Abstract
Background
Gliomas with IDH1/2 mutations without 1p19q codeletion have been identified as the distinct diagnostic entity of IDH mutant astrocytoma (IDHmut astrocytoma). Homozygous deletion of Cyclin-dependent kinase 4 inhibitor A/B (CDKN2A/B) has recently been incorporated in the grading of these tumors. The question of whether histologic parameters still contribute to prognostic information on top of the molecular classification, remains unanswered. Here we evaluated consensus histologic parameters for providing additional prognostic value in IDHmut astrocytomas.
Methods
An international panel of seven neuropathologists scored 13 well-defined histologic features in virtual microscopy images of 192 IDHmut astrocytomas from EORTC trial 22033-26033 (low-grade gliomas) and 263 from EORTC 26053 (CATNON) (1p19q non-codeleted anaplastic glioma). For 192 gliomas the CDKN2A/B status was known. Consensus (agreement ≥ 4/7 panelists) histologic features were tested together with homozygous deletion (HD) of CDKN2A/B for independent prognostic power.
Results
Among consensus histologic parameters, the mitotic count (cut-off of 2 mitoses per 10 high power fields standardized to a field diameter of 0.55 mm and an area of 0.24 mm2) significantly influences PFS (P = .0098) and marginally the OS (P = .07). Mitotic count also significantly affects the PFS of tumors with HD CDKN2A/B, but not the OS, possibly due to limited follow-up data.
Conclusion
The mitotic index (cut-off 2 per 10 40× HPF) is of prognostic significance in IDHmut astrocytomas without HD CDKN2A/B. Therefore, the mitotic index may direct the therapeutic approach for patients with IDHmut astrocytomas with native CDKN2A/B status.
Keywords: astrocytoma, CDKN2A/B, glioma, grading, IDH mutation, molecular pathology, prognosis, proliferation
Key Points.
The mitotic count discriminates PFS in IDH mutant astrocytomas.
CDKN2A/B status and mitoses are relevant for grading of IDH mutant astrocytomas.
Importance of the Study.
In the recent WHO classification of CNS tumors, the question of how to grade IDHmut astrocytomas remains unanswered. While homozygous deletion of CDKN2A/B in this glioma subset has been proven to be associated with poor outcome, the question of how to grade the group of non-HD CDKN2A/B tumors remains unresolved. Here we present the results of panel review of the histology of 455 IDH-mutant astrocytomas from EORTC trial 26053-22054 (CATNON trial) and EORTC trial 22033-26033 (low-grade gliomas). Following multivariate testing of prognostic factors including CDKN2A/B, the (consensus) mitotic count appeared to have independent prognostic value and to discriminate between two survival groups within the CDKN2A/B wild type tumors. The results may impact current therapeutic strategies, and the design of future trials of this important glioma subset.
The classification of diffuse gliomas has lately undergone profound changes. Because the molecular profile of certain tumors correlates closely with clinical behavior, the subtyping of diffuse gliomas is now largely based on their molecular signature.1 Gliomas with IDH mutations are associated with better clinical outcomes than their IDH wild type counterparts.2–4 Currently, the value of histologic grading to delineate useful prognostic subgroups within distinct molecular tumor categories is unclear. In the group of IDHmut astrocytomas, there is still considerable variation in clinical behavior. Homozygous deletion (HD) of the CDKN2A/B locus, which is almost exclusively present in cases with obvious high-grade histology, has been shown to correlate with shorter survival in IDHmut astrocytomas. Therefore, HD of the CDKN2A/B has been included as a grade 4 criterion in the WHO 5th edition.5–8 In the past, the most important histological discriminant between astrocytomas grade 2 and 3 has been the mitotic count.9 However, conclusive data are lacking on the significance of the mitotic count in the context of the IDH status.7 The present investigation aimed to evaluate the prognostic value of consensus histological parameters in IDH mutated astrocytomas. In order to obtain consensus on various histopathologic features including the mitotic count, a panel of seven neuropathologists scored virtual microscopic slides of the 455 IDHmut astrocytomas (1p19q non-codeleted) that were part of EORTC trial 22033-26033 (low-grade gliomas) or EORTC trial 26053 (CATNON, grade 3, 1p19q non-codeleted glioma) (Supplemental Materials). The IDH mutation and 1p/19q codeletion status was known in all cases, while information on HD CDKN2A/B was available for part of the CATNON cohort. The prognostic value of histological parameters was further tested against prognostically relevant molecular parameters. The results show that in IDHmut astrocytoma, consensus mitotic count retains prognostic power when compared to HD CDKN2A/B.
