Abstract
The efficacy of immunotherapy in advanced HER2‐mutated non‐small‐cell lung cancer (NSCLC) remains incomprehensively studied. A total of 107 NSCLC patients with de novo HER2 mutations were retrospectively studied at Guangdong Lung Cancer Institute [GLCI cohort, exon 20 insertions (ex20ins): 71.0%] to compare clinical/molecular features and immune checkpoint inhibitor (ICI)‐based therapy efficacy between patients with ex20ins and non‐ex20ins. Two external cohorts (TCGA, n = 21; META‐ICI, n = 30) were used for validation. In the GLCI cohort, 68.2% of patients displayed programmed death‐ligand 1 (PD‐L1) expression < 1%. Compared with ex20ins patients, non‐ex20ins patients had more concurrent mutations in the GLCI cohort (P < 0.01) and a higher tumour mutation burden in the TCGA cohort (P = 0.03). Under ICI‐based therapy, advanced NSCLC patients with non‐ex20ins had potentially superior progression‐free survival [median: 13.0 vs. 3.6 months, adjusted hazard ratio (HR): 0.31, 95% confidence interval (CI): 0.11–0.83] and overall survival (median: 27.5 vs. 8.1 months, adjusted HR: 0.39, 95% CI: 0.13–1.18) to ex20ins patients, consistent with findings in the META‐ICI cohort. ICI‐based therapy may serve as an option for advanced HER2‐mutated NSCLC, with potentially better efficacy in non‐ex20ins patients. Further investigations are warranted in clinical practice.
Keywords: advanced NSCLC, exon 20 insertion, human epidermal growth factor receptor 2, immune checkpoint inhibitor, non‐exon 20 insertion
HER2‐mutated non‐small‐cell lung cancer (NSCLC) patients with HER2 mutations other than exon 20 insertions (non‐ex20ins) had higher tumour mutation burden than patients with HER2 ex20ins, whereas similar programmed death‐ligand 1 expression was observed. Under immune checkpoint inhibitor‐based therapy, advanced NSCLC patients with HER2 non‐ex20ins had potentially superior progression‐free survival and overall survival compared with patients with HER2 ex20ins.
Abbreviations
- CI
confidence interval
- ex20ins
exon 20 insertions
- FFPE
formalin‐fixed paraffin‐embedded
- GLCI
Guangdong Lung Cancer Institute
- HER2
human epidermal growth factor receptor 2
- HR
hazard ratio
- ICI
immune checkpoint inhibitor
- IHC
immunohistochemistry
- NGS
next‐generation sequencing
- non‐ex20ins
mutations other than ex20ins
- NSCLC
non‐small‐cell lung cancer
- ORR
objective response rate
- OS/mOS
overall survival/median overall survival
- PD‐1
programmed cell death‐receptor 1
- PD‐L1
programmed cell death‐ligand 1
- PFS/mPFS
progression‐free survival/median progression‐free survival
- TCGA
The Cancer Genome Atlas Program
- T‐DM1
ado‐trastuzumab emtansine
- T‐DXd
trastuzumab deruxtecan
- TKI
tyrosine kinase inhibitor
- TMB
tumour mutational burden
- TME
tumour microenvironment
- TPS
tumour proportion score
1. Introduction
Human epidermal growth factor receptor 2 (HER2/ERBB2) is an oncogenic driver of tumour cell proliferation and metastasis [1]. Mutated HER2 genes are rare (prevalence: 2–4%) in non‐small‐cell lung cancer (NSCLC) patients, and are enriched in patients who are females, non‐smokers and younger, and in adenocarcinoma patients [2, 3]. Over 20 types of HER2 exon 20 insertion (ex20ins) mutations have been identified in NSCLC, accounting for 25–50% of NSCLC patients exhibiting HER2 mutations [4, 5, 6]. However, the genomic and immune characteristics of patients with different HER2 mutations have not been comprehensively investigated.
Advanced NSCLC patients harbouring HER2 mutations have a worse prognosis (median survival: 1.9–2.3 years) [3, 7], compared with those harbouring EGFR or ALK mutations. Treatments targeting HER2 mutations in advanced NSCLC include tyrosine kinase inhibitors (TKIs) and HER2‐antibody‐drug conjugates (ADCs). Objective response rates (ORRs) of treatment with TKIs, such as afatinib [8], dacomitinib [9], or pyrotinib [10, 11], are < 30%. The efficacy of TKIs in NSCLC treatment may depend on HER2 mutation subtypes [12, 13, 14]; however, no significant differences in pyrotinib effectiveness were found between patients with HER2 ex20ins and non‐ex20ins mutations. Under ADC treatment, patients receiving ado‐trastuzumab emtansine (T‐DM1) and trastuzumab deruxtecan (T‐DXd) achieved ORRs of 50% and 55%, respectively, and the median progression‐free survival (mPFS) of T‐DXd treatment was 8.2 months [15, 16]. However, the majority of patients in these studies harboured HER2 ex20ins mutations (73–86%), and there was a scarcity of data on patients carrying HER2 non‐ex20ins mutations [17]. The toxicity of T‐DXd remains an important issue; interstitial lung disease is presented by 26% of T‐DXd users [15], despite the recent approval (by the U.S. Food and Drug Administration) of T‐DXd for patients with unresectable or metastatic HER2‐mutated NSCLC. Overall, there is a need to identify effective and tolerable treatments for patients with advanced HER2‐mutated NSCLC.
Because neither TKIs nor ADCs have been approved as first‐line treatments for HER2‐mutated NSCLC patients with advanced disease [18], immune checkpoint inhibitors (ICIs) with/without platinum doublet chemotherapy serve as the standard first‐line therapy [19]. Treatment‐naive patients receiving ICI combination treatment achieved ORR of 52% and mPFS of 6 months [20], whereas other studies have reported ORRs of 7.0–27.0% and mPFS of 2.2–4.0 months under ICI‐based therapy in second‐line or subsequent treatment [21, 22]. Notably, it has been revealed that none of the ICI responders harboured HER2 YVMA mutation [23], suggesting that ICI efficacy in patients with HER2 ex20ins mutations might differ from patients harbouring other HER2 mutations. Moreover, the efficacy of immunotherapy has not been systematically investigated in HER2 non‐ex20ins patients, owing to the low prevalence of HER2 mutations in NSCLC and the focus on HER2 ex20ins. Thus, the association between HER2 mutation subtypes and immunotherapy efficacy remains controversial, and the potential underlying mechanisms have not been comprehensively investigated.
This study aimed to compare the molecular features and tumour microenvironment (TME) characteristics of NSCLC patients harbouring HER2 ex20ins and HER2 non‐ex20ins mutations. The efficacy of ICI‐based therapy was analysed in patients with advanced disease. Two external datasets of HER2‐mutated NSCLC patients were used to validate the results.
2. Materials and methods
2.1. Patients
This retrospective study was performed at Guangdong Lung Cancer Institute (GLCI), Guangdong Provincial People's Hospital; participants in the GLCI cohort were consecutively enrolled between January 2016 and December 2020. The main inclusion criteria were follows: (1) adults having at least 18 years of age; (2) pathologically confirmed NSCLC, according to the 2016 World Health Organization classification; and (3) identified with somatic de novo HER2 mutations in tissue or plasma samples. Patients with other oncogenic drivers, including sensitizing EGFR mutations, BRAF V600E mutation, KRAS G12X/Q61H mutation, ALK/NTRK/RET/ROS1 fusion, and MET exon 14 skipping, were excluded. Insertion mutations in HER2 exon 20 (residues 770–831) were identified as HER2 ex20ins; HER2 mutations outside exon 20 (residues before 770 or after 831) were identified as HER2 non‐ex20ins mutations. Patients harbouring both HER2 exon 20 and non‐ex20ins mutations were grouped into the non‐ex20ins subgroup. The clinical stages and histological subtypes of NSCLC were determined according to the ‘8th edition of the American Joint Committee on Cancer classification system’. Demographics and clinical characteristics of the participants, including age, sex, Eastern Cooperative Oncology Group performance status, and smoking history were obtained from the electronic medical record system of Guangdong Provincial People's Hospital.
ICI‐based therapy was defined as a regimen that included inhibitors of programmed cell death‐1 receptor or its ligand (PD‐1/PD‐L1). Advanced NSCLC patients in the GLCI cohort treated with ICI monotherapy or ICI combination therapy were grouped as the GLCI‐ICI cohort (Fig. 1), and they were followed up until November 2021 or until death. The clinical response to ICI‐based therapy was evaluated using computed tomography according to the ‘Response Evaluation Criteria in Solid Tumours version 1.1’. The study procedures were approved by the Ethics Committee of Guangdong Provincial People's Hospital (2013185H), and written informed consent was obtained from each patient. The study methodologies also conformed to the standards set by the Declaration of Helsinki.
