Abstract
Production of soluble full-length nonstructural protein 5B (NS5B) of hepatitis C virus (HCV) has been shown to be problematic and requires the addition of salts, glycerol, and detergents. In an effort to improve the solubility of NS5B, the hydrophobic C terminus containing 21 amino acids was removed, yielding a truncated NS5B (NS5BΔCT) which is highly soluble and monodispersed in the absence of detergents. Fine deletional analysis of this region revealed that a four-leucine motif (LLLL) in the hydrophobic domain is responsible for the solubility profile of the full-length NS5B. Enzymatic characterization revealed that the RNA-dependent RNA polymerase (RdRp) activity of this truncated NS5B was comparable to those reported previously by others. For optimal enzyme activity, divalent manganese ions (Mn2+) are preferred rather than magnesium ions (Mg2+), whereas zinc ions (Zn2+) inhibit the RdRp activity. Gliotoxin, a known poliovirus 3D RdRp inhibitor, inhibited HCV NS5B RdRp in a dose-dependent manner. Kinetic analysis revealed that HCV NS5B has a rather low processivity compared to those of other known polymerases.
Hepatitis C virus (HCV) is currently the leading etiological agent of non-A non-B hepatitis. According to a press release from a recent World Health Organization meeting, more than 170 million people worldwide may be infected with HCV. About 80% of patients with acute HCV infection will progress to chronic hepatitis; 20% of these will develop cirrhosis, and 1 to 5% of these will develop hepatocellular carcinoma (28a). More than four million individuals in the United States are estimated to be infected with HCV (2).
Current therapies with alpha interferon alone and the combination of alpha interferon-ribavirin have been shown to be effective in a portion of patients with chronic HCV infection (20, 24). Vaccine development has been hampered by the high degree of immune evasion and the lack of protection against reinfection, even with the same inoculum (7, 14, 26, 29). Development of small molecule inhibitors directed against specific viral targets has thus become the focus of anti-HCV research. The determination of crystal structures for NS3 protease (16, 19, 30) and NS3 RNA helicase (15, 31) has provided important structural insights for rational design of specific inhibitors.
One key enzyme encoded by HCV is NS5B, which has been shown to be an RNA-dependent RNA polymerase (1, 4, 6, 17, 32). NS5B is thus believed to be responsible for genome replication of HCV. Cellular localization studies revealed that NS5B is membrane associated and distributed in the perinuclear region (12). This coincides with the distribution of NS5A (27), suggesting that NS5A and NS5B may stay together after proteolytic cleavage at NS5A/NS5B. It has been postulated that the nonstructural proteins of HCV (NS3 to -5B) may assemble into membrane-associated replication complexes which are competent for authentic RNA genome replication.
By itself, HCV NS5B RdRp appears to lack the specificity for HCV RNA and can “copy back” heterologous nonviral RNA (4). This lack of specificity for HCV RNA may reflect the notion that additional viral or cellular factors are required for specific recognition of the replication signal, most likely present at the 3′ untranslated region. Recent studies by Lohmann et al. (17) demonstrated that NS5B alone can replicate the entire HCV genome via a copy-back mechanism initiated from the end of the 3′ untranslated region.
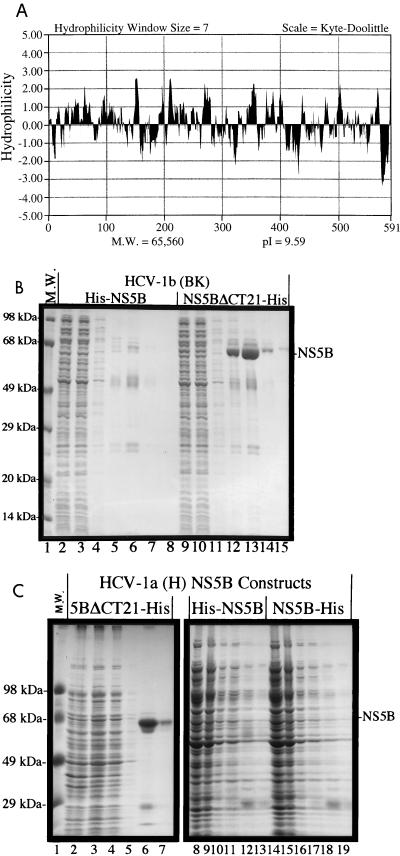
Our earlier attempts to express and purify full-length NS5B were hampered by its poor solubility. Recent reports demonstrated that detergents, salts, and glycerol are required to solubilize the NS5B protein (4, 6, 17). The hydropathy profile of NS5B revealed that there is a highly hydrophobic domain at the C terminus (Fig. 1A), which may affect the solubility of NS5B. In an effort to improve the solubility of NS5B, the C-terminal hydrophobic domain containing 21 amino acids was removed and the truncated protein was compared in parallel with the full-length NS5B for expression and purification.
