Abstract
Unlike the multiple-shelled organization of other Reoviridae members, cytoplasmic polyhedrosis virus (CPV) has a single-shelled capsid. The three-dimensional structures of full and empty CPV by electron cryomicroscopy show identical outer shells but differ inside. The outer surface reveals a T=1 icosahedral shell decorated with spikes at its icosahedral vertices. The internal space of the empty CPV is unoccupied except for 12 mushroom-shaped densities attributed to the transcriptional enzyme complexes. The ordered double-stranded RNA inside the full capsid forms spherical shells spaced 25 Å apart. The RNA-protein interactions suggest a mechanism for RNA transcription and release.
Cytoplasmic polyhedrosis virus (CPV), one of the most widespread insect pathogens (24), belongs to the Cypovirus genus in the Reoviridae family. Infectious CPV capsids are usually embedded in a characteristic crystalline inclusion body composed of viral proteins named polyhedra (18, 27). Insects are infected by ingestion of the polyhedra, followed by the alkaline disruption of the polyhedrin matrix in the gastrointestinal tract. Consequently, the viral capsids are released and penetrate the membranes of the epithelial cells where they replicate. During viral replication, a large number of polyhedrin proteins are produced in the cytoplasm of the infected cells and cell death results because of the metabolic burden.
The infectious CPV capsid contains a 10-segmented double-stranded RNA (dsRNA) genome (12) and five structural proteins (21). When examined by negative-stain electron microscopy, CPV appears as a single-shelled icosahedron, with a diameter of 600 Å and 12 characteristic turret-like spikes. This organization contrasts with the double-shelled (e.g., rice dwarf virus [19]) or triple-shelled (e.g., animal reovirus [4], rotavirus [22], bluetongue virus [13]) arrangements which are typical for viruses in the Reoviridae. The full, infectious CPV capsid resembles that of other reoviruses functionally since it contains an RNA-dependent RNA polymerase and is fully capable of RNA transcription (17). Each transcriptionally active CPV releases newly transcribed mRNA from the intact capsid. The spikes at the icosahedral vertices were suggested to be involved in this unique process (20, 28). Since these proposals lack a structural foundation, we have determined the three-dimensional (3D) structures of the empty and full CPV capsids by using electron cryomicroscopy and computer reconstruction. These studies have revealed new features concerning their structural organization and suggest that viruses in the Reoviridae use a similar mechanism of RNA transcription and release despite the striking differences of their capsid shells.
Purification and electron cryomicroscopy of full and empty particles.
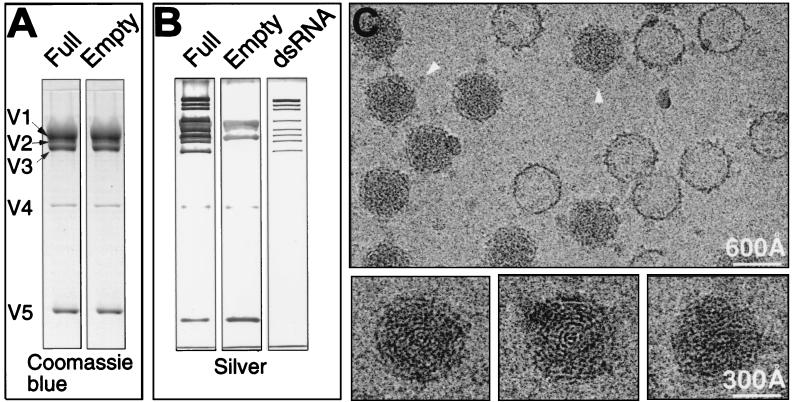
The full and empty CPV particles were purified from infected fifth-instar larvae of Bombyx mori by sucrose gradient centrifugation. Sodium dodecyl sulfate-polyacrylamide gel electrophoretic (SDS-PAGE) analyses were performed to confirm the chemical composition of the particles (Fig. 1). Coomassie blue-stained gels revealed an identical protein pattern for the full and empty CPV capsids (Fig. 1A). A comparison of the silver-stained gel profiles, which stain both the protein and dsRNA contents, showed that the only difference between the full and empty capsids is the 10 segments of dsRNA in the full CPV capsid (Fig. 1B). Subsequently, the full and empty capsids were imaged together to permit a direct structural comparison of the particles recorded under the same conditions (29). Focal pair micrographs were taken at ×30,000 in a JEOL 1200 electron cryomicroscope operated at 100 kv, using standard procedures as described previously (33). As shown in Fig. 1C, the full and empty CPV particles can be unambiguously identified based on the presence of the fingerprint patterns inside the full particles and the lack of them inside the empty particles. Except for this difference, the full and empty capsids appear similar and exhibit prominent protrusions at their vertices (Fig. 1C). Previous negative-stain electron microscopy studies identified these protrusions as spikes consisting of two concentric parts. The portion of the spike extending directly from the shell is called the B spike. Attached to the top of B spike is an extended, narrower mass called the A spike (10). The diameter of the capsid is about 600 Å, excluding the spikes, and increases to ∼800 Å upon their inclusion.
