Abstract
The Vpr protein, encoded by the human immunodeficiency virus type 1 (HIV-1) genome, is one of the nonstructural proteins packaged in large amounts into viral particles. We have previously reported that Vpr associates with the DNA repair enzyme uracil DNA glycosylase (UDG). In this study, we extended these observations by investigating whether UDG is incorporated into virions and whether this incorporation requires the presence of Vpr. Our results, with highly purified viruses, show that UDG is efficiently incorporated either into wild-type virions or into Vpr-deficient HIV-1 virions, indicating that Vpr is not involved in UDG packaging. Using an in vitro protein-protein binding assay, we reveal a direct interaction between the precursor form of UDG and the viral integrase (IN). Finally, we demonstrate that IN-defective viruses fail to incorporate UDG, indicating that IN is required for packaging of UDG into virions.
In addition to the structural gag, pol, and env genes, the human immunodeficiency virus type 1 (HIV-1) genome contains several other open reading frames. Among them, the vpr gene encodes a small protein (14 kDa) specifically packaged within virions through interaction with the Gag C-terminal domain (18, 19, 21, 27). The presence of Vpr in viral particles suggests that it might play a role in the viral life cycle very early in infection. Vpr has been shown to be important for infection of nondividing or slowly dividing cells by facilitating nuclear import of the viral preintegration complex (PIC) in these cells (7, 16, 29). A second function of Vpr is its ability to induce cell cycle arrest, leading Vpr-expressing cells to accumulate in the G2 stage of the cell cycle (17, 22, 30). Other reports have revealed additional functions for Vpr in the stimulation of transcription of the viral long terminal repeat (1, 8, 38) or in the regulation of apoptosis (2, 9, 34).
We have previously reported, in a study using the yeast two-hybrid system, that Vpr binds to the DNA repair enzyme uracil DNA glycosylase (UDG) (4). UDG is an enzyme in the base excision repair pathway required for removal of uracil from DNA (for a review, see reference 31). Another distinct class of proteins, the dUTPases, is also implicated in preventing dUMP incorporation into DNA during DNA synthesis. Both UDG and dUTPase are encoded by some DNA viruses, such as poxviruses and herpesviruses. Genomes of retroviruses encode only dUTPase (11). Lentiviruses from nonprimate species contain in their genome a dUTPase-encoding sequence. Lentiviruses from primate species do not contain in their genome either a dUTPase or UDG encoding sequences. Although dUTPase and UDG activities are mechanistically distinct, both enzymes act to prevent misincorporation of uracil into DNA. In the absence of regulation of uracil misincorporation into genomic DNA, it is expected that DNA will accumulate G-to-A substitutions. Interestingly, dUTPase mutants of the caprine arthritis-encephalitis lentivirus accumulate a high proportion of G-to-A substitutions in their genomes (36). There exists therefore the possibility that Vpr-associated UDG of primate lentiviruses and dUTPase of nonprimate lentiviruses have similar roles in virus replication. dUTPase is encoded by the pol gene and is a virion-associated protein. We hypothesize that UDG, like dUTPase, is present in viral particles. It is therefore of interest to address whether UDG is incorporated into viral particles and whether the requirement for Vpr is related to such a localization.
Packaging of UDG into viral particles is independent of the presence of Vpr.
To investigate the incorporation of UDG into virions and to test whether Vpr is required in this process, we used wild-type NDK and Vpr mutant NDK virions derived from the productively infected H9 T-cell line. Vpr mutant NDK virions contain a stop codon which prematurely terminates the wild-type Vpr protein and removes the last 31 amino acids. Deletions of the C-terminal part of Vpr have been reported to affect the stability and conceivably the protein conformation necessary for virion targeting (37). Virions were pelleted from cell-free supernatant, resuspended in a buffer containing 1 mM CaCl2 and 20 mM Tris-HCl (pH 8.0), and incubated for 18 h at 37°C with 1 mg of subtilisin (Boehringer Mannheim) per ml. This procedure allows the elimination of microvesicles which copurify with HIV-1 virions by sucrose density gradient centrifugation and which are potentially a source of contaminating cellular proteins found in purified virion preparations (3, 15).
