Keywords: cold storage, kidney transplantation, microRNA, NDUFA4, mitochondria, proximal tubular cells
Abstract
Significance Statement
Cold storage-associated transplantation (CST) injury occurs in renal transplant from deceased donors, the main organ source. The pathogenesis of CST injury remains poorly understood, and effective therapies are not available. This study has demonstrated an important role of microRNAs in CST injury and revealed the changes in microRNA expression profiles. Specifically, microRNA-147 (miR-147) is consistently elevated during CST injury in mice and in dysfunctional renal grafts in humans. Mechanistically, NDUFA4 (a key component of mitochondrial respiration complex) is identified as a direct target of miR-147. By repressing NDUFA4, miR-147 induces mitochondrial damage and renal tubular cell death. Blockade of miR-147 and overexpression of NDUFA4 reduce CST injury and improve graft function, unveiling miR-147 and NDUFA4 as new therapeutic targets in kidney transplantation.
Background
Kidney injury due to cold storage–associated transplantation (CST) is a major factor determining the outcome of renal transplant, for which the role and regulation of microRNAs remain largely unclear.
Methods
The kidneys of proximal tubule Dicer (an enzyme for microRNA biogenesis) knockout mice and their wild-type littermates were subjected to CST to determine the function of microRNAs. Small RNA sequencing then profiled microRNA expression in mouse kidneys after CST. Anti–microRNA-147 (miR-147) and miR-147 mimic were used to examine the role of miR-147 in CST injury in mouse and renal tubular cell models.
Results
Knockout of Dicer from proximal tubules attenuated CST kidney injury in mice. RNA sequencing identified multiple microRNAs with differential expression in CST kidneys, among which miR-147 was induced consistently in mouse kidney transplants and in dysfunctional human kidney grafts. Anti–miR-147 protected against CST injury in mice and ameliorated mitochondrial dysfunction after ATP depletion injury in renal tubular cells in intro. Mechanistically, miR-147 was shown to target NDUFA4, a key component of the mitochondrial respiration complex. Silencing NDUFA4 aggravated renal tubular cell death, whereas overexpression of NDUFA4 prevented miR-147–induced cell death and mitochondrial dysfunction. Moreover, overexpression of NDUFA4 alleviated CST injury in mice.
Conclusions
microRNAs, as a class of molecules, are pathogenic in CST injury and graft dysfunction. Specifically, miR-147 induced during CST represses NDUFA4, leading to mitochondrial damage and renal tubular cell death. These results unveil miR-147 and NDUFA4 as new therapeutic targets in kidney transplantation.
Introduction
Renal transplant, including deceased donor and living donor transplantation, is the most effective treatment for ESKD.1,2 Although living donor transplant has a better outcome, its source is very limited, and the majority of renal transplants are from deceased donors.3,4 In deceased donor transplant, the donor kidney experiences cold storage during organ procurement, tissue typing, and transportation before transplantation. Kidney injury due to cold storage–associated transplantation (CST) is known to be a major factor contributing to the pathogenesis of delayed graft function (DGF) and chronic allograft nephropathy.5–7
microRNAs are small noncoding RNAs that are produced endogenously through the transcription of microRNA genes, followed by sequential cleavage by Drosa and Dicer to generate mature microRNAs.8–11 Functionally, microRNAs repress their target gene expression by complementarily binding to the 3′ untranslated region (3′UTR) of their target gene mRNAs. In kidneys, microRNAs play important roles in renal physiology and the pathogenesis of kidney diseases.10,12,13 Targeting microRNAs has been widely explored as a therapeutic strategy,14–16 and microRNA-based drugs have entered clinical trials treating CKDs.17–20 However, therapies targeting microRNAs in AKI or renal transplantation have not yet entered the clinical stage.10, 21–26 In 2010, we established a proximal tubule (PT)-specific Dicer knockout (KO) mouse model with microRNA depletion from kidneys.27 These mice were remarkably resistant to ischemic AKI,27 but they showed similar levels of cisplatin nephrotoxic AKI as their wild-type (WT) littermates,28 indicating a complex role of microRNAs in the pathogenesis of AKI.10,15,21–23,28–30
This study delineates the role and regulation of microRNAs in cold storage–associated kidney transplantation (CST). We show that ablation of Dicer form kidney PTs attenuated CST injury in mice, supporting a pathogenic role of microRNAs. We further identified a robust induction of microRNA-147 (miR-147) in CST mouse kidneys and in human renal grafts with dysfunction. Mechanistically, we identified NDUFA4, a component of mitochondrial respiration complexes, as a novel target of miR-147. We further proved a role of NDUFA4 in miR-147–induced mitochondrial dysfunction and cells death in kidney tubular cells. These findings indicate that miR-147 is induced during CST to mediate kidney tubular cell damage by repressing NDUFA4 and inducing mitochondrial dysfunction.
Results
Dicer Deficiency in Kidney PTs Preserves Graft Function and Attenuates CST Injury
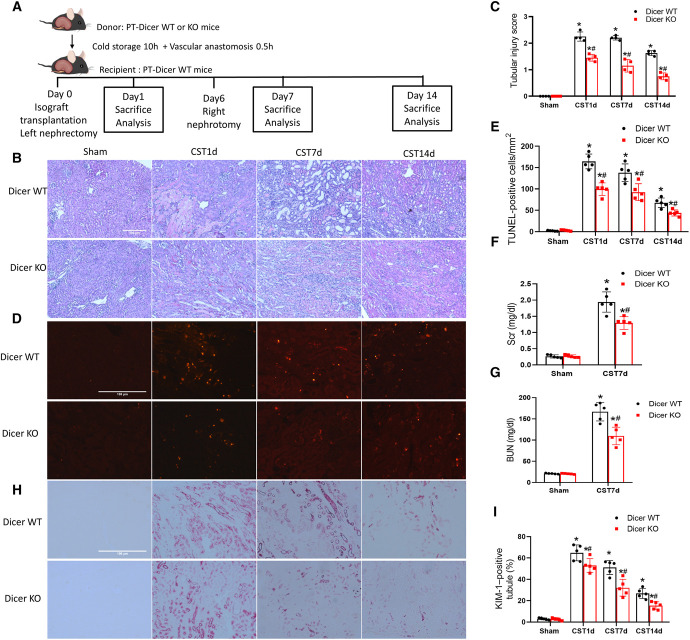
PTs are the main injury site in transplantation-associated kidney injury.31,32 To determine the significance of microRNAs, we subjected the kidneys of PT-Dicer-WT mice and PT-specific Dicer KO (PT-Dicer-KO) mice to cold storage, followed by transplantation into WT recipients according to a well-characterized mouse model of cold storage-associated transplantation (CST)33–36 (Figure 1A). One day after transplantation, WT grafts showed tubular cell loss and necrosis with notable cast formation in the outer medulla and cortex indicating CST injury, whereas Dicer-KO grafts showed better renal histology with fewer lysed tubules and fewer terminal deoxynucleotidyl transferase–mediated digoxigenin-deoxyuridine nick-end labeling (TUNEL)–positive cells (Figure 1, B–E). At day 7, WT grafts had many dilated renal tubules, which were absent in Dicer KO grafts. At day 14, WT grafts showed repair in histology, whereas KO grafts almost fully recovered. Consistently, KO grafts had fewer apoptotic cells in TUNEL staining (Figure 1, D and E). To examine graft function, we allowed the transplanted kidney to recover from initial injury for 6 days and then removed the native kidney, so the transplanted graft became the only life-supporting kidney for excretion. In the mice with WT graft, serum creatinine and BUN markedly increased 1 day after removal of the native kidney (Figure 1, F and G), indicating graft dysfunction. In contrast, the mice with PT-Dicer-KO grafts showed lower levels of serum creatinine and BUN (Figure 1, F and G). Kidney Injury Molecule-1 (KIM-1), a maker for injured PTs,37 increased significantly in WT grafts at day 1 after transplantation and then decreased at day 7 and 14 likely as a result of kidney repair. At all these time points, PT-Dicer-KO grafts showed fewer KIM-1–positive kidney tubules (Figure 1). Dicer is a key enzyme for biogenesis of functional microRNAs,10 and the ablation of PT Dicer led to microRNA depletion in kidneys.27 Therefore, our results indicate that Dicer-associated microRNAs in PTs contribute to CST injury.
Figure 1.
Dicer deficiency in kidney PTs preserves graft function and attenuates cold storage-associated transplantation injury. (A) Diagram showing the experimental design. The left kidney was collected from Dicer-KO or WT mice for 10 hours of cold storage, followed by transplantation into WT mice. On day 6 post-transplantation, the native kidney of the recipient mouse was removed so the transplanted kidney became the life-supporting kidney. On day 1, 7, and 14 the transplanted kidney and blood samples were collected for analysis. The right kidney of donor mice without cold storage-associated transplantation was used as a sham control. Kidneys were fixed for histological examination, and blood was collected for renal function measurement. (B) Representative images of H&E staining of renal histology. Scale bar=100 μm. (C) Pathological score of tubular damage. (D) Representative images of TUNEL staining. Scale bar=100 μm. (E) Quantification of TUNEL staining–positive cells. (F) Serum creatinine. (G) BUN. (H) Representative images of KIM-1 immunohistochemistry. Scale bar=100 μm. (I) Quantification of KIM-1–positive tubules. Quantitative data are expressed as mean±SD (n=4–5). *P < 0.05 versus sham control; #P < 0.05 versus WT graft. KO, knockout, PT, proximal tubule, TUNEL, terminal deoxynucleotidyl transferase–mediated digoxigenin-deoxyuridine nick-end labeling; KIM-1, Kidney Injury Molecule-1; WT, wild-type.
