Abstract
Autophagy is important for cellular homeostasis and to prevent the abnormal accumulation of proteins. While many proteins that comprise the canonical autophagy pathway have been characterized, the identification of new regulators may help understand tissue and/or stress-specific responses. Using an in-silico approach, we identified Striatin interacting protein (Strip), MOB kinase activator 4, and fibroblast growth factor receptor 1 oncogene partner 2 as conserved mediators of muscle tissue maintenance. We performed affinity purification-mass spectrometry (AP-MS) experiments with Drosophila melanogaster Strip as a bait protein and copurified additional Striatin-interacting phosphatase and kinase (STRIPAK) complex members from larval muscle tissue. NUAK family kinase 1 (NUAK) and Starvin (Stv) also emerged as Strip-binding proteins and these physical interactions were verified in vivo using proximity ligation assays. To understand the functional significance of the STRIPAK-NUAK-Stv complex, we employed a sensitized genetic assay combined with RNA interference (RNAi) to demonstrate that both NUAK and stv function in the same biological process with genes that encode for STRIPAK complex proteins. RNAi-directed knockdown of Strip in muscle tissue led to the accumulation of ubiquitinated cargo, p62, and Autophagy-related 8a, consistent with a block in autophagy. Indeed, autophagic flux was decreased in Strip RNAi muscles, while lysosome biogenesis and activity were unaffected. Our results support a model whereby the STRIPAK-NUAK-Stv complex coordinately regulates autophagy in muscle tissue.
INTRODUCTION
Balancing protein synthesis and protein degradation is crucial to maintain normal cellular functions. In addition to the routine turnover of proteins, cells must have the ability to rapidly respond to internal or external signals by modulating the dynamics of protein turnover or rerouting proteins through other degradation pathways. Autophagy is one such degradation pathway that degrades bulk cellular components in the lysosome (Zhao and Zhang, 2019; Xia et al., 2021). Activation of receptors at the cell surface, oxidative stress, and starvation can trigger endocytosis and autophagy to degrade cellular components for energy (Ganesan and Cai, 2021).
Multiple types of autophagy, each with its own subtleties, allow the cell to selectively or nonselectively degrade cellular material. Clearance of organelles or proteins generally occurs through macroautophagy, where a double-membraned structure called the phagophore forms de novo and sequesters cellular components within a closed autophagosome (Parzych and Klionsky, 2014; Hansen et al., 2018). During maturation, autophagosomes can merge with existing late endosomes (LEs)/multivesicular bodies (MVBs) to form a hybrid structure called the amphisome before fusion with lysosomes (Ganesan and Cai, 2021). Selectivity in the degradation process occurs when autophagy receptors recruit and link particular cargo to the autophagy machinery. One example is chaperone-assisted selective autophagy , whereby recognition of the actin crosslinking protein Filamin by the Bcl-2–associated athanogene 3 (Bag3)-Heat-shock cognate 70 complex (Hsc70) recruits p62/Sequestosome-1 (SQSTM1) (Arndt et al., 2010; Kettern et al., 2010; Ulbricht et al., 2013; Höhfeld et al., 2021). This adapter protein then binds to Autophagy-related 8a (Atg8a)/microtubule-associated protein 1 light chain 3 (MAP1LC3/LC3) to coordinate autophagosome formation and eventual lysosomal fusion (Bjørkøy et al., 2005; Komatsu et al., 2007; Pankiv et al., 2007).
Even though autophagy occurs in every cell, the importance of this process in muscle tissue is exemplified by the number of muscle disorders that are linked to mutations in genes that encode for autophagy components and/or the abnormal accumulation of autophagic or lysosomal vacuoles within diseased muscle tissue (Castets et al., 2016; Margeta, 2020; Franco-Romero and Sandri, 2021; Xia et al., 2021). For example, mutations in the lysosomal marker lysosomal Associated Membrane Protein 2 (LAMP2) cause Danon disease, characterized by the accumulation of autophagic vacuoles (Tanaka et al., 2000; Rowland et al., 2016; Kazuma, 2021). Mutations in vacuolar ATPase assembly factor VMA21 result in excessive autophagy and patients with Duchenne muscular dystrophy exhibit lower levels of the microtubule-associated protein 1A/1B-light chain 3-II (LC3 II) and excess p62 (De Palma et al., 2012; Ramachandran et al., 2013; Munteanu et al., 2017). Moreover, mutations in human Filamin C and Bag3 result in Myofibrillar Myopathy (MFM), a progressive disease in which protein aggregates and muscle-tissue disintegration are prevalent in biopsies (Batonnet-Pichon et al., 2017; Fichna et al., 2018; Guglielmi et al., 2018; Luo et al., 2020). Modeling of MFM diseases in zebrafish or mice show that autophagic activity is impaired, thus linking abnormal protein accumulation to a deficiency in protein turnover (Batonnet-Pichon et al., 2017). Identifying additional proteins that mediate autophagy will help understand the etiology and progression of diseases linked to this cellular process.
The striatin-interacting phosphatase and kinase (STRIPAK) multi-subunit signaling complex has emerged as a central component in the coordination of intertwined signaling networks in a wide variety of cellular processes (Hwang and Pallas, 2014; Shi et al., 2016; Kück et al., 2019). Striatin is the founding member of this complex and acts as a scaffolding hub for other components, which together form one of the four families of regulatory subunits for Protein phosphatase 2A (PP2A) (Sents et al., 2013). The core proteins that comprise the STRIPAK complex are evolutionarily conserved across diverse organisms. However, their varied subcellular distribution suggests that STRIPAK proteins may form subcomplexes that target distinct substrates in different intracellular pathways that ultimately influence cell growth, cell division, and actin cytoskeleton organization, among others. Recent papers have uncovered roles for STRIPAK-complex proteins in myofibril assembly in zebrafish and in Drosophila melanogaster (D. melanogaster) adult flight muscles (Kaya-Çopur et al., 2021; Berger et al., 2022), thus highlighting additional functions for members of this protein complex.
Here we uncover components of the STRIPAK complex that affect muscle development through an in-silico screening approach in the flour beetle Tribolium castaneum (T. castaneum). Targeted RNA interference (RNAi) against select STRIPAK proteins in D. melanogaster revealed morphological abnormalities in larval muscles. AP-MS studies confirmed the presence of additional STRIPAK complex members in muscle tissue, including those implicated in autophagy. Detailed analysis of a core STRIPAK member, Striatin interacting protein (Strip), further revealed a role for the STRIPAK complex in the regulation of autophagic flux in muscle tissue.
RESULTS
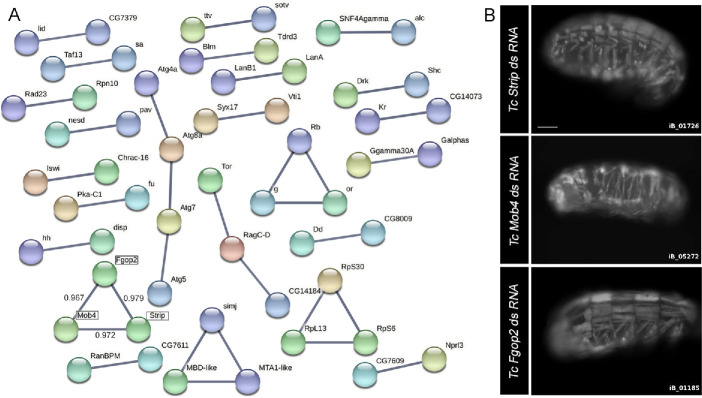
Taking advantage of the evolutionary conservation of genes essential for muscle development and maintenance, we screened iBeetle-Base, a database of Tribolium castaneum (Tc) RNAi phenotypes using the search term “larval muscle irregular” to broadly identify genes that alter muscle morphology (Dönitz et al., 2015; Schmitt-Engel et al., 2015; Dönitz et al., 2017). Out of ∼8500 individual protein-encoding genes targeted using dsRNA injections in the pig-19 line expressing green fluorescent protein (GFP) in all larval somatic muscles (Schultheis et al., 2019), this in-silico approach uncovered 592 genes with abnormal muscle phenotypes (Supplemental Figure S1). After eliminating genes that failed to form Cytoscape interaction networks, we focused on the remaining 55 candidates (Figure 1A). Analysis of representative images for each of these genes in iBeetle-Base revealed similar muscle phenotypes for RNAi knockdown of TC010759 (Tc-Strip), TC012571 (Tc-Mob4), and TC007423 (Tc-Fgop2) (Figure 1B). These abnormalities were primarily characterized by gaps in the stereotypical embryonic muscle pattern, indicative of missing and/or detached muscles. Representative images in the iBeetle database were obtained from a first-pass screen without systematic repetitions (Schmitt-Engel et al., 2015). Despite this limitation, the similarity in phenotypes for three different members of the STRIPAK complex, combined with physical interactions identified by Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) network analysis provided the rationale for further analysis.
FIGURE 1:
Network analysis uncovers interacting genes in T. castaneum. (A) Cytoscape rendering of the 55 genes identified in the iBeetle-base in-silico screen that exhibits one or more known interactions (see Supplemental Figure S1). (B) Representative images of muscle morphology defects in T. castaneum late-stage embryos visualized with GFP after injection of dsRNA for Strip, Mob4, or Fgop2 (https://ibeetle-base.uni-goettingen.de/). Reproduced with permission. Scale bar, 50 µm.
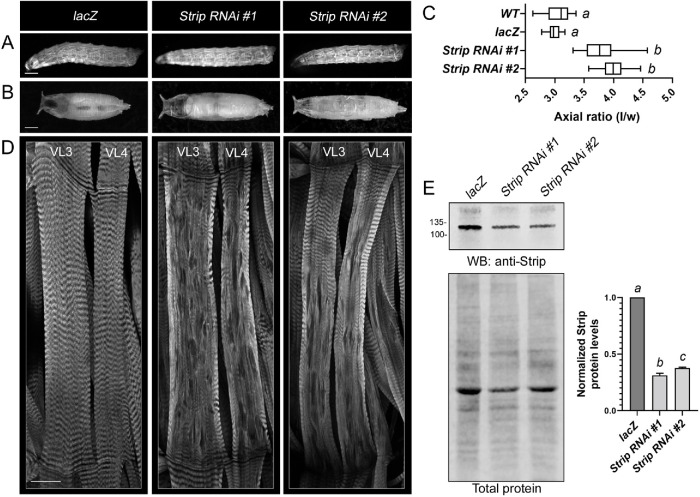
To determine whether these proteins play a conserved role in D. melanogaster muscle development, we induced mRNA knockdown of Strip, MOB kinase activator 4 (Mob4), or Fibroblast growth factor receptor 1 oncogene partner 2 (Fgop2) using previously published upstream activating sequence (UAS)-RNAi lines for each gene under control of the Mef2-Gal4 driver (Brand and Perrimon, 1993). Mef2>lacZ controls showed an overall normal muscle pattern as visualized with Myosin heavy chain (Mhc)-GFP in the late third larval (L3) instar stage (Figure 2A). At this whole organismal level, missing muscles were occasionally present in Strip RNAi L3 larvae. However, a role for Strip in muscle function became more obvious during the L3 to pupal transition when body wall muscles contract to form a stereotypical pupal size. Induction of Strip RNAi using two independent UAS-hairpin lines caused longer pupal cases (Figure 2B), whereby the pupal axial ratio (length/width) was increased by ∼30% compared with Mef2>lacZ controls (Figure 2C).
FIGURE 2:
Induction of Strip RNAi in muscle tissue compromises muscle structure and function. Mef2-Gal4 driven expression of UAS-lacZ control or two independent UAS-Strip RNAi lines. (A) Whole mount images of L3 larvae visualized with MHC-GFP reveal no obvious difference between the control and Strip RNAi muscle patterns. Anterior is to the left. (B) Images of stereotypical pupal cases representative of Mef2>lacZ controls or elongated pupal cases upon a reduction of Strip. Anterior is to the left. (C) Box and whiskers plot showing the relative axial ratio (length/width) of pupal case size in control vs Strip RNAi muscles. N ≥20. Different letters indicate statistical significance between groups. (D) Phalloidin labeling for F-actin reveals normal, repeated sarcomeres in VL3 and VL4 control muscles (left panel). This striated pattern is disrupted in Strip RNAi muscles. All images are maximum intensity projections with anterior towards the top. (E) Representative Western blot of Strip protein levels upon induction of Strip RNAi in L3 muscles (top panel). Total protein loading control (bottom panel). Different letters indicate statistical significance between groups. Scale bars, 1 mm (panels A and B), 50 µm (panel D).