FIG. 1.
SDS-PAGE analyses and electron cryomicroscopic images of purified CPV capsids. Fifth-instar larvae of B. mori were infected with CPV by spraying a suspension of polyhedra (108/ml) onto mulberry leaves. Ten days after infection, midguts were removed from the larvae and polyhedra were purified by a method modified from that of Hayashi and Bird (11). In our procedure, 0.5% SDS and 100 μg of trypsin/ml were used instead of the 1% sodium deoxycholate and an enzyme mixture (RNase A, DNase, trypsin [50 μg/ml each]). We also used 20 mM phosphate-buffered saline (PBS) buffer (pH 7.4) instead of TK buffer (0.03 M tris-HCl buffer, pH 7.5; 0.025M KCl). The polyhedra were resuspended in 0.2 M sodium carbonate-sodium bicarbonate buffer. After 1 h at 30°C, the pH was adjusted to 7.4 with 20 mM NaH2PO4 and the mixture was centrifuged at 10,000 × g for 10 min. The virus particles were pelleted from the supernatant at 90,000 × g for 70 min. The pellet was resuspended in PBS and purified in 4 ml of 10 to 40% (wt/wt) sucrose gradient by centrifugation at 55,000 × g for 1 h. Virus-containing fractions were recovered. After being diluted with PBS, the viral sample was pelleted again at 90,000 × g for 70 min and resuspended in PBS. (A) SDS-PAGE analyses of the purified full and empty capsids were carried out with Coomassie blue staining to identify the differences of protein compositions between the full and empty capsids. Viral proteins V1 to V5 are labeled. (B) The SDS gels stained with silver reveal the nucleic acid compositions as well as protein compositions in full and empty capsids. The right panel shows the profile of the RNA genome extracted from the full capsid. (C) (Upper panel) Typical area of a close-to-focus electron micrograph (1.46 μm defocus) in a focal pair of ice-embedded CPV capsids taken at 100 kV. The micrographs were digitized on a Zeiss SCAI microdensitometer (Carl Zeiss, Inc., Englewood, Colo.) at a step size of 4.67 Å/pixel, and individual virus particles were extracted into individual images of 200 × 200 pixels. Arrowheads indicate the A spikes visible in some capsid views. (Lower panels) three different full capsids are enlarged to show the characteristic fingerprint patterns.
3D reconstruction.
The determination of the center and the orientation parameters of each boxed out particle and their subsequent refinement were carried out using procedures previously described (14, 30, 31), which are based on Fourier common lines (2, 7). The particles from the strongly underfocused micrographs were used as an aid in the determination of the parameters of the particles in the close-to-focus micrographs (31). Prior to the merging of particle images for 3D reconstruction using Fourier-Bessel synthesis (2), the Fourier transform values of individual images were corrected for the contrast transfer function as described elsewhere (33). We merged particle images from five micrographs with different defocus values in order to obtain an even data sampling across a wide range of spatial frequency or resolution zones. The reconstructions for the full and empty capsids were computed from 2,134 and 387 individual particle images, respectively. Using the criterion of the Fourier-ring correlation coefficient between two independent reconstructions (33) being larger than 0.5, the effective resolution of the full and empty capsid reconstructions are 17 and 25 Å, respectively.
