jia HH, Xu YQ, et al. Paracrine and epigenetic control of CAF-induced metastasis: The role of HOTAIR stimulated by TGF-ss1 secretion. Mol Cancer 2018;17:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Chen Q, Xu H, Zhu J, et al. LncRNA MCM3AP-AS1 promotes breast cancer progression via modulating miR-28-5p/CENPF axis. Biomed Pharmacother 2020;128:110289. [DOI] [PubMed] [Google Scholar]
- 120. Cantile M, Di Bonito M, Cerrone M, et al. Long non-coding RNA HOTAIR in breast cancer therapy. Cancers (Basel) 2020;12(5):1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Woolston C. Breast cancer. Nature 2015;527:S101. [DOI] [PubMed] [Google Scholar]
- 122. Augereau P, Patsouris A, Bourbouloux E, et al. Hormonoresistance in advanced breast cancer: A new revolution in endocrine therapy. Ther Adv Med Oncol 2017;9:335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Presti D, Quaquarini E. The PI3K/AKT/mTOR and CDK4/6 pathways in endocrine resistant HR+/HER2− metastatic breast cancer: Biological mechanisms and new treatments. Cancers (Basel) 2019;11(9):1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Ghafouri-Fard S, Fathi M, Zhai T, et al. LncRNAs: Novel biomarkers for pancreatic cancer. Biomolecules 2021;11(11):1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Liu Y, Ao X, Wang Y, et al. Long non-coding RNA in gastric cancer: Mechanisms and clinical implications for drug resistance. Front Oncol 2022;12:841411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Yuan D, Chen Y, Li X, et al. Long non-coding RNAs: Potential biomarkers and targets for hepatocellular carcinoma therapy and diagnosis. Int J Biol Sci 2021;17:220–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Jiang L, Li Z, Wang R. Long non-coding RNAs in lung cancer: Regulation patterns, biologic function and diagnosis implications (Review). Int J Oncol 2019;55:585–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Tan BS, Yang MC, Singh S, et al. LncRNA NORAD is repressed by the YAP pathway and suppresses lung and breast cancer metastasis by sequestering S100P. Oncogene 2019;38:5612–5626. [DOI] [PubMed] [Google Scholar]
- 129. De Troyer L, Zhao P, Pastor T, et al. Stress-induced lncRNA LASTR fosters cancer cell fitness by regulating the activity of the U4/U6 recycling factor SART3. Nucleic Acids Res 2020;48:2502–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Jadaliha M, Gholamalamdari O, Tang W, et al. A natural antisense lncRNA controls breast cancer progression by promoting tumor suppressor gene mRNA stability. PLoS Genet 2018;14:e1007802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Han M, Gu Y, Lu P, et al. Exosome-mediated lncRNA AFAP1-AS1 promotes trastuzumab resistance through binding with AUF1 and activating ERBB2 translation. Mol Cancer 2020;19:26. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 132. Lin HC, Yeh CC, Chao LY, et al. The hypoxia-responsive lncRNA NDRG-OT1 promotes NDRG1 degradation via ubiquitin-mediated proteolysis in breast cancer cells. Oncotarget 2018;9:10470–10482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Calanca N, Paschoal AP, Munhoz EP, et al. The long non-coding RNA ANRASSF1 in the regulation of alternative protein-coding transcripts RASSF1A and RASSF1C in human breast cancer cells: Implications to epigenetic therapy. Epigenetics 2019;14:741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Pang B, Wang Q, Ning S, et al. Landscape of tumor suppressor long noncoding RNAs in breast cancer. J Exp Clin Cancer Res 2019;38:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Li Y, Zhao Z, Liu W, et al. SNHG3 functions as miRNA sponge to promote breast cancer cells growth through the metabolic reprogramming. Appl Biochem Biotechnol 2020;191:1084–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Wang N, Hou M, Zhan Y, et al. LncRNA PTCSC3 inhibits triple-negative breast cancer cell proliferation by downregulating lncRNA H19. J Cell Biochem 2019;120:15083–15088. [DOI] [PubMed] [Google Scholar]
- 137. Tang J, Li Y, Sang Y, et al. LncRNA PVT1 regulates triple-negative breast cancer through KLF5/beta-catenin signaling. Oncogene 2018;37:4723–4734. [DOI] [PubMed] [Google Scholar]
