Abstract
Herpes simplex virus (HSV) DNA is cleaved from concatemers and packaged into capsids in infected cell nuclei. This process requires seven viral proteins, including UL15 and UL28. UL15 expressed alone displays a nuclear localization, while UL28 remains cytoplasmic. Coexpression with UL15 enables UL28 to enter nuclei, suggesting an interaction between the two proteins. Additionally, UL28 copurified with UL15 from HSV-infected cells after ion-exchange and DNA affinity chromatography, and the complex sedimented as a 1:1 heterodimer upon sucrose gradient centrifugation. These findings are evidence of a physical interaction of UL15 and UL28 and a functional role for UL15 in directing UL28 to the nucleus.
At least seven herpes simplex virus (HSV) genes (UL6, UL15, UL17, UL25, UL28, UL32, and UL33) encode proteins required for the cleavage and packaging of HSV DNA into capsids (1–4, 10, 11, 14, 16–18, 21). These proteins act in concert to mediate the cleavage event and the packaging of monomer-length viral genomes into capsids. The coordination of cleavage and packaging events suggests that these proteins interact directly, but there is no direct evidence to support this. The most promising interaction involves UL28 and UL15. Previous work demonstrated that pseudorabies virus (PRV) UL28 was limited to the cytoplasm in the absence of other viral proteins (7), and coexpression with HSV UL15 was sufficient to direct PRV UL28 to nuclei (7).
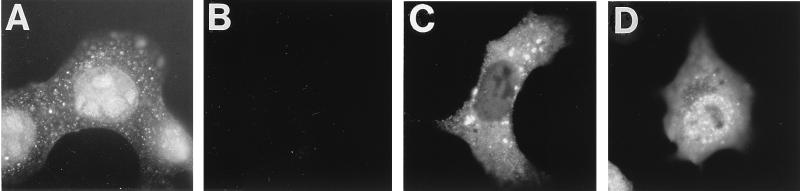
The possibility that HSV UL28 interacts with HSV UL15 was explored by first establishing that HSV UL15 directs the nuclear localization of HSV UL28. In Vero cells infected with HSV(KOS), UL28 accumulated in nuclei (Fig. 1A), consistent with a direct role in the cleavage and packaging reaction (18). UL28 was not detected in Vero cells infected with the UL28-null mutant HSV(gCB) (18) (Fig. 1B). To examine the subcellular distribution of UL28 in the absence of other viral proteins, Vero cells were transfected with plasmid pF1′-CMV-UL28 containing the 3.0-kb BstYI-BstYI fragment from pECH82 (18) in the vector pF1′-CMV (6) as described previously (7). HSV UL28 remained in the cytoplasm (Fig. 1C), consistent with the finding that PRV UL28 was also unable to enter nuclei in the absence of other viral proteins (7). Since UL28 did not efficiently enter nuclei of cells in the absence of HSV proteins, the simplest explanation was that another protein directed UL28 to nuclei. The possibility that UL15 directed UL28 to nuclei was tested. Cells were transfected with pF1′-CMV-UL28 plus pF1′-CMV-UL15 as described previously (7) to express UL28 and UL15, respectively. UL28 was directed to nuclei in the presence of UL15 (Fig. 1D), suggesting that the two proteins interact. Another HSV cleavage and packaging protein, UL25, was also coexpressed with UL28 but did not direct UL28 to nuclei (12). The results suggest that the transport of UL28 to nuclei by UL15 is conserved among alphaherpesviruses, consistent with its playing an important role during infection.
FIG. 1.
Fluorescence photomicrographs of UL28 in Vero cells. Vero cells were infected with HSV(KOS) (A) or HSV(gCB) (B) at an MOI of 2 for 12 h or transfected with plasmids to express UL28 (C) or UL28 plus UL15 (D) for 24 h. Cells were processed to detect UL28 with affinity-purified (13) H85 antiserum (18) as described previously (7).