Materials and Methods
Trials and Clinical Data
Pathology materials of EORTC trial 22033-26033 and trial 26053-22054 were used for this study. For detailed descriptions of EORTC trial 26053-22054 (anaplastic glioma, not 1p/19q codeleted) and EORTC trial 22033-26033 (low-grade gliomas), see Supplemental Materials. EORTC trial 22033-26033, comprising a total of 477 randomized patients, compared standard radiation therapy with temozolomide treatment in high-risk, low-grade gliomas. The first interim analysis revealed no differences in PFS between the two treatment arms.10 EORTC trial 26053-22054 enrolled 745 patients at the time of the interim analysis. In 2017, the first interim analysis showed survival benefit of adjuvant temozolomide in non-codeleted anaplastic gliomas.11 The second interim analysis revealed that IDH mutational status is predictive of the benefit of adjuvant temozolomide.6
Pathology Review Procedure and Scoring
Pathology material of 883 tumors was available for review. For the current study, a total of 455 tumors (namely, 192 fom EORTC 22033-26033 and 263 from CATNON) were IDH mutant and lacked codeletion 1p/19q (Supplementary Table 1). All hematoxylin and eosin stained histologic slides were scanned for virtual microscopy (Hamamatsu scanner; Hamamatsu-shi, Japan). The scanned images were combined, randomly sorted and submitted to the panelists for scoring. A panel of seven neuropathologists, blinded to trial inclusion criteria and clinical course, independently reviewed the histology (AL, AM, MA-H, FB, CM, ER, GM). Thirteen histologic features, namely cell density, calcifications, increased number of blood vessels, microvascular proliferation, neoplastic appearing astrocytes and oligodendrocytes, giant cells, gemistocytes, miniature gemistocytes, nuclear pleomorphism, microcysts and mucoid degeneration, necrosis and the mitotic count, were scored. All features, except the mitotic count and cell density, were scored in an on-off manner. Mitotic activity was scored into three groups, namely: ≤ 1 mitosis, 1–2 mitoses and > 2 mitoses per 10 consecutive (40×) HPF standardized to a field diameter of 0.55 mm and an area of 0.24 mm2.12 Cell density was scored in three groups, viz. low, high and intermediate. Consensus was defined as similar scores by at least four out of the seven neuropathologists. An inter-rater coëfficient (ICC) of < 0.30 was considered poor; 0.30–0.40, fair and > 0.40, good.
Molecular Data
The tumors from CATNON were all tested for CDKN2A/B status. The CDKN2A/B status was obtained from genome-wide methylation profiling data.6 The arrays allow extraction of the CDKN2A/B status using the conumee Bioconductor package.13
Definition of Outcomes and Covariates of Interest
Progression-free survival (PFS; primary) and overall survival (OS; secondary) were considered parameters of outcome. The analysis was carried out by stratifying the data into six treatment groups: two groups for EORTC trial 26053-22054 and four groups for EORTC trial 22033-26033.
Statistical Approaches
All statistical analyses were performed using the SAS version 9.4 and 3.6.0 R version. Cox regression analysis was performed on the consensus histological features as well as combined molecular parameters and histologic features.
Results
Consensus (ie, 60% agreement on scores) was reached by four out of the seven reviewers and their results were used for the analysis.
The ICC for cell density was good; whereas, the presence of giant cells, calcifications, microcystic/mucoid degeneration, gemistocytes and neoplastic oligodendrocytes was fair. The ICCs of all other parameters remained below 0.30 (Supplemental Table 2).
