Abstract
Purpose
Although retinal light injury models have been useful in understanding aspects of retinal degeneration and retinal oxidative stress, information on retinal recovery from oxidative/photoinflammatory retinal injury is scarce. The fundus camera-delivered light-induced retinal degeneration model is a simple and reproducible retinal light injury model developed to recapitulate not only the retinal degeneration aspect, but also the retinal recovery from injury. In this study, we used the fundus camera-delivered light-induced retinal degeneration model to perform cell type-specific analyses of the acute and subacute retinal responses to light injury.
Methods
C57BL/6J eyes were collected before or after light injury (4 hours, 48 hours, and day 5). Retina samples were processed into single-cell suspensions. Droplet-based encapsulation of single cells was performed to generate libraries for sequencing.
Results
Gene expression analysis generated 23 clusters encompassing all known major retinal cell populations. Using unbiased analyses, we identified genes and pathways that were significantly altered in each cell type after light injury, including some cellular processes suggestive of activation of pathways for retinal recovery (e.g., synaptogenesis signaling, ephrin receptor signaling, and Reelin signaling in neurons). More importantly, our data show that a subpopulation of Muller glia cells may play an important role in the cellular recovery process.
Conclusions
This work identifies acute and subacute cell type-specific responses to retinal photo-oxidative injury. A subpopulation of Muller glia seems to initiate the cellular recovery process. A better understanding of these responses may be helpful in identifying therapeutic approaches to minimize retinal damage and maximize recovery after exposure to injury.
Keywords: light injury, retinal recovery, single cell RNA sequencing, scRNA-seq, Muller cells, FCD-LIRD
The goal of developing strategies to protect photoreceptors from acute injury has led to a large number of studies aimed at understanding the acute response of the retina to injury.1–9 However, many human retinal diseases are caused or modulated by sublethal stressors that, by the time of diagnosis, have already induced some damage. Understanding how the retina recovers from sublethal injury may help us to optimize the speed and the extent to which the photoreceptors and RPE overcome insults.
There is clinical evidence of partial retinal recovery in a wide range of retinal diseases, including central serous retinopathy,10–12 phototoxicity,13,14 retinal detachment,15–17 inflammatory diseases,18,19 and even AMD.20 A sublethal retinal light injury model may be helpful in understanding the mechanisms involved. The possible role of light as a stressor in retinal diseases has generated great interest,21–24 and light-induced retinal degeneration (LIRD) has been a useful model to study mechanisms of disease in the retina.25–31 However, data concerning the retinal recovery processes from insults in these mouse models are scarce.32 We have recently developed and characterized a mouse model of LIRD that is easily modulated in terms of severity.33,34 This fundus camera-delivered LIRD (FCD-LIRD) model is a photo-oxidative/photoinflammatory injury model, that is fast, reproducible, and effective in mice carrying the RPE65 450-Met variant and, thus, can be applied to all mouse lines on a C57BL6 background. Using this model, we have shown previously that, in mice treated with a sublethal FCD-LIRD protocol, an initial acute retina injury phase leads to changes in the reflectivity and thinning of the outer retina on optical coherence tomography (OCT). It is followed by a recovery phase in which the retinal volume between Bruch's membrane and the external limiting membrane partially recovers. Because the FCD-LIRD model mimics well the anatomical and functional changes seen in a variety of clinical entities with outer retinal damage, we now have a test system that can be used to understand general pathways of retinal recovery from sublethal injury.
We propose that both the acute response and the subacute recovery phases lead to time-dependent and cell-specific gene expression changes. In this work, we use a low-intensity stimulus to induce FCD-LIRD–mediated sublethal retinal injury in B6J mice and then isolate cells from the central retina for single cell RNA sequencing (RNA-seq) studies at different time points after injury. Our findings shed light on the mechanisms involved in mouse retinal recovery from mild light induced retinal injury. In the current study, we find that 4 hours after the light injury stimulus there is an upregulation of mitochondrial dysfunction signaling and downregulation of oxidative phosphorylation, glycolysis and EIF2 signaling in most cell types of the retina. These stress response metabolic and translational changes continued through 48 hours and up to day 5 after light injury in most cell types except a subpopulation of Muller cells. The data for Muller cells, show that cell signaling pathways such as synaptogenesis signaling and ephrin receptor signaling pathways are upregulated starting at 48 hours in a subpopulation (Muller-1 [Mu-1]) of Muller cells while they are either downregulated or unchanged in the other cell types. The upregulation of these cellular processes and pathways suggests that this subpopulation of Muller cells may play a key role in guiding the retinal recovery mechanism after mild light injury.
Materials and Methods
Animals
Animals were handled in accordance with all applicable international, national, and institutional guidelines for the care and use of animals, including the National Institutes of Health Guide for the Care and Use of Laboratory Animals and the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. All procedures were approved by the UT Southwestern Medical Center Institutional Animal Care and Use Committee. Four-month-old C57BL/6J mice (B6J; Jackson Laboratory, Bar Harbor, ME, USA) were used. Mice were acclimated to our animal facility for at least 1 week before being used for experiments. Mice were bred and kept in a barrier animal facility at UT Southwestern Medical Center under normal lighting conditions with 12-hour-on/12-hour-off cycles. Before performing all procedures, mice were anesthetized with a ketamine-xylazine cocktail (ketamine 100 mg/kg, Xylazine 5mg/kg) one at a time. Mouse eyes were dilated using one drop per eye of tropicamide 1% solution (Alcon Laboratories, Inc., Fort Worth, TX, USA) and phenylephrine hydrochloride 2.5% solution (Alcon, Inc., Lake Forest, IL, USA).
Light Injury
Mice were dark adapted overnight in a darkroom equipped with a red light source during the experiment. Before applying light to each eye, the light intensity from the Micron IV mouse fundus camera was measured using a light meter (Fisher Scientific, Hampton, NH, USA; Cat # S90199) to ensure that equal illumination was provided to all eyes. Each mouse was anesthetized, one at a time, and the pupils dilated as stated above. We applied GenTeal gel to eyes to prevent corneal dryness during the length of the experiment. A single intraperitoneal injection of fluorescein (100 µL of a 1:5 diluted 10% fluorescein solution) was administered just before the camera was centered on the optic disc head and focused on the RPE. The desired light intensity was then applied to the retina as described previously.34 In brief, after a 4-minute delay after the fluorescein injection the light intensity was increased to 45K lux and continued for 3 minutes (a protocol we called FA 3@4) while monitoring that no changes happen in the fundus orientation during this time. Mice were kept under normal lighting conditions after the light procedure.
Sample Collection
For immunohistochemistry of glial fibrillary acidic protein (GFAP), eyes were enucleated from deeply anesthetized control (baseline) and light injured mice at 2, 3, and 5 days after injury and processed by a freeze substitution method as detailed before.35 After freeze substitution fixation, whole globe mouse eyes were paraffin processed, embedded, and sectioned. Paraffin sections were concomitantly prepared and checked by dark-field microscopy to ensure comparative sections through the optic disc and the inclusion of superior to inferior regions of the retina. Resulting serial sections were deparaffinized and immunostained by incubating overnight at 4°C with mouse anti-GFAP antibody (GA5 clone, Biogenex, Fremont, CA, USA; Cat# MU020-UC, 1:800 dilution) followed by chromogenic detection using brown diaminobenzidine-chromogen (ImmPACT DABEqV Substrate Kit, Vector Laboratories, Burlingame, CA, USA; Cat# SK-4103). Nuclei were counterstained lightly with hematoxylin, the slides were then dehydrated, cleared, and cover-slipped with synthetic mounting media. Images of the stained sections were taken at an original magnification of ×20 on either side of the optic nerve head on a Leica DM200 microscope (Leica Microsysytms, Wetzlar, Germany).
For retinal single cell suspension, eyes were collected from deeply anesthetized mice at 4 hours, 48 hours, or 120 hours (5 days) after the application of light injury. Eyes were also collected from mice not exposed to light, which served as control, referred to herein as baseline. After enucleation, the anterior tissues (cornea, lens, iris, and the vitreous body) were removed and the retinas were peeled off from the posterior eyecups. A 2-mm diameter biopsy punch (Robbins Instruments, Houston, TX, USA) was used to carefully cut off the center part of the retina for the preparation of single cell suspension.
Preparation of Single Cells From Mouse Retina
Single-cell suspensions were prepared from the retinas using the papain dissociation system (Worthington Biochemical Corporation, Lakewood, NJ, USA; Cat. No. LK003150), which was based on an originally described method for purifying retinal ganglion cells (RGCs) from rat retina by Barres et al. with slight modifications.36–38 Briefly, retinas were digested/dissociated by incubation with activated papain at 37°C for 45 minutes, followed by gentle trituration (pipetting up and down using a 10-mL pipette for approximately 10–20 times). Papain was removed by aspiration after cells were centrifuged at 300×g for 5 minutes, and the cells were then resuspended in medium containing 10% ovomucoid, a papain inhibitor with 5% DNase I. Intact cells are separated from cell membranes by centrifugation through a single step discontinuous density gradient at 100×g for 6 minutes.
To decrease the acute, endogenous transcriptional alterations, we applied the general transcription inhibitor, actinomycin D, during the dissociation process.39 Actinomycin D inhibits transcription mediated by all three eukaryotic RNA polymerases and provides wide spectrum, fast transcriptional inhibition with little and slow reversibility.40 Actinomycin D (Sigma-Aldrich, St. Louis, O, USA; Cat# A1410) was added at three steps during the dissociation process: to the papain solution at 45 mM, to the papain inhibitor solution at 45 mM, and during trituration at 3 mM.
