Abstract
Late-onset (more than 48 h after ICU admission) acute respiratory distress syndrome (ARDS) is associated with shorter survival time and higher mortality; however, the underlying molecular targets remain unclear. As the WNT gene family is known to drive inflammation, immunity, and tissue fibrosis, all of which are closely related to the pathogenesis and prognosis of ARDS, we aim to investigate the associations of the WNT family with late-onset ARDS and 28-day survival. Genetic (n = 380), epigenetic (n = 185), transcriptional (n = 160), and protein (n = 300) data of patients with ARDS were extracted from the MEARDS (Molecular Epidemiology of ARDS) cohort. We used sure independence screening to identify late onset-related genetic biomarkers and constructed a genetic score on the basis of eight SNPs, which was associated with risk for late-onset ARDS (odds ratio [OR], 2.72; P = 3.81 × 10−14) and survival (hazard ratio [HR], 1.28; P = 0.008). The associations were further externally validated in the iSPAAR (Identification of SNPs Predisposing to Altered Acute Lung Injury Risk) (ORlate onset, 2.49 [P = 0.006]; HRsurvival, 1.87 [P = 0.045]) and MESSI (Molecular Epidemiology of Severe Sepsis in the ICU) (ORlate onset, 4.12 [P = 0.026]; HRsurvival, 1.45 [P = 0.036]) cohorts. Furthermore, we functionally interrogated the six mapped genes of eight SNPs in the multiomics data and noted associations of WNT9A (WNT family member 9A) in epigenetic (ORlate onset, 2.95 [P = 9.91 × 10−4]; HRsurvival, 1.53 [P = 0.011]) and protein (ORlate onset, 1.42 [P = 0.035]; HRsurvival, 1.38 [P = 0.011]) data. The mediation analysis indicated that the effects of WNT9A on ARDS survival were mediated by late onset (HRindirect, 1.12 [P = 0.014] for genetic data; HRindirect, 1.05 [P = 0.030] for protein data). The essential roles of WNT9A in immunity and fibrosis may explain the different trajectories of recovery and dysfunction between early- and late-onset ARDS, providing clues for ARDS treatment.
Keywords: acute respiratory distress syndrome, WNT9A, transomics, onset time, overall survival
Clinical Relevance
Analyses of transomics data reveal that biomarkers of WNT9A at different molecular levels are associated with late-onset acute respiratory distress syndrome, which may contribute to poor survival. This novel susceptibility gene may provide a therapeutic target for the adjuvant treatment of patients with acute respiratory distress syndrome, especially during the coronavirus disease (COVID-19) pandemic.
Acute respiratory distress syndrome (ARDS) is a life-threatening lung injury characterized by acute inflammatory pulmonary disorders and pulmonary fibrosis (1), with an in-hospital mortality rate exceeding 40% (2). Survivors may experience chronic physical, psychosocial, and cognitive sequelae (3). These potentially life-changing conditions have been exacerbated during the coronavirus disease (COVID-19) pandemic, causing severe global concern (4).
Wide heterogeneity in survival has been observed among patients with ARDS, which hampers its management and treatment (5). One factor that has been discussed for decades is the onset time (6–8). In a previous study, we demonstrated that late-onset ARDS (defined as ARDS onset after 48 h of ICU admission but within the first week) is associated with a higher mortality rate and a shorter survival time than early-onset ARDS (9). However, the underlying molecular targets controlling the timing of ARDS development are largely unclear. Moreover, prognosis differs between patients with COVID-19 with early- or late-onset ARDS (10). Therefore, understanding the molecular underpinnings may benefit the treatment of patients with COVID-19.
With interests in the roles of inflammation, immunity, and tissue fibrosis in late-onset ARDS, we focus on the master regulator, the WNT gene family (11–14). WNT signaling can interact with several immune cells, thereby regulating the immune response (15). Moreover, WNT-targeted treatments can interact with the HIF-1α (hypoxia-inducible factor 1 subunit alpha) hypoxia pathway to alleviate severe respiratory diseases (16). Furthermore, WNT family members are crucial regulators for the inflammatory response and are involved in the development of asthma (17). Emerging evidence has unraveled the important role of WNT/β-catenin pathway in tissue development (18). β-Catenin, a central component of the endothelial adherent junctions, engages in microvascular permeability regulation (19), alveolar repair (20), and epithelium regeneration (21), all of which are tied to ARDS recovery. However, it is unknown whether and how the WNT family members and WNT signaling affect the development and progression of ARDS, because of a lack of solid population-level evidence. Moreover, analysis of multiomics data may shed light on the biological dynamics underlying the pathophysiology and identify novel drug targets for the prevention and treatment of many human diseases, such as asthma (22) and COVID-19 (23). To our knowledge, no multiomics study has ever been conducted to screen the underlying molecular targets of ARDS.
To fill this critical gap, we proposed a three-step strategy to evaluate comprehensively the roles of the WNT gene family in late-onset ARDS and its survival, by leveraging multiomics data from the MEARDS (Molecular Epidemiology of ARDS), iSPAAR (Identification of SNPs Predisposing to Altered Acute Lung Injury Risk), and MESSI (Molecular Epidemiology of Sepsis in the ICU) cohorts (9, 24). Our findings revealed the novel molecular targets underlying late-onset ARDS and its prognosis, which might provide therapeutic clues.
Methods
Study Populations
This study was approved by the institutional review boards of the Harvard School of Public Health, Massachusetts General Hospital, and Beth Israel Deaconess Medical Center. All participants gave written informed consent.
The patients with ARDS included in our analysis were recruited primarily from the MEARDS cohort (ClinicalTrials.gov identifier NCT 00006496) and enrolled in the ICUs of Massachusetts General Hospital and Beth Israel Deaconess Medical Center between 1998 and 2014 (9, 24, 25). Briefly, ARDS was defined in accordance with Berlin criteria (26), and eligible patients were defined as critically ill patients with at least one predisposing condition for ARDS and without any of the exclusion criteria (see the data supplement). Also collected were demographics, history, vital signs, hematology, and chemistry; in addition, arterial blood gas analysis was performed frequently, and chest X-ray examinations were conducted within 24 hours of admission. After 24 hours, patients were followed daily for the development of ARDS.
Clinical Outcomes
The primary outcomes of this study are three acute outcomes in patients with ARDS, namely, late onset, 28-day overall survival, and 28-day mortality. ARDS onset time was defined as the interval from ICU admission to the time all Berlin diagnostic criteria (including arterial blood gas, chest X-ray criteria, onset within 1 week, etc.) were met. Therefore, late onset was defined as ARDS onset 48 hours to 1 week after ICU admission (9); 28-day overall survival time was a time-to-event outcome indicating the time interval from ARDS onset to death or loss to follow-up within 28 days after onset, whichever occurred first; and 28-day mortality was a binary outcome indicating vital status (dead or alive) at Day 28 after ARDS onset, an endpoint to complement 28-day overall survival time. For simplicity, we refer to 28-day overall survival time as 28-day survival hereafter.
