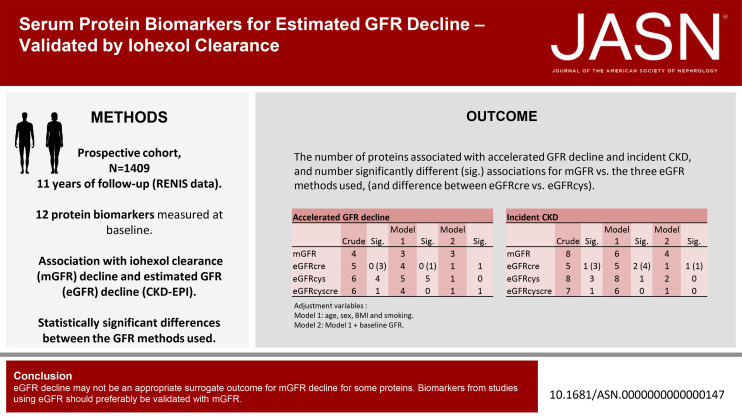
Keywords: CKD, GFR, renal function decline, epidemiology and outcomes, clinical epidemiology, creatinine, iohexol, biomarkers, blood proteins
Abstract
Significance Statement
eGFR from creatinine, cystatin C, or both has been primarily used in search of biomarkers for GFR decline. Whether the relationships between biomarkers and eGFR decline are similar to associations with measured GFR (mGFR) decline has not been investigated. This study revealed that some biomarkers showed statistically significant different associations with eGFR decline compared with mGFR decline, particularly for eGFR from cystatin C. The findings indicate that non–GFR-related factors, such as age, sex, and body mass index, influence the relationship between biomarkers and eGFR decline. Therefore, the results of biomarker studies using eGFR, particularly eGFRcys, should be interpreted with caution and perhaps validated with mGFR.
Background
Several serum protein biomarkers have been proposed as risk factors for GFR decline using eGFR from creatinine or cystatin C. We investigated whether eGFR can be used as a surrogate end point for measured GFR (mGFR) when searching for biomarkers associated with GFR decline.
Methods
In the Renal Iohexol Clearance Survey, GFR was measured with plasma iohexol clearance in 1627 individuals without diabetes, kidney, or cardiovascular disease at baseline. After 11 years of follow-up, 1409 participants had one or more follow-up GFR measurements. Using logistic regression and interval-censored Cox regression, we analyzed the association between baseline levels of 12 serum protein biomarkers with the risk of accelerated GFR decline and incident CKD for both mGFR and eGFR.
Results
Several biomarkers exhibited different associations with eGFR decline compared with their association with mGFR decline. More biomarkers showed different associations with eGFRcys decline than with eGFRcre decline. Most of the different associations of eGFR decline versus mGFR decline remained statistically significant after adjustment for age, sex, and body mass index, but several were attenuated and not significant after adjusting for the corresponding baseline mGFR or eGFR.
Conclusions
In studies of some serum protein biomarkers, eGFR decline may not be an appropriate surrogate outcome for mGFR decline. Although the differences from mGFR decline are attenuated by adjustment for confounding factors in most cases, some persist. Therefore, proposed biomarkers from studies using eGFR should preferably be validated with mGFR.
Introduction
The prevalence of CKD is increasing worldwide.1,2 Several studies have, therefore, investigated biomarkers to identify people at risk for CKD as well as the underlying mechanisms for GFR loss.
Biomarkers related to inflammation, immunity, fibrosis development, cell proliferation, angiogenesis, and apoptosis have been associated with loss of GFR in general population studies.3–13 These studies used equations on the basis of creatinine and/or cystatin C to eGFR and eGFR change rates. This method may introduce confounding and spurious associations between the biomarkers and renal outcomes because eGFR is biased by non–GFR-related factors, such as diet, muscle mass, inflammation, obesity, and cardiometabolic risk factors.14–19 Some of these studies included persons with diabetes, cardiovascular disease (CVD), and other comorbidities, potentially increasing the problem of confounding.
The aim of this study was to investigate whether eGFR can be used as a surrogate end point for mGFR decline when searching for biomarkers for GFR decline. We assessed the validity of associations between protein biomarkers and eGFR decline relative to the decline in mGFR. This was performed by comparing the associations between 12 protein biomarkers measured at baseline and loss of kidney function assessed by eGFR from creatinine and cystatin C versus the mGFR. We analyzed associations of protein biomarkers and risk of accelerated GFR decline and incident CKD using eGFR and mGFR in a representative sample of a middle-aged general population without self-reported diabetes, CVD, or kidney disease at baseline, where iohexol clearance had been measured three times during 11 years of follow-up.
Methods
Subjects
The Renal Iohexol Clearance Survey in Tromsø 6 (RENIS-T6) (2007–2009) was a substudy of the sixth Tromsø Study (T6), a general population survey in the municipality of Tromsø, Northern Norway.20 In total, 2825 participants from T6 between age 50 and 62 years without self-reported diabetes, kidney disease, or CVD were invited to RENIS-T6. The response rate was 75%, and 1627 persons were investigated according to a predetermined study target size (Figure 1).16 Of these 1627, 1324 had a follow-up in the RENIS follow-up (RENIS-FU) (2013–2015) and 1174 in RENIS-3 (2018–2020) after a median of 5.6 (interquartile range [IQR], 4.3–7.0) and 11 (IQR, 10.6–11.5) years, respectively, leaving 1410 participants with at least one follow-up GFR measurement and a total of 4213 GFR measurements.21 Another 210 participants were included in RENIS-3,21 and these were not included in this study because they did not have their GFR or proteins measured at baseline. This study was approved by the Norwegian Data Inspectorate and the Regional Ethics Committee of Northern Norway, and all participants provided written informed consent.
Figure 1.
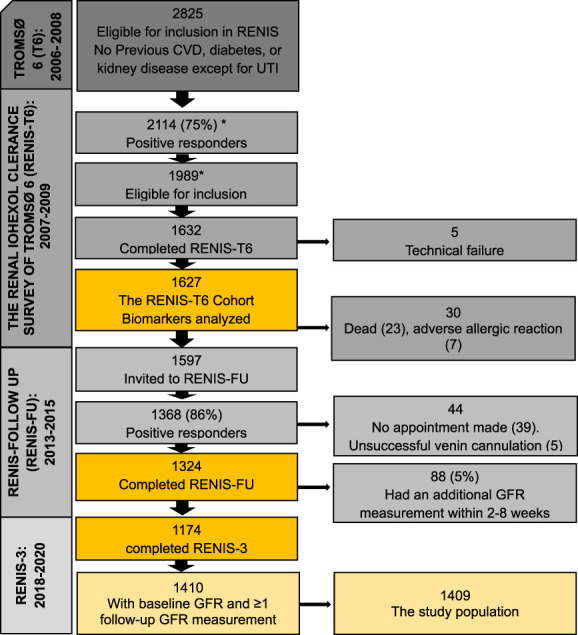
An overview of the Renal Iohexol Clearance Survey with the number (n) of participants included and response rate (%) with relevant remarks. *Miscount in previous publications (earlier numbers are 2107 and 1982, and the correct numbers are 2114 and 1989, respectively). UTI, urinary tract infection. Figure 1 can be viewed in color online at www.jasn.org.
Iohexol Clearance
GFR was measured using single sample plasma clearance of iohexol (Omnipaque, 300 mgI/ml; Amersham Health, London, United Kingdom)22 in all three surveys; details have been reported previously.23–25 The mean coefficient of variation for intraindividual mGFR variation was 4.2% in RENIS-FU (95% confidence interval [CI], 3.4% to 4.9%).24 Iohexol concentration was measured using high-performance liquid chromatography in RENIS-T6 and RENIS-FU and liquid chromatography-mass spectrometry (LC‒MS) in RENIS-3. A calibration equation for the conversion of results between HPLC and LC-MS was developed, and the equation development has been described in detail.21,26
Serum Protein Biomarker Selection and Measurement
We selected serum protein biomarkers that have been associated with an increased risk of accelerated eGFR loss, CKD development, and renal aging in previous studies. The biomarkers were selected based on a structured literature search of published articles during the past 5 years. Two researchers conducted the search using the PubMed database from March to April 2018, applying three slightly different searches to expand the search (listed in Supplemental Table 1). Studies with diabetes type 1 and acute kidney injury patients were excluded. In total, 72 different proteins were identified from original research and review articles, mainly from longitudinal general population studies but also studies involving patients with type 2 diabetes, of which each researcher listed 20 proteins on the basis of relevance in findings. We selected proteins which associated with different pathophysiological pathways to investigate a broad range of biomarkers associated with eGFR loss. Thirteen proteins available for analysis using the Luminex assay were selected (Supplemental Figure 1). Full names of the 13 proteins are listed in Table 1, and abbreviations are used hereinafter.
Table 1.
