Abstract
One of the major structural differences between rabies virus and vesicular stomatitis virus (VSV) is that the nucleoprotein (N) is the major phosphoprotein and the nominal phosphoprotein (P) is less phosphorylated in rabies virus, whereas P is the major phosphoprotein and N is not phosphorylated in VSV. We investigated the function of phosphorylation of rabies virus N after dephosphorylation of N with alkaline phosphatase or after changing the phosphorylated serine at position 389 to alanine by site-directed mutagenesis. The unphosphorylated N, in comparison to the phosphorylated N, was studied for its abilities to encapsidate rabies virus leader RNA and to support transcription and replication of a rabies virus minigenome. We found that unphosphorylated N binds more strongly to leader RNA than the phosphorylated N; however, the rates of transcription and replication of the rabies virus minigenome were significantly lower with the unphosphorylated N than with the phosphorylated N. This indicates that the phosphorylation of rabies virus N plays an important role in the regulation of rabies virus transcription and replication, probably via modulation of leader RNA encapsidation.
Both rabies virus and vesicular stomatitis virus (VSV) belong to the Rhabdoviridae family (7). The two viruses share a number of characteristics, such as the genome organization and replication strategies (15). However, rabies virus and VSV also show marked differences. For example, rabies virus has lower transcriptase activity (8, 12) and replicates more slowly than VSV (4). Furthermore, rabies virus is almost exclusively neurotropic in vivo (7), whereas VSV is pantropic (15). One of the major structural differences between the two viruses is that N is the major phosphoprotein and P is less phosphorylated in rabies virus, whereas P is the major phosphoprotein and N is not phosphorylated in VSV (14).
Previous research has proposed that rabies virus N and VSV N play a crucial role in the transition from viral RNA transcription to replication by encapsidating the positive-strand leader RNA, the first viral products made in an infected cell, and preventing them from further initiating genomic RNA transcription (2, 16–18). In vitro studies have shown that both rabies virus N and VSV N preferentially interact with positive-strand leader RNA over other RNA species (3, 18). Furthermore, Patton et al. (13) showed that VSV N alone can support the transition from transcription to replication. Therefore, N plays important roles in the transcription and replication of rhabdoviruses. The phosphorylation of rabies virus N, but not VSV N, raises questions concerning the function(s) of the phosphorylated N in its interaction with genomic RNA and P to bring about transcription and replication in rabies virus. The phosphorylation site has been mapped to the serine residue at position 389 of rabies virus N (6). In the present study, we investigated the role of phosphorylation of serine 389 of rabies virus N by either dephosphorylation of the N with alkaline phosphatase or mutation of serine 389 to alanine. The dephosphorylated or the unphosphorylated N was studied for its ability to encapsidate rabies virus leader RNA and to support transcription and replication of a rabies virus minigenome (5).
Dephosphorylated N binds more strongly to leader RNA than does phosphorylated N.
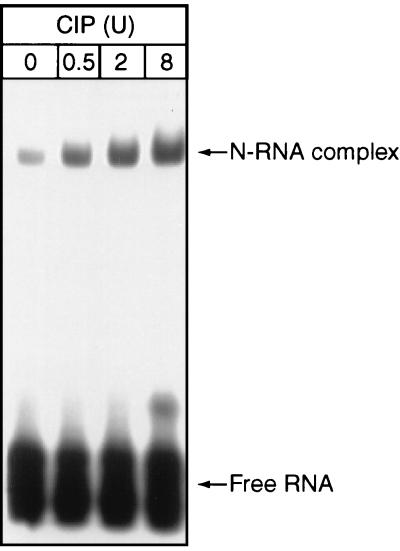
Because rabies virus N plays an important role in the transition from viral RNA transcription to replication by encapsidating leader RNA, we compared the abilities of the phosphorylated and the dephosphorylated N to encapsidate leader RNA. Recombinant rabies virus N was expressed in insect cells and purified by affinity chromatography with anti-N antibodies as described previously (9). It then was subjected to dephosphorylation with calf intestine phosphatase (CIP; Boehringer Mannheim Biochemicals). The dephosphorylated N was used to encapsidate in vitro-transcribed leader RNA (L-70) as described previously (18). RNA transcription and labeling were carried out with a transcription kit (Promega) according to the manufacturer’s specifications. RNA encapsidation was performed as follows. Recombinant N (2 μg) purified from insect cells treated with CIP at 0.5, 2, or 8 U was allowed to react with in vitro-transcribed RNA (106 cpm per reaction) as described previously (18). Purified N without dephosphorylation was included as a control. The RNA-protein mixtures were subjected to digestion with 2 U of micrococcal nuclease and 2 μg of RNase A at 37°C for 30 min. The reaction products were analyzed by electrophoresis on a 4% polyacrylamide gel containing 5% glycerol. As shown in Fig. 1, the dephosphorylated N encapsidated more leader RNA than the phosphorylated N. The intensity of binding of the dephosphorylated N to leader RNA was proportional to the amount of CIP added to the dephosphorylation reaction, showing that RNA encapsidation was regulated by the amount of N phosphorylation.