Transomics Data and Quality Control
The transomics data to be analyzed include genetic, epigenetic, transcriptional, and protein data from MEARDS subjects. We included all 19 WNT family members that were identified from the HUGO Gene Nomenclature Committee database (see Table E1).
-
1.
Genetic data: 380 patients with ARDS (2000–2009) were collected from the MEARDS genome-wide association study, as part of the iSPAAR Consortium. DNA samples were extracted from peripheral white blood cells and genotyped using the Infinium HumanExome BeadChip (Illumina, Inc.). Furthermore, 1,223 SNPs located within ±500-kb windows of 19 WNT family genes were identified using ANNOVAR (https://annovar.openbioinformatics.org/en/latest/) (27, 28) and included in the subsequent analysis. All samples were of European ancestry, and the first five principal components were adjusted to account for the potential population structure.
-
2.
Epigenetic data: peripheral blood was collected from 185 participants between 2000 and 2013 and stored at −80°C. DNA was extracted from whole blood, and methylation was assessed using the Infinium HumanMethylation450 BeadChip (Illumina, Inc.). Focusing on WNT, 2,477 CpG probes located within ±500-kb windows of WNT family members were identified using ANNOVAR (27, 28) and remained in further analysis.
-
3.
Transcriptional data: 160 patients with ARDS (2005–2014) were recruited for RNA sequencing, and total RNA was extracted from blood using the PAXgene Blood RNA Kit (Qiagen) and processed with high-throughput sequencing using the MGISEQ-2000 (MGI Tech) platform.
-
4.
Protein data: 300 patients with ARDS were randomly selected from the MEARDS cohort, which was recruited from 2001 to 2010. Plasma samples also were collected within 24 hours of ICU admission, and plasma protein was quantified using the Olink Target 96 (BGI), including 1,161 proteins; only WNT9A protein was included.
Standard quality control was performed for both biomarkers and samples (see the data supplement). Details on the participants for multiomics data are displayed in Figures E1–E4. After quality control, included for further analyses were 1,025 patients with ARDS (380 with genetic variants, 185 with DNA methylation, 160 with gene expression, and 300 with protein data); demographic and clinical characteristics are detailed in Tables 1 and E2.
Table 1.
Description and Comparison of Demographic and Clinical Characteristics across Patients with Early-Onset and with Late-Onset Acute Respiratory Distress Syndrome in Transomics Data
| Genetic Data |
Epigenetic Data |
Transcriptional Data |
Protein Data |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Early Onset (n = 259) | Late Onset (n = 121) | P Value | Early Onset (n = 124) | Late Onset (n = 61) | P Value | Early Onset (n = 135) | Late Onset (n = 25) | P Value | Early Onset (n = 192) | Late Onset (n = 108) | P Value | |
| Age, yr | 59.02 ± 17.73 | 63.17 ± 16.02 | 0.024 | 64.85 ± 15.39 | 65.51 ± 15.97 | 0.792 | 54.93 ± 18.19 | 52.84 ± 19.50 | 0.623 | 56.58 ± 19.36 | 61.96 ± 17.94 | 0.016 |
| Gender | ||||||||||||
| Female | 99 (38.22) | 46 (38.02) | 1.000 | 39 (31.45) | 26 (42.62) | 0.183 | 46 (34.07) | 9 (36.00) | 1.00 | 74 (38.54) | 40 (37.04) | 0.894 |
| Male | 160 (61.78) | 75 (61.98) | — | 85 (68.55) | 35 (57.38) | — | 89 (65.93) | 16 (64.00) | — | 118 (61.46) | 68 (62.96) | — |
| Smoking status | ||||||||||||
| Never | 69 (31.22) | 25 (26.60) | 0.279 | 30 (28.57) | 16 (30.19) | 0.944 | 52 (42.98) | 5 (20.83) | 0.084 | 49 (30.25) | 31 (35.63) | 0.322 |
| Former | 74 (33.48) | 30 (31.91) | — | 42 (40.00) | 20 (37.74) | — | 31 (25.62) | 9 (37.50) | — | 52 (32.10) | 28 (32.18) | — |
| Current | 78 (35.29) | 39 (41.49) | — | 33 (31.43) | 17 (32.08) | — | 38 (31.40) | 10 (41.67) | — | 61 (37.65) | 28 (32.18) | — |
| ARDS risk factors | ||||||||||||
| Bacteremia | 57 (22.01) | 17 (14.05) | 0.092 | 23 (18.55) | 13 (21.31) | 0.804 | 19 (14.07) | 5 (20.00) | 0.540 | 33 (17.19) | 19 (17.59) | 1.000 |
| Sepsis | 241 (93.05) | 109 (90.08) | 0.427 | 113 (91.13) | 55 (90.16) | 1.000 | 125 (92.59) | 22 (88.00) | 0.430 | 174 (90.63) | 92 (85.19) | 0.216 |
| Septic shock | 223 (86.10) | 92 (76.03) | 0.023 | 107 (86.29) | 47 (77.05) | 0.170 | 109 (80.74) | 15 (60.00) | 0.043 | 135 (70.31) | 65 (60.19) | 0.097 |
| Pneumonia | 196 (75.68) | 78 (64.46) | 0.032 | 98 (79.03) | 41 (67.21) | 0.117 | 113 (83.70) | 18 (72.00) | 0.167 | 153 (79.69) | 56 (51.85) | 9.42 × 10−7 |
| Multiple fractures | 3 (1.16) | 8 (6.61) | 0.006 | 0 (0.00) | 2 (3.28) | 0.108 | 2 (1.48) | 0 (0.00) | 1.00 | 6 (3.13) | 8 (7.41) | 0.161 |
| Pulmonary contusion | 4 (1.54) | 4 (3.31) | 0.272 | 1 (0.81) | 2 (3.28) | 0.253 | 7 (5.19) | 1 (4.00) | 1.00 | 9 (4.69) | 3 (2.78) | 0.547 |
| Aspiration | 27 (10.42) | 8 (6.61) | 0.314 | 13 (10.48) | 3 (4.92) | 0.323 | 15 (11.11) | 2 (8.00) | 1.00 | 23 (11.98) | 5 (4.63) | 0.058 |
| Multiple transfusion | 17 (6.56) | 12 (9.92) | 0.347 | 8 (6.45) | 8 (13.11) | 0.216 | 5 (3.70) | 3 (12.00) | 0.111 | 8 (4.17) | 16 (14.81) | 0.002 |
| APACHE III score | 79.86 ± 22.14 | 76.42 ± 22.75 | 0.168 | 90.29 ± 23.71 | 83.61 ± 24.30 | 0.079 | 76.34 ± 26.24 | 62.29 ± 18.41 | 0.002 | 78.47 ± 23.82 | 78.52 ± 21.42 | 0.987 |
Definition of abbreviations: APACHE = Acute Physiology and Chronic Health Evaluation; ARDS = acute respiratory distress syndrome.
Data are expressed as mean ± SD or as n (%). Sum of frequency numbers may not equal the total sample size because of missing values. ARDS onset within 48 hours after ICU admission was defined as early onset and onset after 48 hours as late onset.