Dicer Deficiency Reduces Inflammation and Fibrosis in CST Kidneys
Immunohistochemistry detected macrophage infiltration in transplanted WT kidneys at day 7 and day 14, which was suppressed in PT-Dicer-KO grafts (statistical power=0.7 approximately 0.8) (Supplemental Figure 1, A and E). Injured tubular cells induce local production of complement C3, which is associated with inflammation and plays a role in the development of chronic allograft nephropathy.38 C3 was rarely seen in sham control kidneys, but granular deposits of C3 around tubular basement membrane were detected in transplanted kidneys at both day 7 and day 14. C3 staining was significantly reduced in PT-Dicer-KO grafts (Supplemental Figure 1, B and F).
Chronic inflammation contributes to the development of renal fibrosis and ultimately graft dysfunction.39,40 We analyzed fibrosis by Sirius red staining and immunostaining of alpha-smooth muscle actin (α-SMA). Sham control kidneys had little Sirius red staining and α-SMA signal, but notable α-SMA and Sirius red staining was detected in transplanted WT kidneys at day 7 and day 14 (Supplemental Figure 1, C and D). Quantitatively, 3.5% and 4.8% of corticomedullary areas were positive for α-SMA and Sirius red staining, respectively, in PT-Dicer-WT grafts on day 14. Sirius red and α-SMA staining was significantly less in PT-Dicer-KO grafts (Supplemental Figure 1, C, D, G, and H).
These results support a role of Dicer-associated microRNAs in chronic inflammation and renal fibrogenesis in CST injury.
miR-147 Is Increased in CST Kidneys and Human DGF Samples
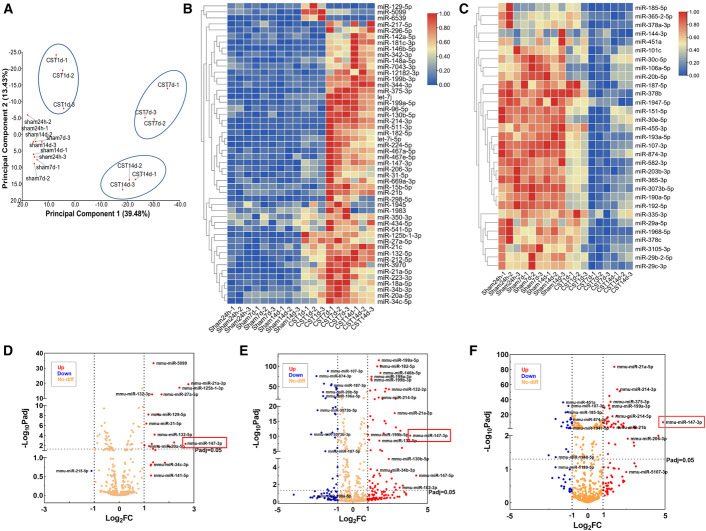
We performed small RNA deep sequencing to profile microRNA expression in CST mouse kidneys. Complete data of the sequencing analysis are presented in Supplemental Datasets 1, 2, and 3. Principal components analysis score plot showed a clear cluster separation between the control and CST samples (Figure 2A). We detected 505 known mouse microRNAs, among which 50 increased significantly after CST (Figure 2B), whereas 31 decreased at least at one time point of day 1, day 7, or day 14 (Figure 2C). Volcano plots further showed the differential expression of microRNAs (Figure 2, D and E). We also verified the increase of miR-132 and the decreases of miR-874 and miR-185 in CST kidneys by Taqman real-time PCR assay (Supplemental Figure 2 and Supplemental Datasets 1, 2, and 3).
Figure 2.
Changes in microRNA expression profile after kidney cold storage-associated transplantation. The left kidney was collected from WT mice for 10 hours of cold storage, followed by transplantation into WT mice for 1, 7, or 14 days. The right kidney of donor mice without cold storage-associated transplantation was used as sham control. Kidney RNA was collected for deep sequencing. (A) PCA of microRNA sequencing data. (B and C) Heatmaps showing upregulated and downregulated microRNAs in indicated groups. The microRNAs that were significantly altered (an average fold changes >1 and P < 0.01) at least one time point (1, 7, or 14 days) after cold storage-associated transplantation are presented. (C–E) Volcano plots showing differentially expressed microRNAs at 1, 7, or 14 days after cold storage-associated transplantation. The threshold in the volcano plot was P < 0.05 and fold changes >1. Upregulated genes are shown in red, whereas downregulated genes are in blue. WT, wild-type; principal components analysis, PCA. Figure 2 can be viewed in color online at www.jasn.org.
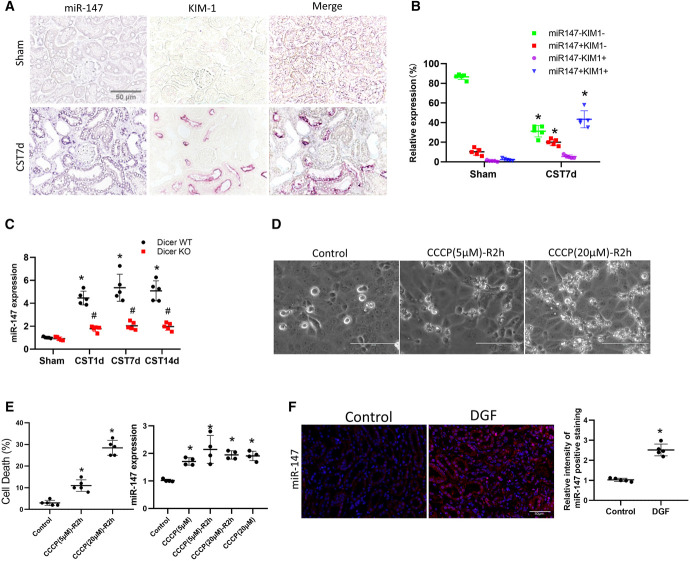
Compared with sham control, CST kidneys showed a robust induction of miR-147 at all time points in small RNA sequencing (Figure 2, D–F: red boxed). In situ hybridization detected miR-147 induction mainly in the cytoplasm of kidney tubular cells, and notably, the induction occurred mainly in KIM-1–positive tubules in CST kidney (Figure 3, A and B). Taqman real-time PCR further confirmed miR-147 induction in WT grafts, and interestingly, the induction was suppressed in Dicer-KO grafts (Figure 3C). In vitro, ATP depletion and recovery treatment induced miR-147 expression in cultured renal proximal tubular cells (RPTCs) (Figure 3, D and E). We further analyzed kidney biopsies of human patients with DGF and para-cancer normal kidney tissues by in situ hybridization. As shown in Figure 3F, miR-147 expression was significantly higher in kidney tubular cells in DGF samples than in normal kidney tissues. Collectively, these data demonstrate the induction of miR-147 in renal PT cells after CST injury.
Figure 3.
miR-147 is increased in cold storage-associated transplantation kidneys and human DGF samples. (A) In situ hybridization analysis of miR-147 and immunostaining of KIM-1 in serial sections of sham control and cold storage-associated transplantation kidneys. Scale bar=50 μm. (B) Quantification of tubules according to miR-147 and KIM-1 staining. (C) qPCR analysis of miR-147 in Dicer WT and KO kidneys at 1, 7, or 14 days after cold storage-associated transplantation or sham surgery. (D) Cellular morphology and in RPTCs with or without CCCP-R treatment for indicated time. (E) Percentage of cell death assessed morphologically and qPCR analysis of miR-147. (F) In situ hybridization showing miR-147 upregulation in human grafts with DGF in comparison with normal (para-cancer) kidney tissues. Scale bar=50 μm. Quantitative data are expressed as mean±SD (n=3–5). Scale bar=100 μm. *P < 0.05 versus control. #P < 0.05 versus WT graft. Carbonyl Cyanide Chlorophenylhydrazone-Recovery, CCCP-R; DGF, delayed graft function; KO, knockout; miR-147, microRNA-147; RPTC, renal proximal tubular cell; WT, wild-type. Figure 3 can be viewed in color online at www.jasn.org.
Anti–miR-147 Improves Graft Function and Reduces Graft Damage and Inflammation
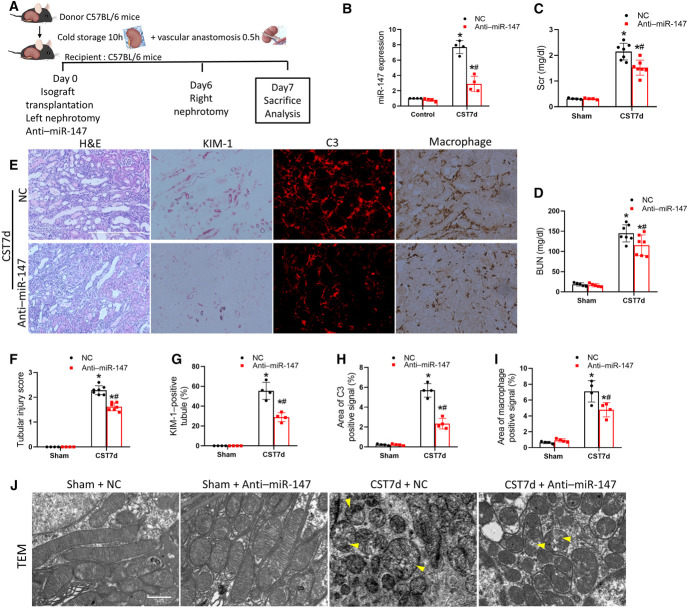
To determine the role of miR-147 in CST injury, we tested the effect of anti–miR-147, a specific antagomir of miR-147. Anti–miR-147 or negative control (NC) oligo was given to recipient mice immediately after cold storage-associated transplantation (CST) (Figure 4A). On day 7, we detected an approximately eight-fold increase of miR-147 in CST kidneys, which was reduced to approximately three-fold by anti–miR-147 (Figure 4B). When the native kidney was removed on day 6, the recipient mouse showed marked increases in serum creatinine and BUN on day 7. Notably, these increases were partially but significantly decreased by anti–miR-147 (Figure 4, C and D). Consistently, anti–miR-147 reduced renal tubular damages and KIM-1 staining in comparison with NC-treated group (Figure 4, E–G). Anti–miR-147 also suppressed C3 (Figure 4, E and H) and macrophage infiltration (Figure 4, E and I) in the graft. Furthermore, electron microscopy showed disrupted mitochondrial structure in CST grafts, which were ameliorated after anti–miR-147 treatment (Figure 4J). Together, these results indicate the protective effect of anti–miR-147, suggesting a pathogenic role of miR-147 in CST injury.
