Abstract
Gummosis is 1 of the most common and destructive diseases threatening global peach (Prunus persica) production. Our previous studies have revealed that ethylene and methyl jasmonate enhance peach susceptibility to Lasiodiplodia theobromae, a virulent pathogen inducing gummosis; however, the underlying molecular mechanisms remain obscure. Here, 2 ethylene response factors (ERFs), PpERF98 and PpERF1, were identified as negative regulators in peach response to L. theobromae infection. Expression of 2 putative paralogs, PpERF98-1/2, was dramatically induced by ethylene and L. theobromae treatments and accumulated highly in the gummosis-sensitive cultivar. Silencing of PpERF98-1/2 increased salicylic acid (SA) content and pathogenesis-related genes PpPR1 and PpPR2 transcripts, conferring peach resistance to L. theobromae, whereas peach and tomato (Solanum lycopersicum) plants overexpressing either of PpERF98-1/2 showed opposite changes. Also, jasmonic acid markedly accumulated in PpERF98-1/2-silenced plants, but reduction in PpPR3, PpPR4, and PpCHI (Chitinase) transcripts indicated a blocked signaling pathway. PpERF98-1 and 2 were further demonstrated to directly bind the promoters of 2 putative paralogous PpERF1 genes and to activate the ERF branch of the jasmonate/ethylene signaling pathway, thus attenuating SA-dependent defenses. The lesion phenotypes of peach seedlings overexpressing PpERF1-1/2 and PpERF98-1/2 were similar. Furthermore, PpERF98-1/2 formed homodimers/heterodimers and interacted with the 2 PpERF1 proteins to amplify the jasmonate/ethylene signaling pathway, as larger lesions were observed in peach plants cooverexpressing PpERF98 with PpERF1 relative to individual PpERF98 overexpression. Overall, our work deciphers an important regulatory network of ethylene-mediated peach susceptibility to L. theobromae based on a PpERF98-PpERF1 transcriptional cascade, which could be utilized as a potential target for genetic engineering to augment protection against L. theobromae-mediated diseases in crops and trees.
Two interacting ethylene response factors (ERFs) negatively regulate resistance by activating the ERF branch of jasmonate/ethylene signaling in Lasiodiplodia theobromae-induced peach gummosis.
Introduction
Peach (Prunus persica) gummosis is 1 of the most prevalent and detrimental diseases in this tree crop and severely impacts economical production of peach orchards (Wang et al. 2011). This disease is mainly caused by Botryosphaeriaceae, including L. theobromae, Botryosphaeria dothidea, and Diplodia seriata, among which L. theobromae is known as a latent and widely distributed hemi-biotrophic pathogen in the tropical and subtropical regions (Paolinelli-Alfonso et al. 2016; Liu et al. 2022). The causal agent colonizes the permanent woody structures of peach trees via wounds and lenticels, causing large amounts of gum exudation, wood necrosis, and stem-bark cracking, eventually leading to poor tree vigor, fruit yield, and quality (Beckman et al. 2003). Due to the severe damage caused by L. theobromae on economically important horticultural crops, such as apple (Malus domestica), grape (Vitis vinifera), and citrus (Citrus sinensis), an increasing number of studies have focused on the modes of plant defense response to tissue invasion by this pathogen (Wang et al. 2011; Travadon et al. 2013; Delgado-Cerrone et al. 2016; Paolinelli-Alfonso et al. 2016). However, the molecular mechanisms influencing the severity of L. theobromae infection, and the final extent of tissue colonization during peach gummosis, are largely unknown.
The phytohormone jasmonates (JAs), ethylene (ET), and salicylic acid (SA) and their elaborate cross talk play important roles in plant response to diverse pathogens. Generally, the activation of the JA/ET pathway results in enhanced defense toward necrotrophic pathogens, whereas the activation of SA signaling leads to increased plants resistance to biotrophic and hemi-biotrophic pathogens (Zhang et al. 2017). The JA/ET and SA pathways have usually been considered as mutually antagonistic in plant response to pathogen attacks, namely in Arabidopsis (Arabidopsis thaliana) (Pieterse et al. 2012). The APETALA2 (AP2)/ET response factor (ERF) superfamily is defined by an AP2/ET responsive element-binding (EREB) factor domain that consists of ∼60 to 70 amino acids and is important for DNA binding (Nakano et al. 2006a). ERF members of the AP2/ERF superfamily of transcription factors regulate the so-called ERF signaling branch of JA/ET signaling and act as integrators between the JA/ET and SA pathways (Lorenzo et al. 2003; Pré et al. 2008). The AP2/ERF transcription factors usually interact with several cis-elements, such as the GCC-box, dehydration responsive elements (DRE), or C-repeat binding factor (CBF) in the promoters of target genes (Nakano et al. 2006a).
Notably, ERF members of the VIII and IX subgroups, such as ERF1, ERF96, and Octadecanoid-responsive Arabidopsis 59 (ORA59) in Arabidopsis, are key regulatory hubs in the cross talk between JA/ET and SA pathways and have been reported as important players in different plant–pathogen interactions (Fujimoto et al. 2000; Nakano et al. 2006b; Catinot et al. 2015). ERF1, a core regulator in the ERF branch of the JA/ET pathway, modulates plant defense responses by activating the expression of the pathogenesis-related (PR) genes CHI (BASIC CHITINASE) and PLANT DEFENSIN 1.2 (PDF1.2) and increases Arabidopsis resistance to necrotrophic fungi, such as Botrytis cinerea and Plectosphaerella cucumerina (Berrocal-Lobo et al. 2002). In wheat (Triticum aestivum), ERF1 overexpression dramatically increases plant resistance to the necrotrophic fungus Rhizoctonia cerealis (Zhu et al. 2014). On the contrary, AtERF1-overexpressing Arabidopsis lines show reduced tolerance against the hemi-biotrophic bacterium Pseudomonas syringae pv. tomato (Pst) DC3000, demonstrating that the role of ERF1 in plant resistance may depend on the pathogen lifestyle (Berrocal-Lobo et al. 2002). Similarly, AtERF96, a small ERF, could modulate the transcription of AtORA59 and of several PRs (e.g. PR3, PR4, and PDF1.2a) by binding to the GCC elements of their promoters, revealing an important role for AtERF96 in the JA/ET-dependent resistance to B. cinerea (Catinot et al. 2015).
Our previous RNA-seq data have shown that ERF members such as putative PpERF1 paralogs, and 2 putative paralogous PpERF98 genes, PpERF98-1 and 2, are sharply and markedly induced in peach shoots inoculated with L. theobromae (Gao et al. 2016; Zhang et al. 2022). ERF98 protein putative orthologs that belong to the subgroup IX of the AP2/ERF superfamily are 128 to 139 amino acid long with an EDLL/CMIX-Ⅰ motif (a potent plant transcriptional activation domain) at the C-terminal region (Tiwari et al. 2012). AtERF98 directly binds to the cis-element GCC-box in the promoters of ascorbic acid biosynthetic genes and activates their transcription, positively regulating plant salt tolerance (Zhang et al. 2012). AtERF98 is also shown to physically interact with AtMED25 (MEDIATOR 25), an integrative hub of JA signaling (Cevik et al. 2012). In addition, the transcript of AtERF98 is induced by chitin, a plant defense elicitor (Libault et al. 2007). However, knowledge on the regulatory network in which ERF98 is embedded during the response to biotic stress remains quite limited. We have additionally shown that suppression of the JA and ET pathway boosts peach resistance to L. theobromae infection (Zhang et al. 2022). Here, we describe a regulatory module composed of physically interacting PpERF98-PpERF1 proteins activates JA/ET signaling. This triggers the ERF branch of the JA/ET-dependent response, which in turn may repress the SA-dependent defense pathway and promote peach susceptibility to L. theobromae. Our findings characterize key molecular players in the cross talk between JA/ET- and SA-related defense pathways, during peach responses to L. theobromae attack.
Results
PpERF98-1/2 transcripts accumulate in the infected shoots of a peach cultivar susceptible to L. theobromae
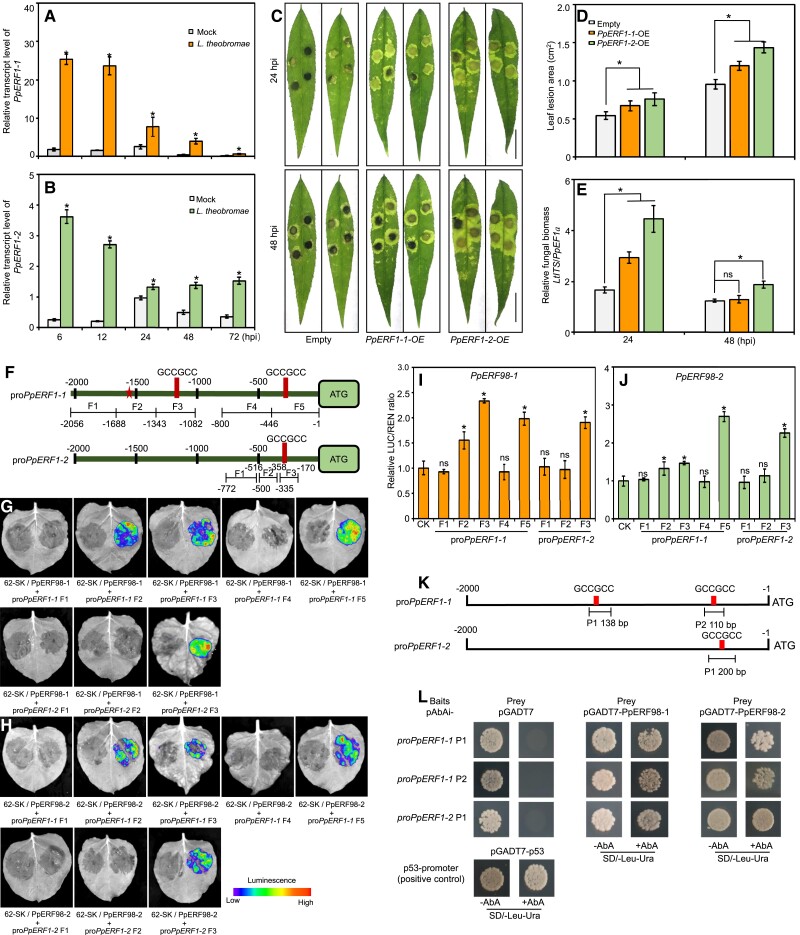
To investigate ET involvement in L. theobromae-induced peach gummosis, we identified 19 differentially expressed ERF members from a previous RNA-seq data set (Zhang et al. 2022). Two putative paralogs belonging to the IX subgroup of ERF family were dramatically upregulated in L. theobromae-infected peach shoots (Supplemental Figure S1A). Based on contig sequences, their coding sequences contained 540 and 492 bp encoding 179 and 163 amino acids, respectively. We designated them PpERF98-1 and 2 (hereafter indicated as PpERF98-1/2 when referring to both members, and likewise for other PpERFs), based on homology in Arabidopsis (Supplemental Figure S1B). The phylogenetic tree showed that PpERF98-1 and 2 are evolutionarily closest to ERF98 putative orthologs in Prunus mume, Prunus dulcis, and M. domestica (Supplemental Figure S1B).
