Abstract
The human immunodeficiency virus type 1 coreceptor activity of CCR5 depends on certain polar and charged residues in its amino-terminal domain. Since studies of chimeric receptors have indicated that the extracellular loops of CCR5 are also involved in viral fusion and entry, we have explored the role of bulky, polar and nonpolar residues in these regions. Selected amino acids in the three extracellular loops were individually changed to alanines, and the coreceptor activities of the mutant CCR5 proteins were tested in a luciferase reporter virus-based entry assay. We found that the cysteines in the extracellular loops of CCR5 are essential for coreceptor activity. However, only minor (two- to threefold) effects on coreceptor function were noted for all of the other alanine substitutions. We also demonstrated that when the first 19 residues of the amino-terminal region were separated from the rest of CCR5, by insertion of glycine/serine spacers between proline 19 and cysteine 20, coreceptor function decreased. Together with our previous studies, these data indicate that both an amino-terminal gp120-binding site and extracellular domain geometry play a role in viral entry.
Proteins belonging to the chemokine receptor family are essential for human immunodeficiency virus (HIV) fusion and entry into target cells. All known viral strains use either CCR5 and/or CXCR4 as coreceptors (3, 4, 8, 9, 14, 20). Other, more recently identified coreceptors can function only with limited subsets of HIV and simian immunodeficiency virus strains and are usually not as efficient as CCR5 and CXCR4 in mediating viral fusion and entry (2, 25, 31). Certain strains of HIV type 1 (HIV-1) are capable of interacting with more than one coreceptor, although not always with the same efficiency (7, 10, 27). This observation has raised a number of questions about the range and flexibility of the coreceptor-binding site on the envelope glycoproteins of HIV and simian immunodeficiency virus. The coreceptor-binding site on HIV-1 gp120 is composed mostly of conserved residues (19, 23, 30), but the V3 loop, which has been shown to influence the gp120-CCR5 interaction (14, 20), may determine which of the multiple coreceptor proteins is used by a particular virus. The envelope-binding site on the coreceptors has not been fully elucidated.
Recent alanine scanning mutagenesis studies performed by us and others show that negatively charged residues and tyrosine residues in the amino-terminal (Nt) region of CCR5 are essential for the entry of macrophage (M)-tropic and dual-tropic HIV-1 isolates (11, 13, 15, 18, 22). These amino acids are involved in gp120 binding. However, initial studies that attempted to elucidate the functionally relevant domains of the coreceptors relied on the study of receptor chimeras (1, 5, 18, 21, 26, 29). Despite some discordance between the different reports, the general conclusion emerged that all of the extracellular domains of CCR5, and not just the Nt, are important for coreceptor function. It was hypothesized, therefore, that gp120 interacts with residues in all four of these domains. Indeed, several studies that analyzed polymorphism of CCR5 genes from different species found that a number of amino acid changes in the extracellular loops (ECLs) impaired HIV-1 entry and/or cell-cell fusion (11, 18, 24). However, systematic and exhaustive mutagenesis of residues in the CCR5 loops was not performed. We have now extended the alanine scanning approach to study the role of bulky, polar and nonpolar residues in the ECLs of CCR5. We sought to identify functionally relevant residues that the early chimera studies predicted might be found in these domains.
Using PCR-based site-directed mutagenesis, we individually changed the polar serine (S), cysteine (C), tyrosine (Y), threonine (T), asparagine (N), and glutamine (Q) residues and the nonpolar proline (P), phenylalanine (F), and tryptophan (W) residues to alanines (A), as described previously (13, 22) (Fig. 1). A similar methodology was used to insert alternating glycine/serine (G/S) coding spacers between P19 and C20 in the Nt domain of CCR5. All CCR5 molecules used in this study had a 9-residue hemagglutinin (HA) tag as a carboxy-terminal extension, to allow detection by dot blotting with anti-HA antibodies (13, 22). We tested the CCR5 mutants for their abilities to mediate the entry of different HIV-1 isolates into U87MG-CD4 cells. This is a human neuronal cell line that does not express CCR5, CCR3, or CXCR4 and is not infectable by any of our test isolates (6, 13, 22). Briefly, cells were lipofected with wild-type or mutant CCR5 genes and then infected with a 200 to 500-ng/ml concentration of p24 from NLluc+env− viruses complemented in trans by envelope glycoproteins from JR-FL (17) or Gun-1 (28). Luciferase activities were measured in cell lysates 72 h postinfection as described previously (13, 22).