Figure 4.
Anti–miR-147 improves graft function and reduces graft damage and inflammation. (A) Diagram showing the experimental design. The left kidney was collected from C57BL/6 mice for 10 hours of cold storage, followed by transplantation into WT recipient mice. Recipient mice were injected with anti–miR-147 or NC oligo intravenously for three consecutive days after transplantation. On day 6 post-transplantation, the native kidney of the recipient mouse was removed so the transplanted kidney became the life-supporting kidney. On day 7, the transplanted kidney and blood samples were collected for analysis. The right kidney of donor mice without cold storage-associated transplantation was used as sham control. (B) miR-147 expression. (C) Serum creatinine. (D) BUN. (E) Representative images of H&E staining, C3 immunohistochemistry, KIM-1 immunohistochemistry, and macrophage staining. Scale bar=100 μm. (F) Quantitative analysis of tubular damage. (G) Quantitative analysis of C3 immunohistochemistry. (H) Quantitative analysis of KIM-1–positive tubules. (I) Quantitative analysis of macrophage staining. (J) Representative electronic microscopy images of proximal tubular cells. Yellow arrowheads indicate swollen or damaged mitochondria. Quantitative data are expressed as mean±SD (n=4–7). *P < 0.05 versus sham control; #P < 0.05 versus NC injected graft. miR-147, microRNA-147; NC, negative control; WT, wild-type. Figure 4 can be viewed in color online at www.jasn.org.
Anti–miR-147 Protects against Cell Death and Mitochondria Dysfunction after ATP Depletion in Renal Tubular Cells
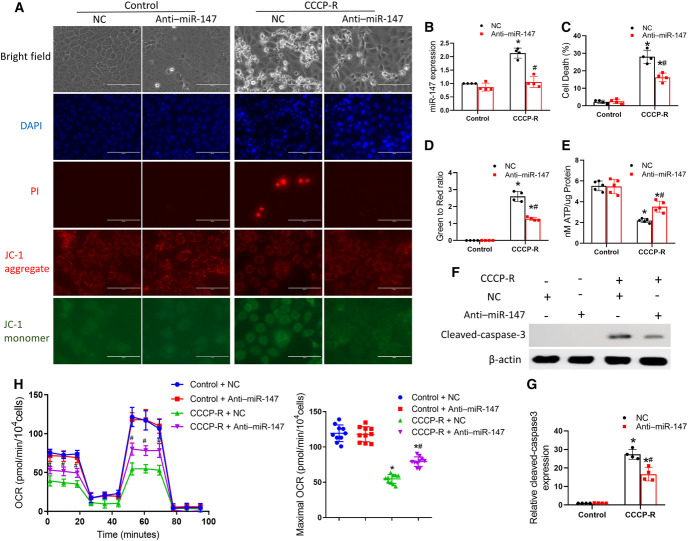
To understand how miR-147 mediates CST injury, we examined the effect of anti–miR-147 on ATP depletion–induced injury in cultured RPTCs. RPTCs were subjected to 3 hours of ATP depletion with CCCP and then returned to normal culture medium for 2 hours of recovery Carbonyl Cyanide Chlorophenylhydrazone-Recovery (CCCP-R) to mimic in vivo renal ischemia/reperfusion. CCCP-R treatment led to apoptosis (Figure 5A). It also induced an approximately two-fold increase of miR-147 expression, which was attenuated by anti–miR-147 (Figure 5B). Cell death was assessed by morphology along with 4′,6-diamidino-2-phenylindole and phosphatidylinositol staining as previously described.41 Compared with NC oligos, anti–miR-147 markedly reduced cell death and caspase 3 activation during CCCP-R treatment (Figure 5, A, C, F, and G). Recently, a clinical study indicated that mitochondrial membrane potential in renal grafts independently predicted DGF, suggesting a pathogenic role of mitochondrial dysfunction.42 CCCP-R treatment resulted in the loss of punctate red JC-1 fluorescence accompanied by the appearance of green JC-1 fluorescence, indicating the loss of mitochondrial membrane potential. Importantly, anti–miR-147 prevented the loss of mitochondrial membrane potential as shown by the preservation of punctate red JC-1 fluorescence without distinguishable green puncta (Figure 5, A and D). Furthermore, CCCP-R treatment reduced cellular ATP, which was preserved by anti–miR-147 (Figure 5E). To determine whether miR-147 regulates mitochondrial oxidative phosphorylation, we measured the oxygen consumption rate (OCR) using a Seahorse XF-24 Extracellular Flux Analyzer. CCCP-R significantly decreased the basal OCR and maximal respiratory capacity, which were preserved by anti–miR-147 treatment (Figure 5H). The protective effects of anti–miR-147 suggest that miR-147 may mediate kidney tubular cell injury and death by inducing mitochondrial damage.
Figure 5.
Anti–miR-147 protects against cell death and mitochondria dysfunction after ATP depletion in renal tubular cells. RPTCs were transfected with 100 nM anti–miR-147 or NC oligo and then subjected to 3 hours of CCCP treatment for ATP depletion with 2 hours of recovery (CCCP-R) or control incubation. (A) Representative images of cell morphology, 4′,6-diamidino-2-phenylindole staining of cell nuclei, PI staining, JC-1 aggregate, and JC-1 monomer staining. Scale bar=100 μm. (B) qPCR analysis of miR-147. (C) Percentage of cell death assessed morphologically. (D) Quantitative analysis of JC-1 green to red signal ratio. (E) Cellular ATP. (F) Immunoblots of cleaved caspase-3 with β-actin as the internal control. (G) Densitometry of cleaved caspase-3. (H) Measurement of OCR using an XF24 Extracellular Flux Analyzer. Quantitative data are expressed as mean±SD (n=4–10). *P < 0.05 versus control; #P < 0.05 versus CCCP-R with NC group. miR-147, microRNA-147; NC, negative control; OCR, oxygen consumption rate; PI, phosphatidylinositol; RPTCs, renal proximal tubular cells.
miR-147 Targets NDUFA4 during Kidney Transplantation and ATP Depletion Injury
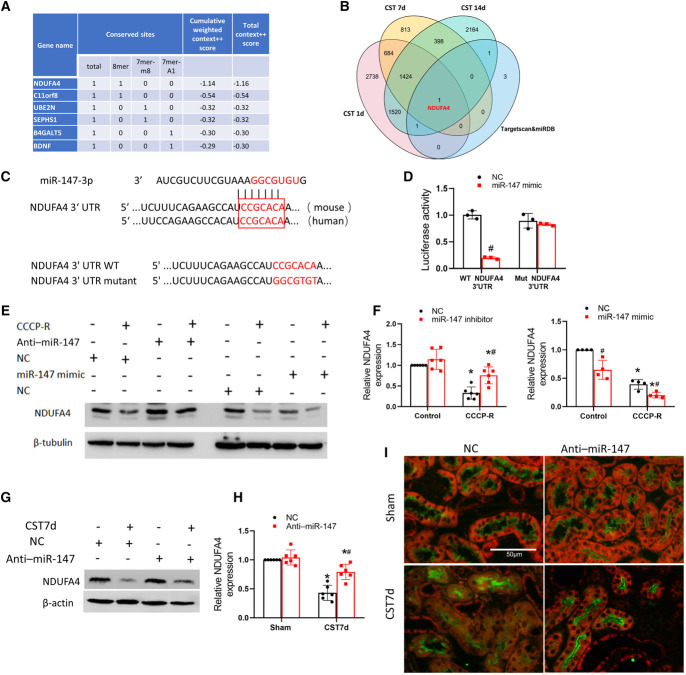
To examine how miR-147 mediates mitochondrial dysfunction and cell death, we sought to identify the target genes of miR-147. Bioinformatics analysis with the TargetScan database (http://www.targetscan.org/vert_72/) ranked NDUFA4, a component of mitochondrial respiratory chain,43,44 as a top target of miR-147 (Figure 6A). NDUFA4 was also one of the six target genes of miR-147 predicted by both TargetScan and miRDB, another microRNA database (http://www.mirbase.org/). Remarkably, NDUFA4 was the only predicted gene that was induced in CST injury (Figure 6B, Supplemental Datasets 4, 5, and 6). A conserved miR-147 targeting site was identified in the 3′UTR of NDUFA4 mRNA (Figure 6C). To confirm that NDUFA4 is a target of miR-147 experimentally, we performed the luciferase microRNA target reporter assay. To this end, NDUFA4-3′UTR or its miR-147 target sequence mutant (Figure 6C) was separately subcloned into the reporter construct for transfection into cells together with miR-147 mimic or NC oligos. As shown in Figure 6D, miR-147 mimic significantly reduced luciferase expression in the cells transfected with WT NDUFA4-3′UTR but not in those transfected with mutant NDUFA4-3′UTR, indicating that miR-147 directly targets NDUFA4-3′UTR. In RPTCs, CCCP-R treatment led to a decrease in NDUFA4 expression (Figure 6, E and F), which was prevented by anti–miR-147 (Figure 6F: lane 2 versus lane 4). In contrast, miR-147 mimic further repressed NDUFA4 during CCCP-R treatment (Figure 6E: lane 8 versus lane 6). Consistently, CST injury significantly decreased proximal tubular NDUFA4 expression in mouse kidneys, and this decrease was partially suppressed by anti–miR-147 (Figure 6, G–I), further proving NDUFA4 as a target of miR-147 in CST injury.