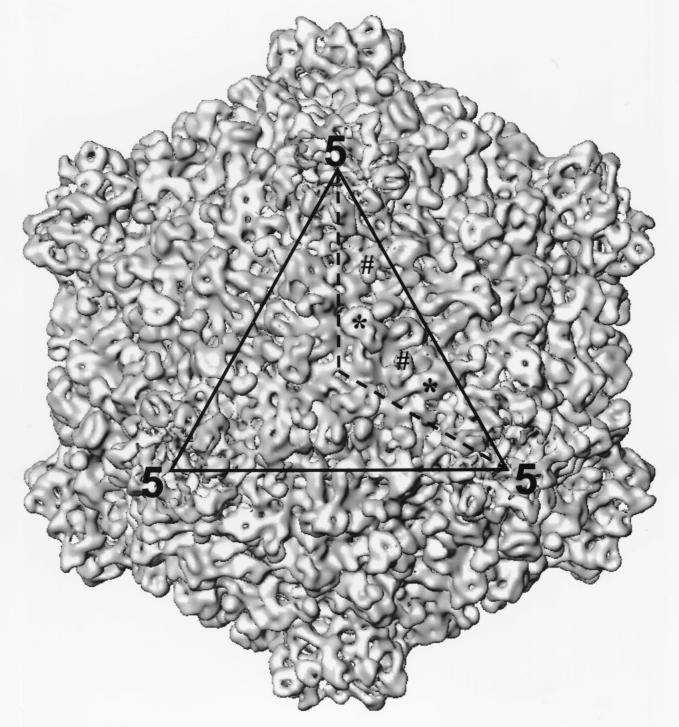
The 17-Å reconstruction of the full capsids shows a T=1 icosahedral shell, with 12 protrusions at the fivefold axes (Fig. 2). These protrusions correspond to the B spike observed in the electron cryomicroscopic images (Fig. 1A). The hollow B spike is 150 Å wide and 50 Å high. Each subunit of the B spike has two discernible, elongated domains that form side-by-side connections at the top of the spike and merge to form a Y-shaped feature at the base of the spike where it interacts with the shell proteins (Fig. 2). The A spike has a lower density and thus is not visible when displayed at the isosurface value of 1ς (standard deviation) (Fig. 2). On the capsid shell, there are 120 large protrusions (LPs) and 120 small protrusions (SPs) (Fig. 2). The SDS gels show that V3 and V5 are the most abundant proteins in CPV capsids (Fig. 1A and B), suggesting that they are the likely candidates for the LPs and SPs.
FIG. 2.
Surface representation of the 3D structure of the full CPV capsid at 17-Å resolution as viewed along the icosahedral threefold axis. One of the three asymmetric units outlined in the icosahedral triangular face (dotted line) contains two SPs and two LPs which are designated by # and ∗, respectively.
Locations of RNA and the transcriptional enzyme complex.
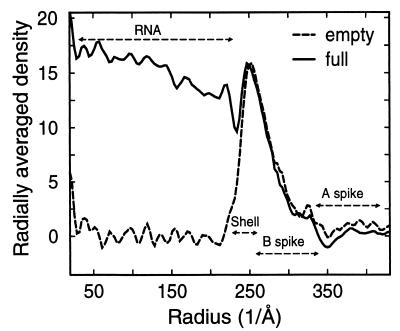
The density distribution from different radii of from 230-Å to 425-Å radii, which encompasses the shells and the spikes, is similar for the full and empty capsids (Fig. 3). Below the radius of 230 Å, the averaged density is significant in the full capsid, which contrasts with the lack of density in the empty capsid (Fig. 3). Since the only difference in the chemical composition in the full and empty capsids is the presence of dsRNA in the full capsid (Fig. 1A and B), it can be concluded that the majority of dsRNA is located internally within a radius of 230 Å.
FIG. 3.
The averaged density distribution of the final 3D reconstructions of the full and empty capsids as a function of particle radius. The density beyond a radius of 230 Å appears almost identical in the empty and full capsids. Below a radius of 230 Å, there are several layers of high density in the full capsid, which are completely absent in the empty capsid. The radial range locations of structural components in full and empty capsids are indicated.