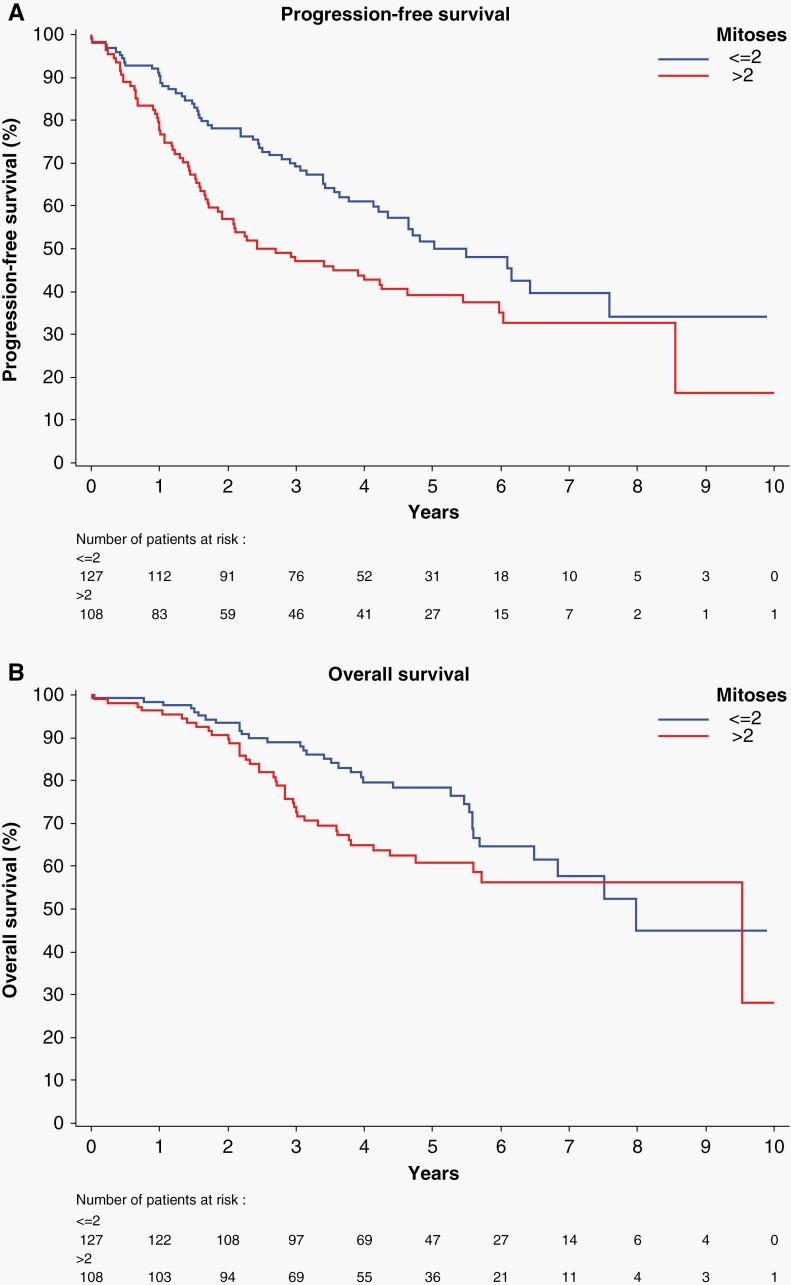
In 91% of cases, the mitotic counts of the consensus reviewers ranged from 1 to 6 mitoses per 10 40× HPF. Univariate analysis showed that mitotic count and the presence of giant cells correlated significantly with PFS (P < .001 and P < .001, respectively) as well as OS (P < .04 and P < .01, respectively) (Supplementary Table 2). For the mitotic count, differences in the discrimination index between step-wise cut-off values was small with the optimal cut-off ≤ 2 and > 2 mitoses (Supplementary Table 3). Univariate analysis revealed that homozygous deletion of CDKN2A/B correlated significantly with PFS and OS (both P < .001) (Supplementary Table 4). The PFS of patients with IDHmut astrocytomas was also significantly influenced by the mitotic count (cut-off 2 mitoses) (P < .001) (Figure 1A). The effect of mitotic count on OS was of marginal significance (P = .07), possibly due to incomplete follow-up (Figure 1B). Multivariate analysis of HD CDKN2A/B, mitotic count and the presence of giant cells showed significant effects of mitotic count and CDKN2A/B status on PFS, whereas OS correlated only with CDKN2A/B (P < .001) (Table 1).