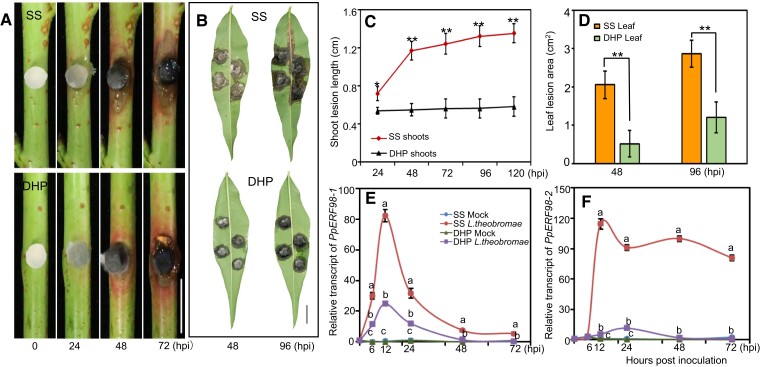
Subsequently, we quantified relative transcripts of PpERF98-1/2 in different peach cultivars infected with L. theobromae. The pathogen inoculation assays in shoots and leaves confirmed the susceptibility of the ‘Spring Snow’ (SS) cultivar to L. theobromae and the tolerance of ‘Da Hongpao’ (DHP) (Fig. 1, A and B). The infected shoots and leaves showed visible necrotic lesions at 48 h post-inoculation (hpi) in both cultivars, but the lesion size on susceptible shoots rapidly and continuously expanded, whereas expansion on tolerant shoots was significantly slower (Fig. 1C). At 48 and 96 hpi, leaf lesions on the susceptible cultivar were 3.9- and 2.4-fold larger than on the tolerant one, respectively (Fig. 1D). The transcripts of PpERF98-1/2 were comparable in the noninfected shoots of the 2 cultivars and highly induced in both after L. theobromae infection (Fig. 1, E and F). The transcript of PpERF98-1 was sharply upregulated at 6 hpi, peaked at 12 hpi, then declined (Fig. 1E), while PpERF98-2 was quickly and significantly upregulated at 12 hpi and remained high in tissues of the susceptible cultivar (Fig. 1F). Noticeably, the transcripts of PpERF98-1/2 were significantly higher in the susceptible cultivar SS than those in the tolerant cultivar DHP.
Figure 1.
PpERF98-1/2 transcripts accumulate in the infected shoots of a peach cultivar susceptible to L. theobromae. A and B) Morphological progression of gummosis in the current-year shoots and fully expanded leaves inoculated with L. theobromae. All leaf images were digitally extracted with the same scale in panels A (bar = 1 cm) and B (bar = 2 cm). C and D) Comparison of lesion size in the infected shoots and leaves between the susceptible cultivar ‘Spring Snow' (SS) and the tolerant cultivar ‘Da Hongpao' (DHP). Asterisks indicate statistical significance between SS and DHP at the same timepoint with *P < 0.05, **P < 0.01 using Student's t test. E and F) Relative transcript levels of PpERF98-1/2 in infected shoots normalized on the reference gene PpTEF2, and displayed as fold changes compared with mock samples at time 0 h. Hpi: hours post inoculation. Different letters show statistical significance of different treatments at the same time point and P < 0.05 based on Duncan’s post hoc test. Error bars represent means ± SD of three biological replicates.
Subsequently, the cis-acting regulatory elements in the promoter regions of PpERF98-1/2 were analyzed, identifying a variety of hormone- and stress-response elements (Supplemental Table S1). In detail, the promoter of PpERF98-1 contains multiple cis-binding elements for ET- (1), abscisic acid- (1), JA- (2), and auxin-related responsive factors (2), while the promoter of PpERF98-2 contains 4 ET-responsive elements, 1 ERF-binding site, 2 abscisic acid-responsive elements, and 2 auxin-responsive elements. We further tested the transcriptional response of PpERF98-1/2 to hormone treatments and ET inhibitors. The transcripts of PpERF98-1/2 were significantly upregulated by SA and ET treatment of shoot segments, whereas methyl JA (MeJA) induced PpERF98-1 but repressed PpERF98-2 transcripts (Supplemental Fig. S2). Transcripts of PpERF98-1/2 in the inoculated shoots treated with inhibitors of ET synthesis (aminoethoxyvinylglycine [AVG]) and perception (1-methylcyclopropene [1-MCP]) were significantly decreased, compared with the L. theobromae-infected shoots alone (Supplemental Fig. S3). These results indicate that PpERF98-1/2 are induced shortly after L. theobromae inoculation in dependence from ET synthesis and signaling and more intensely so in the susceptible cultivar.
PpERF98-1/2 are transcriptional activators that localize in the nucleus
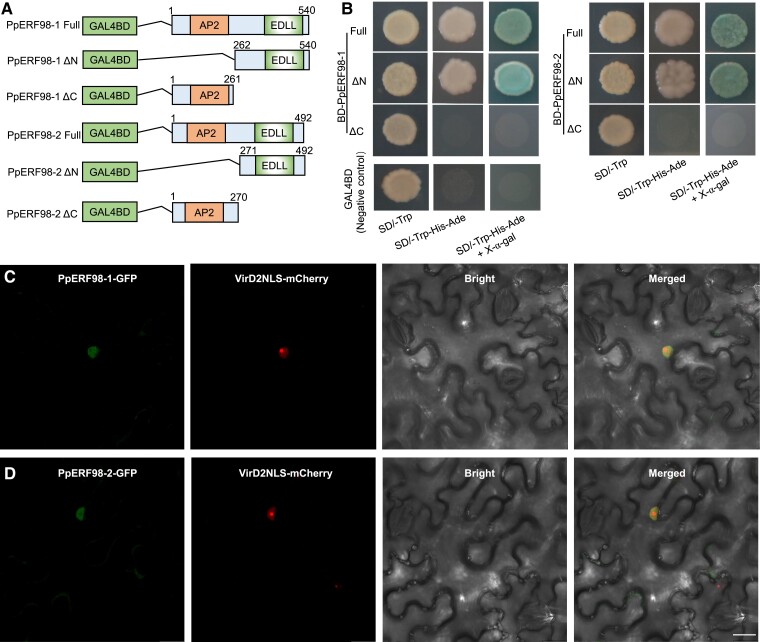
Multiple alignments revealed that PpERF98-1/2 have 1 highly conserved AP2 domain and a short EDLL motif at the N- and C-termini, respectively, and share high sequence similarity to ERF98 proteins of Arabidopsis, rice (Oryza sativa), tomato (S. lycopersicum), apple (M. domestica), plum (P. mume), and citrus (C. sinensis) (Supplemental Fig. S4). To examine the transcriptional activities of PpERF98-1/2, the full-length protein and truncated fragments were tested in a yeast transactivation reporter assay (Fig. 2A). Yeast cells grew well on the synthetic dextrose (SD)/-Trp medium, whereas only those transformed with constructs expressing the full-length PpERF98-1/2 or their C-termini exhibited normal growth on the selective media and displayed α-galactosidase activity (Fig. 2B). To analyze the subcellular localization, the 35S:PpERF98-1/2-GFP constructs and the nuclear marker VirD2NLS-mCherry were coinfiltrated into Nicotiana benthamiana leaves. The microscopic observation of the transiently cotransformed leaves showed the PpERF98-1/2-GFP signal in the nucleus, colocalized with the nuclear marker (Fig. 2, C and D). These results indicate that PpERF98-1/2 have transcription-activating capacity via the activation domain located at the C-terminus and localized in the nucleus. Both features are consistent with a role in transcriptional regulation.
Figure 2.
PpERF98-1 and 2 are transcriptional activators. A) Schematic diagrams of construct vectors used for transcriptional activity assays. The full-length and truncated fragments of PpERF98-1/2 were introduced downstream of the GAL4BD (galactose-specific transcription enhancing factor binding domain) in the pGBKT7 vector. PpERF98-1/2 ΔN and ΔC represent the deletion of the N and C termini of PpERF98-1/2 proteins, respectively. Numbers above shaded boxes indicate the position of nucleotides. AP2, APETALA2-domain; EDLL, a potent plant transcriptional activation domain. B) Growth of the yeast strain AH109 (Saccharomyces cerevisiae) transformed with the vectors, along with the negative control (pGBKT7), on synthetic dextrose (SD)/-Trp, SD/-Trp-His-Ade and SD/-Trp-His-Ade added with X-α-gal. C and D) Subcellular localization of PpERF98-1/2 based on the visualization of the GFP signal. The fusion construct PpERF98-1/2-GFP was co-transformed with the nuclear marker gene VirD2NLS fused to mCherry into Nicotiana benthamiana leaves. Confocal microscopic images of the cells were taken for GFP, mCherry fluorescences or bright field. Scaleb bar = 20 μm.
Knock-down of PpERF98-1/2 enhances peach resistance to L. theobromae
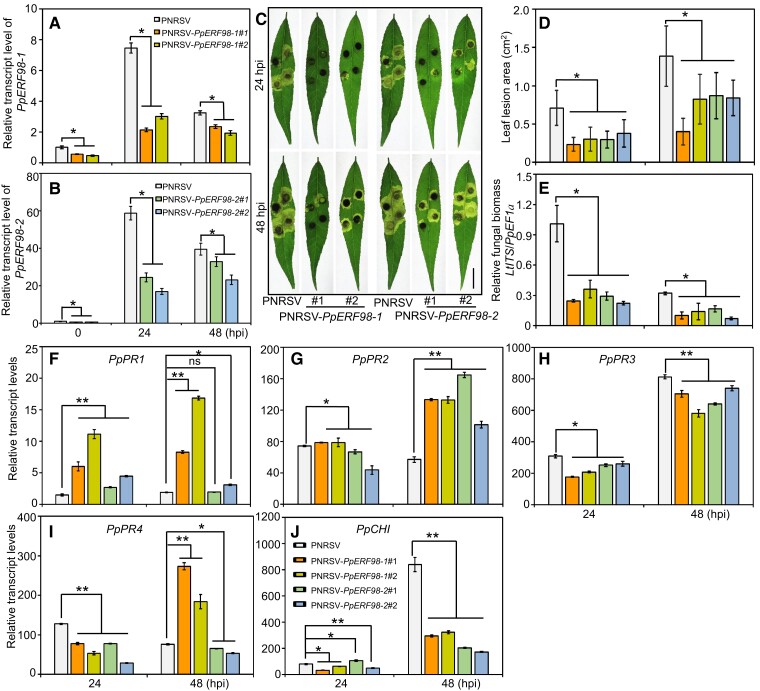
To functionally characterize PpERF98 in peach defense against L. theobromae, we generated peach plants silenced in PpERF98-1/2 expression using a Prunus necrotic ringspot virus (PNRSV)-mediated virus-induced gene silencing (VIGS) system. The top leaves of peach seedlings showed visible bleaching 2 wk after agroinfiltration with PNRSV:Phytoene desaturase (PpPDS) (Supplemental Figure S5, A and B), which verified the VIGS efficiency. The transcripts of PpERF98-1/2 were correspondingly reduced in the silenced plants compared with mock controls (PNRSV; Figs. 3, A and B, and S5, C and D). When infected with L. theobromae, the lesion area of PpERF98-1/2-silenced leaves decreased by ∼40% compared with the mock control (Fig. 3, C and D). Furthermore, the relative fungal biomass in the infected PpERF98-1/2-silenced peach seedlings was significantly lower than that of the infected, mock-transformed controls (Fig. 3E). In the infected plants at 24 or 48 hpi, the transcripts of PpPR1 and PpPR2 were significantly higher in the PpERF98-1/2-silenced plants than the mock control (Fig. 3, F and G). In contrast, the transcripts of PpPR3, PpPR4, and PpCHI were significantly reduced at 24 and/or 48 hpi (Fig. 3, H to J). These results suggest that the silencing of PpERF98-1/2 enhances peach resistance to L. theobromae and that resistance corresponds with PpPR1 and PpPR2 transcripts.
Figure 3.
