Abstract
Data on the performance of blood-based nucleocapsid antigen tests for diagnosing severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection and infectious viral shedding are limited. To address this knowledge gap, we conducted a systematic review to assess the performance of blood-based nucleocapsid (N) antigen tests in diagnosing SARS-CoV-2 infection and identifying infectiousness. This review was registered on PROSPERO (registration no. CRD42022339635). We comprehensively searched PubMed, Embase, Web of Science, and the Coronavirus Research Database for relevant studies published through 27 February 2023. Each study's risk of bias was evaluated using the Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2) tool. Our findings indicate that the performance of the N-antigen test is influenced by factors such as assay type, sampling timing, and illness severity. Sensitive assays provide suitable methods for viable screening and laboratory diagnostic tests in different clinical and research settings during the early phase of illness.
Keywords: SARS-CoV-2, blood, nucleocapsid antigen, sensitivity and specificity, systematic review
The presence of severe acute respiratory syndrome coronavirus 2 nucleocapsid antigen in blood is an early indicator of coronavirus disease 2019 infection and infectious viral shedding. Sensitive assays provide suitable methods for viable screening and laboratory diagnostic tests in different clinical and research settings.
The early detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection has been an important yet challenging issue. Reverse-transcriptase polymerase chain reaction (RT-PCR) testing of nasopharyngeal samples is considered the reference standard for the diagnosis of SARS-CoV-2 infection [1, 2], while virus culture is used as the reference standard for detection of active replicative virus and hence assessment of infectiousness [3, 4]. The nucleocapsid protein (N-antigen) in the nasopharyngeal samples has been identified as one of the predominantly expressed proteins that could have comparable performance with RT-PCR, particularly early in the course of disease [5]. Antigen testing using nasal and saliva specimens has become part of routine clinical practice.
Recent studies suggest that SARS-CoV-2 viremia is a strong predictor of coronavirus disease 2019 (COVID-19) severity and outcome [6–8]. Therefore, detecting viral N-antigen in plasma during the early stages of infection could be a novel approach to improving the screening and diagnosis of SARS-CoV-2 infection and assessing infectious viral shedding, depending on the test performance characteristic in different populations. This test has the potential to be useful in situations where nasopharyngeal swab samples are unavailable or RT-PCR is not feasible. This can be particularly beneficial for individuals who may find the nasal swab uncomfortable or for those who have difficulty providing adequate nasal samples. There has always been interest in identifying a laboratory test that could predict the likelihood of an individual transmitting the virus to others because the current reference standard molecular diagnostic tests cannot differentiate between the active replicating (ie, infectious virus) remnant viral RNAs [9, 10]. For this reason the Centers for Disease Control and Prevention does not recommend using RT-PCR results as guidance for isolation practice [11]. Antigen testing serves as a closer proxy for infectiousness than RT-PCR, although few studies have assessed its ability to measure infectious viral shedding in nasal or blood specimens.
The objective of this article is to assess clinical performance characteristics, such as sensitivity and specificity, while describing commonalities and trends of blood-based N-antigen tests for diagnosis of SARS-CoV-2 infection and assessing infectious viral shedding. This analysis may guide clinicians and policymakers to decide on the value of the plasma N-antigen test in various settings and use cases.
METHODS
Information Sources and Search
This review was performed following the PRISMA (Preferred Reporting Items for Systematic reviews and Meta-Analyses) guidelines [12]. We systematically searched multiple electronic databases, including PubMed, Embase, Web of Science, and Coronavirus Research Database, using both keywords and index terms (Mesh/Emtree). The search was divided into 3 main concepts: COVID-19, N-antigen, blood, and sensitivity/specificity. Multiple synonyms for each concept were included to create comprehensive search strategies. The full search strategies for all databases are provided in the search appendix (Supplementary Table 1). We included articles published before 27 February 2023. Preprints hosted on platforms such as medRxiv, which are not indexed by the aforementioned databases, were not included in this search to focus the discussion of this review on peer-reviewed data. There were no restrictions on time or language, and the citation lists of the articles identified as relevant to the research topic were hand searched for additional articles. This study did not include any factors necessitating patient consent.
Data Extraction and Items
The titles and abstracts of initially found articles were independently screened by 2 reviewers (S. M. and M. S.) using the COVIDence tool. Information was extracted from selected studies, including citation details, country, age range, sex distribution, study design, included population, sample size, timing of sample collection, type of N-antigen test used, comparator test used, sensitivity or positive predictive value, and specificity or negative predictive value. Any discrepancies were resolved through consultation with a third reviewer (J. D. K.) until a consensus was reached.
Eligibility Criteria
Any study that evaluated the sensitivity and specificity of the blood-based N-antigen test was considered eligible. Studies conducted during the acute phase of illness, reporting nasopharyngeal data, and using the following study designs (case-control, cohort, cross-sectional, and clinical trials) were included. Studies assessing seroprevalence, SARS-CoV-1, case reports, and systematic reviews were excluded.
