Abstract
We present detailed studies of human immunodeficiency virus (HIV)-specific cytotoxic T-lymphocyte (CTL) responses to clade A or C HIV type 1 in three donors infected in East Africa. We define several novel non-clade B CTL epitopes, including some restricted by HLA alleles common in Africans. Although cross-clade CTL recognition of these epitopes does occur, recognition can also be highly clade specific.
Of the 30 million people thought to be infected with human immunodeficiency virus (HIV) worldwide, the majority live in sub-Saharan Africa (32). Geographically distinct epidemics have been characterized by the dominance of particular HIV type 1 (HIV-1) subtypes; these have been separated into clades on the basis of their sequences, which differ between clades by approximately 30% for env and 14% for gag (14). In Europe and North America most individuals are infected with clade B virus, which has therefore been the principal focus of virological and immunological investigations. However, a much more extensive diversity of HIV-1 subtypes is found in sub-Saharan Africa, and the majority of these belong to non-B clades (14). Vaccine development has focused almost exclusively on clade B immunogens, which raises the concern that these vaccines may not be effective in the areas of the world where they are most needed. It is now believed that vaccine efficacy will depend on the generation of broadly cross-reactive neutralizing antibody and cytotoxic T-lymphocyte (CTL) responses (1, 4). The extent to which different HIV-1 strains elicit cross-clade cellular immune responses has only recently been addressed. HIV-1-specific CTLs which cross-react with diverse HIV clades were observed in some (but not all) recipients of a clade B Env-based vaccine (9). More recently, several studies have demonstrated that cross-clade CTL activity can be detected both in clade B-infected Caucasians and in African and Thai patients naturally infected with subtypes A, C, D, E, and G (5, 7, 21, 22). Currently, however, no CTL epitopes have been defined for any subtype other than clade B (18). CTL epitope mapping is essential for identifying the immunodominant responses to different HIV strains and for reaching an understanding of how antigenic variation affects the immune response. Furthermore, since the selection of viral peptides for presentation to CTLs is determined by HLA class I alleles, interethnic variation in class I allele frequency should also be considered. Identification of the immunodominant CTL epitopes in the viruses prevailing in different populations should assist greatly in monitoring the immunogenicity of vaccine candidates when they become available.
Here we describe detailed studies of the fine specificity of CTL responses to HIV in three individuals infected with HIV-1 in East Africa, where the dominant subtype is clade A, with some clade C and D infections (14). We identified several new epitopes from these African virus strains using recombinant vaccinia viruses expressing HIV Gag and peptides based on the consensus sequences of HIV-1 clades A, C, and D (Los Alamos database [17]), and we show that while cross-clade CTL responses can often be elicited, recognition of individual peptide epitopes may be highly clade specific. We also demonstrate that when the CTLs were studied by the recently described technique of staining CD8+ T cells with soluble peptide-HLA tetrameric complexes, CTLs from a clade C-infected donor stained only with HLA-A2 complexed with the A or C clade version of the dominant HLA-A2-restricted epitope in p17, which differs by just a single amino acid from the previously defined B clade epitope.
Subjects who were infected with HIV-1 in East Africa (Kenya, Rwanda, or Uganda) were recruited from the HIV clinic in Oxford, United Kingdom (UK). Patient 1 (HLA-A1, -A*0201, -B7, and -B37) is a British man who presented recently with AIDS and is thought to have been infected some years ago in Uganda. Patient 2 (HLA-A*0201, -A24, -B44, and -B55) is a British woman who presented with acute HIV infection on returning from a vacation in Kenya in 1995 and is currently asymptomatic. Patient 3 (HLA-A*2601, -A*6802, -B70, and B*8101) is a Kenyan woman who was diagnosed as HIV positive after presenting with lymphadenopathy to the Oxford clinic and is currently well. Proviral DNA was extracted from peripheral blood mononuclear cells from these donors, and the envelope subtype of their infecting viruses was determined by heteroduplex mobility assays (HMA). Sample DNA was amplified by nested PCR, using first-round primers spanning the first exon of rev to the N-terminal half of gp41 and then second-round primers amplifying env V1 to V5, followed by HMA with reference plasmids from HIV strains A to H, as previously described (8). The patients’ infecting subtypes are as follows: patient 1, subtype C, and patients 2 and 3, subtype A. HLA typing was carried out by amplification refractory mutation system (ARMS)-PCR with sequence-specific primers (19).