VIGS of PpERF98-1 and 2 increases peach seedlings resistance to L. theobromae infection. A and B) Relative transcript levels of PpERF98-1/2 in PNRSV-mediated VIGS seedlings and the negative control (PNRSV). C) Phenotype of PpERF98-1/2-silenced peach leaves at 24 and 48 hpi. Bar = 1 cm. D and E) Lesion size and relative fungal biomass in the PpERF98-1/2-silenced peach leaves inoculated with L. theobromae.F to J) Relative transcript levels of PR genes (PRs; PpPR1, PpPR2, PpPR3, PpPR4, and PpCHI) in the PpERF98-1/2-silenced peach leaves infected with L. theobromae. Error bars represent means ± Sd of 3 independent replicates. Asterisks on top of bars indicate statistical significance with *P < 0.05 and **P < 0.01 using Student's t test, while ns indicates no significance.
OE of PpERF98-1/2 increases peach susceptibility to L. theobromae infection
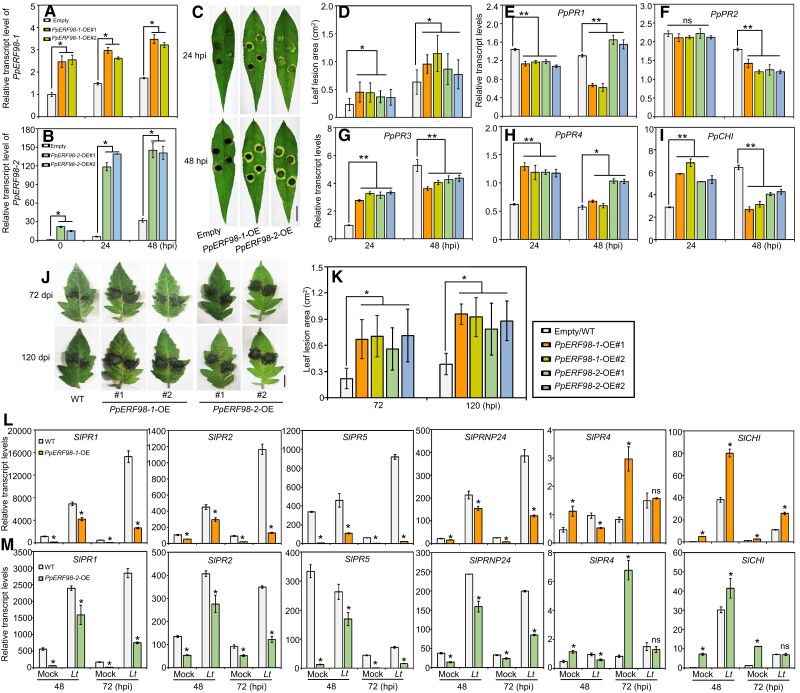
To confirm the hypothesis that PpERF98-1/2 promote susceptibility, PpERF98-1/2 were overexpressed individually in peach leaves, along with empty vector controls (Fig. 4, A and B), and then inoculated with L. theobromae. The lesions of PpERF98-1/2-overexpressing leaves were ∼1.5-fold larger than that of the inoculated, mock-transformed controls (Fig. 4, C and D). Consistently, the transcripts of PpPR1 and PpPR2 were significantly repressed by PpERF98-1/2-overexpression (OE) (Fig. 4, E and F). Notably, the transcripts of PpPR3, PpPR4, and PpCHI were significantly upregulated at 24 hpi, whereas PpPR3 and PpCHI were significantly downregulated at 48 hpi in the infected leaves of the PpERF98-1/2-overexpressing plants compared with the control (Fig. 4, G to I).
Figure 4.
OE of PpERF98-1 and 2 in peach and tomato leaves attenuates resistance to L. theobromae infection. A and B) Relative transcript levels of PpERF98-1/2-overexpressing peach seedling lines (#1 and #2). Peach seedlings transformed with the empty vector and inoculated with L. theobromae were regarded as controls (empty). C) Phenotype of PpERF98-1/2-overexpressing peach leaves inoculated with L. theobromae at 24 and 48 hpi. Leaf images were digitally extracted with a same scale, and bar = 1 cm. D) Lesion areas in L. theobromae-infected leaves of the PpERF98-1/2-overexpressing peach seedlings. E to I) Relative transcript levels of PRs (PpPR1, PpPR2, PpPR3, PpPR4, and PpCHI) in the infected leaves of PpERF98-1/2-overexpressing seedlings. J and K) Leaf symptoms and lesion areas in tomato wild type (WT) and lines (#1 and #2) constitutively overexpressing PpERF98-1/2 and inoculated with L. theobromae. Leaf images were digitally extracted with a same scale, and bar = 1 cm. L and M) Relative transcripts levels of PRs (SlPR1, SlPR2, SlPR5, SlPRNP24, SlPR4, and SlCHI) in the PpERF98-1/2-overexpressing tomato lines infected with L. theobromae (Lt) or treated with a sterile PDA plug (Mock). Relative transcript levels were normalized on the reference genes PpTEF2 in peach and SlActin in tomato and are displayed as fold change compared with mock samples at 0 hpi (which were therefore set to 1). Error bars represent means ± Sd of 3 independent replicates. Asterisks on top of bars indicate statistical significance with *P < 0.05 and **P < 0.01 using Student's t test, whereas ns indicates no significance.
PpERF98-1/2 negatively affect tomato defense against L. theobromae infection
To further consolidate the association between PpERF98-1/2 expression and susceptibility to L. theobromae in peach, we generated analogous tomato lines ectopically overexpressing PpERF98-1/2 in a stable fashion. Totally, 5 and 6 transgenic lines were obtained for the 35S:PpERF98-1 and 2 constructs, respectively (Supplemental Figure S5, E and F). The 2 lines for each gene with the highest transgene expression (labeled #1 and #2) were chosen for phenotypic and transcriptional analyses in comparison with wild type as a control.
The transgenic lines and wild-type plants showed comparable phenotypes in the absence of the pathogen. After inoculation with L. theobromae, leaf lesions of the PpERF98-1/2-overexpressing lines were ∼2-fold larger than those of the wild type (Fig. 4, J and K). Subsequently, we also quantified the transcripts of PR genes in response to L. theobromae. Compared with wild-type plants, the transcripts of SlPR1, SlPR2, SlPR5, and SlPRNP24 (Prion protein 24) were consistently and significantly decreased in noninoculated, PpERF98-1/2-overexpressing lines, indicating that their basal expression was affected (Fig. 4, L and M). When inoculated with L. theobromae, the transcripts of these 4 SlPRs were upregulated in all plants, but their transcripts in the PpERF98-1/2-overexpressing lines were significantly less concentrated than in the wild type. However, the transcripts of SlPR4 were significantly increased in the uninfected PpERF98-1/2-overexpressing plants and were repressed after L. theobromae infection, while transcripts of SlCHI were highly accumulated in the transgenic lines irrespective of inoculation (Fig. 4, L and M). In addition, when tomato plants were challenged by the necrotrophic pathogen B. cinerea, the necrotic lesions on PpERF98-1/2-overexpressing leaves were larger than in the wild type (Supplemental Figure S6, A and B). Transcripts of most SlPRs (except for SlPR4 and SlCHI) in the PpERF98-1/2-overexpressing plants were significantly repressed compared with the wild type after B. cinerea infection (Supplemental Figure S6, C and D). These results confirm, in stably transformed tissues, the role of PpERF98-1/2 also in the interaction of a taxonomically unrelated plant with the same pathogen and with a pathogen with a slightly different lifestyle (necrotrophic vs. hemi-biotrophic).
PpERF98-1/2 activate the transcription of 2 PpERF1 genes and of their shared PR gene targets
Based on the expression patterns of the bona fide marker genes PR1 and PR2 (for the SA-dependent defense pathway) and PR3, PR4, and CHI (for the JA/ET pathway) (Lorenzo et al. 2003) in PpERF98-1/2-overexpressing or silenced plants, we hypothesized that PpERF98-1/2 might be a cross talk node between these 2 pathways. Both our reverse transcription quantitative PCR (RT-qPCR) and RNA-seq data showed that 2 PpERF1 genes (hereafter designated PpERF1-1 and 2) were significantly induced by L. theobromae inoculation (Figs. 5, A and B, and S1A). ERF1, belonging to the IX subcluster of the ERF family together with ERF98, is a central regulator in the JA/ET signaling pathway (Lorenzo et al. 2003). Since ERF1 is a hub gene in the JA/ET and SA-related defense pathways in Arabidopsis, we hypothesized that also for peach and tested if PpERF98-1/2 may interact with PpERF1.
Figure 5.
PpERF1-1/2 negatively regulate peach defense against L. theobromae infection, and are activated by PpERF98-1/2. A and B) Time-course transcripts of PpERF1-1/2 in shoots infected with L. theobromae. C) Lesion phenotype of the infected peach leaves of PpERF1-1/2-overexpressing plants at 24 and 48 hours post inoculation (hpi). Peach seedlings transformed with the empty vector and inoculated with L. theobromae were regarded as controls (Empty). Leaf images were digitally extracted with a same scale, bars = 2 cm. D and E) Lesion size and relative fungal biomass in the infected leaves of peach seedlings transiently overexpressing PpERF1-1/2. Data are means ± SD of three independent replicates and four analytical replicates. F) Schematic diagram of different fragments (Fn) used for reporter constructs for luciferase (LUC) activity assay. The pentagram and rectangle stand for the putative ERF binding site and the GCC-box (GCCGCC), respectively, in the promoters of PpERF1-1/2 (proPpERF1-1/2). Numbers below the fragments indicate the positions of the nucleotides at the 5'-3' end of each fragment relative to the translation start-site in reporters. G and H) Transient expression assays showing that PpERF98-1/2 could activate the expression of PpERF1-1/2. Luminescence imaging of N. benthamiana leaves is shown 48 h after co-infiltration with different pairs of reporter and effector constructs. pGreenII-62-SK(62-SK)+pGreenII-0800-promoter-LUC(proPpERF1-1/2 Fn) was the control on the left, and 62-SK-PpERF98-1/2(PpERF98-1/2)+pGreenII-0800-promoter-LUC(proPpERF1-1/2 Fn) was the treatment on the right in one N. benthamiana leaf. I and J) Activation of proPpERF1-1/2 by PpERF98-1/2 in N. benthamiana leaves, as measured by the transient dual LUC assay. Data are means ± SD of five independent replicates. K) Schematic diagram of fragments (Pn) of proPpERF1-1/2 used as baits in yeast one-hybrid assay. L) Interaction analysis of PpERF98-1/2 with the fragments of proPpERF1-1/2. Transformed yeast cells containing both prey and bait were cultured on the selective medium SD/-Leu-Ura+AbA (aureobasidin A). Co-transformation of pGADT7-53 and pAbAi-p53 was used as positive and of pGADT7 and pAbAi-bait as negative controls, respectively. Asterisks on top of bars show statistical significance between transgenic and control plants at same time point and P < 0.05 using Student's t test, while ns indicates no significance.
