Abstract
Common genetic mutations are absent in neuroblastoma, one of the most common childhood tumours. As a demethylase of 5‐methylcytosine (m5C) modification, TET1 plays an important role in tumourigenesis and differentiation. However, the association between TET1 gene polymorphisms and susceptibility to neuroblastoma has not been reported. Three TET1 gene polymorphisms (rs16925541 A > G, rs3998860 G > A and rs12781492 A > C) in 402 Chinese patients with neuroblastoma and 473 cancer‐free controls were assessed using TaqMan. Multivariate logistic regression analysis was used to evaluate the association between TET1 gene polymorphisms and susceptibility to neuroblastoma. The GTEx database was used to analyse the impact of these polymorphisms on peripheral gene expression. The relationship between gene expression and prognosis was analysed using Kaplan–Meier analysis with the R2 platform. We found that both rs3998860 G > A and rs12781492 A > C were significantly associated with increased neuroblastoma risk. Stratified analysis further showed that rs3998860 G > A and rs12781492 A > C significantly increased neuroblastoma risk in certain subgroups. In the combined risk genotype model, 1–3 risk genotypes significantly increased risk of neuroblastoma compared with the 0 risk genotype. rs3998860 G > A and rs12781492 A > C were significantly associated with increased STOX1 mRNA expression in adrenal and whole blood, and high expression of STOX1 mRNA in adrenal and whole blood was significantly associated with worse prognosis. In summary, TET1 gene polymorphisms are significantly associated with increased neuroblastoma risk; further research is required for the potential mechanism and therapeutic prospects in neuroblastoma.
Keywords: m5C modification, neuroblastoma, polymorphism, susceptibility, TET1
1. INTRODUCTION
Neuroblastoma is an embryonic tumour of the sympathetic nervous system, but its origin is unclear. 1 Currently, it is generally believed that it originates from incomplete precursors of neural crest tissue. 2 , 3 , 4 Neuroblastoma is the most common solid tumour and the second most common extracranial tumour in children. 5 , 6 In developed countries, the incidence of neuroblastoma in children aged 0–14 years is approximately 10.1–15.0 per 1 million. 7 In China, the incidence of neuroblastoma is 7.7 cases per 1 million children. 8 Neuroblastoma exhibits a high degree of heterogeneity, from biological characteristics to clinical processes. 1 Some neuroblastomas can spontaneously subside. 1 Even after a series of intensive treatments, such as surgery, immunotherapy and radiation therapy, high‐risk neuroblastomas still have a relatively poor prognosis and high risk of recurrence. 9 , 10 , 11 Patients with high‐risk neuroblastoma will also face serious psychological problems such as anxiety. 12 Identifying biological characteristics associated with high‐risk neuroblastoma is a research focus. In addition to MYCN amplification, some important other genetic changes have been found in neuroblastomas, such as ATRX, 13 TERT, 14 ALK 15 and RAS 16 mutations. However, data thus far are insufficient to explain the genetic variations associated with neuroblastoma risk.
As the most common genetic variation in DNA, single‐nucleotide polymorphisms (SNPs) are closely related to tumour susceptibility, prognosis and immunity. 17 , 18 , 19 Different tumour types have different associated SNPs. 20 Genome‐wide association studies (GWASs) have enabled large‐scale exploration of SNPs truly related to tumours in the entire genome. With the application of GWAS to neuroblastoma, many SNPs have been proven to be related to its susceptibility. 21 , 22 , 23 , 24 John et al. 24 have shown that rs2168101 G > T located in the LMO1 gene super‐enhancer is closely related to genetic susceptibility to neuroblastoma; the rs2168101 T allele is associated with reduced LMO1 expression and tumour suppression in primary neuroblastoma tumours. However, the genetic variations associated with neuroblastoma susceptibility in different populations are yet to be elucidated. Our previous studies have shown that the BER pathway and ERCC1, XPF, NRAS and ALKBH5 gene polymorphisms affect neuroblastoma risk in Chinese children. 16 , 25 , 26 , 27 However, the genetic polymorphisms associated with the risk of neuroblastoma in Chinese children are not fully understood.
Epigenetic modifications commonly found in genomes and transcriptomes have been shown to play important biological roles in growth and development, aging and various diseases. 28 , 29 , 30 , 31 In recent years, RNA modifications have been shown to play an important role in cancer. 28 , 32 Common RNA modifications include N6‐methyladenosine (m6A), N1‐methyladenosine (m1A), 5‐methylcytosine (m5C), 5‐hydroxymethylcytosine (hm5C), pseudouridine (Ψ) and N7‐methylguanosine (m7G). As the most common DNA modification and important RNA modification, the role of m5C modification in cancer cannot be ignored. 29 , 33 , 34 , 35 As m5C demethylases, the ten‐eleven translocation (TET) family of enzymes oxidize m5C in RNA to form hm5C. 36 TET further oxidizes 5hmC to 5fC (5‐formylcytosine) and 5caC (5‐carboxycytidine) in DNA 37 ; thymidine DNA glycosylase recognizes 5fC and 5caC and converts them to unmethylated cytosine. 38 , 39 , 40 A member of the TET (ten‐eleven translocation) family, TET1 (tet methylcytosine dioxygenase 1) has been shown to play an important biological role in cancer, including neuroblastoma, in recent years. 41 , 42 , 43 , 44 Fragile X mental retardation protein (FMRP) promotes demethylation of m5C by interacting with TET1 and induces transcription‐coupled homologous recombination, which is an important mechanism for mRNA repair and cell survival in cancer. 41 The oncoprotein YAP induces expression of TET1, which promotes transcriptional activation and induces tumourigenesis through epigenetics, such as DNA demethylation caused by its interaction with TEAD. 42 Gao et al. 44 found that TET1 correlated negatively with neuronal differentiation in neuroblastoma cells. TET1 mediates negative regulation of neuronal differentiation by srGAP3 through non‐catalytic action. 44 The TET1 gene is repeatedly mutated in lung cancer, gastrointestinal cancer, skin cancer and urinary system cancer. 45 In cancer patients treated with immune checkpoint inhibitors (ICIs), the TET1 mutation is associated with better therapeutic efficacy. 45 This suggests a potential therapeutic target for the TET1 gene. However, the important role of TET1 gene mutations in neuroblastoma needs to be further explored. Currently, there is no research indicating that TET1 gene polymorphisms are associated with susceptibility to neuroblastoma.