- 138. Li Z, Hou P, Fan D, et al. The degradation of EZH2 mediated by lncRNA ANCR attenuated the invasion and metastasis of breast cancer. Cell Death Differ 2017;24:59–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Gwangwa MV, Joubert AM, Visagie MH. Effects of glutamine deprivation on oxidative stress and cell survival in breast cell lines. Biol Res 2019;52:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Wang K, Li J, Xiong YF, et al. A potential prognostic long noncoding RNA signature to predict recurrence among ER-positive breast cancer patients treated with tamoxifen. Sci Rep 2018;8:3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Wang J, Xie S, Yang J, et al. The long noncoding RNA H19 promotes tamoxifen resistance in breast cancer via autophagy. J Hematol Oncol 2019;12:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Gooding AJ, Zhang B, Gunawardane L, et al. The lncRNA BORG facilitates the survival and chemoresistance of triple-negative breast cancers. Oncogene 2019;38:2020–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Vaidya AM, Sun Z, Ayat N, et al. Systemic delivery of tumor-targeting siRNA nanoparticles against an oncogenic LncRNA facilitates effective triple-negative breast cancer therapy. Bioconjug Chem 2019;30:907–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Nayak S, Herzog RW. Progress and prospects: Immune responses to viral vectors. Gene Ther 2010;17:295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Jin SJ, Jin MZ, Xia BR, et al. Long non-coding RNA DANCR as an emerging therapeutic target in human cancers. Front Oncol 2019;9:1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. D'Mello SR, Cruz CN, Chen ML, et al. The evolving landscape of drug products containing nanomaterials in the United States. Nat Nanotechnol 2017;12:523–529. [DOI] [PubMed] [Google Scholar]
- 147. Fang C, Shi B, Pei YY, et al. In vivo tumor targeting of tumor necrosis factor-alpha-loaded stealth nanoparticles: Effect of MePEG molecular weight and particle size. Eur J Pharm Sci 2006;27:27–36. [DOI] [PubMed] [Google Scholar]
- 148. Immordino ML, Dosio F, Cattel L. Stealth liposomes: Review of the basic science, rationale, and clinical applications, existing and potential. Int J Nanomed 2006;1:297–315. [PMC free article] [PubMed] [Google Scholar]
- 149. Pathak K, Keshri L, Shah M. Lipid nanocarriers: Influence of lipids on product development and pharmacokinetics. Crit Rev Ther Drug Carrier Syst 2011;28:357–393. [DOI] [PubMed] [Google Scholar]
- 150. Perche F, Torchilin VP. Recent trends in multifunctional liposomal nanocarriers for enhanced tumor targeting. J Drug Deliv 2013;2013:705265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Slaby O, Laga R, Sedlacek O. Therapeutic targeting of non-coding RNAs in cancer. Biochem J 2017;474:4219–4251. [DOI] [PubMed] [Google Scholar]
- 152. Ulbrich K, Hekmatara T, Herbert E, et al. Transferrin- and transferrin-receptor-antibody-modified nanoparticles enable drug delivery across the blood-brain barrier (BBB). Eur J Pharm Biopharm 2009;71:251–256. [DOI] [PubMed] [Google Scholar]
- 153. Alvarez-Erviti L, Seow Y, Yin H, et al. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol 2011;29:341–345. [DOI] [PubMed] [Google Scholar]
- 154. Lennox KA, Behlke MA. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res 2016;44:863–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Limoni SK, Moghadam MF, Moazzeni SM, et al. Engineered exosomes for targeted transfer of siRNA to HER2 positive breast cancer cells. Appl Biochem Biotechnol 2019;187:352–364. [DOI] [PubMed] [Google Scholar]
- 156. Milane L, Singh A, Mattheolabakis G, et al. Exosome mediated communication within the tumor microenvironment. J Control Release 2015;219:278–294. [DOI] [PubMed] [Google Scholar]
- 157. Ohno S, Takanashi M, Sudo K, et al. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol Ther 2013;21:185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Quah BJ, O'Neill HC. The immunogenicity of dendritic cell-derived exosomes. Blood Cells Mol Dis 2005;35:94–110. [DOI] [PubMed] [Google Scholar]
- 159. Xie X, Wu H, Li M, et al. Progress in the application of exosomes as therapeutic vectors in tumor-targeted therapy. Cytotherapy 2019;21:509–524. [DOI] [PubMed] [Google Scholar]