Quality Assessment
The quality of each study was assessed using the Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2) tool [13, 14, p 11]. QUADAS-2 comprises 4 key domains: patient selection, index test, reference standard, and flow and timing. We evaluated all domains for potential risk of bias and the first 3 domains for applicability concerns. The risk of bias was classified as “low,” “high,” or “unclear.” Two reviewers (S. M. and M. S.) independently completed the QUADAS-2 assessment, and any discrepancies were resolved through consensus between the reviewers.
Synthesis of Results
Owing to the heterogeneity of the available data in each study, it was challenging to generate meaningful summary measures or synthesize the results. Instead, common findings among multiple studies were identified and described in this article. Summary statistics were calculated, including ranges for continuous variables and percentages for dichotomous variables.
RESULTS
Search Results
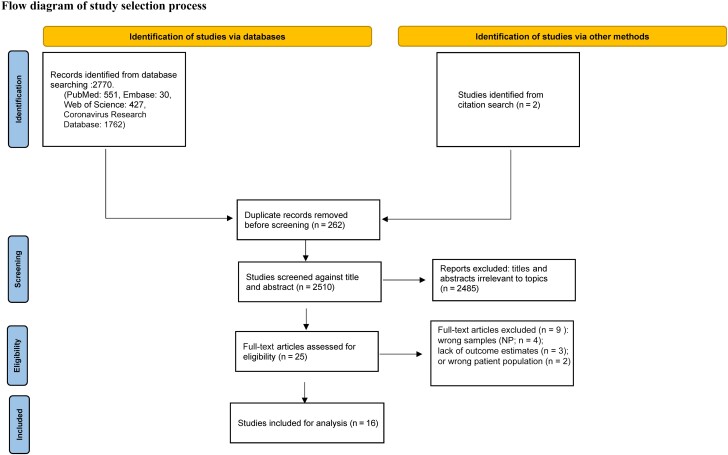
The search initially returned 2770 eligible studies, including 262 duplicates, as of 27 February 2023. After screening of their titles and abstracts, 2485 articles were excluded as not relevant to the topic of interest, and 2 articles were included by citation search. The full text of the remaining 25 articles was reviewed, and 9 were excluded. The reasons for exclusion were as follows: the use of nasopharyngeal samples for N-antigen testing (n = 4), missing test performance characteristics (n = 3), or selection of the wrong population (n = 2; SARS-CoV-1). Ultimately, a total of 16 studies met our inclusion criteria [15–30]. A flow diagram describing the process is shown in Figure 1. These studies were conducted in various countries, including the United States (n = 6); China, France, Denmark, and Germany (n = 2 each); and Finland, and Japan (n = 1 each).
Figure 1.
The Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) flow diagram detailing the number of screened and included abstracts and articles, In total, 16 studies met inclusion criteria and were included in the analysis. Abbreviation: NP, nasopharyngeal.
Characteristics of Included Studies
Of the 16 included studies, 11 did not report the age and/or sex of the participants. Among the remaining 5 studies that provided demographic information, 1 study specifically focused on pediatric patients aged <18 years [24], while the remaining 4 studies included adult participants [19, 20, 28, 29]. Among these 5 studies, female participants accounted for 52.2% of the total sample size (889 of 1703). The timing of blood sample collection was calculated from the day of symptom onset in 11 of 16 studies (68.8%) [16–20, 22, 23, 26–29], from the day of hospitalization in 3 studies (18.8%) [24, 25, 30], and from the day of RT-PCR positivity in 1 study (6.3%) [21]; in 1 study (6.3%) the details were unknown [15]. The majority of studies included hospitalized and/or hospitalized plus outpatient individuals (68.8%), while 1 study [22] focused exclusively on a longitudinal cohort of nonhospitalized community-dwelling individuals.
RT-PCR of nasopharyngeal swab samples was used as the comparator test in 15 of 16 studies. In the remaining study, a pairwise comparison was performed between 3 tests using contingency tables [15]. Of the total 16 studies, 2 assessed infectious viral shedding in addition to the diagnosis of SARS-CoV-2 infection [17, 22]. The type of N-antigen test and the corresponding cutoff used for considering N-antigen positivity varied across the 16 studies. Four studies used SIMOA technology (Quanterix Laboratory). Of these 4 studies, 2 used a cutoff of 1.25 pg/mL, 1 used a cutoff of 0.15 ng/dL, and 1 did not report the cutoff value. One study used the SARS-CoV-2 antigen enzyme-linked immunosorbent assay (ELISA) kit (Solsten Diagnostics International) with a cutoff of 10 pg/mL. Another used the iFlash-2019-nCoV Antigen kits and an iFlash3000 fully automated chemiluminescence assay analyzer with cutoff values of 1 cutoff index and 1.46 pg/mL, respectively.