Bulk CTL cultures were generated by restimulation with autologous virus in phytohemagglutinin blasts in the presence of interleukin-2, as previously described (24), and were tested after 2 weeks for specific lysis of autologous Epstein-Barr virus-transformed B-lymphoblastoid cell lines infected with recombinant vaccinia viruses (multiplicity of infection, 5 PFU/cell) expressing full-length gag sequences from HIV-1 clades A (A1, 28-12 [Ugandan isolate], and A2, 45-5 [Thai isolate]), B (HXB2), and C (45-31 [Ugandan isolate]) (22). A vaccinia virus construct expressing influenza virus polymerase (PB2) was used as a control, and effector/target (E/T) ratios were 50:1 to 100:1.
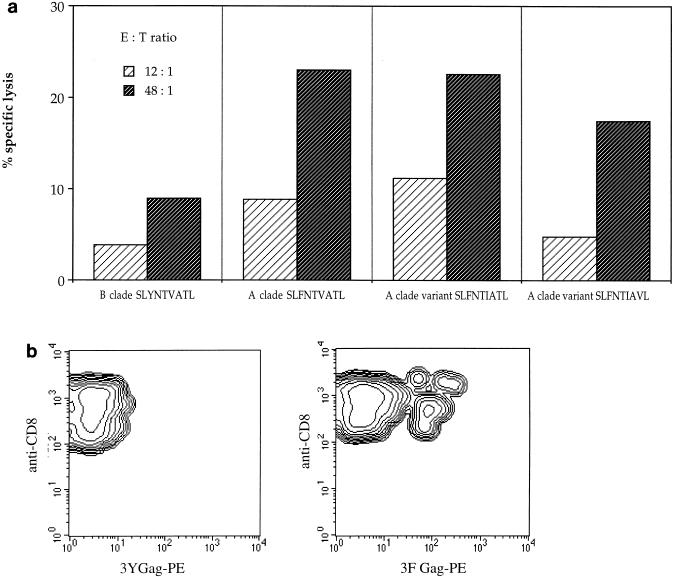
CTL cultures from patient 1, who was infected with C clade HIV-1, recognized A and C but not B clade Gag proteins presented by target cells matched for HLA-A*0201 and -B7 (data not shown). The immunodominant HLA-A2-restricted epitope from clade B HIV-1 p17:77-85 is recognized by approximately two-thirds of HLA-A*0201 HIV-1-seropositive individuals (12); in Caucasian donors with HLA-A*0201, there is an inverse correlation between plasma viral load and the numbers of CTLs responding to this epitope (26). The consensus amino acid sequence of the epitope in clade B from the Los Alamos database (17) is SLYNTVATL: in most A and C clade isolates, this region contains a Y→F substitution at position 3. The F3 variant is only sometimes recognized by CTLs from B clade-infected individuals (6, 30). In contrast, CTLs from patient 1 responded only to the A and C clade variant (F3) of this epitope. The fine specificity of this response was further characterized by using the SLFNTVATL peptide to generate a CTL line, as described previously (20). These CTLs had an absolute requirement for F at position 3 but were able to tolerate other amino acid substitutions (I6 and V8), which are found in some A and C clade isolates (Fig. 1a). The clade specificity of this response was further confirmed by fluorescence-activated cell sorter analysis of a p17-specific CTL line from patient 1, cells of which were stained with HLA-A*0201 tetrameric complexes refolded around either the clade A and C or clade B version of the p17 peptide, as described previously (3, 26). Patient 1 p17-specific CTLs were doubly positive for CD8 and HLA-A*0201–SLFNTVATL tetrameric complexes but not for tetramers refolded around SLYNTVATL (Fig. 1b), thereby demonstrating the sensitivity to a single amino acid substitution of both the CTL response and the tetramer staining.
FIG. 1.
Recognition of an HLA-A2-restricted peptide in A clade p17 by patient 1 CTLs. (a) Gag-specific CTLs from donor 1 specifically lysed HLA-A2-matched target cells pulsed with multiple variants of the A clade version of the previously defined HLA-A2-restricted peptide in p17, but not the B clade index version (all peptides were prepulsed for 1 h at 10 μM). (b) p17-specific CTLs from patient 1 were stained with the HLA-A*0201 tetramer complexed with either the B clade version (Y3; shown as 3Y) of the known p17 epitope peptide or the A clade version (F3; shown as 3F). The plots show the mononuclear cell population costained with tetramer (phycoerythrin [PE]) and anti-CD8 (fluorescein isothiocyanate). A double-positive population is seen only with the F3 tetramer.