Based on the importance of the TET1 gene in neuroblastoma, we hypothesize that TET1 gene polymorphisms have an impact on risk of neuroblastoma. To test our hypothesis, we conducted a case–control study in Jiangsu, China, to explore whether TET1 gene polymorphisms are associated with susceptibility to neuroblastoma.
2. MATERIALS AND METHODS
2.1. Study subjects
This case–control study included 402 children with neuroblastoma and 473 noncancer children (Table S1) recruited from the Children's Hospital of Nanjing Medical University in Jiangsu Province. 46 Epidemiological data were collected using structured questionnaires. The inclusion criterion for patients was neuroblastoma confirmed by biopsy or histology. The control group subjects were matched based on the expected population characteristic (sex, age and race) distribution of the case group and recruited simultaneously with the case group. For our study, all subjects signed an informed consent form. This study was approved by the Institutional Review Committee of Children's Hospital of Nanjing Medical University (Approval No.: 202112141–1).
2.2. Polymorphisms selection and genotyping
We first selected potential functional SNPs located in the 5′ flanking region, 5′ untranslated region (UTR), exon, intron and 3′ UTR regions of the TET1 gene from the dbSNP database and SNPinfo website. The secondary allele frequency of all SNPs in the Han Chinese population reported in 1000 Genomes is >5%. Linkage disequilibrium (LD) between the selected SNPs is less than 0.8. The final SNPs selected were rs16925541, rs3998860 and rs12781492. Both rs16925541 and rs3998860 are missense variants located in the coding region of the TET1 gene. The variant rs12781492 is located in the 3′ UTR of the TET1 gene and is predicted to bind to miRNA. We used TIANamp Blood DNA Kit (TianGen Biotech Co. Ltd.) to extract genomic DNA from tissue or blood samples from all subjects. The concentration and purity of the extracted DNA were measured using a UV spectrophotometer (Nano Drop Technologies, Inc.). The DNA sample was diluted and transferred to a 96‐well plate. For all samples, TaqMan® SNP Genotyping Assays (Applied Biosystems) were used for genotyping. We randomly selected 10% of the samples from the case and the control groups for repeated genotyping, and the results were 100% consistent.
2.3. Statistical analysis
Differences in genotype frequency distribution and demographic characteristics between the case group and the control group were analysed using a bilateral chi‐square test. The goodness of fit χ2 test was used for Hardy–Weinberg equilibrium (HWE). To explore the association between TET1 gene polymorphisms and susceptibility to neuroblastoma, we used multiple logistic regression analysis to analyse the odds ratio (OR) and 95% confidence interval (CI) of TET1 gene polymorphisms in the case group and the control group. The OR calculates the crude OR and adjusted OR before and after adjustment for age and sex, respectively. Subsequently, further stratification analysis was conducted based on age, sex, tumour location and clinical stage for significant SNPs. The above statistical analyses were performed using SAS V9.4 (SAS Institute). Gene expression quantitative trait loci (eQTLs) were analysed through the Genotype‐Tissue Expression (GTEx) official website (https://www.gtexportal.org/home/). Analysis of the relationship between gene expression and prognosis of neuroblastoma cases derived from the GSE62564 dataset using Kaplan–Meier analysis through R2: Genomics Analysis and Visualization Platform (http://r2.amc.nl). All statistics were conducted using a two‐sided test with a significance level of 0.05.
3. RESULTS
3.1. Associations of TET1 gene polymorphisms with neuroblastoma susceptibility
We successfully obtained the genotypes of the three SNPs in 400 cases and 473 controls. As shown in Table 1, all SNPs in the control group conformed to HWE (p > 0.05). After adjusting for age and sex, we found that subjects with the rs3998860 AA genotype had a 1.5‐fold increased neuroblastoma risk compared with those with the GG genotype (AA vs. GG: adjusted OR = 2.51, 95% CI = 1.27–4.95, p = 0.008). We also found that the recessive model of the SNP rs3998860 was significantly associated with increased neuroblastoma risk (AA vs. GG/GA: adjusted OR = 2.69, 95% CI = 1.37–5.27, p = 0.004). Compared with the rs12781492 AA genotype, the rs12781492 CC genotype was significantly associated with increased neuroblastoma risk (CC vs. AA: adjusted OR = 2.04, 95% CI = 1.01–4.10, p = 0.046). Compared to the rs12781492 AA/AC genotypes, the CC genotype also significantly increased neuroblastoma risk (CC vs. AA/AC: adjusted OR = 2.17, 95% CI = 1.08–4.34, p = 0.029). Based on the above statistical analysis results, we defined rs16925541 GG, rs3998860 AA and rs12781492 CC as risk genotypes to further analyse the impact of combined genotypes on susceptibility to neuroblastoma. The results showed that the combination of 1–3 risk genotypes significantly increased neuroblastoma susceptibility compared to the 0 risk genotype (1–3 vs. 0: adjusted OR = 3.01, 95% CI = 1.67–5.40, p = 0.0002).
TABLE 1.
Association of TET1 gene polymorphisms with neuroblastoma risk in children from Jiangsu province.