To determine whether obvious morphological differences may explain the failure of muscle contraction, we examined L3 muscles before the pupal stage. Phalloidin labeling of filleted Mef2>lacZ control larvae revealed a striated pattern of F-actin in the easily imaged ventral longitudinal 3 (VL3) and 4 (VL4) muscles (Figure 2D). Muscle-induced Strip RNAi disrupted this normal F-actin morphology, typified by a loss of the repeated sarcomere pattern and small regions that lacked phalloidin staining. Western blot analysis showed that Strip protein levels were reduced by ∼60-70% in Strip RNAi muscle carcasses (Figure 2E).
Identification and functional characterization of muscle STRIPAK complex components
RNAi knockdown of Mob4 or Fgop2 mRNA in muscle tissue resulted in stereotypical pupal case sizes and largely normal muscle morphology with occasional areas of hypercontraction (Supplemental Figure S2, A–C). Four possibilities, which are not mutually exclusive, may explain the absence of Strip RNAi-like phenotypes. While reduced transcript levels of Mob4 or Fgop2 mRNA are evident by quantitative RT-PCR analysis (Supplemental Figure S3), we cannot rule out the possibility that Mob4 or Fgop2 protein levels are not decreased. Stage-specific and/or species-specific differences may explain the phenotypic variations between beetles and flies. At present, we cannot conclude if Mob4 or Fgop2 are required earlier in development because we did not analyze embryonic or early larval phenotypes. Recent comparisons between T. castaneum and D. melanogaster have revealed more divergence in gene function governing basic developmental processes, including myogenesis, than previously thought (Schultheis et al., 2019; Hakeemi et al., 2022). Thus, species-specific differences may at least partially account for a lack of phenotype in Mob4 or Fgop2 RNAi L3 muscles. Finally, Strip may function independently of the STRIPAK complex in L3 muscle tissue.
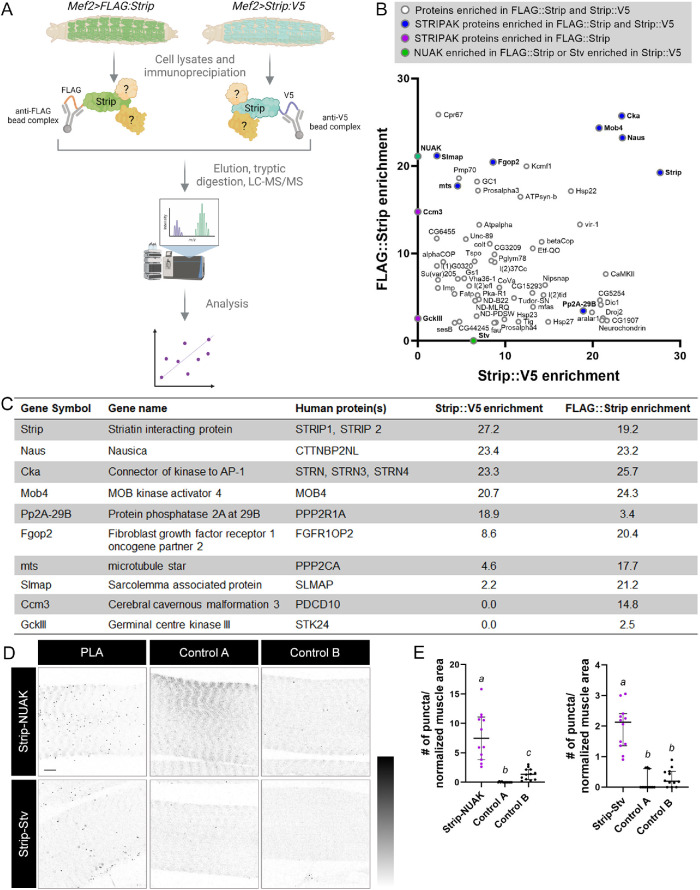
To identify proteins that physically interact with Strip in this context, we developed an AP-MS approach (Figure 3A). N-terminally tagged FLAG::Strip or C-terminally tagged Strip::V5 were independently expressed in muscle under Gal4/UAS control and isolated from L3 larvae using affinity-tag-specific beads. The resulting protein complexes were trypsin digested and subjected to MS analysis. Peptide coverage of the Strip bait proteins varied from 49.6 to 77% in three independent experiments (Supplemental Table S1). Enrichment was calculated by averaging the label-free quantification (LFQ) intensity values for each experimental group (Mef2>FLAG::Strip or Mef2>Strip::V5) minus the average values for control groups (Mef2>lacZ). Using a cutoff of 2.0 as the minimum enrichment value, a total of 195 proteins copurified with FLAG::Strip and 336 proteins copurified with Strip::V5.
FIGURE 3:
Co-purification of STRIPAK complex proteins from larval muscle tissue. (A) Schematic of the AP-MS strategy. Either N-terminally tagged FLAG::Strip or C-terminally tagged Strip::V5 fusion proteins were expressed in larval body wall muscles with Mef2-Gal4. After the preparation of cell lysates and immunoprecipitation with either anti-FLAG or anti-V5 beads, the resulting protein complexes were subjected to MS/MS and quantitative peptide identification. Mef2>lacZ lysates were used for control pulldowns. Created with BioRender.com (B) Enrichment plot showing proteins that copurified with FLAG::Strip vs lacZ (y-axis) or Strip::V5 vs lacZ (x-axis). The enrichment difference was calculated by subtracting the average peptide intensities for each protein in the tagged pulldown minus controls (Supplemental Table S1). (C) Canonical proteins of the STRIPAK complex that copurified with the bait protein Strip from larval muscle tissue. The corresponding human proteins and relative abundance in MS experiments are listed. (D) Representative confocal single-plane images corresponding to PLA experiments characterizing physical interactions of complexes containing Strip-NUAK (top panels) or Strip-Stv (bottom panels) within VL3 muscles. The PLA experiments were performed on Mef2>FLAG::Strip muscles or Mef2>Strip::V5 muscles. Control A includes no primary antibodies and only PLA reagents on either Mef2>FLAG::Strip muscles or Mef2>Strip::V5 muscles. Control B includes both primary and PLA reagents on WT muscles. Images were inverted for visualization. (E) Dot plot depicting the number of PLA puncta/normalized muscle areas in the indicated genotypes. N ≥ 12. Genotypes with different letters are statistically different. Scale bars, 10 µm (panel D).
We next plotted proteins that were present in both datasets based on their relative enrichment values (Figure 3B). Both Mob4 and Fgop2 had enrichment values >20 in the FLAG::Strip pulldowns or >9 in the Strip::V5 pulldowns, thus validating our AP-MS approach and STRING-Cytoscape interactions. We also confirmed the presence of five additional STRIPAK components, Connector of kinase to AP-1 (Cka), Nausicaa (Naus), Sarcolemma Associated Protein (Slmap), microtubule star (mts), and PP2A at 29B (PP2A-29B) in both datasets. Two STRIPAK proteins, Cerebral cavernous malformation 3 (Ccm3) and Germinal center kinase III (GckIII) emerged only in the FLAG::Strip pulldowns, possibly indicating that the C-terminal V5 tag interfered with binding to Strip. This data is summarized in Figure 3C and Supplemental Table S1.
Two additional proteins that caught our interest as possible Strip-binding proteins were NUAK and Starvin (Stv). NUAK was enriched ∼20× in the FLAG::Strip dataset, while Stv was present ∼7× in our V5::Strip samples. Because we previously published a role for the NUAK-Stv complex in autophagy (Brooks et al., 2020), we next verified the presence of Strip-NUAK or Strip-Stv complexes using the proximity ligation assay (PLA) approach. Rabbit antibodies raised against NUAK (Bawa et al., 2020) or Stv (Coulson et al., 2005; Arndt et al., 2010) were used in conjunction with commercially available anti-FLAG or anti-V5 antibodies generated in mice (Figure 3D). The number of puncta corresponding to a Strip-NUAK complex was increased in Mef2>FLAG::Strip muscles compared with two independent controls (Figure 3E). Control experiment A was performed on Mef2>FLAG::Strip muscles with no primary antibody and only PLA reagents. Control experiment B used both primary antibodies and PLA reagents on wild-type (WT) muscles lacking a FLAG-tagged version of Strip. Similar results were obtained for Mef2>Strip::V5 muscles, whereby more puncta corresponding to a Strip-Stv complex were observed than in control experiments (Figure 3E). Together, our AP-MS and PLA results confirm that the STRIPAK complex, together with NUAK and Stv, is expressed in muscles and can be copurified with Strip.
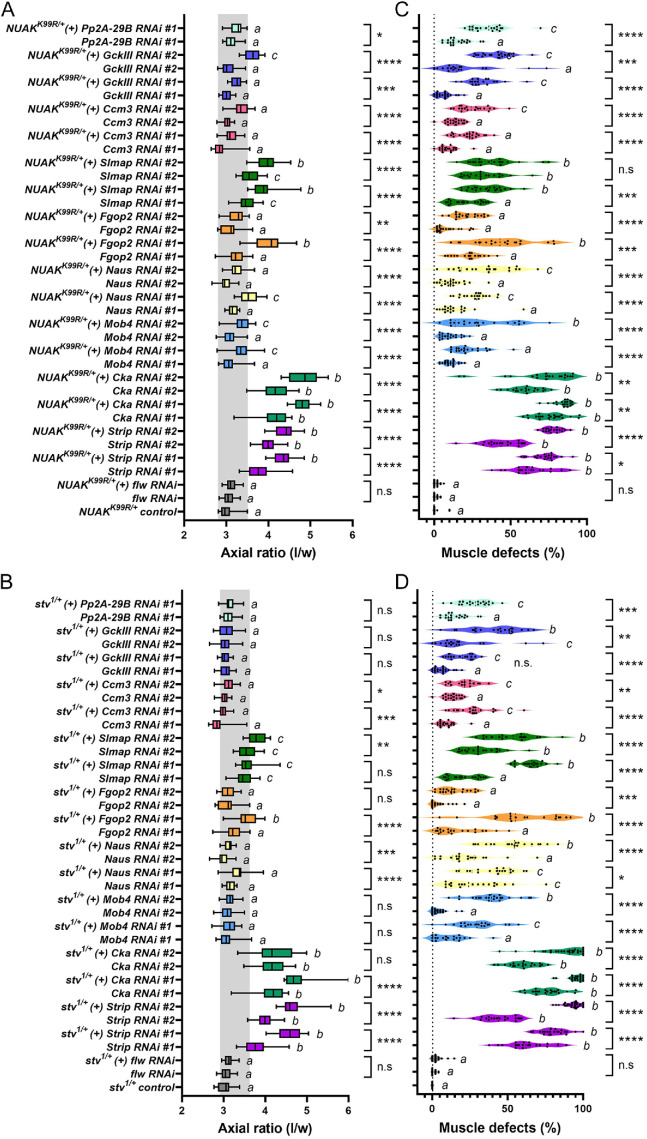
To determine whether individual STRIPAK components function in NUAK and/or Stv-mediated processes, we utilized two separate assays that take advantage of sensitized genetic backgrounds and available RNAi lines. Stable fly stocks of the genotype NUAKK99R±, Mef2-Gal4 and stv1±, Mef2-Gal4 were mated with two independent UAS-RNAi lines for each STRIPAK complex protein, except Pp2A-29B (as other RNAi lines were lethal before the L3 stage). First, we assessed muscle contraction during the larval to pupal transition as described earlier (LaBeau-DiMenna et al., 2012; Green et al., 2016; Brooks et al., 2020). Heterozygous NUAKK99R± or stv1± larvae underwent normal muscle contraction and exhibited a pupal axial ratio comparable with WT and Mef2>lacZ controls (Figure 2C). RNAi-directed knockdown of Strip alone showed decreased muscle contraction (Figure 2C and Figure 4, A and B) and removal of a single copy of NUAK or stv enhanced this phenotype, demonstrating that Strip functions in the same biological process as the NUAK-Stv complex (Figure 4, A and B). The same results were obtained for the knockdown of Cka or Naus, whereby muscle-contraction deficits of either gene alone were exacerbated in the sensitized NUAKK99R± or stv1± backgrounds. While all RNAi lines used in these experiments showed knockdown of mRNA transcripts (Supplemental Figure S3), some of the STRIPAK complex members exhibited milder and more variable results in the phenotype induced by RNAi. All pair-wise combinations of RNAi lines (e.g., Mef2>Strip RNAi #1 vs. NUAKK99R±, Mef2>Strip RNAi #1) showed a statistically significant enhancement of muscle-contraction defects upon removal of a single copy of NUAKK99R (Figure 4A), but only a single RNAi line directed against Fgop2 and Slmap and none against GckIII or Pp2A-29B reached significance values of p < 0.05 (Figure 4B). Neither Mob4 RNAi line enhanced muscle contraction defects in the stv1/+ background.