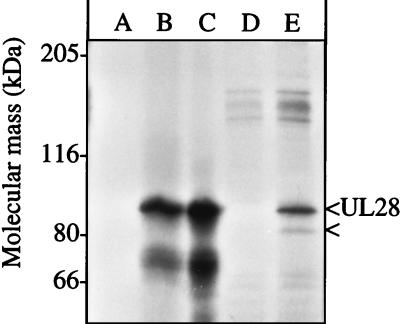
The idea that UL28 interacted directly with another protein was also suggested by coimmunoprecipitation. Radiolabeled infected cell extracts were immunoprecipitated with antiserum, protein A-Sepharose 4B beads (Sigma), separated by sodium dodecyl sulfate (SDS)–8% polyacrylamide gel electrophoresis (PAGE), and visualized by autoradiography as described previously (13) (Fig. 2, lanes D and E). For comparison, [35S]methionine-labeled UL28 was synthesized in reticulocyte lysates (Promega) from UL28-encoding RNA synthesized in vitro by T3 RNA polymerase (Ambion) from linearized pECH82 (18) (Fig. 2, lanes A to C). The H85 anti-UL28 antiserum immunoprecipitated an 87-kDa protein from the infected cell extract (Fig. 2, lane E) as well as the in vitro-synthesized UL28 (Fig. 2, lane B), whereas the preimmune serum did not (Fig. 2, lanes A and D). The band of approximately 75 kDa in the in vitro-expressed reactions is related to UL28 and may result from premature translation termination or protein degradation.
FIG. 2.
In vitro and in vivo expression of UL28. Equivalent amounts of in vitro-translated UL28 were immunoprecipitated with either preimmune (A) or anti-UL28 (B) antisera or left neat (C). Vero cells were infected with HSV(KOS) at an MOI of 5, radiolabeled with [35S]methionine (New England Nuclear) between 6 and 9 h postinfection, and then harvested at 16 h as described previously (13). Cell extracts were immunoprecipitated with either preimmune (D) or anti-UL28 H85 (E) antisera. The mobilities of molecular mass markers are indicated to the left. UL28 and the 81-kDa protein are indicated to the right.
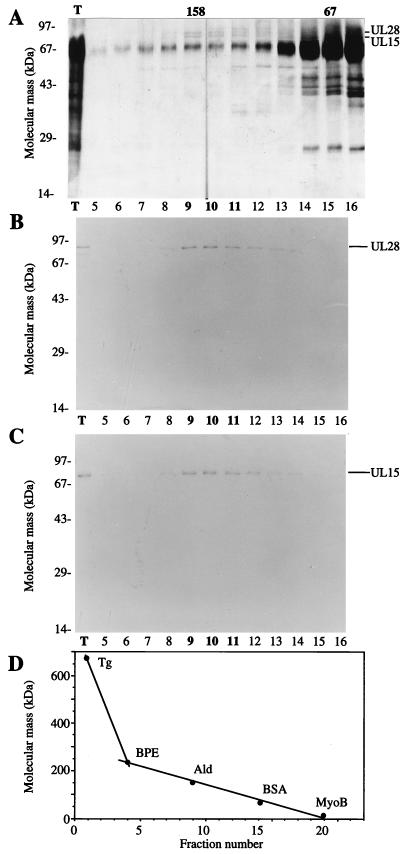
An 81-kDa protein was radiolabeled during infection and immunoprecipitated with UL28 from infected cell extracts (Fig. 2E). The mobility of this protein was consistent with the expected size of UL15 and not with UL28 products seen in Western blots (18). However, in order to conclude that the 81-kDa protein coimmunoprecipitating with HSV UL28 was HSV UL15, a series of protein purification procedures were performed. Approximately 109 Vero cells were infected with HSV(KOS) at a multiplicity of infection (MOI) of 5. At 24 h postinfection the cells were lysed in 10 ml of buffer A (10 mM Tris-Cl [pH 7.8], 1.5 mM MgCl2, 500 μM EDTA, 10 mM KCl, 1 mM dithiothreitol [DTT], and the following protease inhibitors: 1 mM phenylmethylsulfonyl fluoride [PMSF]), 5 μg of leupeptin/ml, 2.3 μg of ubenimex Bestatin/ml, 3.0 μg of aprotinin/ml, 2.7 μg of antipain/ml, and 3.3 μg of pepstatin/ml) by Dounce homogenization. Pelleted nuclei were lysed in 6 ml of buffer B (420 mM NaCl, 25 mM Tris-Cl [pH 7.8], 1.5 mM MgCl2, 25% glycerol, 200 μM EDTA, 1 mM DTT, and the protease inhibitors). The clarified lysate was adjusted to 300 mM NaCl, passed through a DEAE-cellulose column to remove DNA, and directly loaded onto a 5-ml hydroxyapatite column. Proteins were eluted with buffer E (125 mM potassium phosphate [pH 7.4], 100 μM EDTA, 1 mM DTT, 100 μM PMSF, 5% glycerol). The eluate was adjusted to 1 mM DTT and 25 mM potassium phosphate and then chromatographed on a Mono-Q HR5/5 column. Proteins were eluted with a linear gradient of 0 to 500 mM NaCl in buffer F (25 mM Tris-Cl [pH 7.8], 100 μM EDTA, 1 mM DTT, 100 μM PMSF). Fractions were screened with anti-UL15 AS7 and anti-UL28 H85 antisera. AS7 is an anti-glutathione S-transferase-UL15 exon 2 serum similar to AS9 (7). The peak fractions containing UL15 were diluted threefold