| Genotype | Cases (n = 400) | Controls (n = 473) | p a | Crude OR (95% CI) | p | Adjusted OR (95% CI) b | p b |
|---|---|---|---|---|---|---|---|
| rs16925541 A > G (HWE = 0.142) | |||||||
| AA | 308 (77.00) | 365 (77.17) | 1.00 | 1.00 | |||
| AG | 77 (19.25) | 97 (20.51) | 0.94 (0.67–1.32) | 0.722 | 0.94 (0.67–1.32) | 0.722 | |
| GG | 15 (3.75) | 11 (2.33) | 1.62 (0.73–3.57) | 0.235 | 1.63 (0.73–3.60) | 0.231 | |
| Additive | 0.641 | 1.07 (0.82–1.39) | 0.640 | 1.07 (0.82–1.39) | 0.637 | ||
| Dominant | 92 (23.00) | 108 (22.83) | 0.953 | 1.01 (0.74–1.39) | 0.953 | 1.01 (0.74–1.39) | 0.952 |
| AA/AG | 385 (96.25) | 462 (97.67) | 1.00 | 1.00 | |||
| GG | 15 (3.75) | 11 (2.33) | 0.217 | 1.64 (0.74–3.61) | 0.221 | 1.65 (0.75–3.64) | 0.217 |
| rs3998860 G > A (HWE = 0.582) | |||||||
| GG | 276 (69.00) | 319 (67.44) | 1.00 | 1.00 | |||
| GA | 96 (24.00) | 141 (29.81) | 0.79 (0.58–1.07) | 0.124 | 0.79 (0.58–1.07) | 0.128 | |
| AA | 28 (7.00) | 13 (2.75) | 2.49 (1.26–4.90) | 0.008 | 2.51 (1.27–4.95) | 0.008 | |
| Additive | 0.488 | 1.09 (0.86–1.37) | 0.487 | 1.09 (0.86–1.38) | 0.478 | ||
| Dominant | 124 (31.00) | 154 (32.56) | 0.623 | 0.93 (0.70–1.24) | 0.623 | 0.93 (0.70–1.24) | 0.628 |
| GG/GA | 372 (93.00) | 460 (97.25) | 1.00 | 1.00 | |||
| AA | 28 (7.00) | 13 (2.75) | 0.003 | 2.66 (1.36–5.21) | 0.004 | 2.69 (1.37–5.27) | 0.004 |
| rs12781492 A > C (HWE = 0.279) | |||||||
| AA | 303 (75.75) | 348 (73.57) | 1.00 | 1.00 | |||
| AC | 74 (18.50) | 112 (23.68) | 0.76 (0.55–1.06) | 0.103 | 0.76 (0.54–1.06) | 0.103 | |
| CC | 23 (5.75) | 13 (2.75) | 2.03 (1.01–4.08) | 0.046 | 2.04 (1.01–4.10) | 0.046 | |
| Additive | 0.822 | 1.03 (0.80–1.32) | 0.822 | 1.03 (0.80–1.32) | 0.819 | ||
| Dominant | 97 (24.25) | 125 (26.43) | 0.462 | 0.89 (0.66–1.21) | 0.462 | 0.89 (0.66–1.21) | 0.462 |
| AA/AC | 377 (94.25) | 460 (97.25) | 1.00 | 1.00 | |||
| CC | 23 (5.75) | 13 (2.75) | 0.026 | 2.16 (1.08–4.32) | 0.030 | 2.17 (1.08–4.34) | 0.029 |
| Combine risk genotypes c | |||||||
| 0 | 360 (90.00) | 456 (96.41) | 1.00 | 1.00 | |||
| 1–3 | 40 (10.00) | 17 (3.59) | 0.0001 | 2.98 (1.66–5.34) | 0.0002 | 3.01 (1.67–5.40) | 0.0002 |
Note: Values are in bold if the p values are less than 0.05 or the 95% CIs excluded 1.
Abbreviations: CI, confidence interval; HWE, Hardy–Weinberg equilibrium; OR, odds ratio.
χ2 test for genotype distributions between neuroblastoma patients and cancer‐free controls.
Adjusted for age and sex.
Risk genotypes were carriers with rs16925541 GG, rs3998860 AA and rs12781492 CC genotypes.
3.2. Stratification analysis of significant SNPs
We further analysed the impact of TET1 gene polymorphisms on susceptibility to neuroblastoma by stratified analysis of age, sex, site of origin and clinical stage (Table 2). Our results suggested that the rs3998860 AA genotype significantly increases risk of neuroblastoma in children compared to the GG/GA genotype in the >18 month subgroup (AA vs. GG/GA: adjusted OR = 3.07, 95% CI = 1.16–8.12, p = 0.024). Similarly, the rs3998860 AA genotype significantly increased risk of neuroblastoma in male children in sex stratification (AA vs. GG/GA: adjusted OR = 3.57, 95% CI = 1.26–10.14, p = 0.017), the retroperitoneal subgroup of sites in origin stratification (AA vs. GG/GA: adjusted OR = 4.03, 95% CI = 1.91–8.51, p = 0.0003), and I + II + 4 s (AA vs. GG/GA: adjusted OR = 3.00, 95% CI = 1.35–6.64, p = 0.007) and III + IV (AA vs. GG/GA: adjusted OR = 2.69, 95% CI = 1.17–6.17, p = 0.019) subgroups in clinical stage stratification. The recessive model of rs12781492 was significantly associated with increased neuroblastoma risk in the retroperitoneal subgroup (CC vs. AA/AC: adjusted OR = 2.75, 95% CI = 1.23–6.15, p = 0.014) and the III + IV subgroup (CC vs. AA/AC: adjusted OR = 2.64, 95% CI = 1.15–6.04, p = 0.022). In all subgroups stratified by age (≤18 months: adjusted OR = 3.22, 95% CI = 1.32–7.90, p = 0.011; >18 months: adjusted OR = 2.67, 95% CI = 1.23–5.80, p = 0.013), sex (females: adjusted OR = 2.97, 95% CI = 1.33–6.67, p = 0.008; males: adjusted OR = 3.05, 95% CI = 1.30–7.14, p = 0.010) and clinical stage (I + II + 4 s: adjusted OR = 2.98, 95% CI = 1.48–6.01, p = 0.002; III + IV: adjusted OR = 3.03, 95% CI = 1.49–6.19, p = 0.002), a combination of 1–3 risk genotypes was significantly associated with increased neuroblastoma risk compared to the 0 risk genotype. Combination of 1–3 risk genotypes significantly increased neuroblastoma risk in the retroperitoneal subgroup compared to the 0 risk genotype (1–3 vs. 0: adjusted OR = 4.54, 95% CI = 2.37–8.70, p < 0.0001).
TABLE 2.
