Abstract
The α-arrestin ARRDC3 is a recently discovered tumor suppressor in invasive breast cancer that functions as a multifaceted adaptor protein to control protein trafficking and cellular signaling. However, the molecular mechanisms that control ARRDC3 function are unknown. Other arrestins are known to be regulated by posttranslational modifications, suggesting that ARRDC3 may be subject to similar regulatory mechanisms. Here we report that ubiquitination is a key regulator of ARRDC3 function and is mediated primarily by two proline-rich PPXY motifs in the ARRDC3 C-tail domain. Ubiquitination and the PPXY motifs are essential for ARRDC3 function in regulating GPCR trafficking and signaling. Additionally, ubiquitination and the PPXY motifs mediate ARRDC3 protein degradation, dictate ARRDC3 subcellular localization, and are required for interaction with the NEDD4-family E3 ubiquitin ligase WWP2. These studies demonstrate a role for ubiquitination in regulating ARRDC3 function and reveal a mechanism by which ARRDC3 divergent functions are controlled.
INTRODUCTION
Despite advances in treatment, breast cancer remains the second leading cause of cancer death among women in the United States (Giaquinto et al., 2022). Thus, there is an urgent need for identification of new therapeutic targets. Arrestin domain–containing protein 3 (ARRDC3) has emerged as a tumor suppressor for metastatic breast cancer (Adelaide et al., 2007; Draheim et al., 2010; Soung et al., 2014). ARRDC3 inhibits epithelial-to-mesenchymal transition in breast cancer cells (Soung et al., 2019; Zapparoli et al., 2020) and sensitizes invasive breast cancer cells to chemotherapy (Soung et al., 2019). Moreover, ARRDC3 controls endolysosomal trafficking of plasma membrane proteins including protease-activated receptor-1 (PAR1), a G protein–coupled receptor (GPCR), and integrins, thereby suppressing invasive breast cancer cell migration, invasion, and metastasis in vitro and in vivo (Draheim et al., 2010; Arakaki et al., 2018, 2021; Soung et al., 2018). However, the molecular mechanisms that control ARRDC3 tumor suppressor function remain unknown.
ARRDC3 is a member of the mammalian α-arrestin family, which consists of ARRDC1-5 and thioredoxin-interacting protein (TXNIP) (Alvarez, 2008). Although mammalian α-arrestins share little sequence homology with β-arrestins, the two families are structurally similar, with both containing N- and C-domains composed of seven-stranded β-sheet arrestin domains and a finger loop region (Aubry et al., 2009; Polekhina et al., 2013; Hwang et al., 2014; Qi et al., 2014a). However, α-arrestins and β-arrestins differ considerably in the C-tail region. Most α-arrestins, besides TXNIP (Qualls-Histed et al., 2023), are not known to contain a C-tail clathrin-binding motif, and all α-arrestins lack adaptor protein-2 (AP-2) recognition motifs, which are present in β-arrestins. Instead, α-arrestins contain proline-proline-X-tyrosine (PPXY) motifs, which are known to interact with WW domains, particularly of the NEDD4 family of E3 ubiquitin ligases (Nabhan et al., 2010; Rauch and Martin-Serrano, 2011). However, compared with the well-characterized β-arrestins, very little is known about how mammalian α-arrestin function is regulated.
In contrast to mammalian α-arrestins, yeast α-arrestins have been widely studied. Yeast do not express β-arrestins, multifunctional GPCR adaptor proteins, but do express 14 distinct α-arrestin adaptor proteins. In Saccharomyces cerevisiae, where α-arrestins have been most extensively studied, α-arrestins have been shown to function in large part as adaptor proteins that facilitate protein trafficking through recruitment of Rsp5 E3 ubiquitin ligase (the orthologue of human NEDD4) via the Rsp5 WW domains and α-arrestin PPXY motifs to ubiquitinate plasma membrane proteins (Boase and Kelly, 2004; Lin et al., 2008; Nikko and Pelham, 2009; Novoselova et al., 2012). Besides harboring PPXY motifs, some yeast α-arrestins have also been demonstrated to interact with clathrin and clathrin adaptor proteins (O’Donnell et al., 2010). Like mammalian β-arrestins, yeast α-arrestin function is regulated through postranslational modifications—primarily phosphorylation and ubiquitination. Phosphorylation of yeast α-arrestins is suggested to inhibit α-arrestin function; upon dephosphorylation, yeast α-arrestins can promote endocytosis of plasma membrane proteins (O’Donnell and Schmidt, 2019; Zbieralski and Wawrzycka, 2022). Conversely, ubiquitination of yeast α-arrestins appears to promote α-arrestin function through regulated interactions with both cargo proteins and Rsp5 E3 ubiquitin ligase (MacDonald et al., 2020; Zbieralski and Wawrzycka, 2022).
Because previous studies have described potential interactions between ARRDC3 and multiple NEDD4-family E3 ubiquitin ligases (Rauch and Martin-Serrano, 2011; Qi et al., 2014b; Dores et al., 2015; Xiao et al., 2018), we hypothesized that regulation of ubiquitination may be important for controlling mammalian α-arrestin ARRDC3 divergent functions. Here, we report that the ARRDC3 PPXY motifs are required for ARRDC3 ubiquitination and are critical determinants for ARRDC3 function in regulating both PAR1 endolysosomal trafficking and GPCR-induced Hippo pathway signaling. ARRDC3 ubiquitination and the PPXY motifs are required for regulation of ARRDC3 protein stability, ARRDC3 subcellular localization, and ARRDC3 interaction with WWP2 E3 ligase. These findings demonstrate that ubiquitin is an important mediator of mammalian α-arrestin ARRDC3 divergent functions.
RESULTS
ARRDC3 ubiquitination is regulated by PAR1 stimulation and requires WWP2 E3 ubiquitin ligase
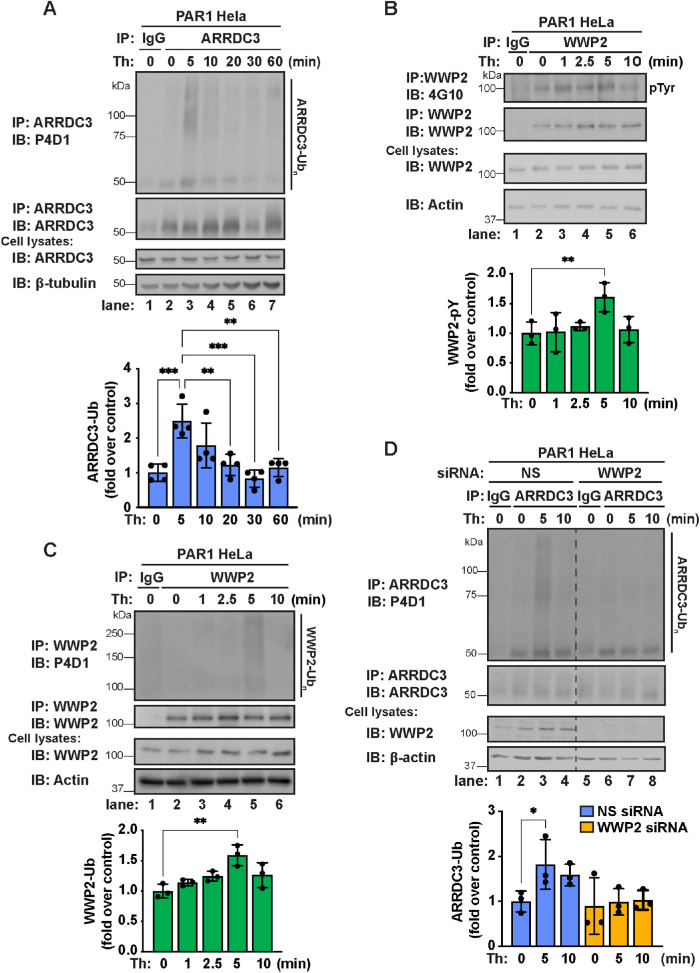
To assess whether ARRDC3 function might be regulated by ubiquitin, we first examined whether ARRDC3 ubiquitination status is modulated by thrombin-induced PAR1 activation. We observed a significant increase in ARRDC3 ubiquitination compared with control cells after 5 min of thrombin stimulation (Figure 1A, lanes 2 and 3), which was significantly decreased by 20 min of thrombin stimulation (Figure 1A, lane 5). These findings suggest that ARRDC3 ubiquitination modulated by thrombin stimulation may regulate ARRDC3 function.
FIGURE 1:
ARRDC3 ubiquitination is modulated by thrombin stimulation and requires WWP2. (A) Top: PAR1 HeLa cells were stimulated with 10 nM thrombin for the indicated times, and endogenous ARRDC3 was immunoprecipitated. Immunoprecipitates were immunoblotted for endogenous ARRDC3 ubiquitination. Bottom: Quantification of ARRDC3 ubiquitination (mean ± SD from four independent experiments; one-way ANOVA; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001). (B) Top: PAR1 HeLa cells were stimulated with 10 nM thrombin for indicated times and then endogenous WWP2 was immunoprecipitated. Immunoprecipitates were immunoblotted for WWP2 tyrosine antibody using an anti–phospho-tyrosine antibody (4G10). Bottom: Quantification of WWP2 tyrosine phosphorylation (mean ± SD from five independent experiments; one-way ANOVA; **, p < 0.01). (C) Top: PAR1 HeLa cells were stimulated with 10 nM thrombin for indicated times and then endogenous WWP2 was immunoprecipitated. Immunoprecipitates were immunoblotted for WWP2 ubiquitination. Bottom: Quantification of ubiquitination (mean ± SD from three independent experiments; one-way ANOVA; **, p < 0.01). (D) Top: PAR1 HeLa cells were treated with nonspecific (NS) or WWP2-specific siRNA, stimulated with 10 nM thrombin for the indicated times, and then endogenous ARRDC3 was immunoprecipitated. Immunoprecipitates were immunoblotted for endogenous ARRDC3 ubiquitination. Bottom: Quantification of ARRDC3 ubiquitination (mean ± SD from three independent experiments; one-way ANOVA; *, p < 0.05).