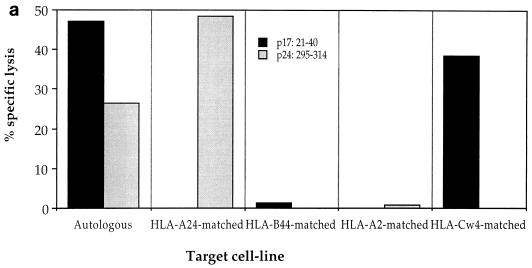
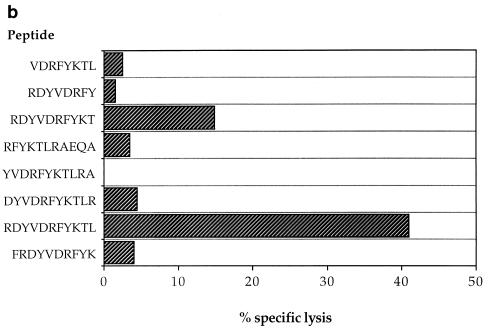
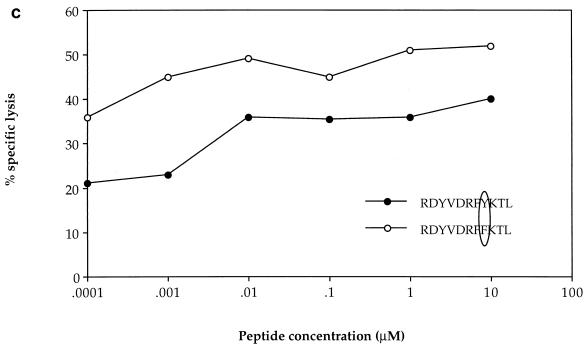
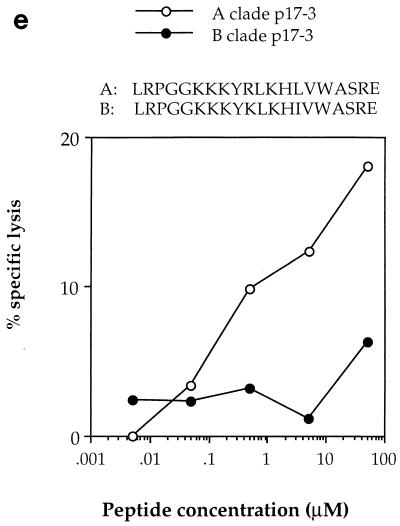
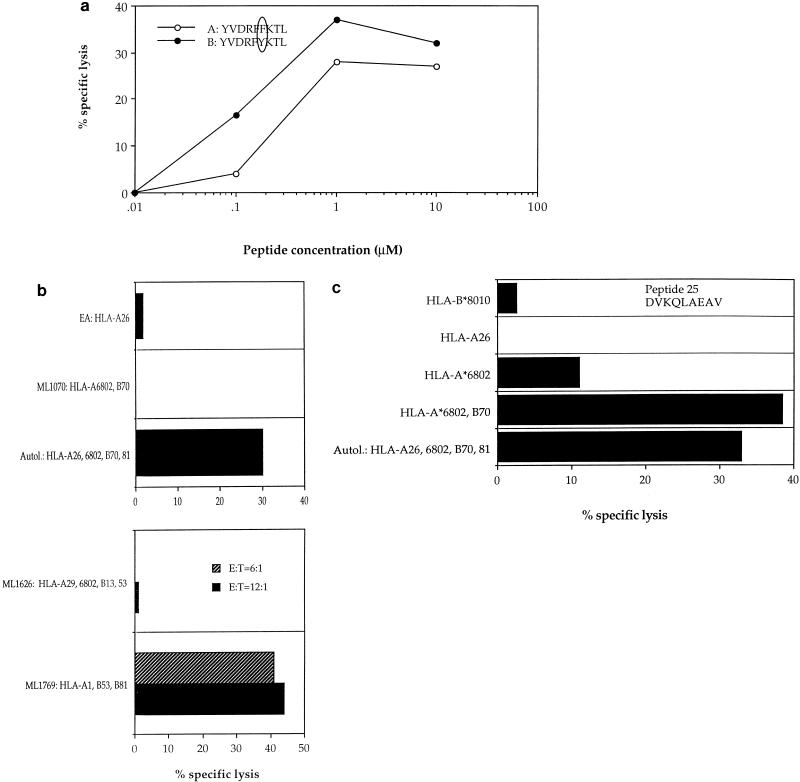
HIV-specific CTLs from patient 2, with A clade infection, had previously been shown to recognize autologous targets expressing gag sequences from clades A and C but not clade B (22). These gag-specific responses were shown to be restricted through HLA-A*0201, -A24, and -B44 (data not shown), and two of the epitopes recognized were mapped by using overlapping 20-mer peptides based on the clade A gag consensus sequence [p17 peptides; Peptide and Protein Research Cons, Exeter, UK, and Genosys Biotechnologies (Europe) Ltd., Pampisford, Cambridge, UK; p24 peptides were kindly donated by the AIDS Reagent Programme of the Medical Research Council (MRC). The HLA restriction of CTL responses to two 20-mer peptides, p17:21-40 and p24:295-314, was mapped to HLA-Cw4 and HLA-A24, respectively (Fig. 2a). The HLA-A24-restricted CTL response to the p24 peptide showed significant cross-reactivity against the equivalent B clade peptide, so the optimal epitope was mapped to an 11-mer, RDYVDRFFKTL, with truncated B clade peptides (which had been made by 9-fluorenylmethoxycarbonyl chemistry with a Zinnser peptide synthesizer and previously used to map other CTL epitopes in this region of p24 [8]) (Fig. 2b). Peptide titrations with a CTL line from donor 2 confirmed significant cross-reactivity between the A and B or D clade versions of the optimal epitope, in which there is an F→Y substitution at position 8 (Fig. 2c). This is one amino acid longer than an A24-restricted epitope, p24:297-306 (DYVDRFYKTL), which was previously identified in HIV-1 clade B-infected individuals (13); however, the shorter peptide was not recognized by CTLs from our donor (Fig. 2b). These epitopes lie in a highly conserved region of p24gag, the major homology region (MHR), where a number of other CTL epitopes have recently been mapped (25). The response to p17 was found to be restricted by HLA-Cw4 (Fig. 2a), although the optimal epitope has not yet been defined. Recognition of the A clade 20-mer was compared with that of the equivalent B clade 20-mer peptide (Fig. 2d and e); no cross-reactivity was observed. This suggests that the optimal epitope may include one or both of the two amino acid substitutions in the B clade peptide, which abrogate recognition by A clade-specific CTLs.
FIG. 2.