FIG. 1.
(A) Hydropathy profile of NS5B. (B) Parallel expression and purification of full-length and truncated NS5B from HCV-1b, the BK isolate. NS5B cDNAs were cloned into the pET-21b vector (Novagen, Inc.) between the NheI and XhoI sites. The resulting plasmids were transformed into the bacterial host, JM109(DE3), for expression driven by T7 polymerase. Induction by 0.2 mM isopropyl-β-thiogalactopyranoside (IPTG) was carried out at 24°C for 4 h. Polyhistidine tags (His−) were engineered into the NS5B clones at either the N terminus or the C terminus to facilitate the purification. All engineered NS5B plasmids were verified by sequencing with the automated sequencer (ABI 377) from Perkin-Elmer (Foster City, Calif.). The purification procedures were similar to those previously reported for the purification of the NS3 protease domain (16) with minor modifications. Protein samples in lanes 2 to 8 are from the full-length His-NS5B, and those in lanes 9 to 15 are from the truncated NS5BΔCT21-His. Lanes 2 and 9, total soluble lysates; lanes 3 and 10, flowthrough unbound proteins; lanes 4 and 11, proteins from high-salt wash; lanes 5 to 8 and 12 to 15, elution fractions off the Ni-NTA resin. (C) Parallel expression and purification of full-length and truncated NS5B from HCV-1a, the Hutchinson (H77) isolate. The NS5B cDNAs were cloned into pET-28a between 14, total soluble lysates; lanes 3, 9, and 15, flowthrough unbound proteins; lanes 4, 10, and 16, high-salt wash; lanes 5 to 7, 11 to 13, and 17 to 19, elution fractions. M.W., molecular weight.
To facilitate the purification, both full-length NS5B and C-terminally truncated NS5B were expressed as polyhistidine (His−)-tagged fusion proteins. The N-terminally tagged proteins are designated His-NS5B or His-NS5BΔCT21, whereas the C-terminally tagged proteins are designated NS5B-His or NS5BΔCT21-His. The results in Fig. 1B show that the C-terminally truncated NS5B, NS5BΔCT21-His, derived from the BK (HCV-1b) isolate, is soluble and yields good purification when applied to a nickel-chelated (Ni-nitrilotriacetic acid [NTA]) column (lanes 12 to 15, elution fractions 1 to 4). On the other hand, the full-length NS5B, His-NS5B, is not soluble under the lysis conditions and thus not purifiable on an Ni-NTA column (lanes 5 to 8). The expression levels for both proteins were comparable in the total lysates as detected by Western blot analysis (data not shown). Further studies using NS5B from a different isolate, H77 (HCV-1a), showed the same results. The full-length NS5Bs (His-NS5B and NS5B-His) are not soluble with little, if any, purification over the Ni-NTA column (Fig. 1C, lanes 8 to 19), whereas the truncated protein (NS5BΔCT21-His) has good solubility and can be purified readily (Fig. 1C, lanes 2 to 7). Attempts to purify the full-length NS5B from the bacterial lysates by the protocols described by Lohmann et al. (17) failed, possible due to the problem that full-length NS5B expressed in bacterial cells accumulated in the inclusion bodies, which are resistant to the solubilization method.
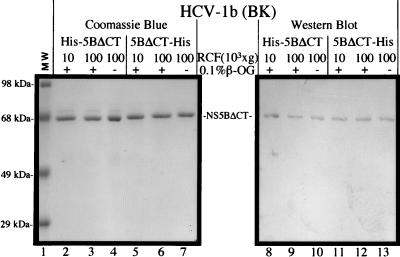
The truncated proteins were further subjected to a more stringent solubility test by ultracentrifugation at 100,000 × g for 30 min. The results (Fig. 2, lanes 3 and 4 and 6 and 7) demonstrated that the truncated proteins remained in the supernatant under these conditions in the presence or absence of detergent (0.1% octyl-β-glucoside). The location of the His tag does not affect the solubility (His-NS5BΔCT21 versus NS5BΔCT21-His), and the His tag can be removed without any loss of solubility (data not shown). In a separate experiment, glycerol (10%) was dialyzed out of the protein samples and no significant loss of solubility was observed. In fact, a high concentration of salt (NaCl at ≥300 mM) is the only essential requirement for solubility. The truncated proteins were well monodispersed in solutions containing high concentrations of salt as detected by light scattering analysis (data not shown). The above results suggest that the entire or part of the 21-amino-acid domain at the C terminus plays an important role in determining the solubility of NS5B. Its removal improves the solubility of NS5B in a detergent-glycerol-free and strain-independent manner.