Next, we sought to identify the E3 ubiquitin ligase that mediates ARRDC3 ubiquitination. In previous studies we showed that WWP2 and ARRDC3 coassociate (Dores et al., 2015), indicating a potential role for WWP2. As a NEDD4-family E3 ubiquitin ligase, the activity of WWP2 is known to be regulated through the release of autoinhibition. This can occur through phosphorylation at Y369 and Y392 within the WW2-WW3 linker region (Chen et al., 2017) or through an allosteric mechanism that involves interaction of WWP2 WW domains and PPXY motifs of other proteins (Rauch and Martin-Serrano, 2011; Mund et al., 2015). Thus, to determine whether WWP2 activity is regulated by thrombin stimulation, we examined WWP2 tyrosine phosphorylation using an anti–phospho-tyrosine 4G10 antibody. We observed a significant increase in WWP2 tyrosine phosphorylation compared with untreated control cells after 5 min of thrombin stimulation (Figure 1B, lanes 2–5), suggesting that WWP2 is activated by thrombin stimulation. To verify WWP2 activation, we also assessed WWP2 autoubiquitination. Upon activation, HECT domain–containing E3 ligases are autoubiquitinated (Grimsey et al., 2018); thus, WWP2 ubiquitination status was used to assess its activity. We observed a significant increase in WWP2 ubiquitination after 5 min of thrombin stimulation compared with untreated control cells (Figure 1C, lanes 2–5), providing further support that WWP2 is activated by thrombin stimulation. Together, these data indicate that WWP2 activity is regulated by thrombin stimulation.
To test whether WWP2 is required for ARRDC3 ubiquitination, we examined thrombin-induced ARRDC3 ubiquitination in cells depleted of WWP2 expression. In cells transfected with nonspecific small interfering RNA (siRNA), thrombin induced a significant increase in ARRDC3 ubiquitination after 5 min compared with untreated cells (Figure 1D, lanes 1–4). However, in cells depleted of WWP2 expression by using siRNA, thrombin treatment failed to induce ARRDC3 ubiquitination (Figure 1D, lanes 5–8). These findings indicate that WWP2 is required for thrombin-induced ARRDC3 ubiquitination and further suggest that ARRDC3 function could be regulated by ubiquitination.
The ARRDC3-WWP2 interaction is predicted to occur through the PPXY motifs and WW1 and WW2 domains of WWP2
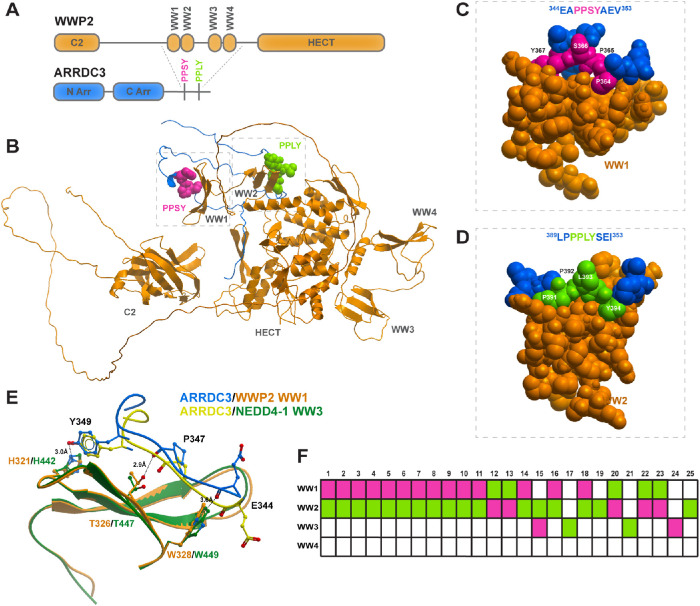
Next, we sought to elucidate the molecular determinants that mediate ARRDC3 and WWP2 interaction. WWP2 consists of a C2 domain, four WW domains, and a HECT ubiquitin ligase domain. WW domains are known to preferentially recognize and bind to short, proline-rich motifs (Macias et al., 2002), and thus we hypothesized that ARRDC3 PPXY motifs (346PPSY motif is shown as pink and the 391PPLY motif as green) may mediate interaction with WWP2 WW domains (Figure 2A). To test this hypothesis, we computationally generated an ensemble of 25 ranked molecular complexes between the C-tail region of ARRDC3 (residues 315–414) and the full-length WWP2 by using Alphafold2 Multimer (Jumper et al., 2021; Varadi et al., 2022). Alphafold2 predicted a direct interaction of the ARRDC3 C-tail region (which contains the PPXY motifs) with WWP2 WW domains. In the highest-ranked model of ARRDC3 C-tail–WWP2 interaction generated with the Alphafold2 multimer, it appears that the ARRDC3 346PPSY motif interacts with the WW1 domain and 391PPLY with the WW2 domain of WWP2 (Figure 2B). As illustrated in the space-filling model (Figure 2, C and D), both 346PPSY and 391PPLY are predicted to pack tightly into the WW domains, consistent with the idea that the ARRDC3 PPXY motifs and WWP2 WW domains are the key determinants that mediate ARRDC3-WWP2 interaction.
FIGURE 2:
The ARRDC3-WWP2 interaction is predicted to occur through the PPXY motifs and preferential WW domains. (A) WWP2 (orange) contains a C2 autoinhibitory domain, four WW domains, and a HECT E3 ligase domain. ARRDC3 (blue) contains an N- and C-arrestin domain and two PPXY motifs (pink and lime green) in the C-tail. (B) Alphafold2 model of interaction between the ARRDC3 C-tail and WWP2. (C, D) Space-filling model of Alphafold2 prediction of ARRDC3 PPSY peptide and WWP2 WW1 or ARRDC3 PPLY and WWP2 WW2. (E) Superposition of ARRDC3 PPSY peptide and NEDD4-1 WW3 (PDB: 4N7H). (F) Graphical depiction of interaction between PPSY (pink) and PPLY (lime green) and the indicated WWP2 WW domain in each of the 25 top-ranked Alphafold2 models of interaction.
Next, we compared our predicted interaction of ARRDC3 346PPSY and WWP2 WW1 domain with the previously published experimental structure of ARRDC3 364PPSY and NEDD4-1 WW3 domain (Figure 2E) (Qi et al., 2014b). Upon superimposition of the WW domains, we observed that the structures of WWP2 WW1 and NEDD4-1 WW3 are highly conserved, with both consisting of a triple-stranded β-sheet (Figure 2E, orange and green), and that the potential sites of hydrogen bonding appear to be similar as well. As was shown for NEDD4-1 and ARRDC3 (Qi et al., 2014b), WWP2 and ARRDC3 appear to be in close enough proximity to make hydrogen bonds between E344, P347, or Y349 of the PPSY peptide and conserved residues W328, T326, or H321 of WWP2, respectively (Figure 2E). The data suggest that the interaction between ARRDC3 and NEDD4-family E3 ligases may be conserved across family members, although the interaction may occur at different WW domains.
To assess whether ARRDC3 may preferentially interact with the individual WW domains of WWP2, we examined the predicted interaction between the ARRDC3 C-tail and the individual WW1, WW2, WW3, and WW4 domains of WWP2 across the top 25 ranked Alphafold2 models. The computational analysis indicates that the ARRDC3 PPXY motifs display a clear preference for interaction with the WW1 and WW2 domains of WWP2, as these domains were predicted to be occupied by the ARRDC3 PPXY motifs in 22 of 25 models (Figure 2F). In only 4 of 25 models, Alphafold2 predicted either ARRDC3 346PPSY or 391PPLY to interact with the WW3 domain of WWP2 (Figure 2F). Taken together, these data suggest that the key determinants of ARRDC3-WWP2 interaction appear to be the ARRDC3 PPXY motifs and the WW1 and WW2 domains of WWP2 E3 ubiquitin ligase.
The ARRDC3 PPXY motifs are required for interaction with WWP2 E3 ligase at WW1 and WW2 domains and mediate thrombin-induced ARRDC3 ubiquitination
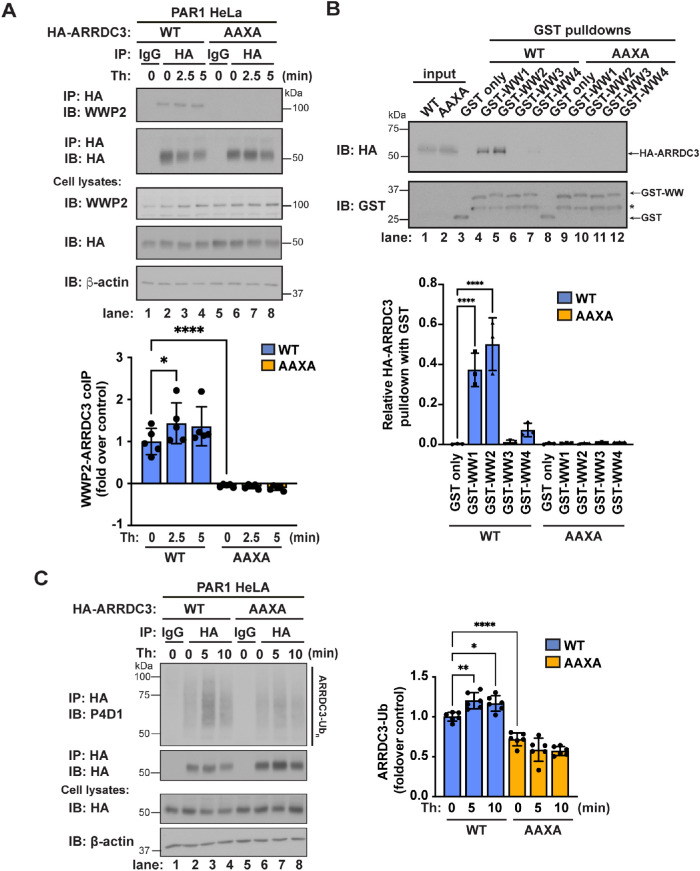
To experimentally examine whether ARRDC3-WWP2 interaction occurs through the ARRDC3 PPXY motifs and to determine whether the ARRDC3-WWP2 interaction is regulated by thrombin stimulation, coassociation of ARRDC3 wild type (WT) and a PPXY mutant was assessed in cells. An HA epitope-tagged ARRDC3 WT and mutant containing PPXY mutations in which both prolines (P) and tyrosines (Y) were converted to alanines (A), yielding a HA-ARRDC3-AAXA mutant (Arakaki et al., 2021), were expressed comparably in cells. The association between HA-ARRDC3-WT or the -AAXA mutant and endogenous WWP2 was then assessed by coimmunoprecipitation. We observed a modest but significant increase in WWP2 and HA-ARRDC3-WT coassociation following thrombin stimulation (Figure 3A, lanes 1–4). In contrast, no association was detected between WWP2 and HA-ARRDC3-AAXA, suggesting that the ARRDC3 PPXY motifs are essential for the interaction between ARRDC3 and WWP2 (Figure 3A, lanes 5–8). These results suggest that the ARRDC3 PPXY motifs are the key determinants of the ARRDC3-WWP2 interaction.
