Abstract
Flock House virus is a small icosahedral insect virus of the family Nodaviridae. Its genome consists of two positive-sense RNA molecules, which are believed to be encapsidated into a single viral particle. However, evidence to support this claim is circumstantial. Here we demonstrate that exposure of nodavirus particles to heat causes the two strands of viral RNA to form a stable complex, directly establishing that both RNAs are copackaged into one virion. The physical properties of the RNA complex, the effect of heat on the particles per se, and the possible relevance of these findings to the nodavirus life cycle are presented.
Flock House virus (FHV) is a nonenveloped, icosahedral insect virus of the family Nodaviridae (for a review, see reference 20). Its genome consists of two messenger-sense RNA molecules: RNA1 (3.1 kb; 1.1 × 106 Da) encodes replicase functions, and RNA2 (1.4 kb; 0.48 × 106 Da) encodes protein alpha, the precursor of the coat protein (5, 8). Studies on FHV assembly in cultured Drosophila melanogaster cells have demonstrated that 180 subunits of alpha protein initially assemble to form a labile precursor particle called a provirion (10, 14). Provirions subsequently mature into infectious virions by cleavage of the coat protein near the C terminus (10, 21). X-ray crystallography of mature virions has provided detailed insights into the atomic structure of the viral protein capsid (7), but little is known about the structure and organization of the encapsidated RNAs.
During particle assembly, both RNA1 and RNA2 are thought to be encapsidated into a single nodavirus particle by an as yet unknown mechanism. The evidence for coencapsidation of both RNAs is indirect and based on the following observations: (i) infectivity of virus preparations rises linearly with increasing particle concentrations, suggesting that a single virion is sufficient for initiating an infectious cycle (22); and (ii) virion populations form a single peak on CsCl gradients, indicating homogeneity with regard to particle density. While homogeneous particle density could be due to virions containing different combinations of RNA1 and RNA2 (e.g., the mass of three copies of RNA2 is roughly equal to the sum of one copy each of RNA1 and RNA2), the fact that extraction of viral RNAs from purified virus consistently yields a 1:1 molar ratio of the two species suggests that each virion carries one strand of RNA1 and one strand of RNA2 (11). More direct evidence indicating a copackaging mechanism comes from earlier studies by Newman and Brown on Nodamura virus (NOV), the prototype member of the nodavirus family (15). Treatment of NOV with 1.5 M guanidine and subsequent centrifugation on a sucrose gradient containing 0.01% sodium dodecyl sulfate (SDS) yielded a component that sedimented at 27S. Heating of this component or extraction with phenol caused it to break down into a 22S (RNA1) and a 15S (RNA2) species (15).
Detection of a noncovalent interaction between FHV RNA1 and RNA2.
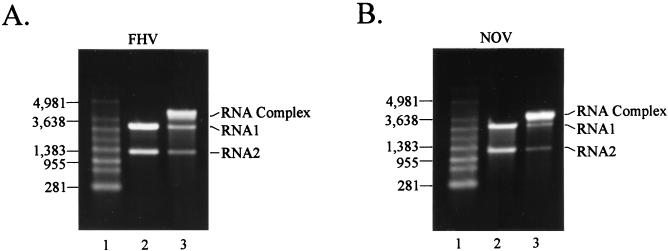
In an attempt to find in vitro conditions that would mimic the release of RNA from virus particles during the infection process, we analyzed the effect of heat on virions. This strategy was based on a report by Cheng et al., who observed that heating of virus particles resulted in extrusion of RNA from a single site on the virion (3). An aliquot of gradient-purified FHV (10 μg) in 50 mM HEPES (pH 7), 5 mM CaCl2, 0.1% 2-mercaptoethanol (2-ME), and approximately 25% (wt/wt) sucrose was heated for 10 min at 65°C. The sample was cooled on ice for several minutes, and viral RNA was extracted with acidic phenol-chloroform after addition of NaCl and SDS to 0.2 M and 0.1%, respectively. RNA in the aqueous phase was precipitated in the presence of sodium acetate and glycogen, pelleted, washed with 70% ethanol, and dried. Following resuspension in nuclease-free water, the entire sample (1 to 1.5 μg) was electrophoresed through a nondenaturing 1% agarose gel in Tris-acetate-EDTA (TAE) buffer at 100 V for 1 h and visualized with ethidium bromide (Fig. 1A). Surprisingly, most of the RNA migrated as a species of 4.5 kb, the combined size of RNA1 and RNA2 (Fig. 1A, lane 3). Small amounts of RNA1 and RNA2 appeared to be present as well, but they were reduced compared to a control preparation of RNA1 and RNA2 extracted from unheated virions (Fig. 1A, lane 2).