with 1 mM DTT and loaded onto a native DNA-cellulose column. Proteins were eluted with a gradient of 0 to 500 mM NaCl in buffer F into tubes containing 5 μg of bovine serum albumin (BSA) to reduce nonspecific protein binding to the tubes. Fractions containing UL15 were pooled and concentrated to 0.5 ml by centrifugation in a Centricon 30 unit (Amicon). The sample was applied to a 5 to 20% sucrose gradient in buffer F plus 100 mM NaCl and 1 mM MgCl2 and then sedimented at 37,000 rpm in a Beckman SW50.1 rotor for 13 h. Fractions were collected, and proteins in these fractions were detected after SDS-PAGE by silver staining (Fig. 3A) and Western blotting (Fig. 3B and C). Silver staining of the SDS-PAGE-separated proteins revealed several bands in fractions 9 through 11. The most prominent band, at 67 kDa, corresponded to exogenous BSA carrier protein. Two other distinct proteins of 87 and 81 kDa were also detected. The 87-kDa protein was confirmed to be UL28 by Western blot analysis with H85 antiserum of a duplicate gel (Fig. 3B). The 81-kDa protein was confirmed to be UL15 by Western blotting with AS7 antiserum of a duplicate gel (Fig. 3C). There were no immunoreactive bands on the AS7 Western blot consistent with the size predicted for UL15.5. The peak fractions containing UL15 and UL28 corresponded to a sedimentation mass near the 157-kDa standard (Fig. 3D) based on the markers for sedimentation. This sedimentation mass was greater than the monomer size of either protein and was most consistent with a UL28-UL15 heterodimer. In contrast, UL25 sedimented as a monomer from whole infected cell extracts (22). Due to the low yield of this complex routinely obtained, it is not possible to accurately estimate its amount or purity.
FIG. 3.
Copurification of UL15 and UL28 from infected cell extracts. Proteins purified from HSV(KOS)-infected nuclear extracts were loaded onto the final sucrose gradient, and the indicated aliquots were separated by SDS-PAGE. Proteins were detected by silver staining (A), Western blotting with anti-UL28 H85 antiserum (B), or Western blotting with anti-UL15 AS7 antiserum (C). An aliquot of the total protein sample (T) loaded onto the sucrose gradient is indicated (A to C). Western blot analyses of proteins on nitrocellulose membranes included alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G (Boehringer-Mannheim) and the colorimetric conversion of 5-bromo-4-chloro-3-indolylphosphate (BCIP)-nitroblue tetrazolium (NBT) (Kirkegaard and Perry Laboratories). The peak fractions of UL15 and UL28 are indicated in bold type. (D) Sedimentation size standards were run in a parallel gradient: thyroglobulin (Tg, 670 kDa), β-phycoerythrin (BPE, 240 kDa), aldolase (Ald, 158 kDa), BSA (67 kDa), and myoglobin (MyoB, 17 kDa). The 158- and 67-kDa markers correspond to the peak fractions of aldolase and BSA.
We have demonstrated that HSV UL28 accumulates within the nuclei of infected cells yet fails to enter nuclei in the absence of other viral proteins. Comparison of UL28 with other proteins required for cleavage and packaging shows considerable variation in their propensities to enter nuclei. Although UL6 and UL15 accumulate in nuclei independently (3, 7, 9, 11, 20, 23), UL25 disperses evenly throughout the cell (7). UL32 is more prominently localized to the cytoplasm even during infection (5, 9). The dependence of UL28 on other proteins to enter the nuclei distinguishes UL28 from the other cleavage and packaging proteins for which information is available.