Recognition of A clade Gag peptides through HLA-A24 and -Cw4 by patient 2 CTLs. (a) Bulk CTL cultures from donor 2 were tested for recognition of autologous and target cell lines matched through a single class I HLA molecule presenting two 20-mer A clade Gag peptides (used at 10 μM; E/T ratio, 50:1). (b) Patient 2 bulk CTL cultures were tested for recognition of target cells matched through HLA-A24 pulsed with peptides representing truncations of the B clade version of the epitope containing residues 295 to 314 (used at 10 μM; E/T ratio, 36:1). (c) A CTL line from donor 2 specific for the HLA-A24-restricted peptide was tested for recognition of HLA-A24-matched targets with various concentrations of the A and B clade peptides in the assay (E/T ratio, 25:1). (d) A CTL line from donor 2 specific for the HLA-Cw-4-restricted p17 peptide was tested for recognition of autologous targets presenting either the A clade 20-mer or the equivalent B clade 20-mer (peptides used at 10 μM). (e) The same p17-specific CTL line from donor 2 was used to study recognition of target cells presenting either the A or B clade 20-mers at different concentrations in the assay.
The class I haplotype of A clade-infected patient 3 (HLA-A*2601, -*6802, -B70, and -*8101) contains HLA molecules which are more typically found in East Africans: the only reported HIV-1 CTL epitope for any of these alleles is an HLA-A26-restricted Gag peptide (11). CTL responses were studied with both overlapping consensus A peptides and 9-mer peptides based on sequences in gag and pol of clades A and D which had previously been used to identify HLA-A*6802-restricted CTLs (29). Initial screening of 49 pooled peptides revealed strong bulk CTL responses to autologous B cells presenting two pools of peptides, one comprising predicted clade A epitopes and the other comprising their equivalents in clade D (data not shown). The responses were mapped to two individual peptides in the pools, Gag residues 298 to 306 and Pol residues 517 to 525. CTL lines were then established by peptide restimulation. The Gag-specific CTL line recognized both the index peptide, which is conserved between clades A and C, and the B or D clade variant, which has an F→Y substitution at position 6 (Fig. 3a). Peptide titrations demonstrated a modest preference for the B or D clade sequence. Truncation of the index peptide at either terminus abrogated the CTL response (data not shown), indicating that the minimal epitope was contained in the sequence YVDRFYKTL, which also lies in the conserved MHR of p24gag (25). CTLs from donor 3 recognized this peptide when presented by targets HLA matched at either HLA-A26 or -B70 and not at HLA-A*6802 (not shown). The presence of valine at position 2 and leucine at the carboxy terminus of the peptide is consistent with the predicted motif for an HLA-A26-restricted epitope, although it does not have a negatively charged residue at position 1 (11). Little is known about the nature of peptides presented by HLA-B70, although it is found frequently in Africans (28).