Validation Data Sets from the iSPAAR Consortium and MESSI Cohort
We further obtained genetic data from the iSPAAR Consortium (except those already in the MEARDS cohort) and the MESSI cohort (29, 30). The iSPAAR Consortium is a multi-institutional cooperative study, including studies from the ARDS Clinical Trials Network and MEARDS. Independent subjects from the ARDS Clinical Trials Network were included for external validation, and DNA was genotyped using the 660 Quad BeadChip (Illumina, Inc.) (29). MESSI is a prospective cohort in the medical ICU of the Hospital of the University of Pennsylvania, an urban tertiary referral center, between 2008 and 2015 (30), and SNPs were genotyped using the Axiom TxArray version 1 (Affymetrix). Standard quality control was conducted for both cohorts using the same process as for MEARDS, and eligible participants were defined using the same criteria. Finally, 219 ARDS cases from iSPAAR and 215 ARDS cases from MESSI were included in the validation analysis.
Three-Step Study Design and Statistical Analysis
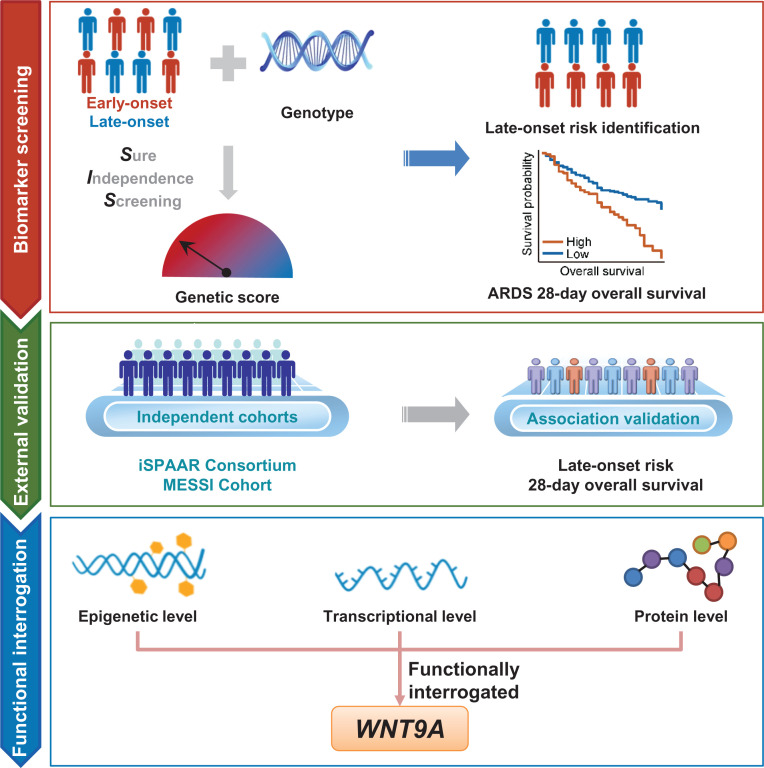
The design of the study, as depicted in Figure 1, features a three-step analytic strategy for evaluating the roles of the WNT gene family in late-onset ARDS and its prognosis.
Figure 1.
Flowchart of the three-step study design. In the biomarker screening step, the late-onset risk-related genetic biomarkers were screened using SIS and combined linearly as a genetic score. Furthermore, the relationships among genetic score, late-onset risk, and 28-day survival were comprehensively evaluated. In the external validation step, the associations were further externally validated in the iSPAAR and MESSI cohorts. In the functional interrogation step, we performed gene annotation for the screened SNPs and further functionally interrogated those mapped genes in the epigenetic, transcriptional, and protein data. ARDS = acute respiratory distress syndrome; iSPAAR = Identification of SNPs Predisposing to Altered Acute Lung Injury Risk; MESSI = Molecular Epidemiology of Severe Sepsis in the ICU; SIS = sure independence screening; WNT9A = WNT family member 9A.
In the biomarker screening step, we used sure independence screening (SIS) to select late onset–related SNPs in the MEARDS cohort using the R (R Foundation for Statistical Computing) package SIS (31, 32). SIS tests the joint effect of SNPs on the basis of a multivariable regression model, which is an efficient variable selection method (for details, see the data supplement). These SNPs were further confirmed using a multivariable logistic regression model, and only those with multivariable P values ⩽0.05 were defined as significant biomarkers related to late-onset ARDS. To understand the cumulative risk effect of variants, we constructed a genetic risk score that was formed using a linear combination of those confirmed SNPs weighted by the coefficients from the multivariable logistic regression model, with adjustment of principal-component analysis and the imbalanced characteristics between early- and late-onset ARDS as covariates. The association between this genetic score and 28-day survival was evaluated using a Cox regression model and illustrated using Kaplan-Meier curves. The cutoff points were derived from the threshold point of the receiver operating characteristic curve with the maximum Youden index. VanderWeele’s mediation analysis was performed to explore the pathway between the genetic score and 28-day survival via late-onset ARDS (33).
In the external validation step, we further confirmed the associations of genetic variants with late-onset ARDS and survival in other existing ARDS genome-wide association study data sets (e.g., independent samples from the iSPAAR Consortium, except those already in the MEARDS cohort, and independent samples from MESSI cohort). Specifically, while focusing on the association, but not prediction, we retested the joint associations between eight screened SNPs and late-onset ARDS in the iSPAAR and MESSI cohorts. Furthermore, genetic scores were constructed with the weights derived from the iSPAAR and MESSI cohorts. In addition, we tested the associations between genetic scores and 28-day survival using the same analytic strategy as in the MEARDS cohort and illustrated using Kaplan-Meier survival curves with cutoff points derived from the threshold point of the receiver operating characteristic curve with the maximum Youden index.
In the functional interrogation step, we performed gene annotation for those selected SNPs and tested the associations of the correspondingly mapped genes at the epigenetic, transcriptional, and protein levels. During the epigenetic data analysis, in which each gene may host numerous CpG probes, we applied SIS to select the top important CpG biomarkers and evaluated their associations with 28-day survival using Cox models. For the transcriptional and protein data analysis, we assessed the associations of mapped genes (mRNA) or proteins with late-onset ARDS and 28-day survival using logistic and Cox regression, respectively.
Continuous variables are summarized as mean ± SD and categorized variables as frequency (n) and proportion (percentage). To understand the overall associations across transomics, the overall associations of WNT genes from multiomics data were assessed by combining P values on the basis of the aggregated Cauchy association test (ACAT) method (34). The covariates adjusted were the imbalanced characteristics between early- and late-onset ARDS that derived from our large clinical cohort study (9). Specifically, in the logistic regression model for late onset, the adjusted covariates were those collected within 24 hours of ICU admission (i.e., Acute Physiology and Chronic Health Evaluation III score, septic shock, pneumonia, multiple fractures, transfusion within 7 d before admission, vasopressors before admission, intubation, ventilation, and highest potassium concentration); in the Cox model for 28-day survival, the adjusted covariates included those just mentioned plus those collected at ARDS onset (i.e., arterial oxygen pressure [PaO2]/fraction of inspired oxygen [FiO2], platelet count, and creatinine concentration), as well as ICU 28-day ventilation-free days. Statistical analyses were performed using R version 3.6.3. Significance was defined by a two-sided P value ⩽0.05, unless otherwise specified.