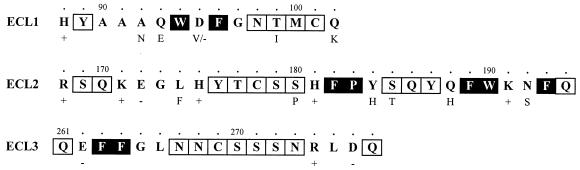
FIG. 1.
Mutagenesis of the predicted ECLs of CCR5. The amino acid sequences of the three human CCR5 ECLs (ECL1 to -3) are shown. The polarity (positive [+] or negative [−]) of charged residues is indicated below the main sequences, as are the identities of residues which differ in murine CCR5. Human CCR5 residues with polar (open squares) and bulky nonpolar side chains (black squares) were independently modified to alanines by PCR-based site-directed mutagenesis. Fidelity was confirmed by sequencing both strands of the entire CCR5 coding region.
Expression levels of mutant and wild-type CCR5 proteins were determined in each experiment (13, 22). Thus, lipofected cells were lysed with detergent, and clarified lysates were diluted (1:1) in 1% sodium dodecyl sulfate–1 mM dithiothreitol. A 70-μl aliquot, corresponding to approximately 105 cells, was loaded onto a Protran nitrocellulose membrane (Schleicher & Schuell) by using a Bio-Dot apparatus (Bio-Rad). Detection of CCR5 expression was carried out with rabbit anti-HA tag antibody diluted 1:103 (Berkeley Antibody Company), followed by horseradish peroxidase-labeled goat anti-rabbit immunoglobulin G, also diluted 1:103 (Amersham). Horseradish peroxidase activity was detected with enhanced chemiluminescence Western blotting reagents (Amersham) in accordance with the manufacturer’s instructions, and autoradiographs were scanned with the IS-1000 digital imaging system (Alpha Innotech Corporation). Integrated density values were used to standardize luciferase activities. In a separate set of experiments, we also analyzed CCR5 mutant protein expression by flow cytometry: cells were incubated with a panel of murine anti-CCR5 monoclonal antibodies (MAbs), washed, and then stained with phycoerythrin-conjugated goat anti-mouse immunoglobulin G (12). All mutants were thus shown to be present on the cell surface, and similar expression levels were measured by dot blotting and flow cytometry (data not shown). The same was true for the CCR5 molecules carrying insertions in the Nt domain (data not shown). Expression levels of all mutants were between 10 and 130% of wild-type-coreceptor expression (Fig. 2 and 3).
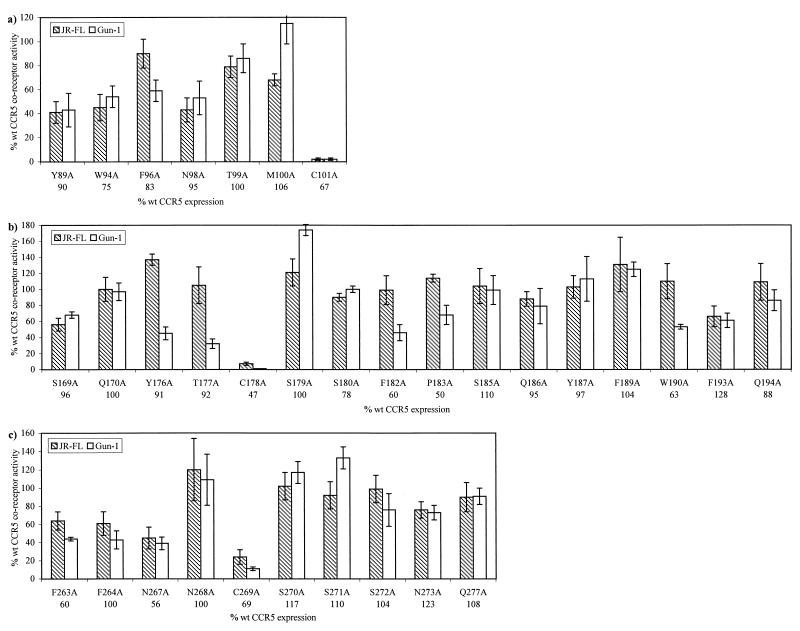
FIG. 2.