Figure 1.
(A) The mitotic count significantly influences PFS (P < .001) in IDHmut astrocytomas. (B) In IDHmut astrocytomas, the mitotic count has a near-significant (P = .07) effect on the OS.
Table 1.
PFS and OS multivariate analyses according to selected consensus histological features and CDKN2A in patients with IDHmut astrocytoma
| Parameter | PFS | OS | |||
|---|---|---|---|---|---|
| P | HR 95% CI |
P | HR 95% CI |
||
| Mitoses | >2 | 0.04 | 1.62 (1.02–2.56) | 0.26 | 1.39 (0.78–2.47) |
| Giant cells | Yes | 0.27 | 1.51 (0.73–3.15) | 0.65 | 1.23 (0.49–3.09) |
| CDKN2A | HD | 0.0006 | 2.57 (1.50–4.42) | 0.0001 | 3.39 (1.81–6.34) |
Based on 90 PFS events and 54 OS events out of 173 patients with complete histological features and CDKN2A data, mitoses (P = .04) and CDKN2A (P = .0006) significantly influenced PFS. Only HD CDKN2A (P = .0001) significantly influenced the OS.
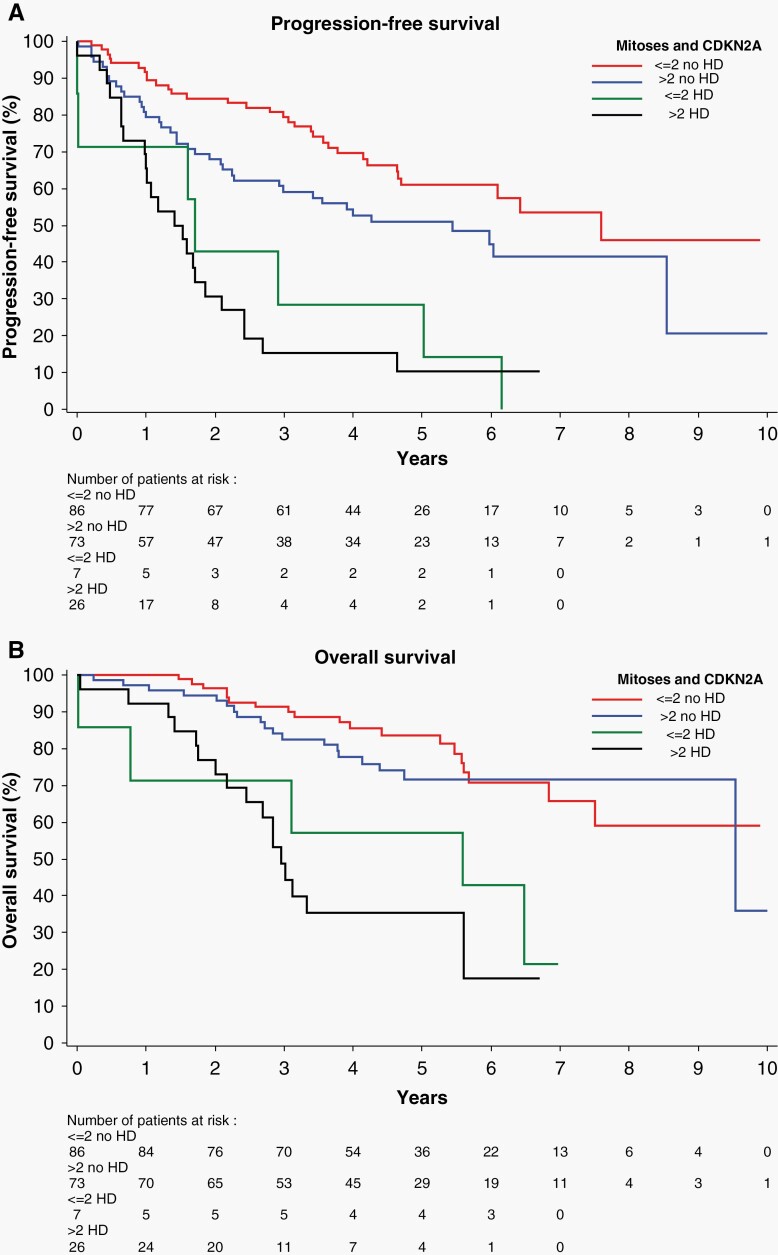
Mitotic index significantly influenced PFS within the subset of tumors without HD CDKN2A/B (P < .001) (Figure 2A). The discriminating effect of mitotic count was absent in gliomas with HD CDKN2A/B (P = .66). The OS was significantly influenced by HD CDKN2A/B status. The mitotic count had no effect on OS, regardless of HD CDKN2A/B status (Figure 2B).