FIG. 1.
In vitro encapsidation of rabies virus positive-strand leader RNA by dephosphorylated N. Rabies virus positive-strand leader RNA (L-70) transcript was made and labeled with 32P by in vitro transcription and interacted with recombinant rabies virus N protein treated with different concentrations of CIP; untreated N protein was included as a control. The complexes formed were subjected to RNase digestion and then electrophoresis on a 4% polyacrylamide gel. The gel was dried and autoradiographed.
Mutation of phosphoserine 389 of rabies virus N to alanine.
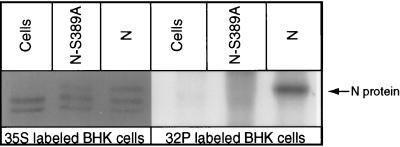
To confirm the observation that phosphorylation of N affects RNA encapsidation, the phosphoserine at position 389 of N (6) was mutated to alanine by site-directed mutagenesis. A mutation primer [5′CCTCGTCGTCAGC(a)GTTGACAGTTCC3′], which changes the serine to alanine, and a selection primer [5′TTTCCCGAAGAAC(t)TCCTCCCAGA3′], which eliminates the EcoRI site but does not change the corresponding amino acid residue, were made, and site-directed mutagenesis was carried out according to the protocol for the Transformer mutagenesis kit (Clontech). The resulting plasmid was sequenced to confirm the mutation, and the plasmid with correct sequence was designated pRN-S389A. Like the wild-type N gene, the gene for the unphosphorylated N in the plasmid is under the control of the T7 promoter. To ascertain that the mutated N is not phosphorylated, plasmid pRN or pRN-S389A was transfected into BHK cells infected with recombinant vaccinia virus vTF7-3 (11) in duplicate sets. One set of the transfected cells was labeled with [35S]methionine (Amersham) and the other was labeled with [32P]phosphoric acid (ICN). Transfected cells were harvested and subjected to immunoprecipitation with anti-N monoclonal antibody (MAb) 377-7 (6). The immunoprecipitate was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis. As shown in Fig. 2, the mutated N, which was immunoprecipitated from the cells labeled with [35S]methionine, had the same mobility as the wild-type N in SDS-PAGE, indicating that the mutated N was synthesized. A phosphorylated N band was observed in SDS-PAGE with cells transfected with the wild-type N gene (pRN) labeled with [32P]phosphoric acid but not from cells transfected with the mutated N gene (pRN-S389A) also labeled with [32P]phosphoric acid, indicating that the wild-type N was phosphorylated in BHK cells but the mutated N was not.
FIG. 2.
Expression of unphosphorylated N. Plasmids expressing wild-type N (pRN) or unphosphorylated N (pRN-S389A) were transfected into BHK cells, which were labeled with [35S]methionine or [32P]phosphoric acid. Cells were lysed and subjected to immunoprecipitation with anti-N MAb 377-7. Lanes: Cells, cells transfected without plasmids; N-S389A, cells transfected with pRN-S389A; N, cells transfected with pRN.