FIGURE 4:
Functional interactions between NUAK and stv with STRIPAK complex members. (A–D) Each RNAi line was expressed alone in muscle tissue using Mef2-Gal4 or in a sensitized NUAKK99R (A and C) or stv1 (B and D) heterozygous background. Two RNAi lines were selected against each STRIPAK complex gene except for PP2A-29B as other PP2A-29B RNAi lines tested were lethal before the L3 stage. (A and B) Box and whisker plots of muscle contractility are depicted by the axial ratio (length/width) of individual pupae of the indicated genotypes. A shaded gray box indicates the min to max values for controls. (C and D) Scatter plots depicting muscle defects in L3 larvae (aberrant F-actin patterning, gaps between myofibrils, and detached muscles). Each data point presents the percentage of muscles with abnormal morphology in individual larvae. N ≥20 for pupal case morphology and N ≥18 for muscle morphology abnormalities. Genotypes with different letters are statistically different between groups (p < 0.05). ****, p < 0.0001; ***, p < 0.001; **, p < 0.01; * p < 0.05; n.s, not significant.
Measurements of muscle contractility during the larval to pupal transition is a relatively stringent assay that reveals the failure of multiple muscles to contract. Thus, we also assessed the morphology of individual L3 muscles with the rationale that minor structural defects may be present, but not severe enough to cause functional deficits in overall contractile ability. Each genotype displayed morphological abnormalities that were enhanced in the NUAKK99R or stv1 sensitized backgrounds, although there was some variability among RNAi lines (Figure 4, C and D). Aside from Strip, muscle-directed RNAi against either Cka line showed strong muscle contraction and sarcomere morphology phenotypes in our quantitative analysis. Indeed, F-actin patterning in Cka RNAi muscles phenocopies Strip phenotypes (Supplemental Figure S4). RNAi knockdown of the phosphatase flapwing (flw) did not exhibit a phenotype on its own or in combination with either the NUAKK99R or stv1 alleles, thus serving as a negative control for all experiments. These data together firmly demonstrate a requirement for the STRIPAK complex in muscle contraction and the maintenance of the integrity of muscle tissue in concert with the NUAK-Stv complex.
Markers associated with autophagy are increased in Strip RNAi muscles
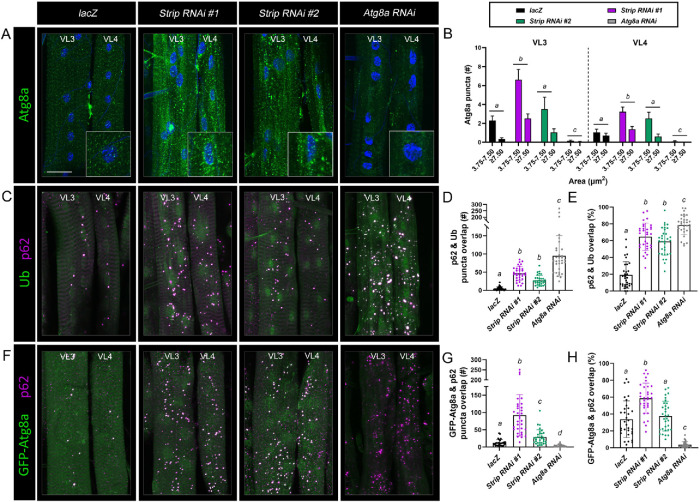
Because the loss of NUAK or Stv impairs autophagy in muscle tissue (Brooks et al., 2020), we next examined the relative distribution and levels of the autophagosome marker Atg8a upon induction of Strip RNAi. In control Mef2>lacZ muscles, fluorescence corresponding to Atg8a puncta was visible at low levels (Figure 5A). RNAi-silencing of Strip with either RNAi line not only increased the overall number of puncta but also shifted the sizes of each Atg8a(+) puncta to greater than (gt) 3.75 μm2 (Figure 5B). While some of these larger puncta were located near nuclei (see inset), a large proportion was broadly distributed throughout the cell in Strip RNAi muscles. The inability to visualize puncta corresponding to autophagosomes in Atg8a RNAi muscles confirmed the specificity of the anti-Atg8a antibody (Figure 5A).
FIGURE 5:
p62-Ub and p62-Atg8a complexes accumulate in Strip RNAi muscles. (A) Mef2-Gal4 flies were mated with UAS-lacZ, UAS-Strip RNAi #1, UAS-Strip RNAi #2, or UAS-Atg8a RNAi and immunostained to assess the relative amount of Atg8a. Maximum intensity projection images of muscles VL3 and VL4 from L3 larvae reveal an increase in the size and number of Atg8a puncta (green). Inset depicts the area around a single nucleus. (B) Bar chart quantification shows an increase in the overall size and number of Atg8a(+) structures in Strip RNAi muscles compared with lacZ controls. N ≥ 15. Genotypes with different letters are statistically different. (C) Maximum intensity projections of VL3 and VL4 muscles immunostained for p62 (magenta) and Ub (green) in lacZ control, Strip RNAi, or Atg8a RNAi muscles. There is a clear increase in both p62 and Ub puncta upon knockdown of Strip or Atg8a. (D and E) Scatter bar graphs reveal an increase in the number of puncta (D) and the percentage of puncta (E) in VL3 and VL4 muscles that show overlap between p62 and Ub in Strip or Atg8a RNAi muscles compared with controls. N ≥32. Genotypes with different letters are statistically different. (F) VL3 or VL4 muscles from L3 larvae expressing GFP-Atg8a (green) and labeled with anti-p62 (magenta) in the indicated genotypes. There are more p62(+) puncta that show an overlap with GFP-Atg8a in Strip knockdown muscles than in lacZ control muscles. Atg8a RNAi shows increased p62, but a loss of GFP-Atg8a staining. (G and H) Scatter bar plots of puncta in VL3 and VL3 muscles. There is an obvious increase in the number of puncta (G) and the percentage of puncta (H) that show overlap between p62 and GFP-Atg8a in Strip RNAi line #1 muscles compared with controls or Atg8a RNAi. N≥32. Genotypes with different letters are statistically different. Scale bars, 25 µm (panels A, C, and F).
Refractory to sigma P (ref(2)p)/p62 is an adapter protein that simultaneously interacts with ubiquitinated cargo and Atg8a to ensure the delivery of autophagic substrates to the autolysosome for degradation (Bjørkøy et al., 2005; Komatsu et al., 2007; Pankiv et al., 2007). Thus, increased p62 protein would be predicted upon a decrease in Strip function. Indeed, the total number and relative size of p62-containing structures were increased in Strip RNAi muscles (Supplemental Figure S5A). Fewer than 10 puncta corresponding to p62 were present within a single VL3 or VL4 muscle and the area of most puncta was between 3.75 and 7.50 μm2 (Supplemental Figure S5B). RNAi-silencing of Strip induced abundant p62 signal within each muscle and a concomitant increase in the sizes of each punctum, of which a substantial proportion was also enlarged gt 3.75 μm2. To confirm this increase in p62, we also examined the quantity and relative size of p62-containing structures in control and Strip RNAi muscles using a p62-GFP fusion protein (Supplemental Figure S5C). Similar increases in puncta number and size were observed for p62-GFP, with a noticeable increase in the area of larger puncta (>7.5 μm2) (Supplemental Figure S5D). Some of the larger, irregular p62-GFP structures (arrows) may correspond to aggregates of p62 due to overexpression (Pircs et al., 2012).
If the clustering and delivery of ubiquitinated proteins to the autophagosome are dependent upon p62, then both Ub and Atg8a should overlap with p62 staining. To test this, we first immunostained L3 muscles with anti-p62 and anti-Ub antibodies. Consistent with anti-p62 staining alone in Supplemental Figure S5C, the number of p62(+) puncta was increased in Strip RNAi muscles (Figure 5C). The total number of puncta that stained positive for both Ub and p62 increased about fivefold in Strip RNAi #1 and ∼2.5fold in Strip RNAi #2 compared with controls (Figure 5D). When quantitated as percentage overlap, the ∼20% observed in lacZ controls increased to ∼60% upon a reduction in Strip (Figure 5E). This trend was even more pronounced upon knockdown of Atg8a, in which Ub-p62 conjugates form but fail to recruit Atg8a thereby blocking delivery to the lysosome.
To examine the overall distribution of Ub-tagged proteins, soluble and insoluble fractions were prepared from L3 muscle-tissue fillets followed by Western blotting with an anti-Ub antibody that recognizes both mono-and polyubiquitin, but not free ubiquitin. Elevated levels of ubiquitinated species were observed in both the soluble and insoluble fractions obtained from Strip RNAi #1 and Atg8a RNAi muscles, with a mild increase in the insoluble portion in Strip RNAi #2 muscles (Supplemental Figure S6A). Ubiquitin chain assembly on outer membrane proteins of damaged mitochondria recruits autophagic machinery for eventual lysosomal degradation (Palikaras et al., 2018; Onishi et al., 2021). To assess if Strip influences mitochondrial turnover, we examined the colocalization of mitochondria and p62-GFP (Supplemental Figure S6B). Overall, mitochondrial morphology was similar in control and Strip RNAi muscles with no indication of swollen mitochondria that fail to undergo autophagic clearance due to their large size (Wang et al., 2016). Moreover, mitochondria and p62-GFP staining did not overlap, as would be expected if ubiquitinated cargo on damaged organelles is tagged for destruction. Thus, it seems likely that the primary substrates of Ub and p62 targeting are not mitochondrial proteins.
We next examined the overlap between p62 and Atg8a. Because antibodies against each protein were generated in the same species and cannot effectively be used in double immunostaining experiments, we took advantage of the GFP-tagged Atg8a fusion protein (GFP-Atg8a). This protein faithfully recapitulates results obtained with anti-Atg8a antibody staining (Figure 5, A and B) as we observed an increase in the number and size of GFP-Atg8a (+) structures corresponding to autophagosomes (Supplemental Figure S5, E and F). Similar to the results for Ub-p62 conjugates, the total number and percentage of puncta that stained positive for p62 and GFP-Atg8a were increased in Strip RNAi muscles (Figure 5F). This result was most striking in RNAi line #1, where the number of puncta that showed overlap increased from an average of 10 puncta in lacZ control muscles to nearly 100 puncta in Strip RNAi #1 (Figure 5G) and the percentage of puncta that overlapped in Strip RNAi was nearly doubled (Figure 5H). Together these data confirm that loss of Strip causes an accumulation in ubiquitinated substrates that get recognized by p62 and recruit Atg8a.