Figure 6.
miR-147 targets NDUFA4 in kidney cold storage-associated transplantation and ATP depletion injury. RPTCs were transfected with 100 nM anti–miR-147 or NC oligo and then subjected to CCCP-induced ATP depletion with recovery (CCCP-R) or control incubation. (A) miR-147 targets predicted by the online TargetScan database. (B) Venn map of the DEGs in kidney cold storage-associated transplantation. (C) The conserved, putative miR-147-3p binding sequence in 3′UTR of mouse and human NDUFA4 mRNAs. Listed at the bottom are the 3′UTR sequence of mouse NDUFA4 and its miR-147-3p binding sequence mutant. (D) Luciferase microRNA target reporter assay. The 3′UTR sequence of mouse NDUFA4 and its miR-147-3p binding sequence mutant were separately cloned into the pMIR-REPORT plasmid and then transfected into HEK cells. The cells were also transfected with miR-147 mimic or NC oligos, along with β-gal reporter plasmid for normalization. Luciferase activity was measured and normalized with β-gal activity. Data are expressed as mean±SD (n=3), #P < 0.05 versus the cells with NC oligo. (E) Effects of anti–miR-147 and miR-147 mimic on NDUFA4 expression during ATP depletion injury. RPTCs were subjected to CCCP-R treatment or control incubation in presence of anti–miR-147, its NC oligo, miR-147 mimic, or its NC oligo to collect lysate for immunoblot analysis of NDUFA4 and β-actin. (F) Densitometry analysis of NDUFA4. Data are expressed as mean±SD (n=4–6). *P < 0.05 versus control; #P < 0.05 versus NC groups. (G) Effect of anti–miR-147 on NDUFA4 expression in cold storage-associated transplantation kidneys. Anti–miR-147 or NC oligo was injected to mice during cold storage transplantation to collect the transplanted kidney at day 7 for immunoblot analysis of NDUFA4 and β−tubulin. (H) Densitometry analysis of NDUFA4 blots. Data are expressed as mean±SD (n=4–6). *P < 0.05 versus respective sham controls; #P < 0.05 versus the grafts with NC oligo. (I) Representative NDUFA4 immunofluorescence and LTL staining. Scale bar=50 μm. 3′UTR, 3′ untranslated region; DEG, differentially expressed gene; LTL, lotus tetragonolobus lectin; miR-147, microRNA-147; NC, negative control; RPTCs, renal proximal tubular cells.
NDUFA4 Preserves Mitochondrial Function and Cell Viability during ATP Depletion Injury in Renal Tubular Cells
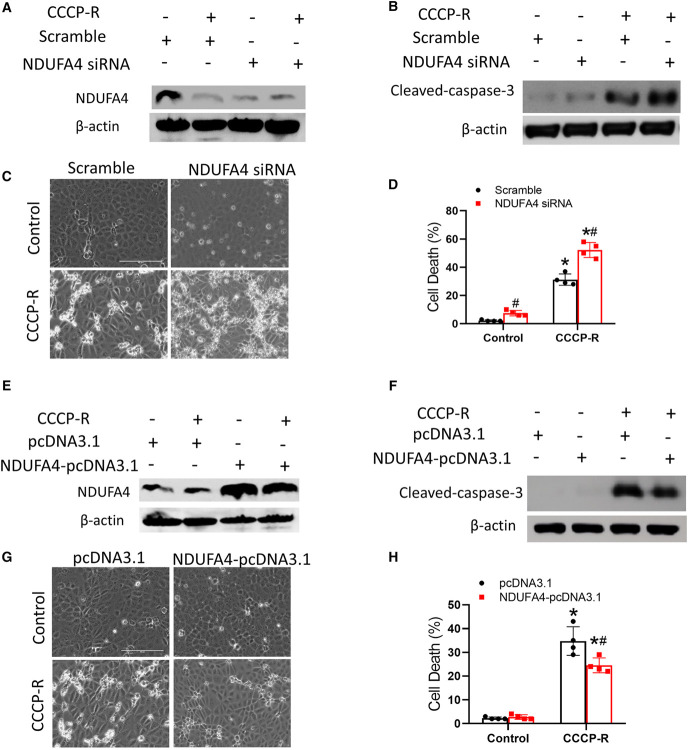
To determine the role of NDUFA4 in miR-147–mediated mitochondrial dysfunction, we transfected lentivirus containing small interfering RNAs (siRNAs) against NDUFA4 or scrambled sequence into RPTCs. Knockdown of NDUFA4 by its siRNA was confirmed by immunoblot analysis (Figure 7A). CCCP-R treatment led to approximately 28% apoptosis in scrambled sequence transfected cells, whereas approximately 40% in NDUFA4-siRNA cells (Figure 7, C and D). NDUFA4-siRNAs cells also had more cleaved/active caspase-3 after CCCP-R treatment (Figure 7B). On the contrary, transfecting NDUFA4-pcDNA3.1 enhanced NDUFA4 expression (Figure 7E) and prevented cell death and caspase activation during CCCP-R treatment, further supporting a protective role of NDUFA4 in renal tubular cells (Figure 7, F–H).
Figure 7.
NDUFA4 preserves mitochondrial function and cell viability during ATP depletion injury in renal tubular cells. RPTCs were stably transfected with lentivirus containing NDUFA4 siRNA or scrambled sequence (A–D), or NDUFA4-pcDNA3.1 or empty pcDNA3.1 (E–H). The cells were then subjected to CCCP-induced ATP depletion treatment or control incubation. (A) Immunoblots showing NDUFA4 downregulation by its siRNA. (B) Immunoblots of cleaved caspase-3 and β-actin. (C) Cell morphology. Bar=100 μm. (D) The percentage of apoptosis evaluated morphologically. (E) Immunoblots verifying NDUFA4 overexpression in the cells transfected with NDUFA4-pcDNA3.1. (F) Immunoblots of cleaved caspase-3 and β-actin. (G) Cell morphology. Scale bar=100 μm. (H) The percentage of apoptosis evaluated morphologically. Quantitative data are expressed as mean±SD (n=4), *P < 0.05 versus control, #P < 0.05 versus CCCP-R treated groups transfected with scramble siRNA or empty pcDNA3.1. RPTCs, renal proximal tubular cells; siRNA, small interfering RNA. Figure 7 can be viewed in color online at www.jasn.org.
Overexpression of NDUFA4 Suppresses miR-147–Induced Mitochondrial Dysfunction and Cell Death
We then determined whether miR-147 mediates mitochondrial dysfunction and cell injury and death by repressing NDUFA4. miR-147 mimic induced cell death in RPTCs, which was prevented by NDUFA4 overexpression (Supplemental Figure 3, A and B). The OCR results showed that the overexpression of miR-147 in RPTCs significantly decreased the respiratory capacity, which were preserved by adenoviral vectors harboring NDUFA4 (Ad-NDUFA4) transfection (Supplemental Figure 3C). miR-147 mimic also induced a decrease in mitochondrial membrane potential as shown by flow cytometry of JC-1 staining and a decrease in ATP production, both of which were partially yet significantly prevented by NDUFA4 overexpression (Supplemental Figure 3, D and E). These results suggest that miR-147 induces mitochondrial dysfunction and cell death by targeting NDUFA4.
NDUFA4 Overexpression in the Donor Kidney Ameliorates CST Injury
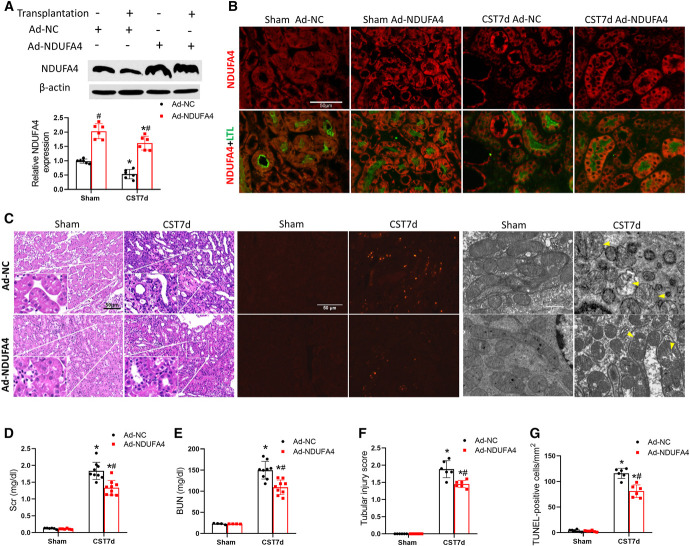
To examine the role of NDUFA4 in CST injury, we overexpressed NDUFA4 in the donor kidney by in situ transducing Ad-NDUFA4 or NC sequence (Ad-NC) as previously described.45 Immunoblot and immunofluorescence analysis confirmed NDUFA4 overexpression in transfected donor kidneys (Figure 8A) and proximal tubular cells (Figure 8B). Importantly, Ad-NDUFA4 transfected grafts had better renal function and histology than Ad-NC transfected grafts (Figure 8, C–E). Ad-NDUFA4 transfected grafts also had fewer TUNEL-positive cells (Figure 8 C and F) and better preservation of mitochondrial structure (Figure 8C).