Firstly, PpERF1-1/2 were successfully overexpressed in peach leaves (Supplemental Fig. S7). The leaf lesions and relative fungal biomass were significantly increased by PpERF1-1/2-OE (Fig. 5, C to E), implying that each of them promotes peach susceptibility to L. theobromae infection. The coding sequences of PpERF98-1/2 were separately used as effectors and promoter fragments of PpERF1-1/2 (proPpERF1-1/2) as reporters fused to luciferase (LUC) in a dual-LUC assay (Fig. 5F). The results indicated that coexpression of either PpERF98-1/2 effector with any proPpERF1-1/2 reporter fragment containing a GCC-box element and putative ERF-binding site (Fig. 5F) dramatically increased LUC activity relative to the control. By contrast, the reporter sequences without the ERF-binding site did not (Fig. 5, G and H). The LUC/Renilla (REN) ratio values corroborate the finding (Fig. 5, I and J). Yeast 1-hybrid (Y1H) assay confirmed physical binding of PpERF98-1/2 and proPpERF1-1/2 fragments containing the GCC-box element (Fig. 5, K and L). Although the dual-LUC assay showed that PpERF98-1/2 could activate fragment F2 in the PpERF1-1 promoter, yeast cells harboring it could grow on selective media even in the absence of exogenous effectors, making it impossible to test physical binding by PpERF98-1/2 via Y1H assay. These results collectively demonstrate that PpERF98-1/2 can function as transcriptional activators of PpERF-1/2 via the GCC-box element in proPpERF1-1/2 sequences.
Given the upregulation of PpPR3, PpPR4, and PpCHI in PpERF98-1/2-overexpressing plants (Fig. 4), we speculated that these PR genes may also be regulated by PpERF98 proteins. The promoters of PpPR3, PpPR4, and PpCHI contain putative ERF-binding sites (ERE) (Supplemental Figure S8A). The coexpression of PpERF98-1/2 effectors, and of promoter fragments for PpPR3, PpPR4, and PpCHI containing the ERE element, showed robust luminescence intensity compared with the control (Supplemental Figure S8B). This confirms that PpERF98-1/2 could promote the transcription of the PR genes PpPR3, PpPR4, and PpCHI, which are induced by the JA/ET pathway based on published findings in Arabidopsis and our own in peach.
PpERF98-1 and 2 form homodimers/heterodimers and interact with PpERF1-1/2 proteins to promote peach susceptibility to L. theobromae
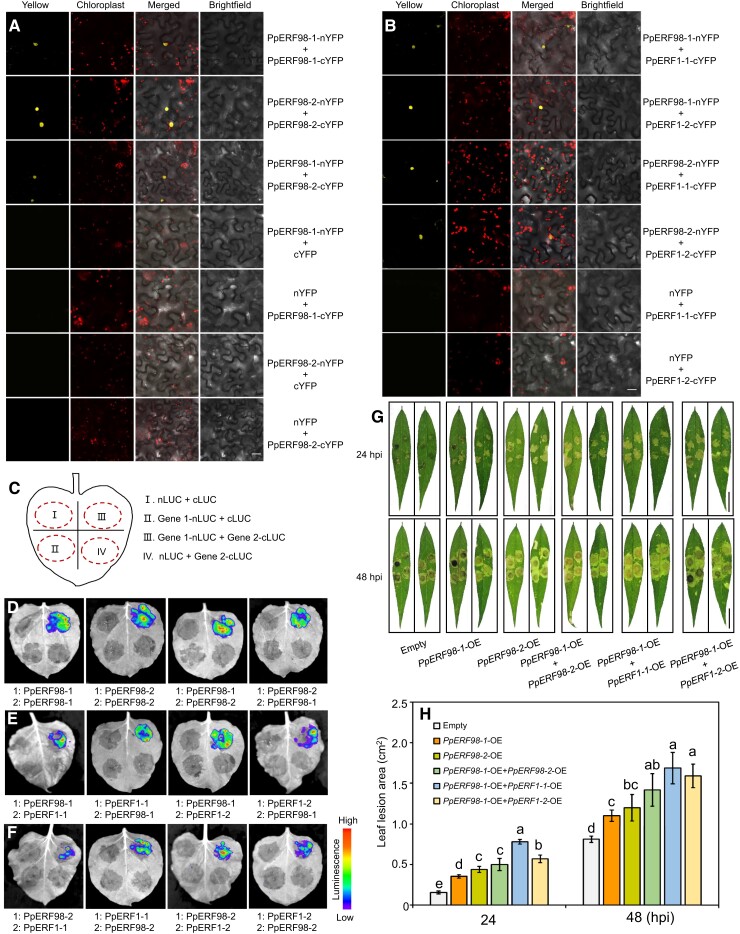
A previous study in Arabidopsis has shown that ERF proteins can form homodimers/heterodimers with themselves or each other in responses to heat stress (Huang et al. 2021). To validate a possible physical interaction between PpERF98-1 and 2; PpERF1-1 and 2; and PpERF98s and PpERF1s, we used the bimolecular fluorescence complementation (BiFC) and LUC complementation imaging (LCI) assays. Coexpression of PpERF98-1-nYFP (N-terminal yellow fluorescent protein) and PpERF98-1-cYFP (C-terminal YFP) in N. benthamiana leaves resulted in strong YFP signals in the nucleus. Similar results were observed with the cotransformation of PpERF98-2-nYFP and PpERF98-2-cYFP (Fig. 6A), demonstrating that both PpERF98-1 and 2 can form homodimers. Coinfiltration of PpERF98-1-nYFP with PpERF98-2-cYFP into N. benthamiana leaves also showed YFP signals in the nucleus, indicating that PpERF98-1 can interact with PpERF98-2 and form heterodimers. Negative controls, i.e. PpERF98-1/2-nYFP coexpressed with cYFP, or nYFP coexpressed with PpERF98-1/2-cYFP, did not develop fluorescence (Fig. 6A). Further, coexpression of PpERF98-1/2-nYFP with PpERF1-1/2-cYFP showed YFP signals in the nucleus, implying that PpERF98-1/2 can interact with PpERF1-1/2 (Fig. 6B). These results were confirmed by LCI assays, for which a simplified layout is shown in Fig. 6C. Here, the reconstitution of LUC expressed as separate C- and N-terminal fragments in fusion with each of the PpERF98-1/2 or PpERF1-1/2 proteins led to the emission of luminescence. As shown in Fig. 6, D to F, the same pairs driving BiFC complementation led to LUC reconstitution. However, PpERF1-1 cannot form dimers with itself or PpERF1-2 (Supplemental Fig. S9).
Figure 6.
PpERF98-1 and 2 form heterodimers and homodimers and interact with PpERF1-1/2 in aggravating peach seedlings susceptibility to L. theobromae. A) In vivo interaction between PpERF98-1 and PpERF98-2 by BiFC assay in N. benthamiana leaves. Bar = 20 μm. B) Heterodimer formation between PpERF98-1/2 and PpERF1-1/2 in vivo by BiFC analysis. Bar = 20 μm. C) Schematic diagram of the construct combinations for LCI analysis, showing 1 experimental group (Gene1-nLUC + Gene2-cLUC) and 3 parallel controls in 1 tobacco leaf. D) Interactions between PpERF98-1 and PpERF98-2 by LCI. E and F) Interactions between PpERF98-1/2 and PpERF1-1/2 as determined by LCI. Combinations of nLUC or cLUC with the corresponding PpERF98-1/2 and PpERF1-1/2 constructs were used as negative controls. G and H) Lesion phenotype and size in peach leaves after coinfiltration with PpERF98-1/2-OE and PpERF1-1/2-OE constructs in Agrobacterium for 48 h and inoculation with L. theobromae at 24 and 48 hpi. Bar = 2 cm. Peach seedlings transformed with the empty vector and inoculated with L. theobromae were regarded as controls (empty). Leaf images were digitally extracted with a same scale in panel G), bar = 2 cm. Different letters show statistical significance of different treatments at the same time point and P < 0.05 based on Duncan's post hoc test. Error bars represent are means ± Sd of 3 independent replicates.
These results drove us to explore whether dimerization by PpERF98s and PpERF1s could increase peach sensitivity to the gummosis pathogen. Thus, peach leaves were coinfiltrated with the OE constructs PpERF98-1/2-OE and PpERF1-1/2-OE in various combinations and inoculated with L. theobromae. All OE combinations of PpERF98-1/2 and PpERF1-1/2 obviously increased lesion areas compared with empty vector controls (Fig. 6G). Lesion areas were significantly increased in leaves cooverexpressing PpERF98-1 and PpERF1-1/2, while only slightly increased in leaves cooverexpressing PpERF98-1 and PpERF98-2, relative to leaves overexpressing either 1 alone (Fig. 6, G and H). These data indicate that PpERF98-1 and 2 can form homodimers with themselves or heterodimers with each other and that they can interact with PpERF1-1/2 proteins, increasing peach susceptibility to L. theobromae.
PpERF98-1/2 modulate the JA/ET- and SA-related defense pathways
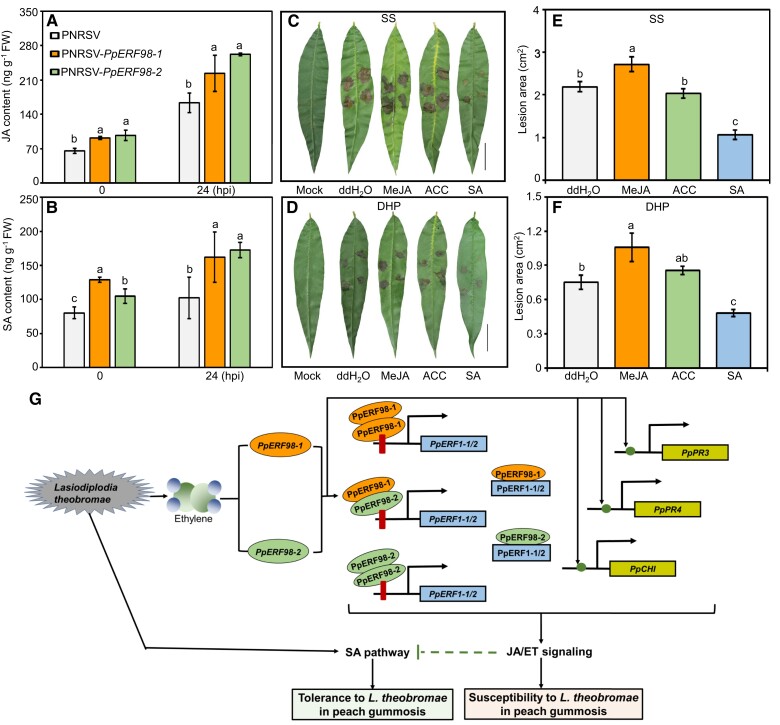
To clarify whether hormone contents corresponded with PpERF98-1/2 levels, we quantified JA and SA after L. theobromae inoculation in peach leaves silenced in PpERF98-1/2 or tomato plants overexpressing them. Both JA and SA contents in PpERF98-1/2-silenced peach leaves were significantly higher than in the controls regardless of L. theobromae inoculation (Fig. 7, A and B). The JA content of the PpERF98-1/2-overexpressing tomato leaves was instead lower than that of the wild type but showed no significant difference when inoculated (Supplemental Figure S10A). The SA content of these plants was also significantly decreased, by at least 42% compared with mock-transformed controls, when infected with L. theobromae (Supplemental Figure S10B). Consistently, pretreatment with exogenous MeJA of both the susceptible and tolerant peach genotypes significantly increased the lesion areas relative to uninoculated, and the water pretreated and inoculated controls, irrespective of peach cultivars; SA treatment decreased them (Fig. 7, C to F). However, the 1-aminocyclopropane-1-carboxylic acid (ACC, a key ET biosynthetic precursor) treatment group showed comparable lesion size relative to L. theobromae inoculation alone.
Figure 7.