Loss of Strip disrupts autophagic flux
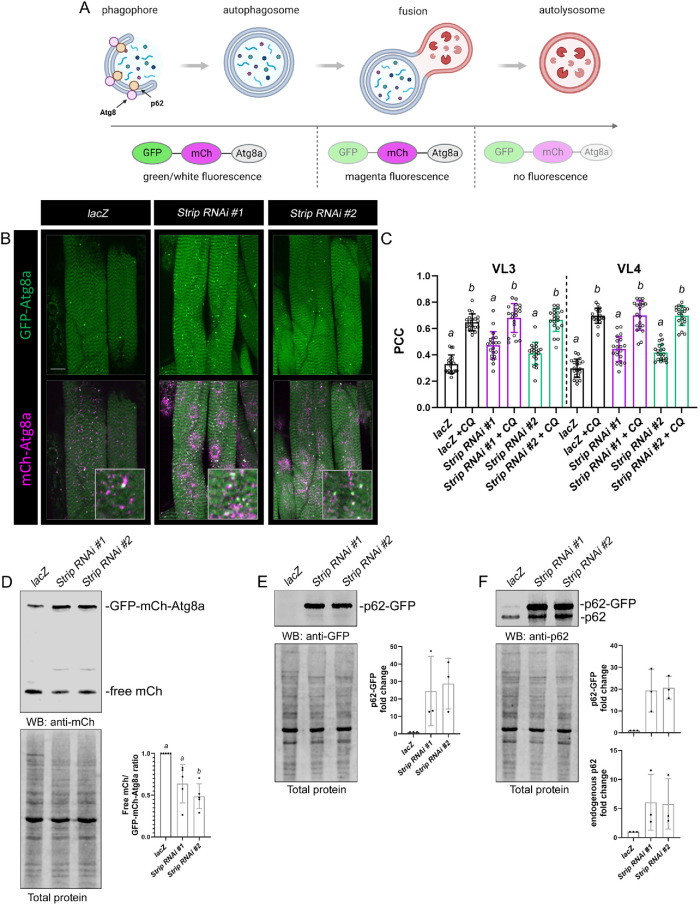
The coordinated increase in Ub, p62, and Atg8a suggests that the autophagy pathway is blocked. To examine autophagic flux, or the overall progression from the formation of an autophagosome to the degradation of cargo in the autolysosome, we next took advantage of the differential pH sensitivity of fluorescent tags in the GFP-mCherry (mCh)-Atg8a tandem fusion protein (Mauvezin et al., 2014; Nagy et al., 2015; Lőrincz et al., 2017a). In autophagosomes, both tags emit fluorescent light resulting in cofluorescence (white puncta) (Figure 6A). However, the fusion of autophagosomes with LEs or lysosomes exposes the fluorescent tags to an acidic environment where the GFP fluorescence signal is quenched. mCh signal persists until all proteins within the autolysosome are degraded. Thus, the GFP signal labels the autophagosome before its fusion with the lysosome.
FIGURE 6:
Knockdown of Strip impedes autophagic flux. (A) Schematic showing how the pH-sensitive GFP-mCh-Atg8a tandem fusion protein is used to monitor autophagy. Created with BioRender.com. (B) Stacked confocal images of the GFP-mCh-Atg8a fusion protein in control or Strip RNAi muscles. Autophagosomes are marked by GFP-Atg8a (green or white puncta) and lysosomal compartments are labeled by mCh-Atg8a (magenta). Note the increase in autophagosomes upon a decrease in Strip. Inset shows the area around a single nucleus. (C) The scatter bar plot depicts the PCC, or the amount of overlap between green and white signals corresponding to autophagosomes. N ≥20. (D) mCh conversion assay assessed by Western blotting in muscles expressing UAS-GFP-mCh-Atg8a. Probing with an antibody against mCh shows a decrease in the amount of liberated mCh in Strip RNAi muscles (top panel). Loading control shows total protein levels (bottom panel). Bar scatter plot quantifying the ratio of free mCh-Atg8a/GFP-mCh-Atg8a normalized to total protein. N = 3 biological replicates. (E and F) The overall level of p62 is increased in Strip RNAi muscles using two independent methods. Anti-GFP detects the p62-GFP fusion protein only (E), while an anti-p62 antibody detects both endogenous and GFP-tagged p62 (F). Total protein loading control (bottom panel). N = 5 biological replicates for each experiment. Scale bars, 25 µm (panel A). Genotypes with different letters are statistically different (p < 0.05).
This GFP-mCh-Atg8a fusion protein was expressed in lacZ control or Strip RNAi muscles. The first observation was an increase in the overall number of GFP-Atg8a puncta corresponding to autophagosomes (Figure 6B). These results are consistent with the elevated numbers of autophagosomes marked by anti-Atg8a or GFP-Atg8a in Figure 5A and Supplemental Figure S4E, respectively. Pearson’s correlation coefficient (PCC) was used to quantitatively assess the overlap of GFP and mCh as a measure of autophagosomes (Supplemental Figure S7). PCC values for control muscles averaged ∼0.3 but increased to ∼0.5 in Strip RNAi muscles (Figure 6C). Next, both control and experimental L3 larvae expressing GFP-mCh-Atg8a were fed the lysosomal fusion inhibitor chloroquine (CQ) for 6 h before analysis (Zirin et al., 2013; Zirin et al., 2015). As expected, the relative amount of autophagosomes (using PCC as a readout) was increased upon CQ feeding in Mef2>lacZ controls, demonstrating an efficient block of autophagosomal-lysosomal fusion. CQ treatment of Mef2>Strip RNAi larvae caused a similar accumulation of autophagosomes as observed in CQ-treated control muscles (Figure 6C). These results together demonstrate that decreasing Strip function impedes autophagy. The additional increase in PCC values upon further blocking the fusion between autophagosomes with lysosomes with CQ may be due to the perdurance of Strip protein in L3 muscles (Figure 2E).
Two additional assays were used to probe autophagic flux with isolated muscle carcasses to eliminate tissues (e.g., fat body) that exhibit high autophagic activity. First, we assessed the conversion of GFP-mCh-Atg8a to ‘free’ mCh to monitor autophagy-dependent delivery into the lysosome (Nagy et al., 2015; Lőrincz et al., 2017a). When autophagy proceeds normally, the GFP-mCh-Atg8a reporter is cleaved within the lysosome and the free mCh tag can be visualized via Western blotting. Our results showed an impairment in the amount of the full-length GFP-mCh-Atg8a fusion protein that is cleaved to produce the free tag in Strip RNAi knockdown muscles compared with control samples (Figure 6D). Because p62 itself is degraded in autolysosomes during normal autophagy, an increase or decrease in the amount of p62 serves as a marker of autophagic flux (Nagy et al., 2015; Lőrincz et al., 2017a; Ding and Hong, 2020). Expression of the p62-GFP fusion protein in lacZ control or Strip RNAi muscles by Western blotting allowed us to assess both tagged and endogenous p62 simultaneously. Using an antibody against GFP, there was a clear increase in the amount of the p62 fusion protein upon induction of Strip RNAi compared with lacZ alone (Figure 6E). Probing with an anti-p62 antibody detected the same increase in p62-GFP, but also showed elevated levels of endogenous p62 protein upon a loss of Strip (Figure 6F). Together, it is clear that autophagic flux is decreased in Strip RNAi muscles.
Lysosomal homeostasis is maintained in Strip RNAi muscles
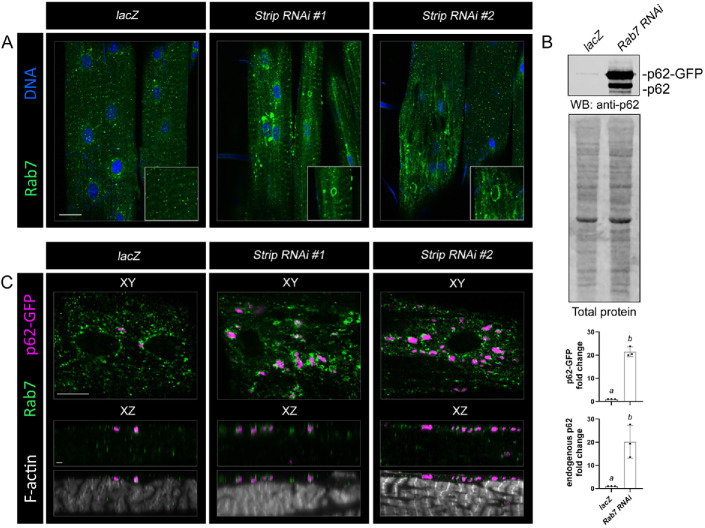
Autophagosomal maturation involves a series of fusion events with endocytic compartments before gaining competency for lysosomal fusion (Zhao and Zhang, 2019). Thus, the intersection between the autophagy and endocytic pathways makes it difficult to distinguish between effects due to altered autophagic flux and/or dysfunction of the endolysosomal pathway. For example, Ras-related protein Rab7 (Rab7) is associated with LEs and lysosomes and plays a role in the fusion of autophagosomes to lysosomes (Kirisako et al., 1999; Hyttinen et al., 2013; Guerra and Bucci, 2016; Fujita et al., 2017; Lőrincz et al., 2017b). To determine whether Strip influences Rab7-dependent events, we first assessed Rab7 localization in control and Strip RNAi muscles. Rab7 was present as puncta throughout control muscles and accumulated in large, circular structures upon induction of Strip RNAi (Figure 7A). Next, we assessed the relationship between Rab7 and p62. Knockdown of Rab7 increased both endogenous p62 and p62-GFP protein levels (Figure 7B), thus supporting the functional link between the endocytic and autophagic pathways. Moreover, p62(+) puncta could be found associated with Rab7 structures in Strip RNAi muscles compared with controls (Figure 7C). These data together raise the possibility that flux through the endolysosomal and/or autolysosomal pathway may be affected.
FIGURE 7:
Endosome morphology is altered upon loss of Strip. (A) VL3 or VL4 muscles of the indicated genotypes were immunostained with Rab7 to visualize LEs. Larger structures labeled with Rab7 were present in Strip RNAi muscles. (B) Mef2-driven p62-GFP was visualized by Western blotting in lacZ control or Rab7 RNAi muscles. Autophagic flux is impeded upon a reduction in Rab7 as visualized with anti-p62 that detects the p62-GFP fusion protein and endogenous p62. (C) High magnification images of L3 control or Strip RNAi muscles expressing UAS-p62-GFP and labeled with anti-Rab7 and phalloidin. Rab7 and p62 accumulate close to one another at the muscle surface. Scale bars, 50 µm (panels A and C), 10 µm (panel C, XY scans), 2 µm (panel C, XZ scans).
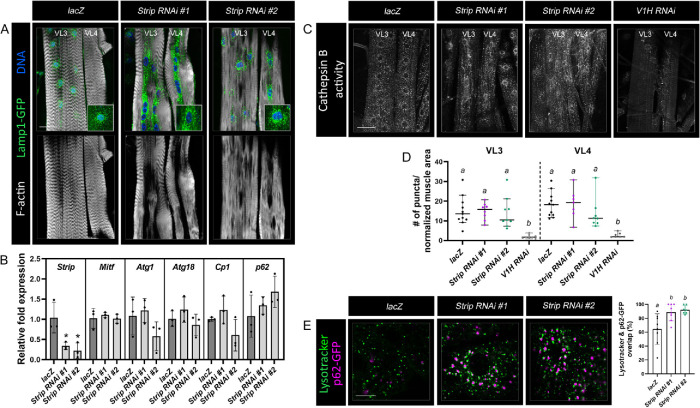
Elevated numbers of autophagosomes and a corresponding reduction in autophagic flux could be due to decreased lysosome biogenesis and/or a block in the lysosomal turnover of cargo. First, we examined the relative amount of lysosomes using lysosomal-associated membrane protein 1 (Lamp1)-GFP as a marker for this organelle. In control muscles, lysosomes were generally distributed throughout the muscle with some accumulation around nuclei (Figure 8A). Enhancement of this perinuclear lysosome clustering in Strip RNAi muscles obscured an accurate quantification of lysosomal number. Therefore, we assessed whether the expression of autophagosome and/or lysosome biogenesis markers is dependent on Strip. First, we verified the utility of the real-time PCR approach in muscle carcasses using the Mef2-Gal4 driver to induce Strip RNAi. Indeed, there was an ∼60% reduction in Strip transcript levels using quantitative RT-PCR (qPCR) (Figure 8B). Autophagy and lysosomal biogenesis is regulated by mammalian transcription factor EB (TFEB), or melanocyte-inducing transcription factor (Mitf) in D. melanogaster (Bouché et al., 2016; Yang and Wang, 2021; Agostini et al., 2022). The relative abundance of Mitf transcripts, as well as its transcriptional targets Autophagy-related 1 (Atg1), Autophagy-related 18 (Atg18a), and Cysteine proteinase-1 (Cp1), were not significantly changed in Strip RNAi muscles (Figure 8B). Thus, it seems unlikely that autophagosome biogenesis (Atg1, Atg18) or lysosomal biogenesis (Cp1) are altered as has been previously shown for mutants of D. melanogaster Myotubularin-related phosphatase 6 (Allen et al., 2020). Levels of p62 transcripts were also not appreciably changed in Strip RNAi muscles, further highlighting that the increase in p62 protein is indeed due to a block in autophagy rather than increased expression levels (Figure 8B).