Figure 8.
NDUFA4 overexpression in the donor kidney ameliorates cold storage-associated transplantation injury. Donor mice kidneys were infected with Ad-NDUFA4 or Ad-NC, stored in cold for 10 hours, and then transplanted into recipient mice. On day 6 post-transplantation, the native kidney of the recipient mice was removed. On day 7, the transplanted kidney and blood samples were collected for analysis. The right kidney of donor mice without cold storage-associated transplantation was used as sham control. (A) Immunoblots verifying NDUFA4 overexpression following Ad-NDUFA4 infection. (B) Representative NDUFA4 immunofluorescence and LTL staining. (C) H&E staining of histology, TUNEL staining of apoptosis, and electronic microscopy images. Yellow arrowheads label swollen or damaged mitochondria. Scale bar=50 or 1 μm. (D) Serum creatinine. (E) BUN. (F) Pathological score of tubular damage. (G) Quantitative analysis of TUNEL-positive cells. Quantitative data are expressed as mean±SD (n=6–9), *P < 0.05 versus respective sham controls; #P < 0.05 versus Ad-NC–infected transplantation groups. Ad-NC, normal control adenoviral vector; Ad-NDUFA4, adenovirus containing NDUFA4; LTL, lotus tetragonolobus lectin; NC, negative control; TUNEL, terminal deoxynucleotidyl transferase–mediated digoxigenin-deoxyuridine nick-end labeling.
Discussion
In human patients, each additional hour of cold storage results in a significant increase in graft failure and mortality after kidney transplantation,6 but the mechanism of CST injury is largely unclear. In this study, we have demonstrated that (1) Dicer deficiency in PT cells ameliorates CST injury and promotes better graft function; (2) CST injury is associated with significant changes in the expression profile of microRNAs, among which miR-147 is consistently induced in mice and in dysfunctional human renal grafts; (3) inhibition of miR-147 protects against CST injury in mice and protects renal tubular cells in vitro; (4) miR-147 mediates tubular cell injury by repressing NDUFA4 and inducing mitochondrial dysfunction. Together, these findings unveil a pathogenic role of the miR-147/NDUFA4 axis in CST injury, suggesting new therapeutic targets in kidney transplantation.
MicroRNAs are critical regulators of gene expression in health and diseases, including renal pathogenesis.10,12,13 Recent work has also implicated microRNAs in kidney transplantation.21,24–26,46–48 Although those studies revealed the changes in microRNA expression suggesting their diagnostic potential, the role of microRNAs as a whole class of molecules in kidney transplantation was not clear. Our current study has filled this knowledge gap by testing kidney PT-Dicer-KO mice, which as shown previously27 depleted more than 80% of microRNAs in kidney tissues. microRNA depletion in donor kidneys because of Dicer KO led to resistance to CST injury (Figure 1). In addition, these grafts had lower levels of C3 complement and macrophage infiltration, suggesting less inflammation. These results indicate that microRNAs as a whole class of molecules play an injurious role in kidney transplantation. This conclusion is consistent with the pathogenic role of microRNAs in renal ischemia/reperfusion injury (IRI).27 In fact, renal ischemia is inevitable during donor kidney procurement and cold storage, and reperfusion occurs after transplantation into the recipient. Renal IRI is also an important pathogenic factor in acute and chronic allograft nephropathy.5–7
Using microRNA sequencing or microRNA arrays, previous studies analyzed microRNA expression and the microRNA gene regulatory networks in renal IRI. In 2010, Godwin et al. profiled microRNA expression in unilateral renal IRI mice by microRNA array, showing that miR-21, miR-17, miR-20a, miR-214, and miR-132 were increased, whereas miR-874 and miR-187 decreased.49 In the same year, we profiled the changes in microRNA expression in bilateral renal IRI mice and demonstrated significant induction of 13 microRNAs and downregulation of 11 microRNAs.27 Interestingly, among the induced microRNAs, miR-68750 was later proved to be injurious, whereas miR-668,41 miR-489,51 and miR-1752 were shown to be protective in renal IRI. In comparison with those early studies of warm renal IRI, our present study has identified some similar changes in microRNA expression profile in the grafts with CST injury. For example, miR-17–92 cluster, miR-21, and miR-132 are upregulated in both warm renal IRI and cold storage transplant kidneys, indicating shared molecular regulations in pathogenesis. Nonetheless, our deep sequencing also unveiled unique microRNA signatures in CST injury. Especially, there was a rapid induction of miR-147 starting from day 1 after transplantation, which persisted to day 7 and day 14. Notably, miR-147 was upregulated in human renal grafts with DGF and in cultured kidney proximal tubular cells after ATP depletion injury. Upregulation of miR-147 was detected in X-linked hereditary nephropathy,53 3-chloro-1, 2-propanediol–induced AKI,54 and sepsis-induced renal injury,55 but the role and regulation of miR-147 in renal pathophysiology have not been investigated. In this study, anti–miR-147 protected against CST injury in mice and renal tubular cells in vitro. Interestingly, the protection was connected to the preservation of mitochondrial membrane potential, mitochondria ultrastructure, and cellular ATP production (Figures 4 and 5). Of note, transplantation was done between syngeneic mice with identical genetic background in this study, so renal inflammation was induced mainly by kidney injury during CST and not by alloimmune reactions. miR-147 likely contributed to inflammation by promoting PT damage, leading to the production of cytokines to attract inflammatory cells, such as macrophages (Figure 4).
To delineate the mechanism whereby miR-147 mediates mitochondrial dysfunction and tubular cell death, we have identified NDUFA4 as a critical target gene of miR-147. NDUFA4 appeared as a top candidate target gene of miR-147 in bioinformatics analysis. The 3′UTR of NDUFA4 contains a conserved miR-147 targeting seed sequence. Importantly, microRNA target luciferase reporter assay proved that mutation of this seed sequence could block the effect of miR-147 on NDUFA4. Consistently, the level of miR-147 expression negatively correlated with that of NDUFA4 (Figure 6). NDUFA4 is a subunit or assembly factor of cytochrome c oxidase (CcO) in the respiration complex IV in mitochondria.43,44 Downregulation of NDUFA4 led to a significant decrease in CcO activities.43 NDUFA4 mutations were reported to underlie CcO dysfunction in neurological disorders in human,56 whereas overexpression of NDUFA4 enhanced the activity and viability of cultured neurons.57,58 In our study, knockdown of NDUFA4 increased the basal level of apoptosis in renal tubular cells and aggravated ATP depletion–associated cell death. Conversely, NDUFA4 overexpression preserved cell viability after ATP depletion treatment in RPTCs and protected against CST injury in mouse kidneys (Figure 8). These results support a critical role of NDUFA4 in mitochondrial function and tubular cell viability. Notably, NDUFA4 overexpression could reduce miR-147 mimic-induced mitochondrial dysfunction and tubular cell death (Supplemental Figure 3), suggesting that miR-147 may mediate kidney tubular cell injury via NDUFA4.
This study has several limitations. First, transplantation was done between syngeneic mice. Although this syngeneic model has the advantage of isolating CST injury for study, it does not have the alloimmune reaction or rejection seen in human patients. In addition, a functioning kidney was kept for the first 6 days to avoid initial animal loss. In these aspects, the experimental model may not fully mimic the clinical situation of renal allograft transplantation. Second, the effect size of interventions shown in the in vivo experiments (Figure 1 and Figure 8, D-F is relatively modest. This is likely due to the complexity of the pathophysiological process of CST injury. Finally, this study only tested renal transplantation between male mice. It is interesting to examine the sex effect on transplantation outcomes and extend the findings of this study to female mice in follow-up studies.
In conclusion, this study demonstrates a pathogenic role of microRNAs in tubular damage and inflammation in CST injury. Specifically, miR-147 is consistently upregulated in CST kidneys and represses NDUFA4, resulting in mitochondrial dysfunction and renal tubular damage. Inhibition of miR-147 and overexpression of NDUFA4 alleviate CST injury, unveiling new therapeutic targets in kidney transplantation.
Methods
Human Samples
Human samples were collected through a protocol (WDRY2021-KS059) approved by the Renmin Hospital of Wuhan University Review Board with written consent from all individual participants. DGF was defined as the need for dialysis in the first week after kidney transplantation. Graft biopsies were performed within 1 month after transplantation. The control group included kidney samples from patients who had undergone nephrectomy for kidney cancer and did not have diabetes or kidney disease. The general patient information is shown in Supplemental Table 1.