In 1 study each, the SARS-CoV-2 antigen quantitative assay kit (Biohit Healthcare) had a cutoff of 2.97 pg/mL, the S-PLEX SARS-CoV-2 N kit (Meso Scale Diagnostics) had a cutoff of 2.80 log10 fg/mL, and the MSD S-PLEX CoV-2 N assay kits (Meso Scale Discovery) had a cutoff of 1.28 pg/mL. A specific cutoff value was not reported for the E-IVD ELISA microplate assay, COVID-Quantigen (AAZ). The Salocor N-antigen ELISA (Salofa) had a cutoff of 2.97 pg/mL, and the iFlash assay had a cutoff of 1 cutoff index. One study used the MAGPIX assay with a cutoff of 1046, along with the BIOPLEX assay with a cutoff of 3683. The COVID-VIRO-LFIA and COV-QUANTO-ELISA (AAZ) had a cutoff of 2.98. The details on the type of N-antigen and the manufacturer's cutoff can be found in Table 1.
Table 1.
Characteristics of the included studies
| Study Count | Author Name | Country | Median Age in Years | Gender Distribution | Study Design | Patient Included | Sample Size | Time of Sample Collection | Type of N-antigen Test | Comparator Test | Manufacturer's Cutoff | Sensitivity (at Manufacturer's Cutoff) | Specificity (at Manufacturer's Cutoff) | Sensitivity (0–7 Days) | Sensitivity (8–14 Days) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Zhang et al | United States of America | 48 | 67% M, 33% F | Cohort | Hospitalized + outpatient | Not reported | Days from symptom onset | SARS-CoV-2 Ag quantitative assay kit (Biohit Healthcare) | RT-PCR | 2.97 | 81.5% | 98.3% (91.1–99.9) | 90.9% (85.1–94.6) | Not reported |
| 2 | Yokoyama et al | Japan | Not reported | Not reported | Case-control | Hospitalized | 487 | Days from symptom onset | iFlash-2019-nCoV Antigen kits and an iFlash3000 fully automated CLIA analyzer (sandwich complex): (Shenzhen, China) | RT-PCR | 1 cutoff Index | 84.8% | 99.0% | Not reported | Not reported |
| 3 | Wang et al | United States of America | 56 | 47% M, 53% F | Case-control | Hospitalized + outpatient | Not reported | Days from symptom onset | S-PLEX SARS-CoV-2 N Kit (Meso ScaleDiagnostics) | RT-PCR | 2.80 log 10 fg/ml | 91.9% ( 83.2–97) | 94.2% (84.1–98.8) | 89.8% (77.8–96.6) | 91.4% (82.3–96.8) |
| 4 | Thudium et al | Denmark | Not reported | 38% M, 62% F | Cohort | Hospitalized + outpatient | 914 | Days from RT-PCR positive | SARS-CoV-2 Antigen ELISA kit (Solsten Diagnostics International, Aarhus, Denmark) | RT-PCR | 10 | 82.8% (73–92.5) | 99.8% (99.4–100) | 92.9% (87.9–98.0) | 91.6% (87.1–96.2) |
| 5 | Sigal et al | United States of America | 12.9 | 56% M, 44% F | Case-Control | Hospitalized | 36 | Days from Hospitalisation | MSD S-PLEX CoV-2 N assay kits (Meso Scale Discovery, Rockville, MD | RT-PCR | 1.28 | 89% (75–96) | 95% (85–99) | Not reported | Not reported |
| 6 | Shan et al | Germany | Not reported | Not reported | Case-Control | Hospitalized | 135 | Days from Hospitalisation | SIMOA technology | RT-PCR | 1.25 | 97.5% | 100% | Not reported | Not reported |
| 7 | Li et al | China | Not reported | Not reported | Case-Control | Hospitalized | SARS-COV-2 RT-PCR+, ab neg: 50 samples SARS-COV-2 RT-PCR+, ab pos: 37 samples |
Days from Hospitalisation | ELISA | RT-PCR | 10 | 76.8% | 100% | Not reported | Not reported |
| 8 | Hingrat et al | France | Not reported | Not reported | Case-Control | Hospitalized | SARS-COV-2 RT-PCR−: 63 samples SARS-COV-2 RT-PCR+: 227 samples |
Days from symptom onset | E-IVD ELISA microplate assay, COVID-Quantigene¬Æ(AAZ) | RT-PCR | … | 79.3% (74–84.6) | 98.4% (95.3–100) | <14 days: 93% (88.7–97.2) | Not reported |
| 9 | Deng et al | China | Not reported | 52% M, 48% F | Case-Control | Hospitalized | 914 | Days from symptom onset | Chemiluminescence immunoassay by iFlash immunoassay analyzer (Shenzhen Yhlo Biotech Co., Ltd,Shenzhen, China) | RT-PCR | 1.46 | Not reported | 98.8% | 76.3% | 62.5% |