FIGURE 8:
Lysosomal fusion appears partially blocked upon a reduction of Strip in muscles. (A) Labeling with Lamp1-GFP (green) shows no overt difference in the overall number of lysosomes between lacZ or Strip RNAi muscles, although the lysosomes appear clustered around nuclei in the latter. (B) mRNA transcripts implicated in autophagosome or lysosome biogenesis were analyzed by qPCR in muscle fillets from lacZ or Strip RNAi individuals. Primers corresponding to Strip were used as a positive control to assure the validity of RNAi knockdown. No statistically significant differences in transcript levels were observed for Atg1, Atg18, Cp1, Mitf, or p62. (C) Monitoring lysosomal activity by Cathepsin B staining reveals no appreciable difference in control muscles or muscles expressing ds hairpins against Strip. V1H RNAi was used as a positive control for a reduction in lysosomal activity (Mauvezin et al., 2015). (D) Quantitation of puncta stained with Cathepsin Red(+)/1000 µm2 represented in a scatter plot. N ≥ 8. Genotypes with different letters are statistically different (p < 0.05). (E) Mef2-Gal4, p62-GFP flies were mated with UAS-lacZ or UAS-Strip RNAi flies. Lysotracker was used to label lysosomes. Muscles deficient for Strip show close proximity between p62 and lysosomes compared with controls. Scale bars, 50 µm (panels A and C), 10 µm (panel E).
To determine if Strip impairs the degradation of cargo, thus leading to autophagosome accumulation, we assayed for lysosomal function using an assay to measure Cathepsin-B activity. Cleavage of a fluorescent substrate by active cathepsins is a general indicator of lysosome function (Barral et al., 2022). However, we observed no obvious difference in the relative number of puncta that stained positive for Cathepsin B activity (Figure 8, C and D). The Vacuolar H+-ATPase SFD subunit (VhaSFD)/V1H served as a negative control because lysosomal acidification does not occur upon RNAi depletion of V1H in fat-body tissue RNAi (Mauvezin et al., 2015). These data together indicate that lysosome biogenesis and lysosome activity are not impaired in Strip RNAi muscles.
An alternative explanation for the increase in autophagosomes is the inability of this organelle to undergo fusion with lysosomes. Because Lysotracker labels lysosomes in live tissues, we colabeled with p62-GFP to mark ubiquitinated cargo that could be captured by autophagosomes for degradation. In control lacZ muscles, ∼60% of lysosomes were found to be associated with p62-GFP (Figure 8E). This value increased to ∼85% in muscles that were expressing either Strip RNAi line, raising the possibility that impaired lysosomal fusion may be one contributing factor to explain the autophagic block observed in Strip RNAi muscles.
DISCUSSION
Our efforts to understand how evolutionarily conserved proteins required for muscle development in T. castaneum also function in D. melanogaster led us to explore how the STRIPAK complex intersects with autophagy. Our previous studies defined a role for the NUAK-Stv complex in mediating the autophagic degradation of Filamin (Brooks et al., 2020). Here we functionally link the STRIPAK complex to NUAK and Stv and show that autophagy is blocked, possibly due to inefficient fusion with lysosomes. Although we did not observe alterations in autophagosome or lysosome biogenesis (assessed by qPCR) or lysosomal activity, we cannot rule out the possibility that yet unidentified factors affect endosomal or endolysosomal flux, thus overloading the pathway.
The search term “larval muscle irregular” used to screen iBeetle-base RNAi phenotypes broadly describes muscles that are mispatterned, thinner, round, degenerative, and more. The descriptor terms primarily used for Tc-strip dsRNA, Tc-Mob4 dsRNA, and Tc-Fgop2 dsRNA larval musculature phenotypes were “pattern irregular” and “pattern not present.” Moreover, RNAi phenotypes for each of these gene products also affected other stages of the T. castaneum lifecycle, including oogenesis, larval cuticle development, and elytra formation, just to name a few (Schmitt-Engel et al., 2015). Thus, the STRIPAK complex has broad functions throughout development and likely even multiple functions within the same tissue, including muscle. Loss of Mob4 or Striatin-3 compromises myofibril assembly in zebrafish (Berger et al., 2022), while the STRIPAK complex family members Slmap, Cka, and Strip are essential for sarcomerogenesis in D. melanogaster indirect flight muscles (Kaya-Çopur et al., 2021). Our identification of STRIPAK proteins copurifying with Strip in muscle tissue combined with the functional involvement of Strip with the NUAK-Stv complex in autophagy demonstrates a role for STRIPAK complex members in both muscle development and maintenance.
One advantageous feature of model organisms is the presence of a single orthologue compared with vertebrates which may possess multiple protein family members. For example, the human genome encodes for two Strip proteins (Strip1/2) and three Striatin proteins (STRN1/3/4), while D. melanogaster possesses one gene of each (Kück et al., 2019). This feature allows for the unambiguous characterization of phenotypes through genetic analysis without complications of overlapping family members exhibiting compensatory roles. In larval body-wall muscles, knockdown of Strip or Cka affected muscle architecture and muscle contraction to a greater extent than a reduction in other STRIPAK complex proteins. It is possible that inefficient protein knockdown of the selected RNAi lines in muscle tissue accounts for the weaker phenotypes. This seems unlikely because many of these RNAi lines, such as those directed against Mob4, were used to assess autophagosome transport defects in neurons (Neisch et al., 2017). Alternatively, some members of the STRIPAK complex function coordinately with other pathways. Mob4 or Fgop2 phenotypes are only visible in the sensitized NUAKK99R/+ or stv1/+ background compared with expression of RNAi alone.
Autophagosome nucleation has been most extensively studied in yeast where phagophore assembly sites are initiated in close proximity to the endoplasmic reticulum (ER) and may directly fuse with nearby vacuoles (Zhao and Zhang, 2018; Hollenstein and Kraft, 2020). Autophagosome biogenesis in eukaryotes can occur simultaneously at multiple locations, including the ER. In both yeast and mammalian cells, specialized subdomains of the ER, termed ER exit sites, serve as an organizing platform for autophagosome formation (Graef et al., 2013). Because ER exit sites are dispersed throughout both mammalian and D. melanogaster multinucleated muscle cells (Ralston et al., 2001; Wang et al., 2015), it stands to reason that autophagosomes can also form at several locations before eventual fusion with endosomes and/or lysosomes. D. melanogaster Cka physically interacts with Dynein and directly binds Atg8a to facilitate the transport of autophagosomes and dense core vesicles within the axons of motoneurons (Neisch et al., 2017). This relatively long-range movement from the site of autophagosome initiation to the site of lysosomal fusion is not a major impediment in muscle cells where the accumulation of stalled autophagosomes appears closer to their lysosomal targets. In Strip RNAi muscles, p62-marked autophagosomes were rarely observed far away from lysosomes, suggesting that transport in this genotype was largely unaffected in muscle tissue.
After the completion of autophagosome formation, additional fusion events with various endolysosomal compartments are required before the final fate of degradation by autolysosomes. This process, referred to as autophagosome maturation, includes the fusion of MVBs or LEs with autophagosomes to form the intermediate or hybrid amphisome structure (Zhao and Zhang, 2019; Ganesan and Cai, 2021). While Rab5 is normally present on early endosomes, Rab7 is predominant on LEs; this Rab5 to Rab 7 switch ensures endosome maturation (Hyttinen et al., 2013; Guerra and Bucci, 2016; Ganesan and Cai, 2021). Rab7/Ypt7 can also be recruited to autophagosomes by the Mon1-Ccz1 guanosine-exchange-factor complex in starved yeast or larval fat cells (Hegedűs et al., 2016; Gao et al., 2018). This nutrition-dependent switch in organelle localization diversifies possible functions of Rab7 but also merits caution in the interpretation of deciphering which step of autophagy may be affected. Roles for Rab7 have been delineated in the movement of LEs toward lysosomes as well as autophagosome maturation (Guerra and Bucci, 2016; Zhao and Zhang, 2019). Autophagosome accumulation in Ypt7-KO yeast or Rab7 knockdown in D. melanogaster cells indicates that the fusion step between autophagosomes and lysosomes is blocked (Cong et al., 2015; Fujita et al., 2017; Lőrincz et al., 2017b). However, CRISPR/Cas9-mediated KO of Rab7 in Madin-Darby canine kidney II cells showed an increase in structures positive for LC3 and the lysosome marker Lamp2, indicating that autophagosome fusion could proceed to generate autolysosomes (Klionsky et al., 2016). More studies need to be performed to determine whether this result is a general feature in mammalian cell types.
Deciphering the molecular role of STRIPAK complex components can be difficult due to subunit heterogeneity in different tissues and/or a functional requirement for core complex members in different stages of the same biological process, such as autophagy. In D. melanogaster, some (Cka, Strip and Mob4), but not all (Ccm3, GckIII), of the core STRIPAK complex components tested were required for axonal autophagosome transport (Neisch et al., 2017), while Strip and Cka (but not Fgop2, GckIII, Mob4, or Slmap) were required for spermatogenesis (La Marca et al., 2019). Because mammalian Ccm3 links Striatin and GckIII proteins (Kean et al., 2011), these data provide support for different STRIPAK subcomplexes dependent upon the biological context. Unc-51 like autophagy activating kinase (Ulk1) is well known for its role in autophagy initiation (Mercer et al., 2018; Zhao and Zhang, 2018; Hollenstein and Kraft, 2020). In a comprehensive set of experiments, Hu et al. (2021) uncovered multiple phosphatase complexes as targets of Ulk1 kinase activity, one of these being Striatin (Hu et al., 2021). Interestingly, many of these phosphorylated substrates are also required for autophagosome maturation and autophagosome-lysosome fusion, thus suggesting that intricate regulation and feedback loops regulate autophagy. Defining targets of STRIPAK-complex activity during each step of autophagy would help clarify these intricate details.
MATERIALS AND METHODS
In-silico screen
The search terms “larval,” “muscle,” and “irregular” were queried using the search function in iBeetle-Base (https://ibeetle-base.uni-goettingen.de/) (Dönitz et al., 2015). The retrieved Tc genes were converted to current Flybase IDs (Fbgn number) and analyzed using Cytoscape (https://cytoscape.org/) (Shannon et al., 2003).