FIG. 3.
Recognition of novel CTL epitopes presented by HLA-A26, -B70, and -B*8101 by CTLs from donor 3. (a) A CTL line from donor 3 was tested for recognition of autologous targets with various concentrations of the A and B clade equivalents of a Gag peptide presented by HLA-A26 and -B70 (E/T ratio, 20:1). (b) Bulk CTL cultures from donor 3 were shown to recognize a 20-mer peptide from A clade p24 only when presented by target cells matched through HLA-B*8101. (c) Peptide-specific CTLs from donor 3 recognized a 9-mer peptide from D clade Pol when presented by target cells matched through HLA-B70 (peptide used at 10 μM; E/T ratio, 20:1).
CTL responses of patient 3 to overlapping A clade Gag peptides on autologous targets revealed responses to another Gag epitope within p24 (positions 168 to 187) (SALSEGATPQDLNMMLNIVG); this was restricted by HLA-B*8101 (Fig. 3b). HLA-B*8101 has only recently been identified as a distinct allele by PCR with sequence-specific primers in individuals of African origin (31), and little is known about its peptide-binding requirements. Therefore, fine-mapping studies will be required to define precisely the minimal epitope in this peptide.
The response of donor 3 to the Pol 9-mer peptides, DVKQLTEVV (A clade, positions 470 to 478) and DVKQLAEAV (D clade), was also restricted by HLA-B70 (Fig. 3c), rather than HLA-A*6802 as anticipated. Bulk CTL cultures from donor 3 recognized the A and D clade variants of the Pol epitope, which differ by two residues, equally well (data not shown). With a CTL line which had been generated by repeated restimulation with autologous B cells pulsed with the D clade peptide, there was no cross-reactive recognition of the B clade equivalent of this epitope, which has the sequence DVKQLTEAV (data not shown).
These studies demonstrate the existence of several novel clade-specific and cross-reactive CTL epitopes in individuals infected with HIV in Africa (Table 1), several of which are expected to be important epitopes in African populations on the basis of class I allele frequency (2). Of particular interest is the identification of new epitopes in the highly conserved MHR of p24gag restricted by HLA-A24, -A26, and -B70, which overlap several other known epitopes (13, 15, 25), confirming the immunogenicity of this region. Predictably, CTL responses to some of these epitopes showed the greatest degree of cross-reactivity to diverse HIV-1 clades. Our data therefore support earlier reports of cross-clade reactivity in individuals infected with diverse HIV-1 subtypes and suggest that an immunogen incorporating such conserved epitopes could be used to vaccinate individuals from different ethnic groups.
TABLE 1.
CTL epitopes identified in patients infected with HIV-1
| HLA restriction | Protein | Amino acid positions | Clade | Consensus sequence |
|---|---|---|---|---|
| HLA-A*201 | p17gag | 77–85 | A | SLFNTVATL |
| B | --Y------ | |||
| C | --?------ (F/H/Y at position 3) | |||
| D | --Y------ | |||
| HLA-Cw4 | p17gag | 21–40 | A | LRPGGKKKYRLKHLVWASRE |
| B | -------K-KL--I------ | |||
| C | -------h-MI--L------ | |||
| D | -------K-?L--I------ | |||
| HLA-A24 | p24gag | 296–306 | A | RDYVDRFFKTL |
| B | -------Y--- | |||
| C | ----------- | |||
| D | -------Y--- | |||
| HLA-A26 or -B70 | p24gag | 298–306 | A | YVDRFFKTL |
| B | -----Y--- | |||
| C | --------- | |||
| D | -----Y--- | |||
| HLA-B*8101 | p24gag | 179–188 | A | SALSEGATPQDLNMMLNIVG |
| B | -------------T---T-- | |||
| C | -------------T---T-- | |||
| D | -------------T---T-- | |||
| HLA-B70 | Pol (RTa) | 470–478 | A | DVKQLTEVV |
| B | -------A- | |||
| C | --?---?-- | |||
| D | -----A-A- |
RT, reverse transcriptase.