Fig. 1.
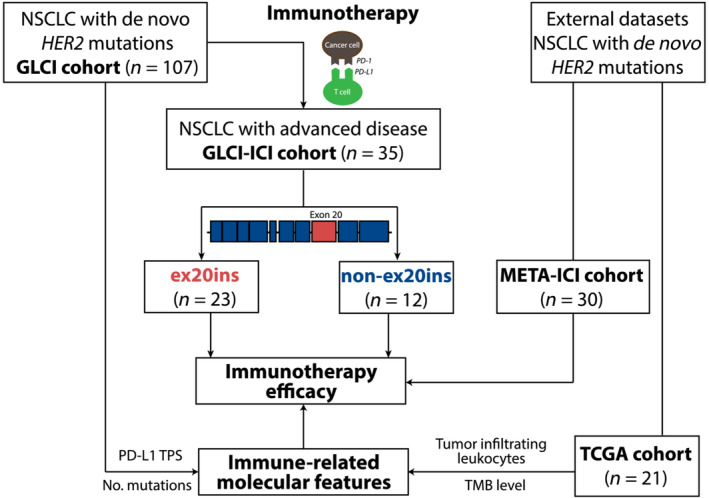
The flow chart of patient enrolment and validation cohorts. A total of 107 NSCLC patients harbouring HER2 mutations were enrolled in the GLCI cohort, including 76 patients with HER2 ex20ins and 31 patients with HER2 non‐ex20ins mutations. The efficacy of ICI‐based therapy and TME features were investigated among 35 of these 107 patients (GLCI‐ICI cohort). Two external datasets of HER2‐mutated NSCLC patients, The Cancer Genome Atlas Program (TCGA, n = 21) and META‐ICI (n = 30) were used for result validation.
2.2. External cohorts
From The Cancer Genome Atlas Program (TCGA), NSCLC patients with de novo HER2 mutations and without other NSCLC oncogenic drivers were included in the TCGA cohort for the validation of genomic features and immune characteristics (Fig. 1). Tumour mutational burden (TMB) of each patient in this cohort was recalculated using a standardized method [24]. The abundance of tumour‐infiltrating leukocytes was estimated by CIBERSORT using RNA‐Seq data, and significantly differentially expressed genes between patients with HER2 ex20ins and HER2 non‐ex20ins mutations were investigated using Gene Set Enrichment Analysis.
From six external data sets, advanced HER2‐mutated NSCLC patients who did not harbour other NSCLC oncogenic drivers and received ICI‐based therapy were grouped into the META‐ICI cohort (Fig. 1), including 10 patients from a study by Rizvi et al. [25], nine patients from a study by Gandara et al. [26], two patients from a study by Miao et al. [27], five patients from a study by Anagnostou et al. [28], and four patients from a study by Samstein et al [29]. Clinicopathological and prognostic data were analysed.
2.3. DNA extraction, library preparation, and next‐generation sequencing data processing
Genomic profiling of tumour tissue or plasma samples before systemic treatment was performed using multiple targeted next‐generation sequencing (NGS) panels, according to the protocol approved by the Ethics Committee of Guangdong Provincial People's Hospital. Tumour genomic DNA was extracted from formalin‐fixed paraffin‐embedded (FFPE) samples using the QIAamp DNA FFPE Tissue Kit (QIAGEN, Dusseldorf, Germany). Genomic DNA was extracted from leukocyte (normal blood controls) using the QIAamp Circulating Nucleic Acid Kit (QIAGEN). Peripheral blood was collected and centrifuged (at 1800 g for 10 min at room temperature) within 2 h to separate the plasma and leukocytes. Cell‐free DNA was extracted from the plasma using a QIAamp Circulating Nucleic Acid Kit (QIAGEN). Sequencing libraries were prepared using the KAPA Hyper Prep Kit (KAPA Biosystems, Wilmington, MA, USA). Briefly, fragmented genomic DNA was subjected to end‐repair, A‐tailing, adapter ligation, size selection, polymerase chain reaction amplification, and purification sequentially. Target enrichment was performed using a customized xGen Lockdown Probes Panel (Integrated DNA Technologies, Coralville, IA, USA), human cot‐1 DNA (Life Technologies) and xGen Universal Blocking Oligos (Integrated DNA Technologies). All procedures were performed according to the manufacturer's instructions. The enriched libraries were sequenced using the Illumina Hiseq4000 NGS platforms (Illumina, San Diego, CA, USA).
trimmomatic was used for quality control of fastq files by removing leading/trailing low quality (reading < 15) or N bases [30]. Sequencing data were then aligned to the reference human genome (build hg19) and processed using the picard suite and the genome analysis toolkit (gatk) [31, 32]. A somatic mutation, filtered for common single nucleotide polymorphisms and germline mutations, was retained when it had at least 1% mutant allele frequency and at least three unique reads on different strands with good quality scores. Gene fusions and copy number variations were analysed using FACTERA and ADTEx [33, 34], respectively, and manually reviewed using integrative genomics viewer Software (igv; Broad Institute, Cambridge, MA, USA). A total of 72 overlapping cancer‐relevant genes from multiple NGS panels were included in the data analysis (Table S1).
2.4. Immunohistochemistry (IHC) for PD‐L1 expression
Formalin‐fixed paraffin‐embedded tumour tissue specimens were stained using PD‐L1 IHC 22C3 pharmDx (Agilent, Santa Clara, CA, USA), and a PD‐L1‐positive cell was defined as complete circumferential or partial cell membrane staining of viable cells with 1+ to 3+ intensity. PD‐L1 protein expression was evaluated using the tumour proportion score (TPS), which was calculated as the percentage of PD‐L1‐positive tumour cells divided by total tumour cells. Patients were categorized into three subgroups according to PD‐L1 TPS: < 1%, 1–49%, and ≥ 50%.
2.5. Statistical analysis
Progression‐free survival was defined as the time from the initiation of ICI‐based therapy to disease progression or death from any cause; overall survival (OS) was defined as the time from the initiation of ICI‐based therapy to death from any cause. The median follow‐up time for the GLCI‐ICI cohort was estimated using the observation time method. Fisher's exact test and two‐sample t‐test were performed to compare the frequencies and means of patients with HER2 ex20ins and HER2 non‐ex20ins mutations, respectively. For survival data, Kaplan–Meier curves for PFS and OS were generated, and log‐rank tests were used to compare differences. Hazard ratios (HR) with 95% confidence intervals (CI) were estimated using Cox proportional hazards models. Multivariable Cox proportional hazards models included clinical and molecular features that were identified as having a potentially strong influence on PFS or OS in univariate analyses or with significantly unbalanced distribution between HER2 ex20ins and non‐ex20ins patients. The proportionality of hazards was assessed using log(−log) survival plots. Individuals with missing data were excluded from analysis. All quoted P‐values were two‐tailed, and P‐values < 0.05 were considered to be statistically significant. Data were analysed using r software (version 4.0.3, Vienna, Austria) and the survival package.
3. Results
3.1. Patient characteristics
A total of 107 eligible patients (76 with HER2 ex20ins and 31 with HER2 non‐ex20ins mutations) were retrospectively enrolled in the GLCI cohort, with only one patient identified to have ex20ins and non‐ex20ins mutations simultaneously. Thirty‐five of patients were identified in the GLCI‐ICI cohort, all of whom received ICI‐based therapy (Fig. 1). The median age of the GLCI cohort was 59 years (range: 24–81). It presented as a majority of adenocarcinoma (99/107, 92.5%), males (60/107, 56.1%), stage IV at initial diagnosis (73/107, 68.2%), and never‐smokers (78/107, 72.9%) (Table 1). Most patients (73/107, 68.2%) had PD‐L1 TPS of < 1%. Compared with patients with HER2 ex20ins mutations, non‐ex20ins patients were older (63 vs. 57 years, P < 0.01), had a greater proportion of smokers (41.9% vs. 21.1%, P = 0.03), and had a lower proportion of adenocarcinoma (83.9% vs. 96.1%, P = 0.02). Two subgroups had similar PD‐L1 expression levels, with over 65% of patients identified with PD‐L1 expression < 1% (< 1%: 71.0% vs. 67.1%, 1–49%: 22.6 vs. 26.3, ≥ 50%: 6.5% vs. 6.6%, P = 0.94).