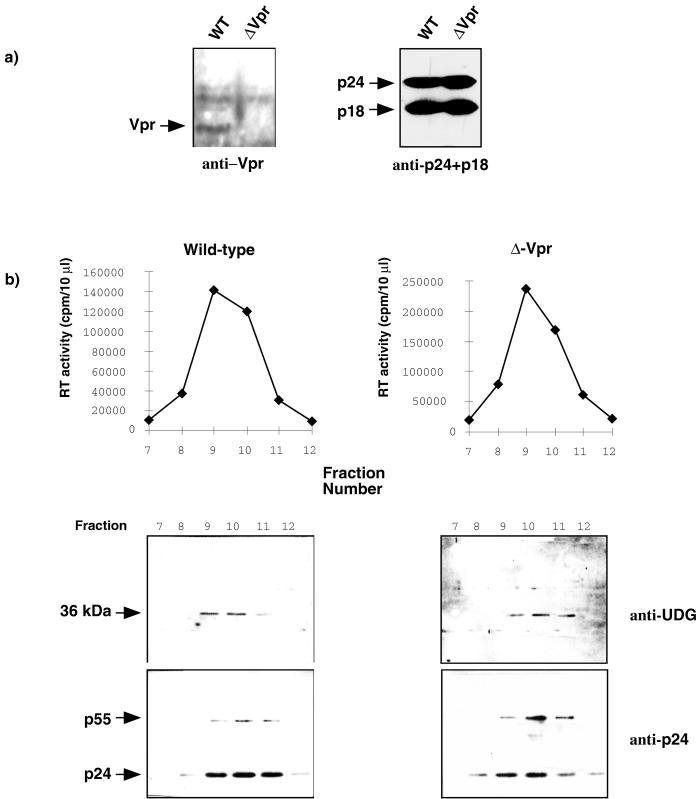
We performed a Western blot analysis using anti-Vpr antibody (kindly provided by N. Landau) on viral lysates before their purification on a sucrose density gradient and confirmed previous data (37) reporting that no detectable Vpr protein was present in viral particles released from cells infected with Vpr-defective viruses (Fig. 1a). Similar amounts of Vpr+ and Vpr− viruses from each stock were resolved on a linear 20 to 60% sucrose density gradient, and aliquots of each fraction were collected and analyzed for reverse transcriptase activity. Virions were harvested from individual gradient fractions, and the content of p24 and UDG was underscored by Western blotting with monoclonal anti-p24+p18 antibody (kindly provided by Q. Sattentau) and rabbit polyclonal anti-UDG antibody (a gift of G. Slupphaug). Bound antibodies were visualized with ECL blotting detection reagents (Amersham). Approximately equivalent amounts of viral Gag proteins were recovered in each gradient (Fig. 1b). For wild-type virus, the presence of UDG was evident in the gradient fraction coinciding with peak of reverse transcriptase activity (Fig. 1b, left panel). Unexpectedly, we observed that gradient fractions from Vpr mutant virions also contain UDG (Fig. 1b, right panel). UDG was also detected in both wild-type and Vpr mutant virions, in two independent experiments, when wild-type AD8 and Vpr mutant AD8 virions derived from productively infected primary macrophages (29) were analyzed (data not shown). These results suggest that UDG is indeed a virion-associated protein and that the incorporation of UDG into viral particles may not depend on the presence of packaged Vpr.
FIG. 1.
Packaging of UDG into viral particles is independent of the presence of Vpr. (a) Cell-free supernatants from wild-type and Vpr-defective virus-infected H9 T cells were treated with subtilisin, pelleted by ultracentrifugation, and monitored for equal amounts of p24. The viral pellet was solubilized in sample buffer, separated on an SDS–12% polyacrylamide gel, and analyzed by Western blotting for the presence of Vpr and Gag products with ECL reagents. (b) Virions were purified on a linear 20 to 60% sucrose density gradient. Aliquots of each gradient fraction were analyzed for reverse transcriptase activity (upper panel). Virus particles in each gradient fraction were pelleted and solubilized in sample buffer, and viral proteins were separated by SDS-PAGE and analyzed by Western blotting for the presence of UDG and Gag products (lower panel).
In addition, our Western blotting analysis indicated that the size of the virion-packaged UDG has an apparent molecular mass of 36 to 38 kDa. It has been reported that cellular UDG could be found in two distinct subcellular compartments, either cytoplasmic or nuclear (32). The cytoplasmic and nuclear forms of UDG differ in their apparent molecular masses. The cytoplasmic form of UDG (38 kDa) contains a presequence of 77 amino acids which is removed to give rise to the mature form of UDG (28 kDa), which is then targeted to the nucleus. Our results indicating that the virion-packaged UDG has a size similar to that found for the precursor form of UDG are consistent with the notion that it is the cytoplasmic form of UDG, and not the nuclear form, which is incorporated into viral particles.
The Pr55Gag protein is not required in UDG packaging.