The ability of HSV UL15 to direct UL28 proteins from HSV and PRV corroborates the substantial conservation of primary sequences among UL15 and UL28 proteins from different herpesviruses, but more importantly it underscores a conservation of function. Biochemical evidence supports the hypothesis that UL28 interacts with UL15. The copurification of UL28 with UL15 through several chromatographic procedures and a sucrose sedimentation gradient strongly supports the hypothesis that UL15 and UL28 form a heterodimer.
Independent evidence regarding interactions of UL28 with UL15 comes from two paths of investigation. Human cytomegalovirus isolates resistant to a compound (TCRB) that inhibits DNA cleavage and packaging (8, 19) show mutations in their UL15 and UL28 homologues. The second line of evidence involves the finding that UL15 (15, 24) and UL28 (24) bind B capsids in infected cells. In cells infected with a UL28-null mutant, at least some forms of UL15 fail to bind capsids (15, 24). UL28 still entered nuclei and bound B capsids in cells infected with UL15 insertion viruses (24), suggesting that truncated UL15 proteins or other proteins transport UL28 to nuclei. In summary, the UL28-UL15 protein interaction can be demonstrated physically in the copurification of a complex and functionally in the transport of UL28 to the nucleus. The direct interaction of UL15 with UL28 is consistent with data from genetic, biochemical, and cellular experiments involving three different herpesviruses. Many questions regarding the role of this complex in the cleavage and packaging reaction remain.
Acknowledgments
This work was supported by Public Health Service grant AI-32501 (N.E.P.).
We thank John Wiley and his group for use of the microscope, Fred Homa for HSV(gCB) and C1 cells, Ann Kwong for pF1′-CMV, and Bernard Roizman for the UL15 antiserum used to initiate these studies.
REFERENCES
- 1.Addison C, Rixon F J, Preston V G. Herpes simplex virus type 1 UL28 gene product is important for the formation of mature capsids. J Gen Virol. 1990;71:2377–2384. doi: 10.1099/0022-1317-71-10-2377. [DOI] [PubMed] [Google Scholar]
- 2.Al-Kobaisi M F, Rixon F J, McDougall I, Preston V G. The herpes simplex virus UL33 gene product is required for the assembly of full capsids. Virology. 1991;180:380–388. doi: 10.1016/0042-6822(91)90043-b. [DOI] [PubMed] [Google Scholar]
- 3.Baines J D, Poon A P W, Rovnak J, Roizman B. The herpes simplex virus 1 UL15 gene encodes two proteins and is required for cleavage of genomic viral DNA. J Virol. 1994;68:8118–8124. doi: 10.1128/jvi.68.12.8118-8124.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavalcoli J D, Baghian A, Homa F L, Kousoulas K G. Resolution of genotypic and phenotypic properties of herpes simplex virus type 1 temperature-sensitive mutant (KOS) tsZ47: evidence for allelic complementation in the UL28 gene. Virology. 1993;197:23–34. doi: 10.1006/viro.1993.1563. [DOI] [PubMed] [Google Scholar]
- 5.Chang Y E, Poon A P W, Roizman B. Properties of the protein encoded by the UL32 open reading frame of herpes simplex virus 1. J Virol. 1996;70:3938–3946. doi: 10.1128/jvi.70.6.3938-3946.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hong Z, Ferrari E, Wright-Minogue J, Chase R, Risano C, Seelig G, Lee C-G, Kwong A D. Enzymatic characterization of hepatitis C virus NS3/4A complexes expressed in mammalian cells by using the herpes simplex virus amplicon system. J Virol. 1996;70:4261–4268. doi: 10.1128/jvi.70.7.4261-4268.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koslowski K M, Shaver P R, Wang X-Y, Tenney D J, Pederson N E. The pseudorabies virus UL28 protein enters the nucleus after coexpression with the herpes simplex virus UL15 protein. J Virol. 1997;71:9118–9123. doi: 10.1128/jvi.71.12.9118-9123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krosky P M, Underwood M R, Turk S R, Feng K W H, Jain R K, Ptak R G, Westerman A C, Biron K K, Townsend L B, Drach J C. Resistance of human cytomegalovirus to benzimidazole ribonucleosides maps to two open reading frames: UL89 and UL56. J Virol. 1998;72:4721–4728. doi: 10.1128/jvi.72.6.4721-4728.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lamberti C, Weller S K. The herpes simplex virus type 1 cleavage/packaging protein, UL32, is involved in efficient localization of capsids to replication compartments. J Virol. 1998;72:2463–2473. doi: 10.1128/jvi.72.3.2463-2473.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamberti C, Weller S K. The herpes simplex virus type 1 UL6 protein is essential for cleavage and packaging but not for genomic inversion. Virology. 1996;226:403–407. doi: 10.1006/viro.1996.0668. [DOI] [PubMed] [Google Scholar]
- 11.Patel A H, Rixon F J, Cunningham C, Davison A J. Isolation and characterization of herpes simplex virus type 1 mutants defective in the UL6 gene. Virology. 1996;217:111–123. doi: 10.1006/viro.1996.0098. [DOI] [PubMed] [Google Scholar]
- 12.Pederson, N. E. Unpublished data.