FIGURE 3:
ARRDC3 ubiquitination requires the ARRDC3 PPXY motifs. (A) Top: PAR1 HeLa cells exogenously expressing HA-ARRDC3-WT or the -AAXA mutant were stimulated with 10 nM thrombin for the indicated times, and HA-ARRDC3 was immunoprecipitated. Immunoprecipitates were immunoblotted for HA-ARRDC3 and coimmunoprecipitation of WWP2. Bottom: Quantification of coimmunoprecipitation of WWP2 with HA-ARRDC3 (mean ± SD from three independent experiments; one-way ANOVA; ****, p < 0.0001). (B) Top: HEK293T cell lysates with transfected HA-ARRDC3-WT or the -AAXA mutant were incubated with GST or the indicated GST-WWP2 WW domain fusion protein. Pull downs were immunoblotted for HA-ARRDC3 and GST fusion proteins. On GST immunoblot, * indicates GST fusion protein degradation product. Bottom: Quantification of pull down of HA-ARRDC3 with GST-WWP2 WW domain fusion protein (mean ± SD from three independent experiments; one-way ANOVA; ****, p < 0.0001). (C) Left: HA-ARRDC3-WT or -AAXA was exogenously expressed in PAR1 HeLa cells, and then the cells were stimulated with 10 nM thrombin for the indicated times and HA-ARRDC3 was immunoprecipitated. Immunoprecipitates were immunoblotted for HA-ARRDC3 ubiquitination. Right: Quantification of HA-ARRDC3 ubiquitination (mean ± SD from six independent experiments; one-way ANOVA; *, p < 0.05; **, p < 0.01; ****, p < 0.0001).
We next examined whether ARRDC3 preferentially interacts with the individual WW domains of WWP2 in cells as predicted by Alphafold2. To test this, we expressed and purified the four individual WW domains tagged with GST (Figure 3B, lanes 3–12). Each GST-WW domain was then incubated with cell lysates prepared from HEK293T cells expressing the comparable HA-ARRDC3-WT or HA-ARRDC3-AAXA mutant (Figure 3B, lanes 1 and 2). We observed a strong association between HA-ARRDC3-WT and WW1 and WW2 domains of WWP2, but not with the WW3 or WW4 domains of WWP2 (Figure 3B, lanes 3–6). Additionally, no association between the HA-ARRDC3-AAXA mutant and any of the WWP2 WW domains was observed (Figure 3B, lanes 8–12). Taken together, these results confirm that the ARRDC3-WWP2 interaction occurs via the ARRDC3 PPXY motifs and WWP2 WW domains 1 and 2.
Because the ARRDC3 PPXY motifs are required for association with WWP2, we hypothesized that the ARRDC3 PPXY motifs are key determinants of ARRDC3 ubiquitination. To test this, we assessed HA-ARRDC3-WT and -AAXA mutant ubiquitination in cells stimulated with or without thrombin. We observed a modest but significant increase in WT HA-ARRDC3 ubiquitination following thrombin treatment compared with that in untreated cells and the immunoglobulin G (IgG) control (Figure 3C, lanes 1–4), like that observed for endogenous ARRDC3 (Figure 1A, lanes 1–4). Conversely, thrombin failed to induce ubiquitination of the HA-ARRDC3-AAXA mutant (Figure 3C, lanes 5–8). These results suggest that the PPXY motifs are a key determinant of ARRDC3 ubiquitination.
Ubiquitin is important for ARRDC3 regulation of PAR1 trafficking and signaling
To examine the possibility that ubiquitination regulates divergent ARRDC3 functions, we first determined whether ubiquitination plays a role in ARRDC3-mediated regulation of PAR1 trafficking and signaling using MDA-MB-231 invasive breast carcinoma cells. JNK1 is an effector of PAR1-stimulated Gα12/13 signaling and a known mediator of PAR1-stimulated breast carcinoma invasion (Kelly et al., 2006; Juneja et al., 2011). In invasive breast cancer cells, ARRDC3 expression is diminished (Adelaide et al., 2007; Soung et al., 2014) and endogenous PAR1 lysosomal trafficking is dysregulated, resulting in increased PAR1 expression and persistent signaling through JNK1 (Booden et al., 2004; Arora et al., 2008; Arakaki et al., 2018). When ARRDC3 was reexpressed in MDA-MB-231 invasive breast carcinoma cells, PAR1 lysosomal trafficking and degradation was rescued and normal JNK1 signaling was restored (Arakaki et al., 2018).
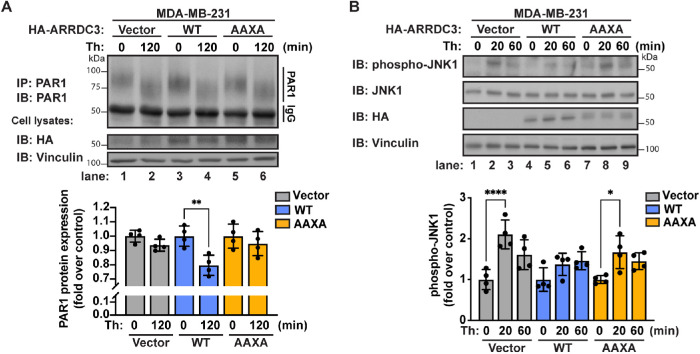
To determine whether ARRDC3 ubiquitination regulates PAR1 endolysosomal trafficking, endogenous PAR1 degradation was measured after prolonged thrombin stimulation. In these experiments, HA-ARRDC3-WT or the -AAXA mutant was reexpressed in MDA-MB-231 cells, which lack endogenous ARRDC3 expression, by lentiviral transduction using HA-ARRDC3-WT or the -AAXA mutant cloned in the pLJM1 vector (Figure 4A). Following thrombin incubation, a shift in PAR1 mobility is observed, indicative of proteolytic cleavage of the PAR1 N-terminus by thrombin. In MDA-MB-231 vector control cells, incubation with thrombin failed to induce PAR1 degradation (Figure 4A, lanes 1–2), whereas reexpression of HA-ARRDC3-WT was sufficient to restore thrombin-induced PAR1 degradation (Figure 4A, lanes 3–4), consistent with our previous study (Arakaki et al., 2018). In contrast to WT HA-ARRDC3, reexpression of the HA-ARRDC3-AAXA mutant was not able to rescue thrombin-stimulated PAR1 degradation (Figure 4A, lanes 5–6). These results suggest that ARRDC3 PPXY motifs and ubiquitination are required for ARRDC3 regulation of PAR1 endolysosomal trafficking.
FIGURE 4:
Ubiquitin differentially regulates ARRDC3 function in controlling PAR1 trafficking and persistent signaling. MDA-MB-231 cells were transduced with vector control, HA-ARRDC3-WT, or -AAXA lentivirus and stimulated with 10 nM thrombin for the indicated times. (A) Top: Endogenous PAR1 was immunoprecipitated and immunoblotted for PAR1 expression. Bottom: Quantification of PAR1 protein expression (mean ± SD from four independent experiments; one-way ANOVA; **, p < 0.01). (B) Top: Lysates were immunoblotted for phospho-JNK1 and total JNK1. Bottom: Quantification of JNK1 phosphorylation (mean ± SD from four independent experiments; one-way ANOVA; *, p < 0.05; ****, p < 0.0001).
In previous studies, we showed that ARRDC3-mediated suppression of PAR1 persistent signaling to JNK1 is linked to restoration of PAR1 endolysosomal sorting (Arakaki et al., 2018, 2021). Thus, we expected that ARRDC3 ubiquitination would have a similar effect on ARRDC3-mediated regulation of PAR1 trafficking and PAR1 persistent signaling to JNK1. To test this, HA-ARRDC3-WT or the -AAXA mutant was reexpressed in MDA-MB-231 cells by lentiviral transduction and then thrombin-induced JNK1 signaling was assessed by immunoblotting using anti–phospho-JNK1 threonine-183 and tyrosine-185 antibody. As expected, persistent signaling to JNK1 was observed in vector control cells following 20 min of thrombin stimulation (Figure 4B, lanes 1–3), and reexpression of WT HA-ARRDC3 was sufficient to suppress thrombin-induced persistent signaling to JNK1 at 20 min, compared with the 0 min control (Figure 4B, lanes 4–6). In contrast, reexpression of the HA-ARRDC3-AAXA mutant failed to suppress thrombin-stimulated JNK1 persistent signaling (Figure 4B, lanes 7–9), consistent with the idea that dysregulated PAR1 trafficking is linked to JNK1 persistent signaling. These results suggest that ARRDC3 regulation of PAR1 signaling is linked to receptor endolysosomal trafficking and that ubiquitin is important for normal regulation of ARRDC3 function.
Ubiquitin regulates ARRDC3 divergent functions in GPCR-mediated Hippo pathway signaling
To further understand the mechanisms by which ubiquitin regulates ARRDC3 function, we also examined the role of ubiquitin in ARRDC3 regulation of GPCR-stimulated Hippo pathway signaling. The Hippo pathway is dysregulated in many cancers and can be regulated by soluble factors that signal through GPCRs (Mo et al., 2012; Yu et al., 2012; Nag et al., 2018). We previously showed that in invasive breast carcinoma, PAR1, PAR2, and lysophosphatidic acid receptor (LPAR) regulate Hippo pathway signaling, leading to nuclear translocation of transcriptional coactivators YAP and TAZ and induction of CTGF expression (Arakaki et al., 2021). We additionally showed that for each of these receptors, reexpression of ARRDC3 significantly suppressed GPCR-promoted Hippo pathway signaling (Arakaki et al., 2021).