D. melanogaster genetics
All stocks were maintained on standard cornmeal-molasses-yeast-agar medium at 25°C unless specified. Gal4/UAS crosses were set up at 25°C and shifted to 31°C at the beginning of the L1 stage. The following RNAi lines stocks were obtained from either the Bloomington D. melanogaster Stock Center (BDSC, Bloomington, IN) or the Vienna D. melanogaster Resource Center (VDRC, Vienna, Austria): UAS-Strip RNAi #1 (v16211); UAS-Strip RNAi #2 (BL34657); UAS-cka RNAi #1 (v106971); UAS-cka RNAi #2 (BL28927); UAS-Mob4 RNAi #1 (v110742); UAS-Mob4 RNAi #2 (v40442); UAS-Naus RNAi #1 (BL34879); UAS-Naus RNAi #2 (v31375); UAS-Fgop2 RNAi #1 (v47389); UAS-Fgop2 RNAi #2 (BL60065); UAS-Slmap RNAi #1 (v8199); UAS-Slmap RNAi #2 (BL32509); UAS-Ccm3 RNAi #1 (v106841); UAS-Ccm3 RNAi #2 (v109453); UAS-GCKIII RNAi #1 (BL35339), UAS-GCKIII RNAi #2 (BL57042); UAS-PP2A-29B RNAi #1 (BL43283); UAS-flw RNAi (BL38336); UAS-Atg8a RNAi (V43097); UAS-NUAK RNAi (BL35194); UAS-stv RNAi (v34408), UAS-Rab7 RNAi (BL27051); and UAS-V1H RNAi (v47471). Additional fly stocks obtained from the BDSC include Mef2-Gal4 (BL27390), da-Gal4 (BL37291), MHC-GFP (BL38462), UAS-lacZ (BL3956), UAS-GFP-Atg8a (BL51656), and UAS-GFP-mCh-Atg8a (BL37749). The YFP::Lamp1CPTI001775 insertion (DGGR115240) is from the Kyoto D. melanogaster Stock Center (DGGR, Kyoto, Japan). Other stocks include the alleles stv1(Coulson et al., 2005) and NUAKK99R (unpublished data) in addition to the transgenic lines UAS-FLAG::Strip (Sakuma et al., 2014), UAS-Strip::V5 (Sakuma et al., 2014); and UAS-p62-GFP (Chang and Neufeld, 2009).
Muscle tissue visualization
Larval whole mounts.
Wandering L3 larvae resulting from parents of the genotype Mef2-Gal4, MHC-GFP x UAS-lacZ or a UAS-RNAi line of interest were individually selected from the sides of a vial or bottle. Larvae were heat-killed and imaged on a Leica M165FC fluorescent stereoscope.
Muscle carcass dissections and immunostaining.
Wandering L3 larvae were placed on a Sylgard plant, pinned near the mouth hooks at the anterior end, and the internal organs were removed. The resulting muscle carcasses were fixed in 4% formaldehyde and subjected to standard immunostaining protocols with normal goat serum used as a blocking agent in PBT (Brooks et al., 2020). Tissues were stained with the following primary antibodies: rabbit anti-p62/ref(2)p (1:200, Abcam, Cambridge, MA), rabbit anti-Atg8a (1:200, Millipore Sigma, Burlington, MA), mouse anti-Rab7 (1:25, Developmental Studies Hybridoma Bank, Iowa City, IA), and rabbit anti-Filamin DZ00502 (1:200, Boster Bio, Pleasanton, CA). Fluorescence was detected using the following secondary antibodies: Alexa Fluor anti-rabbit or anti-mouse 488 or 594 (1:400, Invitrogen, Waltham, MA). F-actin was labeled with phalloidin 488 or 594 (1:400, Molecular Probes, Eugene, OR) and nuclei were stained with Hoechst dye (1:400, Invitrogen, Waltham, MA). Images were captured using a Zeiss 700 confocal microscope. Image processing and analysis were performed using a combination of Zen Black (Zeiss), ImageJ (NIH), and Adobe Photoshop. All images taken at 20× are displayed as maximum intensity projections. Data acquisition at 63× magnifications is presented as single-plane confocal micrographs (Figures 3D, 7C, and 8E) or maximum intensity projections (Figures 2D, 5 A, C and F, 6B, 7A, and 8 A and C).
PLA.
Wandering L3 larvae of the genotypes Mef2>FLAG::Strip, Mef2>Strip::V5, or w1118 were heat-killed, dissected, fixed in 4% formaldehyde, and washed in PBT before following general instructions for the Duolink In Situ Red Starter Kit Mouse/Rabbit (Sigma-Aldrich, St. Louis, MO). Tissues were blocked for 1 h at 37°C in the Duolink blocking solution, incubated with primary antibody diluted in the Duolink antibody diluent overnight at 4°C. Primary antibodies used were anti-FLAG F1804 (1:400, Sigma-Aldrich, St. Louis, MO), anti-NUAK DZ41105 (1:400, Boster Bio, Pleasanton, CA), anti-V5 R960-25 (1:400, Invitrogen, Waltham, MA), and rabbit anti-Stv (1:400; Coulson et al., 2005; Arndt et al., 2010). Tissues were washed twice for 5 min in wash buffer A and then incubated in 40 μl PLA probe solution (8 μl PLUS probe, 8 μl MINUS probe, 24 μl antibody diluent) for 1 h at 37°C. Tissues were washed twice for 5 min in wash buffer A and then incubated for 30 min at 37°C in 40 μl ligation solution (8 μl ligation buffer, 31 μl ddH2O, 1 μl ligase). Tissues were washed twice for 5 min in wash buffer A and incubated for 100 min at 37°C in 40 μl amplification solution (8 μl amplification buffer, 31.5 μl ddH2O, 0.5 μl polymerase). Tissues were washed twice for 10 min in wash buffer B, then rinsed for 1 min with 0.01 × wash buffer B and Phalloidin 488 to label F-actin (1:400, Molecular Probes, Eugene, OR). Tissues were mounted in PLA mounting medium.
Lysosomal dyes.
For LysoTracker red staining, wandering L3 larvae were live dissected in Ca2+-free HL3.1 buffer. After internal organs were removed, the muscle carcasses were incubated in 100-nM LysoTracker Red DND-99 diluted in HL3.1 (Invitrogen, Waltham, MA) for 5 min at room temperature (wrapped in foil to avoid light) and imaged immediately. For Magic red staining, wandering L3 larvae were live dissected in HL3.1, the internal organs were removed, and muscle carcasses were incubated with Magic Red diluted in HL3.1 buffer (1:250, Magic Red Cathepsin B Assay, ImmunoChemistry, Davis, CA) for 20 min at 37°C (wrapped in foil to avoid light). After a quick wash in PBS, the samples were imaged immediately.
Immunoblot analysis
Dissected muscle carcasses were placed into SDS sample buffer, boiled at 95°C for 3 min, homogenized, boiled for an additional 10 min at 95°C, and centrifuged at 20,000 × g for 1 min to pellet debris. The resulting protein samples were separated by SDS–PAGE and transferred to the nitrocellulose blotting membrane (pore size 0.45 µm, Cytiva, Marlborough, MA) using the Trans-Blot® Turbo™ Transfer System (Bio-Rad, Hercules, CA). Membranes were probed with the following primary antibodies: rabbit anti-mCh ab167453 (1:5000, Abcam, Cambridge, UK), rabbit anti-GFP A-21311 (1:5000, Thermo Fisher Scientific, Waltham, MA), rabbit anti-ref(2)p ab178440 (1:2000, Abcam, Cambridge, UK), and rabbit anti-Strip DZ41200 (1:2000, Boster Bio, Pleasanton, CA). The rabbit anti-Strip antibody was generated against a recombinant protein that included amino acids 70–322 of the Strip-PB isoform and affinity-purified. IRDye 800CW secondary antibodies (LI-COR Biosciences, Lincoln, NE) were used at 1:10,000 and Revert 700 Total Protein Stain (LI-COR Biosciences, Lincoln, NE) was used as a loading control. Membranes were developed using the LI-COR Odyssey XF and quantitation of relative protein levels was performed in Empiria Studio Software (LI-COR Biosciences, Lincoln, NE).
Soluble/insoluble fraction preparation.
Muscle carcasses from six dissected larvae were homogenized in 80 μl ice cold Triton X-100 soluble buffer (1% Triton X-100 and 1% Halt Protease Inhibitors Cocktail [ThermoFisher Scientific, Waltham, MA]) in PBS. The homogenate was centrifuged at 21,000 × g for 10 min at 4°C. The supernatant (Triton X-100 soluble sample) was collected as the soluble fraction. The pellet from the Triton X-100 soluble buffer homogenization was washed three times with 600μL ice cold Triton X-100 soluble buffer. 80 μl of room-temperature insoluble buffer (4 M urea, 4% SDS, 10% 2-Mercaptoethanol, 125 mM Tris-HCl, pH 6.8, 50% glycerol, 0.2% (wt/vol) orange G) was added and the pellet was rehomogenized. The resulting mixture represented the insoluble fraction. All the samples were boiled for 10 min and centrifuged before loading to a SDS–PAGE gel. Western blotting was performed using rabbit anti-mono- and polyubiquitinylated conjugates monoclonal antibody FK2 (1:1000, Enzo Life Sciences, Farmingdale, NY).
Quantitative RT-PCR
Total RNA was collected from five wandering L3 larvae which were raised at 31°C, dissected and purified using the RNeasy Mini Kit (Qiagen, Hilden, Germany). For Figure 8B, three RNA samples for each genotype were prepared. cDNA was synthesized from 350 ng of RNA using the qScript XLT cDNA SuperMix kit (Quanta Biosciences, Beverly, MA). For the qPCR reactions, 1:15 dilutions of the cDNA were combined with PowerUp SYBR Green Master Mix (ThermoFisher, Waltham, MA). The following primers were used at a final concentration of 1 µM for the assessment of autophagosome and lysosome biogenesis: Strip forward 5′-CGGAACTTCAAACGCTCCCT, reverse 5′-TGTTGGCATTATTTTGACCCTCG; Atg1 forward 5′-GTCGGGGAATATGAATACAGCTC, reverse 5′-GCATGTGTTTCTTGCGATGAC; Atg18a forward 5′-GTGTTCGTCAACTTCAACCAGA, reverse 5′-TGTCCAGGGTCGAGTCCAC; Cp1 forward 5′-TCAACTACACTCTGCACAAGC, reverse 5′-GCCAGTCCACAGATTTGGG; Mitf forward 5′-AGTATCGGAGTAGATGTGCCAC, reverse 5′-CGCTGAGATATTGCCTCACTTG; p62/ref(2)p forward 5′-GCC CTC CCA GAA TTA CAC CA, reverse 5′-GTT GGC CGA AGA ACC CTC T; and rp49 forward 5′-GCCCAAGGGTATCGACAACA, reverse 5′-GCGCTTGTTCGATCCGTAAC. For validation of RNAi knockdowns in Supplemental Figure S3, eight RNA samples for each genotype were prepared, cDNA was synthesized from 150 ng of RNA, and a 1:10 dilution of cDNA was used for qRT-PCR. Primer sequences used are as follows: Cka forward 5′-CGCCCCAATACACAATACCG, reverse 5′-GGCGCGTCAAATCCGATTT; Mob4 forward 5′-GGCCAGGCACAAAATCCAAG, reverse 5′-TACTTCCACACGCCCTCATC; Naus forward 5′-CGACAGTTCAGCGACGAC, reverse 5′-GATTCCAGCGCTCCGGAT; Fgop2 forward 5′-ATGTCGAACCTATCCGTGGG, reverse 5′-GATTGTTGTTCTCCGCCTCC; Slmap forward 5′-ATGCACTGCGAAACGAGATT, reverse 5′-GAAGTATGCCGCACTCGTTT; Ccm3 forward 5′-TCGTTCTGCCCGTCATACTG, reverse 5′-GGATTGTTCTGCTCCGACTTG; GckIII forward 5′-CAGCTGACCAACACCACATC, reverse 5′-CTTGGGGATGAGGAAGAGCA; and Pp2a-29B forward 5′-AGGATGATGTAGAGCACCTGG, reverse 5′-GCACCAAGTCCACCCTAGTA. All primers were synthesized at Integrated DNA Technologies (IDT, Stokie, IL). Quantitative transcript levels were obtained using a QuantStudio 3 instrument with QuantStudio Design and Analysis software (TheroFisher, Waltham, MA). Normalize fold change was calculated using the 2-ΔΔCt method and graphed as Mean ± SEM using GraphPad 9.4.1.
AP-MS
Experiment.