Results
Eight SNPs Associated with Risk for Late-Onset ARDS in the Biomarker Screening and External Validation Step
SIS identified eight SNPs that have the most predominant contributions to the risk of late-onset ARDS: rs2744728WNT4, rs7971903WNT5B, rs468141WNT7A, rs1356234WNT7A, rs1550160WNT7A, rs2031134WNT9A, rs4944092WNT11, and rs2402574WNT16 (see Table E3). A weighted genetic score of these eight SNPs significantly discriminated patients with early-onset ARDS and those with late-onset ARDS (odds ratio [OR], 2.72 [95% confidence interval (CI), 2.12–3.56]; P = 3.81 × 10−14) (Figure 2A). Furthermore, the associations remained significant in the independent samples from iSPAAR (except for those already in the MEARDS cohort) and MESSI (ORiSPAAR, 2.49 [95% CI, 1.45–6.05; PiSPAAR = 0.006]; ORMESSI, 4.12 [95% CI, 1.21–15.12; PMESSI = 0.026) cohorts (Figure 2B). We also conducted a series of subgroup analyses, with subgroups defined by age, gender, smoking status, and ARDS risk factors. The genetic score remained discriminative in all these subpopulations, with the estimated odd ratios ranging from 2.24 (95% CI, 1.52–3.51; P = 1.41 × 10−4) to 5.65 (95% CI, 2.41–17.99; P = 5.57 × 10−4) (Figures 2C and E5).
Figure 2.
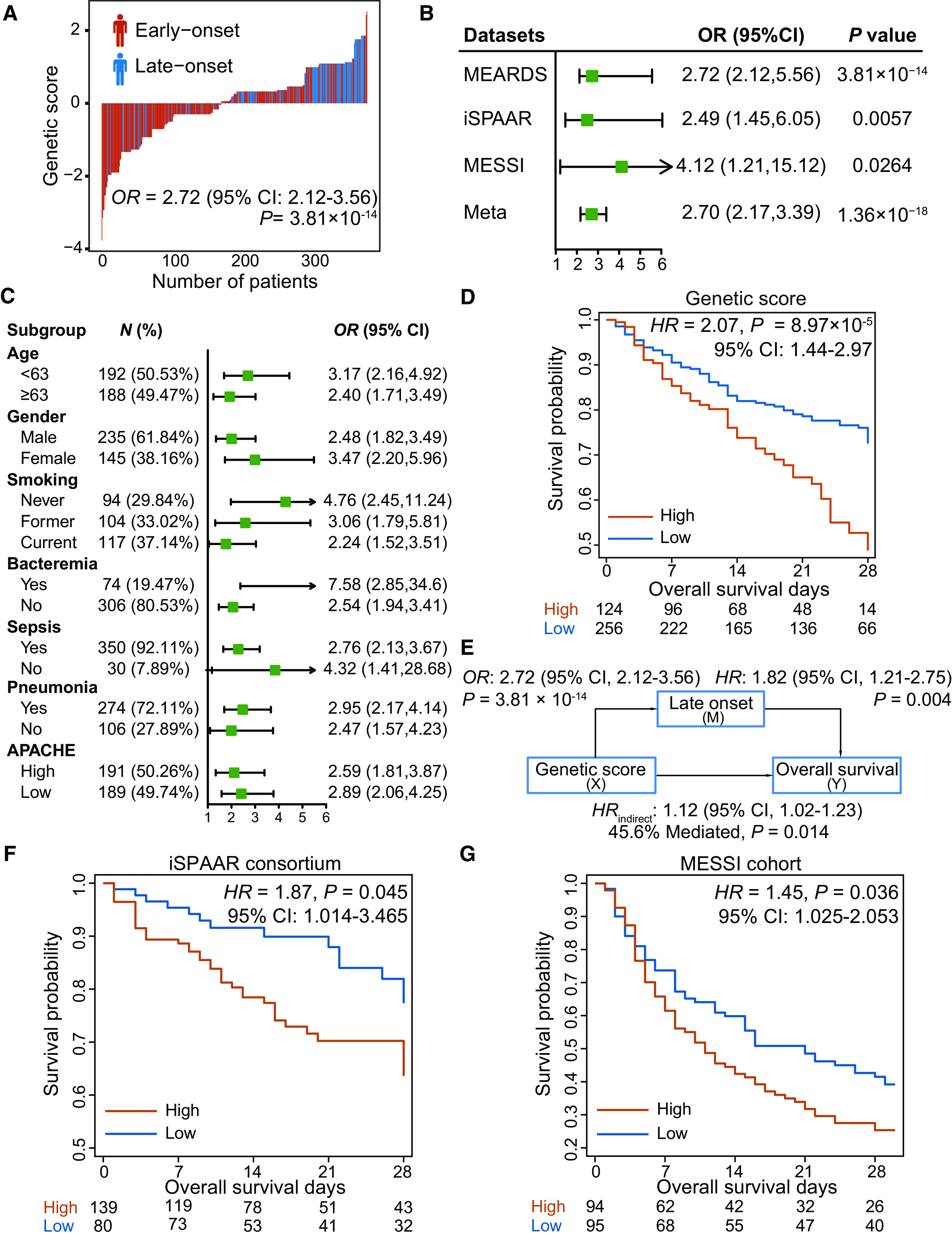
Significant genetic biomarkers screened in the biomarker screening and external validation step. (A) Distribution of genetic score and ARDS onset time. The x-axis is sorted by genetic score. Patients with early-onset ARDS and those with late-onset ARDS are indicated by red and blue color, respectively. (B) External validation of genetic biomarkers in the iSPAAR (except those already in the MEARDS cohort) and MESSI cohorts. (C) Forest plots for subgroup analysis of genetic score stratified by various clinical characteristics. (D) Survival differences between high- and low-risk patients grouped by the genetic score. The optimal cutoff point (0.045) was derived from the threshold point of the receiver operating characteristic curve with the maximum Youden index. (E) Mediation analysis for the indirect effect of genetic score on 28-day survival via late-onset risk. (F and G) External validation of association between genetic score and ARDS survival in other centers of the iSPAAR Consortium (F) and MESSI cohort (G). APACHE = Acute Physiology and Chronic Health Evaluation; CI = confidence interval; HR = hazard ratio; MEARDS = Molecular Epidemiology of ARDS; OR = odds ratio.