The 13 selected protein biomarkers analyzed with the Luminex multiplex method
| Abbreviation | Protein Name | Size (kDa) | Function/Pathways | Type of Marker | Reference |
|---|---|---|---|---|---|
| MCP-1 (CCL2) | Monocyte chemoattractant protein-1 | 13 | Inflammation, immunity | Profibrotic | 27,28 |
| TRAIL-R2 | TNF-related apoptosis-inducing ligand receptor 2 | 48 | Apoptosis, inflammation | — | 29,30 |
| FABP4 | Fatty acid-binding protein 4 | 15 | Binds long-chain fatty acids, fat absorption, transportation, metabolism, inflammation, fibrosis | Filtration and profibrotic | 31–34 |
| TNFR2 | TNF receptor 2 | 40 | Inflammation, antiapoptotic | — | 35,36 |
| CD40Lig | CD40 receptor ligand | 18 | Immunity | Profibrotic | 37,38 |
| GDF-15 | Growth/differentiation factor 15 | 12 (dimer: 25–30) | Cell damage, inflammation, apoptosis | Injury | 39 |
| Tie2 | TEK tyrosine kinase | 75–78 | Embryonic angiogenesis, immunity, anti-inflammatory | — | 40 |
| MMP7 | Matrix metalloproteinase 7 | 19 | Fibrosis, matrix remodulation, wound healing | Profibrotic, injury | 41,42 |
| suPAR | Soluble urokinase-type plasminogen activator receptor | 24–66 | Phosphate metabolism, apoptosis, inflammation | — | 43,44 |
| MMP2 | Matrix metalloproteinase 2 | 72 (64 active) | Fibrosis, matrix remodulation. Neural system | Profibrotic | 45,46 |
| Umod | Uromodulin | 85 | Immunity | Nephron mass | 47,48 |
| Gal-3 | Galectin-3 | 30 | Carbohydrate-binding protein, apoptosis, immunity, antimicrobial, fibrosis | Profibrotic | 49–51 |
| KIM-1 | Kidney injury molecule 1 | 40–80 | Kidney damage marker (proximal tubule), allergies, immunity | Injury | 52 |
Name of the protein, abbreviation, and function/involvement in pathways.
Fasting serum samples collected at baseline and stored at −80°C were thawed for protein measurement. In 2015, TNF receptor 2 (TNFR2) was analyzed using a quantitative sandwich ELISA with a QuantiKine kit from R&D systems, Inc (Minneapolis, MN), as part of a previous project.53 The other 12 proteins were analyzed between June 2018 and January 2019 using Luminex xMap multiplex technology (Bio-Plex 200 systems, BIO-RAD) and human magnetic bead-based assays from R&D Systems (Bio-Teche), consisting of microplates with 96-well plates and magnetic antibody-coated beads. All the microplates and accompanying standard solutions were from the same batch to avoid batch differences.
Owing to differences in the required dilution factor of serum samples before protein analyses with Luminex technology, we used two separate kits. The first nine proteins (CD40 ligand receptor, growth/differentiation factor 15 [GDF-15], monocyte chemoattractant protein-1 [MCP-1], Tie-2, TNF-related apoptosis-inducing ligand receptor 2 [TRAIL-R2], fatty acid binding protein 4 [FABP4], Kidney injury molecule 1 [KIM-1], matrix metalloproteinase 7 [MMP7], and soluble urokinase-type plasminogen activator receptor [suPAR]) were measured at a 1:2 dilution, and the last three (Uromodulin [Umod], Gal-3, and matrix metalloproteinase 2 [MMP2]) were measured at a 1:50 dilution. Protein levels were calculated using a five-parameter logistic standard curve and displayed as the mean of duplicate measurements. KIM-1 was undetectable (below the lower limit of detection) in all but 37 of the baseline samples and was thus excluded from the analysis. Intra-assay and interassay coefficients of variation (CVs) were between 2.9%–6.4% and 4.9%–19.3%, respectively (Supplemental Table 2).
Of the selected biomarkers, two markers of fibrosis (MMP7 and MMP2) as well as TNFR2 have been included in previous publications from the first RENIS-FU after a median of 5.6 years (RENIS-FU).14,53,54
Other Study Variables
Information on medication use and smoking habits (current, previous, or never daily smoker) was collected through a questionnaire. Height and weight were measured to calculate body mass index (BMI: weight in kilogram divided by height in meters squared). Blood pressure (BP) was measured three times at 1-minute intervals after 2 minutes of rest using an automated device (model UA 799; A&D, Tokyo, Japan). The average BP of the last two measurements was used in the analyses. Hypertension was defined as systolic BP (sBP) ≥140 mm Hg, diastolic BP ≥90 mm Hg, or the use of antihypertensive medication. Fasting glucose, creatinine (Cre), and cystatin C (Cys C) were measured using COBAS 8000 (Roche Diagnostics).15 Serum creatinine was measured using an enzymatic assay standardized to the isotope dilution mass spectroscopy method (CREA Plus, Roche Diagnostics, GmbH, Mannheim, Germany). Cystatin C was measured by a particle-enhanced turbidimetric immunoassay (Gentian, Moss, Norway). The interassay coefficient of variation for creatinine and cystatin C was 2.3% and 3.1%, respectively.23 Owing to the lack of an established international standard at baseline, the baseline cystatin C measurements were calibrated to the international reference ERM-DA471/IFCC (as previously described).55 This standard was in use during data collection in RENIS-FU and RENIS-3. eGFR was based on Cre, Cys C, or both using the Chronic Kidney Disease Epidemiology Collaboration equation.56,57 Three samples of first-void morning urine were collected on consecutive days, and fresh samples were analyzed for the urinary albumin-to-creatinine ratio (mg/mmol).20 All measurements and blood samples were collected in the morning between 08:00 and 10:00 am.
Statistical Analyses
Descriptive statistics for normally distributed variables and skewed variables were given as the means (SD) and medians (IQR), respectively. Categorical variables were given as numbers (n) and percentages. Normally distributed proteins were modeled per SD increase in concentration, whereas proteins with skewed distributions were modeled per logarithmic unit increase after log2-transformation.
The association between proteins measured at baseline and a dichotomous variable for accelerated mGFR and eGFR decline was investigated using multiple logistic regression analyses. In studies of CKD, which include many patients with very rapid eGFR decline, accelerated GFR decline has often been defined as loss of GFR > 3.0 ml/min per 1.73 m2 per year. To obtain a reasonable number of persons with accelerated GFR in this relatively healthy population, we instead defined it as the 10% steepest mGFR or eGFR decline slopes, calculated using linear mixed model for each GFR method adjusted for baseline age, sex, BMI, smoking, BP medication, sBP, and fasting glucose, using a within-person centered time variable, as described in previous publications.54,58,59 The slope obtained from a linear mixed model is considered to be more precise than a slope on the basis of linear regression and a better surrogate marker of end-stage kidney disease, especially among those with GFR >60 ml/min per 1.73 m2.60
To test the differences between the odds ratios (ORs) for the GFR methods in the logistic regressions (mGFR versus each eGFR or between eGFRcre and eGFRcys), we used the suest (seemingly unrelated estimation) command in STATA.
Incident CKD was defined as new-onset eGFR or mGFR <60 ml/min per 1.73 m2, a definition used by others.21,61 Since the exact time of the event was not observed, the risk of incident CKD during the study period was assessed using interval-censored Cox regression analysis.62 Censoring was performed at the first time interval incident CKD was assumed to have occurred. Thus, persons with prevalent CKD at baseline were categorized as left censored, i.e., the event occurred before the first visit. Those who developed CKD in between visits were categorized as interval censored, and those who had GFR >60 ml/min per 1.73 m2 at their last visit as right censored. To avoid misclassification if baseline GFR was < 60 ml/min per 1.73 m2 whereas subsequent GFR was >60 ml/min per 1.73 m2, we did not register this as CKD unless GFR fell below 60 ml/min per 1.73 m2 at a later measurement. For each protein, the statistical significance of the differences between the hazard ratio (HR) for mGFR and the other three eGFR methods was calculated with the bootstrap method from 1000 resamples with replacement of the participants in the study population.
Both the analyses of accelerated decline and incident CKD were adjusted for covariates in three different models: unadjusted analysis (protein concentration only); adjustment for factors commonly included in biomarkers studies that may be related to creatinine and/or cystatin C production (non–GFR-related factors influencing creatinine or cystatin C): age, sex, BMI, and tobacco smoking19,63 (model 1). In model 2, we also adjusted for the respective baseline measured or eGFR (model 2), as doing so could reduce confounding of baseline eGFRcre or eGFRcys due to unmeasured non–GFR-related factors.15–17 The association between a protein biomarker and eGFR decline was judged to be dissimilar from the association with mGFR decline if the difference between the associations estimated as OR (Table 2) or HR (Table 3) was statistically significant with alpha set at 0.05 in any of these models.
Table 2.