After centrifugation, the supernatant was removed completely and the cell pellet was resuspended in 100 µL of dead cell removal microbeads (Miltenyi Biotec, USA, Gaithersburg, MD, USA; No.130-090-101). This was followed by passing the cell suspension through MS column (Miltenyi Biotec, USA, No.130-042-201) which was placed in a magnetic field of a suitable MACS Separator (Miltenyi Biotec, USA, No.130-042-109). The MS column was primed by rinsing with 1× binding buffer. The effluent cells were collected as live cell fraction. The live cells are separated by centrifugation at 70×g for 6 minutes at room temperature and the resulting pellet was resuspended in Dulbecco's phosphate-buffered saline containing 0.4% BSA. Finally, the viability of cells was determined on a cellometer (Nexcelom Bioscience LLC, Lawrence, MA, USA) by staining with PI. The viability after dead cell removal was close to 80%.
Droplet-Based Single Cell RNA-seq of Dissociated Cells
The number of viable retinal single cells in each sample was determined and processed through the GemCode Single Cell Platform using the GemCode Gel Bead, chromatin immunoprecipitation, and Library Kits (10X Genomics, Pleasanton, CA, USA) according to a standard platform protocol. In brief, single cells in 0.4% BSA-Dulbecco's phosphate-buffered saline suspension were sorted and approximately 14,000 cells were added to a channel. After partitioning into Gel Beads in Emulsion in the 10x Genomics Chromium Controller, the cells were lysed and primed with barcoded oligo-DT. This was then followed by reverse transcription of poly-adenylated RNA, amplification and shearing of cDNAs for adaptor and sample index attachment. Libraries were sequenced on an Illumina HiSeq 2500.
Analyses of Single Cell Transcriptomes
Identification of Clusters and Analysis of Their Gene Expression
Cell Ranger 3.0.0 (10X Genomics, https://www.10xgenomics.com/) was used to process the raw sequencing data. BCL files were converted to FASTQ files and aligned to mouse (mm10) reference transcriptome. Transcript counts of each cell were quantified using UMI and valid cell barcode. The gene expression matrix from cell ranger was used as input to Seurat R package (v3.0.0) for the downstream analysis.41 Cells with less than 200 genes per cell and very high mitochondrial gene content were filtered out. The global-scaling normalization method “Log Normalize” was used for normalization. A subset of genes exhibiting high variation across the single cells was determined. The highly variable genes were calculated using “FindVariableFeatures” module in Seurat. Average expression and dispersion per gene were calculated and features were divided into bins to get z-scores for dispersion per bin. Data were then scaled, and dimensional reduction was performed with principal component analysis. Seurat integrated analysis was performed across samples at different time points (baseline, 4 hours, 48 hours, and day 5 after light injury). For the sample, a shared nearest neighbor graph was constructed with the “FindNeighbors” module in Seurat by determining the k-nearest neighbors of each cell. The clusters were then identified by optimizing shared nearest neighbor modularity using the “FindClusters” module. This process allowed for a sensitive detection of rare cell types. We obtained 23 clusters with a resolution of 0.5. UMAP plot was generated using the RunUMAP module in Seurat. Each cluster was compared with all other clusters using the Wilcoxon rank-sum test to test for significant differentially expressed genes.
Identities of the clusters representing cell types and subpopulations of retinal cells were determined based on expression profile of known gene markers specific to various cell types found in the retina.42 To find differentially expressed genes between different time point (4 hours vs baseline, 48 hours vs baseline, day 5 vs baseline, 48 hours vs 4 hours and day 5 vs 48 hours) for each cell type, FindMarkers module in Seurat was used. To show a trend of gene expression regulation across four time points, differentially expressed genes with statistical significance of a false discovery rate of less than 0.05 for each of the four time point comparisons were considered. Average expression values of statistically significant genes in each cell type were presented as heat maps and spaghetti. Heatmaps were generated using heatmap.2 function in R (v 3.6.1; The R Foundation, Vienna, Austria). For heatmap and cluster detection, we used a hierarchical clustering approach with the agglomeration method as complete. Spaghetti plots were generated using custom scripts in R (v 3.6.1).
Analysis of Muller Cells Based on Their Expression of GFAP
The Mu-1 cluster was divided into Gfap+ cells (Gfap expression of >0 at 48 hours) and Gfap− cells (Gfap expression of 0 at 48 hours). Differential expression between Gfap+ and Gfap− cells was performed using the “FindMarkers” module in Seurat. The “VlnPlot” module of Seurat was used to generate violin plots. Dot plot was generated using ggplot2 (v3.3.6) in R (v3.6.1).
Pathway Analysis
Gene set enrichment analysis (GSEA) and pathway analysis was performed using the MsigDB database in GSEA (gsea-msigdb.org) and ingenuity pathway analysis (IPA) software (https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/). For GSEA, genes from selected spaghetti plots which showed significant upregulation or downregulation of clusters of genes were used as input data. For IPA, the top 1500 differentially expressed genes (P < 0.05) from comparisons between 48 hours versus baseline and day 5 versus baseline were taken as input. Comparisons of the resulting pathways for these two time points for each cell type are presented as bar plots. The bar plots were generated using ggplot2 (v3.3.6) in R (v3.6.1). The differentially expressed genes between Gfap+ and Gfap− cells with a P value of less than 0.05 were used as input for IPA.
Statistical Analyses
Data are presented as the mean ± standard error of mean. A two-tailed Student t-test or the Mann–Whitney U test was performed when comparing two groups. A P value of less than 0.05 was considered to be statistically significant.
Results
Evidence of Clinical, Structural, and Functional Recovery of the Retina From Photo-Oxidative Injury
The flow chart presented in Figure 1 shows the protocols and steps used in this work starting from the application of light injury to the analysis of single cell RNA-seq (scRNA-seq). Based on our prior studies,33,34,43,44 we had determined that the FCD-LIRD model led to retinal light injury that could modulated be easily by modifying the intensity of the light stimulus. In this model, maximal injury as seen on fundus photos and OCT can be seen at approximately day 5 after injury. More important, if a sublethal stimulus is chosen (Fig. 2), evidence of partial recovery can be documented on both fundus photos and OCT by day 14. Specifically, at day 5, in addition to pigmentary changes on fundus photography (Fig. 2B, compare with Fig. 2A), a very obvious disruption and hyporeflectivity of the ellipsoid zone is seen (white arrows in Fig. 2E, compare with Fig. 2D). Moreover, some thinning of the outer retina and hyper-reflectivity of the photoreceptor outer segment band is seen (yellow arrow in Fig. 2E). Importantly, by day 14, some recovery of the fundus pigmentary changes (Fig. 2C), as well as changes in the ellipsoid and the photoreceptor outer segment bands (Fig. 2F) can be seen in the sublethal injury model. This type of damage and recovery is strongly reminiscent of the changes seen in many retinal diseases.10–20 Because our intention was to analyze the recovery phase, we deliberately chose this level of sublethal light stimulus. We named the protocol FA 3@4 (3 minutes of 45K lux stimulus, starting 4 minutes after intraperitoneal fluorescein injection). Our prior work developing and characterizing the FCD-LIRD model demonstrates that higher levels of light stimulus would lead to more prominent retinal thinning.33,34,43
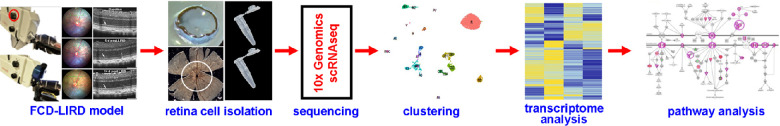
Figure 1.
Schematic diagram showing the experimental protocol. (A) Application of light injury using the FCD-LIRD model. (B) Isolation of central retinal cells from enucleated mouse eyes using a papain dissociation system. (C) Sequencing and clustering using 10× genomics and Cell ranger. (D) Transcriptomics and pathway analysis (IPA).
Figure 2.
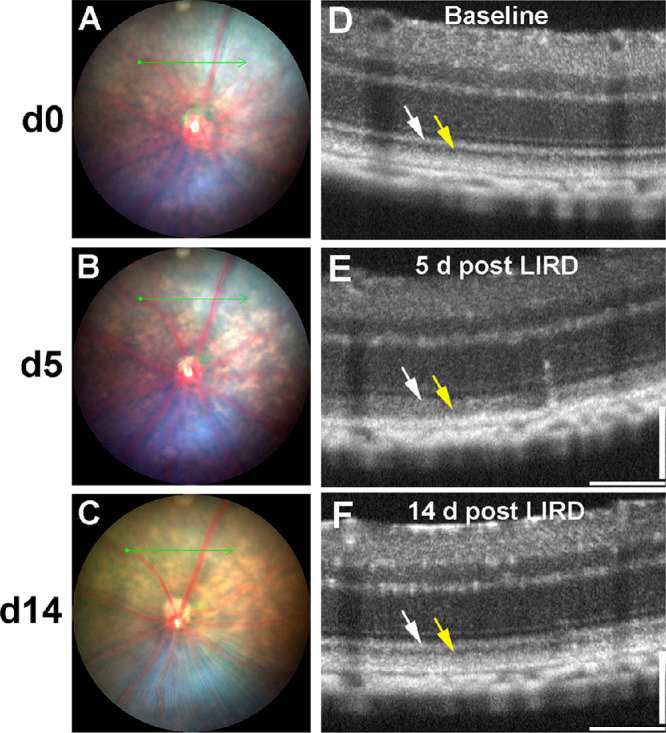
Representative fundus and OCT images showing the results of sublethal retinal injury using the FCD-LIRD model. Baseline fundus (A) and OCT (D) are taken before the injury. At day 5 after light injury, there is a mild RPE discoloration on fundus photography (B). There is also a decrease in reflectivity of the ellipsoid zone and an increase in reflectivity of the photoreceptor outer segments on OCT (E). At day 14 after light injury we could see evidence of recovery of the retina from injury on fundus photography (C) and on OCT (F).
scRNA-seq Identifies Major Cell Populations of the Mouse Retina
Having established an appropriate sublethal injury model, we proceeded to explore the mechanisms of recovery using scRNA-seq. Because in the FCD-LIRD model the stimulus is focused on the posterior retina, we isolated the central retina for our studies (2-mm biopsy punch centered on the disc). We pooled the samples collected from four mice (one eye per mouse) at each of the following timepoints: baseline (no injury), 4 hours, 48 hours, and day 5 after light injury. The retinas were dissociated using a papain-based protocol into single cell suspensions and processed for scRNA-seq analysis using the 10X Genomics platform. We were able to isolate 3400 to 6500 central retinal cells (≥80% viability) from the four eyes per group.