FIG. 2.
Solubility analysis of the truncated NS5B proteins. RCF represents relative centrifugal force (g force). Protein samples were subjected to ultracentrifugation at 100,000 × g for 30 min (at 4°C) with a Beckman Optima TLX Benchtop Ultracentrifuge (in a TLA-45 rotor at a speed of 41,000 rpm), in the presence (+) or absence (−) of a nonionic detergent, 0.1% octyl-β-glucoside. Polyclonal antibodies (α-NS5) raised in rabbits were used for the Western blotting analysis as described previously (11) in lanes 8 to 13.
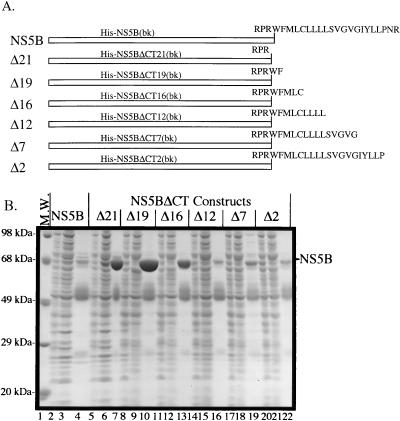
To map the minimum sequence in the 21-amino-acid domain that is responsible for the lack of solubility, amino acids from the hydrophobic domain were added back sequentially to the C terminus of truncated NS5B (Fig. 3A). The resulting proteins were expressed, and the soluble lysates were applied to the Ni-NTA columns for affinity purification. Figure 3B shows that up to 5 amino acids can be added back to the truncated NS5B without significant loss of purifiable proteins (lanes 10 and 13, representing His-NS5BΔCT19 and His-NS5BΔCT16). However, when an LLLL motif was added back, it rendered the protein (His-NS5BΔCT12) virtually unpurifiable (lane 16), indicating a dramatic reduction in solubility. Adding back additional amino acids (in His-NS5BΔCT7 and His-NS5BΔCT2) failed to improve its solubility. These results demonstrate that the LLLL motif-containing proteins, His-NS5B, His-NS5BΔCT2, His-NS5BΔCT7, and His-NS5BΔCT12, have reduced solubility compared to those that lack the motif, suggesting that the LLLL motif contributes to the insolubility of NS5B. Multiple alignments of the C-terminal domains from different genotypes of HCV revealed that this LLLL motif is absolutely conserved.
FIG. 3.
Fine deletional mapping of an LLLL motif in the C-terminal hydrophobic domain. (A) The N-terminally His-tagged NS5B proteins from HCV-1b (BK isolate) were used to map the motif responsible for the insolubility of NS5B. The sequence for the C-terminal 24 amino acids is shown in the full-length protein (NS5B). Removal of the C-terminal 21 amino acids yields a truncated NS5B ending with a C-terminal sequence of RPR. Adding back the C-terminal 21 amino acids in a sequential manner results in NS5B proteins with different C termini. These truncated proteins are designated according to the number of amino acids deleted from the C-terminal end. (B) Purifiability analysis of NS5B deletion mutants by polyacrylamide gel electrophoresis. Only total soluble lysates, flowthrough proteins, and elution fractions are analyzed for NS5B (lanes 2 to 4), CTΔ21 (lanes 5 to 7), ΔCT19 (lanes 8 to 10), ΔCT16 (lanes 11 to 13), ΔCT12 (lanes 14 to 16), ΔCT7 (lanes 17 to 19), and ΔCT2 (lanes 20 to 22). M.W., molecular weight.
Further truncations from the C-terminal end of NS5B were constructed, and the resulting proteins, NS5BΔCT55-His, NS5BΔCT63-His, NS5BΔCT70-His, NS5BΔCT77-His, NS5BΔCT84-His, and NS5BΔCT92-His, were subjected to the same purification procedures as that for NS5BΔCT21-His. Only NS5BΔCT55-His and NS5BΔCT63-His could be purified from the soluble lysates, whereas the further truncated proteins were not soluble and thus unpurifiable (data not shown). We concluded that at least 63 amino acids can be removed from the C terminus without significant loss of solubility, whereas deletions of more than 70 amino acids lead to insoluble products. Similar results were reported by Lohmann et al., who demonstrated that 55 amino acids could be deleted without loss of activity, whereas NS5B with 84 amino acids deleted could not be purified (17).