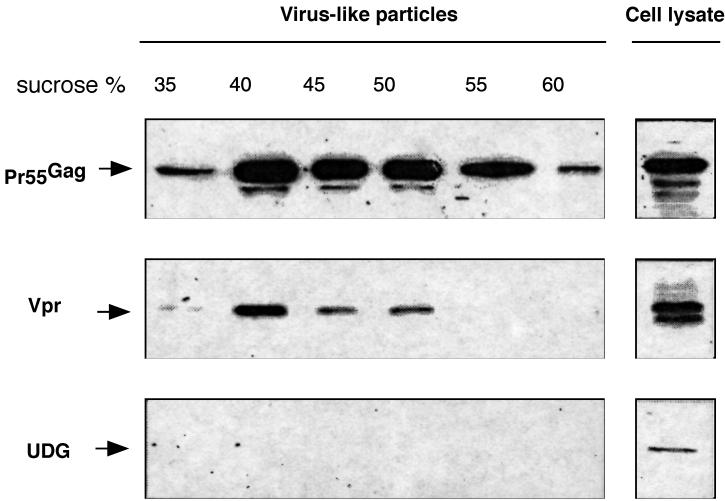
Our above results indicating that UDG is packaged within virions through a Vpr-independent mechanism led us to investigate which viral protein is required to incorporate UDG into viral particles. It has been well documented that reverse transcription occurs, after viral entry and uncoating, in the context of a PIC that includes Vpr in addition to the integrase (IN) and matrix (MA) proteins (7, 12, 16). We thus hypothesized that viral proteins present within the PIC could be a target for UDG. We first investigated whether Pr55Gag could be a target for UDG. The gag gene and the vpr gene were amplified by PCR from the pNL43 molecular clone and then cloned under the control of the T7 polymerase promoter into the Pos7 vector (kindly provided by B. Moss) (39). HeLa cells previously infected for 30 min with 1 PFU of recombinant vaccinia virus (T7-MVA, Ankara strain; a kind gift of G. Sutter) (35) per cell to express T7 polymerase were transfected with Pos7-Gag and Pos7-Vpr plasmids. Cell-free supernatant containing virus-like particles (21) was harvested 24 h after transfection, concentrated by centrifugation, and analyzed on a linear 20 to 60% sucrose density gradient. Individual gradient fractions were then ultracentrifuged and analyzed by Western blotting with anti-p24, anti-Vpr, and anti-UDG antibody (Fig. 2). As a control, Pr55Gag and Vpr overexpressed in cell lysate are shown, as well as endogenous expression of UDG. As expected (21), expression of Pos7-Gag together with Pos7-Vpr resulted in cosedimentation of both Pr55Gag and Vpr. We failed to detect the presence of incorporated UDG in gradient fractions containing both Pr55Gag and Vpr, however. The possibility that the failure to detect packaged UDG into virus-like particles was due to low UDG content seems unlikely since UDG was detected in gradient fractions containing virions (Fig. 1b). These results indicate that the gag gene products are probably not important for the incorporation of UDG within virions and confirm that Vpr is not required in this event.
FIG. 2.
Packaging of UDG into viral particles is independent of the presence of Pr55Gag and Vpr. HeLa cells were cotransfected with expression plasmids encoding Pr55Gag and Vpr. Cell-free supernatant containing virus-like particles was subjected to a linear sucrose density gradient, and individual gradient fractions were collected and analyzed by Western blotting for the presence of Pr55Gag and Vpr, as well as for the presence of packaged UDG. As a control, HeLa cell extracts were analyzed by Western blotting for the presence of Pr55Gag, Vpr, and endogenous UDG.
UDG binds to IN in an in vitro protein-protein binding assay.
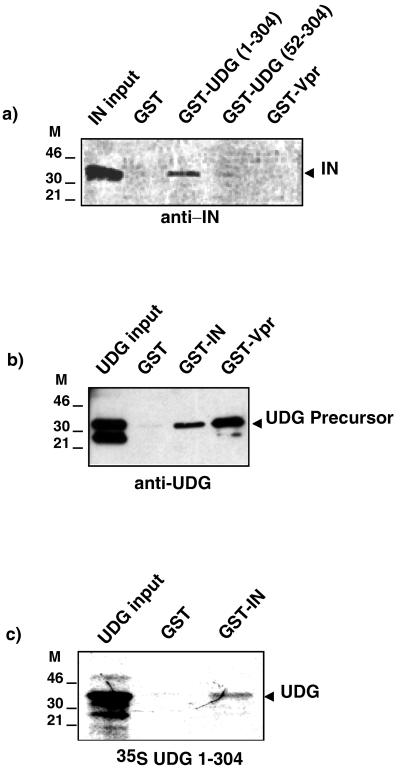
We next investigated whether IN could be a viral partner for UDG. We carried out binding studies between IN and UDG recombinant proteins in vitro. The IN open reading frame was amplified by PCR from the pNL43 molecular clone and then cloned either under the control of the T7 polymerase promoter into the Pos7 vector or in frame with the glutathione S-transferase (GST) sequence. The cDNA encoding the complete UDG protein (residues 1 to 304) was amplified with appropriate primers by PCR from clone p15 (25), kindly provided by G. Slupphaug, and cloned in frame with the GST sequence or cloned into the Pos7 vector. GST, GST-Vpr, and GST-UDG (52–304) fusion proteins were prepared as previously described (4). To overexpress IN or UDG, recombinant vaccinia virus-infected HeLa cells were transfected with Pos7-IN or Pos7-UDG plasmids. Cells overexpressing IN or UDG were harvested 20 h after transfection and lysed as previously described (6). Cell lysate overexpressing IN was incubated in Tris-buffered saline–Tween 20 binding buffer with each of the GST fusion proteins immobilized on glutathione (GSH)-agarose beads. After extensive washes, bound proteins were eluted and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Western blotting analysis with rabbit anti-IN antibody (a gift from D. Trono). As seen in Fig. 3a, IN bound to GST-UDG (1–304) but not to GST-UDG (52–304), indicating that the presence of the UDG presequence is required for the interaction. In contrast, IN failed to associate with GST alone or with GST-Vpr. Cell lysate overexpressing UDG was incubated with equivalent amounts of GST, GST-IN, or GST-Vpr, and bound proteins were revealed by Western blotting with an anti-UDG antibody (Fig. 3b). As shown