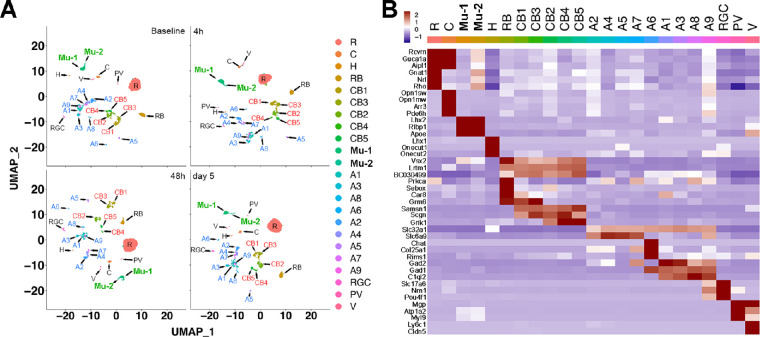
Unbiased analysis of the scRNA-seq data revealed 23 clusters (Fig. 3A) encompassing the major retinal cell populations. Using expression profiles of known cell type-specific markers reported in prior published reports (Fig. 3B), we were able to promptly assign 17 of the 23 clusters to retinal cell types: rod photoreceptors (“R”; markers = Rho, Nrl, Gnat1); cone photoreceptors (“C”; markers = Opn1sw, Opn1mw, Arr3, and Pde6h); rod bipolar cells (“RB”; markers = combination of Prkca, Sebox, and Car8), cone bipolar cells (“CB1–CB5”; markers = Samsn1 and Scgn); Muller glia (“M”; markers = Lhx2, Rlbp1, and Apoe); amacrine cells (“A1–A7”; marker = Slc32a1); and RGC (markers = Slc17a6 and Pou4f1). With further probing using additional cell type markers, we were able to assign the remaining six clusters: horizontal cells (“H”; markers = Lhx1, Onecut1, and Onecut2), additional amacrine subtypes (“A8 and A9”; markers = Slc32a1 plus C1ql2), one additional Muller glia subtype (“Mu-2”; marker = Lhx2 and Rlbp1), vascular endothelial cells (“V”; markers = Ly6c1 and Cldn5), and perivascular cells (“PV”; marker = Myl9). All 23 clusters were present in all 4 time points. We were further able to subclassify cone bipolar cells as ON-bipolars (CB1 and CB3; marker = Grm6) and OFF-bipolars (CB2, CB4, and CB5; marker = Grik1). Similarly, amacrine cells could be subgrouped into GABAergic (“A2, A4, A5, A7”; marker = Slc6a9), glycinergic (“A1, A3, A8, A9”; marker = C1ql2), and Starburst (“A6”; marker = Chat). Of interest, the percentage of retinal cell types in our clusters correlated well with findings from other groups.42,45 The rod photoreceptor cells accounted for approximately one-half of the cells (40%–53% in the different samples). The ratio of rods to cones ranged from 15:1 to 22:1, which was also consistent with previously published data.42,45 Of note, also in line with prior studies, we only found a few cells positive for microglia markers (Tmem119 or Fcrls; Supplementary Fig. S1) or astrocyte markers (GFAP or S100b; Supplementary Fig. S2), and these were not enough to be identified as independent clusters.
Figure 3.
Identification and clustering of retinal cell types at baseline, 4 hours, 48 hours, and day 5 after light injury. (A) UMAP analysis of the mouse retina cells showing 23 different clusters at the 4 time points (top left, baseline; top right, 4 hours; bottom left, 48 hours; bottom right, day 5). (B) The retinal cell types and subtypes were identified using multiple known published cell type-specific marker genes. All the cell type and subtype clusters were present in all the time points.
Cell Type-Specific and Time-Dependent Differential Gene Expression Changes in Response to Light Injury
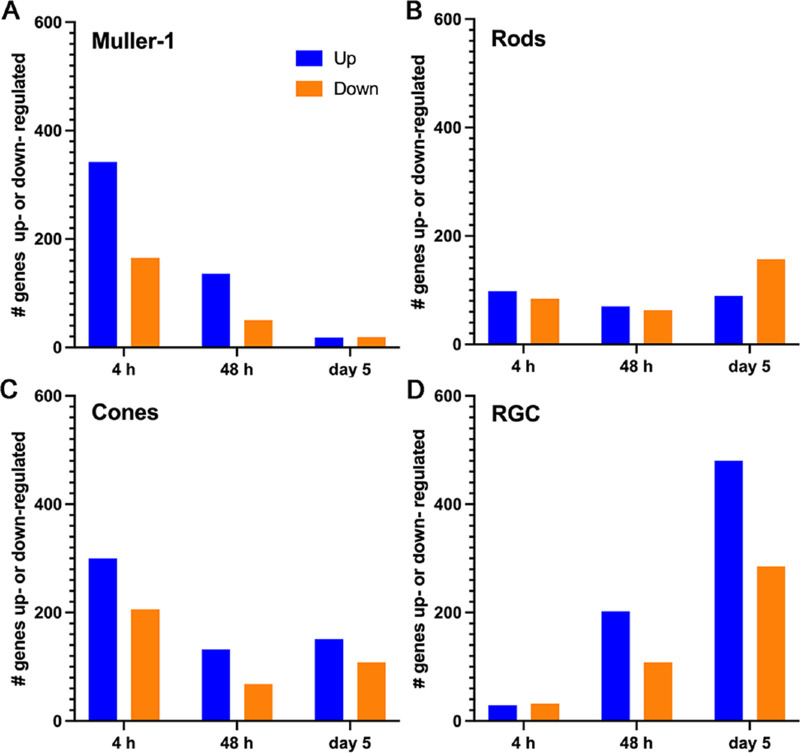
Next, in an unbiased manner, we compared gene expression levels obtained at the different time points after light injury with the baseline levels. Given the choice of a mild light injury stimulus, we expected correspondingly mild gene expression changes. Thus, we graphed the total number of genes with a |log2FC| of greater than 0.25 and a P value of less than 0.05 for each time point in the selected cell types (Mu-1, rods, cones and RGCs) (Fig. 4 and Supplementary Table S1). Using this approach, we uncovered a very interesting temporal pattern of behavior: the largest number of differentially expressed genes in Muller cells (both upregulation and downregulation) occurred at the early time point, whereas most changes in the RGCs were seen at the late time point. In the meantime, the response of photoreceptors seemed to fall between these two extremes; they showed some significant gene expression changes at all timepoints.
Figure 4.
Different cell types show different patterns of up- or down-regulation of genes after light injury. (A) Number of up- or down-regulated genes in Mu-1 cell show a greater number of changes at the early time point of 4 hours, but very few at the late time point. (B, C) Rod and cone cells show a more evenly distributed pattern of gene expression changes in all time points. (D) RGCs show a fewer number of gene expression changes at the early time point, but a greater number of changes at the late time point.
To focus on genes with larger expression changes, we applied more stringent criteria (a |log2FC| > 1, and P < 0.05; shown in bold type in Supplementary Table S1). We found that the greatest proportion of gene expression changes, especially those with upregulation, occurred at the early time point (4 hours). Of interest, most upregulated genes were seen in Muller 1 (51 genes) and Muller 2 (45 genes) cells. At this early time point, a few genes were also upregulated in rods (Samd7, Hmgb2) and cones (Egr1, Ubc, Jund). Downregulation of Hes5 was seen in Mu-1 cells. At the intermediate time point (48 hours), we saw the upregulation of 5 genes (S100a6, Rpsa, Nupr1, Rho, and Rbp3) in Mu-2 cells, and 1 gene (mt-Nd4l) in cones. Downregulated genes at 48 hours included one gene (mt-Nd3) in Mu-1 cells, two genes (mt-Nd3, Ddah2) in Mu-2 cells, three genes (mt-Nd3, mt-Nd1, mt-Nd2) in rods, one gene (mt-Md3) in cones, and two genes (mt-Nd3, mt-Nd1) in RGCs. At the late time point (day 5), two genes (Pcp4, Calb2) were upregulated in rods.
Of interest, a subset of genes in Muller cells even met a more stringent cut-off point of a |log2FC| > 2, with a P value of less than 0.05 at the 4-hour time point (14 genes in Mu-1 and 8 genes in Muller-2 [Mu-2]). Many of these genes are the known immediate early response genes (see genes highlighted in yellow in Supplementary Table S1). They include transcription factors/regulators such as Fos, Cebpb, and Egr1, which regulate cell proliferation and differentiation in response to different stress stimuli. Furthermore, oxidative and environmental stress response genes (Srxn1 and Gadd45b) were also found in this list.
Systematic Analysis of Gene Expression Trends Over Time Confirmed Unique Patterns in Different Cell Types
To better understand the temporal patterns of gene expression changes in the different cell types, we selected those genes with significant differential expression (adjusted P < 0.05) for any pair-wise comparisons (4 hours vs baseline, 48 hours vs baseline, day 5 vs baseline, 48 hours vs 4 hours, and day 5 vs 48 hours). Then, the average expression of these genes was used to generate expression trends across the four time points. The data are depicted as heat maps of average gene expression for all four time points for selected major cell types (Mu-1, rods, cones, and RGCs) (Supplementary Fig. S3).