Stratification analysis for the association between TET1 risk genotypes with neuroblastoma susceptibility in Jiangsu children.
| Variables | rs3998860 (cases/controls) | Adjusted OR a | p a | rs12781492 (cases/controls) | Adjusted OR a | p a | Risk genotypes (cases/controls) | Adjusted OR a | p a | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GG/GA | AA | (95% CI) | AA/AC | CC | (95% CI) | 0 | 1–3 | (95% CI) | ||||
| Age, month | ||||||||||||
| ≤18 | 124/132 | 14/7 | 2.15 (0.84–5.49) | 0.111 | 127/134 | 10/5 | 2.11 (0.70–6.34) | 0.184 | 117/132 | 20/7 | 3.22 (1.32–7.90) | 0.011 |
| >18 | 249/328 | 14/6 | 3.07 (1.16–8.12) | 0.024 | 250/326 | 13/8 | 2.12 (0.86–5.19) | 0.101 | 243/324 | 20/10 | 2.67 (1.23–5.80) | 0.013 |
| Sex | ||||||||||||
| Females | 177/217 | 14/8 | 2.15 (0.88–5.24) | 0.093 | 178/219 | 13/6 | 2.67 (0.99–7.16) | 0.052 | 170/216 | 21/9 | 2.97 (1.33–6.67) | 0.008 |
| Males | 195/243 | 14/5 | 3.57 (1.26–10.14) | 0.017 | 199/241 | 10/7 | 1.75 (0.65–4.69) | 0.267 | 190/240 | 19/8 | 3.05 (1.30–7.14) | 0.010 |
| Sites of origin | ||||||||||||
| Adrenal gland | 89/460 | 4/13 | 1.66 (0.53–5.23) | 0.389 | 90/460 | 3/13 | 1.19 (0.33–4.27) | 0.789 | 88/456 | 5/17 | 1.55 (0.56–4.33) | 0.401 |
| Retroperitoneal | 149/460 | 17/13 | 4.03 (1.91–8.51) | 0.0003 | 154/460 | 12/13 | 2.75 (1.23–6.15) | 0.014 | 142/456 | 24/17 | 4.54 (2.37–8.70) | <0.0001 |
| Mediastinum | 113/460 | 6/13 | 1.91 (0.71–5.17) | 0.202 | 112/460 | 7/13 | 2.23 (0.87–5.72) | 0.096 | 110/456 | 9/17 | 2.21 (0.96–5.11) | 0.063 |
| Others | 18/460 | 0/13 | / | / | 17/460 | 1/13 | 2.05 (0.25–16.63) | 0.502 | 17/456 | 1/17 | 1.55 (0.19–12.36) | 0.680 |
| Clinical stages | ||||||||||||
| I + II + 4 s | 160/460 | 13/13 | 3.00 (1.35–6.64) | 0.007 | 163/460 | 10/13 | 2.20 (0.94–5.14) | 0.068 | 156/456 | 17/17 | 2.98 (1.48–6.01) | 0.002 |
| III + IV | 152/460 | 11/13 | 2.69 (1.17–6.17) | 0.019 | 152/460 | 11/13 | 2.64 (1.15–6.04) | 0.022 | 147/456 | 16/17 | 3.03 (1.49–6.19) | 0.002 |
Abbreviations: CI, confidence interval; OR, odds ratio.
Note: Values are in bold if the p values are less than 0.05 or the 95% CIs excluded 1.
Adjusted for age and sex, omitting the correspondence factor.
3.3. Functional effect of rs3998860 G > A and rs12781492 A > C on surrounding genes
Based on the impact of TET1 rs3998860 G > A and rs12781492 A > C on susceptibility to neuroblastoma, we further explored the impact of these two loci on expression of nearby genes. We conducted cis‐eQTL analysis on rs3998860 (Figure 1) and rs12781492 (Figure 2) using the GTEx database. The results showed that the rs3998860 A allele was significantly associated with increased STOX1 mRNA expression in the adrenal gland (Figure 1A) and whole blood (Figure 1B) compared to the G allele. In cultured fibroblasts (Figure 1C), the A allele was significantly associated with increased KIF1BP mRNA expression compared to the rs3998860 G allele. In the tibial artery, the rs3998860 A allele was significantly associated with increased mRNA expression of SLC25A16 (Figure 1D) and RUFY2 (Figure 1E). In skin not exposed to sun, the rs3998860 A allele was significantly associated with decreased CCAR1 mRNA expression (Figure 1F). In the adrenal gland (Figure 2A) and whole blood (Figure 2B), the rs12781492 C allele was significantly associated with increased STOX1 mRNA expression compared to the rs12781492 A allele. The rs12781492 C allele significantly enhanced mRNA expression of CCAR1 in the oesophageal mucosa (Figure 2C) and skin not exposed to sun (Figure 2D) compared to the A allele.
FIGURE 1.
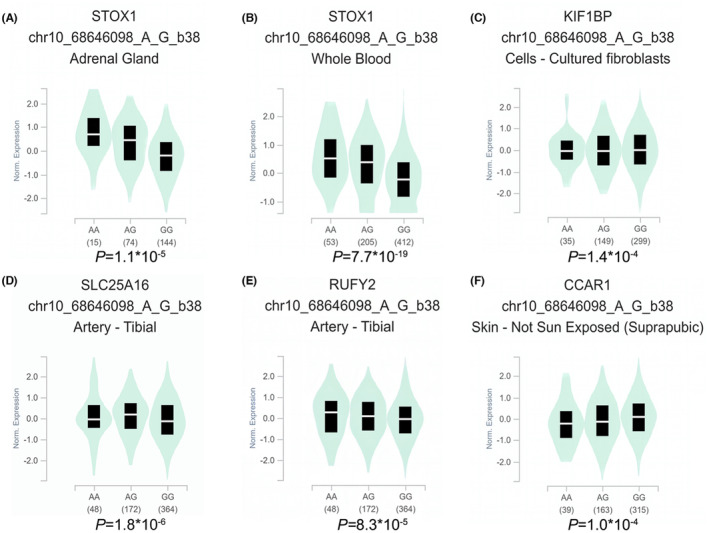
eQTL analysis shows the effect of TET1 rs3998860 G > A on surrounding gene expression. (A, B) In the adrenal gland (p = 1.1 × 10−5) and whole blood (p = 7.7 × 10−19), rs3998860 G was significantly associated with decreased STOX1 mRNA expression. (C) In cultured fibroblasts, rs3998860 G was significantly associated with an increase in KIF1BP mRNA expression (p = 1.4 × 10−4). (D) In the tibial artery, rs3998860 A was significantly associated with increased mRNA expression of SLC25A16 (p = 1.8 × 10−6). (E) In the tibial artery, rs3998860 A was significantly associated with an increase in RUFY2 mRNA expression (p = 8.3 × 10−5). (F) In skin, rs3998860 G was significantly associated with increased mRNA expression of CCAR1 (p = 1.0 × 10−4). Violin diagrams were obtained through the GTEx official website (https://www.gtexportal.org/home/).