However, while cross-clade reactivity may often be elicited with polyclonal CTLs tested against recombinant vaccinia virus-infected targets, studies at the level of individual peptide epitopes revealed that single amino acid substitutions representing sequence variations in different clades may be sufficient to abrogate CTL responses. For example, CTLs can readily distinguish between the A or C (F3) and B clade (Y3) sequences of the immunodominant HLA-A*0201-restricted p17 epitope, even though these peptides bind equally well to HLA-A*0201 (30). This is strikingly demonstrated by the absence of any patient 1 CTL staining with the HLA-A2 tetramer complexed with the B clade peptide (Fig. 1b), which additionally shows that tetramer staining can be sensitive to the level of single amino acid substitutions. Our findings suggest that vaccines based on individual peptide epitopes may be of limited use and that precise definition of CTL epitopes for different HIV-1 subtypes is necessary to determine what sequence variation is biologically significant.
We have focused on CTL responses to Gag and to a lesser extent Pol, as these are known to contain most of the conserved epitopes (23) and CTL responses to Gag have been associated with delayed progression of HIV disease, suggesting that they are an important component of protective immunity (16, 27). More recently, quantification of CTLs by the soluble-peptide–tetramer technique showed an inverse correlation between the numbers of HLA-A2-restricted Gag-specific CTLs and plasma viral load: the correlation was stronger if Pol-specific CTLs were also included (26). Furthermore, recipients of a recombinant vaccine expressing Gag and Env were found to have more extensive cross-clade responses than those vaccinated with a construct expressing Env alone (9). Clade A HIV-1 is the dominant virus strain in East Africa, but these epitopes are likely also to be present in clade E virus, which is now the major strain in southeast Asia, since most of the internal proteins from E clade HIV-1 are from an A clade parent virus (10).
In summary, we have defined both conserved and unique non-B clade CTL epitopes in patients infected in Africa. Further studies should be directed at identifying the CTL responses to the HIV clades prevailing in different populations, taking into account the dominant HLA types of each population, in order to maximize the likelihood of developing a vaccine which could be used and tested globally.
Acknowledgments
Lucy Dorrell and Tao Dong contributed equally to this study.
This work was supported by the MRC of the UK and the Elizabeth Glaser Pediatric AIDS Foundation. L.D. is an MRC Training Fellow, and S.R.-J. is an MRC Senior Fellow and an Elizabeth Glaser Scientist of the Pediatric AIDS Foundation.
We are grateful to Harvey Holmes and Programme EVA for the A clade p24 overlapping peptides, to Kati DeGleria for synthesis of some of the truncated peptides, and to Linda Barber for helpful advice. The HMA kit was supplied by the MRC AIDS Reagent Programme in collaboration with the NIH AIDS Research Reference and Reagent Program.
REFERENCES
- 1.Ada G L, McElrath M J. HIV-1 vaccine-induced cytotoxic T-cell responses: potential role in vaccine efficacy. AIDS Res Hum Retroviruses. 1997;13:205–210. doi: 10.1089/aid.1997.13.205. [DOI] [PubMed] [Google Scholar]
- 2.Allsopp C E, Harding R M, Taylor C, Bunce M, Kwiatkowski D, Anstey N, Brewster D, McMichael A J, Greenwood B M, Hill A V. Interethnic genetic differentiation in Africa: HLA class I antigens in The Gambia. Am J Hum Genet. 1992;50:411–421. [PMC free article] [PubMed] [Google Scholar]
- 3.Altman J, Moss P A H, Goulder P, Barouch D, McHeyzer-Williams M, Bell J I, McMichael A J, Davis M M. Direct visualization and phenotypic analysis of virus-specific T lymphocytes in HIV-infected individuals. Science. 1996;274:94–96. [Google Scholar]
- 4.Bangham C R M, Phillips R E. What is required of an HIV vaccine? Lancet. 1997;350:1617–1621. doi: 10.1016/S0140-6736(97)03258-3. [DOI] [PubMed] [Google Scholar]
- 5.Betts M R, Krowka J, Santamaria C, Balsamo K, Gao F, Mulundu G, Luo C, N’Gandu N, Sheppard H, Hahn B H, Allen S, Frelinger J A. Cross-clade human immunodeficiency virus (HIV)-specific cytotoxic T-lymphocyte responses in HIV-infected Zambians. J Virol. 1997;71:8908–8911. doi: 10.1128/jvi.71.11.8908-8911.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brander C, Hartman K E, Trocha A K, Jones N G, Johnson R P, Korber B, Wentworth P, Buchbinder S P, Wolinsky S, Walker B D, Kalams S A. Lack of strong immune selection pressure by the immunodominant, HLA-A*0201-restricted cytotoxic T lymphocyte response in chronic human immunodeficiency virus-1 infection. J Clin Investig. 