| 10 | Ahava et al | Finland | Panel A: 54, Panel B: 50, Negative Panel: 53 | 41% M, 59% F | Cohort | Unclear | 281 | Days from symptom onset | Salocor N-antigen ELISA (Salofa) | RT-PCR | 2.97 | 91.7% (73–99). | 98% (94.2–99.6). | 96.2% (80.4–99.9) | 91.7% (73–99) |
| 11 | Favresse et al | Germany | 78 | 51% M, 49% F | Cohort | Hospitalized + outpatient | 243 | Days from symptom onset | SIMOA technology + iFlash-2019-nCoV Antigen kits | RT-PCR | Simoa: Unknown; iFlash: >1.0COI | Between day 2–14 in severe patients: Simoa: Sens: 84.6% (57.8–97.3), global sensitivity: 96.9%; IFlash: Sens:76.9% (49.7–91.8). global sensitivity: 96.2% | Between day 2–14 in severe patients: Simoa : 97.2% (90.3–99.5); iFlash: 93.0% (84.6–97.0) | <10 days—Simoa: Severe—100%. Non severe—91.3% (<3 d) to 100% (4 to 10 d); iFlash: Severe—100%. Non severe-93.5% (<3 d) to 96% (4 to 10 d) | 11–20 days—Simoa: severe—88.5%, Non severe: 86.5%; iFlash: severe—80.8%, Non severe—86.5% |
| 12 | Gwyn et al | United States of America & Nigeria | Not reported | Not reported | Cohort | Unclear | 416 | Days from symptom onset | Monoplex and Multiplex testing on MAGPIX and Bioplex | RT-PCR | MAGPIX = 1046, Bio-Plex = 3683 | MAGPIX: Multiplex—96.5% (90.3–99.3); Monoplex- 95.4% (88.6–98.7). Bio-Plex: Multiplex—96.5% (90.3–99.3); Monoplex—96.6% (90.3–99.3) | MAGPIX: Multiplex—98.3 (94.0–99.8); Monoplex—99.2 (95.3–100). Bio-Plex: Multiplex—97.4% (90.3–99.3); Monoplex—98.3 (93.4–100) | Not reported | Not reported |
| 13 | Mathur et al | United States of America | Not reported | 45% M, 55% F | Cohort | Outpatient | 297 | Days from symptom onset | SIMOA technology | RT-PCR | 1.25 | Global Sensitivity not reported | Global Specificity not reported | Sensitivity —Infection-77.6% (64–88.2) Infectious viral shedding: 100%(88.4–100). Specificity—Infection: 100%(84.6–100); Infectious viral shedding—65% (40.8–84.6). | Sensitivity—Infections—43.2(31.1–54.5) infectious viral shedding 70%. Specificity: Infections: 100% (84.6–100), Infectious viral shedding: 64.3% |
| 14 | Oueslati et al | France | Not reported | Not reported | Cohort | Unclear | 289 | Days from symptom onset | COVID-VIRO-LFIA and COV-QUANTO-ELISA (AAZ, Boulogne-Billancourt, France) | RT-PCR | 2.98 | LFIA : Infection: 59% (45–71.6); Infectiousness: 76% (59.4–88). ELISA: Infection—66% (52–77.8) ; Infectiousness : 87% (71.1–95.1) | LFIA: 100% (92–100) | Not reported | Not reported |
| 15 | Verkerke et al | United States of America | Not reported | Not reported | Cohort | Hospitalized + outpatient | 1860 | Days from testing and symptoms | SIMOA technology | RT-PCR | Unclear | 85.80% | 98.60% | Not reported | Not reported |
| 16 | Hiling et al | Denmark | Not reported | Not reported | Cohort | Unclear | 272 | Unclear | SIMOA technology, Solsten ELISA and Elecsys ECLIA | No single comparator test (pairwise comparison between 3 assays) | Solsten ELISA—10 ng/L, SIMOA—0.15 ng/L and Elecsys ECLIA—no cutoff for plasma (used assay linearlity with and without extraction buffer in 10 positive and 10 negative matched heparin and EDTA samples) | SIMOA versus Solsten—For both Tests: 87.8% (81.3–92.6), SIMOA versus Elecsys ECLIA—SIMOA: 75.5% (67.7–82.2); ECLIA: 100% (96.7–100), Solsten ELISA versus Elecsys ECLIA—ELISA: 74.8%(67–81.6); ECLIA 99.1% (95.1–100) | SIMOA versus Solsten—For both Tests: 85.6% (78.2–91.2), SIMOA versus Elecsys ECLIA—SIMOA: 100% (97.1–100); ECLIA: 77.6% (70.4–83.8), Solsten ELISA versus Elecsys ECLIA—ELISA: 99.2%(95.6-100); ECLIA 77% (69.7–83.3) | Not reported | Not reported |
Abbreviations: Ab, antibody; CI, confidence interval; CLIA, chemiluminescence assay; ELISA, enzyme-linked immunosorbent assay; LFIA, lateral flow immunoassay; N, nucleocapsid; NR, not reported; RT-PCR, reverse-transcriptase polymerase chain reaction; SARS-Cov-2, severe acute respiratory syndrome coronavirus 2. MFI-bg, median fluorescence intensity minus background.