Odds ratios for accelerated mGFR and eGFR decline per doubling or SD increase in baseline protein concentration
| Protein | Unadjusted | P Value for Difference | Model 1 | P Value for Difference | Model 2 | P Value for Difference | |||
|---|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | OR | 95% CI | OR | 95% CI | ||||
| MCP-1a | |||||||||
| mGFR | 1.18 | 0.87 to 1.59 | 1.08 | 0.76 to 1.52 | 1.08 | 0.77 to 1.52 | |||
| eGFRcre | 1.11 | 0.80 to 1.55 | 0.80 | 1.11 | 0.79 to 1.57 | 0.90 | 1.10 | 0.78 to 1.56 | 0.94 |
| eGFRcys | 1.31 | 0.98 to 1.75 | 0.61 | 1.28 | 0.91 to 1.79 | 0.99 | 1.00 | 0.62 to 1.63 | 0.80 |
| eGFRcyscre | 1.13 | 0.82 to 1.55 | 0.84 | 1.08 | 0.76 to 1.53 | 0.57 | 1.01 | 0.70 to 1.46 | 0.80 |
| TRAIL-R2a | |||||||||
| mGFR | 1.35b | 1.04 to 1.75b | 1.04 | 0.73 to 1.47 | 1.07 | 0.75 to 1.53 | |||
| eGFRcre | 1.53b | 1.18 to 1.97b | 0.51 | 1.41b | 1.07 to 1.86b | 0.18 | 1.27 | 0.94 to 1.71 | 0.48 |
| eGFRcys | 2.27b | 1.50 to 3.44b | 0.04b | 2.18b | 1.49 to 3.18b | 0.005b | 1.33 | 0.78 to 2.29 | 0.51 |
| eGFRcyscre | 1.52b | 1.17 to 1.97b | 0.52 | 1.34 | 0.99 to 1.83 | 0.28 | 1.01 | 0.63 to 1.63 | 0.85 |
| FABP4a | |||||||||
| mGFR | 1.00 | 0.85 to 1.19 | 1.02 | 0.81 to 1.29 | 1.07 | 0.84 to 1.37 | |||
| eGFRcre | 1.18 | 0.94 to 1.48 | 0.27 | 1.17 | 0.86 to 1.60 | 0.48 | 1.02 | 0.76 to 1.35 | 0.78 |
| eGFRcys | 1.34b | 1.08 to 1.66b | 0.04b | 1.74b | 1.25 to 2.44b | 0.01b | 1.01 | 0.68 to 1.48 | 0.79 |
| eGFRcyscre | 1.32b | 1.04 to 1.68b | 0.07 | 1.44b | 1.00 to 2.06b | 0.12 | 1.11 | 0.79 to 1.55 | 0.87 |
| TNFR2 | |||||||||
| mGFR | 1.40b | 1.20 to 1.64b | 1.21b | 1.02 to 1.43b | 1.28b | 1.07 to 1.53b | |||
| eGFRcre | 1.24b | 1.07 to 1.44b | 0.28c | 1.19b | 1.02 to 1.39b | 0.88d | 1.07 | 0.91 to 1.26 | 0.15 |
| eGFRcys | 2.57b | 2.07 to 3.18b | <0.001b | 2.45b | 1.93 to 3.12b | <0.001b | 1.10 | 0.82 to 1.47 | 0.38 |
| eGFRcyscre | 1.44b | 1.22 to 1.70b | 0.82 | 1.31b | 1.10 to 1.55b | 0.52 | 0.95 | 0.77 to 1.17 | 0.03b |
| CD40Lig | |||||||||
| mGFR | 1.03 | 0.88 to 1.19 | 1.00 | 0.84 to 1.20 | 1.00 | 0.83 to 1.19 | |||
| eGFRcre | 1.10 | 0.96 to 1.26 | 0.49 | 1.07 | 0.93 to 1.24 | 0.56 | 1.09 | 0.94 to 1.26 | 0.45 |
| eGFRcys | 1.15 | 0.98 to 1.35 | 0.29 | 1.18 | 1.00 to 1.39 | 0.20 | 1.20 | 0.96 to 1.50 | 0.21 |
| eGFRcyscre | 1.11 | 0.97 to 1.28 | 0.44 | 1.10 | 0.95 to 1.29 | 0.42 | 1.09 | 0.93 to 1.27 | 0.48 |
| GDF-15 | |||||||||
| mGFR | 1.29 | 0.73 to 2.28 | 1.12b | 1.00 to 1.25b | 1.13b | 1.01 to 1.26b | |||
| eGFRcre | 1.10 | 0.95 to 1.28 | 0.60 | 1.06 | 0.95 to 1.18 | 0.54 | 1.01 | 0.85 to 1.19 | 0.28 |
| eGFRcys | 1.69 | 0.96 to 2.96 | 0.51 | 1.38 | 0.65 to 2.94 | 0.58 | 1.13 | 0.99 to 1.29 | 0.95 |
| eGFRcyscre | 1.19 | 0.83 to 1.71 | 0.81 | 1.11 | 1.00 to 1.25 | 1.00 | 0.96 | 0.75 to 1.24 | 0.28 |
| Tie2 | |||||||||
| mGFR | 1.02 | 0.87 to 1.20 | 0.87 | 0.73 to 1.03 | 0.87 | 0.74 to 1.04 | |||
| eGFRcre | 0.89 | 0.74 to 1.08 | 0.29e | 0.90 | 0.74 to 1.09 | 0.79 | 0.87 | 0.71 to 1.06 | 0.97 |
| eGFRcys | 1.29b | 1.09 to 1.52b | 0.05 | 1.14 | 0.95 to 1.35 | 0.03b | 1.12 | 0.89 to 1.41 | 0.08 |
| eGFRcyscre | 1.04 | 0.88 to 1.23 | 0.86 | 0.99 | 0.83 to 1.17 | 0.31 | 0.94 | 0.78 to 1.12 | 0.59 |
| MMP7 | |||||||||
| mGFR | 1.80b | 1.53 to 2.13 | 1.65b | 1.38 to 1.97b | 1.74b | 1.44 to 2.09b | |||
| eGFRcre | 1.54b | 1.31 to 1.81b | 0.19 | 1.46b | 1.23 to 1.72b | 0.31 | 1.34b | 1.13 to 1.59b | 0.04b |
| eGFRcys | 1.90b | 1.62 to 2.24b | 0.64 | 1.86b | 1.57 to 2.20b | 0.35 | 1.75b | 1.37 to 2.23b | 0.98 |
| eGFRcyscre | 1.83b | 1.56 to 2.15b | 0.89 | 1.71b | 1.45 to 2.02b | 0.79 | 1.52b | 1.27 to 1.81b | 0.08 |
| suPAR | |||||||||
| mGFR | 1.05 | 0.93 to 1.18 | 0.97 | 0.82 to 1.16 | 0.98 | 0.82 to 1.17 | |||
| eGFRcre | 1.05 | 0.94 to 1.18 | 0.95 | 1.02 | 0.90 to 1.15 | 0.69 | 1.00 | 0.86 to 1.16 | 0.86 |
| eGFRcys | 1.14 | 0.98 to 1.31 | 0.40 | 1.13 | 1.00 to 1.27 | 0.20 | 1.04 | 0.85 to 1.27 | 0.64 |
| eGFRcyscre | 1.11 | 0.98 to 1.26 | 0.53 | 1.07 | 0.95 to 1.20 | 0.40 | 1.02 | 0.88 to 1.18 | 0.72 |
| MMP2 | |||||||||
| mGFR | 0.94 | 0.79 to 1.12 | 0.97 | 0.82 to 1.17 | 0.97 | 0.82 to 1.16 | |||
| eGFRcre | 1.03 | 0.88 to 1.21 | 0.46 | 1.02 | 0.88 to 1.19 | 0.67 | 0.99 | 0.84 to 1.17 | 0.89 |
| eGFRcys | 0.98 | 0.82 to 1.17 | 0.78 | 0.95 | 0.79 to 1.14 | 0.84 | 0.84 | 0.66 to 1.07 | 0.33 |
| eGFRcyscre | 1.03 | 0.88 to 1.20 | 0.46 | 1.01 | .87 to 1.18 | 0.75 | 1.00 | 0.85 to 1.17 | 0.85 |
| Umod | |||||||||
| mGFR | 0.76b | 0.64 to 0.90b | 0.94 | 0.78 to 1.14 | 0.92 | 0.76 to 1.12 | |||
| eGFRcre | 0.81b | 0.67 to 0.98b | 0.62f | 0.79b | 0.65 to 0.97b | 0.23 | 0.84 | .69 to 1.03 | 0.52 |
| eGFRcys | 0.52b | 0.43 to 0.63b | 0.003b | 0.61b | 0.49 to 0.75b | 0.002b | 0.85 | 0.62 to 1.16 | 0.64 |
| eGFRcyscre | 0.72b | 0.60 to 0.87b | 0.67 | 0.77b | 0.63 to 0.94b | 0.16 | 0.89 | 0.71 to 1.11 | 0.80 |
| Gal-3 | |||||||||
| mGFR | 0.98 | 0.83 to 1.15 | 1.07 | 0.89 to 1.30 | 1.09 | 0.91 to 1.21 | |||
| eGFRcre | 1.19b | 1.04 to 1.37b | 0.07 | 1.18 | 1.02 to 1.36 | 0.46 | 1.11 | 0.95 to 1.30 | 0.87 |
| eGFRcys | 1.10 | 0.96 to 1.26 | 0.27 | 1.25 | 1.06 to 1.47 | 0.24 | 0.97 | 0.76 to 1.24 | 0.45 |
| eGFRcyscre | 1.21b | 1.06 to 1.38b | 0.04b | 1.25 | 1.08 to 1.45 | 0.22 | 1.16 | 0.99 to 1.36 | 0.62 |
Accelerated GFR decline defined as the 10% with the steepest annual GFR decline slope for the corresponding GFR method.
Model 1: sex, age, body mass index, and smoke (now, previously, and newer).
Model 2: model 1+baseline GFR.
P value for statistically significant differences between mGFR and the respective eGFR from creatinine, cystatine C, or both. OR, odds ratio; CI, confidence interval; mGFR, measured GFR; cre, creatinine; cys, cystatin C; MCP-1, monocyte chemoattractant protein-1; TRAIL-R2, TNF-related apoptosis-inducing ligand receptor 2; FABP4, fatty acid-binding protein 4; TNFR2, TNF receptor 2; CD40Lig, CD40 ligand receptor; GDF-15, growth/differentiation factor 15; Tie2, TEK tyrosine kinase; MMP7, matrix metalloproteinase 7; suPAR, soluble urokinase-type plasminogen activator receptor; MMP2, matrix metalloproteinase 2; Umod, uromodulin; Gal-3, galectin-3.
Log2-transformed.
Statistically significant association between protein and accelerated GFR decline.
A statistically significant difference between eGFRcre and eGFRcys: cP value: <0.001, dP value: <0.001, eP value: 0.004, fP value: 0.001.
Table 3.