Animals
Animals used in this study were housed in the animal facility of Charlie Norwood VA Medical Center (Augusta, GA) or the Experimental Animal Center of Three Gorges University (Yichang, China). Some animal experiments were conducted according to a protocol approved by the Institutional Animal Care and Use Committee of Charlie Norwood VA Medical Center (Augusta) while others were done according to a protocol approved by the Three Gorges University Ethics Committee. Male C57BL/6 mice aged 10–14 weeks were used unless otherwise indicated. Kidney PT-Dicer-KO mice were generated by Cre-Loxp technology, as described previously.27
Cold Storage–Associated Kidney Transplantation
To specifically study kidney injury induced by cold storage-associated transplantation, we conducted transplantation between syngeneic mice that had identical genetic background to avoid the immune reaction/rejection between the donor and recipient. The procedures were previously described with slight modifications.36 In brief, the left kidney of the donor mouse was excised and flushed with cold heparinized University of Wisconsin solution and stored in cold University of Wisconsin solution in an ice water bath for 10 hours. Then, the left kidney of the recipient was removed, and the donor kidney was transplanted. Graft anastomosis time was standardized at 30 minutes. The ureteral implantation was accomplished by fixing to the exterior wall of the bladder. Blood samples and the transplanted kidney were collected at indicated time points after transplantation for analysis. To monitor the life-supporting function of the transplanted kidney, the native kidney was removed on day 6 post-transplantation to let the mouse survive to examine the function and histology of the transplanted kidney on day 7 and day 14. Serum creatinine and BUN levels were measured using commercial kits to indicate the decline of renal function, as previously described.41
Cell Culture and ATP Depletion Injury Model
The RPTC cell line was established by immortalizing rat kidney PT cells with SV40 large T-antigen and originally obtained from Dr. Ulrich Hopfer (Case Western Reserve University).59 This cell line has a kidney proximal tubular phenotype, including a cuboidal morphology, tight junction formation, and transporter function. The cell line was cultured in DMEM/F-12 medium supplemented with 10% fetal bovine serum and growth factors. To induce injury, RPTCs were treated with 20 μM CCCP in glucose-free Krebs-Ringer buffer (in mM: 115 NaCl, 1 KH2PO4, 4 KCl, 1 MgSO4, 1.25 CaCl2, 25 NaHCO3, pH 7.4) for 3 hours, followed by 2 hours of recovery in full culture medium. Cell death was assessed by cell morphology and immunoblot analysis of caspase-3 activation, as previously described.41
Immunoblot Analysis
Immunoblotting was performed using a standard method, as previously described.36,41 The primary antibodies used for immunoblotting included anti-NDUFA4 (catalog number ab182126, Abcam), β-actin (66009-1-Ig, Proteintech), β-tubulin (2146, Cell Signaling Technology), anti–cyclophilin B (43603S, Cell Signaling Technology), and caspase-3 (9664, Cell Signaling Technology).
Immunostaining
Paraffin-embedded kidney sections were rehydrated and steamed in sodium citrate buffer for antigen retrieval. Primary antibodies for immunofluorescence included anti-C3 (ab11862, Abcam) and anti-NDUAF4 (ab129752, Abcam). Cy3-labeled goat anti-rabbit IgG was used as secondary antibodies. Fluorescein-labeled lotus tetragonolobus lectin (FL-1321, Vector Laboratories) was used to mark PTs.60 Primary antibodies for immunohistochemical staining included KIM-1 (AF1817, R&D System, Minneapolis), anti-SMA (ab5694, Abcam), and antimacrophage (ab56297, Abcam). The slides were developed with VECTASTAIN Elite ABC kit and ImmPACT DAB peroxidase substrate (Vector Laboratories).
Renal Histology and TUNEL Assay
For histology, kidney tissues were fixed with 4% paraformaldehyde for paraffin embedding and hematoxylin and eosin staining. Tubular damage was scored by the percentage of renal tubules with cell lysis, loss of brush border, and cast formation (0, no damage; 1, 1%–25%; 2, 25%–50%; 3, 50%–75%; 4, >75%). Ten randomly selected fields per mouse kidney were scored for quantification. For TUNEL staining, paraffin-embedded kidney tissue sections were stained with In Situ Cell Death Detection Kit (Roche Applied Science). The slides were examined with fluorescence microscopy, and TUNEL-positive cells were counted from ten randomly picked images for each specimen in kidney outer medulla and cortex region. All histological examinations were conducted in a blinded manner.
RNA Sequencing
Total RNA was isolated from kidney cortex or cells using mirVana microRNA Isolation Kit (Life Technologies) following the manufacturer's instructions. Isolated RNA sample quality was assessed by BioAnalyzer Nano RNA Assay (Agilent Technologies Inc., CA) and quantified by Qubit 2.0 RNA HS assay (ThermoFisher, MA). Library construction was performed using NEBNext Small RNA Library Prep Set for Illumina according to the manufacturer's recommendation (New England BioLabs Inc., MA). The quantity of final libraries was assessed by Qubit 2.0 (ThermoFisher, MA), and quality was assessed by TapeStation HSD1000 ScreenTape (Agilent Technologies Inc., CA). The average final library size was about 150 bp with an insert size of about 20–30 bp. Illumina 8-nt single indices were used. Equimolar pooling of libraries was performed on the basis of quality control values and sequenced on an Illumina HiSeq (Illumina, CA), with a read length configuration of 150 PE for 20 M total reads per sample (10 M in each direction).
Assembled count values were used as the input of the DESeq2 Bioconductor package. The significantly differentially expressed genes between groups were defined at a cutoff criteria of |log2 fold-change| ≥1 and P value <0.05. Significantly enriched pathways were examined for the identified differentially expressed gene sets.
In Situ Hybridization of miR-147
Tissue sections were incubated with prehybridization solution at 78°C for 3 hours. The hybridization mixture was then prepared by mixing of digoxigenin/Cy3-labeled locked nucleic acid probe of miR-147 (Servicebio, Wuhan, China) and hybridization solution. The hybridization mixture was heated at 65°C for 5 minutes and cooled on ice. After overnight hybridization at 73°C, the specimens were washed with serial dilutions of buffer and incubated with anti-digoxigenin alkaline phosphatase–conjugated antibody. The final signal was revealed by bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium solution and analyzed by microscopy.
Quantitative Real-Time PCR
Total RNAs were extracted from kidney tissues and cells with mirVana microRNA Isolation Kit (Life Technologies) following the manufacturer's instructions. microRNA was quantified by TaqMan microRNA assay (Life Technologies) with small nucleolar RNAs as the internal control for normalization.
Transfection of Cells
Lipofectamine 2000 (Life Technologies) was used for transfection following the manufacturer's instructions. To study miR-147, miR-147 mimic or anti–miR-147 (Ribobio, Guangzhou, China) was transfected into cells. To overexpress NDUFA4, pcDNA3.1-NDUFA4 (Sangon Biotech, Shanghai) was transfected.
In vivo Transfection in Mice
To inhibiting miR-147, anti–miR-147 or NC oligo (Ribobio, Guangzhou, China) was suspended in 200 µl of saline for tail vein injection at a dose of 10 nmol per mouse for three consecutive days after transplantation. To overexpress NDUFA4 in donor kidneys, 100 μl (1×109 PFU/ml) of NDUFA4 adenovirus or NC adenovirus (GeneChem, Shanghai, China) was injected into the temporarily obstructed kidney by using a 30 G needle syringe in donor mice as previously described.45 Five days later, the transfected kidney was harvested for cold storage-associated transplantation.
Luciferase microRNA Target Reporter Assay
DNA sequences containing the predicted miR-147 binding sites in the 3′-UTR region of NDUFA4 or null control insert without the predicted miR-147 binding sites were synthesized (Sangon Biotech, Shanghai, China). The sequences were subcloned downstream of the luciferase gene in the pMIR-REPORT luciferase plasmid (Life Technologies). The luciferase plasmids were cotransfected with pMIR-REPORT β-gal control plasmid and 200 nM miR-147 mimics or NC into HEK293 cells. The luciferase activity and β-galactosidase were examined with the Luciferase Assay System and β-galactosidase Enzyme Assay System, respectively (Beyotime Biotechnology, Shanghai, China). The luciferase activity was normalized with β-galactosidase activity. The ratio of the normalized value between miR-147 mimic and the NC group was used for comparison.
Electron Microscopy
Fresh kidney tissues were fixed in 100 mM sodium cacodylate, 2 mM calcium chloride, 4 mM magnesium sulfate, 4% paraformaldehyde, and 2.5% glutaraldehyde, as previously described.36 The tissue block was examined initially at low magnification (×3000) to identify representative PTs. Cells in these tubules were then examined at high magnification (×15,000) to collect electron micrographs.
Analyses of Cellular Respiration Using a Seahorse XF24 Analyzer
Mitochondrial respiration was measured using a Seahorse XF24 extracellular flux analyzer (Seahorse Bioscience, Copenhagen, Denmark). Cells were plated in Seahorse XF24 cell culture microplates for treatment 24 hours before analysis. Analysis was performed with the appropriate growth media containing 1 mM sodium pyruvate, 1 mM glutamine, and 10 mM glucose. Cells were allowed to acclimate for 1 hour before analysis. Basal oxygen consumption was recorded for 20 minutes, and OCR measurements were performed over time after the successive addition of mitochondrial inhibitors: (1) oligomycin (1.5 μM) to measure the ATP synthesis coupling efficiency; (2) the uncoupler carbonyl cyanide 4-trifluoromethoxy-phenylhydrazone (0.5 μM) to evaluate the spare respiratory capacity; and (3) a mixture of rotenone (0.5 μM) and antimycin A (0.5 μM) to assess the nonmitochondrial consumption of O2.
Mitochondrial Membrane Potential Assay
Mitochondrial membrane potential was detected with JC-1 as previously.61 In brief, RPTCs were incubated with JC-1 (BioVision Inc.) for 35 minutes in dark for examination by fluorescence microscopy. For flow cytometry, cell pellets were suspended in JC-1 working solution and then incubated in the dark at 37°C for 30 minutes. After washing with buffer, the stained cells were analyzed by flow cytometry (BD FACSJazz).
Statistical Analysis
The t test was used to show the difference between two groups, and ANOVA was used for multigroup comparison. The Dunn multiple comparisons and the Fisher least significant difference test were used for one-way ANOVA and two-way ANOVA, respectively. GraphPad Prism 8 and SPSS 25 were used for all calculations. Data were expressed as mean±SD. P values of <0.05 were considered significant. The statistical power of most quantitative data in this study is >80%.