FIG. 1.
Gel electrophoresis of viral RNA extracted from unheated and heated FHV and NOV particles. Aliquots (10 μg) of gradient-purified viral particles were either heated at 65°C for 10 min followed by cooling or kept at 4°C. RNA was extracted with phenol-chloroform and electrophoresed through a nondenaturing 1% agarose-TAE gel. The RNA was visualized with ethidium bromide present in the gel. (A) RNA extracted from FHV particles. Lane 1, RNA size markers; lane 2, RNA extracted from unheated virus; lane 3, RNA extracted from heated virus. (B) RNA extracted from NOV particles. Lane 1, RNA size markers; lane 2, RNA extracted from unheated virus; lane 3, RNA extracted from heated virus. The positions of RNA size markers (in nucleotides), RNA1, RNA2, and the RNA complex are indicated. Note that under these nondenaturing electrophoresis conditions the size markers did not accurately reflect the sizes of the viral RNAs. For example, FHV RNA2 contains 1,400 bases but migrated slightly ahead of the 1,383-base marker.
To determine whether this phenomenon could also be detected with other nodaviruses, we analyzed NOV, an insect virus that is serologically and structurally distantly related to FHV (20). An aliquot of gradient-purified NOV was heated to 65°C for 10 min and RNA was extracted and analyzed by electrophoresis as described above. As with FHV, the RNA molecules were present in the form of a 4.5-kb RNA complex (Fig. 1B, lane 3) while RNA extracted from unheated NOV particles yielded RNA1 and RNA2 as separate molecules (Fig. 1B, lane 2). In contrast, no shift to higher-molecular-weight RNA species was observed for baculovirus-expressed virus-like particles of FHV (data not shown). These particles are crystallographically indistinguishable from native FHV particles (16a), but they contain a mixture of RNA2 and cellular RNAs instead of the normal complement of RNA1 and RNA2 (18). Thus, formation of the RNA complex appeared to be specific for the nodaviral RNAs.
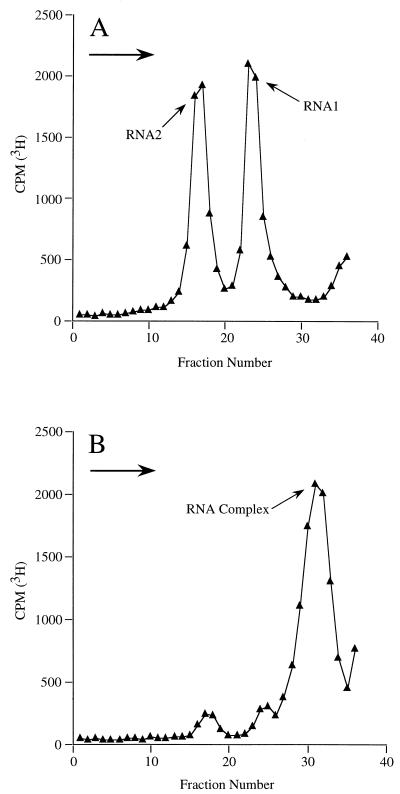
To further analyze the RNA complex, 10 μg (100,000 cpm) of [3H]uridine-labeled FHV particles prepared as previously described (19) were either heated to 65°C followed by cooling on ice or kept at 4°C for 10 min. Viral RNA was extracted from each aliquot, resuspended in 0.2 M sodium acetate (pH 5.0)–0.01 M EDTA, and layered onto an 11-ml 5 to 25% (wt/wt) sucrose gradient prepared in the same buffer. The viral RNAs were sedimented at 36,000 rpm (222,000 × g) in a Beckman SW41Ti rotor for 11 h at 10°C followed by fractionation of the gradient. Radioactivity in an aliquot (100 μl) of each fraction was determined by liquid scintillation counting. As expected, the profile of the gradient containing RNA extracted from unheated particles contained two peaks corresponding to RNA1 and RNA2 (Fig. 2A). However, RNA extracted from the heated particles formed one large peak that sedimented farther down the gradient (Fig. 2B). Correspondingly, the levels of RNA1 and RNA2 in this gradient were significantly reduced. Quantitative analysis revealed that approximately 84% of the RNAs extracted from heated virions was present in the RNA complex (data not shown). Based on the equation Sunknown = Sstandard × (MWunknown/MWstandard)2/3 (where MW is molecular weight) and using RNA1 as the standard, we had previously estimated that a complex of FHV RNA1 and RNA2 would sediment at about 29S (19). Since our sedimentation experiments were performed under near-isokinetic conditions (9), the positions of RNA1 and RNA2 on the gradient and their known sedimentation rates, 15S for RNA2 and 22S for RNA1 (12), could be used to determine the approximate sedimentation rate of the RNA complex. Based on these considerations, the sedimentation rate for the complex was found to be 28 to 29S, which is in good agreement with the predicted value.