FIGURE 2.
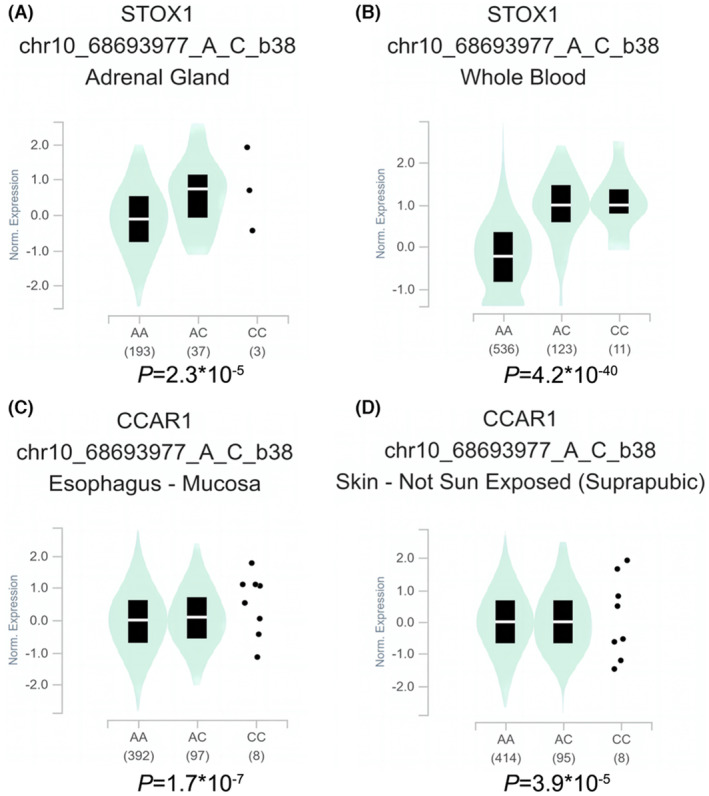
eQTL analysis shows the effect of TET1 rs12781492 A > C on surrounding gene expression. (A, B) In the adrenal gland (p = 2.3 × 10−5) and whole blood (p = 4.2 × 10−40), rs12781492 C was significantly associated with increased STOX1 mRNA expression. (C, D) In the oesophageal mucosa (p = 1.7 × 10−7) and skin (p = 3.9 × 10−5), rs12781492 C was significantly associated with an increase in CCAR1 mRNA expression. Violin diagrams were obtained through the GTEx official website (https://www.gtexportal.org/home/).
3.4. Relationship between STOX1 mRNA expression and prognosis of neuroblastoma
Based on the significant impact of rs3998860 and rs12781492 on STOX1 mRNA expression in the adrenal gland and whole blood, we further analysed the relationship between mRNA expression of STOX1 and prognosis of neuroblastoma through the R2: Genomics Analysis and Visualization Platform (Figure 3). We conducted Kaplan–Meier survival analysis of overall survival (OS) and event‐free survival (EFS) prognostic indicators for neuroblastoma patients in the GSE62564 dataset. Neuroblastoma patients with high STOX1 (n = 249) mRNA expression had significantly (p = 0.039) lower OS than neuroblastoma patients with low STOX1 mRNA expression (n = 249; Figure 3A). Consistent with this, high mRNA expression of STOX1 was significantly (p = 0.016) associated with low EFS (Figure 3B).
FIGURE 3.
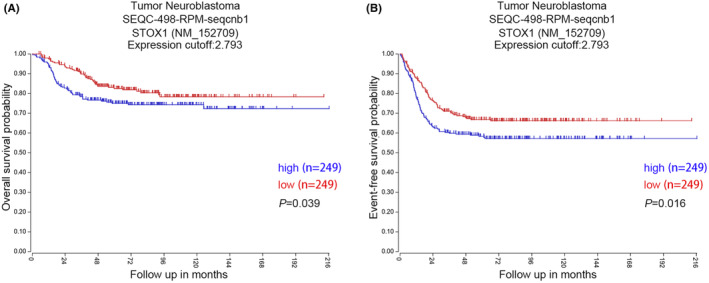
Kaplan–Meier analysis shows the relationship between STOX1 mRNA expression and prognosis. (A) In neuroblastomas, high mRNA expression of STOX1 was significantly associated with low OS (p = 0.039). (B) High mRNA expression of STOX1 was significantly associated with low EFS in neuroblastomas (p = 0.016).
4. DISCUSSION
Neuroblastoma is the most common tumour in children, and its high‐risk type has a high risk of recurrence even after multiple treatments. 3 , 9 , 10 , 11 Fundamentally understanding the genetic variations associated with neuroblastoma is key to early diagnosis and targeted treatment. Based on the important role of m5C modification in cancer, we focus on polymorphisms of m5C modification core genes. Our previous study showed that rs13181449 C > T in the m5C methyltransferase gene NSUN2 confers reduced risk of neuroblastoma. 46 In recent years, the m5C demethylase gene TET1 has been proven to mediate the occurrence and development of some cancers. 41 , 42 , 43 , 44 However, the important role of TET1 gene polymorphisms in cancer, including neuroblastoma, has not been revealed. Therefore, we explored the impact of TET1 gene polymorphisms on risk of neuroblastoma through a case–control study. Our results suggest that TET1 gene polymorphisms (rs3998860 G > A and rs12781492 A > C) are associated with increased neuroblastoma risk.