in the lefthand lane, the two forms of UDG (i.e., precursor and mature) coexist in cell lysate overexpressing UDG. Binding analysis indicated that the precursor form of UDG preferentially associates with GST-IN, while the two forms of UDG associate with GST-Vpr. To test whether the interaction between UDG and IN is direct, we performed an in vitro protein-protein binding assay with equivalent amounts of GST and GST-IN incubated with in vitro-translated, radiolabeled UDG (1–304) (Fig. 3c). Bound labeled proteins were resolved by SDS-PAGE and revealed by autoradiography. Results indicate that the interaction between IN and UDG is direct and does not involve an intermediate bridging protein. Altogether, our results demonstrate that IN associates in an in vitro protein-protein binding assay only with the precursor form of UDG, not with the mature form of UDG. These data are consistent with our above in vivo data revealing that it is the precursor form of UDG which is incorporated within virions and suggest that IN could be the viral protein involved in the packaging of UDG into viral particles.
FIG. 3.
IN associates specifically with the precursor form of UDG. (a) Cell lysate overexpressing IN was incubated with equivalent amounts of GST, GST-UDG (1–304), GST-UDG (52–304), or GST-Vpr fusion proteins affinity purified on GSH-agarose beads. After washes, bound proteins were analyzed by Western blotting with rabbit polyclonal anti-IN antibody. (b) Cell lysate overexpressing UDG was incubated with equivalent amounts of GST, GST-IN, or GST-Vpr fusion proteins affinity purified on GSH-agarose beads, and bound proteins were analyzed by Western blotting with rabbit polyclonal anti-UDG antibody. Lanes marked “input” contain cell lysate overexpressing IN (a) or UDG (b) before binding to GST proteins. (c) Translated, radiolabeled UDG (1–304) was incubated with similar amounts of GST and GST-IN. After washes, bound proteins were separated by SDS-PAGE and revealed by autoradiography. Lane UDG input contains one-fifth of 35S-labeled proteins before binding to GST fusion proteins. M, molecular mass markers (in kilodaltons.).
IN-deficient viruses fail to incorporate UDG.
To demonstrate unequivocally that the IN domain is required for the incorporation of UDG into virions, and that Vpr is not involved in this process, we investigated whether IN mutant viruses or IN-Vpr double mutant viruses are impaired in incorporation of UDG into viral particles. To mutate the IN gene in an NDK or NDK Vpr mutant molecular clone, an NheI digest was performed, cutting at a unique site in the region coding for amino acid 38 of IN. After fill-in with Klenow and autoligation, a frameshift causes a stop codon to be read 8 amino acids downstream. The introduced mutation was confirmed by sequencing.
293 cells were transfected with either wild-type, Vpr-defective, IN-defective, or IN-Vpr double-defective plasmids. Viruses from cell-free supernatant were treated with subtilisin and purified onto a linear sucrose density gradient, as described above. Gradient fractions were pooled, and equivalent amounts of viruses (15 × 106 cpm of reverse transcriptase activity) were analyzed by Western blotting with anti-p24+p18 antibody or anti-UDG antibody. Bound antibodies were visualized with ECL plus reagents (Amersham). Similar amounts of Gag products were observed in each virus preparation (Fig. 4a), indicating that similar amounts of viruses were analyzed. We failed to detect the presence of IN within IN-defective or IN-Vpr double-defective viruses, while IN was detectable within wild-type and Vpr-defective viruses (data not shown). As expected, the incorporation of UDG into viral particles was observed when wild-type as well as Vpr-defective viruses were analyzed. Interestingly, viruses defective for the presence of IN but still expressing Vpr in viral particles did not incorporate detectable amounts of UDG. A similar defect in UDG incorporation into virions was observed when viruses used in the experiments were defective in both IN and Vpr proteins. These results demonstrate that the absence of virion-associated UDG is directly related to the absence of the IN domain of the gag-pro-pol precursor. Our data are consistent with our above in vitro results indicating a direct interaction between the IN domain and UDG. Altogether, our data indicate that the IN domain is sufficient for packaging of UDG into virions and that Vpr is not involved in this process.
FIG. 4.