HIV-1 coreceptor function of CCR5 ECL mutants. U87MG-CD4 transiently expressing wild-type CCR5 or CCR5 with alanine substitutions in ECL1 (a), ECL2 (b), or ECL3 (c) were infected with chimeric reporter viruses carrying M-tropic env–NLluc+env−/JR-FL (shaded bars) or dual-tropic env–NLluc+env−/Gun-1 (open bars). Luciferase (luc) activity (in relative light units [r.l.u.]) was measured 72 h postinfection and standardized for CCR5 expression levels (in integrated density values [i.d.v.]). The coreceptor activity of each mutant, expressed as a percentage of wild-type coreceptor activity, is calculated by using the following formula: [(mutant luc r.l.u./wt luc r.l.u.) × (mutant i.d.v./wt i.d.v.)] × 100%. The expression level of each mutant, expressed as a percentage of wild-type CCR5 expression, is calculated by using the formula (mutant i.d.v./wt i.d.v.) × 100% and is shown below the x-axis. All values are means ± standard deviations of three independent experiments, each performed in quadruplicate. wt, wild type.
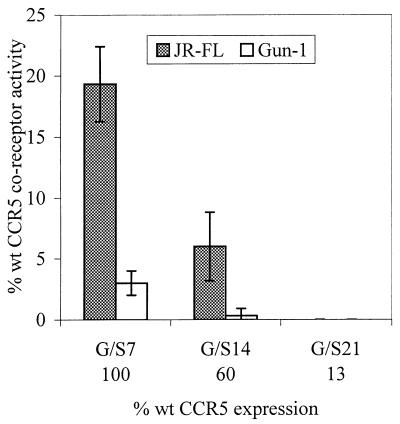
FIG. 3.
HIV-1 coreceptor function of G/S insertions in the Nt of CCR5. Seven, 14, or 21 alternating glycine/serine residues were inserted between P19 and C20 of the Nt domain of CCR5. Coreceptor activity of these insertion mutants was tested in U87MG-CD4 cells by using NLluc+env−/JR-FL (shaded bars) and NLluc+env−/Gun-1 (open bars). The coreceptor activity of each mutant is expressed as a percentage of wild-type (wt) CCR5 activity and is standardized for protein expression levels (see legend to Fig. 1).
We found that only mutation of the three cysteine residues had a significant impact on viral entry (Fig. 2). The coreceptor activity of mutants C101A and C178A was 1 to 5% of that of wild-type CCR5, whereas that of mutant C269A was 10 to 20% of that of wild-type CCR5, depending on the test isolate that was used. It is predicted that the extracellular domains of CCR5 are constrained by two disulfide bridges: one between C101 in ECL1 and C178 in ECL2 and the other between C20 in the Nt and C269 in ECL3 (16). Our results indirectly support this cysteine-pairing pattern since C20A (22) and C269A mutants are phenotypically indistinguishable in that they support similar levels of viral entry; the same is true for the C101A and C178A mutants. None of the other CCR5 mutants exhibited significantly altered coreceptor function; mutants Y89A, W94A, N98A, S169A, F182A, W190A, F263A, F264A, and N267A were the most affected, yet they only lost 40 to 60% of wild-type coreceptor activity, depending on the test isolate (Fig. 2). Our results indicate that the individual side chains of the amino acids that we have studied do not play a significant role in coreceptor function. They do not exclude, however, the possibility that the amino acid main chains have a role in CCR5 coreceptor function (19, 23, 30).