Genetic Score Affects ARDS 28-Day Survival via Late Onset
A higher genetic score was associated with shorter survival time (hazard ratio [HR], 1.28 [95% CI, 1.07–1.54]; P = 0.008) and a higher 28-day mortality rate among patients with ARDS (OR, 1.26 [95% CI, 1.02–1.56]; P = 0.036). Compared with patients with lower genetic scores (i.e., lower than the cutoff 0.045), those with higher scores (i.e., higher than 0.045) tended to have shorter survival time (HRhigh vs. low, 2.07 [95% CI, 1.44–2.97]; P = 8.97 × 10−5) (Figure 2D). Moreover, mediation analysis suggested that 45.6% of the effect of the genetic score on ARDS survival was mediated through late onset in the MEARDS cohort (HRindirect, 1.12 [95% CI, 1.02–1.23]; P = 0.014) (Figure 2E). Furthermore, we confirmed relationships between genetic score and ARDS survival in the iSPAAR Consortium and MESSI cohort and observed that the significant associations remained (HRiSPAAR, 1.87 [95% CI, 1.01–3.47; P = 0.045]; HRMESSI, 1.45 [95% CI, 1.03–2.05; P = 0.036]) (Figures 2F and 2G).
WNT9A Was Identified as a Robust Biomarker in the Functional Interrogation Step
In the functional interrogation step, we verified the six mapped genes, identified in the biomarker screening step, at the epigenetics, transcription, and protein levels (Figures 3A and 3B).
Figure 3.
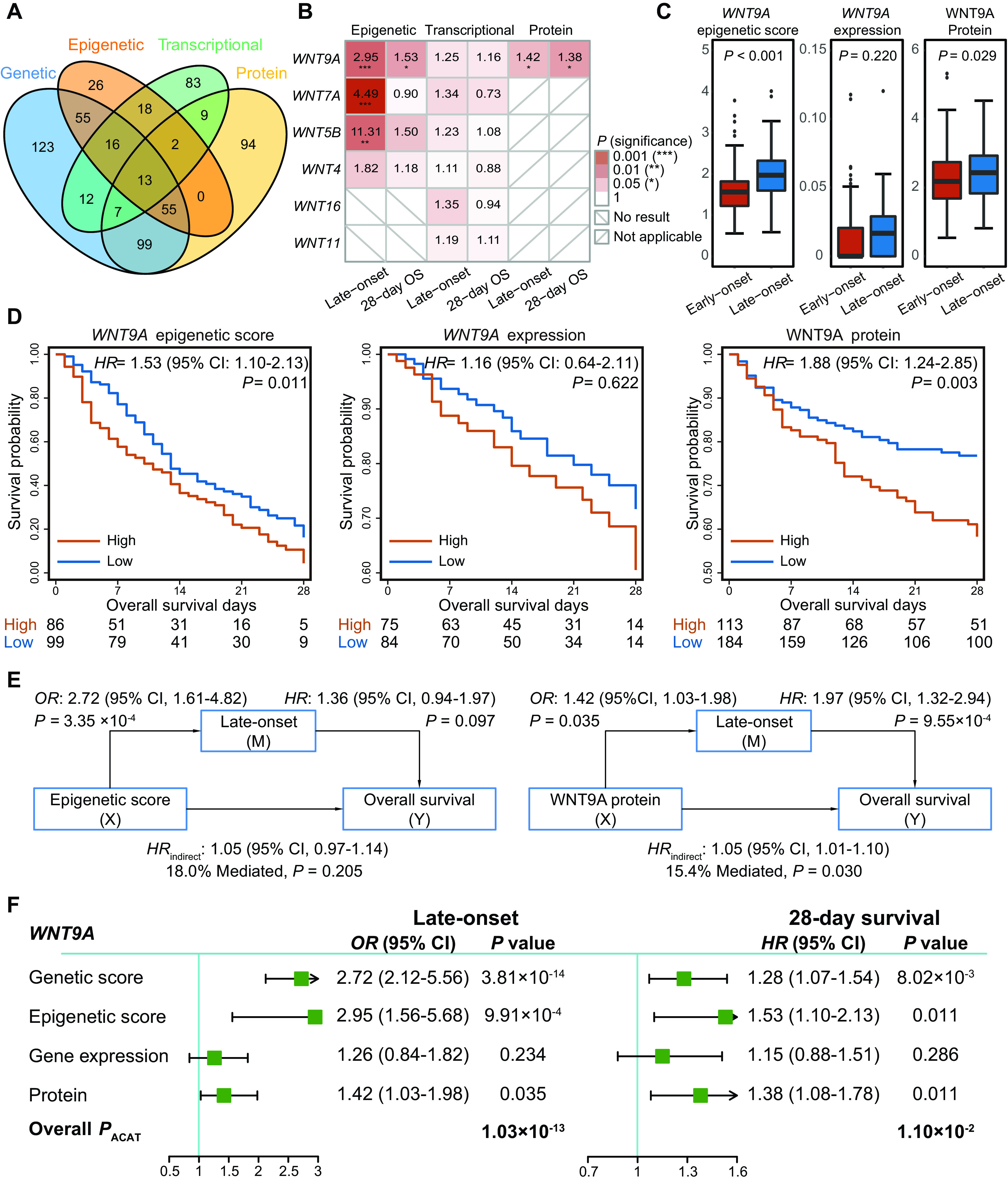
Functional interrogation of screened biomarkers in the epigenetic, transcriptional, and protein data. (A) Venn diagram of overlapping samples across multiomics data. (B) Association pattern for the six mapped genes when validated at the epigenetic, transcriptional, and protein levels. The depth of the color is proportional to the P value. The protein data were quantified using the Olink platform with 1,161 proteins, and only WNT9A in the WNT family was profiled in the platform. (C) Distribution of WNT9A across patients with early-onset ARDS and those with late-onset ARDS at the epigenetic, transcriptional, and protein levels, which were compared using the Wilcoxon test. (D) Survival differences between high- and low-risk patients grouped by WNT9A at the epigenetic, transcriptional, and protein levels. Optimal cutoff points (epigenetic, 1.66; transcriptional, 0.02; protein, 2.51) were derived from the threshold point of the receiver operating characteristic curve with the maximum Youden index. (E) Mediation analyses for indirect effects of WNT9A on 28-day survival via late-onset risk at the epigenetic and protein levels. (F) Forest plots for overall associations of WNT9A with late-onset risk and 28-day survival at transomics levels. ACAT = aggregated Cauchy association test; OS = overall survival.
For the epigenetic analysis, we applied SIS to identify the significant CpG probes within each of these six selected genes; significant CpG probes were identified for WNT9A, WNT7A, WNT5B, and WNT4 but not for WNT11 and WNT16 (Figure 3B). Epigenetic scores were constructed for these four genes (see Table E4). Among them, three epigenetic scores (for WNT9A, WNT7A, and WNT5B) were significantly (false discovery rate ⩽ 0.05) associated with late-onset ARDS (ORhigh vs. low, 2.95 [95% CI, 1.56–5.68; P = 9.91 × 10−4] for WNT9A [cutoff = 1.66]; ORhigh vs. low, 4.49 [95% CI, 2.22–9.64; P = 5.36 × 10−5] for WNT7A [cutoff = 17.78]; ORhigh vs. low, 11.31 [95% CI, 3.26–71.43; P = 1.18 × 10−3] for WNT5B [cutoff = 44.14]) (see Table E5). Only the epigenetic score for WNT9A was observed to be significantly associated with ARDS 28-day survival (HRhigh vs. low, 1.53 [95% CI, 1.10–2.13]; P = 0.011) (Figure 3D; see Table E5).