The presence of IN within the viral particle is required to allow the incorporation of UDG into viral particles. (a) 293 cells were transfected with wild-type, Vpr-defective, IN-defective, or IN-Vpr double-defective molecular clones, and viruses produced in cell-free supernatant were treated with subtilisin and collected by ultracentrifugation. Virions were then resolved on a linear 20 to 60% sucrose density gradient, and gradient fractions coinciding with the peak of reverse transcriptase activity were pooled and normalized for reverse transcriptase activity. Virus pellets were then solubilized in sample buffer, and viral lysates were analyzed by Western blotting for the presence of Gag and UDG products. M, molecular mass markers (in kilodaltons). (b) Evaluation of the enzymatic activity of UDG within virions by using a nicking assay. A 32P-labeled single-stranded 34-mer oligonucleotide containing one uracil in the middle of its sequence was incubated with either recombinant purified UDG (0.005 to 0.1 units) or purified virions (3 × 106 cpm of reverse transcriptase activity). Upon incubation, DNAs were recovered and separated by electrophoresis on denaturing polyacrylamide gels.
We next wondered whether the enzymatic UDG activity could be detected in purified virions. The enzymatic activity of UDG was monitored by means of a “nicking assay” described previously (24). In this procedure, a synthetic 34-mer single-stranded oligonucleotide containing one uracil residue incorporated into the middle of the sequence is 5′ end labeled with 32P. The incubation of 5′-end-labeled 34-mer oligonucleotide with the UDG enzyme induces the excision of the uracil incorporated into the oligonucleotide followed, under standard conditions, by cleavage of the DNA strand, leading to the appearance of a 17-mer oligonucleotide. The labeled oligonucleotide fragments are separated by denaturing PAGE and visualized by autoradiography. The appearance of the 17-mer oligonucleotide followed by the concomitant disappearance of the 34-mer oligonucleotide is the hallmark of the presence of enzymatically active UDG and is proportional to the amount of incubated UDG. Virions treated with subtilisin and purified in a sucrose density gradient were resuspended in a buffer containing 100 mM NaCl, 10 mM Tris-HCl (pH 7.5), 1 mM EDTA and Triton X-100 (0.2%), incubated with the uracil-containing substrate (50 fmol), and analyzed for the presence of enzymatically active UDG. As a control (Fig. 4b), increased amounts (0.005 to 0.1 units) of commercially purified recombinant mature UDG (Perkin-Elmer) led to a progressive conversion from uncleaved to completely cleaved forms of the substrate. When virions (wild type, Vpr defective, IN defective, or Vpr-IN defective) corresponding to 3 × 106 cpm of reverse transcriptase activity were incubated with the uracil-containing substrate, no UDG activity was detected, although the presence of UDG protein is detectable in wild-type and Vpr-defective virions. It is not surprising that the enzymatic activity of the virion-associated UDG, which corresponds to the precursor form of the enzyme, is difficult to detect, because it is noteworthy that the precursor form of UDG exhibits very weak enzymatic activity compared to that of the mature form (33).
In this paper, we report that a cellular protein, namely, the DNA repair enzyme UDG, is specifically incorporated into viral particles. Other host cellular proteins have been reported to be incorporated within virions through interaction with a viral protein. This is the case for cyclophilin A, which is specifically incorporated into HIV-1 particles through association with Gag (13). Ubiquitin has also been reported to be present inside HIV-1 virions via an association with the p6 domain of Gag (26). We also show that intravirion UDG packaging requires a specific interaction with the viral IN protein. Using in vitro studies, we found that the N-terminal domain of UDG (residues 1 to 52) is important for the interaction with IN. Consistent with this, we also demonstrated in an in vivo approach with the yeast double-hybrid system that the N-terminal domain of UDG is required to allow its binding to IN (data not shown). This domain, which corresponds to the presequence of UDG, has been suggested to constitute a separate structural domain (23). Studies to delineate the domain (or domains) of IN which interacts with UDG are in progress. UDG incorporation does not appear to be cell type specific, since we found it in HIV-1 virions produced both from lymphoid T cells and from differentiated macrophages.
We initially reported that UDG has the ability to bind Vpr (4). In this previous study, we failed to detect UDG associated with wild-type virions. We now find that UDG is indeed incorporated into viral particles. It is likely that this discrepancy is due to the amounts of virions used for Western blotting analysis. Although Vpr is incorporated into viral particles, the question arises as to why the intravirion packaging of UDG is not related to the presence of Vpr in virions but to the presence of IN. It has been reported that Vpr is incorporated in virions through interactions with the NC domain of Pr55Gag (10). It is possible that during assembly and budding, the interaction between Vpr and NCp7 impairs the binding of UDG to Vpr via steric hindrance. Experiments to test whether UDG, Vpr, and NCp7 can associate as a trimeric complex would resolve this issue. The other explanation invokes the idea that UDG could bind alternatively Vpr or IN depending on its maturation status. Indeed, UDG is recovered in cells in two forms, either exclusively as a cytoplasmic precursor form or as a mature form (32). Consistent with this, our in vivo and in vitro data indicate that IN has the ability to bind the cytoplasmic precursor form of UDG (this study), while Vpr has the ability to bind preferentially the mature form of UDG, as we previously reported with coimmunoprecipitation experiments (4). It is noteworthy that residues 1 to 52 of UDG are important for binding of IN (this study), while residues 222 to 225 have been reported to be important for binding of Vpr (5). We speculate that the pre-mature form of UDG is cleaved after entry in new cells, and then the mature form may interact with Vpr.