FIG. 2.
Sucrose gradient sedimentation of viral RNA extracted from unheated and heated FHV particles. Ten micrograms (100,000 cpm) of [3H]uridine-labeled FHV particles was either heated to 65°C and cooled or kept at 4°C for 10 min. Viral RNA was extracted from each aliquot, sedimented through an 11-ml 5 to 25% sucrose gradient, and fractionated on an ISCO gradient fractionator. The radioactivity in an aliquot (100 μl) of each fraction was determined by liquid scintillation counting. (A) Plot of RNA extracted from unheated FHV particles. (B) Plot of RNA extracted from heated FHV particles. The peaks corresponding to RNA1, RNA2, and the RNA complex are indicated. The arrows in the top left corners of both graphs indicate the direction of sedimentation.
Gel electrophoresis and sucrose gradient analysis suggested that the RNA complex did not form with 100% efficiency; small amounts of uncomplexed RNA1 and RNA2 were usually detected. This could have been due to a number of reasons. First, it is conceivable that the virus particles were not uniformly heated to 65°C and that the RNA complex therefore did not form in some of the virions. This is unlikely, however, as an increase in the time of incubation at 65°C did not lead to conversion of the remaining monomeric RNA into the complex (data not shown). Alternatively, a small portion of the particles may have been defective in complex formation due to mutations in the viral RNA. A third, and perhaps the most likely, explanation is that the RNA complex was partially disrupted under the conditions of gel electrophoresis or by the physical forces encountered during ultracentrifugation.
We used electrophoretic analysis to further define the conditions that would lead to RNA complex formation (data not shown). Incubation of FHV particles for 10 min at temperatures as low as 50°C resulted in complex formation, although with low efficiency. Optimal conversion was observed between 65 and 75°C. In contrast, at 80°C and above the complex was not detected. Instead, the RNA showed signs of degradation, presumably because the virions were disrupted and the nucleic acid was exposed to RNases in the solution. The pH of the solution could be varied between 5 and 9 without any obvious effects on the efficiency of complex formation. Above pH 9 and below pH 5, however, the complex was not observed.
At this point it was not clear whether the complex was formed during heating of the virions or whether it was generated subsequently during cooling of the preparation, usually to room temperature or 4°C. To test this, virions were heated to 65°C for 10 min and RNA was immediately extracted with hot phenol preequilibrated to the same temperature. RNA purified under these conditions was found to show no signs of complex formation when analyzed by agarose gel electrophoresis (data not shown).
Once formed, the RNA complex could be disrupted again into RNA1 and RNA2 by heating to 85 to 90°C for 10 min (data not shown). This indicated that the interactions between the two RNAs were noncovalent. In line with this notion was the observation that the melting temperature could be reduced to 65°C in the presence of 50% formamide (data not shown).