For better visualization, these gene expression behavior patterns were clustered into behavior subclusters and spaghetti plots for each of these were generated (Supplementary Fig. S4). We then chose the subclusters that demonstrated patterns of gene expression trends that would be most suggestive of a delayed response that could be compatible with a recovery process. These were clusters that showed either progressive increase (green ovals) or decrease (red ovals) in expression up to the late time point (a change in standardized expression of >1.5 units from baseline to day 5). The findings seemed to generally support those shown in Figure 4. Specifically, none of the subclusters in Muller cells showed progressive gene expression changes, but instead included predominantly early and some intermediate changes. In contrast, four of the five subclusters in RGCs met the criteria. Three subclusters (a total of 189 genes) showed a strong gene downregulation trend toward the late time point, and one subcluster (20 genes) showed a progressive upregulation trend. Interestingly, both cone and rod photoreceptors showed a mix of behaviors, but did include some subclusters with upregulation and others with downregulation trends toward the late time point. In rods, out of the nine subclusters, two subclusters (total of 170 genes) were particularly consistent with a downward trend toward the late time point, whereas one subcluster showed some late trending upregulation (37 genes). In cones, the behaviors were less distinct, but out of the five subclusters, only one (22 genes) showed a clear downward trend.
IPA Identifies Pathways Related to Recovery Processes in Muller Cells at Intermediate and Late Time Points After Light Injury
Having identified subclusters of genes with interesting late trending behavior, we decided to investigate whether we could use them to detect enriched pathways related to the recovery process in the four selected cell types. We applied gene ontology analysis (over-representation analysis) to the gene sets resulting from the combination of either subclusters with an upward trend or the combination of subclusters with a downward trend for each individual cell type (Supplementary Table S2). In rod cells, homeostatic processes and pathways related to neurogenesis were identified in the upregulated trends. In contrast, biological processes related to the translation, biosynthetic process, and metabolic process were enriched in downregulated trends. Although there were no enriched processes in upregulated trends in either the cones or the RGCs, the GSEA identified cytoplasmic translation, biosynthetic processes, and oxidative phosphorylation as highly enriched biological processes in the downregulated trends.
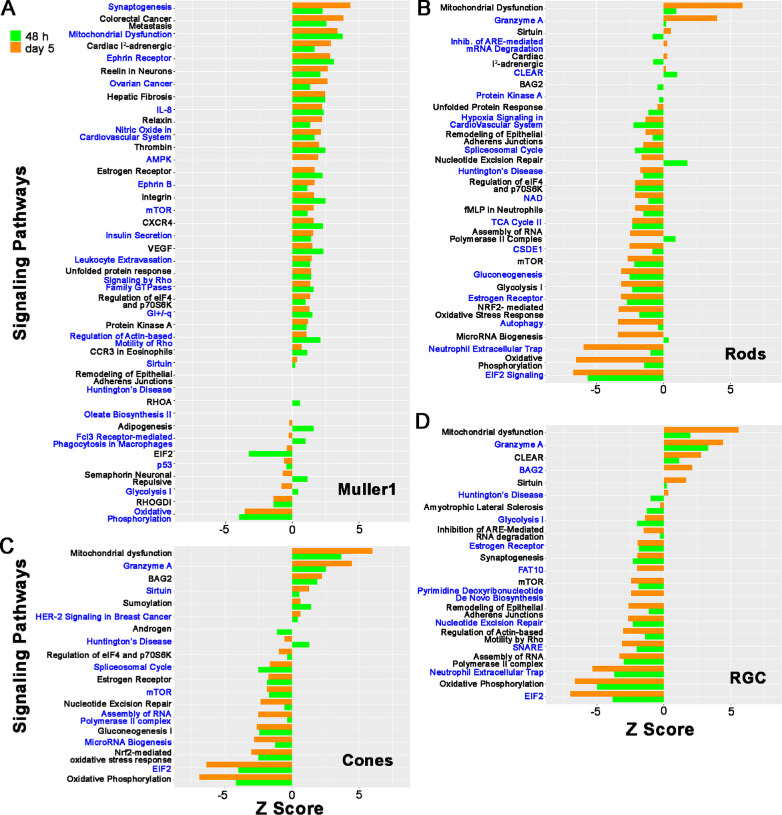
After focusing on late trending gene changes, we decided to do a more inclusive unbiased analysis for each cell type. To this end, we applied IPA. We compared the gene expression profile at 48 hours and day 5 versus baseline for each cell type. (Fig. 5, Supplementary Fig. S5). When considering statistically significant changes (P < 0.05), Muller cells revealed a predominance of upregulated pathways. In contrast, in all other cell types, downregulated pathways were more common, both at 48 hours and day 5. From these pathways, we selected those with a |z-score| > 2 and that are most relevant to our project (Fig. 6) and compared among the different cell types. These include the synaptogenesis pathway, ephrin receptor signaling, Reelin signaling in neurons, IL-8 signaling, EIF2 signaling, mitochondrial dysfunction, granzyme A, and oxidative phosphorylation.
Figure 5.
IPA results at 48 hours and day 5. Results of IPA for the top up- or down-regulated pathways at 48 hours (green bars) and day 5 (orange bars) after light injury are shown as bar plots for selected cell types (Mu-1, rods, cones, and RGCs). The top 1500 statistically significant differentially expressed genes (P < 0.05) between 48 hours and baseline and between day 5 and baseline were used as input for IPA. Pathways were considered to be significantly regulated if they had a P value of <0.05 and a |Z-score| of >2.0.
Figure 6.
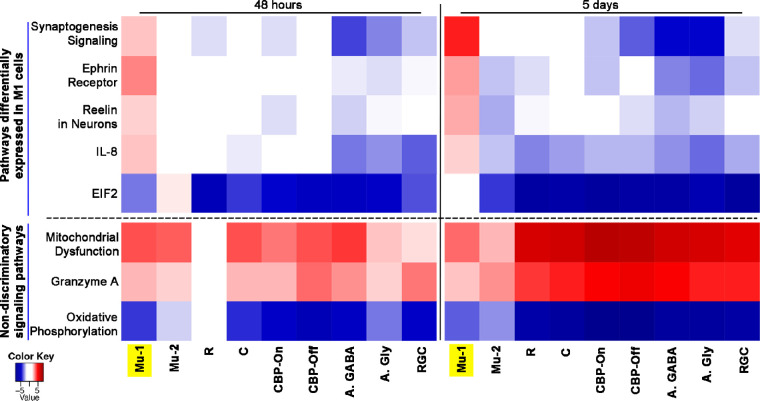
Differential activation of signaling pathways in Mu-1 cells. IPA results of relevant selected up- or down-regulated pathways at 48 hours, and day 5 after light injury compared with baseline are shown as heat map for selected cell types. The cell types shown are Mu-1, Mu-2, rods (R), cones (C), ON-cone bipolar cells (CB-On), OFF-cone bipolar (CB-Off), amacrine GABAergic (A, GABA), amacrine glycinergic (A, Gly), and RGCs. Significantly upregulated (red) or down-regulated (blue) pathways differ for Mu-1 cells compared with other cell types. Pathways are considered as significantly regulated if they meet a P value of <0.05 and a |Z-score| of >2.0.
To better understand the behavior of these selected top upregulated or downregulated pathways in different cell types toward the time frame when we would expect recovery-related gene expression changes, we generated a heat map for the 48-hour and day-5 time points comparing different retinal cell types. This analysis was done on Mu-1 cells, Mu-2 cells, rods, cones, ON-bipolar, OFF-bipolar, glycinergic amacrine, and GABAergic amacrine cells, as well as RGCs. Upregulated pathways with a z-score of greater than 2 are indicated with red, and downregulated pathways with a z-score of less than –2 are shown in blue. Pathways with z-score between 2 and –2 are colored white. Several pathways were regulated similarly in all cell types, including Mu-1. For example, oxidative phosphorylation was downregulated strongly, and the closely related mitochondrial dysfunction and granzyme A pathways were upregulated in all cell types. However, for several of the most significantly affected pathways, the behavior of the Mu-1 cluster was strikingly different from that of Mu-2 and all other cell types. Specifically, although the pathways involving synaptogenesis signaling, ephrin receptor signaling, Reeling signaling, in neurons and IL-8 signaling were mostly downregulated or unchanged in all cell types (including Mu-2), this was not the case for the Mu-1 cluster. For Mu-1 cells, all of these pathways were upregulated both at 48 hours and day 5. Similarly, although the EIF2 Signaling pathway (involved in protein synthesis) was strongly downregulated in all other cell types at day 5, this was not the case for the Mu-1 cluster.