The fact that HIV-1 viruses have evolved by elaborating two viral proteins, IN and Vpr, which both have the ability to bind UDG argues for the importance of these interactions during the viral life cycle. What then are the respective roles, during the viral life cycle, of the Vpr-UDG and IN-UDG interactions? We have previously proposed that the Vpr-UDG interaction might play a role similar to that played by the dUTPases of nonprimate lentiviruses, i.e., the reduction of uracil misincorporation into newly synthesized viral DNA in order to avoid G-to-A substitutions (4). In this regard, the fact that UDG is a virion-packaged protein, through interaction with IN, might be important to ensure the presence of the protein at the site of nascent viral DNA synthesis.
It is well documented that Vpr and IN, in addition to MA, belong to the viral PIC and participate in the nuclear import of viral DNA (7, 14, 16, 28). Although the presence of these viral proteins in the PIC has been demonstrated, whether some of their cellular partners are present remains to be explored. It has been reported that the precursor form of UDG, upon removal of its presequence, is targeted from cytoplasm to nucleus (32). Therefore, one possibility is that the Vpr-UDG and/or IN-UDG interactions participate in the nuclear import of the viral PIC. It would thus be interesting to determine whether UDG can be found, through its association with Vpr and/or IN, to be an integral part of the PIC. In conclusion, future analyses of the role played by interactions between viral and cellular proteins should continue to provide important insights into the biology of HIV-1.
Acknowledgments
We thank N. Landau, B. Moss, Q. Sattentau, G. Slupphaug, G. Sutter, and D. Trono for generous gifts of reagents. We thank Q. Sattentau for careful reading of the manuscript.
K.E.W. was supported by a fellowship from INSERM. This work was supported by INSERM and by grants from the French Agency against AIDS (ANRS).
REFERENCES
- 1.Agostini I, Navarro J M, Rey F, Bouhamdan M, Spire B, Vigne R, Sire J. The human immunodeficiency virus type 1 Vpr transactivator: cooperation with promoter-bound activator domains and binding to TFIIB. J Mol Biol. 1996;261:599–606. doi: 10.1006/jmbi.1996.0485. [DOI] [PubMed] [Google Scholar]
- 2.Ayyavoo V, Mahboubi A, Mahalingam S, Ramalingam R, Kudchodkar S, Williams W V, Green D G, Weiner D B. HIV-1 Vpr suppresses immune activation and apoptosis through regulation of nuclear factor κB. Nat Med. 1997;3:1117–1123. doi: 10.1038/nm1097-1117. [DOI] [PubMed] [Google Scholar]
- 3.Bess J W, Powell P J, Issaq H J, Schumack L J, Krimes M K, Henderson L E, Arthur L O. Microvesicles are a source of contaminating cellular proteins found in purified HIV-1 preparations. Virology. 1997;230:134–144. doi: 10.1006/viro.1997.8499. [DOI] [PubMed] [Google Scholar]
- 4.Bouhamdan M, Benichou S, Rey F, Navarro J M, Agostini I, Spire B, Camonis J, Slupphaug G, Vigne R, Benarous R, Sire J. Human immunodeficiency virus type 1 Vpr protein binds to the uracil DNA glycosylase DNA repair enzyme. J Virol. 1996;70:697–704. doi: 10.1128/jvi.70.2.697-704.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouhamdan M, Xue Y, Baudat Y, Hu B, Sire J, Pomerantz R, Duan L. Diversity of HIV-1 Vpr interactions involves usage of the WxxF motif of host cell proteins. J Biol Chem. 1998;273:8009–8016. doi: 10.1074/jbc.273.14.8009. [DOI] [PubMed] [Google Scholar]
- 6.Bouyac M, Courcoul M, Bertoia G, Baudat Y, Gabuzda D, Blanc D, Chazal N, Boulanger P, Sire J, Vigne R, Spire B. Human immunodeficiency virus type 1 Vif protein binds to the Pr55Gag precursor. J Virol. 1997;71:9358–9365. doi: 10.1128/jvi.71.12.9358-9365.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bukrinsky M, Haggerty S, Dempsey M, Sharova N, Adzhubel A, Spitz L, Lewis P, Goldfarb D, Emerman M, Stevenson M. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature. 1993;365:666–669. doi: 10.1038/365666a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen E A, Terwilliger E F, Jalinoos Y, Proulx J, Sodroski J G, Haseltine W A. Identification of HIV-1 Vpr product and function. J Acquired Immune Defic Syndr. 1990;3:11–18. [PubMed] [Google Scholar]