Performance of Blood-Based N-Antigen Test, Overall and by Subgroups
On analysis of the 16 included studies, the overall performance of blood-based N-antigen test was found to be similar in pediatric and adult populations. Sigal et al [24] reported a sensitivity of 89% (95% confidence interval [CI], 75%–96%) and specificity of 95% (85%–99%) in the pediatric population. In the remaining 15 studies, the reported ranges of sensitivity and specificity were 67%–99% and 77%–100% respectively [15–23, 25–30]. Of these 16 studies, 11 used the receiver operating characteristic (ROC) curve for assessing the diagnostic performance of the N-antigen test. Among these 11 studies, Shan et al [25], Mathur et al [22], Gwyn et al [27], and Verkerke et al [26] did not report the area under the curve (AUC). Ahava et al [19] and Li et al [30] reported similar AUCs of 0.97 when the sample was collected <14 days after symptom onset. Three studies [20, 23, 29] reported AUCs based on the time of sample collection from the day of symptom onset. The AUC ranged from 0.87 to 0.96 during days 0–7 (week 1) and from 0.88 to 0.95 during days 8–14 (week 2). The other 2 studies reported AUCs ranging from 0.92 to 0.99 during the initial 6 days (0–6 days), from 0.93 to 0.98 during the middle 13 days (0–13 days), and from 0.91 to 0.98 during the final 0–20 days. Four studies mentioned that an optimum sensitivity could be achieved at a lower cutoff for N-antigen positivity, while increasing the cutoff to higher levels would optimize specificity [16, 21, 22, 30].
Regardless of the study population (hospitalized and/or nonhospitalized participants), test performance characteristics were similar. However, the tests showed slightly higher sensitivity for diagnosing SARS-CoV-2 infection in symptomatic and severe illness, with sensitivity ranging from 76.6% to 97.5% and specificity from 85% to 100%. In mixed populations of hospitalized and outpatient individuals, the sensitivities ranged from 73% to 97%, and specificities from 84.1% to 100%. Among the 6 studies that stratified results by first and second week after diagnosis [19–23, 28], the point estimates of sensitivity were higher during the first week (76.3%–99.9%) than during the second week (62.5%–99%). The performance of the blood-based N-antigen test also varied according to the type of assay used for testing. Among the studies included in the review, the tests with the highest sensitivities were SIMOA technology, with a global sensitivity of 97.5% [25], followed by MAGPIX and BIO-PLEX multiplex tests with a global sensitivity 96.5% (95% CI, 90.3%–99.3%) [29] and Salocor N-antigen ELISA with a sensitivity of 91.7% (73%–99%) [19]. On the other hand, the ELISA platform had lowest sensitivity, at 76.8% [30]. The test with lowest specificity was S-PLEX SARS-CoV-2 N-Kit (Meso Scale Diagnostics), with a specificity of 94.2% (95% CI, 84.1%–98.8%) [28].
Two studies assessed the performance of plasma N-antigen test for diagnosing infectious viral shedding [17, 22]. Mathur et al [22] reported a sensitivity of 100% (95% CI, 88.4%–100%) for the N-antigen test during the first 7 days after symptom onset, indicating its ability to accurately detect infectious viral shedding during this period. On the other hand, Oueslati et al [17] found that the sensitivity of the lateral flow immunoassay (LFIA) was 76% (95% CI, 59%–88%) and sensitivity of ELISA was 87% (71%–95%) for detecting infectious viral shedding. In addition, Oueslati et al reported a global specificity of 100% (95% CI, 92%–100%) for the LFIA. For a more comprehensive overview of the performance of the tests assessed in the included studies, see Table 1.
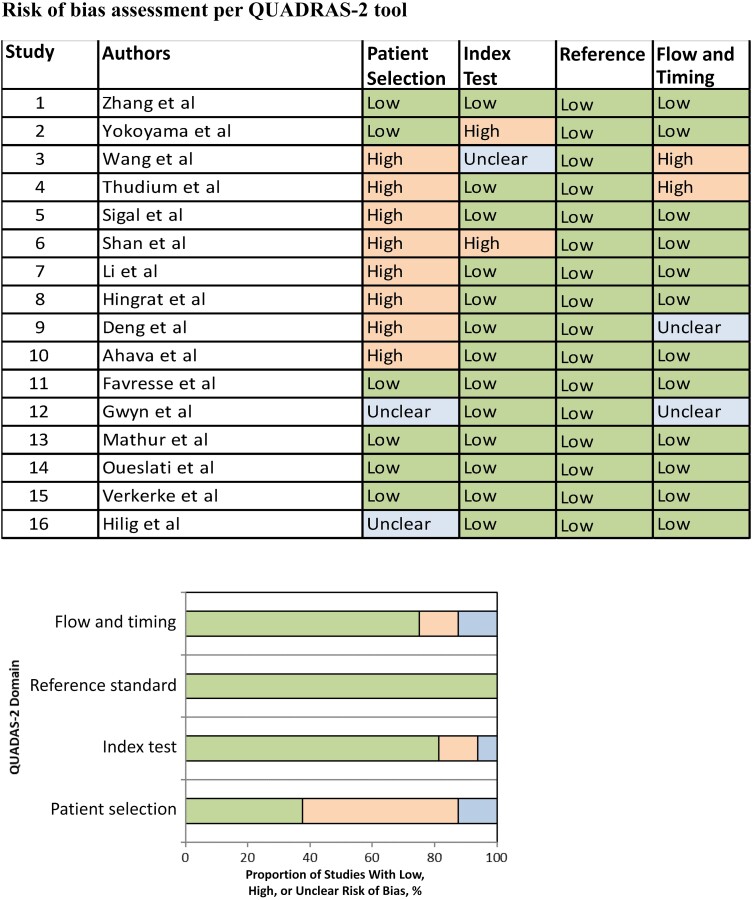
Based on the quality assessment by QUADAS-2, we found that majority of the included studies (68.8% [11 of 16]) included hospitalized individuals and/or those with symptoms of SARS-CoV-2 infection. These participants are more likely to be sicker and represent only a portion of the spectrum of SARS-CoV-2–infected individuals, hence introducing a selection bias. This may limit the generalizability of the findings to the broader population of individuals with SARS-CoV-2 infection. In addition, it is noteworthy that a small proportion of studies included only outpatients (6.3%), and in some studies (29.4%), details regarding the study population were not clear. For a more detailed understanding of the QUADAS-2 bias assessment, see Figure 2.