Differential Gene Expression Changes in a Subgroup of Activated Muller 1 Cells
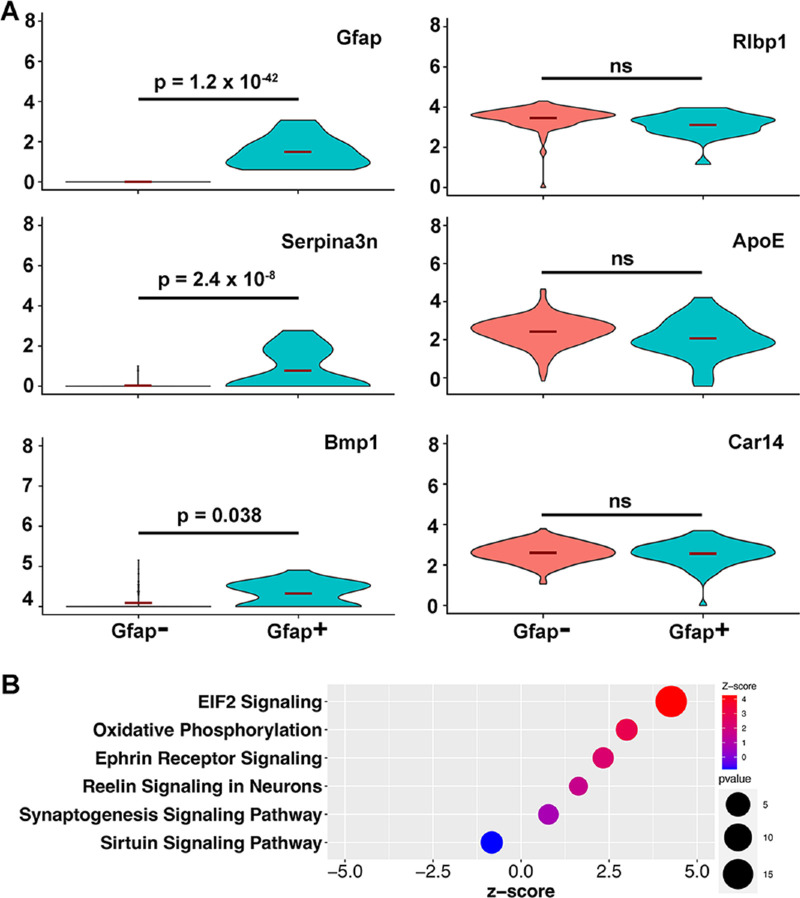
Given our observation that, after light injury, several genes and pathways are expressed differentially in Mu-1 cells compared with other cell types, we wanted to determine if we could find a differentially activated subgroup of Mu-1 cells. Gfap is a known marker of Muller cell activation that is typically maximally expressed 48 hours after a stimulus. We found that a proportion of retinal Mu-1 cells showed Gfap upregulation at 48 hours after light injury (Fig. 7). We confirmed on immunohistochemistry that we could see Muller cell activation starting at 48 hours after light injury (Supplementary Fig. S6). Under normal conditions, GFAP staining does not involve Muller glia and is restricted mostly to astrocytes surrounding ganglion cell axons in the nerve fiber layer and into the optic nerve.46,47 This finding is consistent with what we saw in immunohistochemistry at baseline (Supplementary Fig. S6A): GFAP stains astrocytes surrounding ganglion cell axons in the NFL, but not Muller glia. In contrast, 2 days after the light injury (Supplementary Fig. S6B), vertical lines of staining consistent with Muller glia start appearing. Maximum staining of activated Muller glia is seen at day 3 and is decreased by day 5 (Supplementary Figs. S6C, S6D). Thus, using Gfap as a marker of activation, Mu-1 cells were split into Gfap+ versus Gfap− subgroups to determine if we could find activation-specific gene expression changes. First, to corroborate that both subgroups of cells were indeed Muller cells, we compared their expression of known Muller cell markers and found no difference in their expression (Rlbp1, ApoE, and Car14). Interestingly, despite the relatively low number of cells in the analysis, we were able to identify several activation-related genes including Serpina3n and Bmp1 (Fig. 7A). Serpina3n encodes for a member of the serpin family of proteins and is known to be involved in neurodegenerative diseases including Alzheimer's disease and Parkinson disease. In particular, it seems to play a role in the response to elevated platelet cytosolic Ca2+ and also in the activation of the innate immune system, likely through its inhibition of serine proteases. Bmp1 is a metalloprotease known to have collagen C-protease activity. It is involved in collagen maturation and plays a key role in regulating the formation of extracellular matrix. In addition, Bmp1-related signaling may promote survival of retinal cells and modulate immune cell processes.
Figure 7.
Differential expression of genes and signaling pathways in activated Mu-1 glial cells. (A) Violin plots show that the expression of Gfap, Serpina3n, and Bmp1 are differentially upregulated in Gfap+ cells at 48 hours compared with Gfap– cells. Muller cell-specific markers ApoE, Rlbp1, and Car14 have similar expression levels in both cell populations. (B) Results of pathway analysis (IPA) using all differentially expressed genes between Gfap+ and Gfap– cells show upregulation of EIF2 signaling, oxidative phosphorylation, ephrin receptor signaling, Reelin signaling in neurons, and synaptogenesis signaling pathways in Gfap+ Mu-1 cells.
We then used IPA for pathway analysis comparing Gfap+ cells versus Gfap– cells. Interestingly, the IPA results show that EIF2 signaling is highly upregulated in the Gfap+ cells (Fig. 7B). This finding is striking because this pathway is found to be highly downregulated in all major cell types at all time points after light injury (except in Mu-1 cells at day 5). Of note, EIF2 is thought to be essential for activation of the protein synthesis machinery. Furthermore, Gfap+ cells also show upregulation of the ephrin receptor signaling, Reelin signaling in neurons, and synaptogenesis signaling pathways. This finding is consistent with the finding in Mu-1 cells and still opposite to that of all other cell types.
The Canonical Signaling Pathways Identified by IPA in Muller Cells 48 Hours and 5 Days After Injury Suggest Retinal Recovery Processes
Synaptogenesis Signaling Pathway in Muller Cells and Rod Photoreceptors
Because the IPA analysis of the scRNA-seq data on mouse retina showed upregulation of the synaptogenesis signaling pathway in Muller cells at 48 hours and day 5 after light injury, we explored the potential significance of specific genes in this pathway further (Supplementary Fig. S7). We found upregulation of Neurexin, a gene that is known to be associated with presynaptic terminal differentiation and synapse organization toward the intermediate and late time points. There was also moderate upregulation of several genes affecting neurite outgrowth and synaptic vesicle docking and binding (Camk2d, Pka, Synapsin, Snap25, Complexin, Synuclein, SGTA, and synaptotagmin). Interestingly, several genes relating to the postsynaptic neuron, as well as several genes leading to microtubule stabilization and synaptic spine development, were also upregulated moderately. These genes included Map1b, Mapt, Akt, Rasgrp2, Camkk2d, Pka, and p38Mapk. Furthermore, the increased expression of EphA and EphB may also promote synaptic spine development via a decreased expression of Cdk5 and Arp2 (Supplementary Fig. S7). This result is also interesting because, as discussed elsewhere in this article, increased expression of EphA and EphB play an important role in the upregulation of the ephrin receptor signaling pathway.
The Ephrin Receptor Signaling Pathway Is Induced in Muller Cells at 48 Hours and Day 5
Upregulation of the ephrin receptor signaling pathway was seen only in Muller cells at 48 hours and day 5 after light injury and not in any other cell types (Supplementary Fig. S8). Ephrin receptor signaling through the activation of ephrin receptors (EphA and EphB) mediates a number of biological processes, including cell adhesion, cell proliferation, cell migration, and axon guidance. Upregulation of EphB leads to both dendritic spine morphogenesis and cell morphology changes via cytoskeleton reorganization. Upregulation of EphA leads to cell proliferation through Stat3 or MAP2K/ERK signaling.
Reelin Signaling in Neurons Pathway in Muller Cells
IPA also showed upregulation of the Reelin signaling in neurons pathway only in Muller cells. This was seen both at 48 hours and day 5 after light injury (Supplementary Fig. S9). This pathway is known to be involved in cytoskeletal rearrangement and dendrite outgrowth and neurogenesis. We found increased expression of Jip and Rhogef, and there was also a downregulation of RhoA. These three genes are involved in cytoskeletal rearrangement. We also found an upregulation of Pi3k and Akt, which are involved in dendrite outgrowth and neurogenesis.
Discussion
The strongest evidence for the existence and importance of a retinal recovery response after insults comes from studies in human patients with multiple retinal disorders. These studies document a clinical course characterized by initial anomalies of the photoreceptor outer segments on OCT, followed by partial recovery that, when insufficient, leads to vision loss. Some clinical scenarios include retinal phototoxicity,13,14 postphotodynamic therapy toxicity,10,11 recovery after repair of macula-OFF retinal detachments,15–17 patients with central serous retinopathy,12 AMD,20 and inflammatory diseases like Vogt–Koyanagi–Harada18 and acute idiopathic blind spot enlargement.19 Similar clinical and OCT findings have been described in nonhuman primates after light-induced macular damage.48,49 In their study, Mukai et al.49 found that, shortly after sublethal light injury to the retina, they could document a change in outer retinal reflectivity on OCT, damage to the photoreceptors on light microscopy, and a decrease in the ERG signal. All of these changes demonstrated significant recovery within 2 weeks. Also, in a 2017 publication, Sudharsan et al.50 state that, “Transient alterations in IS/OS [inner segment/outer segment] structure have been previously demonstrated by OCT imaging in RHO T4R dogs exposed to similar low doses of light, suggesting that a repair mechanism of OS may take place over time after a sublethal insult.” Although in vitro studies have shown that ARPE-19 cells are able to recover to some extent from sublethal oxidative injury,51,52 the investigators have concluded that more work needs to be devoted to understanding the mechanisms, particularly using in vivo models.
To study the important and therapeutically exploitable process of retinal recovery from injury, an appropriate test system is needed. We have developed and validated a FCD-LIRD model33,34 that faithfully reproduces the patterns of changes described elsewhere in this article in clinical entities. Like in many of these conditions, oxidative stress and inflammation are important disease mechanisms in the FCD-LIRD model. Also, ellipsoid zone and photoreceptor outer segment changes, accompanied by decreased visual function, are anatomical and functional changes shared by our FCD-LIRD model and these retinal diseases. Finally, as seen in several of these conditions, these anatomic and functional changes can be followed by partial recovery. In contrast, although in FCD-LIRD the insult to the outer retina is transient, this is only the case in some situations in these eye conditions (e.g., acute CSR, retinal detachment after repair, and phototoxicity). But in many clinical scenarios (e.g., AMD, retinal dystrophies, chronic CSR, chronic retinal detachment), the insult is chronic and progressive. Still, the FCD-LIRD model provides us with a test system that can be used to understand general pathways of retinal recovery from sublethal injury.