The defense pathway of JA/ET and SA are regulated by PpERF98-1/2. A and B) Accumulation of JA and SA in the PpERF98-1/2-silenced peach seedlings at 0 and 24 hpi with L. theobromae. C to F) Lesion symptom and size in the hormone-treated leaves of the susceptible cultivar SS and the tolerant cultivar DHP prior to L. theobromae inoculation. Fully expanded leaves were evenly sprayed with 100 µM MeJA, 500 µM SA, 500 µM ACC (ET precursor, 1-aminocyclopropane-1-carboxylate), or ddH2O for 24 h prior to L. theobromae or mock inoculation. Leaf images were digitally extracted with a same scale in panes C) and D), bar = 2 cm. Different letters on top of bars indicate statistical significance at the same time point and P < 0.05 based on Duncan's post hoc test. G) Proposed regulatory model for the role of PpERF98-1 and 2 in peach response to the gummosis fungus L. theobromae. When plants are infected by L. theobromae, large amounts of ET are produced and induce the transcription of PpERF98-1/2. These 2 PpERF98 proteins form heterodimers and homodimers with themselves and directly bind to the GCC-box cis-elements of the promoters of PpERF1-1/2, activating their transcription. Further, these 2 PpERF98 proteins interact with 2 PpERF1 proteins to form heterodimers and amplify JA/ET signaling. Moreover, the PpERF98s can also activate the transcription of the JA/ET-responsive genes PpPR3, PpPR4, and PpCHI through putative ERF-binding cis-elements. Blocking the JA and ET pathways can decrease peach susceptibility to L. theobromae, while SA can promote tolerance (our earlier work; Zhang et al. 2022). Altogether, our data show that 2 interacting PpERF98 genes negatively regulate peach resistance to L. theobromae by activating JA/ET signaling to attenuate the SA-dependent defense pathway. Red bars represent GCC-box, and solid green circles indicate other putative ERF-binding sites in target genes.
Subsequently, we quantified the relative transcripts of JA/ET- and SA-related genes in the PpERF98-1/2-silenced or overexpressing plants. In L. theobromae-infected peach seedlings, the transcripts of PpERF1-1/2 were significantly decreased by 50% in the PpERF98-1-silenced plants compared with mock-transformed, infected controls (Supplemental Figure S11, A and B). This agrees with the data reported above, about the activation of PpERF1-1/2 promoter sequences by PpERF98-1/2. The transcript of PpICS2 (Isochorismate synthase 2), a key SA biosynthetic gene, was upregulated in PpERF98-1-silenced plants irrespective of L. theobromae inoculation (Supplemental Figure S11C). NPR1 (nonexpresser of PR 1) and TGA (TGACG motif-binding factor) are crucial components of SA signaling (Zhang et al. 2003). The transcript of PpNPR1 was downregulated after L. theobromae inoculation but slightly less so in the PpERF98-1-silenced plants than in the mock-silenced, infected control (Supplemental Figure S11D). The silencing of PpERF98-1 significantly also increased the transcript of PpTGA3 (Supplemental Figure S11E). In the PpERF98-1-overexpressing plants, on the other hand, the transcripts of PpERF1-1/2 were significantly upregulated when noninoculated, but transcripts did not increase in the infected leaves as much as in the corresponding controls (infected but not overexpressing; Supplemental Figure S11, F and G). The transcripts of PpICS2, PpNPR1, and PpTGA3 were significantly repressed in the infected PpERF98-1-overexpressing plants relative to the controls (Supplemental Figure S11, H to J). Similar transcriptional patterns were observed for these genes in the peach seedlings altered in PpERF98-2 expression (Supplemental Fig. S12). These results demonstrate that PpERF98-1/2 confer peach susceptibility to L. theobromae infection by positively regulating the ERF branch of the JA/ET pathway and repressing SA-related defense genes, acting at the level of hormonal biosynthesis, and/or in downstream signaling.
Discussion
Gummosis is a destructive trunk fungal disease that causes huge economic losses in peach production and quality. ERF transcription factors play important roles in response to diverse pathogens, and different ERFs have positive and/or negative functions in plant resistance depending on the pathogen lifestyles (Berrocal-Lobo et al. 2002; Müller and Munné-Bosch 2015; Van den Broeck et al. 2017; Feng et al. 2020). However, how individual ERF members participate in peach resistance to gummosis-inducing pathogens remains poorly elucidated. Hereto, we comprehensively deciphered the role of 2 interacting PpERF98 proteins involved in peach defense to L. theobromae-mediated gummosis and shed light on part of their regulatory network (Fig. 7G).
PpERF98-1/2 reduce peach resistance to gummosis caused by the hemi-biotrophic pathogen L. theobromae
In Arabidopsis, 4 small ERFs (ERF95, ERF96, ERF97, and ERF98) are involved in the regulation of plant tolerance to stresses; for example, AtERF98 positively modulates salt tolerance by activating ascorbic acid biosynthesis (Zhang et al. 2012), while AtERF96 and AtERF97 modulate plant defense against biotic attack (Catinot et al. 2015; Wang et al. 2015; Wang et al. 2017; Huang et al. 2021). AtERF98 is identified as a core component of a transcription factor network managing resource allocation decisions between abiotic stress tolerance and leaf growth (Van den Broeck et al. 2017), but a role for ERF98 in biotic stress has not been reported yet.
Most members of the IX subgroup in the ERF family are involved in plant defense responses to pathogens (Nakano et al. 2006a). Two peach ERF98s also belong to this subgroup and contain the EDLL motif at the C-termini (Supplemental Fig. S4), which was identified as a strong plant transcriptional activation domain in AP2/ERF transcription factors (Tiwari et al. 2012). As these putative PpERF98 paralogs are significantly and differentially expressed upon L. theobromae inoculation (Supplemental Fig. S1), we focused on their roles in peach responses to gummosis. The starting hypothesis was that they would increase susceptibility to L. theobromae, since they are significantly more expressed in the susceptibility than in the tolerant peach cultivar (Fig. 1) and are induced by ET, which promotes susceptibility (Zhang et al. 2022). Indeed, the PpERF98-1/2-overexpressing peach and tomato plants were more susceptible to infection by L. theobromae, whereas the PpERF98-silenced peach plants were more tolerant (Figs. 3 and 4). These results confirm that PpERF98-1/2 negatively regulate plant resistance to this hemi-biotrophic causal agent of gummosis. In addition, PpERF98-overexpressing tomato lines were more susceptible to the necrotrophic pathogen B. cinerea than the wild type (Supplemental Fig. S6). While internally consistent, this finding may seem to contradict previous reports, where other members of subgroup IX in the ERF family such as AtERF1, AtORA59, and AtERF96 positively regulate A. thaliana resistance to B. cinerea (Lorenzo et al. 2003; Pré et al. 2008; Catinot et al. 2015). However, previous studies have demonstrated that SA, not JA, plays a major role in plant basal defense to B. cinerea in tomato (Audenaert et al. 2002; Achuo et al. 2004; Li and Zou 2017; Zhang et al. 2018). Therefore, this suggests that the role of each ERF gene will depend on the specific pathosystem and thus will require direct experimental confirmation for each plant–pathogen pair.
PpERF98-1/2 control the expression of PpERF1 and activate the JA/ET-dependent pathway
ERF1 has been known to act as important regulatory hub for the JA/ET pathway (Lorenzo et al. 2003; Huang et al. 2016). OE of AtERF1 positively regulates plant resistance to B. cinerea through binding to the GCC element of the defense-related PR genes PDF1.2a, PR3, PR4, and CHI, whereas it decreases resistance to the hemi-biotrophic bacterium P. syringae (Lorenzo et al. 2003). In our study, OE of PpERF1-1/2 increased peach susceptibility to the hemi-biotrophic fungus L. theobromae (Fig. 5, C and D), consistently with the function of AtERF1 in Arabidopsis response to P. syringae. Notably, the promoters of PpERF1-1/2 contain GCC-boxes, and we could show that PpERF98-1/2 can directly bind them to activate gene transcription (Fig. 5, G and H), implying that PpERF98-1/2 work upstream of PpERF1-1/2. Also AtERF96, another small ERF, could control the transcription of AtORA59 and JA/ET-dependent defense genes by binding to the GCC elements of their promoters (Catinot et al. 2015). Although the GCC-box was not found in the promoters of PpPR3, PpPR4, and PpCHI, PpERF98-1/2 could activate their transcription via other putative ERF-binding elements (Supplemental Fig. S8). Our results suggest that PpER98s, like AtERF1, AtORA59, and AtERF96, could regulate specific subsets of defense-related genes and activate the JA/ET signaling pathway by targeting the GCC-box element or other ERF-binding elements in the promoters of their targets (Lorenzo et al. 2003; Pré et al. 2008; Catinot et al. 2015).
The defense responses activated by the JA/ET-dependent pathways are especially effective against necrotrophic pathogens and insects, whereas SA-dependent defense signaling is thought to be crucial against biotrophic and hemi-biotrophic pathogens; L. theobromae is a typical hemi-biotrophic fungus (Wang et al. 2011; Delgado-Cerrone et al. 2016; Liu et al. 2022). Our study showed that JA treatment significantly increased lesion area in infected peach leaves, while SA treatment alleviated it (Fig. 7), in accordance with our previous results (Zhang et al. 2022). These findings imply that JA/ET promote peach susceptibility to the hemi-biotrophic L. theobromae, while SA positively regulates resistance.
Does L. theobromae hijack the ERF branch of the JA/ET defense pathways, to suppress SA synthesis and signaling and favor tissue colonization?
In PpERF98-1/2-silenced peach seedlings, we observed higher SA content and transcription of the SA-related genes PpICS2, PpTGA3, and PpPR1 (Figs. 3 and 7). JA content was significantly increased in the same plants, but common targets of JA/ET signaling (e.g. PpPR3, PpPR4, and PpCHI) (Naidoo et al. 2013) were downregulated (Figs. 3 and 7). The opposite finding was observed in the PpERF98-1/2-overexpressing plants (Figs. 4, S11, and S12). Therefore, we speculate that PpERF98-1/2 positively regulate JA/ET signaling and thus downregulate SA biosynthesis, signaling, and responses. This hormonal imbalance will increase peach susceptibility to L. theobromae.
In Arabidopsis, the JA/ET-induced defense signaling is antagonistic to the SA-dependent defense pathway, which allows the plant to fine-tune defense strategies against pathogens (Kunkel and Brooks 2002). Several biotrophic and hemi-biotrophic pathogens have developed strategies to activate JA/ET signaling, suggesting this to be beneficial for the attacker due to the attenuation of the SA-dependent defense responses (Doehlemann et al. 2008; Howe et al. 2018; Darino et al. 2021). For instance, several pathogens like L. theobromae and P. syringae can generate JA or JA analogs. These in turn inhibit SA biosynthesis in the infected plants, thus resulting in suppression of SA-mediated defense responses and decreased plant resistance (Uppalapati et al. 2007; Tsukada et al. 2010). In maize (Zea mays) and Arabidopsis, ectopic expression of Jsi1 (encoding the Ustilago maydis effector JA/ET signaling inducer 1) leads to stimulation of the ERF branch of the JA/ET signaling pathway and increases susceptibility to U. maydis (Darino et al. 2021). Thus, it is tempting to speculate that the same may occur in the peach tree–L. theobromae pathosystem and that L. theobromae activates or hijacks the PpERF98-dependent branch of the pathway to attenuate SA biosynthesis/signaling and trigger susceptibility. The characterization of the role and downstream targets of PpERF98-1/2 achieved in this work are important preliminary steps to explore this possibility.