Figure 2.
QUADAS-2 (Quality Assessment of Diagnostic Accuracy Studies 2) criteria for the 16 studies included in this systematic review [15–30]. Orange = High, Blue = Unclear, Green = Low.
DISCUSSION
The studies assessing the performance of blood-based N-antigen reported a wide range of sensitivity and specificity for acute diagnosis of SARS-CoV-2 infection and infectious viral shedding. Our findings reveal that a subgroup of studies had sensitivities approaching those of other tests used in clinical practice to diagnose and evaluate transmission potential, particularly during the first 7 days of illness [17, 22]. Furthermore, these studies highlight the generally very high specificity of blood-based N-antigen tests. According to 1 study we reviewed, the values of N-antigen tend to peak approximately 7 days after the onset of symptoms in patients with severe illness, and there was no peak response for those with nonsevere illness [29]. In nonsevere cases, there was an exponential decay in N-antigen concentration over time [22]. Therefore, N antigens in serum may provide a valuable new marker for COVID-19 diagnosis and evaluation of disease severity. However, given the heterogeneous nature of these studies, clinicians should be aware of the performance characteristics associated with blood-based tests when in use. As these tests enter clinical, public health, and research settings, more research needs to be done to determine best practices.
We observed some degree of heterogeneity among these studies, suggesting that this test should be used with caution in the following settings: patient populations (patients hospitalized with severe illness vs outpatients), testing at different stages of infection (first week of illness vs convalescent phase), use of different testing methods, and implementation of different cutoffs for N-antigen positivity. Some of these differences, particularly those related to selection of cutoff, can be resolved by using ROC curves for the purpose of optimizing sensitivity, specificity and/or accuracy. However, other differences—such as the use of different testing methods (platform, timing) or the severity of illness—cannot be overcome with ROC curves and highlight the need to replicate investigations with these factors in focus.
A variety of detection platforms were used across studies, and the different technologies underpinning antigen testing likely had a strong influence on clinical performance. All the assays detecting N-antigen in blood samples are based on the principle of interaction between precoated antibodies with antigen present in the patient's blood serum sample. This antigen-antibody interaction can be visualized manually or by using an immunofluorescence machine reader. Nucleocapsid protein is mostly expressed during the early stages of SARS-CoV-2 infection and has the least amount of variation in its gene sequence, indicating that it is a stable protein [31]. Our study's findings regarding the impact of timing of testing and severity of illness on the performance of the blood-based N-antigen test are in line with other systematic reviews that have evaluated rapid antigen tests in detecting SARS-CoV-2 infection in the nasopharynx [32]. These findings are expected because the viral load is higher in the early phase of illness and when disease is more severe [33–35].
The blood-based N-antigen test offers a distinct advantage over commonly used, qualitative laboratory tests for SARS-CoV-2 detection because it is quantitative in nature. Blood plasma or serum is homogenous, reproducible, and well-characterized sample material, in contrast to the semiqualitative/qualitative results of upper respiratory swab samples. This minimizes the impact of potential errors or imprecise sample amounts, enhancing the accuracy and reliability of the test results. The ability to precisely measure viral load levels can aid in risk stratification, treatment decision making, and epidemiological investigations. It provides valuable information for assessing the efficacy of antiviral therapies, monitoring disease progression, and evaluating the potential for transmission within a population.