The FCD-LIRD model is effective in mice expressing the 450 Met variant of RPE65, including B6J mice and many genetically modified mice, and is efficient, titratable, and reproducible. As a side note, mice carrying the 450 Met variant of RPE65 have a lower level of susceptibility to light that is more similar to what is seen in humans, when compared with the highly susceptible albino mice (carrying the 450 Leu variant) that have been used in most published LIRD studies. Finally, we have shown that mice simultaneously deficient in SOD1, DJ1, and Parkin demonstrate a pronounced increase in retinal/RPE damage after FCD-LIRD.33 They also have a stronger acute upregulation of oxidative response genes. However, these triple knockout mice still are able to mount a significant outer retinal recovery. Similarly, Nrf2 knockout mice are also able to mount a recovery response.33 These data suggest that the recovery phase is mediated by oxidative stress response/Nrf2-independent mechanisms.
Importantly, we have observed that the severity of the stimulus not only determines the severity of the retinal damage, but also the ability of the retina to recover.33,34,43 In designing our experiments, one of the principal considerations was choosing a mild light stimulus. This factor was not only essential to generating sublethal injury that would allow for recovery, but was also meant to better simulate typical pathophysiological levels of injury in common retinal diseases. We were able to confirm that our sublethal stimulus generated a mild but visible level of injury that we could document on OCT by day 5, and that showed evidence of recovery by day 14. Based on this timeline, we hypothesized that we would be able to find evidence of activation of the biological processes responsible and leading to cellular recovery starting before the time of maximal injury. Hence, we collected retinal cells for scRNA-seq at and before day 5 after light injury. Our scRNA-seq studies generated clustering of cell types that was similar to that described by others.42,45
One intriguing finding seen 5 days after light injury (Fig. 2E) is a hyper-reflective aberration extending from the RPE anteriorly into the ONL, which is strongly reminiscent of the RPE plumes described by Cao D et al. in the setting of AMD, which they suggest represent ectopic RPE that has moved along the path of Muller cells.53 As they explain, there is also a possibility that other cells, such as microglia and/or macrophages, may also be involved. Owing to the unpredictable nature of the occurrence and exact location of this finding, solving this debate would require extensive histological and immunohistochemistry work, which is outside the scope of the current work.
The absence of microglia and astrocyte clusters in our analysis deserves special attention. First, we analyzed the Seurat files and the entire expression profiles across all the time points to identify these cells. Using Tmem119 and Fcrls as microglia markers, we only found a very low number of positive cells (4–9 cells at different time points). This corresponds with less than 0.2% of isolated cells, which was not sufficient for the unbiased clustering algorithm to identify them as a cluster. These cells were included within the vascular endothelium cluster (Supplementary Fig. S1), which we did not analyze. Importantly, our data fit very well with prior studies reporting that microglia only make up approximately 0.2% of isolated retinal cells in snRNA-seq analyses.42 Specific studies of microglia would require enrichment techniques.54 Moreover, the light injury model did not change the clustering profile or the number of microglia cells. This result could perhaps be explained by the fact that only a small percentage of the total retinal microglia become activated after our mild light injury and migrate to the subretinal space. Finally, we know that, when we isolate the retina for dissociation, any activated subretinal microglia remain attached to the RPE and are thus excluded from the analysis.55,56 Regarding astrocytes, we also found very few cells positive for the astrocyte markers GFAP or S100b (only 0%–0.3% of total cells had expression of >1 for either of these markers), which is consistent with other studies (approximately 0.1% was reported by Macosko et al.42). Moreover, most of these cells were GFAP+, S100b– cells detected at the 48-hour time point, which as we discussed elsewhere in this article represent activated Muller glia rather than astrocytes. Although the GFAP+ or S100b+ cells were included in the Muller glia clusters (Supplementary Fig. S2), even within these two clusters they accounted for only 0% to 3% of the cells. Thus, astrocyte gene expression did not seem to influence our findings significantly.
Using our clustering analysis, we were able to find some interesting time-dependent gene expression changes that were specific to the different cell types. We found that gene expression changes in Muller cells were seen predominantly at the earlier time points, whereas responses from RGCs were seen predominantly at the late time point, with photoreceptor responses distributed across all time points. A potential explanation for our observation that many of the pathways of interest that could be related to retinal recovery in Muller cells were seen in the late time points would be that, even though gene expression changes that control the recovery response are of a fairly small magnitude, these small changes are still able to have an impact when they are orchestrated to occur in concert.
Interestingly, in almost all cell types, we find upregulation of the mitochondrial dysfunction and granzyme A signaling pathways and downregulation of oxidative phosphorylation and EIF2 signaling pathways. Mitochondrial dysfunction is considered a core pathological process in neurodegenerative disorders, including retinal diseases such as AMD. It is related intimately to crises of energy production, especially oxidative phosphorylation.57–59 In models such as the FCD-LIRD, which mount oxidative/inflammatory responses, the expression of granzymes, cell death-inducing enzymes, is upregulated and implicated in immune regulation and inflammatory pathways. Granzyme A signaling is involved in several biological processes, including cell cytotoxicity, cell death, and mitochondrial and DNA damage.60
In rods, cones, and RGCs, the combination of GSEA and IPA results suggest a decrease in metabolic activity, perhaps as a survival strategy. The results also suggest that by day 5 some pathways related to neurogenesis, perhaps related to retinal recovery, were starting to be enriched in rods.
Muller cells span the entire width of the retina and are important in metabolic processes supporting the retina.61 When the photoreceptors suffer from stress, the neighboring Muller cells initiate cellular signaling and differentiation cascades. The upregulation of Gfap, a known activation marker of Muller cells, at 48 hours after light injury may possibly correlate with the upregulation of recovery pathways and biological processes. The capacity of Muller cells to mount recovery responses in neurons upon retinal injury has been studied before both in mice62–64 and also in retinal regeneration-competent species such as zebrafish and chick.62 The retinal recovery mechanism, especially after photoreceptor losses, happens concomitant with the activation of Muller cells.65 The reports, however, are conflicting regarding the role Muller cell activation (gliosis) plays in favoring retinal regeneration versus degeneration,66 depending on the experimental setting and the model used. The accumulated data suggest that, when the injury is severe, chronic gliosis favors retinal degeneration, whereas, under mild and acute retinal injury, Muller cell activation guides retinal cell survival. However, to our knowledge, this report is the first that implicates Muller cells as playing an important role in mouse retinal recovery from a mild light injury. We have reported previously retinal recovery from light injury supported by structural and functional analyses33,34,43,44 and now we are starting to understand the mechanisms behind this recovery process.
We were able to identify two different subpopulations of Muller cells: Mu-1 and Mu-2. Intriguingly, Mu-1 cells showed a very distinct pattern of activation of three pathways that may be associated to a recovery response: the synaptogenesis signaling, ephrin receptor signaling, and Reelin signaling in neurons pathways. This was noticed when comparing them with rods, cones, cone bipolar, amacrine, RGCs, and even Mu-2 cells. It was also interesting to see that these pathways were also enriched in activated (Gfaphi) Mu-1 cells. Another pathway of interest was EIF2 signaling, a pathway that has been proposed to be activated when cells need to increase anabolic processes (protein synthesis). It has been proposed previously that the downregulation of EIF2 signaling may facilitate cell survival, perhaps by decreasing the metabolic needs of cells.67 We found that the EIF2 signaling pathway was indeed strongly downregulated in most cell types at day 5. The exception was the Mu-1 cluster, in which EIF2 signaling activation seemed to be similar to baseline. Moreover, both at 48 hours and day 5, Mu-1 cells are the only cell type showing upregulation of the synaptogenesis signaling, ephrin receptor signaling, and Reelin signaling in neurons pathways. These pathways are involved in neuronal synapsis health, affecting both the presynaptic and postsynaptic cells. They also stimulate neurite outgrowth and neurogenesis.
Based on these findings, we decided to look specifically at activated (Gfap+) Muller cells. Accompanying Muller cell activation, we found expression of Serpina3n and Bmp1. Serpina3n is known to be involved in neurodegenerative diseases and in the activation of the immune system. Bmp1 is a metalloprotease involved in collagen maturation. It is known to play a key role in regulating the formation of the extracellular matrix. In addition, Bmp1 may promote survival of retinal cells and modulate immune cell processes. Moreover, IPA results comparing Gfap+ versus Gfap− cells revealed strong activation of EIF2, suggesting that different to other retinal cells undergoing decreased activity (decreased EIF2), the Gfap+ Muller cells have strong activation of the anabolic/protein synthesis pathways. These cells also show enrichment of the ephrin receptor signaling, Reelin signaling in neurons, and synaptogenesis signaling pathways. All these data suggest that a subpopulation of Muller cells are the conductors in the retinal recovery concert.
With regard to RGCs, the finding of strong gene expression changes is interesting for two reasons. First, this activation is happening even though the light injury model tends to affect more of the outer retina. Second, the gene expression changes peak at day 5. Both of these issues suggest that the RGCs activation may not be a direct response to the light stimulus, but perhaps a response triggered by other cells in the retina, perhaps in response to the activated Muller cells. Meanwhile, it was puzzling to note that, although they showed a strong tendency toward gene expression changes at approximately the day 5 time point, we were not able to identify recovery-suggestive pathways in this cluster. A potential explanation would be that the recovery response involving some of the retina cell types may be unique when compared with other tissues and, thus, may not have been reported in the named pathways within the IPA, Gene Ontology, and GSEA databases.
In conclusion, we believe that our findings could be explained by invoking a model in which FCD-LIRD causes photo-oxidative damage. The increased oxidative stress leads to damage of mitochondrial DNA, causing mitochondrial dysfunction (upregulation of the mitochondrial dysfunction pathway). This result is seen in multiple neurodegenerative diseases, glaucoma, and AMD.57–59,68 This dysfunction is expressed by a decrease in the ability of mitochondria to generate energy (downregulation of the oxidative phosphorylation pathway) and by an increase in the expression of granzymes (upregulation of the granzyme A pathway). Meanwhile, the decrease in energy production may trigger a survival response in most cell types, involving the shutdown of protein synthesis (downregulation of the EIF2 pathway).69 However, the injury leads to a different gene expression response pattern in Muller 1 cells that suggests that they may be the orchestrators of the recovery response. Although Mu-1 cells share some of the above responses with other cell types, by day 5 the EIF2 pathway stops being downregulated. Moreover, different from all other cell types, by 48 hours Muller 1 cells are already showing upregulation of several pathways that may be related to a neuronal recovery response (synaptogenesis, ephrin receptor, Reelin in neurons, and IL-8 signaling pathways).