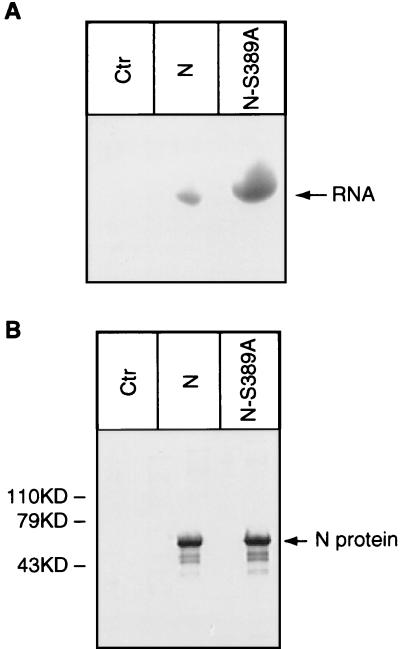
To further study the ability of the unphosphorylated N to encapsidate leader RNA, wild-type N and mutated N were synthesized in the in vitro coupled transcription and translation system (Promega) as described previously (10), and it was found that similar amounts of N and mutant N were synthesized in the system as analyzed by SDS-PAGE (Fig. 3B). For encapsidation of leader RNA, in vitro-labeled leader RNA (L-70) (18) was added to each of the synthesis mixtures. The putative complex was immunoprecipitated with MAb 377-7, and RNA from the immunoprecipitated complex was extracted and analyzed by SDS-PAGE. As shown in Fig. 3A, the mutated (unphosphorylated) N encapsidated more leader RNA than the phosphorylated N, further demonstrating that the unphosphorylated N binds more strongly to leader RNA than the phosphorylated N.
FIG. 3.
Interaction of unphosphorylated N with leader RNA. Plasmids pRN and pRN-S389A were used to synthesize native N and unphosphorylated N, respectively, in the in vitro coupled transcription and translation system. In vitro-transcribed leader RNA (L-70) transcript was added to the reaction mixture. Complexes formed between N and RNA were immunoprecipitated with MAb 377-7. RNA was extracted from the RNA-N complex and analyzed on a 20% polyacrylamide gel containing 6 M urea (A). The amounts of phosphorylated N and unphosphorylated N (N-S389A) synthesized in the in vitro transcription and translation system were compared by SDS-PAGE analysis (B). Ctr, control.
Absence of N phosphorylation results in a decrease of the transcription and replication of a rabies virus minigenome.
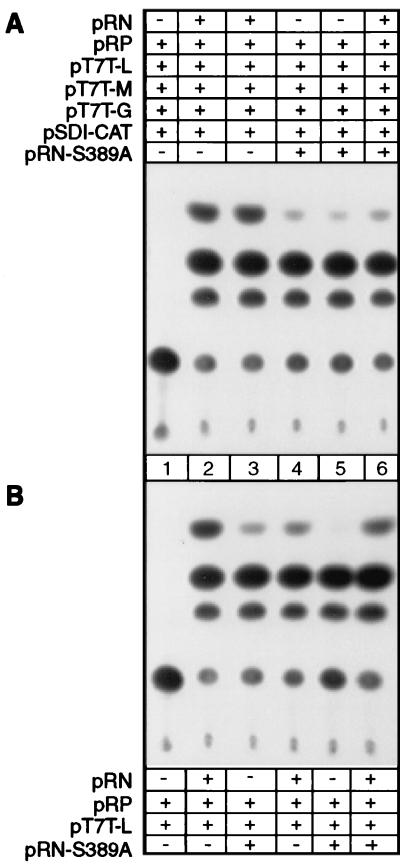
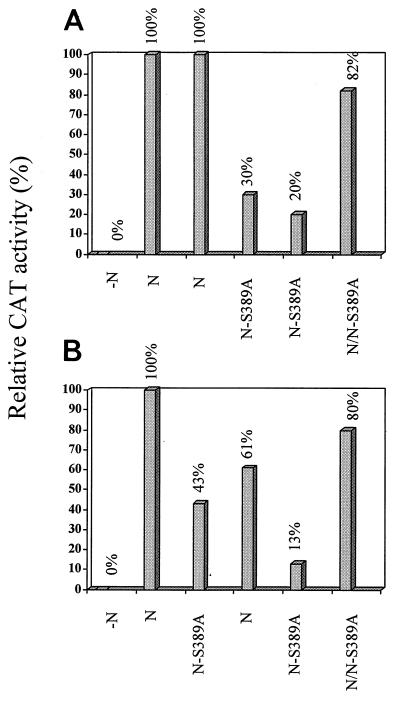
The stronger binding of dephosphorylated N to leader RNA than of phosphorylated N may indicate that the phosphorylation of N affects the encapsidation process and, therefore, may affect viral transcription and replication. To test this hypothesis, we have used the reverse-genetics system described by Conzelmann and Schnell (5) to express a rabies virus minigenome and test directly the effects of unphosphorylated and phosphorylated N on viral RNA transcription and replication. The plasmids for expression of the minigenome (pSDI-CAT), rabies virus L (pT7T-L), glycoprotein (pT7T-G), and matrix protein (pT7T-M) were kindly provided by K. Conzelmann. Plasmids for expression of N (pRN) and P (pRP) were constructed previously in our laboratory (10). These plasmids were transfected into BHK cells infected with recombinant vaccinia virus vTF7-3 (5 to 10 PFU/cell) as described previously (5). Transfected cells were harvested 48 h later and lysed with lysis buffer (0.25 M Tris [pH 7.8]), and chloramphenicol acetyltransferase (CAT) activity was measured in standard CAT assays as described previously (5); the results are presented in Fig. 4A. After autoradiography of the separated acetylated chloramphenicol forms, the spots were quantitated with NIH imaging (National Institutes of Health); the relative CAT activities are presented in Fig. 5A. No CAT activity was detected when N-expressing plasmid was omitted from the transfection (Fig. 4A, lane 1). Strong CAT activity was detected when cells were transfected with all the plasmids, i.e., pT7T-G, pT7T-M, pRN, pRP, pT7T-L, and pSDI-CAT (Fig. 4A, lanes 2 and 3). To study the effects of the unphosphorylated N on viral transcription, plasmid pRN-S389A instead of pRN was