It is important to note that, despite the advantages conferred by its quantitative nature, the blood-based N-antigen test has several limitations. Given that studies on the performance of blood-based N-antigen for diagnosis of SARS-CoV-2 infection and infectious viral shedding have been very limited and heterogeneous, estimates of sensitivity and specificity could not form the basis of a meta-analysis, as the performance of each type of test should be independently considered. Second, there may have been a SARS-CoV-2 variant–specific effect that modified the sensitivity and specificity of the N-antigen test. The reliance on hospitalized and symptomatic patients in the majority of the studies may introduce a selection bias that could affect the overall interpretation of the test's performance. The results obtained from these populations may not accurately reflect the performance of the blood-based N-antigen test in other settings, such as outpatient clinics or community-based screenings. Therefore, caution should be exercised when extrapolating these findings to different populations and clinical scenarios.
To enhance the generalizability of the test's performance, future studies should strive to include a more diverse range of participants, including asymptomatic individuals and those with mild symptoms. This would provide a more comprehensive understanding of the test's accuracy and effectiveness in various clinical and public health settings. By addressing these limitations, the applicability of the blood-based N-antigen test can be better evaluated, and its potential for wider implementation can be more confidently assessed.
Our systematic review indicates that certain platforms have evidence of high sensitivity for detecting SARS-CoV-2 infection and infectious viral shedding in specific populations. In addition to the platform, we found that test performance can vary depending on factors such as the timing of sample collection and the severity of illness. Although more research is needed to evaluate performance in specific settings and determine best practices, blood-based N-antigen tests demonstrate potential in various clinical, public health, and research scenarios, including blood banks, hospitals, public health programs, and surveillance studies.
Supplementary Material
Acknowledgments
Author contributions. S. M. formulated the search strategy, performed the title and abstract screening of extracted articles, performed data analysis, and wrote the first and final drafts of the manuscript. M. S. performed independent title and abstract screening of the extracted articles. P. T. assisted in formulating the search strategy for data extraction. M. J. P., J. N. M., and J. D. K. provided in depth critical review for manuscript revision.
Contributor Information
Sujata Mathur, Department of Epidemiology and Biostatistics, University of California, San Francisco, CA, USA.
Matthew So, Department of Epidemiology and Biostatistics, University of California, San Francisco, CA, USA.
Peggy Tahir, UCSF Library, University of California, San Francisco, CA, USA.
Michael J Peluso, Division of HIV, Infectious Diseases, and Global Medicine, Zuckerberg San Francisco General Hospital, University of California, San Francisco, California, USA.
Jeffrey N Martin, Department of Epidemiology and Biostatistics, University of California, San Francisco, CA, USA.
J Daniel Kelly, Department of Epidemiology and Biostatistics, University of California, San Francisco, CA, USA; Institute for Global Health Sciences, University of California, San Francisco, CA, USA; Francis I. Proctor foundation, University of California, San Francisco, USA; San Francisco Veterans Affairs Medical Centre, San Francisco, CA, USA.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
References
- 1. Chu DKW, Pan Y, Cheng SMS, et al. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin Chem 2020; 66:549–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shen M, Zhou Y, Ye J, et al. Recent advances and perspectives of nucleic acid detection for coronavirus. J Pharm Anal 2020; 10:97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Leland DS, Ginocchio CC. Role of cell culture for virus detection in the age of technology. Clin Microbiol Rev 2007; 20:49–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Berengua C, López M, Esteban M, et al. Viral culture and immunofluorescence for the detection of SARS-CoV-2 infectivity in RT-PCR positive respiratory samples. J Clin Virol 2022; 152:105167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Diao B, Wen K, Zhang J, et al. Accuracy of a nucleocapsid protein antigen rapid test in the diagnosis of SARS-CoV-2 infection. Clin Microbiol Infect 2021; 27:289.e1–e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jacobs JL, Bain W, Naqvi A, et al. Severe acute respiratory syndrome coronavirus 2 viremia is associated with coronavirus disease 2019 severity and predicts clinical outcomes. Clin Infect Dis 2022; 74:1525–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hogan CA, Stevens BA, Sahoo MK, et al. High frequency of SARS-CoV-2 RNAemia and association with severe disease. Clin Infect Dis 2021; 72:e291–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hagman K, Hedenstierna M, Gille-Johnson P, et al. Severe acute respiratory syndrome coronavirus 2 RNA in serum as predictor of severe outcome in coronavirus disease 2019: a retrospective cohort study. Clin Infect Dis 2021; 73:e2995–3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rhee C, Kanjilal S, Baker M, Klompas M. Duration of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infectivity: when is it safe to discontinue isolation? Clin Infect Dis 2021; 72:1467–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Binnicker MJ. Can testing predict SARS-CoV-2 infectivity? the potential for certain methods to be surrogates for replication-competent virus. J Clin Microbiol 2021; 59:e0046921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Centers for Disease Control and Prevention. Ending Isolation and Precautions for People with COVID-19: Interim Guidance. Available at: https://www.cdc.gov/coronavirus/2019-ncov/hcp/infection-control-recommendations.html.
- 12. Page MJ, McKenzie JE, Bossuyt PM, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ 2021; 372:n71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. University of Bristol . QUADAS-2. Available at: https://www.bristol.ac.uk/population-health-sciences/projects/quadas/quadas-2/. Accessed 17 August 2022.