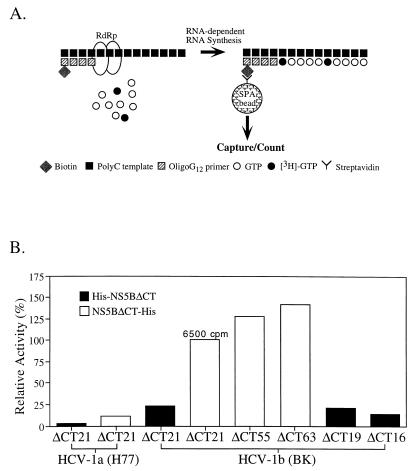
To characterize the activity of these soluble NS5B proteins, an RNA-dependent RNA polymerase assay was developed with poly(C) as the template and biotinylated oligo(G)12 as the primer. Incorporation of [3H]GMP into the primer was measured by the scintillation proximity assay (SPA) after the end products were captured by streptavidin-coated SPA beads (illustrated in Fig. 4A). Under our optimized assay conditions, similar to those described by others (4, 17), NS5BΔCT21-His was shown to catalyze ribonucleotide polymerization (RNA synthesis) in an RNA template-dependent and primer-dependent manner. With the activity of NS5BΔCT21-His normalized to 100% (incorporation of ∼6,500 cpm with 0.25 μg of poly(C)–25 ng of oligo(G)12, 5 μM or 0.05 μCi of GTP, and 50 nM NS5B RdRp at room temperature for 3 h with approximately 10% product formation), the activities of other soluble NS5B proteins were compared (Fig. 4B). The background of this assay was very low at less than 50 cpm. The comparison showed that (i) the truncated NS5Bs with 55 or 63 amino acids deleted (NS5BΔCT55-His and NS5BΔCT63-His, respectively) were consistently more active than NS5BΔCT21-His; (ii) truncations with 19 and 16 amino acids deleted (His-NS5BΔCT19 and His-NS5BΔCT16, respectively) did not affect or slightly reduced activities compared to that of His-NS5BΔCT21; (iii) the C-terminally tagged proteins (NS5BΔCT21-His) are about fourfold more active than the N-terminally tagged proteins (His-NS5BΔCT21); and (iv) NS5Bs from the BK (HCV-1b) isolate are about 5- to 10-fold more active than those corresponding ones derived from the H77 (HCV-1a) isolate. Recent studies by Lohmann et al. also demonstrated that the N terminus of NS5B is important for the RdRp activity and that deletion from the N terminus is detrimental to the RdRp activity (17, 18). Interestingly, the N terminus of poliovirus 3D is also very sensitive to deletion (8, 23) as well as modification: adding two amino acids (alanine and serine) to the N-terminal glycine residue completely abolishes the 3D RdRp activity (10). It has been suggested that the N terminus of poliovirus 3D plays an important role in oligomerization of the polymerase, which is believed to be the functional unit of the viral replicase (8, 21).
FIG. 4.
(A) Illustration of the SPA for RdRp. An SPA (Amersham Life Science) was established for the HCV NS5B RdRp with an RNA homopolymer [poly(C)] as the template complexed with a biotinylated primer [oligo(G)12]. The assay specifically measures the incorporation of tritium-labeled [3H]GMP into the biotinylated RNA products of NS5B which can be captured by the streptavidin-coated SPA beads. The assay conditions were similar to those of a previously described protocol with modifications (4). The assay was carried out at room temperature (≈22°C) for 3 h in a final reaction volume of 50 μl and stopped by adding 50 μl of 100 mM EDTA in phosphate-buffered saline. Unless specified, 50 nM RdRp enzymes were used in the assay containing 250 ng of poly(C)–25 ng of oligo(G)12 and 5 μM or 0.05 μCi of GTP. (B) Comparisons of RdRp activities from different soluble and truncated NS5B proteins. The activity of NS5BΔCT21-His from HCV-1b (BK) is normalized to 100%, which represents a total incorporation of approximately 6,500 cpm. The specific activity is calculated at approximately 870 cpm/pmol of RdRp/h.