transfected along with all the other plasmids into BHK cells. As shown in Fig. 4A (lanes 4 and 5) and Fig. 5A, only 20 to 30% CAT activity was detected in cells transfected with pRN-S389A compared to cells transfected with pRN (Fig. 4A, lanes 2 and 3), indicating that the absence of N phosphorylation results in a decrease of viral transcription. To further study if the unphosphorylated N is a dominant-negative regulator, cells were transfected with both pRN and pRN-S389A. As shown in Fig. 4A (lane 6) and Fig. 5A, 82% CAT activity was detected in the cells transfected with both pRN and pRN-S389A compared to cells transfected with pRN, suggesting that pRN-S389A is not a dominant-negative regulator for viral transcription.
FIG. 4.
Effects of phosphorylation of rabies virus N on viral RNA transcription and replication. Plasmids pRN or pRN-S389A, pRP, pT7T-L, pT7T-G, pT7T-M, and pSDI-CAT were transfected into BHK cells infected with recombinant vaccinia virus vTF7-3 (A). In the passage experiment (B), cells were infected with the supernatant harvested from the transfection (A) and transfected with pRN or pRN-S389A, pRP, and pT7T-L. Cells were harvested for CAT assay.
FIG. 5.
Quantitation of CAT activity. The CAT activities shown in Fig. 4 were quantitated by densitometry with NIH imaging and are expressed as relative CAT activities.
To further examine whether phosphorylation of N also affects viral replication, passage experiments were performed. The supernatants from the initial transfection (containing virus-like particles derived from the rabies virus minigenome) were used to infect fresh BHK cells transfected with pRN or pRN-S389A, pRP, and pT7T-L but not pSDI-CAT. If phosphorylation of N affects only transcription and not replication, viral particles would have been assembled in the initial transfection with pRN-S389A and, therefore, CAT activity would have been detected subsequently in the passage experiment when rescued by the phosphorylated N. The infected cells were harvested for CAT assay after 48 h of incubation, and CAT activity was quantitated. As shown in Fig. 4B and 5B, the highest CAT activity (100%) was detected (Fig. 4B, lane 2), when pRN was used in both the initial transfection and the subsequent passage experiment. When pRN was used in the initial transfection and pRN-S389A was used in the passage experiment, 43% CAT activity was detected (Fig. 4B, lane 3, and Fig. 5B), indicating that phosphorylation of N indeed affects transcription of the minigenome. When pRN-S389A was used in the initial transfection and pRN was used in the passage experiment, 61% CAT activity was detected, indicating that pRN-S389A supported replication of the minigenome in the initial transfection, albeit with somewhat lower efficiency. When pRN-S389A was replaced by pRN in the passage experiment, transcription of the minigenome was partially restored to the level attained by the phosphorylated N. Only 13% CAT activity was detected (Fig. 4B, lane 5, and Fig. 5B) when pRN-S389A was used in both the initial transfection and the passage experiment, suggesting that the absence of N phosphorylation results in decreases of both viral RNA transcription and replication. When both pRN and pRN-S389A were used in the initial transfection and in the passage experiment, 80% CAT activity was detected (Fig. 4B, lane 6), again indicating that unphosphorylated N is not a dominant-negative regulator for viral RNA transcription or replication.
