Dear Editor
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has become a serious emergency to global public health since its outbreak in 2019. SARS-CoV-2 is an enveloped, positive-sense, single-strand RNA virus, belonging to the Betacoronavirus genus of the Coronaviridae family (Wu et al., 2020) including SARS-CoV, SARS-CoV-2, MERS-CoV, etc. After viral entry, the RNA genome first encodes 16 non-structural proteins, including papain-like protease and Nsp5 (also called the main protease), which are essential for viral replication and transcription (Bai et al., 2022). Nsp5 is a 33-kDa cysteine protease whose most frequently found recognition sequence is LQ^(S/A/G). Many protein–protein interactions between the virus and the host have been identified (Gordon et al., 2020a). Only a small number of interactions between host proteins and the viral protein Nsp5 have been experimentally validated (Wenzel et al., 2021). Few reports have linked the role of host protein cleavage by Nsp5 beyond immune evasion. An interaction between catalytically inactive Nsp5 mutant (C145A) and tRNA methyltransferase 1 (TRMT1) has been described previously (Gordon et al., 2020a, b). TRMT1 (659 amino acids) catalyzes the formation of N2-methylguanosine (m2G) or N2,N2-dimethylguanosine (m2,2G) at position 26 of most human tRNAs, which is crucial for tRNA structure and function. TRMT1 is localized in the nucleus, cytoplasm, and mitochondria. Its mitochondrial localization is mediated by an N-terminal mitochondrial targeting sequence (MTS) of ∼35 residues (Dewe et al., 2017). However, it is not known whether TRMT1 is a bone-fide substrate of Nsp5. Moreover, the consequence of the potential cleavage remains unclear.
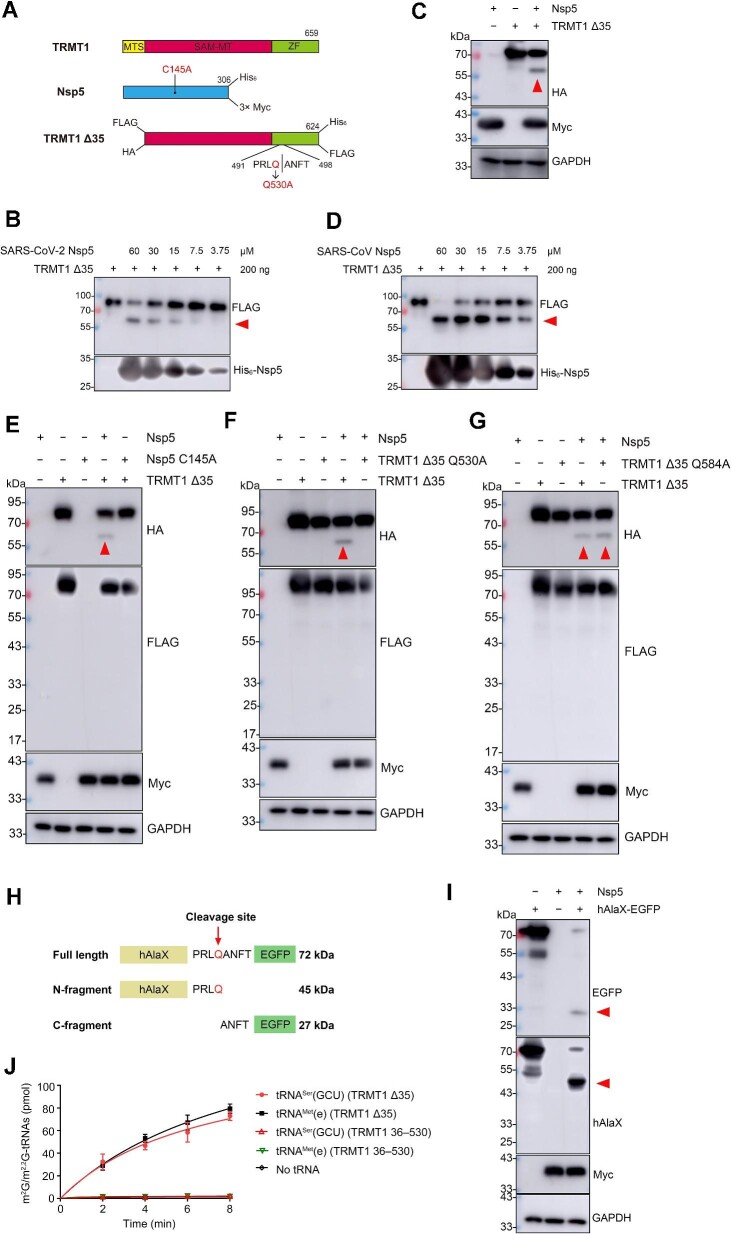
Here, we expressed and purified a recombinant SARS-CoV-2 Nsp5 with a C-terminal His6 tag and an MTS-deleted TRMT1 with an N-terminal FLAG and a C-terminal His6 tag (TRMT1 Δ35) using Escherichia coli (Figure 1A; Supplementary Figure S1A and B). Western blot analysis using anti-FLAG antibodies showed that the amount of TRMT1 Δ35 decreased after incubation with Nsp5 in a dose-dependent manner, accompanied by the appearance of a digested product of ∼58 kDa (Figure 1B), suggesting that the cleavage site is in the latter part of TRMT1. However, the C-terminal cleavage product with a lower molecular weight was not observed using anti-His6 antibodies. Coomassie blue staining using the same protein samples and digestion conditions also showed a clear cleaved fragment after incubation with Nsp5 (Supplementary Figure S1C). To confirm the digestion of TRMT1 by SARS-CoV-2 Nsp5 in vivo, we co-expressed TRMT1 Δ35 (with N-terminal HA and C-terminal FLAG tags) and 3× Myc-tagged Nsp5 (Figure 1A) in HEK293T cells. Indeed, western blot analysis using anti-HA antibodies showed the generation of a digested product of ∼58 kDa, while the cleaved fragment with the lower molecular mass was likewise invisible (Figure 1C). Considering the high sequence identity (∼96%) between SARS-CoV-2 Nsp5 and SARS-CoV Nsp5, we further purified SARS-CoV Nsp5 (Supplementary Figure S1D). Consistently, SARS-CoV Nsp5 was also able to digest TRMT1 Δ35 in vitro with a high efficiency (Figure 1D), indicating that TRMT1 cleavage is a conserved mechanism among different CoVs. To better understand whether TRMT1 digestion is mediated by the protease activity of Nsp5, the C145 residue of SARS-CoV-2 Nsp5 was replaced by alanine, resulting in a C145A mutant that was defective in cleavage (Figure 1E). The C145A mutant is a well-known variant that was constructed to show the contribution of the C145–H41 catalytic dyad to substrate cleavage (Jin et al., 2020). The C145A mutant exhibits the same three-dimensional structure as the wild-type enzyme (Hsu et al., 2005). These data clearly demonstrated that Nsp5 is able to cleave TRMT1 in vitro and in vivo via its protease activity.
Figure 1.
Nsp5 cleaves and inactivates TMRT1 in vitro and in vivo. (A) Diagram of the expression constructs of TRMT1, TRMT1 Δ35, Nsp5, and mutated forms of TRMT1 Δ35 and Nsp5. The SAM methyltransferase (SAM-MT) domain and the zinc finger (ZF) domain were shown in red and green, respectively. For each recombinant protein, tags labelled in the prokaryotic expression system are shown on the top, while tags labelled in the eukaryotic expression system are shown on the bottom. (B) Immunoblotting analysis of the cleavage product of TRMT1 Δ35 by SARS-CoV-2 Nsp5 in vitro. The amount of TRMT1 Δ35 and different concentrations of Nsp5 are indicated. (C) Cleavage of TRMT1 Δ35 by Nsp5 in HEK293T was detected by western blot analysis. (D) Cleavage of TRMT1 Δ35 by SARS-CoV Nsp5 in vitro was detected by western blot analysis. (E) Cleavage of TRMT1 Δ35 by Nsp5 or its inactive mutant (C145A) in HEK293T was detected by western blot analysis. (F and G) Cleavage of TRMT1 Δ35 and its mutants (Q530A and Q584A) by Nsp5 in HEK293T was detected by western blot analysis. (H) Schematic representation of the hAlaX-EGFP reporter with the predicated cleavage site from TRMT1. (I) Cleavage of hAlaX-EGFP by Nsp5 in HEK293T was detected by western blot analysis using anti-hAlaX or anti-EGFP antibodies. (J) Time course curves of m2G/m2,2G modification levels of tRNAMet(e) and tRNASer(GCU) catalyzed by TRMT1 Δ35 and TRMT1 36–530. The data represented mean ± standard deviation (n = 3). In all relevant images, the red arrow indicates the digested product of TRMT1 Δ35 or hAlaX-EGFP.