We used a case–control study to explore the association between risk of neuroblastoma and TET1 gene polymorphisms in children in Jiangsu, China. Each SNP in our control group was found to conform to HWE, indicating that our data source is reliable. We conducted logistic regression analysis of the results of TET1 gene polymorphism genotyping. The results showed that only the GG genotype of TET1 rs3998860 significantly increased neuroblastoma risk compared to the AA genotype. Further model analysis showed significantly increased neuroblastoma risk only for the recessive model, consistent with the above results. In line with our expectations, the OR value in the recessive model also increased (OR: 2.69 > 2.51). Therefore, based on these results, the genetic model of rs3998860 in neuroblastoma is a recessive model, as is the genetic model of rs12781492 A > C in neuroblastoma. Combination of 1–3 risk genotypes significantly increased neuroblastoma risk when compared to the 0 risk genotype, with an OR value reaching the highest of all models. This indicates that risk SNPs can accumulate in neuroblastoma. Considering the effect of confounding factors, we further explored the association between TET1 rs3998860 and susceptibility to neuroblastoma through stratification analysis. The results for the recessive model (AA vs. GG/GA) of TET1 rs3998860 showed significantly increased neuroblastoma risk in subgroups >18 months of age and males. In site of origin stratification, recessive models of rs3998860 and rs12781492 indicated significantly increased risk of neuroblastoma only in the retroperitoneal subgroup, and the OR values were much higher in the other subgroups. This suggests that the variable site of origin might confound the results. rs3998860 G > A and rs12781492 A > C were only significantly associated with increased risk to retroperitoneal neuroblastoma, and the specific mechanisms need to be further studied. The recessive model of TET1 rs3998860 showed significantly increased risk of neuroblastoma in all clinical staging subgroups. The recessive model (CC vs. AA/AC) of rs12781492 was significantly associated with neuroblastoma risk only in the retroperitoneal subgroup and the III + IV subgroup. In all subgroups of age, sex and clinical stage, as well as in the retroperitoneal subgroup, combination of 1–3 risk genotypes was significantly associated with increased neuroblastoma risk compared to the 0 risk genotype. Importantly, all models in the retroperitoneal subgroup were significant, indicating a significant association between TET1 gene polymorphisms and increased risk to retroperitoneal neuroblastoma. These results indicate that there are indeed confounding factors interfering with our research conclusions, and our conclusions should thus be explained by a stratified subgroup.
To further explore the functions of rs3998860 and rs12781492, we used the GTEx database to analyse their impact on nearby gene expression. Importantly, we found that both the rs3998860 A allele and the rs12781492 C allele were significantly associated with increased STOX1 mRNA expression in the adrenal gland and whole blood. Kaplan–Meier analysis showed that high mRNA expression of STOX1 was associated with poor prognosis. Interestingly, our findings precisely indicate that the rs3998860 A allele and the rs12781492 C allele are associated with increased neuroblastoma risk. The STOX1 (storkhead Box 1) gene has been implicated in pre‐eclampsia. 47 Expression of the largest isoform of STOX1 (STOX1A) activates the PI3K‐Akt‐FOX pathway in the nucleus and inhibits the pathway in the cytoplasm. 47 Gao et al. demonstrated that STOX1 can inhibit medulloblastoma by inhibiting Math1 expression and that activation of sonic hedgehog signalling can inhibit STOX1 and restore Math1 expression. 48 Importantly, STOX1A has been shown to directly regulate expression of mitotic cyclin CCNB1 in the SH‐SY5Y neuroblastoma cell line and participate in regulating the cell cycle. 49 Consistent with our findings, knockdown of STOX1A has been shown to inhibit proliferation by neuroblastoma cells. 49 Our study reveals the unique cancer‐promoting effect of STOX1 in neuroblastoma compared to other tumours. In other words, rs3998860 and rs12781492 may affect neuroblastoma susceptibility and prognosis by influencing STOX1 mRNA expression. In skin not exposed to sun, both the rs3998860 G allele and the rs12781492 C allele significantly enhanced mRNA expression of CCAR1. The apoptosis regulatory factor CCAR1 plays an important role in promoting cancer, 50 , 51 and deletion of CCAR1 inhibits the occurrence and development of prostate cancer cells. 50 CCAR1 is an important activator for maintaining the growth of breast cancer. 51 Consistent with our findings, high expression of CCAR1 is associated with cancer occurrence and development. TET1 rs3998860 and rs12781492 may increase neuroblastoma risk by influencing mRNA expression of CCAR1. In cultured fibroblasts, the rs3998860 A allele was significantly associated with increased KIF1BP mRNA expression compared to the G allele. The relationship between the protein family member 1 binding protein KIF1BP and tumours is unclear. Studies have shown that KIF1BP is necessary for the growth and maintenance of nerve axons. 52 In general, neuroblastomas with a lower degree of differentiation are less malignant. 53 This is inconsistent with our research results. The role of KIF1BP in neuroblastoma needs to be further elucidated. In the tibial artery, the rs3998860 A allele was significantly associated with increased mRNA expression of SLC25A16 and RUFY2. There is a lack of research on SLC25A16. The RUN and FYVE domain containing 2 (RUFY2) gene has been found to frequently be mutated in high‐microsatellite instability cancers. 54 RUFY2 expression correlates negatively with risk of glioblastoma. 55 There are almost no relevant studies on the RUFY2 gene in neuroblastoma, and the relationship between the RUFY2 gene and neuroblastoma remains to be elucidated. The GTEx database did not reveal a relationship between expression of the TET1 gene and rs3998860 and rs12781492. Further research is needed to clarify this in the future.
Our study is the first to clarify the association between TET1 gene polymorphisms and susceptibility to neuroblastoma in children in Jiangsu, China, and its possible mechanism. Our sample size was relatively large, and comparability between the case group and the control group was high. Nevertheless, there are shortcomings in this study. First, the subjects included were only recruited from one hospital in Nanjing, China, limiting the extension of the conclusion. Second, the predicted functional SNPs examined may be biased, and functional SNPs that affect neuroblastoma may be missed. Finally, the specific mechanism of TET1 rs3998860 and rs12781492 in neuroblastoma remains to be elucidated.
In summary, our study demonstrates that TET1 gene rs3998860 G > A and rs12781492 A > C significantly increase neuroblastoma risk. The potential mechanisms of TET1 gene polymorphisms in neuroblastoma need to be further elucidated.
AUTHOR CONTRIBUTIONS
Jiaming Chang: Investigation (equal); writing – original draft (equal); writing – review and editing (equal). Lei Lin: Formal analysis (equal); investigation (equal); methodology (equal); writing – original draft (equal); writing – review and editing (equal). Chunlei Zhou: Data curation (equal); investigation (equal); resources (equal); writing – review and editing (equal). Xinxin Zhang: Investigation (equal); writing – review and editing (equal). Tianyou Yang: Investigation (equal); writing – review and editing (equal). Haiyan Wu: Data curation (equal); investigation (equal); resources (equal); writing – review and editing (equal). Yan Zou: Conceptualization (equal); investigation (equal); supervision (equal); writing – review and editing (equal). Jing He: Conceptualization (equal); formal analysis (equal); funding acquisition (equal); investigation (equal); methodology (equal); supervision (equal); writing – review and editing (equal).