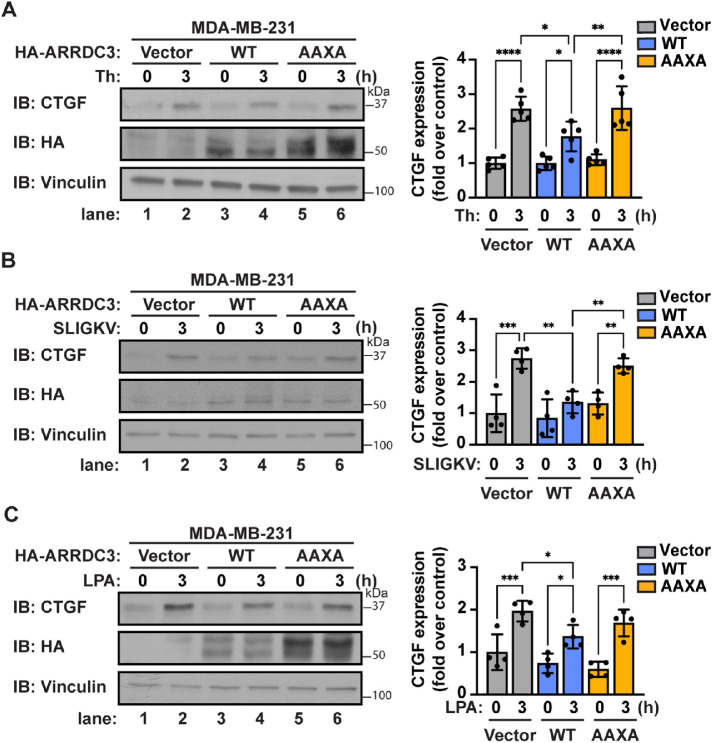
Thus, we examined whether ubiquitination controls ARRDC3 regulation of GPCR-induced Hippo pathway signaling in MDA-MB-231 cells reexpressing HA-ARRDC3-WT or the -AAXA mutant. Cells were stimulated with thrombin or left unstimulated. After agonist incubation, GPCR activation of Hippo pathway signaling was assessed by immunoblotting for induction of CTGF expression. Consistent with our previous study (Arakaki et al., 2021), reexpression of WT HA-ARRDC3 was sufficient to suppress thrombin/PAR1-mediated induction of CTGF expression (Figure 5A, lanes 1-4), whereas reexpression of the HA-ARRDC3-AAXA mutant failed to attenuate PAR1-stimulated CTGF induction (Figure 5A, lanes 5 and 6). To examine whether ARRDC3-mediated suppression of Hippo pathway signaling observed with other GPCRs is dependent on ARRDC3 ubiquitination, we stimulated cells with SLIGKV (PAR2 peptide agonist) or LPA (agonist for LPA receptors). Similar to thrombin/PAR1, activation of PAR2 and LPAR induced CTGF expression (Figure 5, B and C, lanes 1 and 2), which was suppressed by HA-ARRDC3-WT but not by the HA-ARRDC3-AAXA mutant (Figure 5, B and C, lanes 3–6). These results indicate that ARRDC3 ubiquitination mediated by the PPXY motifs is required for suppression of GPCR-stimulated Hippo pathway signaling.
FIGURE 5:
Ubiquitin differentially regulates ARRDC3 function in regulating GPCR signaling to the Hippo pathway. MDA-MB-231 cells were transduced with vector control, HA-ARRDC3-WT, or -AAXA lentivirus and stimulated with (A) 10 nM thrombin, (B) 10 μM SLIGKV, or (C) 100 nM LPA for the indicated times. Left: Lysates were immunoblotted for YAP/TAZ target gene CTGF. Right: Quantifications of CTGF induction (mean ± SD from four or five independent experiments; one-way ANOVA; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001).
The ARRDC3 PPXY motifs regulate protein stability
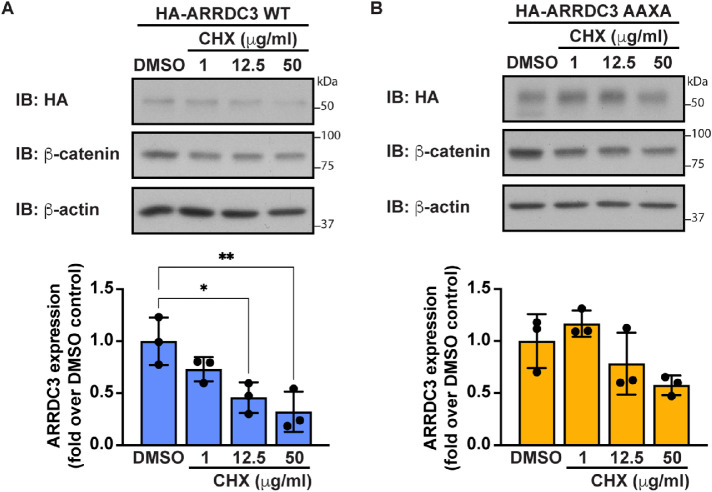
To determine how ubiquitination may modulate ARRDC3 function, we examined ARRDC3 expression. Ubiquitin is known to regulate many different functions; one of the most well-known functions is regulation of protein stability, where ubiquitin acts as a signal for protein degradation. To assess the role of ubiquitination on ARRDC3 protein stability, ARRDC3 protein degradation was measured after prolonged cycloheximide treatment to inhibit protein synthesis. β-Catenin was used as a control because it is known to undergo ubiquitin-induced protein degradation (Aberle et al., 1997). Thus, it was expected that β-catenin protein expression would decrease following cycloheximide treatment. We observed a significant decrease in HA-ARRDC3-WT protein levels following cycloheximide treatment, suggesting that ARRDC3 undergoes protein degradation (Figure 6A). In contrast, no significant loss of HA-ARRDC3-AAXA protein was observed following cycloheximide treatment, suggesting that ubiquitination and the ARRDC3 PPXY motifs regulate ARRDC3 protein expression (Figure 6B). These results indicate that ubiquitin is a key regulator of ARRDC3 protein stability.
FIGURE 6:
Ubiquitin regulates ARRDC3 protein stability. (A) HA-ARRDC3-WT or (B) -AAXA was expressed in HEK293T cells and treated with the indicated concentration of cycloheximide (CHX) for 24 h. Top: Lysates were immunoblotted for HA-ARRDC3. Bottom: Quantification of ARRDC3 expression (mean ± SD from three independent experiments; one-way ANOVA; *, p < 0.05; **, p < 0.01).
Ubiquitin plays a role in regulation of ARRDC3 subcellular localization
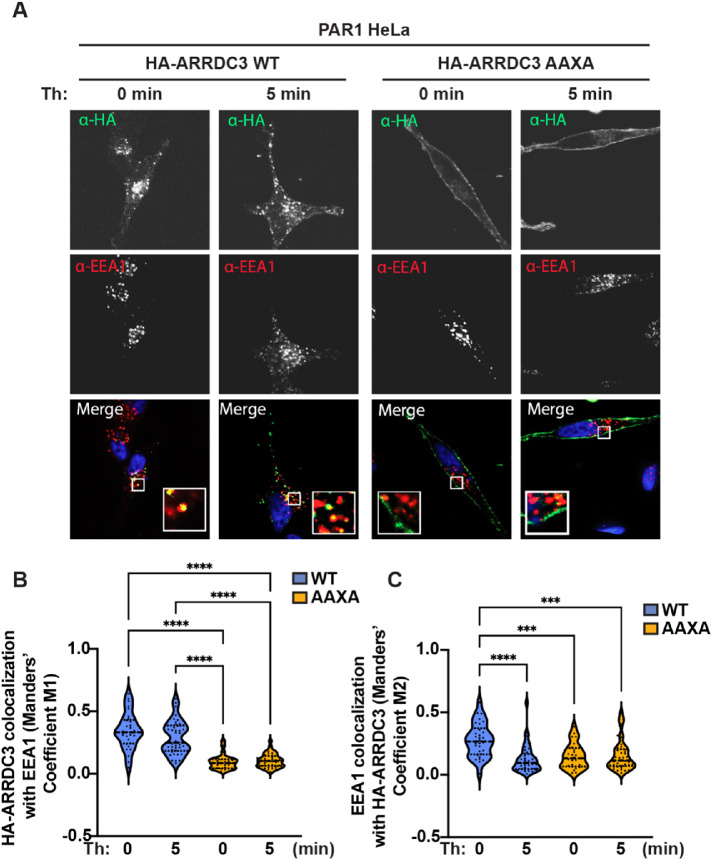
To test whether ubiquitination regulates other facets of ARRDC3 activity, we examined ARRDC3 subcellular localization. ARRDC3 has previously been reported to localize primarily to the early and late endosomal compartments and appears to be mediated by the PPXY motifs (Dores et al., 2015; Tian et al., 2016). We first examined whether thrombin stimulation altered ARRDC3 WT or mutant endosomal subcellular localization by immunofluorescence confocal microscopy. HA-ARRDC3-WT or -AAXA mutant subcellular localization was detected by immunostaining for HA-ARRDC3 (Figure 7A, green) and early endosome antigen 1 (EEA1; Figure 7A, red). Colocalization of HA-ARRDC3 with EEA1 (Figure 7A, yellow) was quantified using the Manders overlap coefficient (MOC) (M1) to determine the proportion of HA-ARRDC3 colocalization with EEA1. The MOC measures the fraction of one protein that colocalizes with a second protein and thus is useful to determine colocalization in situations where there is a difference in the distributions of two proteins (Dunn et al., 2011). As expected, colocalization of HA-ARRDC3-WT with EEA1 was observed (Figure 7A, yellow); however, thrombin failed to induce any significant changes in HA-ARRDC3-WT colocalization with EEA1 (Figure 7B). Consistent with a previous study (Tian et al., 2016), the HA-ARRDC3-AAXA mutant localized primarily at the plasma membrane, in marked contrast to HA-ARRDC3-WT subcellular localization (Figure 7A). HA-ARRDC3-AAXA exhibited significantly reduced colocalization with EEA1 as compared with HA-ARRDC3-WT and was not altered by thrombin stimulation (Figure 7B). These results suggest that while the ARRDC3 PPXY motifs are key determinants for ARRDC3 constitutive association with endosomes, neither the endosomal localization of WT ARRDC3 nor plasma membrane localization of the PPXY mutant is regulated by thrombin signaling.