Figure 2.
(A) CDKN2A/B status was known in 192 tumors. HD CDKN2A/B has a significant overall effect on PFS. Mitotic count discriminates PFS in astrocytomas without CDKN2A/B (P = .006), but there is no effect on PFS of gliomas with HD CDKN2A/B (P = .66). (B) HD CDKN2A/B is a significant discriminator in OS. Mitotic index neither correlates with OS in gliomas without HD CDKN2A/B (P = .42), nor with the OS in cases with HD CDKN2A/B (P = .41).
Post hoc analysis was carried out for the effects of mitotic count on the PFS of the gliomas from the two trials separately. For the IDH mutant astrocytomas from CATNON the mitotic count influenced the PFS significantly (P < .001), while conversely, there was no significant influence on PFS of the tumors from EORTC 26053-22033 (P = .61) (Supplementary Figure1A and B).
Cox regression analysis for the effects of mitoses, CDKN2A/B status and age on PFS and OS revealed that age did not affect the PFS (P = .897) or OS (P = .946) in the subsets of IDH mutant astrocytomas of the two trials (Supplementary Table 5A and B). In the IDH mutant astrocytomas from CATNON necrosis was seen in one case and MVP was present in 170 tumors, while in EORTC 26053-22033 these hallmarks of high-grade glioma were absent. Cox regression analysis in CATNON for the features mitoses and MVP, tested with CDKN2A/B status and extent of surgery, revealed that the mitotic count (P = .05), CDKN2A/B status (P < .001) and extent of surgery (P < .016) had significant effects on the PFS but not MVP (P = .115). Only CDKN2A/B status significantly influenced the OS (P < .001) (Supplementary Table 6A and B).
Discussion
The present study addresses the question of whether classic histological parameters still provide assistance in fine-tuning the prognostication of IDHmut astrocytomas. In order to identify such parameters, it is imperative to evaluate classic histological features in the context of molecular data, preferably in a prospective setting controlled for confounding variables.7,14–16 In a consensus evaluation of virtual pathology slides from two EORTC studies on IDH mutant gliomas harboring all histological grades, we were able to identify mitotic count as the only histological feature associated with progression-free survival (PFS) independent of homozygous CDKN2A/B deletion. However, the impact of mitotic count on overall survival (OS) was not significant, most likely due to incomplete follow-up data.