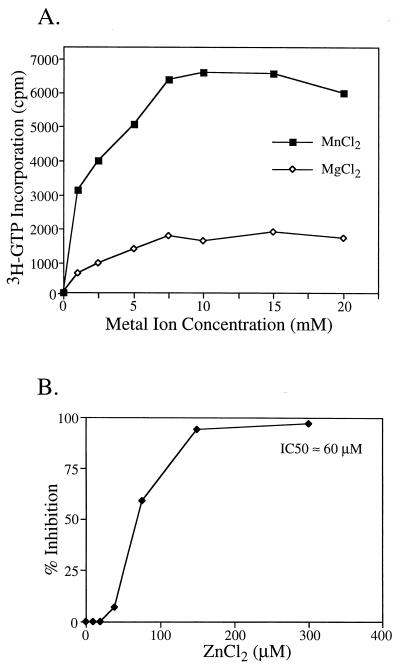
Previous reports showed that divalent magnesium ions (Mg2+) were required for NS5B RdRp activity (4, 17). We further investigated the divalent metal ion requirement of HCV NS5B. Our data showed that manganese ions (Mn2+) were, in fact, approximately four times more effective than Mg2+ in coordinating the catalytic reaction of NS5B RdRp and thus preferred for optimal RdRp activity (Fig. 5A). Zinc ions (Zn2+), on the other hand, not only failed to support the RdRp activity but also inhibited the Mn2+-Mg2+-dependent RdRp with a 50% inhibitory concentration of approximately 60 μM (Fig. 5B). In contrast, the assay conditions for poliovirus 3D RdRp consist of zinc ions (60 μM) (9). Given that polymerases require divalent metal ions for activities (3, 13), inhibition by zinc ions presents several interesting scenarios: (i) is Zn2+ a potent competitor of Mg2+ or Mn2+ for binding to the carboxylate cluster formed by the side chains of three aspartic acids from motif A and C (8), or (ii) does Zn2+ bind to somewhere other than the active site, which allosterically hinders the nucleophilic attack on the α-phosphate by the 3′ hydroxyl group of the primer? If the first scenario is true, Zn2+ has to be supercompetent in competing for binding, since the concentration of Mg2+ or Mn2+ (5 to 10 mM) is much higher than that of Zn2+ (50% inhibitory concentration ≈60 μM). Interestingly, a calcium ion (Ca2+) was found in the crystal structure of poliovirus 3D polymerase coordinated by the carboxylate ligands at the active site (8), even though Ca2+ does not support the RdRp activity of 3D polymerase.
FIG. 5.
(A) Divalent metal ion requirement for NS5B RdRp. (B) Dose-dependent inhibition of NS5B RdRp activity by zinc ion. The RdRp enzyme used here is NS5BΔCT21-His from HCV-1b (BK). The assay conditions are described in Materials and Methods. IC50, 50% inhibitory concentration.
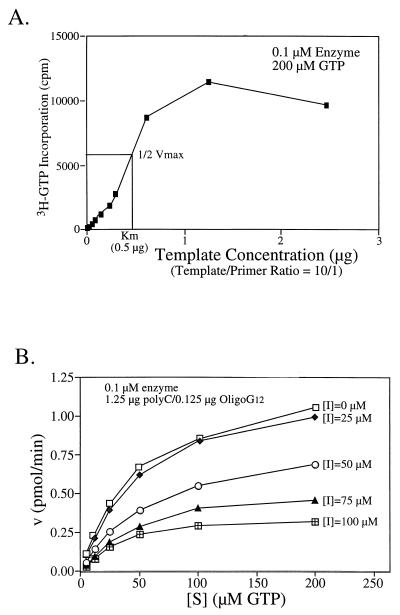
Michaelis-Menten steady-state kinetics were analyzed to characterize the enzymatic activity of NS5B RdRp. In a polymerase reaction, multiple substrates including a template-primer complex and a ribonucleotide are involved. Reaction at each step follows, presumably, a sequential order: the polymerase binds to the template-primer first to form a binary complex which then takes up a nucleotide to form the catalytically competent ternary complex (5, 13). To determine the kinetic parameters for one substrate, the enzyme has to be saturated by the other. Also, the product formation has to be limited to less than 10%. The initial velocities (v) at increasing amounts of each substrate were determined. The data were processed and analyzed with kinetics software (k·cat, version 1.5; BioMetallics, Inc.). To estimate the Km for the template-primer [poly(C)-oligo(G)12] substrate, 200 μM GTP (near saturating amount) was used in the assay in the presence of increasing amounts of poly(C)-oligo(G)12 [0.025 to 2.5 μg of poly(C) with a template/primer ratio of 10). Based on the results from Fig. 6A, the Km for the template is about 0.5 μg, which corresponds to approximately 30 nM primer [oligo(G)12]. To determine the Km and the turnover rate (kcat) for the nucleotide (GTP), saturating amounts of template-primer at 1.25 μg of poly(C)–0.125 μg of oligo(G)12 were used. As shown in Fig. 6B, the Km for GTP was calculated at 52 μM and the kcat is about 0.3 min−1, indicating that NS5B has a rather low processivity. The low kcat value might be in part due to the use of unnatural substrate in an assay measuring both the rate-limiting initiation step and the elongation step (which normally has a higher turnover rate). In a recent report by Lohmann et al., the elongation rate for HCV RdRp was determined at about 150 to 200 nucleotides per min (18). This estimate was based on the size increase of the RNA product as a function of time. However, the number of NS5B molecules required for achieving this elongation rate was not determined. A similar study was also conducted to estimate the elongation rate for poliovirus RdRp at about 1,250 nucleotides per min (28). However, it is difficult to compare such elongation rates (different from kcat values) from these in vitro assays which were carried out under different conditions (for example, the differences in temperatures or the amount of enzymes, etc.). In fact, when we increased the assay temperature from room temperature (∼22°C) to 30 or 37°C, the kcat value for HCV NS5B RdRp was significantly increased (data not shown).