Table 1.
Demographics and clinical characteristics of GLCI cohort.
| Characteristics | Overall (n = 107) | Ex20ins (n = 76) | Non‐ex20ins (n = 31) | P‐value |
|---|---|---|---|---|
| Age, median (range), year | 59 (24–81) | 57 (24–79) | 63 (43–81) | < 0.01* |
| Age, no. (%) | ||||
| < 60 years | 58 (54.2) | 47 (61.8) | 11 (35.5) | 0.01* |
| ≥ 60 years | 49 (45.8) | 29 (38.2) | 20 (64.5) | |
| Sex, no. (%) | ||||
| Female | 47 (43.9) | 35 (46.1) | 13 (41.9) | 0.83 |
| Male | 60 (56.1) | 41 (53.9) | 18 (58.1) | |
| Clinical stage at initial diagnosis, no. (%) | ||||
| I | 10 (9.3) | 7 (9.2) | 3 (9.7) | 0.73 |
| II | 6 (5.6) | 5 (6.6) | 1 (3.2) | |
| III | 18 (16.8) | 11 (14.5) | 7 (22.6) | |
| IV | 73 (68.2) | 53 (69.7) | 20 (64.5) | |
| Histology, no. (%) | ||||
| Adenocarcinoma | 99 (92.5) | 73 (96.1) | 26 (83.9) | 0.02* |
| Squamous cell carcinoma | 5 (4.9) | 1 (1.3) | 4 (12.9) | |
| Adenosquamous carcinoma | 1 (0.9) | 1 (1.3) | 0 (0.0) | |
| Lymphoepithelioma‐like carcinoma | 1 (0.9) | 0 (0.0) | 1 (3.2) | |
| Not otherwise specified | 1 (0.9) | 1 (1.3) | 0 (0.0) | |
| Smoking, no. (%) | ||||
| Ever | 29 (27.1) | 16 (21.1) | 13 (41.9) | 0.03* |
| Never | 78 (72.9) | 60 (78.9) | 18 (58.1) | |
| PD‐L1 expression, no. (%) | ||||
| < 1% | 73 (68.2) | 51 (67.1) | 22 (71.0) | 0.94 |
| 1–49% | 27 (25.2) | 20 (26.3) | 7 (22.6) | |
| ≥ 50% | 7 (6.5) | 5 (6.6) | 2 (6.5) | |
Statistically significant.
3.2. Genomic features and molecular heterogeneity
Of the 107 patients in the GLCI cohort, seven patients had only HER2 alteration records before systemic treatment, without raw sequencing data. The remaining 100 patients (73 with HER2 ex20ins and 27 with HER2 non‐ex20ins mutations) were included in the molecular feature analyses, and their HER2 mutations are summarized in Fig. 2A. The most frequently identified HER2 ex20ins subtype was Y772_A775dup (39/100, 39.0%), followed by G776delinsVC (10/100, 10.0%), 771insAYVM (6/100, 6.0%), G778_P780dup (5/100, 5.0%), and G776delinsLC (5/100, 5.0%) (Fig. 2B). HER2 non‐ex20ins mutations included V659E, L755A, S335C, etc. (Fig. 2B). The most commonly mutated passenger gene was TP53 (ex20ins, 67.6%; non‐ex20ins, 59.3%), followed by LRP1B (ex20ins, 6.8%; non‐ex20ins, 18.5%) and RB1 (ex20ins, 8.2%; non‐ex20ins, 7.4%) (Fig. 2C).
Fig. 2.
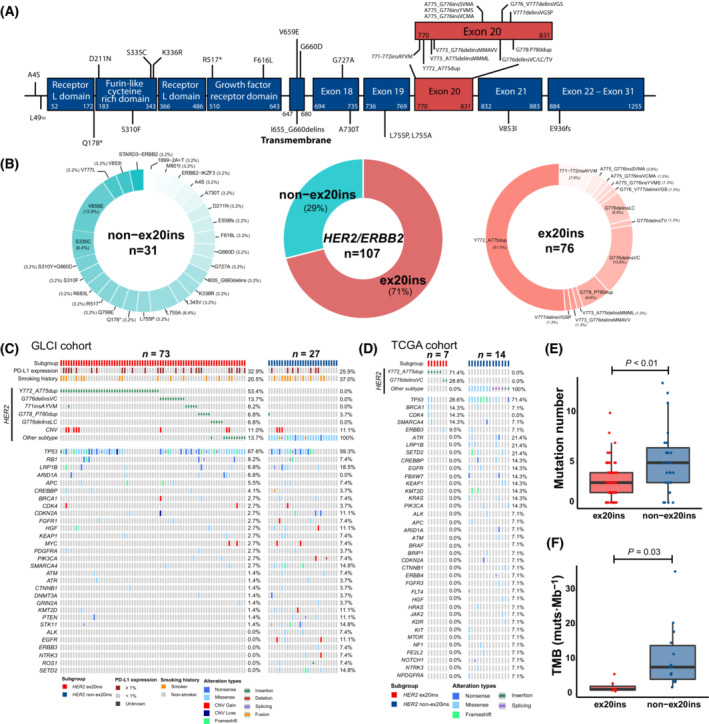
Genomic and immune features in NSCLC patients harbouring HER2 ex20ins and non‐ex20ins mutations. (A) The lollipop plot of HER2 ex20ins and non‐ex20ins mutations in the study cohort. (B) The category and proportion of HER2 ex20ins and non‐ex20ins mutations. (C) Tumour tissue/plasma samples were performed using multiple targeted NGS panels, and 72 overlapping NSCLC relevant genes carried by at least three patients were presented. (D) The genomic profile of The Cancer Genome Atlas Program (TCGA) cohort, including 21 NSCLC patients carrying HER2 mutations. (E) Mutation numbers of NSCLC patients with HER2 ex20ins and HER2 non‐ex20ins mutations. Error bars: interquartile range. P‐value: two‐sample t‐test. (F) TMB of NSCLC patients with HER2 ex20ins and HER2 non‐ex20ins mutations. Error bars: interquartile range. P‐value: two‐sample t‐test.
Compared with patients with HER2 ex20ins mutations, altered SETD2 gene (0.0% vs. 14.8%, P < 0.01) was more frequently observed in non‐ex20ins patients in the GLCI‐ICI cohort (Fig. 2C). In the TCGA cohort [seven with HER2 ex20ins and 14 with HER2 non‐ex20ins mutations, median age: 67 years (range: 52–81), females 66.7%, Table S2], despite a lower TP53 alteration prevalence in HER2 ex20ins patients (28.6%), similar frequently altered genes were observed in comparison with the GLCI cohort, such as BRCA1, CDK4, and LRP1B (Fig. 2D). In the GLCI cohort, the average number of mutations identified in one biopsy was 3.7, and patients with HER2 non‐ex20ins mutations had more concurrent mutations than ex20ins patients (5.2 vs. 2.1, P < 0.01, Fig. 2E). Similarly, in the TCGA cohort, non‐ex20ins patients had higher TMB than ex20ins patients (10.7 vs. 2.2 muts·Mb−1, P = 0.03, Fig. 2F).
3.3. Immune microenvironment features
To gain more insight into the TME of patients with HER2 mutations, transcriptome data from the TCGA cohort were analysed using the CIBERSORT algorithm, and the degree of immune cell infiltration was estimated. Among the 21 patients in the TCGA cohort, HER2 non‐ex20ins patients might have a potentially higher density of resting CD4+ memory T cells than HER2 ex20ins patients, while the difference was not statistically significant (15.6% vs. 12.3%, P = 0.54, Fig. S1A).
3.4. Clinical efficacy of ICI‐based therapy
The genomic and TME features suggested that the efficacy of ICIs in advanced NSCLC patients with HER2 mutations was possibly associated with the subtype of HER2 mutation. Next, we analysed the efficacy of ICIs in the GLCI‐ICI cohort (23 with HER2 ex20ins, 12 with HER2 non‐ex20ins mutations, and one with missing PFS data). The median follow‐up time was 11.6 (range: 0.3–70.0) months. Patients with PD‐L1 TPS < 1% were frequently observed (22/35, 62.9%), and most of the patients received ICI therapy as the second‐line or subsequent treatment (28/35, 80.0%) (Table 2). Of 35 patients in the GLCI‐ICI cohort, 31 had available TMB data, and the percentage of patients with high TMB (≥ 10 muts·Mb−1) appeared to be higher in the non‐ex20ins subgroup than in the ex20ins subgroup (41.7% vs. 21.7%, P = 0.24). The other clinical characteristics (Table 2) and genomic profiles of the GLCI‐ICI cohort were similar to those of the entire study cohort (Fig. S1B).