It is difficult to compare our results with those from other reports on the role of individual residues in the ECLs of CCR5 in coreceptor function. Whereas we have studied the function of single alanine substitutions in the context of human CCR5, others have looked at single or multiple non-alanine mutations in the context of nonhuman or chimeric receptors (11, 18, 24). For example, Kuhmann et al. (18) have found that P183L in the context of the human-murine HHHHloop2M chimera and Q93R in the context of African green monkey (AGM) CCR5 impair HIV-1 coreceptor function; Ross et al. (24) have shown that S180P as well as some double mutations in human-murine CCR5 chimeras yield inefficient coreceptors; Doranz et al. (11) have observed that D11A, D11A/K197A, D11A/D276A, and D11A/K197A/D276A mutations in human CCR5 impair cell-cell fusion. We found that the individual substitution of any of these residues II for alanine other than D11, has no significant effect on human CCR5 coreceptor function. The effect of a given mutation on coreceptor function therefore clearly depends on the context in which it is placed as well as on the kind of substitution that is introduced. Q93R in the context of AGM CCR5 impairs HIV-1 coreceptor function, whereas Q93A in the context of human CCR5 has no significant effect on viral entry (13, 18).
Previously, we found that substitutions of alanine for charged residues in the ECLs of CCR5 had no significant effect on coreceptor function (13). Now we show that other than for the structurally important cysteines, this is also true for residues with polar and bulky nonpolar side chains in the ECLs of CCR5. Altogether, our observations indicate that a gp120-binding site is located exclusively in the Nt domain of CCR5 (13, 22). However, the conformational integrity of the ECLs is important for coreceptor function, since changing cysteine residues to alanines greatly reduces CCR5-mediated viral entry. Also, certain mutations in the ECLs lower viral entry in a weak but reproducible manner. Perhaps multiple, simultaneous substitutions of these residues would create a nonfunctional receptor (and thereby mimic certain chimeras). One interpretation of our results is that either gp120 or the CCR5 Nt domain makes low-affinity contacts with residues in the ECLs. In the latter case, the Nt would be held by these interactions in a conformation that would allow it to interact with gp120.
To determine whether disruption of Nt-to-ECL (or Nt-to-plasma membrane) geometry is important for viral entry, we inserted spacers of 7, 14, or 21 alternating G/S residues between P19 and C20. We assumed that this would impair the formation of any noncovalent bonds that may otherwise form between ECL residues (or the plasma membrane) and Nt residues by increasing the flexibility of the latter. When normalized for protein expression, these insertions do not impair binding of the 2D7 MAb to CCR5 (data not shown), further indicating that CCR5 molecules carrying the G/S spacers are conformationally intact. MAb 2D7 recognizes epitopes in ECL2 (12, 29), and its binding to CCR5 is decreased by ∼50% when either C20 or C269 is changed to alanine (12). Increasing G/S spacer length proportionally decreases NLluc+env−/JR-FL entry: CCR5 with a 7-residue spacer has 19% wild-type activity, a 14-residue spacer lowers activity to 6% of that of the wild-type protein, and CCR5 with a 21-residue spacer no longer supports viral entry (Fig. 3). Entry of dual-tropic NLluc+env−/DH123 was even more severely affected (Fig. 3). Therefore, the greater the degree of flexibility of the Nt domain, the less efficient the mutant coreceptor. This further supports the notion that CCR5 extracellular domain interactions are important for its coreceptor function. Disruption of the tertiary structure might account for the reduced coreceptor function of certain chimeric receptors (1, 5, 18, 21, 26, 29).
We have shown that a gp120-binding site, composed of negatively charged and tyrosine residues, is contained between D2 and E18 of the Nt domain of CCR5 (13, 22). This region needs to be in a specific orientation with regard to the CCR5 ECLs or the plasma membrane for viral entry to occur efficiently. There may be a two-step gp120-binding mechanism: the envelope glycoprotein might first interact with the Nt and then induce a snugger fit by forming a number of weak interactions with residues in the other ECLs. Alternatively, weak interactions between residues in the Nt and ECL1 to -3 or the plasma membrane might maintain the Nt in a conformation that is recognizable by gp120. We are presently exploring these possibilities.
Acknowledgments
We thank John Moore for advice and help with the manuscript.
This study was supported by R01 grant AI43847-01. F.K. is supported by Progenics Pharmaceuticals Inc. Y.G. is supported by ADARC core facility funds.