Subsequently, we intended to identify the cleavage site in TRMT1. Nsp5 digests all protein substrates at conserved glutamine residues. NetCorona 1.0, a website predicting Nsp5 cleavage sites (Kiemer et al., 2004), showed that Q530 was the only confident candidate with a high score of 0.942, while other glutamine residues (such as Q584) were excluded (Supplementary Figure S2A). This prediction was roughly consistent with the molecular mass of the digested product in both in vitro (Figure 1B) and in vivo (Figure 1C) assays. To experimentally validate the predicted cleavage site, we mutated Q530 to alanine in the TRMT1 Δ35 eukaryotic expression construct. In parallel, a Q584A mutant was also constructed for comparison. Immunoblotting with anti-HA antibodies showed that the cleavage of TRMT1 Δ35 Q530A was abolished (Figure 1F). According to the TRMT1 structure predicted by Alphafold, Q530 is spatially located in an unstructured peripheral loop with its side chain protruding into the solvent. The substitution to alanine, with a small uncharged side chain, is not expected to alter the conformation of the loop, suggesting that the inability of TRMT1 Δ35 Q530A to be cleaved by Nsp5 is due to the modification of the recognition sequence rather than a local conformational change in the protein. On the other hand, TRMT1 Δ35 Q584A mutant was clearly digested with a similar efficiency to wild-type TRMT1 (Figure 1G), confirming that Q584 is not an Nsp5 cleavage site as predicted by NetCorona 1.0 and that the Q584A mutation does not influence the local structure. To further confirm that Q530 serves as the cleavage site, we constructed an artificial cleavage reporter by inserting a peptide containing Q530 (PRLQANFT) between the human tRNA trans-editing factor hAlaX and EGFP (Figure 1H). It was expected that, after cleavage of the protein by Nsp5, the N-terminal hAlaX fragment (45 kDa) and the C-terminal EGFP fragment (27 kDa) would be released from the full-length hAlaX-EGFP fusion protein (72 kDa). Indeed, after co-expression of hAlaX-EGFP and Nsp5 in HEK293T cells, both predicted cleaved constituents were detected by anti-hAlaX or anti-EGFP antibodies (Figure 1I). Moreover, we further analyzed the primary sequences of TRMT1 from various mammals and found that Q530 is not evolutionarily conserved. For example, it is a lysine residue in TRMT1 from mice (Mus musculus) and hamsters (Mesocricetus auratus) (Supplementary Figure S2B). Therefore, we expressed Nsp5 together with either mouse Trmt1 (MmTrmt1 Δ13) or hamster Trmt1 (MaTrmt1 Δ13) (MTS deleted, with N-terminal HA and C-terminal FLAG tags). Western blot analysis showed that Trmt1 from these two species was not a substrate of Nsp5 (Supplementary Figure S3A and B). These additional data further confirm that Nsp5 cleaves TRMT1 at the Q530 residue.
To further explore and investigate the consequences of Nsp5 cleavage on the tRNA modification activity of TRMT1, we expressed and purified the cleavage product extending from A36 to Q530 (TRMT1 36–530) in E. coli (Supplementary Figure S4A). The tRNA methylation assays showed that TRMT1 36–530 was unable to catalyze the formation of m2,2G in the tRNASer(GCU) transcript (Figure 1J; Supplementary Figure S4B and C) or m2G in the tRNAMet(e) transcript (Figure 1J; Supplementary Figure S4B and D). Thus, the in vitro data demonstrated that the tRNA modification activity of TRMT1 was abolished by Nsp5 proteolysis.