1998;101:2559–2566. doi: 10.1172/JCI2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao H, Kanki P, Sankalé J-L, Dieng-Sarr A, Mazzara G P, Kalams S A, Korber B, Mboup S, Walker B D. Cytotoxic T-lymphocyte cross-reactivity among different human immunodeficiency virus type 1 clades: implications for vaccine development. J Virol. 1997;71:8615–8623. doi: 10.1128/jvi.71.11.8615-8623.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delwart E L, Shpaer E G, Louwagie J, McCutchan F E, Grez M, Rubsamen Waigmann H, Mullins J I. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science. 1993;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- 9.Ferrari G, Humphrey W, McElrath M J, Excler J L, Duliege A M, Clements M L, Corey L C, Bolognesi D P, Weinhold K J. Clade B-based HIV-1 vaccines elicit cross-clade cytotoxic T lymphocyte reactivities in uninfected volunteers. Proc Natl Acad Sci USA. 1997;94:1396–1401. doi: 10.1073/pnas.94.4.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao F, Robertson D L, Morrison S G, Hui H, Craig S, Decker J, Fultz P N, Girard M, Shaw G M, Hahn B H, Sharp P M. The heterosexual human immunodeficiency virus type 1 epidemic in Thailand is caused by an intersubtype (A/E) recombinant of African origin. J Virol. 1996;70:7013–7029. doi: 10.1128/jvi.70.10.7013-7029.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goulder P, Conlon C, McIntyre K, McMichael A. Identification of a novel human leukocyte antigen A26-restricted epitope in a conserved region of Gag. AIDS. 1996;10:1441–1443. doi: 10.1097/00002030-199610000-00025. [DOI] [PubMed] [Google Scholar]
- 12.Goulder P J R, Sewell A K, Lalloo D G, Price D A, Whelan J A, Evans J, Taylor G P, Luzzi G, Giangrande P, Phillips R E, McMichael A J. Patterns of immunodominance in HIV-1 specific cytotoxic T lymphocyte responses in two human histocompatibility leukocyte antigens (HLA)-identical siblings with HLA-A*0201 are influenced by epitope mutation. J Exp Med. 1997;185:1423–1433. doi: 10.1084/jem.185.8.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikeda-Moore Y, Tomiyama H, Miwa K, Oka S, Iwamoto A, Kaneko Y, Takiguchi M. Identification and characterisation of multiple HLA-A24-restricted HIV-1 CTL epitopes: strong epitopes are derived from V regions of HIV-1. J Immunol. 1997;159:6242–6252. [PubMed] [Google Scholar]
- 14.Janssens W, Buve A, Nkengasong J N. The puzzle of HIV-1 subtypes in Africa. AIDS. 1997;11:705–712. doi: 10.1097/00002030-199706000-00002. [DOI] [PubMed] [Google Scholar]
- 15.Johnson R P, Trocha A, Yang L, Mazzara G P, Panicali D L, Buchanan T M, Walker B D. HIV-1 gag-specific cytotoxic T lymphocytes recognize multiple highly conserved epitopes. Fine specificity of the gag-specific response defined by using unstimulated peripheral blood mononuclear cells and cloned effector cells. J Immunol. 1991;147:1512–1521. [PubMed] [Google Scholar]
- 16.Klein M R, van Baalen C A, Holwerda A M, Kerkhof Garde S R, Bende R J, Keet I P, Eeftinck-Schattenkerk J K, Osterhaus A D, Schuitemaker H, Miedema F. Kinetics of Gag-specific cytotoxic T lymphocyte responses during the clinical course of HIV-1 infection: a longitudinal analysis of rapid progressors and long-term asymptomatics. J Exp Med. 1995;181:1365–1372. doi: 10.1084/jem.181.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korber B, Hahn B, Foley B, Mellors J W, Leitner T, McCutchan M G F, Kuiken C. Human retroviruses and AIDS. Los Alamos, N. Mex: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 1997. [Google Scholar]
- 18.Korber B, Moore J, Brander C, Walker B, Haynes B, Koup R. HIV molecular immunology database. Los Alamos, N. Mex: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 1997. [Google Scholar]
- 19.Krausa P, Bodmer J G, Browning M J. Defining the common subtypes of HLA A9, A10, A28 and A19 by use of ARMS/PCR. Tissue Antigens. 1993;42:91–99. doi: 10.1111/j.1399-0039.1993.tb02243.x. [DOI] [PubMed] [Google Scholar]
- 20.Lalvani A, Dong T, Ogg G, Patham A A, Newell H, Hill A V, McMichael A J, Rowland-Jones S. Optimization of a peptide-based protocol employing IL-7 for in vitro restimulation of human cytotoxic T lymphocyte precursors. J Immunol Methods. 1997;210:65–77. doi: 10.1016/s0022-1759(97)00177-4. [DOI] [PubMed] [Google Scholar]
- 21.Lynch J A, de Souza M, Robb M D, Markowitz L, Nitayaphan S, Sapan C V, Mann D L, Birx D L, Cox J H. Cross-clade cytotoxic T-cell response to HIV-1 proteins among HLA disparate North Americans and Thais. J Infect Dis. 1998;178:1040–1046. doi: 10.1086/515652. [DOI] [PubMed] [Google Scholar]
- 22.McAdam S, Kaleebu P, Krausa P, Goulder P, French N, Collin B, Blanchard T, Whitworth J, McMichael A, Gotch F M. Cross-clade recognition of p55 by CTL in HIV-1 infection. AIDS. 1998;12:571–579. doi: 10.1097/00002030-199806000-00005. [DOI] [PubMed] [Google Scholar]
- 23.McMichael, A. J., and B. D. Walker. 1994. Cytotoxic T lymphocyte epitopes: implications for HIV vaccines. AIDS 8(Suppl. 1):S155–S173.