In addition to the major molecular categories of diffuse gliomas, a number of mutations of additional prognostic relevance have been recently identified, basically replacing classic histopathological grading. Importantly, the significance of some of the mutations appears to depend on the molecular context of the tumors. For example, TERT promoter mutations are associated with more favorable outcomes in IDHmut gliomas (independent of 1p/19q codeletion), while the association with poor outcomes in IDHwt gliomas is variable.17,18 In IDHmut astrocytomas, mutations in PIK3R1, PIK3CA, genes of the RB1 pathway, amplification of EGFR and PDGFRA, higher copy number variance (CNV), and homozygous deletions in CDKN2A/B correlate with an unfavorable outcome.6,14,19–22
Although, there is no dispute on the adverse effects of HD CDKN2A/B on survival in IDHmut astrocytomas,5,7,23–25 the effects of HD CDKN2A/B are reportedly dependent on concomitant mutations in TP53 or ATRX.23 Because HD CDKN2A is not found in low-grade IDHmut astrocytomas, it seems to replace the traditional histopathological features in high-grade glioma.5 However, discrepancies regarding the prognostic significance of histopathological parameters in the IDH mutant astrocytomas persist. The presence of necrosis and microvacular proliferation in tumors without HD CDKN2A/B was found to be prognostically significant in one study,5 while only the influence of necrosis was confirmed in another report.7 The presence of necrosis in IDHmut astrocytomas lacking HD CDKN2A/B dispute the significance of this traditional high-grade histological feature.7 Conversely, we found low mitotic counts in seven tumors with HD CDKN2A/B (Figure 2A, green curve). This discrepancy may well be due to sampling errors in the available histologic slides, and may concur with the lack of significant differences in PFS between the two mitotic groups of tumors with HD CDKN2A/B. In the tumors of the present study neither necrosis nor MVP were identified as independent prognostic parameters in IDHmut astrocytomas. Necrosis was seen in only one tumor disqualifying this featute from analysis. MVP was present in 170 IDH mutated astrocytomas (all in the tumors from the CATNON trial) but lost significance to PFS when tested with the mitotic count (Supplementary Table 6A and B). It would be interesting to see how this feature would hold up against the mitotic count in IDH wild type gliomas.
To date, the mitotic count has served as most important discriminator of WHO grade 2 from grade 3 astrocytomas.9 The WHO adopted the grading scheme of Daumas-Duport but unfortunately, clear cut-off values were not specified and for this reason it has remained controversial in prognosticating gliomas.9,26 In retrospective studies the mitotic index has not been found to correlate with survival in IDHmut tumors, even after testing various cut-offs, ranging from 1 to 6 mitoses per 10 HPF.16,27,28 The present finding that consensus mitotic count is prognostic in non-HD CDKN2A/B tumors reflects the importance of obtaining results in controlled settings. Moreover, acceptable inter-observer concordance on mitotic counts can only be reached in a setting of precisely delineated counting conditions.29–31 Our results also confirm the relevance of the mitotic count as opposed to other histologic parameters in the historic grading schemes to delineate grades 2 and 3 astrocytomas.
In conclusion, we identified the mitotic index (cut-off 2 per 10 40× HPF) as an independent prognostic parameter in IDHmut astrocytomas, which therefore should be retained for WHO grading and fine-tuning the prognosis in these patients.
Supplementary Material
Acknowledgments
Mr. A. Nigg, Dept. of Pathology, Erasmus Medical Center, Rotterdam (scanning pathology slides).
Contributor Information
Johan M Kros, Department of Pathology, Laboratory for Tumor Immunopathology, Erasmus Medical Center, Dr. Molewaterplein 40, 3015 GD Rotterdam, The Netherlands.
Elisabeth Rushing, Department of Neuropathology, University Hospital Zurich, University of Zurich, Switzerland.
Aimé L Uwimana, European Organization for Research and Treatment of Cancer Headquarters, Brussels, Belgium.
Aurelio Hernández-Laín, Department of Pathology (Neuropathology), Hospital Universitario 12 de Octubre Research Institute, Madrid, Spain.
Alex Michotte, Medische Oncologie, Oncologisch Centrum, Academisch Ziekenhuis Vrije Universiteit Brussel (AZ-VUB), Brussel, Belgium.
Maysa Al-Hussaini, Department of Pathology and Laboratory Medicine, King Hussein Cancer Centre, Amman, Jordan.
Franck Bielle, Sorbonne Université, AP-HP, Institut du Cerveau, Paris Brain Institute, ICM, Inserm, CNRS, Hôpitaux Universitaires La Pitié Salpêtrière, Charles Foix, Service de Neuropathologie, Paris, France.
Christian Mawrin, Department of Neuropathology, Otto-von-Guericke University, 39120 Magdeburg, Germany.