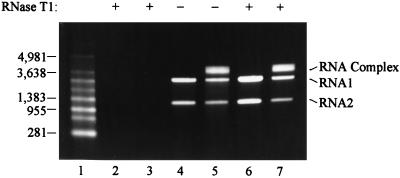
It was important to determine whether the conformational changes observed for FHV RNA during heating of virus particles occurred inside the virion or whether heating resulted in partial or even complete exposure of the nucleic acids. To address this question, aliquots containing 10 μg of heated or unheated FHV particles were treated with 200 U of RNase T1 at room temperature for 15 min. As a positive control 1 μg of viral RNA already extracted from heated and unheated virions was treated with the same amount of RNase, and as a negative control identical aliquots of heated and unheated FHV did not receive nuclease. All samples were subsequently incubated with 50 μg of proteinase K/ml for 30 min at 37°C to inactivate the RNase. RNA was then extracted from the samples and analyzed on a nondenaturing 1% agarose gel. As expected, the naked RNAs were digested by the RNase (Fig. 3, lanes 2 and 3), whereas RNA extracted from unheated and heated FHV particles that had not received RNase T1 was intact (Fig. 3, lanes 4 and 5, respectively). Viral RNAs purified from unheated and heated particles that had been incubated with RNase were also completely protected from degradation, suggesting that the heating and cooling regimen did not result in exposure of the viral genome to the exterior of the virion (Fig. 3, lanes 6 and 7). Thus, formation of the RNA complex had to occur inside the virus particle.
FIG. 3.
Effect of RNase T1 on viral RNA associated with heated and unheated FHV particles. FHV particles were kept at 4°C or heated to 65°C for 10 min and cooled. Each sample was then divided into three aliquots. To the first aliquot RNase T1 was added, and the particles were incubated at room temperature for 15 min. The second aliquot was incubated at room temperature without addition of RNase. The third aliquot was used for extraction of RNA from the virions followed by incubation of the purified RNA with RNase T1. All samples were subsequently treated with proteinase K for 30 min at 37°C to inactivate the RNase. RNA was then extracted from the samples and electrophoresed on a nondenaturing 1% agarose-TAE gel. The RNA was visualized with ethidium bromide. Lane 1, RNA size markers (in nucleotides); lane 2, RNA extracted from unheated virions prior to RNase treatment; lane 3, RNA extracted from heated virions prior to RNase treatment; lane 4, RNA extracted from unheated virions not treated with RNase; lane 5, RNA extracted from heated virions not treated with RNase; lane 6, RNA extracted from unheated virions treated with RNase; lane 7, RNA extracted from heated virions treated with RNase. The positions of RNA size markers, RNA1, RNA2, and the RNA complex are indicated.
Attempts to generate the RNA complex in solution by using purified FHV RNA1 and RNA2.
Our data described so far were somewhat similar to observations that have been made for the retroviruses. The retroviral genome consists of two identical strands of mRNA, which form a noncovalent dimer by hydrogen bonding of sequences located near the 5′ end of the RNAs (1). It has previously been shown that under certain conditions the two strands of genomic RNA spontaneously dimerize in vitro (6). Based on these observations, we attempted to generate the FHV RNA1/RNA2 complex by using similar conditions. Specifically, 1 μg of purified viral RNA1 and RNA2 was incubated in 250 mM NaCl, 1 mM MgCl2, and 10 mM Tris (pH 7.0) at either −20, 4, 37, 55, or 65°C for various time intervals. However, no complex was detected when the RNAs were analyzed by gel electrophoresis (data not shown). It is conceivable that other conditions are required for its formation in vitro. In addition, it is possible that limiting amounts of coat protein are necessary for interaction of the two RNAs.
Physical properties of heated FHV particles.