- 14. Whiting P. QUADAS-2 background document. Available at: https://www.bristol.ac.uk/media-library/sites/quadas/migrated/documents/background-doc.pdf.
- 15. Hillig T, Kristensen JR, Brasen CL, et al. Sensitivity and performance of three novel quantitative assays of SARS-CoV-2 nucleoprotein in blood. Sci Rep 2023; 13:2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yokoyama R, Kurano M, Nakano Y, et al. Association of the serum levels of the nucleocapsid antigen of SARS-CoV-2 with the diagnosis, disease severity, and antibody titers in patients with COVID-19: a retrospective cross-sectional study. Front Microbiol 2021; 12:791489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Oueslati S, Manai Bouokazi M, Ramdhani I, et al. Clinical added value of SARS-CoV-2 antigen detection in blood samples. Diagnostics 2022; 12:2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hingrat QL, Visseaux B, Laouenan C, et al. Detection of SARS-CoV-2 N-antigen in blood during acute COVID-19 provides a sensitive new marker and new testing alternatives. Clin Microbiol Infect 2020; 27:789.e1–e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ahava MJ, Kurkela S, Kuivanen S, Lappalainen M, Jarva H, Jääskeläinen AJ. Detection of SARS-CoV-2 nucleocapsid antigen from serum can aid in timing of COVID-19 infection. J Virol Methods 2022; 302:114469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang Y, Ong CM, Yun C, et al. Diagnostic value of nucleocapsid protein in blood for SARS-CoV-2 infection. Clin Chem 2021; 68:240–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thudium RF, Stoico MP, Høgdall E, et al. Early laboratory diagnosis of COVID-19 by antigen detection in blood samples of the SARS-CoV-2 nucleocapsid protein. J Clin Microbiol 2021; 59:e0100121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mathur S, Davidson MC, Anglin K, et al. Evaluation of severe acute respiratory syndrome coronavirus 2 nucleocapsid antigen in the blood as a diagnostic test for infection and infectious viral shedding. Open Forum Infect Dis 2022; 9:ofac563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Deng Q, Ye G, Pan Y, et al. High performance of SARS-Cov-2N protein antigen chemiluminescence immunoassay as frontline testing for acute phase COVID-19 diagnosis: a retrospective cohort study. Front Med 2021; 8:676560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sigal GB, Novak T, Mathew A, et al. Measurement of severe acute respiratory syndrome coronavirus 2 antigens in plasma of pediatric patients with acute coronavirus disease 2019 or multisystem inflammatory syndrome in children using an ultrasensitive and quantitative immunoassay. Clin Infect Dis 2022; 75:1351–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shan D, Johnson JM, Fernandes SC, et al. N-protein presents early in blood, dried blood and saliva during asymptomatic and symptomatic SARS-CoV-2 infection. Nat Commun 2021; 12:1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Verkerke HP, Damhorst GL, Graciaa DS, et al. Nucleocapsid antigenemia is a marker of acute SARS-CoV-2 infection. J Infect Dis 2022; 226:1577–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gwyn S, Abubakar A, Akinmulero O, et al. Performance of SARS-CoV-2 antigens in a multiplex bead assay for integrated serological surveillance of neglected tropical and other diseases. Am J Trop Med Hyg 2022; 107:260–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang H, Hogan CA, Verghese M, et al. SARS-CoV-2 nucleocapsid plasma antigen for diagnosis and monitoring of COVID-19. Clin Chem 2021; 68:204–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Favresse J, Bayart JL, David C, et al. Serum SARS-CoV-2 antigens for the determination of COVID-19 severity. Viruses 2022; 14:1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li T, Wang L, Wang H, et al. Serum SARS-COV-2 nucleocapsid protein: a sensitivity and specificity early diagnostic marker for SARS-COV-2 infection. Front Cell Infect Microbiol 2020; 10:470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Surjit M, Lal SK. The SARS-CoV nucleocapsid protein: a protein with multifarious activities. Infect Genet Evol 2008; 8:397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ghasemi S, Harmooshi NN, Rahim F. Diagnostic utility of antigen detection rapid diagnostic tests for COVID-19: a systematic review and meta-analysis. Diagn Pathol 2022; 17:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. World Health Organization . Antigen-detection in the diagnosis of SARS-CoV-2 infection using rapid immunoassays: interim guidance. 11 September 2020. Available at: https://apps.who.int/iris/bitstream/handle/10665/334253/WHO-2019-nCoV-Antigen_Detection-2020.1-eng.pdf. Accessed 18 September 2022.
- 34. Wick KD, Leligdowicz A, Willmore A, et al. Plasma SARS-CoV-2 nucleocapsid antigen levels are associated with progression to severe disease in hospitalized COVID-19. Crit Care 2022; 26:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rogers AJ, Wentworth D, Philips A, et al. ; ACTIV-3/TICO Study Group . The association of baseline plasma SARS-CoV-2 nucleocapsid antigen level and outcomes in patients hospitalized with COVID-19. Ann Intern Med 2022; 175:1401–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.