FIGURE 7:
Ubiquitin contributes to regulation of ARRDC3 endosomal localization. (A) HA-ARRDC3-WT or -AAXA was exogenously expressed in PAR1 HeLa cells, and the cells were processed for immunofluorescence and stained for HA-ARRDC3, EEA1, and DAPI. (B, C) Quantification of colocalization using the Manders coefficient of (B) HA-ARRDC3 with EEA1 or (C) EEA1 with HA-ARRDC3 (mean ± SD from three independent experiments, 8–12 cells per experiment; one-way ANOVA; ***, p < 0.001; ****, p < 0.0001).
We also quantified the MOC (M2) to determine the relative proportion of EEA1-positive puncta that overlap with HA-ARRDC3. As expected, in cells expressing HA-ARRDC3-WT, colocalization of EEA1 with HA-ARRDC3-WT was observed, but surprisingly, thrombin stimulation led to a significant decrease in colocalization (Figure 7C). As expected, we observed a significant decrease in EEA1 colocalization with HA-ARRDC3-AAXA compared to HA-ARRDC3-WT (Figure 7C). These results suggest that while thrombin-induced ubiquitination does not regulate HA-ARRDC3 endosomal subcellular localization (Figure 7B), thrombin signaling and ARRDC3 ubiquitination may regulate the proportion of EEA1-positive endosomes that contain HA-ARRDC3 (Figure 7C). Together, these data suggest that ARRDC3 PPXY motifs and ubiquitination are the major determinants for regulation of ARRDC3 subcellular localization.
DISCUSSION
The α-arrestin ARRDC3 has been demonstrated to display tumor suppressor activity in invasive breast carcinoma (Draheim et al., 2010; Arakaki et al., 2018, 2021; Soung et al., 2018). However, unlike for yeast α-arrestins, which have been demonstrated to be regulated by ubiquitination and phosphorylation (O’Donnell and Schmidt, 2019; MacDonald et al., 2020; Zbieralski and Wawrzycka, 2022), the mechanisms that control mammalian α-arrestin function are not known. Here, we show that ubiquitin is a key regulator of ARRDC3 function. We found that WWP2 E3 ubiquitin ligase is activated by thrombin stimulation, interacts with ARRDC3 via the ARRDC3 PPXY motifs and WW1 and WW2 domains of WWP2, and is required for ARRDC3 ubiquitination. In addition, we show that the ARRDC3 PPXY motifs and ubiquitination are required for ARRDC3 divergent functions in regulating GPCR trafficking and signaling. We further demonstrate that ARRDC3 ubiquitination plays a role in regulating ARRDC3 protein stability and endosomal localization.
Thrombin-induced activation of WWP2 E3 ligase leads to ARRDC3 ubiquitination mediated by the ARRDC3 PPXY motifs
We found that the ARRDC3 PPXY motifs are the key determinants for interaction with WWP2 E3 ligase (Figures 2 and 3). Our Alphafold2 model of interaction and biochemical analysis demonstrates that the ARRDC3 PPXY motifs preferentially interact with the WW1 and WW2 domains of WWP2 (Figures 2 and 3). This is also consistent with other studies that have identified an interaction between WWP2 WW domains 1 and 2 and PPXY motifs of nonarrestins such as Dishevelled 2 (Mund et al., 2015). In contrast, when interacting with NEDD4-1, the ARRDC3 PPXY motifs were demonstrated to prefer WW domains 2 and 3 (Qi et al., 2014b). We also found that the structural basis of the interaction of the ARRDC3 346PPSY motifs with NEDD4-1 WW domain 3 was conserved when this motif bound the WWP2 WW domain 1 (Figure 2). For yeast α-arrestins, this interaction is also conserved: a previous study found that the yeast α-arrestin Art1 interacts with Rsp5 via the Art1 PPXY motifs and Rsp5 WW domains; however, Art1 preferentially interacts with Rsp5 WW domains 2 and 3 (Lin et al., 2008). Together, these studies and ours suggest that the specificity of interaction between NEDD4-family E3 ligases and PPXY motif–containing proteins may be dictated by the E3 ligase.
Our results also indicate that WWP2 is required for thrombin-induced ARRDC3 ubiquitination (Figure 1). However, it remains unknown whether this interaction regulates WWP2 activation. The activation of WWP2 and other NEDD4-family E3 ligases is known to occur via tyrosine phosphorylation of an autoinhibitory linker region located between WW domains (Chen et al., 2017; Grimsey et al., 2018) and is often allosterically regulated by WW domain binding to PPXY motifs of substrate proteins (Mund et al., 2015; Mund and Pelham, 2018). Similar to other PPXY motif–containing proteins, ARRDC3 may allosterically promote the activation of WWP2 or function as an adaptor to facilitate WWP2 tyrosine phosphorylation, but direct proof is so far lacking. Furthermore, as we previously demonstrated, activation of the closely related NEDD4-2 E3 ligase following activation of PAR1 occurs through c-Src–mediated tyrosine phosphorylation (Grimsey et al., 2018). Interestingly, our recent proteomics study identified Src family kinases c-Src, Fyn, and Lyn among the top hits of ARRDC3-interacting proteins (Wedegaertner et al., 2022). These studies suggest the possibility that ARRDC3 could function as a mediator of WWP2 activation through c-Src, as well. Thus, ARRDC3, an adaptor protein, may act as a hub to regulate WWP2 activity and substrate specificity, but future studies are necessary to test this idea.
ARRDC3 ubiquitination is required for regulation of PAR1 endolysosomal trafficking and persistent signaling from the plasma membrane
Previous studies showed that both ARRDC3 and ALG-interacting protein X (ALIX) are required for targeting PAR1 to the endolysosomal sorting pathway, preventing recycling of activated PAR1 and suppressing persistent signaling to JNK1 (Dores et al., 2015; Arakaki et al., 2018). We also showed that ARRDC3 mediates ALIX ubiquitination, which facilitates ALIX-mediated PAR1 endolysosomal trafficking (Dores et al., 2015). In addition, in MDA-MB-231 invasive breast cancer cells that were depleted of ALIX by siRNA, reexpression of HA-ARRDC3-WT was not sufficient to suppress PAR1-persistent signaling to JNK1 (Arakaki et al., 2018, 2021). The observation that the ARRDC3 PPXY motifs are required for both ARRDC3-mediated regulation of PAR1 endolysosomal trafficking and suppression of PAR1 persistent signaling to JNK1 (Figure 4) is consistent with the idea that PAR1 dysregulated trafficking and persistent signaling are linked, as expected.
ARRDC3 ubiquitination is required for suppression of GPCR-induced Hippo pathway signaling
We previously demonstrated that ARRDC3-mediated suppression of thrombin-induced Hippo pathway signaling is independent of ARRDC3 trafficking function and requires the ARRDC3 PPXY motifs (Arakaki et al., 2021). However, it was unknown whether this observation was broadly applicable to other GPCRs implicated in breast cancer progression. Only PAR1, and not PAR2 or LPAR, is expected to require ARRDC3 for endolysosomal trafficking (Dores et al., 2012); thus it was not clear whether the PPXY motifs would also be required for ARRDC3-mediated suppression of PAR2- and LPAR-induced Hippo pathway signaling. Here, we found that the ARRDC3 PPXY motifs are also required for suppression of PAR2- and LPAR-mediated Hippo pathway signaling (Figure 5). This is consistent with the idea that ARRDC3-mediated suppression of GPCR-induced Hippo pathway signaling is independent of ARRDC3 trafficking function (Arakaki et al., 2021). Additionally, ARRDC3 regulation of Hippo pathway signaling has been suggested to occur primarily through interaction of the ARRDC3 PPXY motifs with the WW domains of transcriptional coactivators YAP and TAZ (Shen et al., 2018; Xiao et al., 2018; Arakaki et al., 2021). Thus, it appears that the ARRDC3 PPXY motifs and ubiquitination regulate divergent ARRDC3 functions.
The ARRDC3 PPXY motifs are key regulators of ARRDC3 function
Here, we further demonstrate that the PPXY motifs are key determinants of ARRDC3 protein stability, endosomal subcellular localization, and interaction with WWP2 E3 ubiquitin ligase. We showed that ARRDC3 may be both monoubiquitinated and polyubiquitinated (Figure 1A). For yeast α-arrestins, which have been extensively studied, monoubiquitination has been shown to promote α-arrestin function in regulating plasma membrane protein endocytosis (Herrador et al., 2013; Hager et al., 2018), whereas polyubiquitination has been shown to promote α-arrestin degradation (Ho et al., 2017; Hovsepian et al., 2017). The ARRDC3 PPXY motifs appear to mediate ARRDC3 protein degradation, presumably through facilitating ARRDC3 polyubiquitination (Figure 6). This suggests that ubiquitin might control ARRDC3 activity through regulation of ARRDC3 expression levels. However, although the HA-ARRDC3-AAXA mutant appears to be more stable than HA-ARRDC3-WT, we observed that HA-ARRDC3-AAXA is unable to restore PAR1 endolysosomal trafficking and suppress persistent signaling (Figures 4 and 5). This suggests that ARRDC3 function is not solely regulated through ubiquitin-mediated protein degradation, and thus, ubiquitination may regulate other facets of ARRDC3 activity.
To this end, we further showed that the PPXY motifs are the key determinants for ARRDC3 endosomal subcellular localization (Figure 7), consistent with previous reports (Nabhan et al., 2010; Han et al., 2013; Tian et al., 2016). A previous study suggested that ubiquitinated ARRDC3 may interact with Hrs and STAM-1, two endosome-localized proteins that contain ubiquitin-binding motifs, to facilitate ARRDC3 endosomal subcellular localization (Tian et al., 2016). However, it is unclear whether ARRDC3 endosomal localization specifically requires ubiquitination or whether the PPXY motifs mediate interactions at the endosome independent of ubiquitination status.
Interestingly, although we did not observe any change in ARRDC3 endosomal localization following thrombin stimulation, we did observe a change in the relative proportion of EEA1-positive puncta that colocalize with ARRDC3 puncta. Other studies have posited that the relative proportion of EEA1-positive endosomes may change following agonist stimulation (Perini et al., 2014; Kamentseva et al., 2020); however, it is unknown whether this is also true for thrombin stimulation. These results indicate a potential role for thrombin/PAR1-mediated regulation of endosomal subpopulations.