CONFLICT OF INTEREST STATEMENT
None declared.
Supporting information
Table S1.
ACKNOWLEDGEMENTS
This study was supported by grants from the National Natural Science Foundation of China (No: 82173593), Guangzhou Science and Technology Project (No: 202201020622), Guangdong Basic and Applied Basic Research Foundation (Nos: 2021A1515111116, 2023A1515010534), China Postdoctoral Science Foundation (No: 2021M691649) and Postdoctoral Science Foundation of Jiangsu Province (No: 2021K524C).
Chang J, Lin L, Zhou C, et al. Functional polymorphisms of the TET1 gene increase the risk of neuroblastoma in Chinese children. J Cell Mol Med. 2023;27:2239‐2248. doi: 10.1111/jcmm.17820
Jiaming Chang, Lei Lin, and Chunlei Zhou contributed equally to this work.
Contributor Information
Yan Zou, Email: monknut@126.com.
Jing He, Email: hejing198374@gmail.com.
DATA AVAILABILITY STATEMENT
All the data are available upon request from the corresponding authors.
REFERENCES
- 1. Qiu B, Matthay KK. Advancing therapy for neuroblastoma. Nat Rev Clin Oncol. 2022;19:515‐533. [DOI] [PubMed] [Google Scholar]
- 2. Johnsen JI, Dyberg C, Fransson S, Wickstrom M. Molecular mechanisms and therapeutic targets in neuroblastoma. Pharmacol Res. 2018;131:164‐176. [DOI] [PubMed] [Google Scholar]
- 3. Maris JM. Recent advances in neuroblastoma. N Engl J Med. 2010;362:2202‐2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. London WB, Castleberry RP, Matthay KK, et al. Evidence for an age cutoff greater than 365 days for neuroblastoma risk group stratification in the Children's oncology group. J Clin Oncol. 2005;23:6459‐6465. [DOI] [PubMed] [Google Scholar]
- 5. Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer. 2003;3:203‐216. [DOI] [PubMed] [Google Scholar]
- 6. Schwab M, Westermann F, Hero B, Berthold F. Neuroblastoma: biology and molecular and chromosomal pathology. Lancet Oncol. 2003;4:472‐480. [DOI] [PubMed] [Google Scholar]
- 7. Tas ML, Reedijk AMJ, Karim‐Kos HE, et al. Neuroblastoma between 1990 and 2014 in the Netherlands: increased incidence and improved survival of high‐risk neuroblastoma. Eur J Cancer. 2020;124:47‐55. [DOI] [PubMed] [Google Scholar]
- 8. Bao PP, Li K, Wu CX, et al. Recent incidences and trends of childhood malignant solid tumors in Shanghai, 2002–2010. Zhonghua Er Ke Za Zhi. 2013;51:288‐294. [PubMed] [Google Scholar]
- 9. Pinto NR, Applebaum MA, Volchenboum SL, et al. Advances in risk classification and treatment strategies for neuroblastoma. J Clin Oncol. 2015;33:3008‐3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Coughlan D, Gianferante M, Lynch CF, Stevens JL, Harlan LC. Treatment and survival of childhood neuroblastoma: evidence from a population‐based study in the United States. Pediatr Hematol Oncol. 2017;34:320‐330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tumino N, Weber G, Besi F, et al. Polymorphonuclear myeloid‐derived suppressor cells impair the anti‐tumor efficacy of GD2.CAR T‐cells in patients with neuroblastoma. J Hematol Oncol. 2021;14:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zheng DJ, Krull KR, Chen Y, et al. Long‐term psychological and educational outcomes for survivors of neuroblastoma: a report from the childhood cancer survivor study. Cancer. 2018;124:3220‐3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cheung NK, Zhang J, Lu C, et al. Association of age at diagnosis and genetic mutations in patients with neuroblastoma. Jama. 2012;307:1062‐1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Peifer M, Hertwig F, Roels F, et al. Telomerase activation by genomic rearrangements in high‐risk neuroblastoma. Nature. 2015;526:700‐704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Reshetnyak AV, Rossi P, Myasnikov AG, et al. Mechanism for the activation of the anaplastic lymphoma kinase receptor. Nature. 2021;600:153‐157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lin L, Miao L, Lin H, et al. Targeting RAS in neuroblastoma: is it possible? Pharmacol Ther. 2022;236:108054. [DOI] [PubMed] [Google Scholar]
- 17. Decock J, Long JR, Laxton RC, et al. Association of matrix metalloproteinase‐8 gene variation with breast cancer prognosis. Cancer Res. 2007;67:10214‐10221. [DOI] [PubMed] [Google Scholar]
- 18. Safonov A, Jiang T, Bianchini G, et al. Immune gene expression is associated with genomic aberrations in breast cancer. Cancer Res. 2017;77:3317‐3324. [DOI] [PubMed] [Google Scholar]
- 19. Fanfani V, Citi L, Harris AL, Pezzella F, Stracquadanio G. The landscape of the heritable cancer genome. Cancer Res. 2021;81:2588‐2599. [DOI] [PubMed] [Google Scholar]
- 20. Song H, Koessler T, Ahmed S, et al. Association study of prostate cancer susceptibility variants with risks of invasive ovarian, breast, and colorectal cancer. Cancer Res. 2008;68:8837‐8842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Diskin SJ, Hou C, Glessner JT, et al. Copy number variation at 1q21.1 associated with neuroblastoma. Nature. 2009;459:987‐991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Capasso M, Devoto M, Hou C, et al. Common variations in BARD1 influence susceptibility to high‐risk neuroblastoma. Nat Genet. 2009;41:718‐723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Diskin SJ, Capasso M, Schnepp RW, et al. Common variation at 6q16 within HACE1 and LIN28B influences susceptibility to neuroblastoma. Nat Genet. 2012;44:1126‐1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Oldridge DA, Wood AC, Weichert‐Leahey N, et al. Genetic predisposition to neuroblastoma mediated by a LMO1 super‐enhancer polymorphism. Nature. 2015;528:418‐421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhuo Z, Zhou C, Fang Y, et al. Correlation between the genetic variants of base excision repair (BER) pathway genes and neuroblastoma susceptibility in eastern Chinese children. Cancer Commun (Lond). 2020;40:641‐646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhuo ZJ, Liu W, Zhang J, et al. Functional polymorphisms at ERCC1/XPF genes confer neuroblastoma risk in Chinese children. EBioMedicine. 2018;30:113‐119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guan Q, Lin H, Hua W, et al. Variant rs8400 enhances ALKBH5 expression through disrupting miR‐186 binding and promotes neuroblastoma progression. Chin J Cancer Res. 2023;35:140‐162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Barbieri I, Kouzarides T. Role of RNA modifications in cancer. Nat Rev Cancer. 2020;20:303‐322. [DOI] [PubMed] [Google Scholar]