Hazard ratios for incident CKD per doubling or SD increase in baseline protein concentration
| Protein | Unadjusted | P Value for Difference | Model 1 | P Value for Difference | Model 2 | P Value for Difference | |||
|---|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | HR | 95% CI | HR | 95% CI | ||||
| MCP-1a | |||||||||
| mGFR | 0.89 | 0.58 to 1.37 | 0.87 | 0.55 to 1.37 | 0.81 | 0.52 to 1.27 | |||
| eGFRcre | 1.00 | 0.58 to 1.73 | 0.55 | 0.95 | 0.53 to 1.70 | 0.67 | 0.93 | 0.54 to 1.59 | 0.76 |
| eGFRcys | 1.24 | 0.86 to 1.78 | 0.04b | 1.17 | 0.78 to 1.75 | 0.19 | 1.06 | 0.74 to 1.51 | 0.43 |
| eGFRcyscre | 1.06 | 0.64 to 1.75 | 0.40 | 1.00 | 0.59 to 1.68 | 0.56 | 0.87 | 0.51 to 1.46 | 0.91 |
| TRAIL-R2a | |||||||||
| mGFR | 1.64b | 1.37 to 1.96b | 1.54b | 1.27 to 1.87b | 1.48b | 1.18 to 1.87b | |||
| eGFRcre | 1.67b | 1.20 to 2.31b | 0.87 | 1.58b | 1.08 to 2.31b | 0.78 | 1.37 | 0.81 to 2.30 | 0.57 |
| eGFRcys | 1.62b | 1.44 to 1.83b | 0.98 | 1.44b | 1.25 to 1.67b | 0.71 | 1.00 | 0.63 to 1.58 | 0.09 |
| eGFRcyscre | 1.57b | 1.26 to 1.96b | 0.69 | 1.39b | 1.08 to 1.80b | 0.50 | 0.82 | 0.46 to 1.44 | 0.05 |
| FABP4a | |||||||||
| mGFR | 2.00b | 1.58 to 2.53b | 1.69b | 1.28 to 2.23b | 1.10 | 0.82 to 1.48 | |||
| eGFRcre | 1.91b | 1.39 to 2.64b | 0.84 | 1.86b | 1.27 to 2.73b | 0.98 | 1.19 | 0.77 to 1.83 | 0.72 |
| eGFRcys | 2.37b | 1.87 to 3.01b | 0.16 | 1.70b | 1.28 to 2.26b | 0.51 | 0.98 | 0.72 to 1.34 | 0.36 |
| eGFRcyscre | 2.31b | 1.75 to 3.06b | 0.31 | 1.89b | 1.37 to 2.61b | 0.63 | 0.99 | 0.67 to 1.46 | 0.54 |
| TNFR2 | |||||||||
| mGFR | 1.47b | 1.37 to 1.57b | 1.33b | 1.25 to 1.42b | 1.15b | 1.05 to 1.26b | |||
| eGFRcre | 1.37b | 1.20 to 1.57b | 0.36c | 1.27b | 1.12 to 1.44b | 0.52d | 1.09 | .88 to 1.36 | 0.64 |
| eGFRcys | 1.69b | 1.63 to 1.75b | <0.001b | 1.63b | 1.55 to 1.72b | <0.001b | 1.01 | 0.85 to 1.21 | 0.40 |
| eGFRcyscre | 1.58b | 1.47 to 1.71b | 0.08 | 1.46b | 1.34 to 1.58b | 0.13 | 0.95 | 0.80 to 1.14 | 0.16 |
| CD40Lig | |||||||||
| mGFR | 1.08 | 0.91 to 1.29 | 1.05 | 0.89 to 1.24 | 1.04 | 0.89 to 1.23 | |||
| eGFRcre | 0.97 | 0.72 to 1.32 | 0.31 | 0.95 | 0.70 to 1.29 | 0.36 | 0.93 | 0.96 to 1.27 | 0.43 |
| eGFRcys | 1.08 | 0.89 to 1.31 | 0.93 | 0.99 | 0.86 to 1.15 | 0.46 | 0.91 | 0.77 to 1.08 | 0.17 |
| eGFRcyscre | 1.04 | 0.82 to 1.32 | 0.61 | 0.97 | 0.78 to 1.20 | 0.31 | 0.96 | 0.75 to 1.22 | 0.34 |
| GDF-15a | |||||||||
| mGFR | 1.67b | 1.26 to 2.21b | 1.55b | 1.11 to 2.15b | 1.25 | 0.90 to 1.74 | |||
| eGFRcre | 1.27 | 0.91 to 1.76 | 0.35 | 1.09 | 0.72 to 1.64 | 0.31e | 0.76 | 0.48 to 1.20 | 0.09 |
| eGFRcys | 2.18b | 1.72 to 2.77b | 0.04b | 2.18b | 1.59 to 2.99b | 0.05 | 1.13 | 0.76 to 1.68 | 0.67 |
| eGFRcyscre | 2.07b | 1.47 to 2.91 | 0.19 | 1.83b | 1.23 to 2.73b | 0.36 | 0.99 | 0.60 to 1.62 | 0.31 |
| Tie2 | |||||||||
| mGFR | 1.03 | 0.85 to 1.25 | 1.06 | 0.87 to 1.29 | 0.95 | 0.77 to 1.17 | |||
| eGFRcre | 1.12 | 0.87 to 1.46 | 0.50 | 1.14 | 0.85 to 1.53 | 0.56 | 1.08 | 0.82 to 1.42 | 0.49 |
| eGFRcys | 1.11 | 0.93 to 1.33 | 0.45 | 1.05 | 0.88 to 1.27 | 0.97 | 1.12 | 0.92 to 1.36 | 0.33 |
| eGFRcyscre | 1.09 | 0.87 to 1.37 | 0.59 | 1.09 | 0.86 to 1.40 | 0.73 | 1.00 | 0.79 to 1.26 | 0.81 |
| MMP7 | |||||||||
| mGFR | 1.85b | 1.62 to 2.11b | 1.75b | 1.53 to 1.99b | 1.53b | 1.34 to 1.75b | |||
| eGFRcre | 1.71b | 1.47 to 1.99b | 0.48 | 1.65b | 1.39 to 1.96b | 0.61 | 1.42b | 1.13 to 1.79b | 0.53 |
| eGFRcys | 1.88b | 1.69 to 2.09b | 0.81 | 1.80b | 1.58 to 2.06b | 0.71 | 1.65b | 1.35 to 2.02b | 0.48 |
| eGFRcyscre | 1.89b | 1.67 to 2.14b | 0.81 | 1.75b | 1.51 to 2.03b | 0.98 | 1.48b | 1.23 to 1.80b | 0.74 |
| suPAR | |||||||||
| mGFR | 1.11b | 1.06 to 1.15b | 1.08 | 0.94 to 1.24 | 1.11 | 0.98 to 1.25 | |||
| eGFRcre | 0.95 | 0.67 to 1.35 | 0.04f | 0.81 | 0.54 to 1.23 | 0.04g | 0.61b | 0.41 to 0.90b | 0.01h |
| eGFRcys | 1.14b | 1.10 to 1.17b | 0.14 | 1.13b | 1.04 to 1.22b | 0.30 | 1.08 | 0.90 to 1.30 | 0.71 |
| eGFRcyscre | 1.11b | 1.04 to 1.19b | 0.81 | 1.06 | 0.85 to 1.33 | 0.77 | 0.81 | 0.61 to 1.10 | 0.08 |
| MMP2 | |||||||||
| mGFR | 1.12 | 0.93 to 1.35 | 1.04 | 0.87 to 1.24 | 1.08 | 0.91 to 1.30 | |||
| eGFRcre | 1.27 | 1.02 to 1.60 | 0.09 | 1.19 | 0.95 to 1.48 | 0.06 | 1.20 | 0.88 to 1.64 | 0.27 |
| eGFRcys | 1.18 | 1.01 to 1.38 | 0.47 | 1.12 | 0.98 to 1.27 | 0.29 | 1.24b | 1.03 to 1.49b | 0.24 |
| eGFRcyscre | 1.18 | 0.93 to 1.48 | 0.50 | 1.09 | 0.89 to 1.35 | 0.43 | 1.01 | 0.81 to 1.27 | 0.71 |
| Umod | |||||||||
| mGFR | 0.74b | 0.58 to 0.94b | 0.72b | 0.56 to 0.93b | 0.82 | 0.64 to 1.06 | |||
| eGFRcre | 0.91 | 0.66 to 1.26 | 0.09i | 0.92 | 0.65 to 1.30 | 0.08j | 1.13 | 0.84 to 1.52 | 0.07 |
| eGFRcys | 0.67b | 0.50 to 0.89b | 0.30 | 0.71b | 0.53 to 0.95b | 0.81 | 0.98 | 0.70 to 1.37 | 0.26 |
| eGFRcyscre | 0.74 | 0.53 to 1.05 | 0.94 | 0.75 | 0.53 to 1.07 | 0.81 | 1.04 | 0.73 to 1.48 | 0.20 |
| Gal-3 | |||||||||
| mGFR | 1.25b | 1.07 to 1.45b | 1.14 | 0.97 to 1.35 | 1.09 | 0.90 to 1.31 | |||
| eGFRcre | 1.38b | 1.15 to 1.64b | 0.06 | 1.32b | 1.09 to 1.60b | 0.03b | 1.18 | 0.98 to 1.43 | 0.36 |
| eGFRcys | 1.30b | 1.13 to 1.50b | 0.34 | 1.18b | 1.01 to 1.38b | 0.64 | 1.09 | 0.97 to 1.22 | 0.95 |
| eGFRcyscre | 1.38b | 1.18 to 1.63b | 0.02b | 1.28b | 1.07 to 1.52b | 0.06 | 1.12 | 0.96 to 1.32 | 0.61 |
Incident CKD is defined as new-onset GFR <60 ml/min per 1.73 m2 during follow-up.
A total of 1409 individuals are included in the analysis; the respective numbers of baseline CKD and incident CKD for the GFR methods are mGFR; n=8 and, n=95. eGFRcre; n=4 and n=51. eGFRcys; n=5 and n=96. eGFRcyscr; n=3 and n=62.