Supplementary Material
Disclosures
Z. Dong reports Consultancy: DILIsym/RenaSym and Mitobridge; Honoraria: DILIsym/RenaSym and Mitobridge; and Advisory or Leadership Role: Associate Editor for American Journal of Physiology-Renal Physiology, Associate Editor for Kidney Diseases, Board of Directors: Adjunct Professorship at The Second Xiangya Hospital of Central South University in China and Chinese American Society of Nephrology, and Editorial board member for American Journal of Physiology-Cell Physiology, JASN, and Kidney International. All remaining authors have nothing to disclose.
Funding
This work was supported by the National Institutes of Health (DK058831, DK087843) of USA, the Department of Veterans Administration of United States (BX000319), the National Science Foundation of China (grant number 82100803), and Natural Science Foundation of Hubei Province (grant number 2021CFB379/2021CFB101). Z. Dong is a recipient of The Senior Research Career Scientist award from the Department of Veterans Affairs of United States.
Author Contributions
Conceptualization: Zheng Dong, Jiefu Zhu.
Data curation: Xiaoru Hu, Chenglong Li, Zhixia Song, Xiaohong Xiang, Jiefu Zhu.
Formal analysis: Zheng Dong, Xiaoru Hu, Chenglong Li, Jiefu Zhu.
Funding acquisition: Zheng Dong.
Investigation: Zheng Dong, Zhixia Song, Xiaohong Xiang.
Methodology: Chenglong Li, Jiefu Zhu.
Project administration: Zheng Dong.
Resources: Zhixia Song.
Software: Jiefu Zhu.
Supervision: Zheng Dong.
Validation: Zheng Dong, Xiaohong Xiang.
Visualization: Jiefu Zhu.
Writing – original draft: Zhixia Song, Jiefu Zhu.
Writing – review & editing: Zheng Dong, Zhixia Song.
Data Sharing Statement
The complete RNA-Seq dataset is available from the Gene Expression Omnibus repository (Supplemental Datasets 1, 2, 3, 4, 5, and 6).
Supplemental Material
This article contains the following supplemental material online at http://links.lww.com/JSN/E440, http://links.lww.com/JSN/E434, http://links.lww.com/JSN/E435, http://links.lww.com/JSN/E436, http://links.lww.com/JSN/E437, http://links.lww.com/JSN/E438, and http://links.lww.com/JSN/E439.
Supplemental Figure 1. Dicer deficiency reduces inflammation and fibrosis in cold storage-associated transplantation kidneys.
Supplemental Figure 2. Taqman real-time PCR analysis of microRNA expression in cold storage-associated transplantation kidneys.
Supplemental Figure 3. Overexpression of NDUFA4 suppresses miR-147–induced mitochondrial dysfunction and cell death.
Supplemental Table 1. Clinical data of DGF and non-DGF patients examined.
Supplemental Datasets 1. The microRNA sequencing data for the comparison between sham and CST at 1 day (.rar file).
Supplemental Datasets 2. The microRNA sequencing data for the comparison between sham and CST at 7 days.
Supplemental Datasets 3. The microRNA sequencing data for the comparison between sham and CST at 14 days.
Supplemental Datasets 4. The mRNA sequencing data for the comparison between sham and CST at 1 day.
Supplemental Datasets 5. The mRNA sequencing data for the comparison between sham and CST at 7 days.
Supplemental Datasets 6. The mRNA sequencing data for the comparison between sham and CST at 14 days.
References
- 1.Tonelli M, Wiebe N, Knoll G, et al. Systematic review: kidney transplantation compared with dialysis in clinically relevant outcomes. Am J Transplant. 2011;11(10):2093–2109. doi: 10.1111/j.1600-6143.2011.03686.x [DOI] [PubMed] [Google Scholar]
- 2.Tullius SG, Rabb H. Improving the supply and quality of deceased-donor organs for transplantation. N Engl J Med. 2018;378(20):1920–1929. doi: 10.1056/NEJMra1507080 [DOI] [PubMed] [Google Scholar]
- 3.Lentine KL, Smith JM, Hart A, et al. OPTN/SRTR 2020 annual data report: kidney. Am J Transplant. 2022;22(suppl 2):21–136. doi: 10.1111/ajt.16982 [DOI] [PubMed] [Google Scholar]
- 4.Mudiayi D, Shojai S, Okpechi I, et al. Global estimates of capacity for kidney transplantation in world countries and regions. Transplantation 2022;106(6):1113–1122. doi: 10.1097/tp.0000000000003943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith SF, Hosgood SA, Nicholson ML. Ischemia-reperfusion injury in renal transplantation: 3 key signaling pathways in tubular epithelial cells. Kidney Int. 2019;95(1):50–56. doi: 10.1016/j.kint.2018.10.009 [DOI] [PubMed] [Google Scholar]
- 6.Debout A, Foucher Y, Trebern-Launay K, et al. Each additional hour of cold ischemia time significantly increases the risk of graft failure and mortality following renal transplantation. Kidney Int. 2015;87(2):343–349. doi: 10.1038/ki.2014.304 [DOI] [PubMed] [Google Scholar]
- 7.Langewisch E, Mannon RB. Chronic allograft injury. Clin J Am Soc Nephrol. 2021;16(11):1723–1729. doi: 10.2215/CJN.15590920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148(6):1172–1187. doi: 10.1016/j.cell.2012.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo C, Dong G, Liang X, Dong Z. Epigenetic regulation in AKI and kidney repair: mechanisms and therapeutic implications. Nat Rev Nephrol. 2019;15(4):220–239. doi: 10.1038/s41581-018-0103-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cerqueira TCF, de Cerqueira Neto ML, Cacau LdAP, et al. Effect of neuromuscular electrical stimulation on functional exercise capacity in patients undergoing cardiac surgery: a randomized clinical trial. Clin Rehabil. 2022;36(6):789–800. doi: 10.1177/02692155211070945 [DOI] [PubMed] [Google Scholar]
- 12.Bhatt K, Kato M, Natarajan R. Mini-review: emerging roles of microRNAs in the pathophysiology of renal diseases. Am J Physiol Renal Physiol. 2016;310(2):F109–F118. doi: 10.1152/ajprenal.00387.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cerqueira DM, Tayeb M, Ho J. MicroRNAs in kidney development and disease. JCI Insight. 2022;7(9):e158277. doi: 10.1172/jci.insight.158277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–222. doi: 10.1038/nrd.2016.246 [DOI] [PubMed] [Google Scholar]
- 15.Mahtal N, Lenoir O, Tinel C, Anglicheau D, Tharaux PL. MicroRNAs in kidney injury and disease. Nat Rev Nephrol. 2022;18(10):643–662. doi: 10.1038/s41581-022-00608-6 [DOI] [PubMed] [Google Scholar]
- 16.Thompson ER, Sewpaul A, Figuereido R, et al. MicroRNA antagonist therapy during normothermic machine perfusion of donor kidneys. Am J Transplant. 2022;22(4):1088–1100. doi: 10.1111/ajt.16929 [DOI] [PubMed] [Google Scholar]
- 17.Patel V, Williams D, Hajarnis S, et al. miR-17∼92 miRNA cluster promotes kidney cyst growth in polycystic kidney disease. Proc Natl Acad Sci U S A. 2013;110(26):10765–10770. doi: 10.1073/pnas.1301693110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hajarnis S, Lakhia R, Yheskel M, et al. microRNA-17 family promotes polycystic kidney disease progression through modulation of mitochondrial metabolism. Nat Commun. 2017;8(1):14395. doi: 10.1038/ncomms14395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomez IG, MacKenna DA, Johnson BG, et al. Anti-microRNA-21 oligonucleotides prevent Alport nephropathy progression by stimulating metabolic pathways. J Clin Invest. 2015;125(1):141–156. doi: 10.1172/JCI75852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lakhia R, Ramalingam H, Chang CM, et al. PKD1 and PKD2 mRNA cis-inhibition drives polycystic kidney disease progression. Nat Commun. 2022;13(1):4765. doi: 10.1038/s41467-022-32543-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ledeganck KJ, Gielis EM, Abramowicz D, Stenvinkel P, Shiels PG, Van Craenenbroeck AH. MicroRNAs in AKI and kidney transplantation. Clin J Am Soc Nephrol. 2019;14(3):454–468. doi: 10.2215/CJN.08020718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Z, Wang Y, Shu S, Cai J, Tang C, Dong Z. Non-coding RNAs in kidney injury and repair. Am J Physiol Cell Physiol. 2019;317(2):C177–C188. doi: 10.1152/ajpcell.00048.2019 [DOI] [PubMed] [Google Scholar]
- 23.Scian MJ, Maluf DG, Mas VR. MiRNAs in kidney transplantation: potential role as new biomarkers. Expert Rev Mol Diagn. 2013;13(1):93–104. doi: 10.1586/Erm.12.131 [DOI] [PubMed] [Google Scholar]
- 24.Tinel C, Lamarthee B, Anglicheau D. MicroRNAs: small molecules, big effects. Curr Opin Organ Transplant. 2021;26(1):10–16. doi: 10.1097/MOT.0000000000000835 [DOI] [PubMed] [Google Scholar]