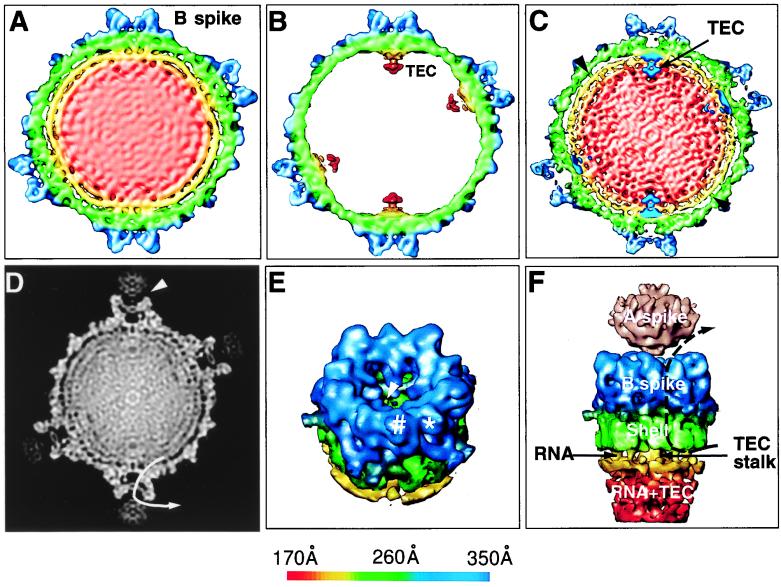
In order to compare the 3D structures of the full and empty capsids, the reconstructions were filtered to a comparable 25-Å resolution. At this resolution, their structural features beyond the radius of 230 Å are indistinguishable. However, the corresponding 50-Å-thick slices extracted from the 3D reconstructions of the full and empty capsids, respectively, show significant structural differences inside the capsids (Fig. 4A and B). The internal space of the full capsid map is densely packed, which is attributed to the dsRNA genome of CPV, whereas no density is visible inside the empty capsid (Fig. 4A and B). When the reconstruction of the empty capsids was displayed at 0.5ς, a flower-shaped feature was seen attached to the inner surface of each fivefold axis from a radius of 160 to 230 Å (Fig. 4B). This structure is connected by a stalk to the inner surface of the capsid underneath each B spike (Fig. 4B). Similar structures have been seen and attributed to be the viral transcription complex in rotavirus (23) and reovirus (4). We propose that the structure seen here is the transcriptional enzyme complex (TEC) in CPV. The lower density of this structure suggests that there are less than 60 copies of the constituent protein in each capsid. It has been shown that the RNA transcriptase has a molecular size of ∼140 kDa and is the most conservative protein in the Reoviridae (25, 26). The SDS gel analyses (Fig. 1A and B) showed that V2 has a molecular size of about 140 kDa, is one of the least abundant proteins in the CPV capsid, and is thus the most likely candidate for the RNA transcriptase of CPV. This suggestion is consistent with the observation that antibody against V2 could block the RNA transcriptase activity of the isolated enzyme complex (3).
FIG. 4.
Visualization of protein-RNA interactions in CPV. (A and B) Structural comparison of full and empty capsids. A central 50-Å thick slice was extracted from the 3D reconstructions of the full (A) and empty (B) capsids at 25-Å resolution. Both maps were contoured at 1ς above the mean except for the internal complexes of the empty capsid, where 0.5ς was used to reveal the TEC. (C) A 50-Å thick central slice from the 17-Å full capsid reconstruction was superimposed with the TECs extracted from the reconstruction of the empty capsids. The interactions between the capsid proteins and the RNA were strongest at the base of the spike. Minor interactions were also observed close to the twofold axis (arrowhead). The densities attributed to dsRNA inside the full capsid formed two shells between the 160- to 230-Å radii, with a distance of 25 Å between adjacent RNA shells. (D) A 3-Å-thick slice extracted from the center of the 3D reconstruction of the full capsid is displayed in gray scale. The A spike is loosely connected to the B spike. The A spike has a globular structure, whereas the B spike is a hollow protrusion from the shell. The pathway of one of the five channels, which connects the inside and outside the capsid, is depicted by the arrow line. (E) The B spike and a portion of the viral shell were extracted to reveal the hole at the top cover of the B spike cavity and the terminal end of the channel connecting the inside and outside of the capsid (arrowhead). Each of the five subunits of the spike has two discernible subdomains (∗ and #). (F) A portion of the full map at the icosahedral vertex was extracted and viewed from the side to illustrate the interactions among the RNA, the B spike, and the shell. The A spike, visible only when displayed at 0.5ς above the mean, is shown in pink. The channel connecting the inside with the outside indicated in panel D is illustrated by the dotted line. Except for the A spike in panel F, the maps are color coded according to the radius of the capsid, using the scheme shown at the bottom.