Model 1: sex, age, body mass index, and smoke (now, previously, and newer).
Model 2: model 1+baseline GFR. HR, hazard ratio; CI, confidence interval; mGFR, measured GFR; cre, creatinine; cys, cystatin C; MCP-1, monocyte chemoattractant protein-1; TRAIL-R2, TNF-related apoptosis-inducing ligand receptor 2; FABP4, fatty acid-binding protein 4; TNFR2: TNF receptor 2; CD40Lig, CD40 ligand receptor; GDF-15, growth/differentiation factor 15; Tie2, TEK tyrosine kinase; MMP7, matrix metalloproteinase 7; suPAR, soluble urokinase-type plasminogen activator receptor; MMP2, matrix metalloproteinase 2; Umod, uromodulin; Gal-3, galectin-3.
Log2 transformed.
Statistically significant association between protein and incident CKD.
Difference between eGFrcre and eGFRcys: cP value: <0.001, dP value: <0.001, eP value: 0.04, fP value: 0.004, gP value: 0.002, hP value: 0.004, iP value: 0.008, jP value: 0.04.
All 1410 participants who attended the baseline examination and had one or more follow-up GFR measurements were included in this study. There was equal number of observations regardless of whether eGFR or mGFR was used. However, we excluded one participant with an extreme outlier in the Cys C measurement at RENIS-3, leaving 1409 participants included in the analysis. The protein biomarkers were measured at baseline only.
All statistical analyses were performed with STATA version 17.0 (StataCorp, College Station, TX). A P value of <0.05 was considered as statistically significant. Owing to the number of multiple analyses with different outcomes, adjustment of the P values for multiple comparisons was considered. This has been a controversial issue in epidemiological research.64 We selected all proteins on the basis of earlier studies with a high probability of being associated with the outcome of interest, thus having high pretest probability, which would not have been considered in commonly used methods for multiple comparisons adjustment. Accordingly, we decided not to adjust for multiple comparisons.
Results
Study Characteristics
The baseline characteristics of the study cohort are presented in Table 4. There were only small differences between the subgroup with one or more follow-up examinations (N=1409; included in this study) and the total RENIS baseline cohort (N=1627) (Supplemental Table 3).
Table 4.
Baseline characteristics of the study cohort
| Baseline Characteristics of RENIS Participants with at Least One Follow-Up GFR Measurement | |
|---|---|
| Participants (n) | 1409 |
| Male sex (n) | 696 (49%) |
| Age (yr) | 58.5 (54.5–61.4) |
| Height (cm) | 170.7 (8.7) |
| Weight (kg) | 79.6 (14.1) |
| BMI (kg/m2) | 27.2 (3.9) |
| mGFR (ml/min per 1.73 m2) | 94.0 (14.3) |
| eGFRcre (ml/min per 1.73 m2) | 94.9 (9.4) |
| eGFRcys (ml/min per 1.73 m2) | 105.7 (12.1) |
| eGFRcyscre (ml/min per 1.73 m2) | 103.2 (11.2) |
| uACR (mg/mmol)a | 0.2 (0.1–0.5) |
| sBP (mm Hg) | 129 (17) |
| dBP (mm Hg) | 83 (10) |
| BP medication (n) | 250 (18%) |
| RAS inhibitors | 143 (10%) |
| Fasting blood glucose (mmol/L) | 5.3 (0.5) |
| LDL cholesterol (mmol/L) | 3.7 (0.8) |
| HDL cholesterol (mmol/L) | 1.5 (0.4) |
| Triglycerides (mmol/L) | 1.2 (0.7) |
| Daily smoker (n) | 949 (67%) |
| Current smoker (n) | 278 (20%) |
| Previously smoker (n) | 670 (48%) |
| Never smoker (n) | 456 (32%) |
| Serum protein biomarkers | |
| CD40Lig (ng/ml) | 6.8 (2.1) |
| GDF-15 (ng/ml) | 0.8 (0.4) |
| MCP-1 (ng/ml) | 0.5 (0.4–0.6) |
| Tie2 (ng/ml) | 16.2 (5.7) |
| TRAIL-R2 (ng/ml) | 0.03 (0.03–0.04) |
| FABP4 (ng/ml) | 9.5 (6.6–13.8) |
| MMP7 (ng/ml) | 1.9 (0.80) |
| suPAR (ng/ml) | 0.3 (0.2) |
| MMP2 (ng/ml) | 315.3 (58.7) |
| Umod (ng/ml) | 237.8 (96.0) |
| Gal-3 (ng/ml) | 7.5 (1.9) |
| TNFR2 (ng/ml) | 2.7 (0.6) |
Mean (SD) for normally distributed variables, median (interquartile range) for skewed variables, and number and percentages (%) for categorical variables.
Variables with missing baseline values (n): smoke (5), TNF-related apoptosis-inducing ligand receptor 2 (1), fatty acid-binding protein 4 (3), TNFR2, TNF receptor 2 (3), matrix metalloproteinase 7 (14), and soluble urokinase-type plasminogen activator receptor (18). RENIS, Renal Iohexol Clearance Survey; BMI, body mass index; mGFR, measured GFR; eGFRcre/cys/cyscre, eGFR based on the CKD-EPI equation for creatinine, cystatin C or both; uACR, urinary albumin-to-creatinine ratio; sBP, systolic BP; dBP, diastolic BP; RAS, renin-angiotensin system; CD40Lig, CD40 ligand receptor; GDF-15, growth/differentiation factor 15; MCP-1, monocyte chemoattractant protein-1; Tie2; TEK tyrosine kinase; TRAIL-R2, TNF-related apoptosis-inducing ligand receptor 2; FABP4, fatty acid-binding protein 4; MMP7, matrix metalloproteinase 7; suPAR, soluble urokinase-type plasminogen activator receptor; MMP2, matrix metalloproteinase 2; Umod, uromodulin; Gal-3, galectin-3; TNFR2, TNF receptor 2.
To convert albumin-to-creatinine ratio in mg/mmol to mg/g, multiply by 8.84.
Accelerated GFR Decline
The mean (SD) mGFR decline rate was 1.07 ml/min per 1.73 m2 per year (0.5). The 140 persons who had an accelerated mGFR decline defined as the 10% with the steepest mGFR decline slope had an mGFR decline rate of ≥1.63 ml/min per 1.73 m2 per year. As reported in a previous publication from the RENIS study, the mean (SD) eGFRcre decline rate was similar to that of mGFR, but the mean eGFRcys decline rate was steeper, and the distribution (SD) wider, using cystatin C-based eGFR.21 Using the eGFR equations with the 10% steepest eGFR decline slopes, the cutoffs for eGFRcre, eGFRcys, and eGFRcyscre were ≥1.49 ml/min per 1.73 m2 per year, ≥2.64 ml/min per 1.73 m2 per year, and ≥2.13 ml/min per 1.73 m2 per year, respectively.
In the unadjusted analysis, higher concentrations of four proteins were associated with accelerated mGFR decline, five with eGFRcre decline, and six with eGFRcys and eGFRcyscre decline (Table 2). For four proteins, there were statistically significant differences in associations between eGFRcys decline and mGFR decline (TRAIL-R2, FABP4, and TNFR2 showed higher OR and Umod lower OR for eGFRcys versus mGFR). No proteins showed significantly different associations between eGFRcre decline and mGFR decline in unadjusted analysis. For Gal-3, it was a statistically significant difference between mGFR and eGFRcyscre. For three proteins, the associations with eGFR decline were statistically significant different when using eGFRcre versus eGFRcys (TNFR2, Tie2, and Umod).
Most associations and differences between the GFR methods remained after adjustment for age, sex, BMI, and smoking (model 1), but many associations changed in the model that included baseline eGFR or mGFR, respectively (model 2). Only one of four proteins for eGFRcre and one of five proteins for eGFRcys in model 1 remained associated with eGFR decline after additional adjustment for baseline eGFR, while three of three remained associated with mGFR decline after adjusting for baseline mGFR. Five and two proteins showed statistically significant different results between eGFR and mGFR in model 1 and model 2, respectively (model 2, Table 2).
Incident CKD
At baseline, eight participants had CKD using mGFR, 95 developed CKD during follow-up, and the rest (n=1307) never developed CKD during the study period. For eGFRcre, eGFRcys, and eGFRcyscre: 4, 5, and 3 had eGFR <60 ml/min per 1.73 m2 per year at baseline and 51, 96, and 62 developed CKD, respectively.
In the interval-censored Cox regression analysis using mGFR, eight proteins were associated with an increased risk of incident CKD in the unadjusted model, five with eGFRcre, eight with eGFRcys, and seven with eGFRcyscre. In unadjusted analyses, there were statistically significant differences in the association of one protein for incident CKD using eGFRcre (suPAR), three for eGFRcys (MCP-1, TNFR2 and GDF-15), and one (Gal-3) for eGFRcyscre, compared with the results using mGFR. For three proteins (TNFR2, suPAR, and Umod), there were also differences in the associations with incident CKD using eGFRcre versus eGFRcys.
Most of the associations between proteins and incident CKD, and the different results compared with mGFR, remained similar after adjustment for age, sex, and BMI (Table 3, model 1). However, after additional adjustment for the corresponding measured or estimated baseline GFR, most of the associations between baseline protein concentration and incident CKD changed. Four proteins remained associated with increased risk of incident CKD using mGFR (TRAIL-R2, TNFR2, GDF-15, and MMP7). For eGFR, only two proteins remained associated with increased risk (GDF-15 with eGFRcys and MMP7 with all eGFRs). Only one protein (suPAR) remained statistically significantly differently associated with incident CKD compared with the association with incident CKD using mGFR (Table 3).