In summary, our study reveals a role for ubiquitin-mediated regulation of divergent mammalian α-arrestin ARRDC3 tumor suppressor functions. Specifically, ubiquitin differentially regulates ARRDC3 function in regulating GPCR trafficking and signaling in cancer. This study further indicates that the ARRDC3 PPXY motifs are the key regulators of ARRDC3 function, likely through recruitment of WWP2 or other E3 ubiquitin ligases. This work provides the basis for future studies to identify druggable targets upstream of ARRDC3 ubiquitination with the aim of developing therapeutic treatments to prevent metastatic breast cancer.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Reagents and antibodies
Human α-thrombin was obtained from Enzyme Research Technologies (South Bend, IN). SLIGKV peptide agonist was synthesized at the Tufts University Core Facility (Boston, MA). Lysophosphatidic acid (LPA) was purchased from Tocris Bioscience (Bristol, UK). Protein A-Sepharose CL-4B beads were from GE Healthcare. Glutathione Sepharose 4B (catalogue no. 17075601) was purchased from Cytiva (Marlborough, MA). Mouse IgG (catalogue no. 010-0102) was purchased from Rockland Immunochemicals (Gilbertsville, PA). Rabbit monoclonal anti-ARRDC3 antibody (catalogue no. ab64187) was purchased from Abcam (Cambridge, UK). Polyclonal anti-GST conjugated to horseradish peroxidase (HRP) (catalogue no. RPN1236V) was purchased from Cytiva (Marlborough, MA). Mouse monoclonal anti-PAR1 WEDE15 was purified from hybridoma as previously described (Brass et al., 1992, 1994). Rabbit polyclonal C5433 anti-PAR1 antibody was described previously (Paing et al., 2006). Rabbit monoclonal β-catenin (catalogue no. 8480), rabbit polyclonal anti-HA (catalogue no. 3742), mouse monoclonal anti-HA conjugated to Alexa Fluor 488 (catalogue no. 2350), rabbit anti–phospho-JNK1/2 (catalogue no. 9251), mouse anti-JNK1 (catalogue no. 3708), and rabbit IgG (catalogue no. 2729) antibodies were purchased from Cell Signaling Technology (Danvers, MA). Mouse 4G10 platinum anti–phospho-tyrosine antibody (catalogue no. 170-6515) was purchased from Millipore (Burlington, MA). Rat monoclonal anti-HA conjugated to HRP (catalogue no. 11667475001) was purchased from Roche (Manheim, Germany). Mouse monoclonal anti-CTGF (catalogue no. sc-365970), mouse monoclonal P4D1 anti-ubiquitin (catalogue no. sc-8017), and mouse monoclonal anti-WWP2 (catalogue no. sc-166240) antibodies were purchased from Santa Cruz Biotechnology (Dallas, TX). Mouse monoclonal anti–β-actin (catalogue no. A5316) and mouse monoclonal anti-vinculin (catalogue no. V9131) antibodies were purchased from Sigma-Aldrich (Irvine, CA). Goat anti-mouse secondary antibody conjugated to Alexa Fluor 594 (catalogue no. A-11001) was purchased from Thermo Fisher Scientific (Waltham, MA). Anti-tubulin hFAB conjugated to rhodamine (catalogue no.12004166) and goat anti-mouse (catalogue no. 170-6516) and goat anti-rabbit (catalogue no. 170-6515) secondary antibodies conjugated to HRP were purchased from BioRad.
Cell culture, plasmids, and transfections
HeLa cells stably expressing FLAG-PAR1 were generated as previously described (Trejo et al., 2000) and maintained in DMEM supplemented with 10% fetal bovine serum (FBS) and 250 μg/ml hygromycin. HEK293T cells were purchased from the American Type Culture Collection (ATCC) and maintained in DMEM supplemented with 10% FBS (vol/vol). MDA-MB-231 cells were purchased from ATCC and maintained without CO2 in Leibowitz-15 medium supplemented with 10% FBS (vol/vol). siRNA transfections were performed using Oligofectamine (Life Technologies) per the manufacturer’s instructions. All single siRNAs were purchased from Qiagen: nonspecific siRNA sequence, 5′-CUACGUCCAGGAGCGCACC-3′; WWP2 target sequence, 5′-AACAGGATGGGAGATGAAATA-3′. cDNA transfections were performed using 1 mg/ml polyethyleneimine (PEI) at a 3:1 ratio (3 μl of PEI:1 μg of plasmid). The HA-ARRDC3 WT cDNA cloned into the pEF6 vector was provided by Q. Lu (Harvard University, Cambridge, MA). The HA-ARRDC3-AAXA plasmid was generated by site-directed mutagenesis. pHH0103 WWP2 WW domain #1 was a gift from Sachdev Sidhu and Marius Sudol (Addgene plasmid #104212; http://n2t.net/addgene:104212; RRID:Addgene_#104212). pHH0103 WWP2 WW domain #2 was a gift from Sachdev Sidhu and Marius Sudol (Addgene plasmid #104213; http://n2t.net/addgene:104213; RRID:Addgene_#104213). pHH0103 WWP2 WW domain #3 was a gift from Sachdev Sidhu and Marius Sudol (Addgene plasmid #104214; http://n2t.net/addgene:104214; RRID:Addgene #104214). pHH0103 WWP2 WW domain #4 was a gift from Sachdev Sidhu and Marius Sudol (Addgene plasmid #104215; http://n2t.net/addgene:104215; RRID:Addgene_#104215). Ubiquitination, stability, coimmunoprecipitation, and immunofluorescence assays described were performed 48–72 h after transfection.
ARRDC3 lentiviral constructs and transduction
pLJM1-eGFP (Addgene plasmid #19319; http://n2t.net/addgene:19319; RRID:Addgene_#19319) was used as backbone to subclone HA-tag, HA-ARRDC3-WT, and -AAXA using NheI and EcoRI to replace eGFP. Briefly, N-terminal HA-tagged ARRDC3-WT or -AAXA cDNAs were cloned into the pLJM vector to generate pLJM-HA-ARRDC3 vectors. Lentivirus was produced by transfecting HEK293T cells with the pLJM-HA-ARRDC3 vector along with packaging plasmids VSV-G and pCMVΔ8.2R using PEI. Lentivirus-containing supernatant was harvested at 48 and 72 h posttransfection, filtered, and used to transduce MDA-MB-231 cells overnight with 8 μg/ml polybrene (Millipore, Burlington, MA). MDA-MB-231 cells expressing pLJM-ARRDC3 vectors were selected using 0.75 μg/ml puromycin.
Immunoblotting
Cell lysates were collected in 2× Laemmli sample buffer containing 200 mM DTT. Samples were resolved by SDS–PAGE, transferred to polyvinylidene fluoride (PVDF) membranes, immunoblotted with appropriate antibodies, and then developed by chemiluminescence. Immunoblots were quantified by densitometry using ImageJ software (National Institutes of Health).
Immunoprecipitations
HeLa cells stably expressing FLAG-PAR1 were grown in 100 mm plates (1.5 × 106 cells/plate) overnight at 37°C and then transfected with either HA-ARRDC3-WT or the -AAXA mutant plasmid or endogenous protein was assessed. Cells were incubated for 48 h, washed, serum-starved for 2 h, and then stimulated for the indicated times with thrombin.
For ubiquitination assays, cells were washed and then lysed with RIPA buffer (5 mM EDTA, 50 mM Tris-HCl, pH 8.0, 0.5% [wt/vol] Na deoxycholate, 1% [vol/vol] NP-40, 100 mM NaCl, 1% SDS) supplemented with protease inhibitors, phosphatase inhibitors (50 mM NaF and 10 mM sodium pyrophosphate), 3 mg/ml N-ethylmaleimide, and 20 μM PR-619 (Boston Biochem, Boston, MA). Lysates were sonicated for 10 s at 10% amplitude (Branson Model 450 sonifier) and then clarified by centrifugation at 14,000 rpm for 30 min at 4°C.
For tyrosine phosphorylation assay, cells were washed and lysed with Triton lysis buffer (50 mM Tris-HCl, pH 7.4, 100 mM NaCl, 1% Triton X-100, 10 mM NaF, 10 mM β-glycerophosphate, 10 mM NaPP, 2 mM NaVO4) supplemented with protease inhibitors, 20 mM N-ethylmaleimide, and 20 µM PR-619. Lysates were sonicated for 10 s at 10% amplitude (Branson Model 450 sonifier) and then clarified by centrifugation at 14,000 rpm for 30 min at 4°C.
For coimmunoprecipitations, cells were washed and lysed with NP-40 buffer (0.5% [vol/vol] NP40, 20 mM Tris-HCl, pH 7.4, 150 mM NaCl) supplemented with protease inhibitors, phosphatase inhibitors (50 mM NaF and 10 mM sodium pyrophosphate), and 3 mg/ml N-ethylmaleimide. Lysates were passed through a 22 GA needle and then clarified by centrifugation at 14,000 rpm for 20 min at 4°C.
The protein concentration was normalized after quantification by bicinchoninic acid (BCA) assay (Thermo Fisher Scientific, Waltham, MA). Clarified lysates were incubated with protein A–Sepharose beads (GE Healthcare, Pittsburgh, PA) and indicated antibodies overnight at 4°C. Beads were washed and resuspended in 2× Laemmli sample buffer. Immunoprecipitates were resolved on 7% SDS–PAGE gels and then transferred onto PVDF and immunoblotted using anti-ubiquitin antibody, anti-HA antibody, and anti-WWP2 antibody.
Molecular modeling
The ARRDC3 C-tail interaction with WWP2 was modeled using a standalone installation of Alphafold2 Multimer version 2.2.0 (Jumper et al., 2021; Varadi et al., 2022). Modeling was run using the Triton Shared Computer Cluster (University of California, San Diego, La Jolla, CA). Structural models were analyzed using ICM v3.8-7a (Molsoft LLC, San Diego, CA).