To visualize the details of the internal RNA, a 50-Å thick section of the 17-Å reconstruction of the full CPV is displayed (Fig. 4C). The mass density close to the inner surface of the capsid forms two clearly distinguishable shells (shown in yellow and red). The distance between the adjacent shells is 25 Å, which corresponds to the distance between close-packed dsRNA revealed by low-angle X-ray diffraction (8). Careful examination of the outermost RNA shell indicates that it has a dodecahedron shape, similar to that reported for rotavirus (23). The density below the radius of 180 Å appeared featureless (Fig. 4C). In order to evaluate the presence of icosahedral symmetry in the dsRNA shells, we computed the disagreement factor between two independent reconstructions of the full capsid (32), which was 10% for the capsid shell and less than 20% for the RNA core. Furthermore, the effective resolution of the RNA core was 30 Å, which contrasts with the 17 Å for the capsid shell alone, indicating that the icosahedral symmetry of the RNA core is not preserved to the same extent as the protein shell.
Icosahedrally organized RNA has been observed in small viral structures by X-ray crystallography (6, 15). The presence of the symmetrically organized dsRNA genome in the full CPV capsid gives a characteristic fingerprint pattern in the electron cryomicroscopic image (Fig. 1), similar to those observed for the reovirus (5) and rotavirus (23). However, how the dsRNA is packaged into this symmetrical form is yet to be determined.
Interactions among RNA, TEC, and the spikes.
By examining the contact points between the RNA and the protein shells, the strongest interaction between the RNA and the capsid proteins was identified at the base of the spike where the TECs connect the shell proteins with the RNA (Fig. 4C). It appeared that the presence of TEC at this location caused the mass density of RNA shells to deviate from its characteristic spherical organization. In addition, some weak interactions could also be observed adjacent to the icosahedral twofold axes. The relative strength of these interactions was revealed in a 3-Å thick central slice of the full capsid reconstruction, using a gray level display (Fig. 4D). These interactions may provide the structural basis for the symmetrical organization of RNA genome and the RNA transcription inside CPV.
The mass density of one B spike was extracted from the reconstruction and viewed in a slightly tilted angle to reveal additional features (Fig. 4E). The two subdomains of each B spike subunit were indicated by * and # in Fig. 4E. Each B spike has an internal cylinder-shaped cavity 60 Å wide and 30 Å high that connects to the shell of the capsid. The subunits of the B spike extend centrally and downward to form a bowl-shaped cover on the cavity, leaving a central hole that is 23 to 28 Å wide. Surrounding the TEC are five elongated channels with a diameter of 18 Å that penetrate the shell and terminate peripherally in the cavity of the B spike. The terminal of one of these channels is indicated in the cavity by an arrowhead (Fig. 4E).
As is apparent from Fig. 4D, the A spike, which is occasionally visible in the electron cryomicroscopic images (Fig. 1C), can be seen loosely attached to the top of the B spike. A 25-Å-wide cleft exists between the two structural components. Due to its much lower density than the rest of the capsid, the A spike can only be seen in a surface representation when the density threshold value is lowered to 0.5ς (Fig. 4F). This indicates that the A spike is relatively flexible and/or less than five symmetry-related sites at each vertex are occupied by this component. Each A spike has a globular structure, about 110 Å wide and 90 Å high. The bottom of the A spike has a cone shape that fits into the opening at the top of the B spike.
Comparison with other dsRNA viruses and functional implications.
The single-shelled capsid of CPV is unique and contrasts with the double- or triple-shelled capsids of other members in the Reoviridae. In the mammalian reovirus system, the two outer layers of the capsid have two functional roles. First, they protect the viral RNA genome from the unfavorable environment outside the virus. Second, they mediate the specific viral attachment to the cells that are infected. Devoid of the outer protein layers, CPV particles are instead embedded in the characteristic polyhedra. The polyhedra mediate the transmission of the CPV and thus may play roles similar to those of the outer layers of other reoviruses. However, it remains unclear how CPV attaches and penetrates the host cell.