Discussion
In this cohort from the general population without diabetes, self-reported kidney disease, or CVD at baseline, biomarkers showed divergent associations with the three eGFR methods compared with mGFR and among the eGFR methods themselves. Several of the differences between accelerated decline of mGFR and eGFR or between eGFRcre and eGFRcys were statistically significant in unadjusted models and when adjusting for age, sex, and BMI. Two differences remained significant in the model that included baseline GFR. More biomarkers showed different associations with eGFRcys decline than with eGFRcre decline. Only MMP7 showed statistically significant associations with GFR decline and incident CKD using all GFR methods, regardless of adjustment.
These findings indicate the influence of non–GFR-related factors when assessing the association of proteins with eGFR decline. We have previously shown that eGFRcre and eGFRcys are influenced by inflammation, as assessed by TNFR2 and C-reactive protein (CRP) and by CKD risk factors.14,15 The current results, showing a statistically significant difference between the GFR methods used and, e.g., associations with TNFR2, indicate that different associations can be found for some biomarkers when assessing GFR change using eGFR or mGFR. For example, muscle mass, obesity, and low-grade inflammation may affect the level of several biomarkers and at the same time influence the production rate of creatinine and/or cystatin C. Muscle wasting (reduced creatinine), increased body fat, and increased inflammation (increased cystatin C) during follow-up, which are commonly seen during aging, may lead to a stronger association with a decline in eGFRcys and a weaker association with the eGFRcre decline rate. Adjustment for factors that have been associated with non–GFR-related influence on creatinine and cystatin C (e.g., age, sex, obesity, and smoking in model 1) at baseline would be expected to reduce such influences. However, for most of the investigated proteins, the differences to mGFR persisted in model 1. Other nontraditional cardiovascular risk markers, such as fasting insulin levels, muscle mass, and dimethylarginines, have also been found to influence creatinine and/or cystatin C levels along non–GFR-related pathways.16 Since these risk factors are typically not available as covariates for adjustment in many studies, residual confounding may persist when using eGFR.
Several of the associations between proteins and eGFR decline, and the different associations compared with mGFR decline in this study, were attenuated and no longer significant when adjusting for baseline eGFR or mGFR, respectively.65 There may be several explanations for this, and one possibility is that the inclusion of baseline eGFR in the model partly blocks the non–GFR-related effects on eGFR change, thereby also reducing the longitudinal confounding. Accordingly, adjusting for baseline GFR may adjust for some of the confounding factors so that change in eGFR is more reflective of true change in mGFR.
In line with the results of the present longitudinal study, several previous cross-sectional studies found cystatin C to be influenced by more non–GFR-related factors than creatinine.14,16,17 We recently reported that eGFRcys overestimates GFR change rates compared with mGFR.21 The distribution of the GFR decline rates was wider with eGFRcys than with mGFR, possibly due to the influence of non–GFR-related factors.
Ten protein biomarkers in this study have previously been shown to be associated with eGFR in general population studies, of which nine were associated with eGFR decline.3–13,53,66–68 Some of these biomarkers were not validated in separate cohorts, whereas others were validated but with mixed results. MMP7 has been associated with GFR decline in patients with diabetes using eGFRcre69,70 and was also associated with mGFR decline over a median of 5.6 years in the RENIS cohort.54 Several of the proteins that have shown associations with eGFR decline in previous studies did not show any associations in our study, regardless of the method used to measure or estimate GFR. The different age distributions, population characteristics of the included persons in the studies, and follow-up times, as well as different methods to measure proteins (ELISA, Luminex/proteomic) and statistical methods, could explain some of the diverging findings. Our cohort was relatively healthy at baseline, without self-reported CVD, kidney disease, or diabetes, with mean estimated and measured baseline GFRs well within the normal range.3–6,8,9 Thus, some of the observed GFR decline may represents age-related GFR decline rather than a pathological decline due to underlying disease. Although age-related GFR decline is an important driver of the high prevalence of CKD in the aging population, some proteins may be associated with specific disease processes in subgroups of people, which were not prevalent in this study. However, our main objective was to validate associations found using eGFR with mGFR. We have no reason to believe that differences between methods will not hold in other more diseased populations. Similar non–GFR-related determinants of cystatin C and creatinine, such as inflammation, obesity, and CVD risk factors, have been found in different populations,16,17,63 and their influence on eGFR may even be larger in patients than in healthy persons.
A study with older Swedish participants from two separate general population cohorts found associations between FABP4, suPAR, GDF-15, TRAIL-R2, and TNFR2 and annual eGFR decline and incident CKD.3 In these Swedish cohorts, 74% and 40% of the participants had diabetes, which may explain the different results compared with this study. However, among 20 proteins that were associated with eGFR decline in both cohorts, none were consistently associated with eGFR decline after adjustment for baseline eGFR. There may be several reasons for these findings, and it is controversial whether it is correct to adjust for the baseline value when investigating change rates of that same variable.65,71 Nevertheless, our study shows that the results of eGFR decline are more similar to mGFR decline in models that adjust for the corresponding baseline GFR.
The results of this study need to be interpreted in the context of some strengths and limitations. The main strength is the longitudinal design with repeated GFR measurements by iohexol clearance during 11 years of follow-up. In addition, we investigated the association with biomarkers that represent different pathways likely involved in kidney function decline and CKD development and used different statistical methods to assess the relationship with different kidney outcomes using both mGFR and eGFR.
There are also limitations. Generalizability to other age groups and ethnicities is limited by only including North European study participants between 50 and 64 years. Misclassification of incident CKD could have occurred because censoring was performed at the first visit with mGFR/eGFR <60 ml/min per 1.73 m2 without repeated eGFR/mGFR after 3 months. However, others have found similar risk patterns using this definition compared with CKD confirmed with later follow-up measurements, but with lower risk estimates.61 The linear mixed model used to calculate the accelerated GFR decline outcome assumes a linear decline in GFR. Although this assumption may not hold for some participants, the nonlinearity of the GFR trajectory in RENIS is modest and fairly similar for eGFR compared with mGFR.21 Most proteins were measured using a Luminex assay with varying intra-assay and interassay CVs between 2.9%–6.4% and 4.9%–19.3% (four >10%), respectively. Thus, misclassification may have occurred. However, our mean intra-assay and interassay CVs were lower than or equal to those reported by others using the Luminex method.3,72,73 In addition, in a previous publication, we explored the high interassay CV of MMP7, where one assay was found to have standards which deviated from the expected concentrations by 7.5%–28%. By excluding the 30 participants on this assay, the interassay CV for the study population was reduced from 19.3% to 13.7%, and the association between the GFR outcomes and MMP7 even became slightly stronger.
To conclude, some associations between biomarkers and eGFR decline were different from the associations with mGFR decline. Thus, spurious associations with eGFR decline may be caused by the influence of non-GFR factors. Although the differences between the methods were attenuated after adjustment, particularly after adjustment for baseline eGFR, persisting differences for some biomarkers may be a problem in research on CKD pathophysiology and risk factors. The results of studies using eGFR to identify biomarkers for GFR decline should therefore be interpreted with caution and preferably be validated with mGFR.
Supplementary Material
Disclosures
T. Melsom reports Honoraria: Novo Nordisk Norway AS, lecture at a local meeting. M.D. Solbu reports Honoraria: AstraZeneca; Advisory or Leadership Role: Baxter and Vifor Pharma; and Other Interests or Relationships: ERA-EDTA, ISN, Norwegian Society of Hypertension, and Norwegian Society of Nephrology. All remaining authors have nothing to disclose.
Funding
The RENIS studies were funded by the Northern Norway Regional Health Authorities (SFP 1100-13), and RENIS-FU was also supported by an unrestricted grant obtained from Boehringer Ingelheim (1235.104 IIS).
Acknowledgments
We are grateful to the staff at the Clinical Research Unit, University Hospital of North Norway, who made it possible to conduct this study by assisting in planning, conducting procedures, and collecting data. We thank Gro Bolstad at the Metabolic Laboratory of UiT–The Arctic University of Norway, who analyzed baseline protein biomarker levels using the Luminex assay. We thank all the participants in the RENIS cohort for their contributions to this investigation.
Author Contributions
Conceptualization: Bjørn O. Eriksen, Toralf Melsom.
Data curation: Bjørn O. Eriksen, Toralf Melsom.
Investigation: Inger T.T. Enoksen, Bjørn O. Eriksen, Toralf Melsom.
Methodology: Inger T.T. Enoksen, Bjørn O. Eriksen, Toralf Melsom, Nikoline B. Rinde, Dmitri Svistounov.
Project administration: Bjørn O. Eriksen, Toralf Melsom.
Supervision: Bjørn O. Eriksen, Toralf Melsom.
Validation: Bjørn O. Eriksen, Toralf Melsom.
Visualization: Inger T.T. Enoksen.
Writing – original draft: Inger T.T. Enoksen.
Writing – review & editing: Inger T.T. Enoksen, Bjørn O. Eriksen, Toralf Melsom, Jon V. Norvik, Nikoline B. Rinde, Marit D. Solbu, Dmitri Svistounov.
Data Sharing Statement
The data underlying this article cannot be shared publicly since this was not included in the research permission due to ethical considerations and the privacy of individuals who participated in the study. The data can be shared upon request as part of the research collaboration.
Supplemental Material
This article contains the following supplemental material online at http://links.lww.com/JSN/E428.
Supplemental Table 1. Literature searches 3.
Supplemental Table 2. Protein intra-assay and interassay % CV 4.
Supplemental Table 3. Baseline characteristics 5.
Supplemental Figure 1. Literature search 7.