In summary, we found that SARS-CoV-2 Nsp5 can cleave human tRNA methyltransferase TRMT1 at the Q530 residue in vitro and in vivo. The N-terminal cleavage product, TRMT1 36–530, can stably exist in vitro and in vivo but is defective in mediating tRNA m2G and m2,2G biogenesis. However, the C-terminal fragment is undetectable, probably due to rapid degradation. The structure of TRMT1 has not yet been determined. According to the primary sequence analysis and structure prediction by Alphafold, TRMT1 36–530 contains the S-adenosylmethionine (SAM) binding site, and the cleaved C-terminal portion is probably involved in tRNA binding. During the preparation and submission of this manuscript, two independent studies have been released on bioRxiv (Oliviera et al., 2023; Zhang et al., 2023), both revealing the cleavage of TRMT1 by SARS-CoV-2 Nsp5 at Q530, which are in full agreement with our results. Very early after the virus enters human cells, the virus genome generates several non-structural proteins, among which Nsp1 has been widely reported to inhibit mRNA translation by blocking the mRNA entry channel (Tidu et al., 2020). TRMT1 is a critical tRNA-modifying enzyme that ensures proper tRNA folding, and its malfunction severely impairs overall protein synthesis (Dewe et al., 2017). We suggest that the cleavage and subsequent inactivation of TRMT1 by Nsp5 likely act in synergy with Nsp1-mediated ribosome dysfunction to inhibit host mRNA translation. The in vivo significance of the Nsp5–TRMT1 interaction should be further investigated at the cellular and viral infection levels.
[Supplementary material is available at Journal of Molecular Cell Biology online. We thank Dr Haitao Yang (ShanghaiTech University) for providing SARS-CoV and SARS-CoV-2 Nsp5 constructs and Dr Ligang Wu (Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences) for providing hamster cDNA. We also thank Dr Gilbert Eriani (University of Strasbourg, France) for carefully reading the manuscript. This work was supported by the National Key Research and Development Program of China (2021YFA1300800 and 2021YFC2700903), the Natural Science Foundation of China (32271300), the Committee of Science and Technology in Shanghai (22ZR1481300 and 22JC1400503), and CAS Project for Young Scientists in Basic Research (YSBR-075). X.-L.Z. conceived the study, designed the experiments, and interpreted the data. J.-L.L. performed the experiments. X.-L.Z. and J.-L.L. wrote the manuscript.]
Contributor Information
Jia-Li Lu, State Key Laboratory of Molecular Biology, Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, University of Chinese Academy of Sciences, Shanghai 200031, China.
Xiao-Long Zhou, State Key Laboratory of Molecular Biology, Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, University of Chinese Academy of Sciences, Shanghai 200031, China; Key Laboratory of Systems Health Science of Zhejiang Province, School of Life Science, Hangzhou Institute for Advanced Study, University of Chinese Academy of Sciences, Hangzhou 310024, China.
References
- Bai C., Zhong Q., Gao G.F. (2022). Overview of SARS-CoV-2 genome-encoded proteins. Sci. China Life Sci. 65, 280–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewe J.M., Fuller B.L., Lentini J.M.et al. (2017). TRMT1-catalyzed tRNA modifications are required for redox homeostasis to ensure proper cellular proliferation and oxidative stress survival. Mol. Cell. Biol. 37, e00214–e00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Hiatt J., Bouhaddou M.et al. (2020a). Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science 370, eabe9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M.et al. (2020b). A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583, 459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu M.F., Kuo C.J., Chang K.T.et al. (2005). Mechanism of the maturation process of SARS-CoV 3CL protease. J. Biol. Chem. 280, 31257–31266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Du X., Xu Y.et al. (2020). Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 582, 289–293. [DOI] [PubMed] [Google Scholar]
- Kiemer L., Lund O., Brunak S.et al. (2004). Coronavirus 3CLpro proteinase cleavage sites: possible relevance to SARS virus pathology. BMC Bioinformatics 5, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliviera A.D., Dai X., Mottaghinia S.et al. (2023). Recognition and cleavage of human tRNA methyltransferase TRMT1 by the SARS-CoV-2 main protease. bioRxiv, 10.1101/2023.02.20.529306 [DOI] [Google Scholar]
- Tidu A., Janvier A., Schaeffer L.et al. (2020). The viral protein NSP1 acts as a ribosome gatekeeper for shutting down host translation and fostering SARS-CoV-2 translation. RNA 27, 253–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel J., Lampe J., Müller-Fielitz H.et al. (2021). The SARS-CoV-2 main protease Mpro causes microvascular brain pathology by cleaving NEMO in brain endothelial cells. Nat. Neurosci. 24, 1522–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B.et al. (2020). A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Ciesla J.H., Eldin P.et al. (2023). Proteolytic cleavage and inactivation of the TRMT1 tRNA modification enzyme by SARS-CoV-2 main protease. bioRxiv, 10.1101/2023.02.10.527147 [DOI] [PMC free article] [PubMed] [Google Scholar]