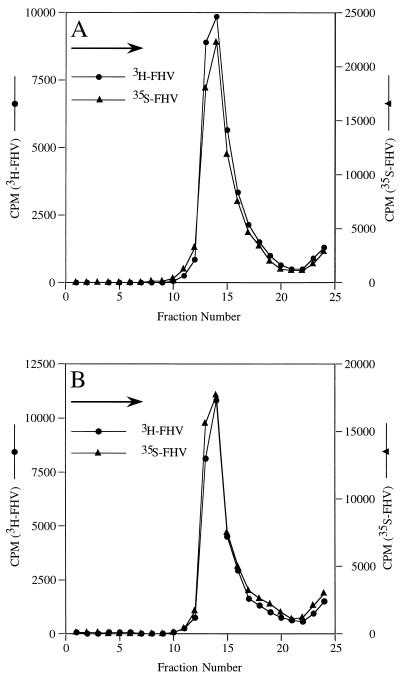
As shown above, heat treatment of FHV particles did not allow exogenously added RNase access to the viral genome. This indicated that particle integrity was not severely affected by heating at 65°C for 10 min. Nonetheless, in order to detect more subtle changes, we performed sucrose gradient sedimentation analysis. Specifically, [35S]methionine-labeled FHV (200,000 cpm) (19), either heated for 10 min at 65°C or unheated, was layered onto 5-ml 5 to 20% (wt/wt) sucrose gradients in 50 mM HEPES (pH 7.0)–0.1% 2-ME. Untreated [3H]uridine-labeled FHV (100,000 cpm) was included in the sample as an internal control. Centrifugation was in a Beckman SW50.1 rotor at 45,000 rpm (243,000 × g) for 25 min at 20°C. Gradients were fractionated, and an aliquot (100 μl) of the radioactivity in each fraction was determined by liquid scintillation counting. As shown in Fig. 4, both unheated and heated FHV cosedimented with the differentially labeled FHV marker, indicating that there were no significant changes in the size or overall morphology of the heated particles. This was confirmed by electron microscopy, which revealed no differences between negatively stained heated and unheated particles (data not shown). Analysis of heat-treated virus by plaque assay on monolayers of Drosophila melanogaster cells (21), however, showed that the specific infectivity of heated virus was somewhat lower than that of untreated virus. On the average (two experiments), the titer of heated particles dropped by 1 log unit to 2.09 × 109 PFU/mg compared to that (2.13 × 1010 PFU/mg) for the untreated virus. This drop in titer, while small, might have been a result of the altered interactions between RNA1 and RNA2 within the particle. Alternatively, heat treatment may have had a subtle effect that could not be detected by sucrose gradient sedimentation or electron microscopy on the structure of the viral capsid.
FIG. 4.
Sedimentation analysis of unheated and heated FHV particles. [35S]methionine-labeled FHV (200,000 cpm) was either left unheated or heated for 10 min at 65°C and cooled. Each sample was then mixed with unheated [3H]uridine-labeled FHV (100,000 cpm) and sedimented through a 5-ml 5 to 25% (wt/wt) sucrose gradient for 25 min at 243,000 × g. The gradients were fractionated, and the radioactivity in each fraction was determined by liquid scintillation counting. (A) Sedimentation profile of unheated FHV particles. (B) Sedimentation profile of heated FHV particles. The arrows in the top left corners of both graphs indicate the direction of sedimentation.
The finding that FHV RNA1 and RNA2 are capable of forming a stable complex may lead to important new insights into nodaviral biology. First, the observation that complex formation occurred within the context of the viral capsid unequivocally demonstrated that both RNAs are packaged into a single particle. Although other results had previously implied that this was the case, direct evidence had not been obtained until now. Newman and Brown’s observation (15) that gentle dissociation of NOV in the presence of guanidine and SDS resulted in the release of the genomic RNAs as a single 27S component could not be reproduced with FHV. Rather, the RNAs were liberated as separate species (data not shown). Previous dissociation experiments involving alkaline pH, high salt, and freezing at −20°C also revealed release of FHV RNAs as individual strands (19). Thus, it was not clear whether NOV represented a unique case or whether the complex in FHV and other nodaviruses was much more labile.
It is not yet known which sequences of the genomic RNAs are involved in complex formation. Regions of complementarity between the two RNAs do not span more than seven bases if standard Watson-Crick base pairing is assumed. This makes it improbable that a single site of interaction would be sufficient for maintaining a stable complex. It is more likely that several regions involving short stretches of primary sequence are required. Alternatively, or additionally, the interacting regions may contain non-Watson-Crick base pairs.
Particles had to be heated to at least 50°C for the complex to be detected. Presumably, heating of the virions resulted in reorganization of the encapsidated RNAs, such that stable interactions between RNA1 and RNA2 could be established. It is possible that the complex already preexisted in a labile form and that it was merely stabilized during the heating and cooling procedure. The notion of a preexisting complex would be in line with the results of Newman and Brown, who detected the NOV complex without heat treatment of virus particles (15). NOV can be dissociated under very mild conditions, such as incubation in 0.1 M NaCl (15). This property may allow release of a labile complex in intact form. In contrast, FHV particles are exceptionally stable, such that extraction of viral RNA is usually performed in the presence of 1% SDS, 0.2 M NaCl, and hot phenol-chloroform (12). Under these conditions only separate strands of RNA1 and RNA2 are normally detected.