References
- 1.Bikbov B, Purcell CA, Levey AS, et al. Global, regional, and national burden of chronic kidney disease, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2020;395(10225):709–733. doi: 10.1016/s0140-6736(20)30045-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hill NR, Fatoba ST, Oke JL, et al. Global prevalence of chronic kidney disease – a systematic review and meta-analysis. PLoS One. 2016;11(7):e0158765. doi: 10.1371/journal.pone.0158765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlsson AC, Ingelsson E, Sundström J, et al. Use of proteomics to investigate kidney function decline over 5 years. Clin J Am Soc Nephrol. 2017;12(8):1226–1235. doi: 10.2215/CJN.08780816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Seaghdha CM, Hwang S-J, Ho JE, Vasan RS, Levy D, Fox CS. Elevated galectin-3 precedes the development of CKD. J Am Soc Nephrol. 2013;24(9):1470–1477. doi: 10.1681/ASN.2012090909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rebholz CM, Selvin E, Liang M, et al. Plasma galectin-3 levels are associated with the risk of incident chronic kidney disease. Kidney Int. 2018;93(1):252–259. doi: 10.1016/j.kint.2017.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu G, Deng Y, Sun L, et al. Elevated plasma tumor necrosis factor-α receptor 2 and resistin are associated with increased incidence of kidney function decline in Chinese adults. Endocrine. 2016;52(3):541–549. doi: 10.1007/s12020-015-0807-3 [DOI] [PubMed] [Google Scholar]
- 7.Shankar A, Sun L, Klein BEK, et al. Markers of inflammation predict the long-term risk of developing chronic kidney disease: a population-based cohort study. Kidney Int. 2011;80(11):1231–1238. doi: 10.1038/ki.2011.283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bao X, Xu B, Borné Y, et al. Growth differentiation factor-15 and incident chronic kidney disease: a population-based cohort study. BMC Nephrol. 2021;22(1):351. doi: 10.1186/s12882-021-02558-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho JE, Hwang S-J, Wollert KC, et al. Biomarkers of cardiovascular stress and incident chronic kidney disease. Clin Chem. 2013;59(11):1613–1620. doi: 10.1373/clinchem.2013.205716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayek SS, Sever S, Ko Y-A, et al. Soluble urokinase receptor and chronic kidney disease. New Engl J Med. 2015;373(20):1916–1925. doi: 10.1056/nejmoa1506362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayek SS, Ko Y-A, Awad M, et al. Cardiovascular disease biomarkers and suPAR in predicting decline in renal function: a prospective cohort study. Kidney Int Rep. 2017;2(3):425–432. doi: 10.1016/j.ekir.2017.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schulz C-A, Persson M, Christensson A, et al. Soluble urokinase-type plasminogen activator receptor (suPAR) and impaired kidney function in the population-based malmö diet and cancer study. Kidney Int Rep. 2017;2(2):239–247. doi: 10.1016/j.ekir.2016.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Then C, Then HL, Lechner A, et al. Serum uromodulin and decline of kidney function in older participants of the population-based KORA F4/FF4 study. Clin Kidney J. 2021;14(1):205–211. doi: 10.1093/ckj/sfaa032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schei J, Stefansson VTN, Mathisen UD, et al. Residual associations of inflammatory markers with eGFR after accounting for measured GFR in a community-based cohort without CKD. Clin J Am Soc Nephrol. 2016;11(2):280–286. doi: 10.2215/CJN.07360715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Melsom T, Fuskevåg OM, Mathisen UD, et al. Estimated GFR is biased by non-traditional cardiovascular risk factors. Am J Nephrol. 2015;41(1):7–15. doi: 10.1159/000371557 [DOI] [PubMed] [Google Scholar]
- 16.Mathisen UD, Melsom T, Ingebretsen OC, et al. Estimated GFR associates with cardiovascular risk factors independently of measured GFR. J Am Soc Nephrol. 2011;22(5):927–937. doi: 10.1681/ASN.2010050479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rule AD, Bailey KR, Lieske JC, Peyser PA, Turner ST. Estimating the glomerular filtration rate from serum creatinine is better than from cystatin C for evaluating risk factors associated with chronic kidney disease. Kidney Int. 2013;83(6):1169–1176. doi: 10.1038/ki.2013.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stevens LA, Coresh J, Greene T, Levey AS. Assessing kidney function—measured and estimated glomerular filtration rate. N Engl J Med. 2006;354(23):2473–2483. doi: 10.1056/nejmra054415 [DOI] [PubMed] [Google Scholar]
- 19.Knight EL, Verhave JC, Spiegelman D, et al. Factors influencing serum cystatin C levels other than renal function and the impact on renal function measurement. Kidney Int. 2004;65(4):1416–1421. doi: 10.1111/j.1523-1755.2004.00517.x [DOI] [PubMed] [Google Scholar]
- 20.Eggen AE, Mathiesen EB, Wilsgaard T, Jacobsen BK, Njolstad I. The sixth survey of the Tromso Study (Tromso 6) in 2007-08: collaborative research in the interface between clinical medicine and epidemiology: study objectives, design, data collection procedures, and attendance in a multipurpose population-based health survey. Scand J Public Health. 2013;41(1):65–80. doi: 10.1177/1403494812469851 [DOI] [PubMed] [Google Scholar]
- 21.Melsom T, Norvik JV, Enoksen IT, et al. Sex differences in age-related loss of kidney function. J Am Soc Nephrol. 2022;33(10):1891–1902. doi: 10.1681/ASN.2022030323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delanaye P, Ebert N, Melsom T, et al. Iohexol plasma clearance for measuring glomerular filtration rate in clinical practice and research: a review. Part 1: how to measure glomerular filtration rate with iohexol? Clin kidney J. 2016;9(5):682–699. doi: 10.1093/ckj/sfw070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eriksen BO, Mathisen UD, Melsom T, et al. Cystatin C is not a better estimator of GFR than plasma creatinine in the general population. Kidney Int. 2010;78(12):1305–1311. doi: 10.1038/ki.2010.321 [DOI] [PubMed] [Google Scholar]
- 24.Eriksen BO, Stefansson VTN, Jenssen TG, et al. Elevated blood pressure is not associated with accelerated glomerular filtration rate decline in the general non-diabetic middle-aged population. Kidney Int. 2016;90(2):404–410. doi: 10.1016/j.kint.2016.03.021 [DOI] [PubMed] [Google Scholar]
- 25.Jacobsson L. A method for the calculation of renal clearance based on a single plasma sample. Clin Physiol. 1983;3(4):297–305. doi: 10.1111/j.1475-097x.1983.tb00712.x [DOI] [PubMed] [Google Scholar]
- 26.Nilsson-Ehle P. Iohexol clearance for the determination of glomerular filtration rate: 15 Years' experience in clinical practice. EJIFCC. 2001;13(2):48–52. https://pubmed.ncbi.nlm.nih.gov/30429722/ [PMC free article] [PubMed] [Google Scholar]
- 27.Deshmane SL, Kremlev S, Amini S, Sawaya BE. Monocyte chemoattractant protein-1 (MCP-1): an overview. J Interferon Cytokine Res. 2009;29(6):313–326. doi: 10.1089/jir.2008.0027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lloyd CM, Minto AW, Dorf ME, et al. RANTES and monocyte chemoattractant protein-1 (MCP-1) play an important role in the inflammatory phase of crescentic nephritis, but only MCP-1 is involved in crescent formation and interstitial fibrosis. J Exp Med. 1997;185(7):1371–1380. doi: 10.1084/jem.185.7.1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walczak H, Degli-Esposti MA, Johnson RS, et al. TRAIL-R2: a novel apoptosis-mediating receptor for TRAIL. EMBO J. 1997;16(17):5386–5397. doi: 10.1093/emboj/16.17.5386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaudhary PM, Eby M, Jasmin A, Bookwalter A, Murray J, Hood L. Death receptor 5, a new member of the TNFR family, and DR4 induce FADD-dependent apoptosis and activate the NF-κB pathway. Immunity. 1997;7(6):821–830. doi: 10.1016/s1074-7613(00)80400-8 [DOI] [PubMed] [Google Scholar]
- 31.Furuhashi M, Saitoh S, Shimamoto K, Miura T. Fatty acid-binding protein 4 (FABP4): pathophysiological insights and potent clinical biomarker of metabolic and cardiovascular diseases. Clin Med Insights Cardiol. 2015;8(suppl 3):23–33. doi: 10.4137/cmc.s17067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ebert T, Hopf LM, Wurst U, et al. Circulating adipocyte fatty acid binding protein is increased in chronic and acute renal dysfunction. Nutr Metab Cardiovasc Dis. 2014;24(9):1027–1034. doi: 10.1016/j.numecd.2014.03.006 [DOI] [PubMed] [Google Scholar]
- 33.Shrestha S, Sunaga H, Hanaoka H, et al. Circulating FABP4 is eliminated by the kidney via glomerular filtration followed by megalin-mediated reabsorption. Sci Rep. 2018;8(1):16451. doi: 10.1038/s41598-018-34902-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng Y, Guo F, Xia Z, et al. Inhibition of fatty acid-binding protein 4 attenuated kidney fibrosis by mediating macrophage-to-myofibroblast transition. Front Immunol. 2020;11:566535. doi: 10.3389/fimmu.2020.566535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kohno T, Brewer MT, Baker SL, et al. A second tumor necrosis factor receptor gene product can shed a naturally occurring tumor necrosis factor inhibitor. Proc Natl Acad Sci U S A. 1990;87(21):8331–8335. doi: 10.1073/pnas.87.21.8331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Romero X, Cañete JD, Engel P. Determination of soluble tumor necrosis factor receptor 2 produced by alternative splicing. In: Bayry J, editor. The TNF Superfamily: Methods and Protocols. Springer New York; 2014:187–199. [DOI] [PubMed] [Google Scholar]