It is interesting that the CPV core is structurally similar to the core of animal reovirus (4) and to that of the bacteriophage φ6, another segmented, dsRNA virus (1). The cores of both these viruses contain prominent turret-like, hollow projections extending outward at the fivefold axes and have a similar diameter when excluding the spikes. Their shells have a T=1 icosahedral symmetry. However, unlike CPV, these viruses do not contain a structure corresponding to the A spike in CPV. The structure organization of CPV is very similar to the model proposed for the subviral core of Fiji virus (9), another member of the Reoviridae. Fiji virus possesses a double-shelled icosahedron that causes plant diseases. Its A spike extends distally from the surface of the outer shell. Interestingly, the A spike only exists in viruses that lack the outer third of the capsid layer. Therefore, it is possible that the A spike provides extra protection for the viral genome by functioning as a cover for the hollow B spike.
In contrast to the capsid of CPV, the double-shelled particle of rotavirus has a relatively smooth surface without prominent projections at the fivefold vertices. Lawton et al. (16) have suggested that the mRNA is synthesized at the base of the icosahedral vertices and then released through the small channels around the fivefold vertices, without the lysis of the viral capsid. The TECs in CPV are also located at the base of the fivefold axes. Therefore, the observed structure of the RNA inside our CPV capsid may represent a snapshot of a dynamic process of the RNA genome. The RNA segments may slide through the TEC at the fivefold axes to act as templates for RNA synthesis. The mRNA may be released through the five channels at the periphery of the TEC to the hollow cavity of the B spike and then exit through the central hole of the B spike (Fig. 4E and F). Since no channels have been observed in the A spike, the RNA has to be released through the cleft between the A and the B spikes. This sliding process may be driven by the continuous release of the transcribed mRNA to the outside. When the CPV capsid is actively being transcribed, the release of mRNA may be delayed in the hollow cavity, causing the spike to swell (28). This mechanism of RNA synthesis in CPV agrees with the negative-stain electron microscopy observation that partially disrupted viral particles release the thread-like RNA from the spike complex (20, 28). It also accords well with the mechanism proposed for rotavirus (16). Therefore, mammalian rotavirus and insect CPV appear to share a similar mechanism of RNA synthesis and release, despite striking differences in their capsid structures; this mechanism may be common among members of the Reoviridae.
Acknowledgments
This project was supported partly by K. C. Wong Education Foundation, Hong Kong, and the National Natural Science Foundation of China.
We are in debt to K. H. Kuo for encouragement and support. We thank the National Center for Macromolecular Imaging directed by W. Chiu at Baylor College of Medicine for the use of their resources, and J. Stoops, S. Kolodziej, U. Qazi, and S. Boatman for helpful discussions and editorial assistance.
REFERENCES
- 1.Butcher S J, Dokland T, Ojala P M, Bamford D H, Fuller S D. Intermediates in the assembly pathway of the double-stranded RNA virus phi6. EMBO J. 1997;16:4477–4487. doi: 10.1093/emboj/16.14.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crowther R A. Procedures for three-dimensional reconstruction of spherical viruses by Fourier synthesis from electron micrographs. Phil Trans R Soc Lond B. 1971;261:221–230. doi: 10.1098/rstb.1971.0054. [DOI] [PubMed] [Google Scholar]