How heating and subsequent cooling of virus particles might result in stabilization of an RNA complex is currently not clear. However, our data are reminiscent of observations made for retroviruses. Retroviruses package two identical copies of genomic RNA that are linked by base pairing of regions located near the 5′ end (1). The dimer is initially labile but is stabilized upon proteolytic maturation of virus particles. The stabilization is due to the action of the nucleocapsid protein, which has been referred to as a “nucleic acid chaperone” (13). Interestingly, the RNA dimer is also stabilized by heating rapidly harvested virions, i.e., immature virions, to 40°C (2).
Like for the retroviruses, synthesis of FHV particles proceeds through a noninfectious precursor, the provirion. Provirions mature by autocatalytic cleavage of coat protein subunits near the C terminus between residues Asn363 and Ala364 (14). In contrast to retroviruses, however, both mature and immature FHV particles yielded the same results with regard to RNA complex formation. Extraction of RNA without heat treatment yielded the two RNAs as separate molecules, whereas heating and cooling resulted in formation of the heterodimer (data not shown). This difference might be due to the fact that maturation of FHV particles causes only subtle conformational adjustments in the viral capsid (16a), whereas retroviral particles undergo substantial structural rearrangements during the maturation process (4).
In retroviruses, formation of the RNA dimer is believed to be a prerequisite for RNA packaging (16). It is not known whether this is also the case for the nodaviruses, although it would represent an elegant mechanism by which RNA1 and RNA2 could be packaged into a single particle. If RNA1 and RNA2 are indeed packaged as a complex, it is possible that this complex is destabilized as coat protein-RNA interactions are established during the assembly process. This destabilization may make it difficult to detect the complex by using standard RNA extraction procedures. More experiments are required to confirm these hypotheses. It would be particularly important to show that the FHV RNA complex can form spontaneously in vitro or that it exists in infected cells prior to packaging.
Alternatively, the complex might form de novo upon heating, and it would be reasonable to assume that the heterodimeric RNA molecules generated in each particle are identical. This in turn would imply that the organization of RNA1 and RNA2 within the assembled virion follows a defined pattern such that the same regions are brought into close proximity in each particle. Unfortunately, current techniques used for high-resolution structural analysis of icosahedral viruses, such as X-ray crystallography and cryoelectron microscopy combined with image reconstruction, do not allow visualization of encapsidated nucleic acid unless it conforms to the icosahedral symmetry of the virus particle. Moreover, the electron density obtained represents only an average of the sequences present at the symmetrically related site. Thus, further insights regarding the conformation of genomic RNA and the precise spatial arrangement of the individual nucleotides within the particle will have to await the development of approaches that eliminate these constraints.
Formation of the RNA complex may also have reflected changes that occur in the particle during viral uncoating. This assumption is based on observations made by Cheng et al. (3), who found that heating of FHV for 10 min at 65°C results in extrusion of RNA from a single site on the virion. This phenomenon was thought to mirror an event of the uncoating process based on analogies with the picornaviruses. For instance, heating of poliovirus particles for 2 to 3 min at 56°C results in release of RNA and formation of 80S particles which are normally detected as intermediates during cell entry (17). Heating for shorter time intervals does not lead to loss of RNA but to conversion into A particles, another intermediate in the cell entry process. Thus, while we did not detect any exposure of viral RNAs from heated FHV particles, we cannot exclude the possibility that the apparent reorganization of the RNA is a prerequisite for its release from the virus particle.
Acknowledgments
We thank J. E. Johnson for critical reading of the manuscript.
This work was supported by NIH grant GM53491 to A.S.
Footnotes
Manuscript no. 11896MB from The Scripps Research Institute.