- 13.Pederson N E, Enquist L W. Overexpression in bacteria and identification in infected cells of the pseudorabies virus protein homologous to herpes simplex virus type 1 ICP18.5. J Virol. 1991;65:3746–3758. doi: 10.1128/jvi.65.7.3746-3758.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poon A P W, Roizman B. Characterization of a temperature-sensitive mutant of the UL15 open reading frame of herpes simplex virus 1. J Virol. 1993;67:4497–4503. doi: 10.1128/jvi.67.8.4497-4503.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salmon B, Baines J D. Herpes simplex virus DNA cleavage and packaging: association of multiple forms of UL15-encoded proteins with B capsids requires at least the UL6, UL17, and UL28 genes. J Virol. 1998;72:3045–3050. doi: 10.1128/jvi.72.4.3045-3050.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salmon B, Cunningham C, Davison A J, Harris W J, Baines J D. The herpes simplex virus type 1 UL17 gene encodes virion tegument proteins that are required for cleavage and packaging of viral DNA. J Virol. 1998;72:3779–3788. doi: 10.1128/jvi.72.5.3779-3788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sherman G, Bachenheimer S L. DNA processing in temperature-sensitive morphogenic mutants of HSV-1. Virology. 1987;158:427–430. doi: 10.1016/0042-6822(87)90214-5. [DOI] [PubMed] [Google Scholar]
- 18.Tengelsen L A, Pederson N E, Shaver P R, Wathen M W, Homa F L. Herpes simplex virus type 1 DNA cleavage and encapsidation require the product of the UL28 gene: isolation and characterization of two UL28 deletion mutants. J Virol. 1993;67:3470–3480. doi: 10.1128/jvi.67.6.3470-3480.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Underwood M R, Harvey R J, Stanat S C, Hemphill M L, Miller T, Drach J C, Townsend L B, Biron K K. Inhibition of human cytomegalovirus DNA maturation by a benzimidazole ribonucleoside is mediated through the UL89 gene product. J Virol. 1998;72:717–725. doi: 10.1128/jvi.72.1.717-725.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ward P L, Ogle W O, Roizman B. Assemblons: nuclear structures defined by aggregation of immature capsids and some tegument proteins of herpes simplex virus 1. J Virol. 1996;70:4623–4631. doi: 10.1128/jvi.70.7.4623-4631.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weller S K, Carmichael E P, Aschman D P, Goldstein D J, Schaffer P A. Genetic and phenotypic characterization of mutants in four essential genes that map to the left half of HSV-1 UL DNA. Virology. 1987;161:198–210. doi: 10.1016/0042-6822(87)90186-3. [DOI] [PubMed] [Google Scholar]
- 22.Yamanaka, G., and T. Wilson. Unpublished data.
- 23.Yu D, Weller S K. Genetic analysis of the UL15 gene locus for the putative terminase of herpes simplex virus type 1. Virology. 1998;243:32–44. doi: 10.1006/viro.1998.9041. [DOI] [PubMed] [Google Scholar]
- 24.Yu D, Weller S K. Herpes simplex virus type 1 cleavage and packaging proteins UL15 and UL28 are associated with B but not C capsids during packaging. J Virol. 1998;72:7428–7439. doi: 10.1128/jvi.72.9.7428-7439.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]