The formation of homodimers/heterodimers by PpERF98-1/2 and PpERF1-1/2 could enhance their functionality
Some studies have demonstrated that most regulatory proteins physically interact with other proteins, to form complexes participating in stress defense (Chi et al. 2013). Noticeably, their interaction with transcription factors could enhance DNA binding (Alves et al. 2014). WRKY proteins, for example, can interact with themselves and with each other to form homo/hetero complexes, which affect the DNA binding and transcription-regulatory activity, thereby modulating WRKY-regulated gene expression (Chi et al. 2013). For instance, cotton (Gossypium hirsutum) GhWRKY41 physically interacts with itself and directly activates its own transcription and the expression of the phenylpropanoid pathway genes, thereby increasing lignin and flavonoids biosynthesis and conferring enhanced resistance against Verticillium dahliae (Xiao et al. 2023). Recently, Huang et al. (2021) showed AtERF95 and AtERF97 to form homodimers or heterodimers with themselves or each other and this interaction to be heat inducible. Here, PpERF98-1/2 proteins also could form homodimers and heterodimers with themselves and each other (Fig. 6, A and D). Combined PpERF98-1 and 2 OE increased peach susceptibility compared to the individual OE of either gene (Fig. 6, G and H). Furthermore, PpERF98-1/2 could form heterodimers with the PpERF1-1 and 2 proteins (Fig. 6, B, E and F). Cooverexpression of PpERF98-1 and PpERF1-1 or 2 further increased peach susceptibility to L. theobromae (Fig. 6H). However, PpERF1-1 and 2 could not form dimers with themselves (Supplemental Fig. S9). Therefore, we speculate that dimer formation between ERF98 and ERF1 proteins may promote binding to the cis-elements of downstream targets, thus maximizing the activation of the JA/ET signaling pathways.
Conclusions
We propose a working model on the role and regulatory network of PpERF98-1/2 in peach resistance to L. theobromae (Fig. 7G). Our previous study has shown that L. theobromae infection induces the accumulation of ET and JA, which in turn reduce the resistance of peach trees to L. theobromae during gummosis progression (Zhang et al. 2022). In this work, ET and infection activate the transcription of PpERF98-1/2. Homodimers/heterodimers of PpERF98-1/2 directly bind the promoters of 2 PpERF1 genes activating their transcription. These encode core regulatory proteins in the ERF branch of JA/ET signaling, which in turn can form heterodimers with PpERF98-1/2, thus likely modulating their activity. PpERF98-1/2 could also activate directly a common subset of target genes downstream of JA/ET signaling: PpPR3, PpPR4, and PpCHI. Dimer formation between PpERF98s and PpERF1s could maximize the activation of the ERF branch in JA/ET signaling and thus suppress the SA-dependent defense pathway at the biosynthesis and signaling level; this is likely the key mechanism by which they decrease peach resistance to gummosis. All findings advance our understanding of the interactions among ERF proteins in plant responses to biotic stresses and of the molecular underpinnings of peach susceptibility to the devastating gummosis pathogen.
Materials and methods
Biological materials and growth conditions
The ‘SS’ and ‘DHP’ peach (P. persica Batsch) cultivars grafted on wild peach rootstocks, showing susceptibility and tolerance to gummosis, respectively, were used in this study. Lignified, current-year shoots were collected from 5-yr-old trees in the peach orchard of Huazhong Agricultural University (Wuhan, China) between July and August. For seedlings generation, mature wild peach seeds were stratified in moist vermiculite and kept at 4 °C for 3 mo to sprout and then transplanted into plastic pots containing commercial nursery substrate: perlite:vermiculite:nutrient soil 2:1:1:1 (Shangdao Biotech, Shandong, China). The tomato (S. lycopersicum) A57 and N. benthamiana seeds were also potted into the same substrate after germination. All seedlings were cultivated in a growth room at 25 °C, 65% relative humidity with 16-h light (300 μmol s−1 m−2) and watered with full-strength Hoagland solution every 4 d.
The L. theobromae strain was originally isolated from an infected peach tree in the Hubei Province, China (Wang et al. 2011). B. cinerea strain B05 was used to infect tomato leaves. The pathogens were cultured on potato dextrose agar (PDA) medium at 28 °C under a 12-h dark/12-h light for 3 d before inoculation.
Pathogen inoculation and chemical treatment
Peach shoots were inoculated as described previously, with slight modifications (Zhang et al. 2022). Leaves were stripped off current-year shoots, which were cut into 15-cm segments. After surface wash and sterilization, the segments were wounded and inoculated with a 5-mm diameter PDA plug covered with L. theobromae mycelium. In parallel, sterile PDA plugs were placed onto the wound as a mock. The inoculated/mock peach shoots were separately placed in plastic boxes with moist filter paper at the bottom and covered with transparent plastic film to maintain humidity; the shoot segments were sprayed with sterilized water every 12 h. Two days after inoculation, the shoots were individually placed in an upright position into plastic bottles containing 200-mL sterilized water, which was refreshed daily, and nursed as above. Unless specified otherwise, the shoots were incubated in a growth chamber at 28 °C, with 90% relative humidity and 12-h light (300 μmol s−1 m−2). For leaf inoculation assays, 5-mm agar plugs covered with mycelium were placed onto the abaxial side of freshly detached, fully expanded peach or tomato leaves, on both sides of the main rib. In parallel, sterile PDA plugs acted as mock inocula.
To evaluate the effect of hormones on transcription, peach shoots prepared as above were pretreated with 100 µM MeJA, 500 µM SA, 10 µL L−1 ET, 15 ng mL−1 AVG, or 0.1 mg mL−1 1-MCP (Zhang et al. 2022). For leaf treatment, fully expanded peach leaves were sprayed with 100 µM MeJA, 500 µM SA, 500 µM ACC, or ddH2O. After 24 h, the pretreated leaves were inoculated with L. theobromae or not (mock) and nursed as aforementioned.
The time of pathogen inoculation was set as the starting point in all treatments. Each treatment and time point was run in 3 biological replicates made of 15 pooled shoot segments or leaves. For lesion phenotyping, 40 individual peach shoot segments, and 20 peach or tomato leaves per replicate were used in each treatment. At harvest, the shoot and leaf tissues within 0.5 to 1.0 cm of lesion were sampled at 0, 6, 12, 24, 48, and 72 hpi for peach shoots and at 0, 24, and 48 hpi for leaves. Tomato leaves were sampled at 0, 48, 72, and 120 hpi (with L. theobromae) or at 0, 48, and 72 hpi (with B. cinerea), instantly frozen in liquid nitrogen and stored at −80 °C for further analyses.
Gene cloning and RT-qPCR assays
Total RNA was isolated using the EASY spin Plus RNA kit (Aidlab, Beijing, China), followed by DNase digestion. Single-strand cDNA was synthesized using the PrimeScript RT reagent Kit with gDNA Eraser (TaKaRa, Kusatsu, Japan). The coding sequences of PpERF98-1 and 2 (Prupe. 8G224700 and Prupe. 1G037800) and PpERF1-1 and 2 (Prupe. 6G348700 and Prupe. 8G224600) were downloaded from the GDR database (https://www.rosaceae.org/) and amplified with specific primers (Supplemental Table S2).
The RT-qPCR assays were performed as previously reported (Zhang et al. 2022). The putative gene sequences were retrieved from NCBI, and the primers are shown in Supplemental Table S2. The gene transcript levels were normalized on the values of PpTEF2 (translation elongation factor 2) in peach (Gao et al. 2016) or SlActin in tomato (Yang et al. 2018). Genomic DNA of the pathogen and host was extracted with cetyltrimethylammonium bromide and quantified in the infected tissues using qPCR (Zhang et al. 2021) with PpEF1α (elongation factor 1α) and LtITS as plant and fungus internal control genes, respectively. The fold-change values were calculated using the 2−ΔΔCT method, and RT-qPCR data are presented as means of 3 biological and 4 analytical replicates.
Plant transformation
For the stable transformation in tomato, PpERF98-1/2 coding sequences were amplified using Phanta Max Super-Fidelity DNA Polymerase (Vazyme, Nanjing, China) and cloned into the pK7WG2D vector by a Gateway technology. The recombinant vectors were transformed into Agrobacterium tumefaciens GV3101 and the transgenic lines were generated and identified as described previously (Horsch et al. 1985), selected under 100 mg L−1 kanamycin, and raised to T2 generation for further analysis, while wild-type plants grown under the same conditions were regarded as controls.
Transient OE in peach leaves was achieved as previously described (Cui and Wang 2017; Zhao et al. 2020), with minor changes. Briefly, PpERF98-1/2 and PpERF1-1/2 were separately subcloned into the OE vector pK7WG2D, the constructs transformed into A. tumefaciens GV3101, and the bacterial cultures resuspended in the infiltration medium (10 mM MgCl2, 10 mM MES, 200 µM acetosyringone, and pH 5.8) to OD600 = 0.8. Equal volumes of 2 Agrobacterium cultures were mixed to obtain various combinations. After incubating for 1 h in the dark, the resuspension was gently infiltrated into fully expanded leaves of peach seedlings at 4 wk after transplantation using a sterile, needleless syringe. In parallel, the bacterial culture harboring the empty pK7WG2D vector acted as the control. At least 20 peach seedlings per treatment (4 leaves per plant) were infiltrated; after 48 h, infiltrated leaves were inoculated with L. theobromae, and the disease index evaluated.
For VIGS in peach seedlings, specific fragments of PpERF98-1/2 and PpPDS cDNA were separately cloned into the XbaI-digested pCaRNA3 vector of the PNRSV system to construct the PNRSV-PpERF98-1/2 and PNRSV-PpPDS vectors under the control of the CaMV 35S promoter. The primers used for plasmid construction are listed in Supplemental Table S2. The pCaRNA1&2, pCaRNA3, and the generated constructs were transformed into A. tumefaciens GV3101 to transfect leaves of peach seedlings 2 wk after transplantation as previously reported (Cui and Wang 2017), with slight modifications. Briefly, equal volumes of 2 Agrobacterium cultures, harboring pCaRNA1&2 and PNRSV-PpERF98-1/2 vectors, were mixed and agroinfiltrated as aforementioned. In parallel, a combination of the pCaRNA1&2 and pCaRNA3 empty vectors was taken as negative controls (marked PNRSV), whereas a combination of the pCaRNA1&2 and PNRSV-PpPDS, which causes an albino phenotype when knocked down, was used as positive control. Four weeks after infiltration, the fully expanded leaves at the top of the silenced peach seedlings were sampled for transcript quantification and then L. theobromae inoculation as described above. A total of 20 plants (3 to 4 leaflets per plant) per treatment were used to evaluate the disease index.
Subcellular localization
The coding sequences of PpERF98-1/2 without the stop codon were separately amplified and inserted into the pRI101 vector fused with the GFP under the control of CaMV 35S promoter. The recombinant plasmids pRI101-PpERF98-1/2-GFP were transformed into A. tumefaciens GV3101 and infiltrated into leaves of 4-wk-old N. benthamiana plants together with the nuclear marker 35S:VirD2NLS-mCherry (Kumar and Kirti 2010). Two days later, the subcellular localization of the target proteins was visualized under a confocal laser scanning microscope (Leica TCS SP8, Wetzlar, Germany). The excitation/emission wavelengths during acquisition were 488 nm/495 to 539 nm for GFP, intensity 4.9%, and gain 800 and 552 nm/560 to 610 nm for RFP, intensity 4.9%, and gain 735.
Transactivation and Y1H assays
The transactivation and Y1H assays were conducted as previously (Ming et al. 2020). The coding sequences of PpERF98-1/2 or their truncated fragments (coding for the N- or C-terminal regions) were individually inserted into the pGBKT7 vector and transformed into the yeast strain AH109, plated on SD/-Trp, SD/-Trp-His-Ade, and SD/-Trp-His-Ade supplemented with 4 mg mL−1 X-α-Gal (Sigma-Aldrich, MO, USA), and cultured at 30 °C for 3 d. The pGBKT7 vector was used as the negative control.