GST pull-down assay
GST-WW1, GST-WW2, GST-WW3, and GST-WW4 fusion proteins were transformed into BL21 CodonPlus-competent (DE3) Escherichia coli (Stratagene, San Diego, CA). Fusion protein expression was induced with 0.5 M isopropyl β-d-1-thiogalactopyranoside (IPTG) at 30°C for 3 h. Cell pellets were resuspended in phosphate-buffered saline (PBS) containing 0.1 M dithiothreitol (DTT) and 1% Triton X-100 and lysed by sonication. GST fusion proteins were affinity purified from bacterial lysates by using Glutathione Sepharose 4B (Cytiva, Marlborough, MA) according to the manufacturer’s protocols.
HEK293T cells were grown in 100 mm plates (3 × 106 cells/plate) overnight at 37°C and then transfected with either HA-ARRDC3-WT or the -AAXA mutant plasmid. Cells were washed and lysed with GST binding buffer (50 mM Tris-HCl, pH 7.0, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 0.05% Triton X-100, 2 mM DTT, 1 mM CaCl2, and 0.5 mM MgCl2) supplemented with protease inhibitors cocktail tablet (Roche, Manheim, Germany). GST fusion proteins bound to Glutathione Sepharose were incubated with equivalent amounts of cell lysate overnight at 4°C and washed. WW domain–interacting proteins were eluted in 2× Laemmli sample buffer. Eluates were resolved on 7% SDS–PAGE gels and then transferred onto PVDF and immunoblotted using anti–HA-HRP antibody and anti-GST-HRP antibody.
PAR1 degradation assays
PAR1 degradation assays were performed essentially as described previously (Grimsey et al., 2015). Briefly, MDA-MB-231 cells expressing pLJM-ARRDC3 WT or mutants were plated in six-well plates (6 × 105 cells/well) and incubated for 72 h at 37°C. Cells were washed and serum-starved for 1 h at 37°C and stimulated with thrombin for the indicated time. Cells were then placed on ice, washed with PBS, and lysed in Triton X-100 lysis buffer (50 mM Tris-HCl, pH 7.4, 100 mM NaCl, 5 mM EDTA, 50 mM NaF, 10 mM sodium pyrophosphate, and 1% [vol/vol] Triton X-100) supplemented with protease inhibitors. Cell lysates were collected and sonicated for 10 s at 10% amplitude (Branson Model 450 sonifier), and the protein concentration was normalized after quantification by BCA assay (Thermo Fisher Scientific, Waltham, MA). Lysates were incubated with protein A–Sepharose beads (GE Healthcare, Pittsburgh, PA) and anti-WEDE antibodies overnight at 4°C. Beads were washed and resuspended in 2× Laemmli sample buffer. Immunoprecipitates were resolved by SDS–PAGE gels immunoblotted using anti-PAR1 C5433 antibody.
Signaling assays
Signaling assays were performed essentially as described previously (Arakaki et al., 2018, 2021). Briefly, MDA-MB-231 cells expressing pLJM-ARRDC3 were starved for 1 h or overnight at 37°C and stimulated with 10 nM thrombin, 10 μM SLIGKV agonist peptide, or 100 nM LPA for the indicated times at 37°C, and cell lysates were collected in Triton X-100 lysis buffer (50 mM Tris-HCl, pH 7.4, 100 mM NaCl, 5 mM EDTA, 50 mM NaF, 10 mM sodium pyrophosphate, and 1% [vol/vol] Triton X-100) supplemented by protease inhibitors and quantified by BCA analysis. Equivalent amounts were diluted in 2× Laemmli sample buffer containing 200 mM DTT. Samples were resolved by SDS–PAGE and immunoblotted with anti–phospho-JNK antibody, anti-JNK1 antibody, or CTGF antibody.
Protein stability assays
HEK293T cells were grown in 60 mm plates (1 × 106 cells/well) overnight at 37°C and then transfected with either HA-ARRDC3-WT or the -AAXA mutant. After 24 h, cells were collected and replated in 24-well plates (1 × 105 cells/well) and incubated overnight at 37°C. Cells were then treated with the indicated concentrations of cycloheximide for 24 h and collected in Triton X-100 lysis buffer (50 mM Tris-HCl, pH 7.4, 100 mM NaCl, 5 mM EDTA, 50 mM NaF, 10 mM sodium pyrophosphate, and 1% [vol/vol] Triton X-100) supplemented by protease inhibitors and quantified by BCA analysis. Equivalent amounts were diluted in 2× Laemmli sample buffer containing 200 mM DTT. Samples were resolved by SDS–PAGE and immunoblotted with anti-HA antibody.
Immunofluorescence microscopy
HeLa cells stably expressing FLAG-PAR1 were plated on coverslips in 12-well plates (9 × 104 cells/well) and incubated overnight at 37°C. Cells were transfected with the indicated plasmids and then incubated for 48 h. Cells were washed, serum-starved for 1 h, and stimulated with 10 nM thrombin for the indicated times at 37°C. Cells were fixed in 4% paraformaldehyde, permeabilized in 100% methanol, and treated with the indicated primary and secondary antibodies. Coverslips were mounted with ProLong Gold reagent (Thermo Fischer Scientific, Waltham, MA). Confocal images of 0.28-μm x–y sections were collected sequentially using an Olympus IX81 DSU spinning confocal microscope fitted with a Plan Apo 60×1.4 NA oil objective and a Hamamatsu ORCA-ER digital camera using Metamorph version 7.7.4.0 software (Molecular Devices). Co-localization was quantified in individual cells using the MOC.
Statistics
Data were analyzed using Prism software (version 9.4.1; GraphPad Software). Statistical analysis was determined, as indicated, by performing either a Student’s t test or a one-way analysis of variance (ANOVA).
Acknowledgments
We thank all members of the Trejo laboratory for comments and advice. We especially thank Norton Cheng and Olga Meiri Chaim for constructive discussions and advice in experimental design. This research was funded by National Institutes of Health (NIH) R35 GM127121 (J.T.), R01 GM136202 (I.K.), R21 AI156662 (I.K.), R01 AI161880 (I.K), NIH T32 GM007752 Pharmacological Sciences Training Grant (H.W.), and University of California, Tobacco-related Disease Research Program Predoctoral Fellowship T31DT1550 (H.W.). Computer cluster expenses were partially covered by NHMRC 2012579 (I.K).
Abbreviations used:
- ALIX
ALG-interacting protein X
- ARRDC
arrestin domain–containing protein
- CHX
cycloheximide
- CTGF
connective tissue growth factor
- FBS
fetal bovine serum
- GPCR
G protein-coupled receptor
- JNK
Jun N-terminal kinase
- LPA
lysophosphatidic acid
- NEDD4
neuronal precursor cell–expressed developmentally down-regulated 4
- PAR1
protease-activated receptor 1
- PAR2
protease-activated receptor 2
- TAZ
WW domain–containing transcription regulator protein 1
- TUBE
tandem ubiquitin binding entity
- TXNIP
thioredoxin-interacting protein
- WWP2
WW domain–containing protein-2
- YAP
yes-associated protein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E23-02-0055) on May 24, 2023.
REFERENCES
- Aberle H, Bauer A, Stappert J, Kispert A, Kemler R (1997). Beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J 16, 3797–3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelaide J, Finetti P, Bekhouche I, Repellini L, Geneix J, Sircoulomb F, Charafe-Jauffret E, Cervera N, Desplans J, Parzy D, et al. (2007). Integrated profiling of basal and luminal breast cancers. Cancer Res 67, 11565–11575. [DOI] [PubMed] [Google Scholar]
- Alvarez CE (2008). On the origins of arrestin and rhodopsin. BMC Evol Biol 8, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakaki AKS, Pan WA, Lin H, Trejo J (2018). The alpha-arrestin ARRDC3 suppresses breast carcinoma invasion by regulating G protein-coupled receptor lysosomal sorting and signaling. J Biol Chem 293, 3350–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakaki AKS, Pan WA, Wedegaertner H, Roca-Mercado I, Chinn L, Gujral TS, Trejo J (2021). α-Arrestin ARRDC3 tumor suppressor function is linked to GPCR-induced TAZ activation and breast cancer metastasis. J Cell Sci 134, jcs254888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora P, Cuevas BD, Russo A, Johnson GL, Trejo J (2008). Persistent transactivation of EGFR and ErbB2/HER2 by protease-activated receptor-1 promotes breast carcinoma cell invasion. Oncogene 27, 4434–4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubry L, Guetta D, Klein G (2009). The arrestin fold: variations on a theme. Curr Genomics 10, 133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boase NA, Kelly JM (2004). A role for creD, a carbon catabolite repression gene from Aspergillus nidulans, in ubiquitination. Mol Microbiol 53, 929–940. [DOI] [PubMed] [Google Scholar]
- Booden MA, Eckert LB, Der CJ, Trejo J (2004). Persistent signaling by dysregulated thrombin receptor trafficking promotes breast carcinoma cell invasion. Mol Cell Biol 24, 1990–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass LF, Pizarro S, Ahuja M, Belmonte E, Blanchard N, Stadel JM, Hoxie JA (1994). Changes in the structure and function of the human thrombin receptor during receptor activation, internalization, and recycling. J Biol Chem 269, 2943–2952. [PubMed] [Google Scholar]
- Brass LF, Vassallo RR Jr, Belmonte E, Ahuja M, Cichowski K, Hoxie JA (1992). Structure and function of the human platelet thrombin receptor. Studies using monoclonal antibodies directed against a defined domain within the receptor N terminus. J Biol Chem 267, 13795–13798. [PubMed] [Google Scholar]
- Chen Z, Jiang H, Xu W, Li X, Dempsey DR, Zhang X, Devreotes P, Wolberger C, Amzel LM, Gabelli SB, Cole PA (2017). A tunable brake for HECT ubiquitin ligases. Mol Cell 66, 345–357.e346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dores MR, Chen B, Lin H, Soh UJ, Paing MM, Montagne WA, Meerloo T, Trejo J (2012). ALIX binds a YPX(3)L motif of the GPCR PAR1 and mediates ubiquitin-independent ESCRT-III/MVB sorting. J Cell Biol 197, 407–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dores MR, Lin H, Grimsey NJ, Mendez F, Trejo J (2015). The alpha-arrestin ARRDC3 mediates ALIX ubiquitination and G protein-coupled receptor lysosomal sorting. Mol Biol Cell 26, 4660–4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draheim KM, Chen HB, Tao Q, Moore N, Roche M, Lyle S (2010). ARRDC3 suppresses breast cancer progression by negatively regulating integrin beta4. Oncogene 29, 5032–5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn KW, Kamocka MM, McDonald JH (2011). A practical guide to evaluating colocalization in biological microscopy. Am J Physiol Cell Physiol 300, C723–C742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaquinto AN, Sung H, Miller KD, Kramer JL, Newman LA, Minihan A, Jemal A, Siegel RL (2022). Breast Cancer Statistics, 2022. CA Cancer J Clin 72, 524–541. [DOI] [PubMed] [Google Scholar]