- 25.Bostjancic E, Veceric-Haler Z, Kojc N. The role of immune-related miRNAs in the pathology of kidney transplantation. Biomolecules. 2021;11(8):1198. doi: 10.3390/biom11081198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mas VR, Le TH, Maluf DG. Epigenetics in kidney transplantation: current evidence, predictions, and future Research directions. Transplantation. 2016;100(1):23–38. doi: 10.1097/TP.0000000000000878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei Q, Bhatt K, He HZ, Mi QS, Haase VH, Dong Z. Targeted deletion of Dicer from proximal tubules protects against renal ischemia-reperfusion injury. J Am Soc Nephrol. 2010;21(5):756–761. doi: 10.1681/ASN.2009070718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hao J, Lou Q, Wei Q, et al. MicroRNA-375 is induced in cisplatin nephrotoxicity to repress hepatocyte nuclear factor 1-β. J Biol Chem. 2017;292(11):4571–4582. doi: 10.1074/jbc.M116.754929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Z, Tang C, He L, et al. The negative feedback loop of NF-κB/miR-376b/NFKBIZ in septic acute kidney injury. JCI Insight. 2020;5(24):e142272. doi: 10.1172/jci.insight.142272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Z, Wang S, Mi QS, Dong Z. MicroRNAs in pathogenesis of acute kidney injury. Nephron. 2016;134(3):149–153. doi: 10.1159/000446551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scholz H, Boivin FJ, Schmidt-Ott KM, et al. Kidney physiology and susceptibility to acute kidney injury: implications for renoprotection. Nat Rev Nephrol. 2021;17(5):335–349. doi: 10.1038/s41581-021-00394-7 [DOI] [PubMed] [Google Scholar]
- 32.Chevalier RL. The proximal tubule is the primary target of injury and progression of kidney disease: role of the glomerulotubular junction. Am J Physiol Renal Physiol. 2016;311(1):F145–F161. doi: 10.1152/ajprenal.00164.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lo S, Jiang L, Stacks S, Lin H, Parajuli N. Aberrant activation of the complement system in renal grafts is mediated by cold storage. Am J Physiol Renal Physiol. 2021;320(6):F1174–F1190. doi: 10.1152/ajprenal.00670.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parajuli N, Shrum S, Tobacyk J, Harb A, Arthur JM, MacMillan-Crow LA. Renal cold storage followed by transplantation impairs expression of key mitochondrial fission and fusion proteins. PLoS One. 2017;12(10):e0185542. doi: 10.1371/journal.pone.0185542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L, Wei J, Jiang S, et al. Effects of different storage solutions on renal ischemia tolerance after kidney transplantation in mice. Am J Physiol Renal Physiol. 2018;314(3):F381–F387. doi: 10.1152/ajprenal.00475.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu J, Zhang G, Song Z, et al. Protein kinase C-delta mediates kidney tubular injury in cold storage-associated kidney transplantation. J Am Soc Nephrol. 2020;31(5):1050–1065. doi: 10.1681/ASN.2019101060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Timmeren MM, van den Heuvel MC, Bailly V, Bakker SJ, van Goor H, Stegeman CA. Tubular kidney injury molecule-1 (KIM-1) in human renal disease. J Pathol. 2007;212(2):209–217. doi: 10.1002/path.2175 [DOI] [PubMed] [Google Scholar]
- 38.Varagunam M, Yaqoob MM, Dohler B, Opelz G. C3 polymorphisms and allograft outcome in renal transplantation. N Engl J Med. 2009;360(9):874–880. doi: 10.1056/NEJMoa0801861 [DOI] [PubMed] [Google Scholar]
- 39.Ferenbach DA, Bonventre JV. Mechanisms of maladaptive repair after AKI leading to accelerated kidney ageing and CKD. Nat Rev Nephrol. 2015;11(5):264–276. doi: 10.1038/nrneph.2015.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Fijter JW. Rejection and function and chronic allograft dysfunction. Kidney Int. 2010;(119):S38–S41. doi: 10.1038/ki.2010.421 [DOI] [PubMed] [Google Scholar]
- 41.Wei Q, Sun H, Song S, et al. MicroRNA-668 represses MTP18 to preserve mitochondrial dynamics in ischemic acute kidney injury. J Clin Invest. 2018;128(12):5448–5464. doi: 10.1172/JCI121859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garonzik-Wang JM, Lonze BE, Ruck JM, et al. Mitochondrial membrane potential and delayed graft function following kidney transplantation. Am J Transplant. 2019;19(2):585–590. doi: 10.1111/ajt.15174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Balsa E, Marco R, Perales-Clemente E, et al. NDUFA4 is a subunit of complex IV of the mammalian electron transport chain. Cell Metab. 2012;16(3):378–386. doi: 10.1016/j.cmet.2012.07.015 [DOI] [PubMed] [Google Scholar]
- 44.Kadenbach B. Regulation of mammalian 13-subunit cytochrome c oxidase and binding of other proteins: role of NDUFA4. Trends Endocrinol Metab. 2017;28(11):761–770. doi: 10.1016/j.tem.2017.09.003 [DOI] [PubMed] [Google Scholar]
- 45.Ding H, Li J, Li Y, et al. MicroRNA-10 negatively regulates inflammation in diabetic kidney via targeting activation of the NLRP3 inflammasome. Mol Ther. 2021;29(7):2308–2320. doi: 10.1016/j.ymthe.2021.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schauerte C, Hubner A, Rong S, et al. Antagonism of profibrotic microRNA-21 improves outcome of murine chronic renal allograft dysfunction. Kidney Int. 2017;92(3):646–656. doi: 10.1016/j.kint.2017.02.012 [DOI] [PubMed] [Google Scholar]
- 47.Hartono C, Muthukumar T, Suthanthiran M. Noninvasive diagnosis of acute rejection of renal allografts. Curr Opin Organ Transplant. 2010;15(1):35–41. doi: 10.1097/mot.0b013e3283342728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van de Vrie M, Deegens JK, Eikmans M, van der Vlag J, Hilbrands LB. Urinary MicroRNA as biomarker in renal transplantation. Am J Transplant. 2017;17(5):1160–1166. doi: 10.1111/ajt.14082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Godwin JG, Ge X, Stephan K, Jurisch A, Tullius SG, Iacomini J. Identification of a microRNA signature of renal ischemia reperfusion injury. Proc Natl Acad Sci U S A. 2010;107(32):14339–14344. doi: 10.1073/pnas.0912701107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bhatt K, Wei Q, Pabla N, et al. MicroRNA-687 induced by hypoxia-inducible factor-1 targets phosphatase and tensin homolog in renal ischemia-reperfusion injury. J Am Soc Nephrol. 2015;26(7):1588–1596. doi: 10.1681/ASN.2014050463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wei Q, Liu Y, Liu P, et al. MicroRNA-489 induction by hypoxia-inducible factor-1 protects against ischemic kidney injury. J Am Soc Nephrol. 2016;27(9):2784–2796. doi: 10.1681/ASN.2015080870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hao J, Wei Q, Mei S, et al. Induction of microRNA-17-5p by p53 protects against renal ischemia-reperfusion injury by targeting death receptor 6. Kidney Int. 2017;91(1):106–118. doi: 10.1016/j.kint.2016.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chu CP, Liu S, Song W, Xu EY, Nabity MB. Small RNA sequencing evaluation of renal microRNA biomarkers in dogs with X-linked hereditary nephropathy. Sci Rep. 2021;11(1):17437. doi: 10.1038/s41598-021-96870-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang G, Xue J, Sun X, Wang J, Yu LL: Necroptosis in 3-chloro-1, 2-propanediol (3-MCPD)-dipalmitate-induced acute kidney injury in vivo and its repression by miR-223-3p. Toxicology. 2018;406-407:33–43. doi: 10.1016/j.tox.2018.05.015 [DOI] [PubMed] [Google Scholar]
- 55.Kim B, Guaregua V, Chen X, et al. Characterization of a murine model system to study MicroRNA-147 during inflammatory organ injury. Inflammation. 2021;44(4):1426–1440. doi: 10.1007/s10753-021-01427-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pitceathly RD, Rahman S, Wedatilake Y, et al. NDUFA4 mutations underlie dysfunction of a cytochrome c oxidase subunit linked to human neurological disease. Cell Rep. 2013;3(6):1795–1805. doi: 10.1016/j.celrep.2013.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fu F, Li Y, Li R, et al. NDUFA4 enhances neuron growth by triggering growth factors and inhibiting neuron apoptosis through Bcl-2 and cytochrome C mediated signaling pathway. Am J Transl Res. 2018;10(1):164–174. https://pubmed.ncbi.nlm.nih.gov/29423002/ [PMC free article] [PubMed] [Google Scholar]
- 58.Fu F, Chen C, Du K, et al. Ndufa4 regulates the proliferation and apoptosis of neurons via miR-145a-5p/homer1/ccnd2. Mol Neurobiol. 2023;60(6):2986–3003. doi: 10.1007/s12035-023-03239-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Woost PG, Kolb RJ, Finesilver M, et al. Strategy for the development of a matched set of transport-competent, angiotensin receptor-deficient proximal tubule cell lines. In Vitro Cell Dev Biol Anim. 2006;42(7):189–200. doi: 10.1290/0511076.1 [DOI] [PubMed] [Google Scholar]
- 60.Baisantry A, Bhayana S, Rong S, et al. Autophagy induces prosenescent changes in proximal tubular S3 segments. J Am Soc Nephrol. 2016;27(6):1609–1616. doi: 10.1681/ASN.2014111059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Livingston MJ, Wang J, Zhou J, et al. Clearance of damaged mitochondria via mitophagy is important to the protective effect of ischemic preconditioning in kidneys. Autophagy. 2019;15(12):2142–2162. doi: 10.1080/15548627.2019.1615822 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The complete RNA-Seq dataset is available from the Gene Expression Omnibus repository (Supplemental Datasets 1, 2, 3, 4, 5, and 6).