The Y1H assay was performed following the manufacturer's protocol (Matchmaker One-Hybrid System; Clontech, CA, USA). To create the prey constructs, the coding sequences of PpERF98-1/2 were separately cloned into the pGADT7 vector containing a GAL4 transcriptional activation domain. The promoter fragments of PpERF1-1/2 containing GCC-boxes were separately cloned into the pAbAi vector. After confirming the integration of each bait vector into the yeast strain Y1H gold, the prey vectors were added. The cotransformed yeast cells were cultured on SD/-Leu-Ura and SD/-Leu-Ura media with aureobasidin A (AbA; TaKaRa, Kusatsu, Japan) at 30 °C for 7 d.
Binding tests
The dual-LUC assay was conducted following Zhang et al. (2022). The coding sequences of PpERF98-1/2 were separately cloned into the pGreenII-62-SK vector to obtain the effector constructs, while the reporter constructs included individual promoter fragments of PpERF1-1/2 in the vector pGreenII-0800-LUC. The reporter and effector constructs, together with the pSoup19 vector, were transformed into A. tumefaciens GV3101. The suspensions of cells containing either effector or reporter constructs were mixed equally and coinfiltrated into leaves of 4-wk-old N. benthamiana seedlings. After 2 d, LUC activity was measured using the VivoGlo Luciferin, In Vivo Grade Kit (Promega, WI, USA) and imaged under a Night SHADE LB985 system (Berthold Technologies, Stuttgart, Germany). Meanwhile, transient expression was evaluated by the activities of firefly LUC and REN LUC using the Dual-Luciferase Reporter Assay System (Promega) on a VICTOR Nivo Multimode Plate Reader (PerkinElmer, Waltham, USA). Six biological replicates were measured for each sample.
For LCI assays, Agrobacterium-mediated transient expression in N. benthamiana leaves was performed as described previously (Huang et al. 2021). The coding regions of PpERF98-1/2 and PpERF1-1/2 were fused into the N-terminus LUC-fusion vector JW771-nLUC and C-terminus LUC-fusion vector JW772-cLUC, respectively. Different combinations of recombinant vectors were transformed into A. tumefaciens GV3101 and coinfiltrated into 4-wk-old N. benthamiana leaves. After 2 d in the dark, the infiltrated leaf regions were injected with 100 μM VivoGlo luciferin solution (Promega) and kept in the dark for 5 min for LUC activity detection using the NightSHADE LB985 In Vivo Plant Imaging system.
For BiFC assay, the coding sequences of PpERF98-1/2 and PpERF1-1/2 were individually cloned into the pCL112-nYellow and the pCL113-cYellow vectors, respectively, and transformed into A. tumefaciens GV3101, which was then infiltrated into the leaves of 4-wk-old N. benthamiana plants. After 2 d of incubation in the dark, fluorescence was visualized under a confocal laser scanning microscope. The excitation/emission wavelengths during acquisition were 514 nm/545 to 580 nm for YFP and 552 nm/650 to 700 nm for chloroplast, intensity 8%, and gain 800. The relevant primers are listed in Supplemental Table S2.
JA and SA measurement
The leaves of PpERF98-1/2-overexpressing and silenced plant seedlings were collected, homogenized in cold extraction buffer (methanol:water:acetic acid, 80:19:1, v:v:v), and extracted following a published method (Zhang et al. 2022). Both JA and SA were quantified by Triple Quadrupole Liquid Chromatography–Mass Spectrometry (TSQ Altis; Thermo Fisher Scientific, MA, USA) using labeled internal standards, in 3 biological replicates at each sampling point of each treatment.
Statistical analysis
All experiments were run independently with at least 3 biological replicates for each treatment and time point. All data sets are shown as means ± Sd of 3 independent replicates (each in 4 analytical replicates for RT-qPCR) and were processed using the SPSS Statistics 25.0. Statistical significance was analyzed using Student's t-test and 1-way ANOVA based on Duncan's post hoc test at P < 0.05 and 0.01 levels.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers: OQ876620 for PpERF98-1 and OQ876621 for PpERF98-2.
Supplementary Material
Acknowledgments
The authors thank Dr. Taotao Wang (Huazhong Agricultural University) and Dr. Hongguang Cui (Hainan University) for gifting A57 tomato and PNRSV-based vectors, respectively. Also, thank Dr. Pengfei Cao (Michigan State University), Dr. Fei Zhang, and Dr. Pengwei Wang (Huazhong Agricultural University) for critical reading of the manuscript.
Contributor Information
Dongmei Zhang, National Key Lab for Germplasm Innovation and Utilization of Horticultural Crops, College of Horticulture and Forestry Sciences, Huazhong Agricultural University, Wuhan, Hubei Province 430070, China.
Kaijie Zhu, National Key Lab for Germplasm Innovation and Utilization of Horticultural Crops, College of Horticulture and Forestry Sciences, Huazhong Agricultural University, Wuhan, Hubei Province 430070, China.
Xingyi Shen, National Key Lab for Germplasm Innovation and Utilization of Horticultural Crops, College of Horticulture and Forestry Sciences, Huazhong Agricultural University, Wuhan, Hubei Province 430070, China.
Jian Meng, National Key Lab for Germplasm Innovation and Utilization of Horticultural Crops, College of Horticulture and Forestry Sciences, Huazhong Agricultural University, Wuhan, Hubei Province 430070, China.
Xue Huang, National Key Lab for Germplasm Innovation and Utilization of Horticultural Crops, College of Horticulture and Forestry Sciences, Huazhong Agricultural University, Wuhan, Hubei Province 430070, China.
Yuqi Tan, National Key Lab for Germplasm Innovation and Utilization of Horticultural Crops, College of Horticulture and Forestry Sciences, Huazhong Agricultural University, Wuhan, Hubei Province 430070, China.
Francesca Cardinale, PlantStressLab, Department of Agriculture, Forestry and Food Science DISAFA, Turin University, Grugliasco, Torino10095, Italy.
Jihong Liu, National Key Lab for Germplasm Innovation and Utilization of Horticultural Crops, College of Horticulture and Forestry Sciences, Huazhong Agricultural University, Wuhan, Hubei Province 430070, China.
Guohuai Li, National Key Lab for Germplasm Innovation and Utilization of Horticultural Crops, College of Horticulture and Forestry Sciences, Huazhong Agricultural University, Wuhan, Hubei Province 430070, China.
Junwei Liu, National Key Lab for Germplasm Innovation and Utilization of Horticultural Crops, College of Horticulture and Forestry Sciences, Huazhong Agricultural University, Wuhan, Hubei Province 430070, China.
Author contributions
D.Z., G.L., and J.W.L. designed the experiments; D.Z., X.S., and Y.T. performed the experiments; D.Z., K.Z., X.H., M.J., and J.W.L. analyzed the data; and D.Z., K.Z., F.C., J.H.L., and J.W.L. wrote the manuscript. All authors read and approved the final manuscript.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Characterization of PpERF98-1 and 2 genes in P. persica.
Supplemental Figure S2. Transcriptional response of PpERF98-1 and 2 to hormonal treatment of peach shoots.
Supplemental Figure S3. Relative transcripts of PpERF98-1 and 2 in the inoculated peach shoots pretreated with ET inhibitors.
Supplemental Figure S4. Amino acid alignment of PpERF98-1 and 2 and their homologs from other plant species.
Supplemental Figure S5. Phenotype of 2-wk-old PpPDS-silenced peach seedlings created by VIGS and molecular characterization of the transgenic peach plants and tomato lines.
Supplemental Figure S6. Constitutive heterologous OE of PpERF98-1/2 increases the susceptibility of tomato plants to B. cinerea.
Supplemental Figure S7. Relative transcript levels in peach seedlings transiently overexpressing PpERF1-1 and 2.
Supplemental Figure S8. Binding test of PpERF98-1/2 with the promoters of PR genes (proPpPRs).
Supplemental Figure S9. PpERF1-1 and 2 cannot form heterodimers and homodimers with each other.
Supplemental Figure S10. Contents of JA and SA in the PpERF98-1/2-overexpressing tomato lines inoculated with L. theobromae at 0 and 24 hpi.
Supplemental Figure S11. Transcripts of PpERF1-1/2 and SA-related genes are affected by PpERF98-1/2.
Supplemental Figure S12. Transcriptional response of PpERF1-1, PpERF1-2, and SA-related genes to L. theobromae invasion in peach leaves transiently modified in PpERF98-2 expression.
Supplemental Table S1. Cis-acting regulatory elements present in the promoter regions of PpERF98-1/2.
Supplemental Table S2. Primer sequences used in this study.
Funding
This work was financially supported by the Natural Science Foundation of China (grant nos. 32202410 and 32172516) and the China Agriculture Research System of MOF and MARA (grant no. CARS-30).
Data availability
The authors confirm that all experimental data are available and accessible via the main text and/or the supplemental data.