Table 2.
Demographics and clinical characteristics of GLCI‐ICI cohort.
| Characteristics | Overall (n = 35) | Ex20ins (n = 23) | Non‐ex20ins (n = 12) | P‐value |
|---|---|---|---|---|
| Age, median (range), year | 56 (35–72) | 54 (35–72) | 58 (51–72) | 0.11 |
| Age, no. (%) | ||||
| < 60 years | 23 (65.7) | 16 (69.6) | 7 (58.3) | 0.71 |
| ≥ 60 years | 12 (34.3) | 7 (30.4) | 5 (41.7) | |
| Sex, no. (%) | ||||
| Female | 12 (34.3) | 8 (34.8) | 4 (33.3) | > 0.99 |
| Male | 23 (65.7) | 15 (65.2) | 8 (66.7) | |
| Clinical stage at initial diagnosis, no. (%) | ||||
| IIIb | 2 (5.7) | 0 (0.0) | 2 (16.7) | 0.20 |
| IVa | 15 (42.9) | 10 (43.5) | 5 (41.7) | |
| IVb | 18 (51.4) | 13 (56.5) | 5 (41.7) | |
| Histology, no. (%) | ||||
| Adenocarcinoma | 32 (91.4) | 22 (95.7) | 10 (83.3) | 0.27 |
| Squamous cell carcinoma | 3 (8.6) | 1 (4.3) | 2 (16.7) | |
| Smoking, no. (%) | ||||
| Ever | 13 (37.1) | 7 (30.4) | 6 (50.0) | 0.30 |
| Never | 22 (62.9) | 16 (69.6) | 6 (50.0) | |
| ECOG performance status, no. (%) | ||||
| 1 | 33 (94.3) | 21 (91.3) | 12 (100.0) | > 0.99 |
| 2 | 1 (2.9) | 1 (4.3) | 0 (0.0) | |
| 3 | 1 (2.9) | 1 (4.3) | 0 (0.0) | |
| PD‐L1 expression, no. (%) | ||||
| < 1% | 22 (62.9) | 12 (52.2) | 9 (75.0) | 0.65 |
| 1–49% | 12 (34.3) | 9 (39.1) | 3 (25.0) | |
| ≥ 50% | 1 (3.9) | 1 (4.3) | 0 (0.0) | |
| TMB, no. (%) | ||||
| < 10 muts·Mb−1 | 19 (54.3) | 15 (65.2) | 4 (33.3) | 0.24 |
| ≥ 10 muts·Mb−1 | 10 (28.6) | 5 (21.7) | 5 (41.7) | |
| Unknown | 6 (17.1) | 3 (13.0) | 3 (25.0) | |
| ICI‐based therapy, no. (%) | ||||
| Monotherapy | 15 (42.9) | 8 (34.8) | 7 (58.3) | 0.28 |
| Combination therapy | 20 (57.1) | 15 (65.2) | 5 (41.7) | |
| ICI lines, no. (%) | ||||
| 1st | 7 (20.0) | 3 (13.0) | 4 (33.3) | 0.20 |
| ≥ 2nd | 28 (80.0) | 20 (87.0) | 8 (66.7) | |
Fifteen patients in the GLCI‐ICI cohort were treated with ICI monotherapy (Fig. 3A), and the remaining 20 patients received ICI combination therapy (Fig. 3B). The overall ORR was 20.0% (95% CI: 8.0–41.2%); the mPFS and median OS (mOS) of the GLCI‐ICI cohort were 5.2 (95% CI: 3.5–9.9) months and 14.8 (95% CI: 8.1–not reached) months, respectively. Patients carrying HER2 non‐ex20ins appeared to achieve relatively high disease control rates compared with those carrying HER2 ex20ins mutations (monotherapy, 85.7% vs. 28.6%, P = 0.10; ICI combination therapy, 100% vs. 66.7%, P = 0.27). Also, patients with HER2 non‐ex20ins mutations exhibited a relatively high durable clinical benefit (complete/partial response or stable disease lasting over 6 months) compared with those with HER2 ex20ins (66.7% vs. 34.8%, P = 0.07, Fig. 3C); however, they did not have a significantly higher ORR (25.0% vs. 17.4%, P = 0.67). Details of treatment and ICI efficacy in the GLCI‐ICI cohort are shown in Table 3.
Fig. 3.
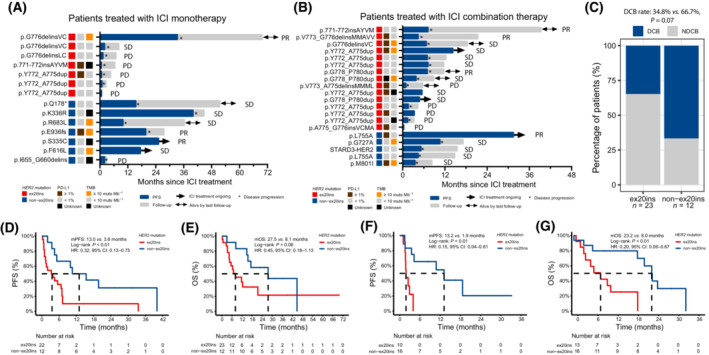
Swimmer plots, best of response, and survival of HER2‐mutated NSCLC patients receiving ICI‐based therapy. (A) A total of 15 HER2‐mutated NSCLC patients were treated with ICI monotherapy, including seven harbouring HER2 ex20ins and eight harbouring HER2 non‐ex20ins mutations (one patient with missing PFS data was not presented here). (B) Other 20 HER2‐mutated NSCLC patients were treated with ICI combination therapy, including 15 ex20ins patients and five patients harbouring HER2 non‐ex20ins mutations. (C) Patients with HER2 non‐ex20ins mutations had a relatively high durable clinical benefit (complete/partial response or stable disease lasting over 6 months) rate in comparison with patients harbouring HER2 ex20ins (66.7% vs. 34.8%, P = 0.07, Fisher's exact test). (D, E) In the GLCI‐ICI cohort, HER2‐mutated NSCLC patients with HER2 non‐ex20ins displayed potentially better PFS and OS than those with HER2 ex20ins mutations. (F, G) In the META‐ICI cohort, similar results were obtained that non‐ex20ins patients had significantly superior PFS and OS than ex20ins patients.
Table 3.
Treatment plan and efficacy of GLCI‐ICI cohort.