- 37.Fitzgerald KA, O'Neill LAJ, Gearing AJH, Callard RE. CD40L. In: Fitzgerald KA, O'Neill LAJ, Gearing AJH, Callard RE, editors. The Cytokine FactsBook and Webfacts, 2nd ed. Academic Press; 2001:183–187. [Google Scholar]
- 38.Zhang S, Breidenbach JD, Khalaf FK, et al. Renal fibrosis is significantly attenuated following targeted disruption of Cd40 in experimental renal ischemia. J Am Heart Assoc. 2020;9(7):e014072. doi: 10.1161/jaha.119.014072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bootcov MR, Bauskin AR, Valenzuela SM, et al. MIC-1, a novel macrophage inhibitory cytokine, is a divergent member of the TGF-beta superfamily. Proc Natl Acad Sci U S A. 1997;94(21):11514–11519. doi: 10.1073/pnas.94.21.11514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reusch P, Barleon B, Weindel K, et al. Identification of a soluble form of the angiopoietin receptor TIE-2 released from endothelial cells and present in human blood. Angiogenesis. 2001;4(2):123–131. doi: 10.1023/a:1012226627813 [DOI] [PubMed] [Google Scholar]
- 41.Liu Z, Tan RJ, Liu Y. The many faces of matrix metalloproteinase-7 in kidney diseases. Biomolecules. 2020;10(6):960. doi: 10.3390/biom10060960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matrisian LM. Chapter 161 - matrix metallopeptidase-7/matrilysin. In: Rawlings ND, Salvesen G, editors. Handbook of Proteolytic Enzymes, 3rd ed. Academic Press; 2013:786–795. [Google Scholar]
- 43.Thunø M, Macho B, Eugen-Olsen J. suPAR: the molecular crystal ball. Dis Markers. 2009;27(3):157–172. doi: 10.1155/2009/504294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Enocsson H, Wirestam L, Dahle C, et al. Soluble urokinase plasminogen activator receptor (suPAR) levels predict damage accrual in patients with recent-onset systemic lupus erythematosus. J Autoimmun. 2020;106:102340. doi: 10.1016/j.jaut.2019.102340 [DOI] [PubMed] [Google Scholar]
- 45.Visse R, Nagase H. Matrix metalloproteinases and tissue inhibitors of metalloproteinases. Circ Res. 2003;92(8):827–839. doi: 10.1161/01.res.0000070112.80711.3d [DOI] [PubMed] [Google Scholar]
- 46.Klein T, Bischoff R. Physiology and pathophysiology of matrix metalloproteases. Amino Acids. 2011;41(2):271–290. doi: 10.1007/s00726-010-0689-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lau W-H, Leong W-S, Ismail Z, Gam L-H. Qualification and application of an ELISA for the determination of tamm horsfall protein (THP) in human urine and its use for screening of kidney stone disease. Int J Biol Sci. 2008;4(4):215–222. doi: 10.7150/ijbs.4.215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Youhanna S, Weber J, Beaujean V, Glaudemans B, Sobek J, Devuyst O. Determination of uromodulin in human urine: influence of storage and processing. Nephrol Dial Transplant. 2014;29(1):136–145. doi: 10.1093/ndt/gft345 [DOI] [PubMed] [Google Scholar]
- 49.Dumic J, Dabelic S, Flögel M. Galectin-3: an open-ended story. Biochim Biophys Acta. 2006;1760(4):616–635. doi: 10.1016/j.bbagen.2005.12.020 [DOI] [PubMed] [Google Scholar]
- 50.Liu F-T, Patterson RJ, Wang JL. Intracellular functions of galectins. Biochim Biophys Acta. 2002;1572(2-3):263–273. doi: 10.1016/s0304-4165(02)00313-6 [DOI] [PubMed] [Google Scholar]
- 51.Henderson NC, Sethi T. The regulation of inflammation by galectin-3. Immunological Rev. 2009;230(1):160–171. doi: 10.1111/j.1600-065x.2009.00794.x [DOI] [PubMed] [Google Scholar]
- 52.Karmakova ТА, Sergeeva NS, Kanukoev КY, Alekseev B, Kaprin A. Kidney injury molecule 1 (KIM-1): a multifunctional glycoprotein and biological marker (review). Sovrem Tekhnologii Med. 2021;13(3):64–78. doi: 10.17691/stm2021.13.3.08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schei J, Stefansson VT, Eriksen BO, et al. Association of TNF receptor 2 and CRP with GFR decline in the general nondiabetic population. Clin J Am Soc Nephrol. 2017;12(4):624–634. doi: 10.2215/CJN.09280916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Enoksen IT, Svistounov D, Norvik JV, et al. Serum matrix metalloproteinase 7 and accelerated glomerular filtration rate decline in a general non-diabetic population. Nephrol Dial Transplant. 2021;37(9):1657–1667. doi: 10.1093/ndt/gfab251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eriksen BO, Løchen ML, Arntzen KA, et al. Estimated and measured GFR associate differently with retinal vasculopathy in the general population. Nephron. 2015;131(3):175–184. doi: 10.1159/000441092 [DOI] [PubMed] [Google Scholar]
- 56.Inker LA, Schmid CH, Tighiouart H, et al. Estimating glomerular filtration rate from serum creatinine and cystatin C. N Engl J Med. 2012;367(1):20–29. doi: 10.1056/nejmoa1114248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Levey AS, Stevens LA, Schmid CH, et al. A new equation to estimate glomerular filtration rate. Ann Intern Med. 2009;150(9):604–612. doi: 10.7326/0003-4819-150-9-200905050-00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Norvik JV, Harskamp LR, Nair V, et al. Urinary excretion of epidermal growth factor and rapid loss of kidney function. Nephrol Dial Transplant. 2021;36(10):1882–1892. doi: 10.1093/ndt/gfaa208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leffondre K, Boucquemont J, Tripepi G, Stel VS, Heinze G, Dunkler D. Analysis of risk factors associated with renal function trajectory over time: a comparison of different statistical approaches. Nephrol Dial Transplant. 2014;30(8):1237–1243. doi: 10.1093/ndt/gfu320 [DOI] [PubMed] [Google Scholar]
- 60.Grams ME, Sang Y, Ballew SH, et al. Evaluating glomerular filtration rate slope as a surrogate end point for ESKD in clinical trials: an individual participant meta-analysis of observational data. J Am Soc Nephrol. 2019;30(9):1746–1755. doi: 10.1681/ASN.2019010008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nelson RG, Grams ME, Ballew SH, et al. Development of risk prediction equations for incident chronic kidney disease. JAMA. 2019;322(21):2104–2114. doi: 10.1001/jama.2019.17379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zeng D, Mao L, Lin DY. Maximum likelihood estimation for semiparametric transformation models with interval-censored data. Biometrika. 2016;103(2):253–271. doi: 10.1093/biomet/asw013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stevens LA, Schmid CH, Greene T, et al. Factors other than glomerular filtration rate affect serum cystatin C levels. Kidney Int. 2009;75(6):652–660. doi: 10.1038/ki.2008.638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rothman KJ. No adjustments are needed for multiple comparisons. Epidemiology. 1990;1(1):43–46. doi: 10.1097/00001648-199001000-00010 [DOI] [PubMed] [Google Scholar]
- 65.Glymour MM, Weuve J, Berkman LF, Kawachi I, Robins JM. When is baseline adjustment useful in analyses of change? An example with education and cognitive change. Am J Epidemiol. 2005;162(3):267–278. doi: 10.1093/aje/kwi187 [DOI] [PubMed] [Google Scholar]
- 66.Hennings A, Hannemann A, Rettig R, et al. Circulating angiopoietin-2 and its soluble receptor tie-2 concentrations are related to renal function in two population-based cohorts. PLoS One. 2016;11(11):e0166492. doi: 10.1371/journal.pone.0166492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Steubl D, Block M, Herbst V, et al. Plasma uromodulin correlates with kidney function and identifies early stages in chronic kidney disease patients. Medicine. 2016;95(10):e3011. doi: 10.1097/md.0000000000003011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Scherberich JE, Gruber R, Nockher WA, et al. Serum uromodulin—a marker of kidney function and renal parenchymal integrity. Nephrol Dial Transplant. 2018;33(2):284–295. doi: 10.1093/ndt/gfw422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pena MJ, Heinzel A, Heinze G, et al. A panel of novel biomarkers representing different disease pathways improves prediction of renal function decline in type 2 diabetes. PLoS One. 2015;10(5):e0120995. doi: 10.1371/journal.pone.0120995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ihara K, Skupien J, Kobayashi H, et al. Profibrotic circulating proteins and risk of early progressive renal decline in patients with type 2 diabetes with and without albuminuria. Diabetes Care. 2020;43(11):2760–2767. doi: 10.2337/dc20-0630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tu YK, Gilthorpe MS. Revisiting the relation between change and initial value: a review and evaluation. Stat Med. 2007;26(2):443–457. doi: 10.1002/sim.2538 [DOI] [PubMed] [Google Scholar]
- 72.O’Seaghdha CM, Hwang S-J, Larson MG, Meigs JB, Vasan RS, Fox CS. Analysis of a urinary biomarker panel for incident kidney disease and clinical outcomes. J Am Soc Nephrol. 2013;24(11):1880–1888. doi: 10.1681/ASN.2013010019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schrauben SJ, Shou H, Zhang X, et al. Association of multiple plasma biomarker concentrations with progression of prevalent diabetic kidney disease: findings from the chronic renal insufficiency cohort (CRIC) study. J Am Soc Nephrol. 2021;32(1):115–126. doi: 10.1681/ASN.2020040487 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data underlying this article cannot be shared publicly since this was not included in the research permission due to ethical considerations and the privacy of individuals who participated in the study. The data can be shared upon request as part of the research collaboration.