Gianluca Marucci, Neuropathology Unit, Fondazione IRCCS Istituto Neurologico Carlo Besta, Milan, Italy.
C Mircea S Tesileanu, Department of Neurology, Brain Tumor Center, Erasmus MC Cancer Institute, Rotterdam, The Netherlands.
Roger Stupp, Northwestern University Feinberg School of Medicine, Chicago, Illinois, USA.
Brigitta Baumert, Department of Radiation Oncology, MediClin Robert Janker Clinic and Clinical Cooperation Unit Neurooncology, University of Bonn Medical Centre, Bonn, Germany.
Martin van den Bent, Neurooncology Unit, Erasmus MC Cancer Institute, Rotterdam, The Netherlands.
Pim J French, Neurooncology Unit, Erasmus MC Cancer Institute, Rotterdam, The Netherlands.
Thierry Gorlia, European Organization for Research and Treatment of Cancer Headquarters, Brussels, Belgium.
Funding
No funding specific for this work has been received.
Conflicts of interest
None.
Authorship
Experimental design: JMK; TG; AU; PF; MJvdB. Data production: JMK; ER; AH-L; AM; MA-H; FB; CM; GM; CMST; RS; BM; MJvdB; PF. Data analysis/ interpretation: JMK; AU; TG; PF; MJvdB. Writing the Ms: JMK; ER; PJF.
References
- 1. Louis DN, Perry A, Wesseling P, et al. . The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol. 2021;23(8):1231–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. van den Bent MJ, Tesileanu CMS, Wick W, et al. . Adjuvant and concurrent temozolomide for 1p/19q non-co-deleted anaplastic glioma (CATNON; EORTC study 26053-22054): second interim analysis of a randomised, open-label, phase 3 study. Lancet Oncol. 2021;22(6):813–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Parsons DW, Jones S, Zhang X, et al. . An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ceccarelli M, Barthel FP, Malta TM, et al. . Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164(3):550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Appay R, Dehais C, Maurage CA, et al. . CDKN2A homozygous deletion is a strong adverse prognosis factor in diffuse malignant IDH-mutant gliomas. Neuro-oncology. 2019;21(12):1519–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tesileanu CMS, van den Bent MJ, Sanson M, et al. . Prognostic significance of genome-wide DNA methylation profiles within the randomized, phase 3, EORTC CATNON trial on non-1p/19q deleted anaplastic glioma. Neuro Oncol. 2021;23(9):1547–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shirahata M, Ono T, Stichel D, et al. . Novel, improved grading system(s) for IDH-mutant astrocytic gliomas. Acta Neuropathol. 2018;136(1):153–166. [DOI] [PubMed] [Google Scholar]
- 8. Wijnenga MMJ, French PJ, Dubbink HJ, et al. . Prognostic relevance of mutations and copy number alterations assessed with targeted next generation sequencing in IDH mutant grade II glioma. J Neurooncol. 2018;139(2):349–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Daumas-Duport C, Scheithauer B, O’Fallon J, Kelly P. Grading of astrocytomas. A simple and reproducible method. Cancer. 1988;62(10):2152–2165. [DOI] [PubMed] [Google Scholar]
- 10. Baumert BG, Hegi ME, van den Bent MJ, et al. . Temozolomide chemotherapy versus radiotherapy in high-risk low-grade glioma (EORTC 22033-26033): a randomised, open-label, phase 3 intergroup study. Lancet Oncol. 2016;17(11):1521–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van den Bent MJ, Baumert B, Erridge SC, et al. . Interim results from the CATNON trial (EORTC study 26053-22054) of treatment with concurrent and adjuvant temozolomide for 1p/19q non-co-deleted anaplastic glioma: a phase 3, randomised, open-label intergroup study. Lancet. 2017;390(10103):1645–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cree IA, Tan PH, Travis WD, et al. . Counting mitoses: SI(ze) matters!. Mod Pathol. 2021;34(9):1651–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hovestadt V, Zapatka M. conumee: Enhanced copy-number variation analysis using Illumina DNA methylation arrays: R package version 1.6.0. 2015. http://bioconductor.org/packages/conumee/.