| Patient ID | HER2 mutation | Subtype | Sex | Age over 60 | Smoking | Adenocarcinoma | TMB over 10 muts·Mb−1 | PD‐L1 over 1% | Clinical stage | ICI line | ICI treatment plan | Clinical response | PFS status | PFS (months) | OS status | OS (months) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HER2‐106 | Exon 20ins | p.Y772_A775dup | Female | Yes | Never | Yes | No | Yes | IVA | 3rd | Pembrolizumab | PD | 1 | 0.7 | 1 | 4.57 |
| HER2‐111 | Non‐exon 20ins | p.Q178* | Male | No | Ever | No | No | No | IVA | 5th | Tislelizumab (BGB‐A317) | SD | 1 | 15.4 | 0 | 22.77 |
| HER2‐125 | Non‐exon 20ins | p.L755A | Female | No | Never | Yes | No | No | IVA | 1st | Pemetrexed + Carboplatin + Pembrolizumab | SD | 1 | 4.4 | 1 | 14.83 |
| HER2‐136 | Non‐exon 20ins | STARD3‐ERBB2 | Female | No | Never | Yes | No | No | IVA | 2nd | Pembrolizumab + Docetaxel | SD | 1 | 5.2 | 1 | 15.50 |
| HER2‐144 | Exon 20ins | p.G776delinsVC | Male | No | Never | Yes | Yes | Yes | IVB | 2nd | Pemetrexed + Carboplatin + Pembrolizumab | SD | 1 | 6.7 | 0 | 18.33 |
| HER2‐148 | Non‐exon 20ins | p.L755A | Female | Yes | Never | Yes | No | Yes | IVA | 1st | Nab‐Pacilitaxel + Carboplatin + Pembrolizumab | PR | 0 | 31.5 | 0 | 31.50 |
| HER2‐159 | Exon 20ins | p.Y772_A775dup | Female | No | Never | Yes | No | No | IVA | 3rd | Camrelizumab + Bevacizumab | SD | 1 | 7.0 | 1 | 12.37 |
| HER2‐164 | Non‐exon 20ins | p.K336R | Female | Yes | Never | Yes | NA | No | IVA | 2nd | Camrelizumab | SD | 1 | 40.0 | 1 | 44.73 |
| HER2‐18 | Non‐exon 20ins | p.R683L | Male | Yes | Ever | Yes | Yes | No | IVB | 1st | Nivolumab | SD | 1 | 9.9 | 0 | 36.30 |
| HER2‐181 | Exon 20ins | p.Y772_A775dup | Male | Yes | Never | Yes | No | No | IVA | 3rd | Pemetrexed + Carboplatin + Pembrolizumab | SD | 1 | 7.2 | 1 | 11.70 |
| HER2‐187 | Non‐exon 20ins | p.M801I | Male | No | Ever | No | Yes | Yes | IIIB | 2nd | Docetaxel + Carboplatin + Pembrolizumab | SD | 1 | 3.5 | 1 | 8.30 |
| HER2‐194 | Exon 20ins | p.V773_G776delinsMMAVV | Male | No | Ever | Yes | No | Yes | IVB | 1st | Pemetrexed + Carboplatin + Pembrolizumab | PR | 1 | 4.4 | 1 | 21.40 |
| HER2‐2 | Exon 20ins | p.Y772_A775dup | Male | No | Never | Yes | No | No | IVB | 3rd | Pembrolizumab | PD | 1 | 0.9 | 1 | 2.60 |
| HER2‐200 | Exon 20ins | p.Y772_A775dup | Male | Yes | Ever | Yes | NA | Yes | IVA | 2nd | Pemetrexed + Carboplatin + Pembrolizumab | SD | 1 | 5.5 | 1 | 5.47 |
| HER2‐202 | Exon 20ins | p.Y772_A775dup | Female | Yes | Never | Yes | Yes | Yes | IVB | 2nd | Pemetrexed + Bevacizumab + Sintilimab | SD | 0 | 14.3 | 0 | 14.33 |
| HER2‐241 | Non‐exon 20ins | p.F616L | Male | No | Ever | Yes | Yes | No | IVB | 2nd | Nivolumab | SD | 0 | 17.4 | 0 | 17.40 |
| HER2‐244 | Exon 20ins | p.G778_P780dup | Male | No | Never | Yes | Yes | NA | IVA | 2nd | Nab‐Pacilitaxel + Carboplatin + Pembrolizumab | SD | 1 | 3.8 | 0 | 8.80 |
| HER2‐264 | Exon 20ins | p.Y772_A775dup | Female | No | Never | Yes | No | Yes | IVB | 2nd | Pemetrexed + Carboplatin + Camrelizumab | PD | 1 | 3.3 | 1 | 3.33 |
| HER2‐280 | Exon 20ins | p.G778_P780dup | Male | No | Ever | Yes | No | No | IVA | 2nd | Pacilitaxel + Pembrolizumab | PR | 1 | 7.0 | 0 | 11.63 |
| HER2‐283 | Exon 20ins | p.G778_P780dup | Male | Yes | Never | Yes | No | No | IVA | 3rd | Pemetrexed + Sintilimab | SD | 0 | 4.8 | 0 | 4.77 |
| HER2‐286 | Exon 20ins | p.A775_G776insVCMA | Male | No | Ever | Yes | No | Yes | IVB | 2nd | Pemetrexed + Carboplatin + ICI (unknown) | PD | 1 | 0.3 | 1 | 0.27 |
| HER2‐287 | Exon 20ins | p.V773_A775delinsMMML | Male | No | Never | Yes | No | Yes | IVB | 2nd | Pemetrexed + Carboplatin + Pembrolizumab | PD | 1 | 0.9 | 0 | 5.73 |
| HER2‐32 | Non‐exon 20ins | p.I655_G660delins | Male | No | Never | Yes | NA | No | IVB | 5th | Nivolumab | PD | 1 | 2.0 | 1 | 2.97 |
| HER2‐33 | Non‐exon 20ins | p.E936fs | Male | Yes | Ever | Yes | Yes | Yes | IVB | 3rd | Pembrolizumab | PR | 1 | 19.7 | 1 | 27.53 |
| HER2‐35 | Non‐exon 20ins | p.G727A | Male | Yes | Ever | Yes | Yes | No | IIIB | 1st | Pacilitaxel + Carboplatin + Durvalumab | SD | 1 | 10.6 | 1 | 17.27 |
| HER2‐4 | Exon 20ins | p.Y772_A775dup | Male | Yes | Ever | Yes | No | No | IVB | 3rd | Nivolumab | PD | 1 | 0.7 | 1 | 0.70 |
| HER2‐41 | Exon 20ins | p.G776delinsVC | Male | No | Ever | Yes | No | No | IVB | 4th | Pembrolizumab | SD | 1 | 2.0 | 1 | 8.07 |
| HER2‐55 | Exon 20ins | p.Y772_A775dup | Female | No | Never | Yes | NA | No | IVB | 4th | Nivolumab + Bevacizumab | PD | 1 | 1.4 | 1 | 3.27 |
| HER2‐60 | Non‐exon 20ins | p.S335C | Male | No | Never | Yes | NA | No | IVB | 3rd | Nivolumab | PR | 0 | 25.2 | 0 | 25.17 |
| HER2‐62 | Exon 20ins | p.G776delinsVC | Female | No | Never | Yes | Yes | No | IVA | 3rd | Nivolumab | PR | 1 | 33.5 | 0 | 70.00 |
| HER2‐68 | Exon 20ins | p.Y772_A775dup | Male | Yes | Ever | Yes | No | No | IVB | 1st | Pemetrexed + Carboplatin + Pembrolizumab | PD | 1 | 1.6 | 1 | 4.30 |
| HER2‐72 | Exon 20ins | p.771‐772insAYVM | Male | No | Never | No | No | No | IVA | 3rd | Pacilitaxel + Pembrolizumab | PR | 1 | 7.2 | 0 | 39.27 |
| HER2‐74 | Exon 20ins | p.G776delinsLC | Female | No | Never | Yes | No | No | IVA | 4th | KL‐A167 | PD | 1 | 1.3 | 1 | 6.90 |
| HER2‐82 | Exon 20ins | p.771‐772insAYVM | Male | No | Never | Yes | NA | Yes | IVB | 1st | Pembrolizumab | PD | 1 | 0.6 | 1 | 6.43 |
| HER2‐83 | Exon 20ins | p.771‐772insAYVM | Female | No | Never | Yes | Yes | Yes | IVB | 4th | Nivolumab | – | – | – | 1 | 4.37 |
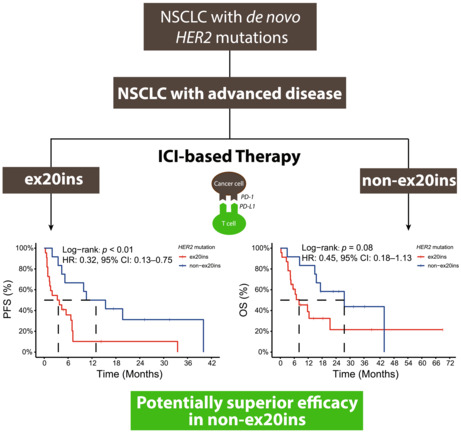
After excluding one patient with unavailable PFS data, 12 patients with HER2 non‐ex20ins mutations had an mPFS of 13.0 months, and they might have superior PFS to ex20ins patients (HR: 0.32, 95% CI: 0.13–0.75, Fig. 3D). Non‐ex20ins patients might also have longer mOS (27.5 vs. 8.1 months) and relatively good OS in comparison with patients carrying HER2 ex20ins (HR: 0.45, 95% CI: 0.18–1.13, Fig. 3E). The influence of potential confounders on prognosis was also investigated (Table 4). Patients whose TMB ≥ 10 muts·Mb−1 had a relatively low risk of disease progression than those with TMB < 10 muts·Mb−1 (HR: 0.40, 95% CI: 0.16–1.00), and a potential association of TMB with OS was also observed (HR: 0.35, 95% CI: 0.11–1.09). None of the patients' age, sex, histologic type, smoking history, PD‐L1 expression, ICI‐based therapy regimen, and whether ICI‐based therapy served as first‐line treatment, showed a strong association with PFS or OS. In the multivariable Cox regression model controlling for age, histologic type, smoking history, and TMB, HER2 non‐ex20ins mutations remained associated with potentially better PFS (adjusted HR: 0.31, 95% CI: 0.11–0.83) and OS (adjusted HR: 0.39, 95% CI: 0.13–1.18).