∼120 wandering L3 larvae resulting from parents of the genotype Mef2-Gal4 ×x UAS-3xFLAG::Strip, Mef2-Gal4 × UAS-Strip::V5, or Mef2-Gal4 × UAS-lacZ were selected from the sides of media bottles. In total, three biological replicates per genotype were collected. The larvae were gently washed using 0.7% NaCl + 0.04% Triton-100 solution to remove food debris, followed by a rinse with double-distilled water. The washed larvae were transferred to a dounce homogenizer with 1-ml lysis buffer (100-mM NaCl, 50-mM Tris-HCl pH 7.3, 1% NP-40, 1× Halt protease inhibitor cocktails, 1× Halt Phosphatase inhibitor cocktails, 10µM MG132). After homogenization with a pestle, the lysate was transferred to 1.5-ml microcentrifuge tubes. The supernatant was collected after centrifugation at 4°C for 5 min at 12,000 × g. Next, anti-FLAG® M2 Magnetic Beads m8823 (MilliporeSigma, Burlington, MA) or the anti-V5 mAb-Magnetic Beads (MBL, Woburn, MA) were incubated with the supernatant at 4°C for 2 h on a Barnstead Labquake Rotating Shaker. For the anti-FLAG immunoprecipitation, beads were washed three times in 400 µl cold wash buffer A (100-mM NaCl, 50-mM Tris-HCl pH 7.3, 0.05% NP-40) followed by two more times in 400 µl cold wash buffer B (100-mM NaCl, 50-mM Tris-HCl pH 7.3). The NaCl concentration was 150 mM for wash and lysis buffers for the anti-V5 pulldown. All washed samples were flash frozen in liquid nitrogen and kept at –80°C until they were sent to the Recombinant DNA/Protein Resource Facility at Oklahoma State University for MS analysis where each biological replicate was performed in triplicate (Brooks et al., 2021; Kawakami et al., 2022).
Analysis.
Peptides were identified and quantified by using MaxQuant v1.5.3.8 (Cox and Mann, 2008). Data analysis and statistical analysis were carried out in the freely available Perseus software platform (https://maxquant.net/perseus/) (Tyanova et al., 2016). LFQ intensities were imported into Perseus and the raw data was filtered to remove potential contaminants or incorrect protein identifications. Expression values were converted to log2 for data normalization before statistical analysis (Cox et al., 2014). For each biological replicate, proteins that passed the statistical threshold after being subjected to a two-sample t-test (Permutation-based FDR p < 0.05) were kept for further analysis. Imputation was performed to replace missing values from a normal distribution. The values for each genotype across all experiments were averaged and the difference between experiments and controls was defined as the enrichment value (Supplemental Table S1).
Quantitation
Assessment of muscle contractility during the L3 to pupal transition.
Pupae of the indicated genotypes were removed from vials, oriented dorsal side up, and attached to slides using a small drop of nail polish. Images were taken with a Leica M165 FC Stereomicroscope. Length and width measurements for each pupa were performed in ImageJ using the line and measure functions. Values were put into an Excel spreadsheet and the axial ratio (length/width) was calculated for each individual. The raw data were imported into Graphpad Prism 9.4.1 and graphed as a box and whiskers plot. N ≥20 for each genotype.
Muscle defect quantification.
Embryos from parents of the genotype Mef2-Gal4 × UAS-RNAi at 25°C were collected and transferred to 31°C at the L1 stage. Wandering L3 larvae were dissected in PBS. The resulting muscle carcasses were fixed in 4% methanol-free formaldehyde (Polysciences) for 15 min at room temperature and washed three times in PBST before incubating with phalloidin 488 (1:400) and Hoechst (1:400). For each individual larva, seven distinct muscle fibers (VL1, VL2, VL3, VL4, LO1, LL1, and SBM) from eight randomly selected abdominal hemisegments were examined under the Zeiss 700 confocal microscope. The number of normal or defective muscles for each larva was divided by the total number of muscles to calculate the percentage of muscle defects. N ≥18 for each genotype.
Puncta quantification.
The Analyze-Particles function in ImageJ (NIH) was used to identify and measure GFP-Atg8a, p62-GFP, Atg8a, and p62 puncta. The colocalization between the GFP channel and the mCh channel using the GFP-mCh-Atg8a construct was determined using the Fiji/ImageJ Just Another Colocalization Plugin (JACoP) plugin. Quantitative analysis is represented by the PCC. N ≥ 20 for each genotype.
Statistical analysis.
Statistical analyses were performed in GraphPad 9.4.1. The unpaired student t-test was used to evaluate the significance between pairwise comparisons followed by the Mann-Whitney test. All other data sets that compared three or more unmatched groups that did not conform to a Gaussian distribution were subjected to one-way ANOVA analysis using the nonparametric Kruskal-Wallis test. For groups of data with the same letter (e.g., a, b), the difference between the means is not statistically significant (p > 0.05). Different letters indicate statistical significance between different data groups (p < 0.05 – p < 0.001). See Supplemental Table S2 for a complete description of statistical tests and p-values.
Supplementary Material
Acknowledgments
We are grateful to Takahiro Chihara for sharing the UAS-FLAG::Strip and UAS-Strip::V5 transgenic flies, Gabor Juhász for providing the UAS-p62-GFP flies, and Jörg Höhfeld for the generous contribution of anti-Stv antibody. Stocks obtained from the BDSC are supported by a grant from the Office of the Director of the National Institutes of Health (NIH) under award number P40OD018537. A portion of the transgenic RNAi stocks was obtained from the VDRC ( www.vdrc.at) and a YFP-tagged insertion was obtained from KYOTO D. melanogaster Stock Center in Kyoto Institute of Technology. Thanks to the Kansas State University Johnson Cancer Research Center (JCRC) for travel funds to Y.G. This study was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) of the NIH under award number RO1AR060788 to E.R.G.
Abbreviations used:
- AP-MS
affinity purification mass spectrometry
- Atg1
Autophagy-related 1
- Atg18a
Autophagy-related 18
- Atg8a
Autophagy-related 8a
- Bag3
Bcl-2–associated athanogene 3
- CASA
chaperone-assisted selective autophagy
- Ccm3
Cerebral cavernous malformation 3
- Cka
Connector of kinase to AP-1
- Cp1
Cysteine proteinase-1
- D. melanogaster
Drosophila melanogaster
- ER
endoplasmic reticulum
- Fgop2
Fibroblast growth factor receptor 1 oncogene partner 2
- flw
flapwing
- GckIII
Germinal centre kinase III
- GFP
green fluorescent protein
- gt
greater than
- Hsc70
Heat-shock cognate 70
- LAMP2
lysosomal Associated Membrane Protein 2
- LE
late endosomes
- LFQ
label-free quantification
- MAP1LC3/LC3
microtubule-associated protein 1 light chain 3
- mCh
mCherry
- MFM
Myofibrillar Myopathy
- Mhc
Myosin heavy chain
- Mitf
melanocyte-inducing transcription factor
- Mob4
MOB kinase activator 4
- mts
microtubule star
- MVBs
multivesicular bodies
- Naus
Nausicaa
- NUAK
NUAK family kinase 1
- PLA
proximity ligation assays
- PP2A
protein phosphatase 2A
- PP2A-29B
protein phosphatase 2A at 29B
- ref(2)p
refractory to sigma P
- RNAi
RNA interference
- Slmap
sarcolemma associated protein
- SQSTM1
sequestosome-1
- Strip
striatin interacting protein
- STRIPAK
striatin interacting phosphatase and kinase
- STRN
striatin
- T. castaneum
Tribolium castaneum
- TFEB
transcription factor EB
- UAS
upstream activating sequence
- VhaSFD
Vacuolar H+-ATPase SFD subunit
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E23-01-0006) on June 28, 2023.
REFERENCES
- Agostini F, Agostinis R, Medina DL, Bisaglia M, Greggio E, Plotegher N (2022). The regulation of MiTF/TFE transcription factors across model organisms: from brain physiology to implication for neurodegeneration. Mol Neurobiol 59, 5000–5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen EA, Amato C, Fortier TM, Velentzas P, Wood W, Baehrecke EH (2020). A conserved myotubularin-related phosphatase regulates autophagy by maintaining autophagic flux. J Cell Biol 219, e201909073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt V, Dick N, Tawo R, Dreiseidler M, Wenzel D, Hesse M, Fürst DO, Saftig P, Saint R, Fleischmann BK, et al. (2010). Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr Biol 20, 143–148. [DOI] [PubMed] [Google Scholar]
- Barral DC, Staiano L, Guimas Almeida C, Cutler DF, Eden ER, Futter CE, Galione A, Marques ARA, Medina DL, Napolitano G, et al. (2022). Current methods to analyze lysosome morphology, positioning, motility and function. Traffic 23, 238–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batonnet-Pichon S, Behin A, Cabet E, Delort F, Vicart P, Lilienbaum A (2017). Myofibrillar myopathies: new perspectives from animal models to potential therapeutic approaches. J Neuromuscul Dis 4, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bawa S, Gameros S, Baumann K, Brooks DS, Kollhoff JA, Zolkiewski M, David Re Cecconi A, Panini N, Russo M, Piccirillo R, et al. (2020). Costameric integrin and sarcoglycan protein levels are altered in a Drosophila model for Limb-girdle muscular dystrophy type 2H. Mol Biol Cell 3, 260-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger J, Berger S, Currie PD (2022). Mob4-dependent STRIPAK involves the chaperonin TRiC to coordinate myofibril and microtubule network growth. PLoS Genet 18, e1010287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T (2005). p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 171, 603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché V, Espinosa AP, Leone L, Sardiello M, Ballabio A, Botas J (2016). Drosophila Mitf regulates the V-ATPase and the lysosomal-autophagic pathway. Autophagy 12, 484–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415. [DOI] [PubMed] [Google Scholar]
- Brooks D, Naeem F, Stetsiv M, Goetting SC, Bawa S, Green N, Clark C, Bashirullah A, Geisbrecht ER (2020). Drosophila NUAK functions with Starvin/BAG3 in autophagic protein turnover. PLoS Genet 16, e1008700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks DS, Vishal K, Bawa S, Alder A, Geisbrecht ER (2021). Integration of proteomic and genetic approaches to assess developmental muscle atrophy. J Exp Biol 224, jeb242698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castets P, Frank S, Sinnreich M, Rüegg MA (2016). “Get the balance right”: pathological significance of autophagy perturbation in neuromuscular disorders. J Neuromuscul Dis 3, 127–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YY, Neufeld TP (2009). An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol Biol Cell 20, 2004–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong J, Fan T, Yang X, Squires JW, Cheng G, Zhang L, Zhang Z (2015). Structural and functional changes in maternal left ventricle during pregnancy: a three-dimensional speckle-tracking echocardiography study. Cardiovasc Ultrasound 13, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulson M, Robert S, Saint R (2005). Drosophila starvin encodes a tissue-specific BAG-domain protein required for larval food uptake. Genetics 171, 1799–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M (2014). Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics 13, 2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26, 1367–1372. [DOI] [PubMed] [Google Scholar]
- De Palma C, Morisi F, Cheli S, Pambianco S, Cappello V, Vezzoli M, Rovere-Querini P, Moggio M, Ripolone M, Francolini M, et al. (2012). Autophagy as a new therapeutic target in Duchenne muscular dystrophy. Cell Death Dis 3, e418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S, Hong Y (2020). The fluorescence toolbox for visualizing autophagy. Chem Soc Rev 49, 8354–8389. [DOI] [PubMed] [Google Scholar]
- Dönitz J, Gerischer L, Hahnke S, Pfeiffer S, Bucher G (2018). Expanded and updated data and a query pipeline for iBeetle-Base. Nucleic Acids Res 46, D831–D835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dönitz J, Schmitt-Engel C, Grossmann D, Gerischer L, Tech M, Schoppmeier M, Klingler M, Bucher G (2015). iBeetle-Base: a database for RNAi phenotypes in the red flour beetle Tribolium castaneum. Nucleic Acids Res 43, D720–D725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichna JP, Maruszak A, Żekanowski C (2018). Myofibrillar myopathy in the genomic context. J Appl Genet 59, 431–439. [DOI] [PubMed] [Google Scholar]
- Franco-Romero A, Sandri M (2021). Role of autophagy in muscle disease. Mol Aspects Med 82, 101041. [DOI] [PubMed] [Google Scholar]