FIG. 6.
Kinetic analysis of NS5B RdRp activity. (A) Titration of the template-primer at various concentrations; (B) titration of GTPs in the absence and presence of an RdRp-specific inhibitor, gliotoxin. Enzyme HCV-1b NS5BΔCT21-His (0.1 μM) was used in the kinetic study.
Finally, a panel of known polymerase inhibitors was screened for inhibition of HCV NS5B RdRp. Gliotoxin, a natural product (fungal metabolite), was found to inhibit HCV NS5B RdRp in a dose-dependent manner (Fig. 6B). Interestingly, gliotoxin also inhibited poliovirus 3D RdRp with a similar potency (25), suggesting that gliotoxin may be an RdRp-specific inhibitor. A recent report by Paul et al. further demonstrated that gliotoxin inhibited both priming (uridylylation of VPg) and elongation mediated by 3D polymerase (22). The kinetic study demonstrated that gliotoxin is a mixed noncompetitive inhibitor with a Ki value of approximately 230 μM (Fig. 6B).
The major finding in this report is the identification of a hydrophobic domain at the C terminus of NS5B and mapping of a conserved LLLL motif in this domain which significantly affects the solubility on NS5B. Although we have characterized the solubility of the truncated NS5B extensively, a similar solubility test for the full-length NS5B has not been performed due to our inability to purify the products from bacterial lysates. In a previous report, full-length NS5B products were solubilized from insect cell lysates in the presence of high concentrations of salt, detergent, and glycerol (17). We failed to solubilize similar full-length NS5B products expressed in bacterial cells. This may be due to the problem that insoluble NS5Bs produced in bacterial cells accumulate in the inclusion bodies and can be purified only under denaturing conditions. Interestingly, in a recently published report, the full-length NS5B solubilized in salt, detergent, and glycerol appeared to form high-molecular-weight aggregates (18) which were unlikely to remain in solution upon ultracentrifugation.
Also, a highly sensitive and quantitative SPA was developed to characterize the RdRp activity. We have demonstrated that the truncated NS5B from Escherichia coli is as active as those NS5Bs produced in insect cells reported previously (4, 17), based on a comparison of radioactivity incorporation under each assay condition. Another important finding is that Mn2+, rather than Mg2+, is preferred for optimal RdRp activity, whereas Zn2+ is inhibitory. Whether there are any differences in RdRp activities among NS5Bs expressed in various systems will be the subject of a future study.
Finally, NS5B RdRp activity is sensitive to the presence of gliotoxin, a fungal metabolite. It is a known inhibitor of poliovirus 3D RdRp (22, 25). Based on our kinetic analysis, gliotoxin is a mixed noncompetitive inhibitor and is not potent. More importantly, it is a very toxic compound with limited pharmaceutical uses. However, a better understanding of its mechanism of action against RdRp may shed light on the structural and enzymatic characterization of RdRp.
Acknowledgments
We thank Gregory Reyes and Patricia Weber for support and Charles Lesburg and Michael Cable for helpful discussions. We also appreciate the excellent technical assistance of Lin Cheng and Michele Beaudet-Miller.