Table 4.
Hazard ratios estimated by univariate and multivariable Cox regression models in GLCI‐ICI cohort. Ref, reference.
| Characteristics | No. of patients (%) | PFS | OS | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Univariate | Multivariable | Univariate | Multivariable | ||||||
| HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | ||
| ERBB2 mutation | |||||||||
| Ex20ins | 12 (34.3) | Ref | Ref | Ref | Ref | ||||
| Non‐ex20ins | 23 (65.7) | 0.32 (0.13–0.75) | < 0.01* | 0.31 (0.11–0.83) | 0.02* | 0.45 (0.18–1.13) | 0.09 | 0.39 (0.13–1.18) | 0.10 |
| Age | |||||||||
| < 60 years | 23 (65.7) | Ref | Ref | Ref | Ref | ||||
| ≥ 60 years | 12 (34.3) | 0.49 (0.21–1.12) | 0.09 | 0.55 (0.23–1.32) 0.18 | 0.93 (0.38–2.23) | 0.87 | 1.06 (0.40–2.83) 0.90 | ||
| Sex | |||||||||
| Female | 12 (34.3) | Ref | – a | Ref | – a | ||||
| Male | 23 (65.7) | 1.60 (0.67–3.80) | 0.29 | – a | 0.91 (0.38–2.20) | 0.83 | – a | ||
| Histology | |||||||||
| Squamous cell carcinoma | 32 (91.4) | Ref | Ref | Ref | Ref | ||||
| Adenocarcinoma | 3 (8.6) | 1.02 (0.31–3.40) | 0.98 | 1.61 (0.41–6.26) 0.49 | 2.91 (0.39–21.78) | 0.30 | 3.60 (0.44–29.28) 0.23 | ||
| Smoking | |||||||||
| Ever | 13 (37.1) | Ref | Ref | Ref | Ref | ||||
| Never | 22 (62.9) | 0.83 (0.39–1.78) | 0.63 | 0.54 (0.24–1.18) 0.12 | 0.85 (0.36–2.01) | 0.70 | 0.61 (0.25–1.50) 0.28 | ||
| PD‐L1 expression | |||||||||
| < 1% | 22 (62.9) | Ref | – a | Ref | – a | ||||
| ≥ 1% | 12 (34.3) | 1.41 (0.64–3.313) | 0.39 | – a | 1.41 (0.59–3.36) | 0.43 | – a | ||
| Unknown | 1 (3.9) | 2.62 (0.29–17.77) | 0.44 | – a | – b | – a | |||
| TMB | |||||||||
| < 10 muts·Mb−1 | 19 (54.3) | Ref | Ref | Ref | Ref | ||||
| ≥ 10 muts·Mb−1 | 10 (28.6) | 0.40 (0.16–1.00) | 0.05 | 0.39 (0.14–1.09) | 0.07 | 0.35 (0.11–1.09) | 0.07 | 0.32 (0.10–1.05) | 0.06 |
| Unknown | 6 (17.1) | 0.47 (0.15–1.47) | 0.20 | 0.82 (0.24–2.86) | 0.76 | 1.04 (0.35–3.07) | 0.94 | 1.37 (0.41–4.53) | 0.61 |
| ICI‐based treatment | |||||||||
| Monotherapy | 15 (42.9) | Ref | – a | Ref | – a | ||||
| Immunochemotherapy | 20 (57.1) | 1.49 (0.65–3.40) | 0.34 | – a | 1.11 (0.90–2.67) | 0.82 | – a | ||
| ICI line | |||||||||
| 1st | 7 (20.0) | Ref | – a | Ref | – a | ||||
| ≥ 2nd | 28 (80.0) | 0.96 (0.39–2.39) | 0.93 | – a | 1.13 (0.41–3.09) | 0.82 | – a | ||
Not included in multivariable Cox regression models.
Insufficient patient number for model fitting.
Statistically significant.
To validate the findings in the GLCI‐ICI cohort, 30 eligible patients from external data sets of ICI‐based therapy studies were grouped into the META‐ICI cohort (13 with HER2 ex20ins and 17 with HER2 non‐ex20ins mutations, four with unavailable PFS data, and one with unavailable OS data, Table S3). Notably, the majority of patients in the META‐ICI cohort were treated with ICI monotherapy. The mPFS and mOS of the META‐ICI cohort were 4.0 (95% CI: 1.9–not reached) months and 18.9 (95% CI: 8.0–24.9) months, respectively. The demographic and clinical characteristics of the META‐ICI cohort are summarized in Table S3. The mPFS and mOS of patients harbouring HER2 ex20ins were 1.9 and 8.0 months, respectively, and patients with HER2 non‐ex20ins mutations presented a longer mPFS of 13.2 months and a longer mOS of 23.3 months. HER2 non‐ex20ins mutations were associated with favourable PFS (HR: 0.15, 95% CI: 0.04–0.61, Fig. 3F) and OS (HR: 0.20, 95% CI: 0.06–0.67, Fig. 3G).
4. Discussion
In this study, we analysed the molecular and TME characteristics of NSCLC patients with de novo HER2 mutations, as well as their responses to immunotherapy. HER2 non‐ex20ins patients had higher mutation numbers than HER2 ex20ins patients, whereas similar low‐level PD‐L1 expression was detected. HER2 non‐ex20ins mutations were potentially associated with superior PFS and OS under ICI‐based therapy, which was consistent with the findings in external cohorts.
Based on the GLCI cohort, we provided a landscape view of the genomic and clinical features of 107 HER2‐mutated NSCLC patients without common driver mutations. The diversity of HER2 mutations suggested potentially uniform responses to the same treatment. In this study, HER2 ex20ins mutations were detected in 76 (71.0%) patients, similar to the results of another East Asian cohort study by Tan et al. (72.7%) [35]. In contrast, a study based on a western cohort (n = 84) revealed that HER2 ex20ins accounted for 34.4% HER2 mutations [6]. For concurrent TP53 mutations, the prevalence was similar between patients harbouring HER2 ex20ins and non‐ex20ins mutations in our study (67.6% vs. 59.3%), whereas the prevalence in the TCGA cohort appeared to be relatively high in the non‐ex20ins subgroup (28.6% vs. 71.4%). Consistent with the TMB results reported by Tan et al. [35], HER2 non‐ex20ins patients in our study had higher mutation numbers than patients with HER2 ex20ins mutations.
Our data provide preliminary insights into the TME heterogeneity in HER2‐mutated NSCLC. Similar to the results of a previous study [36], a generally immunosuppressed TME was observed, whereas a heterogeneous TME was observed in HER2‐mutated NSCLC. In our study, patients with HER2 ex20ins and non‐ex20ins mutations showed similar PD‐L1 TPS, with over 60% of the patients having negative expression levels (< 1%), which aligned with the findings of previous studies [5, 20, 35]. In the TCGA cohort, non‐ex20ins patients might be enriched with resting CD4+ memory T cells, which should be further investigated in a large cohort of HER2‐mutated NSCLC patients.
Our findings suggest that ICI‐based therapy might serve as a treatment option for advanced NSCLC patients with HER2 mutations, and that those harbouring HER2 non‐ex20ins mutations could potentially benefit more. In the GLCI‐ICI cohort, the mPFS and ORR of ICI‐based therapy were 5.2 months and 20.0%, respectively, consistent with the result of previous studies [20, 21, 22, 23]. In the German nNGM lung cancer cohort, a 26.2% ORR was reported in 61 HER2‐mutated NSCLC patients receiving ICI‐based therapy as the first‐line and later lines of treatment [20]. In the French Lung Cancer Group 01‐2018 and MSKCC research, ICI‐based therapy showed ORRs of 27.3% and 11.5%, respectively [21, 22]. In our study, a potentially better response to ICI was observed in patients harbouring HER2 non‐ex20ins mutations than in those harbouring HER2 ex20ins, which was similar to the findings of previous studies [5, 37, 38, 39, 40]. For instance, as reported in Fan's study, none of the six HER2 ex20ins patients treated with PD‐1 inhibitors achieved an objective response [38]. In a study by Tan et al. [35], none of the four ex20ins patients achieved a response after receiving pembrolizumab monotherapy. Our study also revealed that non‐ex20ins patients were more likely to achieve a better response to ICI‐based therapy than ex20ins patients; however, further research with a larger sample size is warranted. The relatively poor prognosis of ex20ins patients could be partially explained by the low TMB levels in patients harbouring HER2 ex20ins [29, 41], as advanced NSCLC patients with higher TMB (above 50% percentile) could achieve better PFS than patients with lower TMB (below 50% percentile) [25]. Mutated SETD2, enriched in non‐ex20ins NSCLC, was identified as a favourable predictive biomarker for immunotherapy, associated with higher TMBs and better OS (HR: 0.55, 95% CI: 0.46–0.65) [42]. LRP1B mutations, being relatively frequent in HER2 non‐ex20ins patients, were also associated with prolonged survival (HR: 0.63, 95% CI: 0.40–0.97) [43]. In our study, we did not detect a strong association between PD‐L1 expression and PFS/OS, which might have resulted from the similar proportions of patients with at least 1% PD‐L1 expression between the ex20ins and non‐ex20ins subgroups.