References
- Achuo EA, Audenaert K, Meziane H, Hofte M. The salicylic acid-dependent defence pathway is effective against different pathogens in tomato and tobacco. Plant Pathol. 2004:53(1):65–72. 10.1111/j.1365-3059.2004.00947.x [DOI] [Google Scholar]
- Alves MS, Dadalto SP, Goncalves AB, de Souza GB, Barros VA, Fietto LG. Transcription factor functional protein-protein interactions in plant defense responses. Proteomes 2014:2(1):85–106. 10.3390/proteomes2010085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audenaert K, De Meyer GB, Höfte MM. Abscisic acid determines basal susceptibility of tomato to Botrytis cinerea and suppresses salicylic acid-dependent signaling mechanisms. Plant Physiol. 2002:128(2):491–501. 10.1104/pp.010605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckman TG, Pusey PL, Bertrand PF. Impact of fungal gummosis on peach trees. HortScience 2003:38(6):1141–1143. 10.21273/HORTSCI.38.6.1141 [DOI] [Google Scholar]
- Berrocal-Lobo M, Molina A, Solano R. Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 2002:29(1):23–32. 10.1046/j.1365-313x.2002.01191.x [DOI] [PubMed] [Google Scholar]
- Catinot J, Huang JB, Huang PY, Tseng MY, Chen YL, Gu SY, Lo WS, Wang LC, Chen YR, Zimmerli L. ETHYLENE RESPONSE FACTOR 96 positively regulates Arabidopsis resistance to necrotrophic pathogens by direct binding to GCC elements of jasmonate- and ethylene-responsive defence genes. Plant Cell Environ. 2015:38(12):2721–2734. 10.1111/pce.12583 [DOI] [PubMed] [Google Scholar]
- Cevik V, Kidd B, Zhang P, Hill C, Kiddle S, Denby K, Holub E, Cahill D, Manners J, Schenk P, et al. MEDIATOR25 Acts as an integrative hub for the regulation of jasmonate-responsive gene expression in Arabidopsis. Plant Physiol. 2012:160(1):541–555. 10.1104/pp.112.202697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi YJ, Yang Y, Zhou Y, Zhou J, Fan BF, Yu JQ, Chen ZX. Protein–protein interactions in the regulation of WRKY transcription factors. Mol Plant. 2013:6(2):287–300. 10.1093/mp/sst026 [DOI] [PubMed] [Google Scholar]
- Cui HG, Wang AM. An efficient viral vector for functional genomic studies of Prunus fruit trees and its induced resistance to Plum pox virus via silencing of a host factor gene. Plant Biotechnol J. 2017:15(3):344–356. 10.1111/pbi.12629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darino M, Chia KS, Marques J, Aleksza D, Soto-Jimenez LM, Saado I, Uhse S, Borg M, Betz R, Bindics J, et al. Ustilago maydis effector Jsi1 interacts with topless corepressor, hijacking plant jasmonate/ethylene signaling. New Phytol. 2021:229(6):3393–3407. 10.1111/nph.17116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Cerrone L, Mondino-Hintz P, Alaniz-Ferro S. Botryosphariaceae species associated with stem canker, die-back and fruit rot on apple in Uruguay. Eur J Plant Pathol. 2016:146(3):637–655. 10.1007/s10658-016-0949-z [DOI] [Google Scholar]
- Doehlemann G, Wahl R, Horst RJ, Voll LM, Usadel B, Poree F, Stitt M, Pons-Kuhnemann J, Sonnewald U, Kahmann R, et al. Reprogramming a maize plant: transcriptional and metabolic changes induced by the fungal biotroph Ustilago maydis. Plant J. 2008:56(2):181–195. 10.1111/j.1365-313X.2008.03590.x [DOI] [PubMed] [Google Scholar]
- Feng K, Hou XL, Xing GM, Liu JX, Duan AQ, Xu ZS, Li MY, Zhuang J, Xiong AS. Advances in AP2/ERF super-family transcription factors in plant. Crit Rev Biotechnol. 2020:40(6):750–776. 10.1080/07388551.2020.1768509 [DOI] [PubMed] [Google Scholar]
- Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M. Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 2000:12(3):393–404. 10.1105/tpc.12.3.393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Wang YT, Li Z, Zhang H, Ye JL, Li GH. Gene expression changes during the gummosis development of peach shoots in response to Lasiodiplodia theobromae infection using RNA-Seq. Front Physiol. 2016:7:170. 10.3389/fphys.2016.00170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, Fraley RT. A simple and general method for transferring genes into plants. Science 1985:227(4691):1229–1231. 10.1126/science.227.4691.1229 [DOI] [PubMed] [Google Scholar]
- Howe GA, Major IT, Koo AJ. Modularity in jasmonate signaling for multistress resilience. Annu Rev Plant Biol. 2018:69(1):387–415. 10.1146/annurev-arplant-042817-040047 [DOI] [PubMed] [Google Scholar]
- Huang PY, Catinot J, Zimmerli L. Ethylene response factors in Arabidopsis immunity. J Exp Bot. 2016:67(5):1231–1241. 10.1093/jxb/erv518 [DOI] [PubMed] [Google Scholar]
- Huang JY, Zhao XB, Burger M, Wang YR, Chory J. Two interacting ethylene response factors regulate heat stress response. Plant Cell 2021:33(2):338–357. 10.1093/plcell/koaa026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar KRR, Kirti PB. A mitogen-activated protein kinase, AhMPK6 from peanut localizes to the nucleus and also induces defense responses upon transient expression in tobacco. Plant Physiol Biochem. 2010:48(6):481–486. 10.1016/j.plaphy.2010.03.010 [DOI] [PubMed] [Google Scholar]
- Kunkel BN, Brooks DM. Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol. 2002:5(4):325–331. [DOI] [PubMed] [Google Scholar]
- Li LL, Zou Y. Induction of disease resistance by salicylic acid and calcium ion against Botrytis cinerea in tomato (Lycopersicon esculentum). Emir J Food Agric. 2017:29(1):78–82. 10.9755/ejfa.2016-10-1515 [DOI] [Google Scholar]
- Libault M, Wan J, Czechowski T, Udvardi M, Stacey G. Identification of 118 Arabidopsis transcription factor and 30 ubiquitin-ligase genes responding to chitin, a plant-defense elicitor. Mol Plant Microbe Interact. 2007:20(8):900–911. 10.1094/MPMI-20-8-0900 [DOI] [PubMed] [Google Scholar]
- Liu YS, Gao YX, Yu ZM, Zhang Y. Study on infection behavior and characteristics of poplar wood dyed by Lasiodiplodia theobromae. Eur J Wood Prod. 2022:80(5):1151–1163. 10.1007/s00107-022-01832-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Piqueras R, Sanchez-Serrano JJ, Solano R. ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 2003:15(1):165–178. 10.1105/tpc.007468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming R, Zhang Y, Wang Y, Khan M, Dahro B, Liu JH. The JA-responsive MYC2-BADH-like transcriptional regulatory module in Poncirus trifoliata contributes to cold tolerance by modulation of glycine betaine biosynthesis. New Phytol. 2020:229(5):2730–2750. 10.1111/nph.17063 [DOI] [PubMed] [Google Scholar]
- Müller M, Munné-Bosch S. Ethylene response factors: a key regulatory hub in hormone and stress signaling. Plant Physiol. 2015:169(1):32–41. 10.1104/pp.15.00677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidoo R, Ferreira L, Berger D, Myburg A, Naidoo S. The identification and differential expression of Eucalyptus grandis pathogenesis-related genes in response to salicylic acid and methyl jasmonate. Front Plant Sci. 2013:4:43. 10.3389/fpls.2013.00043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006a:140(2):411–432 doi: 10.1104/pp.105.073783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Ohtsuki N, Tsujimoto Y, Fujimura T, Shinshi H. Identification of genes of the plant-specific transcription-factor families cooperatively regulated by ethylene and jasmonate in Arabidopsis thaliana. J Plant Res. 2006b:119(4):407–413. 10.1007/s10265-006-0287-x [DOI] [PubMed] [Google Scholar]
- Paolinelli-Alfonso M, Villalobos-Escobedo JM, Rolshausen P, Herrera-Estrella A, Galindo-Sanchez C, Lopez-Hernandez JF, Hernandez-Martinez R. Global transcriptional analysis suggests Lasiodiplodia theobromae pathogenicity factors involved in modulation of grapevine defensive response. BMC Genomics 2016:17(1):615. 10.1186/s12864-016-2952-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CM, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SC. Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol. 2012:28(1):489–521. 10.1146/annurev-cellbio-092910-154055 [DOI] [PubMed] [Google Scholar]
- Pré M, Atallah M, Champion A, De Vos M, Pieterse CMJ, Memelink J. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 2008:147(3):1347–1357. 10.1104/pp.108.117523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Belachew A, Ma SF, Young M, Ade J, Shen Y, Marion CM, Holtan HE, Bailey A, Stone JK, et al. The EDLL motif: a potent plant transcriptional activation domain from AP2/ERF transcription factors. Plant J. 2012:70(5):855–865. 10.1111/j.1365-313X.2012.04935.x [DOI] [PubMed] [Google Scholar]
- Travadon R, Rolshausen PE, Gubler WD, Cadle-Davidson L, Baumgartner K. Susceptibility of cultivated and wild Vitis spp. to wood infection by fungal trunk path ogens. Plant Dis. 2013:97(12):1529–1536. 10.1094/PDIS-05-13-0525-RE [DOI] [PubMed] [Google Scholar]
- Tsukada K, Takahashi K, Nabeta K. Biosynthesis of jasmonic acid in a plant pathogenic fungus, Lasiodiplodia theobromae. Phytochemistry 2010:71(17–18):2019–2023. 10.1016/j.phytochem.2010.09.013 [DOI] [PubMed] [Google Scholar]
- Uppalapati SR, Ishiga Y, Wangdi T, Kunkel BN, Anand A, Mysore KS, Bender CL. The phytotoxin coronatine contributes to pathogen fitness and is required for suppression of salicylic acid accumulation in tomato inoculated with Pseudomonas syringae pv. tomato DC3000. Mol Plant Microbe Interact. 2007:20(8):955–965. 10.1094/MPMI-20-8-0955 [DOI] [PubMed] [Google Scholar]
- Van den Broeck L, Dubois M, Vermeersch M, Storme V, Matsui M, Inzé D. From network to phenotype: the dynamic wiring of an Arabidopsis transcriptional network induced by osmotic stress. Mol Syst Biol. 2017:13(12):961. 10.15252/msb.20177840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hou C, Zheng K, Li Q, Chen S, Wang S. Overexpression of ERF96, a small ethylene response factor gene enhances salt tolerance in Arabidopsis. Biol Plant. 2017:61(4):693–701. 10.1007/s10535-017-0734-7 [DOI] [Google Scholar]
- Wang XP, Liu SD, Tian HN, Wang SC, Chen JG. The small ethylene response factor ERF96 is involved in the regulation of the abscisic acid response in Arabidopsis. Front Plant Sci. 2015:6:1064. 10.3389/fpls.2015.01064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Zhao L, Li G, Huang J, Hsiang T. Identification and characterization of Botryosphaeria spp. causing gummosis of peach trees in Hubei Province, Central China. Plant Dis. 2011:95(11):1378–1384. 10.1094/PDIS-12-10-0893 [DOI] [PubMed] [Google Scholar]
- Xiao SH, Ming YQ, Hu Q, Ye ZX, Si H, Liu SM, Zhang XJ, Wang WR, Yu Y, Kong J, et al. GhWRKY41 forms a positive feedback regulation loop and increases cotton defence response against Verticillium dahliae by regulating phenylpropanoid metabolism. Plant Biotechnol J. 2023:21(5):961–978. 10.1111/pbi.14008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CY, Liang YB, Qiu DW, Zeng HM, Yuan JJ, Yang XF. Lignin metabolism involves Botrytis cinerea BcGs1-induced defense response in tomato. BMC Plant Biol. 2018:18(1):103. 10.1186/s12870-018-1319-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Shen X, Zhang H, Huang X, He H, Ye J, Cardinale F, Liu J, Liu J, Li G. Integrated transcriptomic and metabolic analyses reveal that ethylene enhances peach susceptibility to Lasiodiplodia theobromae-induced gummosis. Hortic Res. 2022:9:uhab019. 10.1093/hr/uhab019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Shen W, Zhang D, Shen X, Wang F, Hsiang T, Liu J, Li G. The bZIP transcription factor LtAP1 modulates oxidative stress tolerance and virulence in the peach gummosis fungus Lasiodiplodia theobromae. Front Microbiol. 2021:12:741842. 10.3389/fmicb.2021.741842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Tessaro MJ, Lassner M, Li X. Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell 2003:15(11):2647–2653 10.1105/tpc.014894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZJ, Wang J, Zhang RX, Huang RF. The ethylene response factor AtERF98 enhances tolerance to salt through the transcriptional activation of ascorbic acid synthesis in Arabidopsis. Plant J. 2012:71(2):273–287. 10.1111/j.1365-313X.2012.04996.x [DOI] [PubMed] [Google Scholar]
- Zhang SJ, Wang L, Zhao RR, Yu WQ, Li R, Li YJ, Sheng JP, Shen L. Knockout of SIMAPK3 reduced disease resistance to Botrytis cinerea in tomato plants. J Agric Food Chem. 2018:66(34):8949–8956. 10.1021/acs.jafc.8b02191 [DOI] [PubMed] [Google Scholar]
- Zhang L, Zhang F, Melotto M, Yao J, He SY. Jasmonate signaling and manipulation by pathogens and insects. J Exp Bot. 2017:68(6):1371–1385. 10.1093/jxb/erw478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Dong W, Zhu Y, Allan AC, Lin-Wang K, Xu C. PpGST1, an anthocyanin-related glutathione S-transferase gene, is essential for fruit coloration in peach. Plant Biotechnol J. 2020:18(5):1284–1295. 10.1111/pbi.13291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Qi L, Liu X, Cai S, Xu H, Huang R, Li J, Wei X, Zhang Z. The wheat ethylene response factor transcription factor pathogen-induced ERF1 mediates host responses to both the necrotrophic pathogen Rhizoctonia cerealis and freezing stresses. Plant Physiol. 2014:164(3):1499–1514. 10.1104/pp.113.229575 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that all experimental data are available and accessible via the main text and/or the supplemental data.