Previously, we studied the mode and specificity of leader RNA encapsidation by N and found that rabies virus N preferentially encapsidates rabies virus leader RNA over other RNA species (18). In the present study, we investigated the role of phosphorylation of rabies virus N in leader RNA encapsidation and in rabies virus RNA transcription and replication.
Serine 389 of rabies virus N was identified as the only phosphorylated residue by Dietzschold et al. (6). Recently, however, Anzai et al. (1) proposed that a minor phosphorylation site may occur at threonine 375. The molar ratio of phosphoserine to phosphothreonine is about 4 to 1. In the present study, no phosphorylation was detected when the serine residue at position 389 was mutated to alanine. This could have been due to only a small portion of the N molecules being phosphorylated at threonine 375 and therefore not being detected by our assay. Alternatively, the phosphorylation at threonine 375 may have been influenced by phosphorylation at serine 389 as suggested by Anzai et al. (1). Once the phosphorylation at the serine residue at 389 is abolished, the threonine at 375 is not phosphorylated.
Rabies virus, like other single-stranded negative-sense RNA viruses, uses its genomic RNA as a template for both transcription and replication. During the transcription process, a positive-strand leader RNA and five monocistronic mRNAs are synthesized, whereas during the RNA replication process, a full-length, positive-strand RNA (replicative intermediate) is synthesized. The replicative intermediate RNA serves as the template for the synthesis of progeny negative-strand genomic RNA (17). This differential regulation of viral RNA transcription and replication is thought to be due to the ability of N to encapsidate the nascent RNA produced, as proposed for VSV and rabies virus (2, 16–18). Therefore, the ability of N to encapsidate leader RNA is a prerequisite for viral RNA replication. In the present study, we found that dephosphorylated or unphosphorylated N encapsidated more RNA. Furthermore, the absence of N phosphorylation resulted in a dramatic decrease of RNA transcription as measured by CAT assay in both the initial transfection and subsequent passage experiments. If unphosphorylated N binds more RNA, it would be reasonable to assume that unphosphorylated N would favor replication. Surprisingly, the absence of N phosphorylation also resulted in reduction of replication, although this was not as dramatic as the reduction of transcription. This may be due to the fact that the unphosphorylated N may affect the interaction between polymerase and the RNP template and consequently result in reduction of replication. In the present study, however, only the passage experiment was carried out to assess the rate of replication. The unphosphorylated N may also influence N-N or N-matrix protein interaction and therefore affect the efficiency of virus assembly. More sensitive assays, such as the RNase protection assay, to directly distinguish viral transcripts from replication products are being developed.
The stronger RNA binding activity of unphosphorylated or dephosphorylated N may be due to the fact that the unphosphorylated N is too basic in charge and, therefore, binds too tightly to the acidic genomic RNA. Mutation of the phosphoserine to aspartic acid or glutamic acid is under way in the laboratory to confirm this hypothesis. Alternatively, the unphosphorylated N may have a configuration different from that of the phosphorylated N, which binds more RNA. The tight binding of RNA by the unphosphorylated N makes it more difficult for other viral proteins to unwind the encapsidated RNA for transcription and replication by viral RNA polymerase L. In summary, we conclude that phosphorylation of rabies virus N supports viral RNA transcription and replication, possibly by modulating the encapsidation of genomic RNA.
Acknowledgments
This work was supported by Public Health Service grants AI-33029 (Z.F.F.) and AI-09706 (H.K.) from the National Institute of Allergy and Infectious Diseases.
We thank K. Conzelmann for supplying plasmids for the reverse genetics, B. Moss for the recombinant vaccinia virus vTF7-3, and W. H. Wunner for critically reading the manuscript.