- 14. Aoki K, Nakamura H, Suzuki H, et al. . Prognostic relevance of genetic alterations in diffuse lower-grade gliomas. Neuro Oncol. 2018;20(1):66–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cimino PJ, Holland EC. The molecular landscape of adult diffuse gliomas and relevance to clinical trials. Oncotarget. 2019;10(19):1758–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yoda RA, Marxen T, Longo L, et al. . Mitotic index thresholds do not predict clinical outcome for IDH-mutant astrocytoma. J Neuropathol Exp Neurol. 2019;78(11):1002–1010. [DOI] [PubMed] [Google Scholar]
- 17. Arita H, Matsushita Y, Machida R, et al. . TERT promoter mutation confers favorable prognosis regardless of 1p/19q status in adult diffuse gliomas with IDH1/2 mutations. Acta Neuropathol Commun. 2020;8(1):201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tesileanu CMS, Dirven L, Wijnenga MMJ, et al. . Survival of diffuse astrocytic glioma, IDH1/2 wildtype, with molecular features of glioblastoma, WHO grade IV: a confirmation of the cIMPACT-NOW criteria. Neuro-oncology. 2020;22(4):515–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang RR, Shi ZF, Zhang ZY, et al. . IDH mutant lower grade (WHO Grades II/III) astrocytomas can be stratified for risk by CDKN2A, CDK4 and PDGFRA copy number alterations. Brain Pathol. 2020;30(3):541–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brat DJ, Aldape K, Colman H, et al. . cIMPACT-NOW update 5: recommended grading criteria and terminologies for IDH-mutant astrocytomas. Acta Neuropathol. 2020;139(3):603–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li KK, Shi ZF, Malta TM, et al. . Identification of subsets of IDH-mutant glioblastomas with distinct epigenetic and copy number alterations and stratified clinical risks. Neurooncol Adv. 2019;1(1):vdz015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Draaisma K, Wijnenga MM, Weenink B, et al. . PI3 kinase mutations and mutational load as poor prognostic markers in diffuse glioma patients. Acta Neuropathol Commun. 2015;3(1):88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reis GF, Pekmezci M, Hansen HM, et al. . CDKN2A loss is associated with shortened overall survival in lower-grade (World Health Organization Grades II-III) astrocytomas. J Neuropathol Exp Neurol. 2015;74(5):442–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Carstam L, Corell A, Smits A, et al. . WHO Grade loses its prognostic value in molecularly defined diffuse lower-grade gliomas. Front Oncol. 2021;11(10):803975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fortin Ensign SP, Jenkins RB, Giannini C, et al. . Translational significance of CDKN2A/B homozygous deletion in IDH-mutant astrocytoma. Neuro Oncol. 2022:noac205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Louis DN, Perry A, Reifenberger G, et al. . The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 27. Duregon E, Bertero L, Pittaro A, et al. . Ki-67 proliferation index but not mitotic thresholds integrates the molecular prognostic stratification of lower grade gliomas. Oncotarget. 2016;7(16):21190–21198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Olar A, Wani KM, Alfaro-Munoz KD, et al. . IDH mutation status and role of WHO grade and mitotic index in overall survival in grade II-III diffuse gliomas. Acta Neuropathol. 2015;129(4):585–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Scolyer RA, Shaw HM, Thompson JF, et al. . Interobserver reproducibility of histopathologic prognostic variables in primary cutaneous melanomas. Am J Surg Pathol. 2003;27(12):1571–1576. [DOI] [PubMed] [Google Scholar]
- 30. Coons SW, Pearl DK. Mitosis identification in diffuse gliomas: implications for tumor grading. Cancer. 1998;82(8):1550–1555. [PubMed] [Google Scholar]
- 31. Kros JM, Huizer K, Hernandez-Lain A, et al. . Evidence-based diagnostic algorithm for glioma: analysis of the results of pathology panel review and molecular parameters of EORTC 26951 and 26882 trials. J Clin Oncol. 2015;33(17):1943–1950. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.