REFERENCES
- 1.Berkowitz R, Fisher J, Goff S P. RNA packaging. In: Krausslich H G, editor. Current topics in microbiology and immunology. Vol. 214. Berlin, Germany: Springer; 1996. pp. 177–218. [DOI] [PubMed] [Google Scholar]
- 2.Canaani E, Helm K V D, Duesberg P. Evidence for the 30-40S RNA as precursor of the 60-70S RNA of Rous sarcoma virus. Proc Natl Acad Sci USA. 1973;72:401–405. doi: 10.1073/pnas.70.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng R H, Reddy V S, Olson N H, Fisher A J, Baker T S, Johnson J E. Functional implications of quasi-equivalence in a T=3 icosahedral animal virus established by cryo-electron microscopy and X-ray crystallography. Structure. 1994;2:271–282. doi: 10.1016/s0969-2126(00)00029-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coffin J M. Retroviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Virology. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1767–1847. [Google Scholar]
- 5.Dasmahapatra B, Dasgupta R, Ghosh A, Kaesberg P. Structure of the black beetle virus genome and its functional implications. J Mol Biol. 1985;182:183–189. doi: 10.1016/0022-2836(85)90337-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng Y X, Copeland T D, Henderson L E, Gorelick R J, Bosche W J, Levin J G, Rein A. HIV-1 nucleocapsid protein induces “maturation” of dimeric retroviral RNA in vitro. Proc Natl Acad Sci USA. 1996;93:7577–7581. doi: 10.1073/pnas.93.15.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisher A J, Johnson J E. Ordered duplex RNA controls capsid architecture in an icosahedral animal virus. Nature. 1993;361:176–179. doi: 10.1038/361176a0. [DOI] [PubMed] [Google Scholar]
- 8.Friesen P D, Rueckert R R. Synthesis of black beetle virus proteins in cultured Drosophila cells: differential expression of RNAs 1 and 2. J Virol. 1981;37:876–886. doi: 10.1128/jvi.37.3.876-886.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fritsch A. Properties of various rotors used for zone centrifugation. Anal Biochem. 1973;55:57–71. doi: 10.1016/0003-2697(73)90290-x. [DOI] [PubMed] [Google Scholar]
- 10.Gallagher T, Rueckert R R. Assembly-dependent maturation cleavage in provirions of a small icosahedral insect ribovirus. J Virol. 1988;62:3399–3406. doi: 10.1128/jvi.62.9.3399-3406.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallagher T M. Ph.D. thesis. Madison: University of Wisconsin—Madison; 1987. [Google Scholar]
- 12.Gallagher T M, Friesen P D, Rueckert R R. Autonomous replication and expression of RNA1 from black beetle virus. J Virol. 1983;46:481–489. doi: 10.1128/jvi.46.2.481-489.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herschlag D. RNA chaperones and the RNA folding problem. J Biol Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 14.Hosur M V, Schmidt T, Tucker R C, Johnson J E, Gallagher T M, Selling B H, Rueckert R R. Structure of an insect virus at 3.0 Å resolution. Proteins. 1987;2:167–176. doi: 10.1002/prot.340020302. [DOI] [PubMed] [Google Scholar]
- 15.Newman J F E, Brown F. Further physicochemical characterization of Nodamura virus. Evidence that the divided genome occurs in a single component. J Gen Virol. 1977;38:83–95. doi: 10.1099/0022-1317-38-1-83. [DOI] [PubMed] [Google Scholar]
- 16.Paillart J C, Marquet R, Skripkin E, Ehresmann C, Ehresmann B. Dimerization of retroviral genomic RNAs: structural and functional implications. Biochimie. 1996;78:639–653. doi: 10.1016/s0300-9084(96)80010-1. [DOI] [PubMed] [Google Scholar]
- 16a.Reddy, V., and J. E. Johnson. Personal communication.
- 17.Rueckert R R. Picornaviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Virology. Vol. 1. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 609–654. [Google Scholar]
- 18.Schneemann A, Dasgupta R, Johnson J, Rueckert R. Use of recombinant baculoviruses in synthesis of morphologically distinct virus-like particles of flock house virus, a nodavirus. J Virol. 1993;67:2756–2763. doi: 10.1128/jvi.67.5.2756-2763.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneemann A, Gallagher T M, Rueckert R R. Reconstitution of flock house provirions: a model system for studying structure and assembly. J Virol. 1994;68:4547–4556. doi: 10.1128/jvi.68.7.4547-4556.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneemann A, Reddy V, Johnson J E. The structure and function of nodavirus particles: a paradigm for understanding chemical biology. Adv Virus Res. 1998;50:381–446. doi: 10.1016/s0065-3527(08)60812-x. [DOI] [PubMed] [Google Scholar]
- 21.Schneemann A, Zhong W, Gallagher T M, Rueckert R R. Maturation cleavage required for infectivity of a nodavirus. J Virol. 1992;66:6728–6734. doi: 10.1128/jvi.66.11.6728-6734.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Selling B H, Rueckert R R. Plaque assay for black beetle virus. J Virol. 1984;51:251–253. doi: 10.1128/jvi.51.1.251-253.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]