Limitations, Prospects, and Future Directions
Based on our study design, the main predicted caveat of choosing a mild, sublethal stimulus was that we would expect less dramatic gene expression changes. As expected, rather than a few dramatic changes in gene expression, we saw low, but statistically significant, changes in gene expression in a cluster of genes, which we found to be driving the important cell signaling pathways discussed elsewhere in this article. A second limitation was that we likely only detected a portion of all the genes expressed in any given cell type. Quality control measures showed that we achieved a mean of 32,519 to 62,875 reads per cell for the different timepoints. We also observed a median number of genes per cell ranging from 1039 to 1774. These statistics are consistent with prior reports applying the 10X Genomics platform to retinal samples, but may be lower than for other platforms.54,70 Our data may indicate a need to add a later time point to detect activation of the recovery phase in other cell types like photoreceptors and RGCs. Perhaps different levels of light intensity in the FCD-LIRD model should also be studied. Promoting retinal/RPE recovery from injury may result in new therapeutic approaches to retinal diseases. This work has two important implications. First, treatments promoting retinal recovery may be helpful to many retinal diseases that cause sublethal injury, independent of the mechanism of injury. Second, drugs promoting retinal recovery from injury would work on different pathways when compared with disease-specific agents, and thus may have added benefits.
Supplementary Material
Acknowledgments
Disclosure: B. Aredo, None; A. Kumar, None; B. Chen, None; C. Xing, None; R.L. Ufret-Vincenty, None
References
- 1. Chen Y, Palczewska G, Masuho I, et al.. Synergistically acting agonists and antagonists of G protein-coupled receptors prevent photoreceptor cell degeneration. Sci Signal. 2016; 9(438): ra74. PMID 27460988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen Y, Sawada O, Kohno H, et al.. Autophagy protects the retina from light-induced degeneration. J Biol Chem. 2013; 288(11): 7506–7518. PMID 23341467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Coyner AS, Ryals RC, Ku CA, et al.. Retinal neuroprotective effects of flibanserin, an FDA-approved dual serotonin receptor agonist-antagonist. PLoS One. 2016; 11(7): e0159776. PMID 27447833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Grimm C, Reme CE. Light damage models of retinal degeneration. Methods Mol Biol. 2019; 1834: 167–178. PMID 30324444. [DOI] [PubMed] [Google Scholar]
- 5. Kunchithapautham K, Coughlin B, Lemasters JJ, Rohrer B. Differential effects of rapamycin on rods and cones during light-induced stress in albino mice. Invest Ophthalmol Vis Sci. 2011; 52(6): 2967–2975. PMID 21273550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li GY, Fan B, Jiao YY. Rapamycin attenuates visible light-induced injury in retinal photoreceptor cells via inhibiting endoplasmic reticulum stress. Brain Res. 2014; 1563: 1–12. PMID 24607296. [DOI] [PubMed] [Google Scholar]
- 7. Narimatsu T, Negishi K, Miyake S, et al.. Blue light-induced inflammatory marker expression in the retinal pigment epithelium-choroid of mice and the protective effect of a yellow intraocular lens material in vivo. Exp Eye Res. 2015; 132: 48–51. PMID 25576667. [DOI] [PubMed] [Google Scholar]
- 8. Rapp LM, Smith SC. Morphologic comparisons between rhodopsin-mediated and short-wavelength classes of retinal light damage. Invest Ophthalmol Vis Sci. 1992; 33(12): 3367–3377. PMID 1428709. [PubMed] [Google Scholar]
- 9. Song D, Song J, Wang C, Li Y, Dunaief JL. Berberine protects against light-induced photoreceptor degeneration in the mouse retina. Exp Eye Res. 2016; 145: 1–9. PMID 26475979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fujita K, Shinoda K, Imamura Y, et al.. Correlation of integrity of cone outer segment tips line with retinal sensitivity after half-dose photodynamic therapy for chronic central serous chorioretinopathy. Am J Ophthalmol. 2012; 154(3): 579–585. PMID 22818904. [DOI] [PubMed] [Google Scholar]
- 11. Gutierrez-Hernandez JC, Martinez-Camarillo JC, Sadda SR. Long-term follow-up of photoreceptor loss and recovery after half-fluence photodynamic therapy for chronic central serous chorioretinopathy. Retin Cases Brief Rep. 2015; 9(2): 109–113. PMID 25383847. [DOI] [PubMed] [Google Scholar]
- 12. Ojima Y, Tsujikawa A, Yamashiro K, Ooto S, Tamura H, Yoshimura N. Restoration of outer segments of foveal photoreceptors after resolution of central serous chorioretinopathy. Jpn J Ophthalmol. 2010; 54(1): 55–60. PMID 20151277. [DOI] [PubMed] [Google Scholar]
- 13. Costagliola C, Menzione M, Chiosi F, Romano MR, Della Corte M, Rinaldi M. Retinal phototoxicity induced by hydrochlorothiazide after exposure to a UV tanning device. Photochem Photobiol. 2008; 84(5): 1294–1297. PMID 18673326. [DOI] [PubMed] [Google Scholar]
- 14. Cartwright K, Tole DM, Haynes RJ, Males JJ, Dick AD, Mayer EJ. Recovery from macular phototoxicity after corneal triple procedure. Cornea. 2007; 26(1): 102–104. PMID 17198023. [DOI] [PubMed] [Google Scholar]
- 15. dell'Omo R, Viggiano D, Giorgio D, et al.. Restoration of foveal thickness and architecture after macula-off retinal detachment repair. Invest Ophthalmol Vis Sci. 2015; 56(2): 1040–1050. PMID 25613940. [DOI] [PubMed] [Google Scholar]
- 16. Nakanishi H, Hangai M, Unoki N, et al.. Spectral-domain optical coherence tomography imaging of the detached macula in rhegmatogenous retinal detachment. Retina. 2009; 29(2): 232–242. PMID 18997641. [DOI] [PubMed] [Google Scholar]
- 17. Ra E, Ito Y, Kawano K, et al.. Regeneration of photoreceptor outer segments after scleral buckling surgery for rhegmatogenous retinal detachment. Am J Ophthalmol. 2017; 177: 17–26. PMID 28189482. [DOI] [PubMed] [Google Scholar]
- 18. Bae SS, Forooghian F. Optical coherence tomography-based quantification of photoreceptor injury and recovery in Vogt-Koyanagi-Harada uveitis. Ocul Immunol Inflamm. 2017; 25(3): 338–343. PMID 26903383. [DOI] [PubMed] [Google Scholar]
- 19. Horton JC, Parker AB, Botelho JV, Duncan JL. Spontaneous regeneration of human photoreceptor outer segments. Sci Rep. 2015; 5: 12364. PMID 26213154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Oishi A, Shimozono M, Mandai M, Hata M, Nishida A, Kurimoto Y. Recovery of photoreceptor outer segments after anti-VEGF therapy for age-related macular degeneration. Graefes Arch Clin Exp Ophthalmol. 2013; 251(2): 435–440. PMID 22576370. [DOI] [PubMed] [Google Scholar]
- 21. Delcourt C, Cougnard-Grégoire A, Boniol M, et al.. Lifetime exposure to ambient ultraviolet radiation and the risk for cataract extraction and age-related macular degeneration: the Alienor Study. Invest Ophthalmol Vis Sci. 2014; 55(11): 7619–7627. PMID 25335979. [DOI] [PubMed] [Google Scholar]
- 22. Paskowitz DM, LaVail MM, Duncan JL. Light and inherited retinal degeneration. Br J Ophthalmol. 2006; 90(8): 1060–1066. PMID 16707518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tomany SC, Cruickshanks KJ, Klein R, Klein BE, Knudtson MD. Sunlight and the 10-year incidence of age-related maculopathy: the Beaver Dam Eye Study. Arch Ophthalmol. 2004; 122(5): 750–757. PMID 15136324. [DOI] [PubMed] [Google Scholar]
- 24. White DA, Fritz JJ, Hauswirth WW, Kaushal S, Lewin AS. Increased sensitivity to light-induced damage in a mouse model of autosomal dominant retinal disease. Invest Ophthalmol Vis Sci. 2007; 48(5): 1942–1951. PMID 17460245. [DOI] [PubMed] [Google Scholar]
- 25. Hadziahmetovic M, Kumar U, Song Y, et al.. Microarray analysis of murine retinal light damage reveals changes in iron regulatory, complement, and antioxidant genes in the neurosensory retina and isolated RPE. Invest Ophthalmol Vis Sci. 2012; 53(9): 5231–5241. PMID 22736611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Joly S, Francke M, Ulbricht E, et al.. Cooperative phagocytes: resident microglia and bone marrow immigrants remove dead photoreceptors in retinal lesions. Am J Pathol. 2009; 174(6): 2310–2323. PMID 19435787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. O'Koren EG, Mathew R, Saban DR. Fate mapping reveals that microglia and recruited monocyte-derived macrophages are definitively distinguishable by phenotype in the retina. Sci Rep. 2016; 6: 20636. PMID 26856416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rohrer B, Guo Y, Kunchithapautham K, Gilkeson GS. Eliminating complement factor D reduces photoreceptor susceptibility to light-induced damage. Invest Ophthalmol Vis Sci. 2007; 48(11): 5282–5289. PMID 17962484. [DOI] [PubMed] [Google Scholar]
- 29. Song D, Song Y, Hadziahmetovic M, Zhong Y, Dunaief JL. Systemic administration of the iron chelator deferiprone protects against light-induced photoreceptor degeneration in the mouse retina. Free Radic Biol Med. 2012; 53(1): 64–71. PMID 22579919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wenzel A, Grimm C, Samardzija M, Reme CE. Molecular mechanisms of light-induced photoreceptor apoptosis and neuroprotection for retinal degeneration. Prog Retin Eye Res. 2005; 24(2): 275–306. PMID 15610977. [DOI] [PubMed] [Google Scholar]