- 29. Greenberg MVC, Bourc'his D. The diverse roles of DNA methylation in mammalian development and disease. Nat Rev Mol Cell Biol. 2019;20:590‐607. [DOI] [PubMed] [Google Scholar]
- 30. Unnikrishnan A, Freeman WM, Jackson J, Wren JD, Porter H, Richardson A. The role of DNA methylation in epigenetics of aging. Pharmacol Ther. 2019;195:172‐185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Koch A, Joosten SC, Feng Z, et al. Author correction: analysis of DNA methylation in cancer: location revisited. Nat Rev Clin Oncol. 2018;15:467. [DOI] [PubMed] [Google Scholar]
- 32. Xue C, Chu Q, Zheng Q, et al. Role of main RNA modifications in cancer: N(6)‐methyladenosine, 5‐methylcytosine, and pseudouridine. Signal Transduct Target Ther. 2022;7:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Niu X, Peng L, Liu W, et al. A cis‐eQTL in NSUN2 promotes esophageal squamous‐cell carcinoma progression and radiochemotherapy resistance by mRNA‐m(5)C methylation. Signal Transduct Target Ther. 2022;7:267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen X, Li A, Sun BF, et al. 5‐methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat Cell Biol. 2019;21:978‐990. [DOI] [PubMed] [Google Scholar]
- 35. Rong D, Sun G, Wu F, et al. Epigenetics: roles and therapeutic implications of non‐coding RNA modifications in human cancers. Mol Ther Nucleic Acids. 2021;25:67‐82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fu L, Guerrero CR, Zhong N, et al. Tet‐mediated formation of 5‐hydroxymethylcytosine in RNA. J Am Chem Soc. 2014;136:11582‐11585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ito S, Shen L, Dai Q, et al. Tet proteins can convert 5‐methylcytosine to 5‐formylcytosine and 5‐carboxylcytosine. Science. 2011;333:1300‐1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156:45‐68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. He YF, Li BZ, Li Z, et al. Tet‐mediated formation of 5‐carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303‐1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5‐formylcytosine and 5‐carboxylcytosine: potential implications for active demethylation of CpG sites. J Biol Chem. 2011;286:35334‐35338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yang H, Wang Y, Xiang Y, et al. FMRP promotes transcription‐coupled homologous recombination via facilitating TET1‐mediated m5C RNA modification demethylation. Proc Natl Acad Sci U S A. 2022;119:e2116251119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wu BK, Mei SC, Chen EH, Zheng Y, Pan D. YAP induces an oncogenic transcriptional program through TET1‐mediated epigenetic remodeling in liver growth and tumorigenesis. Nat Genet. 2022;54:1202‐1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mariani CJ, Vasanthakumar A, Madzo J, et al. TET1‐mediated hydroxymethylation facilitates hypoxic gene induction in neuroblastoma. Cell Rep. 2014;7:1343‐1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gao J, Ma Y, Fu HL, et al. Non‐catalytic roles for TET1 protein negatively regulating neuronal differentiation through srGAP3 in neuroblastoma cells. Protein Cell. 2016;7:351‐361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wu HX, Chen YX, Wang ZX, et al. Alteration in TET1 as potential biomarker for immune checkpoint blockade in multiple cancers. J Immunother Cancer. 2019;7:264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lin L, Deng C, Zhou C, et al. NSUN2 gene rs13181449 C>T polymorphism reduces neuroblastoma risk. Gene. 2023;854:147120. [DOI] [PubMed] [Google Scholar]
- 47. van Dijk M, Mulders J, Poutsma A, et al. Maternal segregation of the Dutch preeclampsia locus at 10q22 with a new member of the winged helix gene family. Nat Genet. 2005;37:514‐519. [DOI] [PubMed] [Google Scholar]
- 48. Zhang C, Ji Z, Wang M, et al. Stox1 as a novel transcriptional suppressor of Math1 during cerebellar granule neurogenesis and medulloblastoma formation. Cell Death Differ. 2016;23:2042‐2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Abel D, Abdul‐Hamid O, Dijk M, Oudejans CB. Transcription factor STOX1A promotes mitotic entry by binding to the CCNB1 promotor. PLoS One. 2012;7:e29769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Seo WY, Jeong BC, Yu EJ, et al. CCAR1 promotes chromatin loading of androgen receptor (AR) transcription complex by stabilizing the association between AR and GATA2. Nucleic Acids Res. 2013;41:8526‐8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yu EJ, Kim SH, Heo K, Ou CY, Stallcup MR, Kim JH. Reciprocal roles of DBC1 and SIRT1 in regulating estrogen receptor alpha activity and co‐activator synergy. Nucleic Acids Res. 2011;39:6932‐6943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lyons DA, Naylor SG, Mercurio S, Dominguez C, Talbot WS. KBP is essential for axonal structure, outgrowth and maintenance in zebrafish, providing insight into the cellular basis of Goldberg‐Shprintzen syndrome. Development. 2008;135:599‐608. [DOI] [PubMed] [Google Scholar]
- 53. Lam WA, Cao L, Umesh V, Keung AJ, Sen S, Kumar S. Extracellular matrix rigidity modulates neuroblastoma cell differentiation and N‐myc expression. Mol Cancer. 2010;9:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shin N, You KT, Lee H, et al. Identification of frequently mutated genes with relevance to nonsense mediated mRNA decay in the high microsatellite instability cancers. Int J Cancer. 2011;128:2872‐2880. [DOI] [PubMed] [Google Scholar]
- 55. Lai W, Li D, Kuang J, Deng L, Lu Q. Integrated analysis of single‐cell RNA‐seq dataset and bulk RNA‐seq dataset constructs a prognostic model for predicting survival in human glioblastoma. Brain Behav. 2022;12:e2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Data Availability Statement
All the data are available upon request from the corresponding authors.