In the era of HER2‐ADC, comprehensive studies including considerable HER2 non‐ex20ins patients were warranted to investigate how HER2 mutation subtype affects ADC efficacy and whether immunotherapy or HER2‐ADC would be the optimal first‐line regimen for NSCLC patients with specific subtypes of HER2 mutations. In DESTINY‐Lung01, HER2 non‐ex20ins patients did not achieve as good response to T‐DXd as ex20ins patients (ORR: 33% vs. 73%, P = 0.02) [15], suggesting that the efficacy of T‐DXd might depend on HER2 mutation subtype. For HER2‐mutated patients in East Asia, Tan et al. reported that three of four (75.0%) ex20ins patients responded to T‐DXd; however, the efficacy of ADCs in non‐ex20ins patients remained unclear. In our study, HER2 non‐ex20ins patients achieved ORR of 25.0% and mPFS of 13.0 months. Further studies comparing the treatment efficacy of HER2‐ADC and immunotherapy might be useful for the treatment of advanced NSCLC patients with HER2 non‐ex20ins mutations.
Our study has some limitations. As this was a retrospective study, the timing of the disease progression assessment was not standardized. The potentially superior PFS in non‐ex20ins patients might be influenced by the line of therapy, which could not be well controlled, owing to the limited sample size of the GLCI‐ICI cohort. In addition, multiple NGS panels were performed on participants' tumour tissue or plasma samples, and only overlapping cancer‐relevant genes were included for data analyses, resulting in the failure to accurately calculate TMB in the GLCI cohort and compare key ICI treatment‐related signalling pathways. We presented the number of mutations per person instead of the TMB value in the GLCI cohort, even though TMB data were available for patients in the GLCI‐ICI cohort. Another limitation is the limited sample size of the GLCI‐ICI cohort, resulting in an inability to comprehensively analyse the prognostic data for patients treated with ICI monotherapy or combination therapy, separately. Additionally, the results of the TME comparison were not comprehensive, owing to the lack of RNA‐Seq data in the GLCI‐ICI cohort.
5. Conclusion
Our study revealed that HER2‐mutated lung cancers present with a high‐level molecular heterogeneity. Patients harbouring HER2 non‐ex20ins mutations had higher mutation numbers than patients harbouring HER2 ex20ins mutations, whereas similar PD‐L1 expression levels were detected. HER2 non‐ex20ins mutations could potentially be considered positive predictors of ICI efficacy in advanced HER2‐mutated NSCLC patients, and ICI‐based therapy might be a good option for patients with HER2 non‐ex20ins mutations.
Conflict of interest
Yi‐Long Wu has received honoraria from AstraZeneca, Eli Lilly, Roche, Pierre Fabre, Pfizer, and Sanofi; consulting or advisory role with AstraZeneca, Roche, Merck and Boehringer Ingelheim, and Roche outside the submitted work. Xiaotian Zhao, Dongqin Zhu, Lingling Yang, and Qiuxiang Ou are employees of Nanjing Geneseeq Technology Inc., China. The remaining authors have nothing to disclose.
Author contributions
H‐YT, KY, and Y‐LW designed the study. Y‐LS, J‐XL, X‐YB, and Y‐CZ were responsible for patient recruitment and sample collection. E‐EK, S‐PW, Y‐SL, M‐MZ, S‐YML, C‐RX, QZ, J‐JY, W‐ZZ, B‐CW, and X‐CZ were responsible for patient clinical and survival data collection and results interpretation. H‐YT, KY, XZ, DZ, LY, and QO analysed data and interpreted results. All authors wrote and reviewed the manuscript, and approved the submitted version.
Supporting information
Fig. S1. Transcriptomic data of The Cancer Genome Atlas Program (TCGA) cohort and the genomic profile of the Guangdong Lung Cancer Institute‐immune checkpoint inhibitor (GLCI‐ICI) cohort.
Table S1. 72 Overlapping cancer‐relevant genes.
Table S2. Demographics and clinical characteristics of TCGA cohort.
Table S3. Demographics and clinical characteristics of META‐ICI cohort.
Acknowledgements
This study was supported by The Key Lab System Project of Guangdong Science and Technology Department—Guangdong Provincial Key Lab of Translational Medicine in Lung Cancer (Grant No. 2017B030314120 to Yi‐Long Wu), GDPH Scientific Research Funds for Leading Medical Talents in Guangdong Province (Grant No. KJ012019426 to Yi‐Long Wu), and Beijing Health Alliance Charitable Foundation (Grant No. TB219010 to Hai‐Yan Tu). The authors thank all the patients who participated in this study.
Hai‐Yan Tu and Kai Yin contributed equally to this work
Data accessibility
The data sets used and/or analysed in the current study are available from the corresponding author on reasonable request.
References
- 1. Spector NL, Blackwell KL. Understanding the mechanisms behind trastuzumab therapy for human epidermal growth factor receptor 2–positive breast cancer. J Clin Oncol. 2009;27:5838–47. [DOI] [PubMed] [Google Scholar]
- 2. Arcila ME, Chaft JE, Nafa K, Roy‐Chowdhuri S, Lau C, Zaidinski M, et al. Prevalence, clinicopathologic associations, and molecular spectrum of ERBB2 (HER2) tyrosine kinase mutations in lung adenocarcinomas ERBB2 (HER2) mutations in lung carcinoma. Clin Cancer Res. 2012;18:4910–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pillai RN, Behera M, Berry LD, Rossi MR, Kris MG, Johnson BE, et al. HER2 mutations in lung adenocarcinomas: a report from the lung cancer mutation consortium. Cancer. 2017;123:4099–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ou S‐HI, Madison R, Robichaux JP, Ross JS, Miller VA, Ali SM, et al. Characterization of 648 non‐small cell lung cancer (NSCLC) cases with 28 unique HER2 exon 20 insertions. J Clin Oncol. 2019;37(15_suppl):9063–3. [Google Scholar]
- 5. Tian P, Zeng H, Ji L, Ding Z, Ren L, Gao W, et al. Lung adenocarcinoma with ERBB2 exon 20 insertions: comutations and immunogenomic features related to chemoimmunotherapy. Lung Cancer. 2021;160:50–8. [DOI] [PubMed] [Google Scholar]
- 6. Wei XW, Gao X, Zhang XC, Yang JJ, Chen ZH, Wu YL, et al. Mutational landscape and characteristics of ERBB2 in non‐small cell lung cancer. Thorac Cancer. 2020;11:1512–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Eng J, Hsu M, Chaft JE, Kris MG, Arcila ME, Li BT. Outcomes of chemotherapies and HER2 directed therapies in advanced HER2‐mutant lung cancers. Lung Cancer. 2016;99:53–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dziadziuszko R, Smit EF, Dafni U, Wolf J, Wasąg B, Biernat W, et al. Afatinib in NSCLC with HER2 mutations: results of the prospective, open‐label phase II NICHE trial of European Thoracic Oncology Platform (ETOP). J Thorac Oncol. 2019;14:1086–94. [DOI] [PubMed] [Google Scholar]
- 9. Kris M, Camidge D, Giaccone G, Hida T, Li B, O'connell J, et al. Targeting HER2 aberrations as actionable drivers in lung cancers: phase II trial of the pan‐HER tyrosine kinase inhibitor dacomitinib in patients with HER2‐mutant or amplified tumors. Ann Oncol. 2015;26:1421–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Song Z, Li Y, Chen S, Ying S, Xu S, Huang J, et al. Efficacy and safety of pyrotinib in advanced lung adenocarcinoma with HER2 mutations: a multicenter, single‐arm, phase II trial. BMC Med. 2022;20:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhou C, Li X, Wang Q, Gao G, Zhang Y, Chen J, et al. Pyrotinib in HER2‐mutant advanced lung adenocarcinoma after platinum‐based chemotherapy: a multicenter, open‐label, single‐arm, phase II study. J Clin Oncol. 2020;38:2753–61. [DOI] [PubMed] [Google Scholar]
- 12. Fang W, Zhao S, Liang Y, Yang Y, Yang L, Dong X, et al. Mutation variants and co‐mutations as genomic modifiers of response to Afatinib in HER2‐mutant lung adenocarcinoma. Oncologist. 2020;25:e545–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu Z, Wu L, Cao J, Yang Z, Zhou C, Cao L, et al. Clinical characterization of ERBB2 exon 20 insertions and heterogeneity of outcomes responding to afatinib in Chinese lung cancer patients. Onco Targets Ther. 2018;11:7323–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ou S‐HI, Schrock AB, Bocharov EV, Klempner SJ, Haddad CK, Steinecker G, et al. HER2 transmembrane domain (TMD) mutations (V659/G660) that stabilize homo‐and heterodimerization are rare oncogenic drivers in lung adenocarcinoma that respond to afatinib. J Thorac Oncol. 2017;12:446–57. [DOI] [PubMed] [Google Scholar]
- 15. Li BT, Smit EF, Goto Y, Nakagawa K, Udagawa H, Mazières J, et al. Trastuzumab deruxtecan in HER2‐mutant non–small‐cell lung cancer. N Engl J Med. 2022;386:241–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mack PC, Redman MW, Moon J, Goldberg SB, Herbst RS, Melnick MAC, et al. Residual circulating tumor DNA (ctDNA) after two months of therapy to predict progression‐free and overall survival in patients treated on S1403 with afatinib+/−cetuximab. J Clin Oncol. 2020;38(15_suppl):9532–2. [Google Scholar]
- 17. Subbiah V. Optimizing anti–body drug conjugates and radiopharmaceuticals for precision therapy: the next frontier in precision oncology. Curr Probl Cancer. 2021;45:100799. [DOI] [PubMed] [Google Scholar]
- 18. Halliday PR, Blakely CM, Bivona TG. Emerging targeted therapies for the treatment of non‐small cell lung cancer. Curr Oncol Rep. 2019;21:1–12. [DOI] [PubMed] [Google Scholar]
- 19. Network NCC . Non‐small cell lung cancer (version 3.2022).