REFERENCES
- 1.Atchison R E, Gosling J, Monteclaro F S, Franci C, Digilio L, Charo I F, Goldsmith M A. Multiple extracellular elements of CCR-5 and HIV-1 entry: dissociation from response to chemokines. Science. 1996;274:1924–1926. doi: 10.1126/science.274.5294.1924. [DOI] [PubMed] [Google Scholar]
- 2.Balter M. AIDS researchers negotiate tricky slopes of science. Science. 1998;280:825–826. doi: 10.1126/science.280.5365.825. [DOI] [PubMed] [Google Scholar]
- 3.Berger, E. A. 1997. HIV entry and tropism: the chemokine receptor connection. AIDS 11(Suppl. A):S3–S16. [PubMed]
- 4.Bieniasz P D, Cullen B R. Chemokine receptors and human immunodeficiency virus infection. Front Biosci. 1998;3:44–58. doi: 10.2741/a265. [DOI] [PubMed] [Google Scholar]
- 5.Bieniasz P D, Fridell R A, Aramori I, Ferguson S S G, Caron M C, Cullen B R. HIV-1-induced cell fusion is mediated by multiple regions within both the viral envelope and the CCR5 co-receptor. EMBO J. 1997;16:2599–2609. doi: 10.1093/emboj/16.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chesebro B, Buller R, Portis J, Wehrly K. Failure of human immunodeficiency virus entry and infection in CD4-positive human brain and skin cells. J Virol. 1990;64:215–221. doi: 10.1128/jvi.64.1.215-221.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. Change in co-receptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dimitrov D S. How do viruses enter cells? The HIV co-receptors teach us a lesson of complexity. Cell. 1997;91:721–730. doi: 10.1016/s0092-8674(00)80460-2. [DOI] [PubMed] [Google Scholar]
- 9.Doms R W, Peiper S C. Unwelcome guests with master keys: how HIV uses chemokine receptors for cellular entry. Virology. 1997;235:179–190. doi: 10.1006/viro.1997.8703. [DOI] [PubMed] [Google Scholar]
- 10.Doranz B, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S, Parmentier M, Collman R G, Doms R W. A dual-tropic, primary HIV-1 isolate that uses fusion and the β-chemokine receptors CKR-5, CKR-3 and CKR-2b as fusion cofactors. Cell. 1996;86:1149–1159. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 11.Doranz B J, Lu Z-H, Rucker J, Zhang T-Y, Sharron M, Cen Y-H, Wang Z-X, Guo H-H, Du J-G, Accavitti M A, Doms R W, Peiper S C. Two distinct CCR5 domains can mediate coreceptor usage by human immunodeficiency virus type 1. J Virol. 1997;71:6305–6314. doi: 10.1128/jvi.71.9.6305-6314.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dragic, T., G. E. Rabut, K. A. Nagashima, P. J. Maddon, J. P. Moore, and W. C. Olson. Unpublished data.
- 13.Dragic T, Trkola A, Lin X W, Nagashima K A, Kajumo F, Zhao L, Olson W C, Wu L, Mackay C R, Allaway G P, Sakmar T P, Moore J P, Maddon P J. Amino-terminal substitutions in the CCR5 coreceptor impair gp120 binding and human immunodeficiency virus type 1 entry. J Virol. 1998;72:279–285. doi: 10.1128/jvi.72.1.279-285.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dragic T, Trkola A, Moore J P. HIV co-receptors: gateways to the cell. Adv Res Ther. 1997;7:2–13. [Google Scholar]
- 15.Farzan M, Choe H, Vaca L, Martin K, Sun Y, Desjardins E, Ruffing N, Wu L, Wyatt R, Gerard N, Gerard C, Sodroski J. A tyrosine-rich region in the N-terminus of CCR5 is important for human immunodeficiency virus type 1 entry and mediates an association between gp120 and CCR5. J Virol. 1998;72:1160–1164. doi: 10.1128/jvi.72.2.1160-1164.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horuk R. Molecular properties of the chemokine receptor family. TIPS. 1994;15:159–165. doi: 10.1016/0165-6147(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 17.Koyanagi Y, Miles S, Mitsuyasu R T, Merrill J E, Vinters H V, Chen I S Y. Dual infection of the central nervous system by AIDS viruses with distinct cellular tropisms. Science. 1987;236:819–822. doi: 10.1126/science.3646751. [DOI] [PubMed] [Google Scholar]