REFERENCES
- 1.Al R H, Xie Y, Wang Y, Hagedorn C H. Expression of recombinant hepatitis C virus non-structural protein 5B in Escherichia coli. Virus Res. 1998;53:141–149. doi: 10.1016/s0168-1702(97)00147-0. [DOI] [PubMed] [Google Scholar]
- 2.Alter M J, Mast E E. The epidemiology of viral hepatitis in the United States. Gastroenterol Clin N Am. 1994;23:437–455. [PubMed] [Google Scholar]
- 3.Beese L S, Steitz T A. Structural basis for the 3′-5′ exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 1991;10:25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behrens S-E, Tomei L, De Francesco R. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 1996;15:12–22. [PMC free article] [PubMed] [Google Scholar]
- 5.Benkovic S J, Cameron C E. Kinetic analysis of nucleotide incorporation and misincorporation by Klenow fragment of Escherichia coli DNA polymerase I. Methods Enzymol. 1995;262:257–269. doi: 10.1016/0076-6879(95)62022-2. [DOI] [PubMed] [Google Scholar]
- 6.De Francesco R, Behrens S E, Tomei L, Altamura S, Jiricny J. RNA-dependent RNA polymerase of hepatitis C virus. Methods Enzymol. 1996;275:58–67. doi: 10.1016/s0076-6879(96)75006-1. [DOI] [PubMed] [Google Scholar]
- 7.Farci P, Alter H J, Govindarajan S, Wong D C, Engle R, Lesniewski R R, Mushahwar I K, Desai S M, Miller R H, Ogata N, Purcell R H. Lack of protective immunity against reinfection with hepatitis C virus. Science. 1992;258:135–140. doi: 10.1126/science.1279801. [DOI] [PubMed] [Google Scholar]
- 8.Hansen J L, Long A M, Schultz S C. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure. 1997;5:1109–1122. doi: 10.1016/s0969-2126(97)00261-x. [DOI] [PubMed] [Google Scholar]
- 9.Hey T D, Richards O C, Ehrenfeld E. Synthesis of plus- and minus-strand RNA from poliovirion RNA template in vitro. J Virol. 1986;58:790–796. doi: 10.1128/jvi.58.3.790-796.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hong, Z. Unpublished data.
- 11.Hong Z, Beaudet-Miller M, Durkin J, Zhang R, Kwong A D. Identification of a minimal hydrophobic domain in the herpes simplex virus type 1 scaffolding protein which is required for interaction with the major capsid protein. J Virol. 1996;70:533–540. doi: 10.1128/jvi.70.1.533-540.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hwang S B, Park K J, Kim Y S, Sung Y C, Lai M M C. Hepatitis C virus NS5B protein is a membrane-associated phosphoprotein with a predominantly perinuclear localization. Virology. 1997;227:439–446. doi: 10.1006/viro.1996.8357. [DOI] [PubMed] [Google Scholar]
- 13.Joyce C M, Steitz T A. Function and structure relationships in DNA polymerases. Annu Rev Biochem. 1994;63:777–822. doi: 10.1146/annurev.bi.63.070194.004021. [DOI] [PubMed] [Google Scholar]
- 14.Kao J H, Chen P J, Wang J T, Yang P M, Lai M Y, Wang T H, Chen D S. Superinfection by homotypic virus in hepatitis C virus carriers: studies on patients with post-transfusion hepatitis. J Med Virol. 1996;50:303–308. doi: 10.1002/(SICI)1096-9071(199612)50:4<303::AID-JMV4>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 15.Kim J L, Morgenstern K A, Griffith J P, Dwyer M D, Thomson J A, Murcko M A, Lin C, Caron P R. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure. 1998;6:89–100. doi: 10.1016/s0969-2126(98)00010-0. [DOI] [PubMed] [Google Scholar]
- 16.Kim J L, Morgenstern K A, Lin C, Fox T, Dwyer M D, Landro J A, Chambers S P, Markland W, Lepre C A, O’Malley E T, Harbeson S L, Rice C M, Murcko M A, Caron P R, Thomson J A. Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell. 1996;87:343–355. doi: 10.1016/s0092-8674(00)81351-3. [DOI] [PubMed] [Google Scholar]
- 17.Lohmann V, Korner F, Herian U, Bartenschlager R. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J Virol. 1997;71:8416–8428. doi: 10.1128/jvi.71.11.8416-8428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lohmann V, Roos A, Korner F, Koch J O, Bartenschlager R. Biochemical and kinetic analyses of NS5B RNA-dependent RNA polymerase of the hepatitis C virus. Virology. 1998;249:108–118. doi: 10.1006/viro.1998.9311. [DOI] [PubMed] [Google Scholar]