No significant results were observed in the transcriptional analysis, possibly because of the small sample size with the transcriptional data (see Table E6; Figures 3C and 3D).
For the protein analysis, the WNT9A plasma protein concentration was significantly lower among the patients with early-onset ARDS (P = 0.029) (Figure 3C), which was confirmed by a covariate-adjusted logistic regression model (OR, 1.42 [95% CI, 1.03–1.98]; P = 0.035) (see Table E7). Moreover, higher WNT9A protein was significantly related to shorter survival time (HR, 1.38 [95% CI, 1.08–1.78]; P = 0.011) and higher mortality rate (OR, 1.64 [95% CI, 1.14–2.39]; P = 0.009) (see Table E7). Kaplan-Meier survival curves showed that a higher concentration of WNT9A protein (cutoff = 2.51) was associated with shorter survival time (HRhigh vs. low, 1.88 [95% CI, 1.24–2.85]; P = 0.003) (Figure 3D). Also observed was a significant indirect effect of WNT9A protein on ARDS 28-day survival mediated by late onset (HRindirect, 1.05 [95% CI, 1.01–1.10]; 15.4% mediated; P = 0.030) (Figure 3E). Although the overlapped samples across multiomics data were limited, we performed an exploratory analysis for methylation quantitative trait locus (QTL), expression QTL, and protein QTL. No significant regulation was observed after adjustment for multiple hypothesis tests (see Table E8).
In summary, WNT9A genetic variants, epigenetic marks, and plasma protein concentrations all differed across early- and late-onset ARDS (overall PACAT = 1.03 × 10−13) and were associated with ARDS 28-day survival (overall PACAT = 0.011) (Figure 3F; see Table E9).
Discussion
For decades, research on the association between ARDS onset time and prognosis has produced controversial conclusions (6–8). Our previous two-stage large-scale population study verified 48 hours after ICU admission as a cutoff for categorizing early- and late-onset ARDS on the basis of a data-driven method and demonstrated the association between late-onset ARDS and poor prognosis (9). However, previous studies did not investigate molecular biomarkers. This three-step multiomics study screens the molecular targets (WNT family members) that are involved in a master regulator of inflammation, immunity, and tissue fibrosis. We reveal that the genetic variants, DNA methylation, and protein concentrations of WNT9A are associated with both late-onset ARDS and 28-day prognosis.
According to the Berlin definition of ARDS, respiratory failure should develop within 1 week of a known clinical insult (35). Therefore, the onset time of ARDS is defined as Day 0 (within 24 h), Day 1 (24–48 h), and so forth. Thus, the value of onset time ranges from 0 to 6 days. Hence, the difference in onset time among patients is not very large. In the MEARDS cohort, the proportion of late-onset ARDS is about 30% (9), which accounts for a minority of patients with ARDS. On the basis of our previous study (9), compared with patients with early-onset ARDS, those with late-onset ARDS have less proportion of ventilation within 24 hours of ICU admission, as they have higher PaO2/FiO2 ratios and lower lung injury scores and Acute Physiology and Chronic Health Evaluation III scores, indicating milder lung injury. In any case, they have greater decreases in PaO2/FiO2 ratio from the time of admission to the time of ARDS onset. Patients with early-onset ARDS and those with late-onset ARDS may have different trajectories of recovery and dysfunction after ARDS onset (36). Patients with late onset may be on a slow-burn trajectory, in which patients have slight loss of function at the beginning but develop more rapid and persistent functional decline later, resulting in worse survival. In contrast, patients with early onset may be on a big-hit trajectory, in which patients have acute loss of function during their critical illness, after which they may gradually recover, survive longer, and have lower mortality. Combined with the results of the present study, WNT signaling and immune inflammatory response might play essential roles in the trajectories of recovery and dysfunction of patients with ARDS (37, 38). WNT signaling is highly specific for macrophage activation, which exists in a dynamic balance between proinflammatory M1 and antiinflammatory M2 macrophages, and further influence the occurrence of pulmonary disorders (39). Alveolar macrophages resident during the early course of pathogenic invasion, detecting pathogens and secreting inflammatory factors through M1 polarization, and causing cytokine storm result in early-onset ARDS (16, 40). In contrast, for patients with late-onset ARDS, higher WNT signaling might inhibit M1 polarization, promote antiinflammatory M2 states, and relieve the inflammatory response (41). However, sustained activation of WNT signaling in response to tissue damage would promote fibroblast activation and collagen release, leading to pulmonary fibrosis and irreversible remodeling of pulmonary tissue (42, 43), resulting in a poor prognosis of late-onset ARDS. In addition, mounting evidence suggests that the physical well-being of a patient in the ICU declines as hospitalization drags on (44). Patients with early onset, although enduring a big hit early during their ICU stays, may gradually recover and then be transferred to general wards and survive longer. In contrast, with a slow-burn trajectory, patients with late onset cannot sustain their physical condition with a constant “hit”; thus they remain in the ICU and ultimately experience a worse prognosis (45, 46).
Our three-step multiomics study provides multilevel evidence that WNT9A is associated with late-onset ARDS, through which it mediates a large proportion of the effect on 28-day survival. Moreover, early- and late-onset ARDS might vary by triggers and have different trajectories, indicating that precision treatment is warranted for patients with ARDS with different onset times. This mechanism might be explained by the essential roles of WNT signaling in immune responses and tissue fibrosis. Nevertheless, further biological experiments are warranted to validate the results of the present exploratory research.
Our study has several strengths. First, we integrate transomics data of patients with ARDS and systematically evaluate and verify associations between WNT9A and phenotypes of ARDS. Second, the associations of WNT9A with late-onset ARDS and 28-day survival are discovered in genetic data using the MEARDS cohort, validated using the iSPAAR and MESSI cohorts, and further confirmed using epigenetic and protein data, which may bolster reliability and usability. Finally, the mediation analysis establishes the path of the impact of WNT9A on ARDS survival via late onset, which may facilitate understanding of the biological mechanism of the disease.
We also acknowledge some limitations. First, because of the high cost of omics studies, we have a small sample size of multiomics data in MEARDS, yielding limited statistical power to identify biomarkers with small to moderate effects. To boost the statistical power in a genetic study with a small sample size, we use SIS, an efficient statistical feature selection method, to select large-effect biomarkers on the basis of their marginal effects on late-onset ARDS. Besides, we again confirm their significance in the independent iSPAAR and MESSI cohorts. By using all available ARDS genomic data, we make an effort to maximize the balance between significance of and confidence in our results. In any case, to our knowledge, our ARDS cohort is probably unique, with multiomics data in a relatively large sample size, providing a rare opportunity to evaluate the relationships between the WNT gene family and late-onset ARDS, as well as overall survival of patients with ARDS from different molecular perspectives. Second, as this was a retrospective study, we can evaluate only the associations between phenotypes of ARDS and the measured domains of biomarkers, including SNPs, DNA methylation, gene expression, and protein. For unmeasured domains (e.g., metabolites), we are looking forward to future studies. Third, as our study hints at WNT9A as a therapeutic target for treating patients with ARDS with various onset times, further biological experiments or clinical trials are warranted. Fourth, the majority of our samples are of European ancestry, limiting the generalizability of the results to other ethnic groups. Finally, the overlapping sample size across multiomics data is limited, hampering the statistical power in the QTL analysis.