- 9.Conti L, Rainaldi G, Matarrese P, Varano B, Rivabene R, Columba S, Sato A, Bellardelli F, Malorni W, Gessani S. The HIV-1 Vpr protein acts as a negative regulator of apoptosis in a human lymphoblastoid T cell line: possible implications for the pathogenesis of AIDS. J Exp Med. 1998;187:403–413. doi: 10.1084/jem.187.3.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Rocquigny H, Petitjean P, Tanchou V, Decimo D, Drouot L, Delaunay T, Darlix J L, Rocques B P. The zinc fingers of HIV nucleocapsid protein NCp7 direct interactions with the viral regulatory protein Vpr. J Biol Chem. 1997;272:30753–30759. doi: 10.1074/jbc.272.49.30753. [DOI] [PubMed] [Google Scholar]
- 11.Elder J H, Lerner D L, Hasseikus-Light C S, Fontenot D J, Hunter E, Luciw P A, Montelaro R C, Phillips T R. Distinct subsets of retroviruses encode dUTPase. J Virol. 1992;66:1791–1794. doi: 10.1128/jvi.66.3.1791-1794.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farnet C M, Haseltine W A. Determination of viral proteins present in the human immunodeficiency virus type 1 preintegration complex. J Virol. 1991;65:1910–1915. doi: 10.1128/jvi.65.4.1910-1915.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franke E K, En Hui Yuan H, Luban J. Specific incorporation of cyclophilin A into HIV-1 virions. Nature. 1994;372:359–362. doi: 10.1038/372359a0. [DOI] [PubMed] [Google Scholar]
- 14.Gallay P, Hope T, Chin D, Trono D. HIV-1 infection of non-dividing cells through the recognition of integrase by the importin//karyopherin pathway. Proc Natl Acad Sci USA. 1997;94:9825–9830. doi: 10.1073/pnas.94.18.9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gluschankof P, Mondor I, Gelderblom H R, Sattentau Q. Cell membrane vesicles are a major contaminant of gradient-enriched human immunodeficiency virus type 1 preparations. Virology. 1997;230:125–133. doi: 10.1006/viro.1997.8453. [DOI] [PubMed] [Google Scholar]
- 16.Heinzenberg N K, Bukrinsky M I, Haggerty S A, Ragland A M, Kewalramani V, Lee M-A, Gendelman H E, Ratner L, Stevenson M, Emerman M. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc Natl Acad Sci USA. 1994;91:7311–7315. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jowett J B M, Planelles V, Poon B, Shah N P, Chen M-L, Chen I S Y. The human immunodeficiency virus type 1 vpr gene arrests infected T cells in the G2 + M phase of the cell cycle. J Virol. 1995;69:6304–6313. doi: 10.1128/jvi.69.10.6304-6313.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kondo E, Mammano F, Cohen E A, Gottlinger H G. The p6Gag domain of human immunodeficiency virus type 1 is sufficient for the incorporation of Vpr into heterologous viral particles. J Virol. 1995;69:2759–2764. doi: 10.1128/jvi.69.5.2759-2764.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lavallée C, Yao X J, Ladha A, Göttlinger H, Haseltine W A, Cohen E A. Requirement of the Pr55Gag precursor for incorporation of the Vpr product in human immunodeficiency virus type 1 viral particles. J Virol. 1994;68:1926–1934. doi: 10.1128/jvi.68.3.1926-1934.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Layne S P, Merges M J, Dembo M, Spouge J L, Conley S R, Moore J P, Raina J L, Renz H, Genderblom H R, Nara P L. Factors underlying spontaneous inactivation and susceptibility to neutralisation of human immunodeficiency virus. Virology. 1992;189:695–714. doi: 10.1016/0042-6822(92)90593-e. [DOI] [PubMed] [Google Scholar]
- 21.Lu Y L, Spearman P, Ratner L. Human immunodeficiency virus type 1 viral protein R localization in infected cells and virions. J Virol. 1993;67:6542–6550. doi: 10.1128/jvi.67.11.6542-6550.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahalingam S, Ayyavoo V, Patel M, Kieber-Emmons T, Weiner D B. Nuclear import, virion incorporation, and cell cycle arrest/differentiation are mediated by distinct functional domains of human immunodeficiency virus type 1 Vpr. J Virol. 1997;71:6339–6347. doi: 10.1128/jvi.71.9.6339-6347.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagelhus T A, Haug T, Singh K K, Keshav K F, Skorpen F, Otterlei M, Bhareti S, Lindmo T, Benichou S, Benarous R, Krokan H E. A sequence in the N-terminal region of human uracil-DNA glycosylase with homology to XPA interacts with the C-terminal part of the 34-kDa subunit of replication protein. A. J Biol Chem. 1997;272:6561–6566. doi: 10.1074/jbc.272.10.6561. [DOI] [PubMed] [Google Scholar]