- 31. Zheng L, Anderson RE, Agbaga MP, Rucker EB 3rd, Le YZ. Loss of BCL-XL in rod photoreceptors: increased susceptibility to bright light stress. Invest Ophthalmol Vis Sci. 2006; 47(12): 5583–5589. PMID 17122152. [DOI] [PubMed] [Google Scholar]
- 32. Redfern WS, Storey S, Tse K, et al.. Evaluation of a convenient method of assessing rodent visual function in safety pharmacology studies: effects of sodium iodate on visual acuity and retinal morphology in albino and pigmented rats and mice. J Pharmacol Toxicol Methods. 2011; 63(1): 102–114. PMID 20619348. [DOI] [PubMed] [Google Scholar]
- 33. Ding Y, Aredo B, Zhong X, Zhao CX, Ufret-Vincenty RL. Increased susceptibility to fundus camera-delivered light-induced retinal degeneration in mice deficient in oxidative stress response proteins. Exp Eye Res. 2017; 159: 58–68. PMID 28336262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhong X, Aredo B, Ding Y, et al.. Fundus camera-delivered light-induced retinal degeneration in mice with the RPE65 Leu450Met variant is associated with oxidative stress and apoptosis. Invest Ophthalmol Vis Sci. 2016; 57(13): 5558–5567. PMID 27768794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Aredo B, Zhang K, Chen X, et al.. Differences in the distribution, phenotype and gene expression of subretinal microglia/macrophages in C57BL/6N (Crb1 rd8/rd8) versus C57BL6/J (Crb1 wt/wt) mice. J Neuroinflammation. 2015; 12: 6. PMID 25588310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Barres BA, Silverstein BE, Corey DP, Chun LL. Immunological, morphological, and electrophysiological variation among retinal ganglion cells purified by panning. Neuron. 1988; 1(9): 791–803. PMID 2908449. [DOI] [PubMed] [Google Scholar]
- 37. Chen B, Aredo B, Ding Y, et al.. Forward genetic analysis using OCT screening identifies Sfxn3 mutations leading to progressive outer retinal degeneration in mice. Proc Natl Acad Sci USA. 2020; 117(23): 12931–12942. PMID 32457148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Feodorova Y, Koch M, Bultman S, Michalakis S, Solovei I. Quick and reliable method for retina dissociation and separation of rod photoreceptor perikarya from adult mice. MethodsX. 2015; 2: 39–46. PMID 26150970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu YE, Pan L, Zuo Y, Li X, Hong W. Detecting activated cell populations using single-cell RNA-seq. Neuron. 2017; 96(2): 313–329.e6. PMID 29024657. [DOI] [PubMed] [Google Scholar]
- 40. Bensaude O. Inhibiting eukaryotic transcription: Which compound to choose? How to evaluate its activity? Transcription. 2011; 2(3): 103–108. PMID 21922053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stuart T, Butler A, Hoffman P, et al.. Comprehensive integration of single-cell data. Cell. 2019; 177(7): 1888–1902.e21. PMID 31178118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Macosko EZ, Basu A, Satija R, et al.. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015; 161(5): 1202–1214. PMID 26000488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen B, Aredo B, Zhu Y, Ding Y, Xin-Zhao C, Ufret-Vincenty RL. A Mouse model of retinal recovery from photo-oxidative/photo-inflammatory injury: Nrf2, SOD1, DJ-1, and Parkin are not essential to recovery. Invest Ophthalmol Vis Sci. 2019; 60(4): 1165–1174. PMID 30908580. [DOI] [PubMed] [Google Scholar]
- 44. Peng H, Ramadurgum P, Woodard DR, et al.. Utility of the DHFR-based destabilizing domain across mouse models of retinal degeneration and aging. iScience. 2022; 25(5): 104206. PMID 35521529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jeon CJ, Strettoi E, Masland RH. The major cell populations of the mouse retina. J Neurosci. 1998; 18(21): 8936–8946. PMID 9786999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tuccari G, Trombetta C, Giardinelli MM, Arena F, Barresi G. Distribution of glial fibrillary acidic protein in normal and gliotic human retina. Basic Appl Histochem. 1986; 30(4): 425–432. PMID 3548695. [PubMed] [Google Scholar]
- 47. Verardo MR, Lewis GP, Takeda M,et al. Abnormal reactivity of muller cells after retinal detachment in mice deficient in GFAP and vimentin. Invest Ophthalmol Vis Sci. 2008; 49(8): 3659–3665. PMID 18469190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jaffe GJ, Irvine AR, Wood IS, Severinghaus JW, Pino GR, Haugen C. Retinal phototoxicity from the operating microscope. The role of inspired oxygen. Ophthalmology. 1988; 95(8): 1130–1141. PMID 3068607. [DOI] [PubMed] [Google Scholar]
- 49. Mukai R, Akiyama H, Tajika Y, Shimoda Y, Yorifuji H, Kishi S. Functional and morphologic consequences of light exposure in primate eyes. Invest Ophthalmol Vis Sci. 2012; 53(10): 6035–6044. PMID 22807297. [DOI] [PubMed] [Google Scholar]
- 50. Sudharsan R, Simone KM, Anderson NP, Aguirre GD, Beltran WA. Acute and protracted cell death in light-induced retinal degeneration in the canine model of rhodopsin autosomal dominant retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2017; 58(1): 270–281. PMID 28114588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Baek SM, Yu SY, Son Y, Hong HS. Substance P promotes the recovery of oxidative stress-damaged retinal pigmented epithelial cells by modulating Akt/GSK-3beta signaling. Mol Vis. 2016; 22: 1015–1023. PMID 27582624. [PMC free article] [PubMed] [Google Scholar]
- 52. Strunnikova N, Zhang C, Teichberg D, et al.. Survival of retinal pigment epithelium after exposure to prolonged oxidative injury: a detailed gene expression and cellular analysis. Invest Ophthalmol Vis Sci. 2004; 45(10): 3767–3777. PMID 15452088. [DOI] [PubMed] [Google Scholar]
- 53. Cao D, Leong B, Messinger JD, et al.. Hyperreflective foci, optical coherence tomography progression indicators in age-related macular degeneration, include transdifferentiated retinal pigment epithelium. Invest Ophthalmol Vis Sci. 2021; 62(10): 34. PMID 34448806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hammond TR, Dufort C, Dissing-Olesen L, et al.. Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity. 2019; 50(1): 253–271.e6. PMID 30471926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ng TF, Streilein JW. Light-induced migration of retinal microglia into the subretinal space. Invest Ophthalmol Vis Sci. 2001; 42(13): 3301–3310. PMID 11726637. [PubMed] [Google Scholar]
- 56. Rashid K, Akhtar-Schaefer I, Langmann T. Microglia in retinal degeneration. Front Immunol. 2019; 10: 1975. PMID 31481963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Johri A, Beal MF. Mitochondrial dysfunction in neurodegenerative diseases. J Pharmacol Exp Ther. 2012; 342(3): 619–630. PMID 22700435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kaarniranta K, Uusitalo H, Blasiak J, et al.. Mechanisms of mitochondrial dysfunction and their impact on age-related macular degeneration. Prog Retin Eye Res. 2020; 79: 100858. PMID 32298788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wu Y, Chen M, Jiang J. Mitochondrial dysfunction in neurodegenerative diseases and drug targets via apoptotic signaling. Mitochondrion. 2019; 49: 35–45. PMID 31288090. [DOI] [PubMed] [Google Scholar]
- 60. Ewen CL, Kane KP, Bleackley RC. A quarter century of granzymes. Cell Death Differ. 2012; 19(1): 28–35. PMID 22052191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Willbold E, Layer PG. Muller glia cells and their possible roles during retina differentiation in vivo and in vitro. Histol Histopathol. 1998; 13(2): 531–552. PMID 9589907. [DOI] [PubMed] [Google Scholar]
- 62. Hoang T, Wang J, Boyd P, et al.. Gene regulatory networks controlling vertebrate retinal regeneration. Science. 2020; 370(6519): eabbb8598. PMID 33004674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kang S, Larbi D, Andrade M, et al.. A Comparative analysis of reactive muller glia gene expression after light damage and microRNA-Depleted Muller glia-focus on microRNAs. Front Cell Dev Biol. 2020; 8: 620459. PMID 33614628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tomita Y, Qiu C, Bull E, et al.. Muller glial responses compensate for degenerating photoreceptors in retinitis pigmentosa. Exp Mol Med. 2021; 53(11): 1748–1758. PMID 34799683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hippert C, Graca AB, Barber AC, et al.. Muller glia activation in response to inherited retinal degeneration is highly varied and disease-specific. PLoS One. 2015; 10(3): e0120415. PMID 25793273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yoo HS, Shanmugalingam U, Smith PD. Harnessing astrocytes and Muller glial cells in the retina for survival and regeneration of retinal ganglion cells. Cells. 2021; 10(6): 1339. PMID 34071545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM. The integrated stress response. EMBO Rep. 2016; 17(10): 1374–1395. PMID 27629041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Eells JT. Mitochondrial dysfunction in the aging retina. Biology (Basel). 2019; 8(2): 31. PMID 31083549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wek RC. Role of eIF2alpha kinases in translational control and adaptation to cellular stress. Cold Spring Harb Perspect Biol. 2018; 10(7): a032870. PMID 29440070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. See P, Lum J, Chen J, Ginhoux F. A single-cell sequencing guide for immunologists. Front Immunol. 2018; 9: 2425. PMID 30405621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.