Conclusions
By integrating multiomics data, we reveal that biomarkers of WNT9A at multiple molecular levels are associated with late-onset ARDS, which may contribute to poor survival.
Acknowledgments
Acknowledgment
The authors thank all participants in the MEARDS, iSPAAR, and MESSI cohorts. The iSPAAR Consortium and MEARDS cohort data sets are deposited in the Database of Genotypes and Phenotypes under accession code phs000631.v1.p1. The Database of Genotypes and Phenotypes submission for the MESSI data set is forthcoming, in accordance with the National Institutes of Health genomic data sharing policy.
Footnotes
Supported by National Natural Science Foundation of China grants 82220108002 (F.C.), 82273737 (R.Z.), 81973142 (Y.W.), and 82173620 (Y.Z.); National Institutes of Health grants CA209414, HL060710, and ES000002 (D.C.C.) and CA209414 and CA249096 (Y.L.); and Priority Academic Program Development of Jiangsu Higher Education Institutions. R.Z. was partially supported by the Qing Lan Project of the Higher Education Institutions of Jiangsu Province and the Outstanding Young Level Academic Leadership Training Program of Nanjing Medical University.
Author Contributions: J.C., J.T., D.C.C., F.C., and R.Z. contributed to the study design. M.M.W., N.J.M., A.J.F., B.T.T., and D.C.C. contributed to data collection. J.C., J.T., and Y.W. performed statistical analysis and interpretation and drafted the manuscript. M.N. performed data cleaning and statistical analysis and drafted the response. Y.L., R.Z., Y.Z., and D.C.C. revised the manuscript. All authors contributed to critical revision of the manuscript and approved its final version.
This article has a data supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1165/rcmb.2022-0416OC on April 24, 2023
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1. Meyer NJ, Gattinoni L, Calfee CS. Acute respiratory distress syndrome. Lancet . 2021;398:622–637. doi: 10.1016/S0140-6736(21)00439-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beitler JR, Thompson BT, Baron RM, Bastarache JA, Denlinger LC, Esserman L, et al. Advancing precision medicine for acute respiratory distress syndrome. Lancet Respir Med . 2022;10:107–120. doi: 10.1016/S2213-2600(21)00157-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sasannejad C, Ely EW, Lahiri S. Long-term cognitive impairment after acute respiratory distress syndrome: a review of clinical impact and pathophysiological mechanisms. Crit Care . 2019;23:352. doi: 10.1186/s13054-019-2626-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fan E, Beitler JR, Brochard L, Calfee CS, Ferguson ND, Slutsky AS, et al. COVID-19-associated acute respiratory distress syndrome: is a different approach to management warranted? Lancet Respir Med . 2020;8:816–821. doi: 10.1016/S2213-2600(20)30304-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Khan YA, Fan E, Ferguson ND. Precision medicine and heterogeneity of treatment effect in therapies for ards. Chest . 2021;160:1729–1738. doi: 10.1016/j.chest.2021.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vincent JL, Sakr Y, Groeneveld J, Zandstra DF, Hoste E, Malledant Y, et al. ARDS of early or late onset: does it make a difference? Chest . 2010;137:81–87. doi: 10.1378/chest.09-0714. [DOI] [PubMed] [Google Scholar]
- 7. Liao KM, Chen CW, Hsiue TR, Lin WC. Timing of acute respiratory distress syndrome onset is related to patient outcome. J Formos Med Assoc . 2009;108:694–703. doi: 10.1016/S0929-6646(09)60392-2. [DOI] [PubMed] [Google Scholar]
- 8. Shari G, Kojicic M, Li G, Cartin-Ceba R, Alvarez CT, Kashyap R, et al. Timing of the onset of acute respiratory distress syndrome: a population-based study. Respir Care . 2011;56:576–582. doi: 10.4187/respcare.00901. [DOI] [PubMed] [Google Scholar]
- 9. Zhang R, Wang Z, Tejera P, Frank AJ, Wei Y, Su L, et al. Late-onset moderate to severe acute respiratory distress syndrome is associated with shorter survival and higher mortality: a two-stage association study. Intensive Care Med . 2017;43:399–407. doi: 10.1007/s00134-016-4638-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li X, Ma X. Acute respiratory failure in COVID-19: is it “typical” ARDS? Crit Care . 2020;24:198. doi: 10.1186/s13054-020-02911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene . 2017;36:1461–1473. doi: 10.1038/onc.2016.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Staal FJ, Luis TC, Tiemessen MM. WNT signalling in the immune system: WNT is spreading its wings. Nat Rev Immunol . 2008;8:581–593. doi: 10.1038/nri2360. [DOI] [PubMed] [Google Scholar]
- 13. Hu HH, Cao G, Wu XQ, Vaziri ND, Zhao YY. Wnt signaling pathway in aging-related tissue fibrosis and therapies. Ageing Res Rev . 2020;60:101063. doi: 10.1016/j.arr.2020.101063. [DOI] [PubMed] [Google Scholar]
- 14. Ahmad S, Manzoor S, Siddiqui S, Mariappan N, Zafar I, Ahmad A, et al. Epigenetic underpinnings of inflammation: connecting the dots between pulmonary diseases, lung cancer and COVID-19. Semin Cancer Biol . 2022;83:384–398. doi: 10.1016/j.semcancer.2021.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Patel S, Alam A, Pant R, Chattopadhyay S. Wnt signaling and its significance within the tumor microenvironment: novel therapeutic insights. Front Immunol . 2019;10:2872. doi: 10.3389/fimmu.2019.02872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhu B, Wu Y, Huang S, Zhang R, Son YM, Li C, et al. Uncoupling of macrophage inflammation from self-renewal modulates host recovery from respiratory viral infection. Immunity . 2021;54:1200–1218.e9. doi: 10.1016/j.immuni.2021.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bartel S, Carraro G, Alessandrini F, Krauss-Etschmann S, Ricciardolo FLM, Bellusci S. miR-142-3p is associated with aberrant WNT signaling during airway remodeling in asthma. Am J Physiol Lung Cell Mol Physiol . 2018;315:L328–L333. doi: 10.1152/ajplung.00113.2018. [DOI] [PubMed] [Google Scholar]
- 18. Nusse R, Clevers H. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell . 2017;169:985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 19. Thibeault S, Rautureau Y, Oubaha M, Faubert D, Wilkes BC, Delisle C, et al. S-nitrosylation of beta-catenin by eNOS-derived NO promotes VEGF-induced endothelial cell permeability. Mol Cell . 2010;39:468–476. doi: 10.1016/j.molcel.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 20. Skronska-Wasek W, Mutze K, Baarsma HA, Bracke KR, Alsafadi HN, Lehmann M, et al. Reduced frizzled receptor 4 expression prevents WNT/beta-catenin-driven alveolar lung repair in chronic obstructive pulmonary disease. Am J Respir Crit Care Med . 2017;196:172–185. doi: 10.1164/rccm.201605-0904OC. [DOI] [PubMed] [Google Scholar]