- 24.Neidermann P, Jiricny J. The purification of a mismatch-specific thymine-DNA glycosylase from HeLa cells. J Biol Chem. 1993;268:21218–21224. [PubMed] [Google Scholar]
- 25.Olsen L C, Aasland R, Wittwer C U, Krokan H E, Helland D E. Molecular cloning of human uracil-DNA glycosylase, a highly conserved DNA repair enzyme. EMBO J. 1989;8:3121–3125. doi: 10.1002/j.1460-2075.1989.tb08464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ott D E, Coren L V, Copeland T D, Kane B P, Johnson D G, Sowder II R C, Yoshinaka Y, Oroszlan S, Arthur L O, Henderson L E. Ubiquitin is covalently attached to the p6Gag proteins of human immunodeficiency virus type 1 and simian immunodeficiency virus and to the P12Gag protein of Moloney murine leukemia virus. J Virol. 1998;72:2962–2968. doi: 10.1128/jvi.72.4.2962-2968.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paxton W, Connor R I, Landau N L. Incorporation of Vpr into human immunodeficiency virus type 1 virions: requirement for the p6 region of Gag and mutational analysis. J Virol. 1993;67:7229–7237. doi: 10.1128/jvi.67.12.7229-7237.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Popov S, Rexach M, Zybarth G, Reiling N, Lee M A, Ratner L, Lane C M, Moore M S, Blobel G, Bukrinsky M. Viral protein R regulates nuclear import of the HIV-1 preintegration complex. EMBO J. 1998;17:909–917. doi: 10.1093/emboj/17.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rey F, Bouhamdan M, Navarro J M, Agostini I, Willetts K, Bouyac M, Tamalet C, Spire B, Vigne R, Sire J. A role for HIV-1 Vpr during infection of peripheral blood mononuclear cells. J Gen Virol. 1998;79:1083–1087. doi: 10.1099/0022-1317-79-5-1083. [DOI] [PubMed] [Google Scholar]
- 30.Rogel M E, Wu L I, Emerman M. The human immunodeficiency virus type 1 vpr gene prevents cell proliferation during chronic infection. J Virol. 1995;69:882–888. doi: 10.1128/jvi.69.2.882-888.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shapiro R. In: Chromosome damage and repair. Seeberg E, Kleppe K, editors. New York, N.Y: Plenum; 1981. pp. 13–18. [Google Scholar]
- 32.Slupphaug G, Markussen F H, Olsen L C, Aaslande R, Aarsaether N, Bakke O, Krokan H E, Helland D E. Nuclear and mitochondrial forms of human uracil-DNA glycosylase are encoded by the ame gene. Nucleic Acids Res. 1993;11:2579–2584. doi: 10.1093/nar/21.11.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slupphaug G, Eftedal I, Kavli B, Bharati S, Helle N M, Haug T, Levine D W, Krokan H E. Properties of a recombinant human uracil-DNA glycosylase from the UNG gene and evidence that UDG encodes the major uracil-DNA glycosylase. Biochemistry. 1995;34:128–138. doi: 10.1021/bi00001a016. [DOI] [PubMed] [Google Scholar]
- 34.Stewart S A, Poon B, Jowett J B M, Chen I S Y. Human immunodeficiency virus type 1 Vpr induces apoptosis following cell cycle arrest. J Virol. 1997;71:5576–5592. doi: 10.1128/jvi.71.7.5579-5592.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sutter G, Ohlmann M, Erfle V. Non-replicating vaccinia vector efficiently expresses T7 RNA polymerase. FEBS Lett. 1995;371:9–12. doi: 10.1016/0014-5793(95)00843-x. [DOI] [PubMed] [Google Scholar]
- 36.Turelli P, Guiguen F, Mornex J-F, Vigne R, Querat G. dUTPase-minus caprine arthritis-encephalitis virus is attenuated for pathogenesis and accumulates G-to-A substitutions. J Virol. 1997;71:4522–4530. doi: 10.1128/jvi.71.6.4522-4530.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yao X J, Subramanian R A, Rougeau N, Boisvert F, Bergeron D, Cohen E A. Mutagenic analysis of human immunodeficiency virus type 1 Vpr: role of a predicted N-terminal alpha-helical structure in Vpr nuclear localization and virion incorporation. J Virol. 1995;69:7032–7044. doi: 10.1128/jvi.69.11.7032-7044.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L, Mukherjee S, Jia F, Narayan O, Zhao L J. Interaction of virion protein Vpr of human immunodeficiency virus type 1 with cellular transcription factor Sp1 and trans-activation of viral long terminal repeat. J Biol Chem. 1995;270:25564–25569. doi: 10.1074/jbc.270.43.25564. [DOI] [PubMed] [Google Scholar]
- 39.Wyatt L S, Moss B, Rozenblatt S. Replication-deficient vaccinia virus encoding bacteriophage T7 RNA polymerase for transient gene expression in mammalian cells. Virology. 1995;210:202–205. doi: 10.1006/viro.1995.1332. [DOI] [PubMed] [Google Scholar]