- 19.Love R A, Parge H E, Wickersham J A, Hostomsky Z, Habuka N, Moomaw E W, Adachi T, Hostomska Z. The crystal structure of hepatitis C virus NS3 proteinase reveals a trypsin-like fold and a structural zinc binding site. Cell. 1996;87:331–342. doi: 10.1016/s0092-8674(00)81350-1. [DOI] [PubMed] [Google Scholar]
- 20.Marcellin P, Boyer N, Gervais A, Martinot M, Pouteau M, Castelnau C, Kilani A, Areias J, Auperin A, Benhamou J P, Degott C, Erlinger S. Long-term histologic improvement and loss of detectable intrahepatic HCV RNA in patients with chronic hepatitis C and sustained response to interferon-alpha therapy. Ann Intern Med. 1997;127:875–881. doi: 10.7326/0003-4819-127-10-199711150-00003. [DOI] [PubMed] [Google Scholar]
- 21.Pata J D, Schultz S C, Kirkegaard K. Functional oligomerization of poliovirus RNA-dependent RNA polymerase. RNA. 1995;1:466–477. [PMC free article] [PubMed] [Google Scholar]
- 22.Paul A V, van Boom J H, Filippov D, Wimmer E. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature. 1998;393:280–284. doi: 10.1038/30529. [DOI] [PubMed] [Google Scholar]
- 23.Plotch S J, Palant O, Gluzman Y. Purification and properties of poliovirus RNA polymerase expressed in Escherichia coli. J Virol. 1989;63:216–225. doi: 10.1128/jvi.63.1.216-225.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reichard O, Norkrans G, Fryden A, Braconier J H, Sonnerborg A, Weiland O, Group T S S. Randomised, double-blind, placebo-controlled trial of interferon alpha-2b with and without ribavirin for chronic hepatitis C. Lancet. 1998;351:83–87. doi: 10.1016/s0140-6736(97)06088-1. [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez P L, Carrasco L. Gliotoxin: inhibitor of poliovirus RNA synthesis that blocks the viral RNA polymerase 3Dpol. J Virol. 1992;66:1971–1976. doi: 10.1128/jvi.66.4.1971-1976.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimizu Y K, Hijikata M, Iwamoto A, Alter H J, Purcell R H, Yoshikura H. Neutralizing antibodies against hepatitis C virus and the emergence of neutralization escape mutant viruses. J Virol. 1994;68:1494–1500. doi: 10.1128/jvi.68.3.1494-1500.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanji Y, Hijikata M, Satoh S, Kaneko T, Shimotohno K. Hepatitis C virus-encoded nonstructural protein NS4A has versatile functions in viral protein processing. J Virol. 1995;69:1575–1581. doi: 10.1128/jvi.69.3.1575-1581.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Dyke T A, Rickles R J, Flanegan J B. Genome-length copies of poliovirion RNA are synthesized in vitro by the poliovirus RNA-dependent RNA polymerase. J Biol Chem. 1982;257:4610–4617. [PubMed] [Google Scholar]
- 28a.World Health Organization. Lancet. 1998;351:1415. [Google Scholar]
- 29.Wyatt C A, Andrus L, Brotman B, Huang F, Lee D-H, Prince A M. Immunity in chimpanzees chronically infected with hepatitis C virus: role of minor quasispecies in reinfection. J Virol. 1998;72:1725–1730. doi: 10.1128/jvi.72.3.1725-1730.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan Y, Li Y, Munshi S, Sardana V, Cole J L, Sardana M, Steinkuehler C, Tomei L, De Francesco R, Kuo L C, Chen Z. Complex of NS3 protease and NS4A peptide of BK strain hepatitis C virus: a 2.2 A resolution structure in a hexagonal crystal form. Protein Sci. 1998;7:837–847. doi: 10.1002/pro.5560070402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yao N, Hesson T, Cable M, Hong Z, Kwong A D, Le H V, Weber P C. Structure of the hepatitis C virus RNA helicase domain. Nat Struct Biol. 1997;4:463–467. doi: 10.1038/nsb0697-463. [DOI] [PubMed] [Google Scholar]
- 32.Yuan Z H, Kumar U, Thomas H C, Wen Y M, Monjardino J. Expression, purification, and partial characterization of HCV RNA polymerase. Biochem Biophys Res Commun. 1997;232:231–235. doi: 10.1006/bbrc.1997.6249. [DOI] [PubMed] [Google Scholar]