- 21. Conlon TM, John-Schuster G, Heide D, Pfister D, Lehmann M, Hu Y, et al. Inhibition of LTβR signalling activates WNT-induced regeneration in lung. Nature . 2020;588:151–156. doi: 10.1038/s41586-020-2882-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Abdel-Aziz MI, Vijverberg SJH, Neerincx AH, Brinkman P, Wagener AH, Riley JH, et al. U-BIOPRED Study Group A multi-omics approach to delineate sputum microbiome-associated asthma inflammatory phenotypes. Eur Respir J . 2022;59:2102603. doi: 10.1183/13993003.02603-2021. [DOI] [PubMed] [Google Scholar]
- 23. Su Y, Chen D, Yuan D, Lausted C, Choi J, Dai CL, et al. ISB-Swedish COVID19 Biobanking Unit Multi-omics resolves a sharp disease-state shift between mild and moderate COVID-19. Cell . 2020;183:1479–1495.e20. doi: 10.1016/j.cell.2020.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wei Y, Tejera P, Wang Z, Zhang R, Chen F, Su L, et al. A missense genetic variant in lrrc16a/carmil1 improves acute respiratory distress syndrome survival by attenuating platelet count decline. Am J Respir Crit Care Med . 2017;195:1353–1361. doi: 10.1164/rccm.201605-0946OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen J, Gao X, Shen S, Xu J, Sun Z, Lin R, et al. Association of longitudinal platelet count trajectory with ICU mortality: a multi-cohort study. Front Immunol . 2022;13:936662. doi: 10.3389/fimmu.2022.936662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ranieri VM, Rubenfeld GD, Thompson BT, Ferguson ND, Caldwell E, Fan E, et al. ARDS Definition Task Force Acute respiratory distress syndrome: the Berlin definition. JAMA . 2012;307:2526–2533. doi: 10.1001/jama.2012.5669. [DOI] [PubMed] [Google Scholar]
- 27. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res . 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gusev A, Ko A, Shi H, Bhatia G, Chung W, Penninx BW, et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet . 2016;48:245–252. doi: 10.1038/ng.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dong X, Zhu Z, Wei Y, Ngo D, Zhang R, Du M, et al. Plasma insulin-like growth factor binding protein 7 contributes causally to ards 28-day mortality: evidence from multistage Mendelian randomization. Chest . 2021;159:1007–1018. doi: 10.1016/j.chest.2020.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guillen-Guio B, Lorenzo-Salazar JM, Ma SF, Hou PC, Hernandez-Beeftink T, Corrales A, et al. Sepsis-associated acute respiratory distress syndrome in individuals of European ancestry: a genome-wide association study. Lancet Respir Med . 2020;8:258–266. doi: 10.1016/S2213-2600(19)30368-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ouyang B, Wang J, He T, Bartel CJ, Huo H, Wang Y, et al. Synthetic accessibility and stability rules of NASICONs. Nat Commun . 2021;12:5752. doi: 10.1038/s41467-021-26006-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fan J, Song R. Sure independence screening in generalized linear models with np-dimensionality. Ann Stat . 2010;38:3567–3604. [Google Scholar]
- 33. Valeri L, Vanderweele TJ. Mediation analysis allowing for exposure-mediator interactions and causal interpretation: theoretical assumptions and implementation with SAS and SPSS macros. Psychol Methods . 2013;18:137–150. doi: 10.1037/a0031034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu Y, Chen S, Li Z, Morrison AC, Boerwinkle E, Lin X. ACAT: a fast and powerful p value combination method for rare-variant analysis in sequencing studies. Am J Hum Genet . 2019;104:410–421. doi: 10.1016/j.ajhg.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ferguson ND, Fan E, Camporota L, Antonelli M, Anzueto A, Beale R, et al. The Berlin definition of ARDS: an expanded rationale, justification, and supplementary material. Intensive Care Med . 2012;38:1573–1582. doi: 10.1007/s00134-012-2682-1. [DOI] [PubMed] [Google Scholar]
- 36. Iwashyna TJ. Trajectories of recovery and dysfunction after acute illness, with implications for clinical trial design. Am J Respir Crit Care Med . 2012;186:302–304. doi: 10.1164/rccm.201206-1138ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Janssens S, Pulendran B, Lambrecht BN. Emerging functions of the unfolded protein response in immunity. Nat Immunol . 2014;15:910–919. doi: 10.1038/ni.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fajgenbaum DC, June CH. Cytokine storm. N Engl J Med . 2020;383:2255–2273. doi: 10.1056/NEJMra2026131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tigue ML, Loberg MA, Goettel JA, Weiss WA, Lee E, Weiss VL. Wnt signaling in the phenotype and function of tumor-associated macrophages. Cancer Res . 2023;83:3–11. doi: 10.1158/0008-5472.CAN-22-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shapouri-Moghaddam A, Mohammadian S, Vazini H, Taghadosi M, Esmaeili SA, Mardani F, et al. Macrophage plasticity, polarization, and function in health and disease. J Cell Physiol . 2018;233:6425–6440. doi: 10.1002/jcp.26429. [DOI] [PubMed] [Google Scholar]
- 41. Liu C, Xiao K, Xie L. Advances in the regulation of macrophage polarization by mesenchymal stem cells and implications for ali/ards treatment. Front Immunol . 2022;13:928134. doi: 10.3389/fimmu.2022.928134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hudson LD, Hough CL. Therapy for late-phase acute respiratory distress syndrome. Clin Chest Med . 2006;27:671–677. doi: 10.1016/j.ccm.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 43. Chilosi M, Poletti V, Zamò A, Lestani M, Montagna L, Piccoli P, et al. Aberrant Wnt/beta-catenin pathway activation in idiopathic pulmonary fibrosis. Am J Pathol . 2003;162:1495–1502. doi: 10.1016/s0002-9440(10)64282-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Guidet B, de Lange DW, Boumendil A, Leaver S, Watson X, Boulanger C, et al. VIP2 Study Group The contribution of frailty, cognition, activity of daily life and comorbidities on outcome in acutely admitted patients over 80 years in European ICUs: the VIP2 study. Intensive Care Med . 2020;46:57–69. doi: 10.1007/s00134-019-05853-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fleiss N, Coggins SA, Lewis AN, Zeigler A, Cooksey KE, Walker LA, et al. Evaluation of the neonatal sequential organ failure assessment and mortality risk in preterm infants with late-onset infection. JAMA Netw Open . 2021;4:e2036518. doi: 10.1001/jamanetworkopen.2020.36518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Giantsou E, Liratzopoulos N, Efraimidou E, Panopoulou M, Alepopoulou E, Kartali-Ktenidou S, et al. Both early-onset and late-onset ventilator-associated pneumonia are caused mainly by potentially multiresistant bacteria. Intensive Care Med . 2005;31:1488–1494. doi: 10.1007/s00134-005-2697-y. [DOI] [PubMed] [Google Scholar]