- 160. Xiu B, Chi Y, Liu L, et al. LINC02273 drives breast cancer metastasis by epigenetically increasing AGR2 transcription. Mol Cancer 2019;18:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Vidovic D, Huynh TT, Konda P, et al. ALDH1A3-regulated long non-coding RNA NRAD1 is a potential novel target for triple-negative breast tumors and cancer stem cells. Cell Death Differ 2020;27:363–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Arun G, Diermeier SD, Spector DL. Therapeutic targeting of long non-coding RNAs in cancer. Trends Mol Med 2018;24:257–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Bass BL. Double-stranded RNA as a template for gene silencing. Cell 2000;101:235–238. [DOI] [PubMed] [Google Scholar]
- 164. Elmen J, Lindow M, Schutz S, et al. LNA-mediated microRNA silencing in non-human primates. Nature 2008;452:896–899. [DOI] [PubMed] [Google Scholar]
- 165. Hua Y, Sahashi K, Rigo F, et al. Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature 2011;478:123–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Monia BP, Lesnik EA, Gonzalez C, et al. Evaluation of 2’-modified oligonucleotides containing 2′-deoxy gaps as antisense inhibitors of gene expression. J Biol Chem 1993;268:14514–14522. [PubMed] [Google Scholar]
- 167. Setten RL, Rossi JJ, Han SP. The current state and future directions of RNAi-based therapeutics. Nat Rev Drug Discov 2019;18:421–446. [DOI] [PubMed] [Google Scholar]
- 168. Stein CA, Hansen JB, Lai J, et al. Efficient gene silencing by delivery of locked nucleic acid antisense oligonucleotides, unassisted by transfection reagents. Nucleic Acids Res 2010;38:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Valerio LG Jr. Tenth anniversary of expert opinion on drug metabolism & toxicology. Exp Opin Drug Metab Toxicol 2014;10:767–768. [DOI] [PubMed] [Google Scholar]
- 170. Lin LC, Lee HT, Chien PJ, et al. NAD(P)H:quinone oxidoreductase 1 determines radiosensitivity of triple negative breast cancer cells and is controlled by long non-coding RNA NEAT1. Int J Med Sci 2020;17:2214–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Li M, Li X, Zhuang Y, et al. Induction of a novel isoform of the lncRNA HOTAIR in Claudin-low breast cancer cells attached to extracellular matrix. Mol Oncol 2017;11:1698–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Elmen J, Thonberg H, Ljungberg K, et al. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res 2005;33:439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Hall AH, Wan J, Shaughnessy EE, et al. RNA interference using boranophosphate siRNAs: Structure-activity relationships. Nucleic Acids Res 2004;32:5991–6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Liao Y, Tang L. Inducible RNAi system and its application in novel therapeutics. Crit Rev Biotechnol 2016;36:630–638. [DOI] [PubMed] [Google Scholar]
- 175. Martinez J, Patkaniowska A, Urlaub H, et al. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 2002;110:563–574. [DOI] [PubMed] [Google Scholar]
- 176. Nie L, Das Thakur M, Wang Y, et al. Regulation of U6 promoter activity by transcriptional interference in viral vector-based RNAi. Genom Proteom Bioinform 2010;8:170–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Turner JJ, Jones SW, Moschos SA, et al. MALDI-TOF mass spectral analysis of siRNA degradation in serum confirms an RNAse A-like activity. Mol Biosyst 2007;3:43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Yang J. Patisiran for the treatment of hereditary transthyretin-mediated amyloidosis. Expert Rev Clin Pharmacol 2019;12:95–99. [DOI] [PubMed] [Google Scholar]
- 179. Singh N, Padi SKR, Bearss JJ, et al. PIM protein kinases regulate the level of the long noncoding RNA H19 to control stem cell gene transcription and modulate tumor growth. Mol Oncol 2020;14:974–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Li Y, Ren Y, Wang Y, et al. A compound AC1Q3QWB selectively disrupts HOTAIR-mediated recruitment of PRC2 and enhances cancer therapy of DZNep. Theranostics 2019;9:4608–4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Hao Q, Wang P, Dutta P, et al. Comp34 displays potent preclinical antitumor efficacy in triple-negative breast cancer via inhibition of NUDT3-AS4, a novel oncogenic long noncoding RNA. Cell Death Dis 2020;11:1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Chandra Gupta S, Nandan Tripathi Y. Potential of long non-coding RNAs in cancer patients: From biomarkers to therapeutic targets. Int J Cancer 2017;140:1955–1967. [DOI] [PubMed] [Google Scholar]