- 3.Dai R M, Wu A Z, Sun Y K. The protein subunits of the double-stranded RNA dependent RNA polymerase and methyltransferase of the cytoplasmic polyhedrosis virus of silkworm, Bombyx mori. Sci Sin Ser B (Chem Biol Agric Med Earth Sci) 1986;29:1267–1272. [PubMed] [Google Scholar]
- 4.Dryden K A, Farsetta D L, Wang G, Keegan J M, Fields B N, Baker T S, Nibert M L. Internal structures containing transcriptase-related proteins in top component particles of mammalian orthoreovirus. Virology. 1998;245:33–46. doi: 10.1006/viro.1998.9146. [DOI] [PubMed] [Google Scholar]
- 5.Dryden K A, Wang G J, Yeager M, Nibert M L, Coombs K M, Furlong D B, Fields B N, Baker T S. Early steps in reovirus infection are associated with dramatic changes in supramolecular structure and protein conformations. J Cell Biol. 1993;122:1023–1041. doi: 10.1083/jcb.122.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fisher A J, Johnson J E. Ordered duplex RNA controls capsid architecture in an icosahedral animal virus. Nature (London) 1993;361:176–179. doi: 10.1038/361176a0. [DOI] [PubMed] [Google Scholar]
- 7.Fuller S D, Butcher S J, Cheng R H, Baker T S. Three-dimensional reconstruction of icosahedral particles—the uncommon line. J Struct Biol. 1996;116:48–55. doi: 10.1006/jsbi.1996.0009. [DOI] [PubMed] [Google Scholar]
- 8.Harvey J D, Bellamy A R, Earnshaw W C, Schutt C. Biophysical studies of reovirus type 3. IV. Low-angle X-ray diffraction studies. Virology. 1981;112:240–249. doi: 10.1016/0042-6822(81)90629-2. [DOI] [PubMed] [Google Scholar]
- 9.Hatta T, Francki R I B. Morphology of Fiji disease virus. Virology. 1977;76:797–807. doi: 10.1016/0042-6822(77)90260-4. [DOI] [PubMed] [Google Scholar]
- 10.Hatta T, Francki R I B. Similarity in the structure of cytoplasmic polyhedrosis virus, leafhopper A virus and Fiji disease virus particles. Intervirology. 1982;18:203–208. doi: 10.1159/000149326. [DOI] [PubMed] [Google Scholar]
- 11.Hayashi Y, Bird F T. The isolation of cytoplasmic polyhedrosis virus from the white-marked tussock moth, Orgyia leucostigma (Smith) Can J Microbiol. 1970;16:695–701. doi: 10.1139/m70-119. [DOI] [PubMed] [Google Scholar]
- 12.Hayashi Y, Kawase S. Base pairing in ribonucleic acid extracted from the cytoplasmic polyhedra of the silkworm. Virology. 1964;23:611–614. doi: 10.1016/0042-6822(64)90247-8. [DOI] [PubMed] [Google Scholar]
- 13.Hewat E A, Booth T F, Roy P. Structure of correctly self-assembled bluetongue virus-like particles. J Struct Biol. 1994;112:183–191. doi: 10.1006/jsbi.1994.1019. [DOI] [PubMed] [Google Scholar]
- 14.Johnson O, Govindan V, Park Y, Zhou Z H. Proceedings of the 4th International Conference on High Performance Computing. Los Alamitos, Calif: IEEE Computer Society Press; 1997. Custom virtual memory policy for an image reconstruction application; pp. 517–521. [Google Scholar]
- 15.Larson S B, Koszelak S, Day J, Greenwood A, Dodds J A, McPherson A. Double-helical RNA in satellite tobacco mosaic virus. Nature (London) 1993;361:179–182. doi: 10.1038/361179a0. [DOI] [PubMed] [Google Scholar]
- 16.Lawton J A, Estes M K, Prasad B V V. Three-dimensional visualization of mRNA release from actively-transcribing rotavirus particles. Nat Struct Biol. 1997;4:118–121. doi: 10.1038/nsb0297-118. [DOI] [PubMed] [Google Scholar]
- 17.Lewandowski L J, Kalmakoff J, Tanada Y. Characterization of a ribonucleic acid polymerase activity associated with purified cytoplasmic polyhedrosis virus of the silkworm Bombyx mori. J Virol. 1969;4:857–865. doi: 10.1128/jvi.4.6.857-865.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewandowski L J, Traynor B L. Comparison of the structure and polypeptide composition of three double-stranded ribonucleic acid-containing viruses (diplornaviruses): cytoplasmic polyhedrosis virus, wound tumor virus, and reovirus. J Virol. 1972;10:1053–1070. doi: 10.1128/jvi.10.5.1053-1070.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu G Y, Zhou Z H, Baker M, Jakana J, Cai D Y, Wei X C, Chen S X, Gu X C, Chiu W. Structure of double-shelled rice dwarf virus. J Virol. 1998;72:8541–8549. doi: 10.1128/jvi.72.11.8541-8549.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishimura A, Hosaka Y. Electron microscopic study on RNA of cytoplasmic polyhedrosis virus of the silkworm. Virology. 1969;38:550–557. doi: 10.1016/0042-6822(69)90175-5. [DOI] [PubMed] [Google Scholar]