- Fujita N, Huang W, Lin TH, Groulx JF, Jean S, Nguyen J, Kuchitsu Y, Koyama-Honda I, Mizushima N, Fukuda M, Kiger AA (2017). Genetic screen in Drosophila muscle identifies autophagy-mediated T-tubule remodeling and a Rab2 role in autophagy. eLife 6, e23367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan D, Cai Q (2021). Understanding amphisomes. Biochem J 478, 1959–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Langemeyer L, Kümmel D, Reggiori F, Ungermann C (2018). Molecular mechanism to target the endosomal Mon1-Ccz1 GEF complex to the pre-autophagosomal structure. Elife 7, e31145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graef M, Friedman JR, Graham C, Babu M, Nunnari J (2013). ER exit sites are physical and functional core autophagosome biogenesis components. Mol Biol Cell 24, 2918–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green N, Odell N, Zych M, Clark C, Wang ZH, Biersmith B, Bajzek C, Cook KR, Dushay MS, Geisbrecht E (2016). A common suite of coagulation proteins function in Drosophila muscle attachment. Genetics 204, 1075-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra F, Bucci C (2016). Multiple roles of the small GTPase Rab7. Cells 5, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmi V, Tomelleri G, Vattemi G (2018). Myofibrillar myopathies through the microscope: from diagnosis to molecular pathogenesis. Microscopie 29. [Google Scholar]
- Hakeemi MS, Ansari S, Teuscher M, Weißkopf M, Großmann D, Kessel T, Dönitz J, Siemanowski J, Wan X, Schultheis D, et al. (2022). Screens in fly and beetle reveal vastly divergent gene sets required for developmental processes. BMC Biol 20, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Rubinsztein DC, Walker DW (2018). Autophagy as a promoter of longevity: insights from model organisms. Nat Rev Mol Cell Biol 19, 579–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegedűs K, Takáts S, Boda A, Jipa A, Nagy P, Varga K, Kovács AL, Juhász G (2016). The Ccz1-Mon1-Rab7 module and Rab5 control distinct steps of autophagy. Mol Biol Cell 27, 3132–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenstein DM, Kraft C (2020). Autophagosomes are formed at a distinct cellular structure. Curr Opin Cell Biol 65, 50–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Sankar DS, Vu B, Leytens A, Vionnet C, Wu W, Stumpe M, Martínez-Martínez E, Stork B, Dengjel J (2021). ULK1 phosphorylation of striatin activates protein phosphatase 2A and autophagy. Cell Rep 36, 109762. [DOI] [PubMed] [Google Scholar]
- Hwang J, Pallas DC (2014). STRIPAK complexes: structure, biological function, and involvement in human diseases. Int J Biochem Cell Biol 47, 118–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyttinen JM, Niittykoski M, Salminen A, Kaarniranta K (2013). Maturation of autophagosomes and endosomes: a key role for Rab7. Biochim Biophys Acta 1833, 503–510. [DOI] [PubMed] [Google Scholar]
- Höhfeld J, Benzing T, Bloch W, Fürst DO, Gehlert S, Hesse M, Hoffmann B, Hoppe T, Huesgen PF, Köhn M, et al. (2021). Maintaining proteostasis under mechanical stress. EMBO Rep 22, e52507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami J, Brooks D, Zalmai R, Hartson SD, Bouyain S, Geisbrecht ER (2022). Complex protein interactions mediate Drosophila Lar function in muscle tissue. PLoS One 17, e0269037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya-Çopur A, Marchiano F, Hein MY, Alpern D, Russeil J, Luis NM, Mann M, Deplancke B, Habermann BH, Schnorrer F (2021). The Hippo pathway controls myofibril assembly and muscle fiber growth by regulating sarcomeric gene expression. Elife 10, e63726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazuma S (2021). Autophagic vacuolar myopathy: Danon disease and related myopathies. Neurol Clin Neurosci 10, 271–347. [Google Scholar]
- Kean MJ, Ceccarelli DF, Goudreault M, Sanches M, Tate S, Larsen B, Gibson LC, Derry WB, Scott IC, Pelletier L, et al. (2011). Structure-function analysis of core STRIPAK Proteins: a signaling complex implicated in Golgi polarization. J Biol Chem 286, 25065–25075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettern N, Dreiseidler M, Tawo R, Höhfeld J (2010). Chaperone-assisted degradation: multiple paths to destruction. Biol Chem 391, 481–489. [DOI] [PubMed] [Google Scholar]
- Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y (1999). Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol 147, 435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, et al. (2016). Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12, 1-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, et al. (2007). Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131, 1149–1163. [DOI] [PubMed] [Google Scholar]
- Kück U, Radchenko D, Teichert I (2019). STRIPAK, a highly conserved signaling complex, controls multiple eukaryotic cellular and developmental processes and is linked with human diseases. Biol Chem 400, 1005–1022. [DOI] [PubMed] [Google Scholar]
- La Marca JE, Diepstraten ST, Hodge AL, Wang H, Hart AH, Richardson HE, Somers WG (2019). Strip and Cka negatively regulate JNK signalling during Drosophila spermatogenesis. Development 146. [DOI] [PubMed] [Google Scholar]
- LaBeau-DiMenna EM, Clark KA, Bauman KD, Parker DS, Cripps RM, Geisbrecht ER (2012). Thin, a Trim32 ortholog, is essential for myofibril stability and is required for the integrity of the costamere in Drosophila. Proc Natl Acad Sci USA 109, 17983–17988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo YB, Peng Y, Lu Y, Li Q, Duan H, Bi F, Yang H (2020). Expanding the clinico-genetic spectrum of myofibrillar myopathy: experience from a chinese neuromuscular center. Front Neurol 11, 1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lőrincz P, Mauvezin C, Juhász G (2017). Exploring autophagy in drosophila. Cells 6, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lőrincz P, Tóth S, Benkő P, Lakatos Z, Boda A, Glatz G, Zobel M, Bisi S, Hegedűs K, Takáts S, et al. (2017). Rab2 promotes autophagic and endocytic lysosomal degradation. J Cell Biol 216, 1937–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margeta M (2020). Autophagy defects in skeletal myopathies. Annu Rev Pathol 15, 261–285. [DOI] [PubMed] [Google Scholar]
- Mauvezin C, Ayala C, Braden CR, Kim J, Neufeld TP (2014). Assays to monitor autophagy in Drosophila. Methods 68, 134–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauvezin C, Nagy P, Juhász G, Neufeld TP (2015). Autophagosome-lysosome fusion is independent of V-ATPase-mediated acidification. Nat Commun 6, 7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TJ, Gubas A, Tooze SA (2018). A molecular perspective of mammalian autophagosome biogenesis. J Biol Chem 293, 5386–5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munteanu I, Kalimo H, Saraste A, Nishino I, Minassian BA (2017). Cardiac autophagic vacuolation in severe X-linked myopathy with excessive autophagy. Neuromuscul Disord 27, 185–187. [DOI] [PubMed] [Google Scholar]
- Nagy P, Varga Á, Kovács AL, Takáts S, Juhász G (2015). How and why to study autophagy in Drosophila: it’s more than just a garbage chute. Methods 75, 151–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neisch AL, Neufeld TP, Hays TS (2017). A STRIPAK complex mediates axonal transport of autophagosomes and dense core vesicles through PP2A regulation. J Cell Biol 216, 441–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi M, Yamano K, Sato M, Matsuda N, Okamoto K (2021). Molecular mechanisms and physiological functions of mitophagy. EMBO J 40, e104705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palikaras K, Lionaki E, Tavernarakis N (2018). Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat Cell Biol 20, 1013–1022. [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Øvervatn A, Bjørkøy G, Johansen T (2007). p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282, 24131–24145. [DOI] [PubMed] [Google Scholar]
- Parzych KR, Klionsky DJ (2014). An overview of autophagy: morphology, mechanism, and regulation. Antioxid Redox Signal 20, 460–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pircs K, Nagy P, Varga A, Venkei Z, Erdi B, Hegedus K, Juhasz G (2012). Advantages and limitations of different p62-based assays for estimating autophagic activity in Drosophila. PLoS One 7, e44214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston E, Ploug T, Kalhovde J, Lomo T (2001). Golgi complex, endoplasmic reticulum exit sites, and microtubules in skeletal muscle fibers are organized by patterned activity. J Neurosci 21, 875–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran N, Munteanu I, Wang P, Ruggieri A, Rilstone JJ, Israelian N, Naranian T, Paroutis P, Guo R, Ren ZP, et al. (2013). VMA21 deficiency prevents vacuolar ATPase assembly and causes autophagic vacuolar myopathy. Acta Neuropathol 125, 439–457. [DOI] [PubMed] [Google Scholar]
- Rowland TJ, Sweet ME, Mestroni L, Taylor MR (2016). Danon disease - dysregulation of autophagy in a multisystem disorder with cardiomyopathy. J Cell Sci 129, 2135–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma C, Kawauchi T, Haraguchi S, Shikanai M, Yamaguchi Y, Gelfand VI, Luo L, Miura M, Chihara T (2014). Drosophila Strip serves as a platform for early endosome organization during axon elongation. Nat Commun 5, 5180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt-Engel C, Schultheis D, Schwirz J, Ströhlein N, Troelenberg N, Majumdar U, Dao VA, Grossmann D, Richter T, Tech M, et al. (2015). The iBeetle large-scale RNAi screen reveals gene functions for insect development and physiology. Nat Commun 6, 7822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheis D, Weißkopf M, Schaub C, Ansari S, Dao VA, Grossmann D, Majumdar U, Hakeemi MS, Troelenberg N, Richter T, et al. (2019). A Large Scale Systemic RNAi Screen in the Red Flour Beetle Tribolium castaneum Identifies Novel Genes Involved in Insect Muscle Development. G3 (Bethesda) 9, 1009–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sents W, Ivanova E, Lambrecht C, Haesen D, Janssens V (2013). The biogenesis of active protein phosphatase 2A holoenzymes: a tightly regulated process creating phosphatase specificity. FEBS J 280, 644–661. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Jiao S, Zhou Z (2016). STRIPAK complexes in cell signaling and cancer. Oncogene 35, 4549–4557. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Guhde G, Suter A, Eskelinen EL, Hartmann D, Lüllmann-Rauch R, Janssen PM, Blanz J, von Figura K, Saftig P (2000). Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature 406, 902–906. [DOI] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J (2016). The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13, 731–740. [DOI] [PubMed] [Google Scholar]
- Ulbricht A, Arndt V, Höhfeld J (2013). Chaperone-assisted proteostasis is essential for mechanotransduction in mammalian cells. Commun Integr Biol 6, e24925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZH, Clark C, Geisbrecht ER (2016). Drosophila clueless is involved in Parkin-dependent mitophagy by promoting VCP-mediated Marf degradation. Hum Mol Genet 25, 1946–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZH, Rabouille C, Geisbrecht ER (2015). Loss of a Clueless-dGRASP complex results in ER stress and blocks Integrin exit from the perinuclear endoplasmic reticulum in Drosophila larval muscle. Biol Open 4, 636–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Q, Huang X, Huang J, Zheng Y, March ME, Li J, Wei Y (2021). The role of autophagy in skeletal muscle diseases. Front Physiol 12, 638983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Wang X (2021). Lysosome biogenesis: regulation and functions. J Cell Biol 220, e202102001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YG, Zhang H (2018). Formation and maturation of autophagosomes in higher eukaryotes: a social network. Curr Opin Cell Biol 53, 29–36. [DOI] [PubMed] [Google Scholar]
- Zhao YG, Zhang H (2019). Autophagosome maturation: an epic journey from the ER to lysosomes. J Cell Biol 218, 757–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirin J, Nieuwenhuis J, Perrimon N (2013). Role of autophagy in glycogen breakdown and its relevance to chloroquine myopathy. PLoS Biol 11, e1001708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirin J, Nieuwenhuis J, Samsonova A, Tao R, Perrimon N (2015). Regulators of autophagosome formation in Drosophila muscles. PLoS Genet 11, e1005006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.