- 18.Kuhmann K E, Platt E J, Kozak S L, Kabat D. Polymorphism in the CCR5 genes of African green monkeys and mice implicate specific amino acids in infections by simian and human immunodeficiency viruses. J Virol. 1997;71:8642–8656. doi: 10.1128/jvi.71.11.8642-8656.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore J P, Trkola A, Dragic T. Co-receptors for HIV-1 entry. Curr Opin Immunol. 1997;9:551–562. doi: 10.1016/s0952-7915(97)80110-0. [DOI] [PubMed] [Google Scholar]
- 21.Picard L, Simmons G, Power C A, Meyer A, Weiss R A, Clapham P R. Multiple extracellular domains of CCR-5 contribute to human immunodeficiency virus type 1 entry and fusion. J Virol. 1997;71:5003–5011. doi: 10.1128/jvi.71.7.5003-5011.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rabut G E E, Konner J A, Kajumo F, Moore J P, Dragic T. Alanine substitutions of polar and nonpolar residues in the amino-terminal domain of CCR5 differently impair entry of macrophage- and dual-tropic isolates of the human immunodeficiency virus type 1. J Virol. 1998;72:3464–3468. doi: 10.1128/jvi.72.4.3464-3468.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rizzuto C, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong P, Hendrickson W, Sodroski J. Identification of a conserved human immunodeficiency virus gp120 glycoprotein structure important for chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 24.Ross T M, Bieniasz P D, Cullen B R. Multiple residues contribute to the inability of murine CCR5 to function as a coreceptor for macrophage-tropic human immunodeficiency virus type 1 isolates. J Virol. 1998;72:1918–1924. doi: 10.1128/jvi.72.3.1918-1924.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rucker J, Edinger A L, Sharron M, Samson M, Lee B, Berson J F, Yi Y, Margulies B, Collman R G, Doranz B J, Parmentier M, Doms R W. Utilization of chemokine receptors, orphan receptors, and herpesvirus-encoded receptors by diverse human and simian immunodeficiency viruses. J Virol. 1997;71:8999–9007. doi: 10.1128/jvi.71.12.8999-9007.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rucker J, Samson M, Doranz B J, Libert F, Berson J F, Yi Y, Smyth R J, Collman R G, Broder C C, Vassart G, Doms R W, Parmentier M. Regions in the β-chemokine receptors CCR-5 and CCR-2b that determine HIV-1 cofactor specificity. Cell. 1996;87:437–446. doi: 10.1016/s0092-8674(00)81364-1. [DOI] [PubMed] [Google Scholar]
- 27.Simmons G, Wilkinson D, Reeves J D, Dittmar M T, Beddows S, Weber J, Carnegie G, Desselberger U, Gray P W, Weiss R A, Clapham P R. Primary, syncytium-inducing human immunodeficiency virus type 1 isolates are dual-tropic and most can use either Lestr or CCR5 as coreceptors for virus entry. J Virol. 1996;70:8355–8360. doi: 10.1128/jvi.70.12.8355-8360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takeuchi Y, Akutsu M, Murayama K, Shimizu N, Hoshino H. Host range mutant of human immunodeficiency virus type 1: modification of cell tropism by a single point mutation at the neutralization epitope in the env gene. J Virol. 1991;65:1710–1718. doi: 10.1128/jvi.65.4.1710-1718.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu L, LaRosa G, Kassam N, Gordon C J, Heath H, Ruffing N, Chen H, Humblias J, Samson M, Parmentier M, Moore J P, Mackay C R. Interaction of chemokine receptor CCR5 with its ligands: multiple domains for HIV-1 gp120 binding and a single domain for chemokine binding. J Exp Med. 1997;186:1373–1381. doi: 10.1084/jem.186.8.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wyatt R, Kwong P D, Desjardins E, Sweet R, Robinson J, Hendrickson W, Sodroski J. The antigenic structure of the human immunodeficiency virus gp120 envelope glycoprotein. Nature. 1998;393:705–710. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, Y., T. Dragic, Y. Cao, L. Kostrikis, D. S. Kwon, D. R. Littman, V. N. KewalRamani, and J. P. Moore. Use of coreceptors other than CCR5 by non-syncytium-inducing adult and pediatric isolates of human immunodeficiency virus type 1 is rare in vitro. J. Virol. 72:9337–9344. [DOI] [PMC free article] [PubMed]