- 21.Payne C C, Rivers C F. A provisional classification of cytoplasmic polyhedrosis viruses based on the sizes of the RNA genome segments. J Gen Virol. 1976;33:71–85. doi: 10.1099/0022-1317-33-1-71. [DOI] [PubMed] [Google Scholar]
- 22.Prasad B V V, Burns J W, Marietta E, Estes M K, Chiu W. Localization of VP4 neutralization sites in rotavirus by three-dimensional cryo-electron microscopy. Nature (London) 1990;343:476–479. doi: 10.1038/343476a0. [DOI] [PubMed] [Google Scholar]
- 23.Prasad B V V, Rothnagel R, Zeng C Q-Y, Jakana J, Lawton J A, Chiu W, Estes M K. Visualization of ordered genomic RNA and localization of the transcription enzymes in rotavirus. Nature (London) 1996;382:471–473. doi: 10.1038/382471a0. [DOI] [PubMed] [Google Scholar]
- 24.Smith K M. The cytoplasmic virus diseases. In: Steinhaus E A, editor. Insect pathology. Vol. 1. New York, N.Y: Academic Press, Inc.; 1963. pp. 457–497. [Google Scholar]
- 25.Valenzuela S, Pizarro J, Sandino A M, Vasquez M, Fernandez J, Hernandez O, Patton J, Spencer E. Photoaffinity labeling of rotavirus VP1 with 8-azido-ATP: identification of the viral RNA polymerase. J Virol. 1991;65:3964–3967. doi: 10.1128/jvi.65.7.3964-3967.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wiener J R, Joklik W K. The sequence of the reovirus serotype 1, 2 and 3 L1 genome segments and analysis of the mode of divergence of the reovirus serotypes. Virology. 1989;169:194–203. doi: 10.1016/0042-6822(89)90055-x. [DOI] [PubMed] [Google Scholar]
- 27.Xia D, Sun Y-K, McCrae M A, Rossmann M G. X-ray powder pattern analysis of cytoplasmic polyhedrosis virus inclusion bodies. Virology. 1991;180:153–158. doi: 10.1016/0042-6822(91)90018-7. [DOI] [PubMed] [Google Scholar]
- 28.Yazaki K, Miura K-I. Relation of the structure of cytoplasmic polyhedrosis viruses and the synthesis of ets messenger RNA. Virology. 1980;105:467–479. doi: 10.1016/0042-6822(80)90047-1. [DOI] [PubMed] [Google Scholar]
- 29.Yeager M, Berriman J A, Baker T S, Bellamy A R. Three-dimensional structure of the rotavirus haemagglutinin VP4 by cryo-electron microscopy and difference map analysis. EMBO J. 1994;13:1011–1018. doi: 10.1002/j.1460-2075.1994.tb06349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Z H, Chiu W, Haskell K, Spears H J, Jakana J, Rixon F J, Scott L R. Refinement of herpesvirus B-capsid structure on parallel supercomputers. Biophys J. 1998;74:576–588. doi: 10.1016/S0006-3495(98)77816-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou Z H, He J, Jakana J, Tatman J D, Rixon F J, Chiu W. Assembly of VP26 in herpes simplex virus-1 inferred from structures of wild-type and recombinant capsids. Nat Struct Biol. 1995;2:1026–1030. doi: 10.1038/nsb1195-1026. [DOI] [PubMed] [Google Scholar]
- 32.Zhou Z H, Macnab S J, Jakana J, Scott L R, Chiu W, Rixon F J. Identification of the sites of interaction between the scaffold and outer shell in herpes simplex virus-1 capsids by difference electron imaging. Proc Natl Acad Sci USA. 1998;95:2778–2783. doi: 10.1073/pnas.95.6.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou Z H, Prasad B V V, Jakana J, Rixon F, Chiu W. Protein subunit structures in the herpes simplex virus A-capsid determined from 400 kV spot-scan electron cryomicroscopy. J Mol Biol. 1994;242:458–469. doi: 10.1006/jmbi.1994.1594. [DOI] [PubMed] [Google Scholar]