- Grimsey NJ, Aguilar B, Smith TH, Le P, Soohoo AL, Puthenveedu MA, Nizet V, Trejo J (2015). Ubiquitin plays an atypical role in GPCR-induced p38 MAP kinase activation on endosomes. J Cell Biol 210, 1117–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimsey NJ, Narala R, Rada CC, Mehta S, Stephens BS, Kufareva I, Lapek J, Gonzalez DJ, Handel TM, Zhang J, Trejo J (2018). A tyrosine switch on NEDD4-2 E3 ligase transmits GPCR inflammatory signaling. Cell Rep 24, 3312–3323.e3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager NA, Krasowski CJ, Mackie TD, Kolb AR, Needham PG, Augustine AA, Dempsey A, Szent-Gyorgyi C, Bruchez MP, Bain DJ, et al. (2018). Select alpha-arrestins control cell-surface abundance of the mammalian Kir2.1 potassium channel in a yeast model. J Biol Chem 293, 11006–11021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SO, Kommaddi RP, Shenoy SK (2013). Distinct roles for beta-arrestin2 and arrestin-domain-containing proteins in beta2 adrenergic receptor trafficking. EMBO Rep 14, 164–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrador A, Leon S, Haguenauer-Tsapis R, Vincent O (2013). A mechanism for protein monoubiquitination dependent on a trans-acting ubiquitin-binding domain. J Biol Chem 288, 16206–16211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho HC, MacGurn JA, Emr SD (2017). Deubiquitinating enzymes Ubp2 and Ubp15 regulate endocytosis by limiting ubiquitination and degradation of ARTs. Mol Biol Cell 28, 1271–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovsepian J, Defenouillere Q, Albanese V, Vachova L, Garcia C, Palkova Z, Leon S (2017). Multilevel regulation of an alpha-arrestin by glucose depletion controls hexose transporter endocytosis. J Cell Biol 216, 1811–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J, Suh HW, Jeon YH, Hwang E, Nguyen LT, Yeom J, Lee SG, Lee C, Kim KJ, Kang BS, et al. (2014). The structural basis for the negative regulation of thioredoxin by thioredoxin-interacting protein. Nat Commun 5, 2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juneja J, Cushman I, Casey PJ (2011). G12 signaling through c-Jun NH2-terminal kinase promotes breast cancer cell invasion. PLoS One 6, e26085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamentseva R, Kosheverova V, Kharchenko M, Zlobina M, Salova A, Belyaeva T, Nikolsky N, Kornilova E (2020). Functional cycle of EEA1-positive early endosome: direct evidence for pre-existing compartment of degradative pathway. PLoS One 15, e0232532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly P, Moeller BJ, Juneja J, Booden MA, Der CJ, Daaka Y, Dewhirst MW, Fields TA, Casey PJ (2006). The G12 family of heterotrimeric G proteins promotes breast cancer invasion and metastasis. Proc Natl Acad Sci USA 103, 8173–8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, MacGurn JA, Chu T, Stefan CJ, Emr SD (2008). Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell 135, 714–725. [DOI] [PubMed] [Google Scholar]
- MacDonald C, Shields SB, Williams CA, Winistorfer S, Piper RC (2020). A cycle of ubiquitination regulates adaptor function of the Nedd4-family ubiquitin ligase Rsp5. Curr Biol 30, 465–479.e465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias MJ, Wiesner S, Sudol M (2002). WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. FEBS Lett 513, 30–37. [DOI] [PubMed] [Google Scholar]
- Mo JS, Yu FX, Gong R, Brown JH, Guan KL (2012). Regulation of the Hippo-YAP pathway by protease-activated receptors (PARs). Genes Dev 26, 2138–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mund T, Graeb M, Mieszczanek J, Gammons M, Pelham HR, Bienz M (2015). Disinhibition of the HECT E3 ubiquitin ligase WWP2 by polymerized Dishevelled. Open Biol 5, 150185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mund T, Pelham HR (2018). Substrate clustering potently regulates the activity of WW-HECT domain-containing ubiquitin ligases. J Biol Chem 293, 5200–5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabhan JF, Pan H, Lu Q (2010). Arrestin domain-containing protein 3 recruits the NEDD4 E3 ligase to mediate ubiquitination of the beta2-adrenergic receptor. EMBO Rep 11, 605–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag JK, Rudina T, Maoz M, Grisaru-Granovsky S, Uziely B, Bar-Shavit R (2018). Cancer driver G-protein coupled receptor (GPCR) induced beta-catenin nuclear localization: the transcriptional junction. Cancer Metastasis Rev 37, 147–157. [DOI] [PubMed] [Google Scholar]
- Nikko E, Pelham HR (2009). Arrestin-mediated endocytosis of yeast plasma membrane transporters. Traffic 10, 1856–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoselova TV, Zahira K, Rose RS, Sullivan JA (2012). Bul proteins, a nonredundant, antagonistic family of ubiquitin ligase regulatory proteins. Eukaryot Cell 11, 463–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell AF, Apffel A, Gardner RG, Cyert MS (2010). Alpha-arrestins Aly1 and Aly2 regulate intracellular trafficking in response to nutrient signaling. Mol Biol Cell 21, 3552–3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell AF, Schmidt MC (2019). AMPK-mediated regulation of alpha-arrestins and protein trafficking. Int J Mol Sci 20, 515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paing MM, Johnston CA, Siderovski DP, Trejo J (2006). Clathrin adaptor AP2 regulates thrombin receptor constitutive internalization and endothelial cell resensitization. Mol Cell Biol 26, 3231–3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perini ED, Schaefer R, Stoter M, Kalaidzidis Y, Zerial M (2014). Mammalian CORVET is required for fusion and conversion of distinct early endosome subpopulations. Traffic 15, 1366–1389. [DOI] [PubMed] [Google Scholar]
- Polekhina G, Ascher DB, Kok SF, Beckham S, Wilce M, Waltham M (2013). Structure of the N-terminal domain of human thioredoxin-interacting protein. Acta Crystallogr D Biol Crystallogr 69, 333–344. [DOI] [PubMed] [Google Scholar]
- Qi S, O’Hayre M, Gutkind JS, Hurley JH (2014a). Insights into beta2-adrenergic receptor binding from structures of the N-terminal lobe of ARRDC3. Protein Sci 23, 1708–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi S, O’Hayre M, Gutkind JS, Hurley JH (2014b). Structural and biochemical basis for ubiquitin ligase recruitment by arrestin-related domain-containing protein-3 (ARRDC3). J Biol Chem 289, 4743–4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qualls-Histed SJ, Nielsen CP, MacGurn JA (2023). Lysosomal trafficking of the glucose transporter GLUT1 requires sequential regulation by TXNIP and ubiquitin. iScience 26, 106150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch S, Martin-Serrano J (2011). Multiple interactions between the ESCRT machinery and arrestin-related proteins: implications for PPXY-dependent budding. J Virol 85, 3546–3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Sun X, Sun B, Li T, Wu G, Li Y, Chen L, Liu Q, Cui M, Zhou Z (2018). ARRDC3 suppresses colorectal cancer progression through destabilizing the oncoprotein YAP. FEBS Lett 592, 599–609. [DOI] [PubMed] [Google Scholar]
- Soung YH, Chung H, Yan C, Ju J, Chung J (2019). Arrestin domain containing 3 reverses epithelial to mesenchymal transition and chemo-resistance of TNBC cells by up-regulating expression of miR-200b. Cells 8, 692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soung YH, Ford S, Yan C, Chung J (2018). The role of arrestin domain-containing 3 in regulating endocytic recycling and extracellular vesicle sorting of integrin beta4 in breast cancer. Cancers (Basel) 10, 507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soung YH, Pruitt K, Chung J (2014). Epigenetic silencing of ARRDC3 expression in basal-like breast cancer cells. Sci Rep 4, 3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, Irannejad R, Bowman SL, Du Y, Puthenveedu MA, von Zastrow M, Benovic JL (2016). The alpha-arrestin ARRDC3 regulates the endosomal residence time and intracellular signaling of the beta2-adrenergic receptor. J Biol Chem 291, 14510–14525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trejo J, Altschuler Y, Fu HW, Mostov KE, Coughlin SR (2000). Protease-activated receptor-1 down-regulation: a mutant HeLa cell line suggests novel requirements for PAR1 phosphorylation and recruitment to clathrin-coated pits. J Biol Chem 275, 31255–31265. [DOI] [PubMed] [Google Scholar]
- Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, Yuan D, Stroe O, Wood G, Laydon A, et al. (2022). AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res 50, D439–D444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedegaertner H, Pan WA, Gonzalez CC, Gonzalez DJ, Trejo J (2022). The alpha-arrestin ARRDC3 is an emerging multifunctional adaptor protein in cancer. Antioxid Redox Signal 36, 1066–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Shi Q, Li W, Mu X, Peng J, Li M, Chen M, Huang H, Wang C, Gao K, Fan J (2018). ARRDC1 and ARRDC3 act as tumor suppressors in renal cell carcinoma by facilitating YAP1 degradation. Am J Cancer Res 8, 132–143. [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Zhao B, Panupinthu N, Jewell JL, Lian I, Wang LH, Zhao J, Yuan H, Tumaneng K, Li H, et al. (2012). Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell 150, 780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapparoli E, Briata P, Rossi M, Brondolo L, Bucci G, Gherzi R (2020). Comprehensive multi-omics analysis uncovers a group of TGF-beta-regulated genes among lncRNA EPR direct transcriptional targets. Nucleic Acids Res 48, 9053–9066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zbieralski K, Wawrzycka D (2022). alpha-arrestins and their functions: from yeast tohuman health. Int J Mol Sci 23, 4988. [DOI] [PMC free article] [PubMed] [Google Scholar]