REFERENCES
- 1.Anzai J, Takamatsu F, Takeuchi K, Kohno T, Morimoto K, Goto H, Minamoto N, Kawai A. Identification of a phosphatase-sensitive epitope of rabies virus nucleoprotein which is recognized by a monoclonal antibody 5-2-26. Microbiol Immunol. 1997;41:229–240. doi: 10.1111/j.1348-0421.1997.tb01195.x. [DOI] [PubMed] [Google Scholar]
- 2.Banerjee A K, Barik S. Gene expression of vesicular stomatitis virus genome RNA. Virology. 1992;188:417–428. doi: 10.1016/0042-6822(92)90495-b. [DOI] [PubMed] [Google Scholar]
- 3.Blumberg B M, Giorgi C, Kolakofsky D. N protein of vesicular stomatitis virus selectively encapsidates leader RNA in vitro. Cell. 1983;32:559–567. doi: 10.1016/0092-8674(83)90475-0. [DOI] [PubMed] [Google Scholar]
- 4.Clark H F. Systems for assay and growth of rhabdoviruses. In: Bishop D H L, editor. Rhabdoviruses. Vol. 1. Boca Raton, Fla: CRC Press, Inc.; 1980. pp. 23–41. [Google Scholar]
- 5.Conzelmann K-L, Schnell M. Rescue of synthetic genomic RNA analogs of rabies virus by plasmid-encoded proteins. J Virol. 1994;68:713–719. doi: 10.1128/jvi.68.2.713-719.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dietzschold B, Lafon M, Wang H, Otvos L, Celis E, Wunner W H, Koprowski H. Localization and immunological characterization of antigenic domains of rabies virus internal N and NS proteins. Virus Res. 1987;8:103–125. doi: 10.1016/0168-1702(87)90023-2. [DOI] [PubMed] [Google Scholar]
- 7.Dietzschold B, Rupprecht C E, Fu Z F, Koprowski H. Rhabdoviruses. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1137–1159. [Google Scholar]
- 8.Flamand A, Delagneau J F, Bussereau F. An RNA polymerase activity in purified rabies virions. J Gen Virol. 1978;40:233–238. doi: 10.1099/0022-1317-40-1-233. [DOI] [PubMed] [Google Scholar]
- 9.Fu Z F, Dietzschold B, Schumacher C L, Wunner W H, Ertl H C J, Koprowski H. Rabies virus nucleoprotein expressed in and purified from insect cells is efficacious as a vaccine. Proc Natl Acad Sci USA. 1991;88:2001–2005. doi: 10.1073/pnas.88.5.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu Z F, Zheng Y, Wunner W H, Koprowski H, Dietzschold B. Both the N- and the C-terminal domains of the nominal phosphoprotein of rabies virus are involved in binding to the nucleoprotein. Virology. 1994;200:590–597. doi: 10.1006/viro.1994.1222. [DOI] [PubMed] [Google Scholar]
- 11.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawai A. Transcriptase activity associated with rabies virion. J Virol. 1977;24:826–835. doi: 10.1128/jvi.24.3.826-835.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patton J, Davies N L, Wertz G W. N protein alone satisfies the requirement for protein synthesis during RNA replication of vesicular stomatitis virus. J Virol. 1984;49:303–309. doi: 10.1128/jvi.49.2.303-309.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sokol F, Clark H F. Phosphoproteins, structural components of rhabdoviruses. Virology. 1973;52:246–263. doi: 10.1016/0042-6822(73)90413-3. [DOI] [PubMed] [Google Scholar]
- 15.Wagner R R, Rose J K. Rhabdoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1121–1136. [Google Scholar]
- 16.Wertz G W, Davies N L, Patton J. The role of proteins in vesicular stomatitis virus RNA replication. In: Wagner R R, editor. The rhabdoviruses. New York, N.Y: Plenum Press; 1987. pp. 271–296. [Google Scholar]
- 17.Wunner W H. The chemical composition and molecular structure of rabies viruses. In: Baer G M, editor. Natural history of rabies. 2nd ed. Boca Raton, Fla: CRC Press, Inc.; 1991. pp. 31–67. [Google Scholar]
- 18.Yang J, Hooper D C, Wunner W H, Koprowski H, Dietzschold B, Fu Z F. The specificity of rabies virus RNA encapsidation by nucleoprotein. Virology. 1998;242:107–117. doi: 10.1006/viro.1997.9022. [DOI] [PubMed] [Google Scholar]