- 20. Saalfeld FC, Wenzel C, Christopoulos P, Merkelbach‐Bruse S, Reissig TM, Laßmann S, et al. Efficacy of immune checkpoint inhibitors alone or in combination with chemotherapy in NSCLC harboring ERBB2 mutations. J Thorac Oncol. 2021;16:1952–8. [DOI] [PubMed] [Google Scholar]
- 21. Guisier F, Dubos‐Arvis C, Viñas F, Doubre H, Ricordel C, Ropert S, et al. Efficacy and safety of anti–PD‐1 immunotherapy in patients with advanced NSCLC with BRAF, HER2, or MET mutations or RET translocation: GFPC 01‐2018. J Thorac Oncol. 2020;15:628–36. [DOI] [PubMed] [Google Scholar]
- 22. Mazieres J, Drilon A, Lusque A, Mhanna L, Cortot A, Mezquita L, et al. Immune checkpoint inhibitors for patients with advanced lung cancer and oncogenic driver alterations: results from the IMMUNOTARGET registry. Ann Oncol. 2019;30:1321–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lai W‐CV, Feldman DL, Buonocore DJ, Brzostowski EB, Rizvi H, Plodkowski AJ, et al. PD‐L1 expression, tumor mutation burden and response to immune checkpoint blockade in patients with HER2‐mutant lung cancers. J Clin Oncol. 2018;36(15_suppl):9060. [Google Scholar]
- 24. Wang VG, Kim H, Chuang JH. Whole‐exome sequencing capture kit biases yield false negative mutation calls in TCGA cohorts. PLoS One. 2018;13:e0204912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rizvi H, Sanchez‐Vega F, La K, Chatila W, Jonsson P, Halpenny D, et al. Molecular determinants of response to anti–programmed cell death (PD)‐1 and anti–programmed death‐ligand 1 (PD‐L1) blockade in patients with non–small‐cell lung cancer profiled with targeted next‐generation sequencing. J Clin Oncol. 2018;36:633–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gandara DR, Paul SM, Kowanetz M, Schleifman E, Zou W, Li Y, et al. Blood‐based tumor mutational burden as a predictor of clinical benefit in non‐small‐cell lung cancer patients treated with atezolizumab. Nat Med. 2018;24:1441–8. [DOI] [PubMed] [Google Scholar]
- 27. Miao D, Margolis CA, Vokes NI, Liu D, Taylor‐Weiner A, Wankowicz SM, et al. Genomic correlates of response to immune checkpoint blockade in microsatellite‐stable solid tumors. Nat Genet. 2018;50:1271–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Anagnostou V, Niknafs N, Marrone K, Bruhm DC, White JR, Naidoo J, et al. Multimodal genomic features predict outcome of immune checkpoint blockade in non‐small‐cell lung cancer. Nat Cancer. 2020;1:99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Samstein RM, Lee C‐H, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51:202–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next‐generation DNA sequencing data. Nat Genet. 2011;43:491–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Amarasinghe KC, Li J, Hunter SM, Ryland GL, Cowin PA, Campbell IG, et al. Inferring copy number and genotype in tumour exome data. BMC Genomics. 2014;15:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Newman AM, Bratman SV, Stehr H, Lee LJ, Liu CL, Diehn M, et al. FACTERA: a practical method for the discovery of genomic rearrangements at breakpoint resolution. Bioinformatics. 2014;30:3390–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tan AC, Saw SP, Chen J, Lai GG, Oo HN, Takano A, et al. Clinical and genomic features of HER2 exon 20 insertion mutations and characterization of HER2 expression by immunohistochemistry in east Asian non–small‐cell lung cancer. JCO Precis Oncol. 2022;6:e2200278. [DOI] [PubMed] [Google Scholar]
- 36. Kirchner M, Kluck K, Brandt R, Volckmar A‐L, Penzel R, Kazdal D, et al. The immune microenvironment in EGFR‐and ERBB2‐mutated lung adenocarcinoma. ESMO Open. 2021;6:100253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Catania C, Passaro A, Rocco EG, Spitaleri G, Barberis M, Noberasco C, et al. Dramatic antitumor activity of nivolumab in advanced HER2‐positive lung cancer. Clin Lung Cancer. 2016;17:e179–83. [DOI] [PubMed] [Google Scholar]
- 38. Chen K, Pan G, Cheng G, Zhang F, Xu Y, Huang Z, et al. Immune microenvironment features and efficacy of PD‐1/PD‐L1 blockade in non‐small cell lung cancer patients with EGFR or HER2 exon 20 insertions. Thorac Cancer. 2021;12:218–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xia L, Yu Y, Lan F, Yan J, Li J, Li W, et al. Case report: tumor microenvironment characteristics in a patient with HER2 mutant lung squamous cell carcinoma harboring high PD‐L1 expression who presented hyperprogressive disease. Front Oncol. 2021;11:760703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhao S, Xian X, Tian P, Li W, Wang K, Li Y. Efficacy of combination chemo‐immunotherapy as a first‐line treatment for advanced non‐small‐cell lung cancer patients with HER2 alterations: a case series. Front Oncol. 2021;1230:633522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Goodman AM, Kato S, Bazhenova L, Patel SP, Frampton GM, Miller V, et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther. 2017;16:2598–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lu M, Zhao B, Liu M, Wu L, Li Y, Zhai Y, et al. Pan‐cancer analysis of SETD2 mutation and its association with the efficacy of immunotherapy. NPJ Precis Oncol. 2021;5:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen H, Chong W, Wu Q, Yao Y, Mao M, Wang X. Association of LRP1B mutation with tumor mutation burden and outcomes in melanoma and non‐small cell lung cancer patients treated with immune check‐point blockades. Front Immunol. 2019;10:1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Transcriptomic data of The Cancer Genome Atlas Program (TCGA) cohort and the genomic profile of the Guangdong Lung Cancer Institute‐immune checkpoint inhibitor (GLCI‐ICI) cohort.
Table S1. 72 Overlapping cancer‐relevant genes.
Table S2. Demographics and clinical characteristics of TCGA cohort.
Table S3. Demographics and clinical characteristics of META‐ICI cohort.
Data Availability Statement
The data sets used and/or analysed in the current study are available from the corresponding author on reasonable request.