- 24.Nixon D F, Townsend A R M, Elvin J G, Rizza C R, Gallwey J, McMichael A J. HIV-1 gag-specific cytotoxic T lymphocytes defined with recombinant vaccinia virus and synthetic peptides. Nature. 1988;336:484–487. doi: 10.1038/336484a0. [DOI] [PubMed] [Google Scholar]
- 25.Ogg G S, Dong T, Hansasuta P, Dorrell L, Clarke J, Coker R, Luzzi G, Conlon C, McMichael A J, Rowland-Jones S L. Four novel CTL epitopes in the highly-conserved MHR region of HIV-1 gag, restricted by B*4402, B*1401, A*2601 and B*70. AIDS. 1998;12:1561–1563. doi: 10.1097/00002030-199812000-00026. [DOI] [PubMed] [Google Scholar]
- 26.Ogg G S, Xin J, Bonhoeffer S, Dunbar P R, Nowak M A, Monard S, Segal J P, Cao Y, Rowland-Jones S L, Cerundolo V, Hurley A, Markowitz M, Ho D D, Nixon D F, McMichael A J. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma viral RNA load. Science. 1998;279:2103–2106. doi: 10.1126/science.279.5359.2103. [DOI] [PubMed] [Google Scholar]
- 27.Rinaldo C, Huang X-L, Fan Z, Ding M, Beltz L, Logar A, Panicali D, Mazzara G, Liebmann J, Cottrill M, Gupta P. High levels of anti-human immunodeficiency virus type 1 (HIV-1) memory cytotoxic T-lymphocyte activity and low viral load are associated with lack of disease in HIV-1-infected long-term nonprogressors. J Virol. 1995;69:5838–5842. doi: 10.1128/jvi.69.9.5838-5842.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez S G, Bei M, Inamdar A, Stewart D, Johnson A H, Hurley C K. Molecular and serological characterization of HLA-B71 in association with different class I haplotypes or in different ethnic groups. Tissue Antigens. 1996;47:58–62. doi: 10.1111/j.1399-0039.1996.tb02514.x. [DOI] [PubMed] [Google Scholar]
- 29.Rowland-Jones S L, Dong T, Fowke K R, Kimani J, Krausa P, Newell H, Blanchard T, Ariyoshi K, Oyugi J, Ngugi E, Bwayo J, MacDonald K S, McMichael A J, Plummer F A. Cytotoxic T-cell responses to multiple conserved epitopes in HIV-resistant prostitutes in Nairobi. J Clin Investig. 1998;102:1758–1765. doi: 10.1172/JCI4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sewell A K, Harcourt G C, Goulder P J, Price D A, Phillips R E. Antagonism of cytotoxic T lymphocyte-mediated lysis by natural HIV-1 altered peptide ligands requires simultaneous presentation of agonist and antagonist peptides. Eur J Immunol. 1997;27:2323–2329. doi: 10.1002/eji.1830270929. [DOI] [PubMed] [Google Scholar]
- 31.Vilches C, Sanz L, de Pablo R, Moreno M E, Puente S, Kreisler M. Molecular characterization of the new alleles HLA-B*8101 and B*4407. Tissue Antigens. 1996;47:139–142. doi: 10.1111/j.1399-0039.1996.tb02527.x. [DOI] [PubMed] [Google Scholar]
- 32.World Health Organization. HIV/AIDS: the global epidemic. World Health Forum. 1997;18:369–372. [PubMed] [Google Scholar]