Abstract
High-throughput sequencing has become a standard first-tier approach for both diagnostics and research-based genetic testing. Consequently, this hypothesis-free testing manner has revealed the true breadth of clinical features for many established genetic disorders, including Meier-Gorlin syndrome (MGORS). Previously known as ear-patella short stature syndrome, MGORS is characterized by growth delay, microtia, and patella hypo/aplasia, as well as genital abnormalities, and breast agenesis in females. Following the initial identification of genetic causes in 2011, a total of 13 genes have been identified to date associated with MGORS. In this review, we summarise the genetic and clinical findings of each gene associated with MGORS and highlight molecular insights that have been made through studying patient variants. We note interesting observations arising across this group of genes as the number of patients has increased, such as the unusually high number of synonymous variants affecting splicing in CDC45 and a subgroup of genes that also cause craniosynostosis. We focus on the complicated molecular genetics for DONSON, where we examine potential genotype-phenotype patterns using the first 3D structural model of DONSON. The canonical role of all proteins associated with MGORS are involved in different stages of DNA replication and in addition to summarising how patient variants impact on this process, we discuss the potential contribution of non-canonical roles of these proteins to the pathophysiology of MGORS.
Subject terms: Disease genetics, Medical genetics
Introduction
Meier-Gorlin syndrome (MGORS) is a rare form of microcephalic primordial dwarfism (MPD) with less than 100 cases reported in the literature. MPD is an umbrella term for a group of rare disorders defined by global growth restriction including microcephaly. The reduced head size is often proportional to the height reduction but can be more severe for some subcategories of MPD, such as Seckel syndrome. Generally, these disorders are inherited in an autosomal recessive manner. Associated genes encode proteins involved in processes required for timely cell cycle progression such as cell cycle checkpoint, centrosome duplication and coordination, DNA replication, and DNA repair [1]. The molecular pathophysiology suggests reduced cellular proliferation (or increased apoptosis) during development ultimately underlies the restricted growth [1]. MGORS is diagnosed clinically by the triad of short stature, microtia, and patella hypo/aplasia, although not all MGORS patients present with all three (Table 1) [2]. There is a common facial gestalt of downslanting palpebral fissures, a fuller bottom lip, and with age, the nose becomes more prominent. Additional features are variably present between individuals (Table 1), with the exception of mammary hypoplasia which is completely penetrant in post-pubescent females [2].
Table 1.
Frequency of features in individuals with a clinical and genetic diagnosis of Meier-Gorlin syndrome.
 |
Information was derived from the reported cases in the literature and clinical information provided, so the number of cases in the literature likely underestimates the true number of affected individuals, especially for established disease genes. na information not available. IUGR intra-uterine growth retardation, ID intellectual disability, DD developmental delay.
MGORS is caused by disruption to DNA replication and is typically associated with early DNA replication processes [3–12]. DNA replication is a complex multi-protein process that is tightly regulated to ensure accurate duplication of the genome (reviewed in refs. [13, 14]). The initiation of DNA replication involves the formation of the pre-replication complex (pre-RC). This occurs during the M/G1 phases of the cell cycle and is initiated by localisation of the origin recognition complex (ORC, composed of subunits ORC1-6) to origins of replication. Recruitment of CDC6 and interaction with CDT1 facilitates the stepwise addition of two inactive MCM2-7 hexamers (MCM) onto chromatin in a process called replication origin licensing. Inhibition of CDT1 by GMNN during S and G2 phases ensures that this licensing step only occurs once during each cell cycle. Origin activation occurs during the S phase transition and first requires the formation of the pre-initiation complex (pre-IC). The pre-IC unites multiple replication factors to chromatin-bound MCM, including CDC45 and the GINS complex (GINS1-4), which associate together with MCM to form the CMG complex. The CMG complex facilitates the activation of MCM helicase activity, which is responsible for unwinding double-stranded DNA during S-phase. Further replication factors are then recruited to form the replisome, a large multi-protein complex that couples the unwinding of double-stranded DNA with active DNA synthesis by DNA polymerases. Multiple progression and cell-cycle checkpoint proteins, including the replication fork stabilisation factor DONSON [13, 15], function at the replisome to maintain genome integrity by ensuring continued and efficient progression of the replisome.
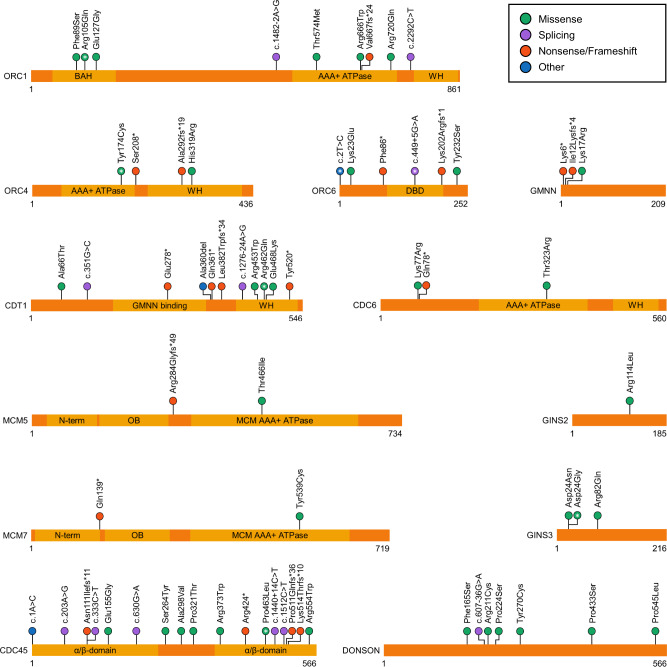
Variants in 13 DNA replication genes have been associated with MGORS, these are ORC1, ORC4, ORC6, CDT1, CDC6, GMNN, MCM3, MCM5, MCM7, CDC45, GINS2, GINS3, and DONSON (Fig. 1) [3–12]. Twelve of these have essential roles in early DNA replication, including nine which are specifically involved in pre-RC formation. Consequently, MGORS has been defined by the reduced ability to load early replicative machinery onto replication origins [13]. The identification of replication fork stabiliser DONSON as a new MGORS gene challenges this idea [9, 16, 17], since DONSON has no known role during early DNA replication processes.
Fig. 1. Meier-Gorlin syndrome variants and their locations.
Schematics for each MGORS protein detailing published MGORS variants and known protein domains. Circles containing a white asterisk (*) represent variants that are recurrent in three or more families. Domain name abbreviations: BAH (bromo-adjacent homology), WH (winged helix), DBD (DNA binding domain), OB (oligonucleotide/oligosaccharide binding). RefSeq IDs: ORC1 (NM_004153.3), ORC4 (NM_181741.3), ORC6 (NM_014321.3), GMNN (NM_015895.4), CDT1 (NM_030928.3), CDC6 (NM_001254.3), MCM5 (NM_006739.3), MCM7 (NM_005916.4), GINS2 (NM_016095.2), GINS3 (NM_022770.3), CDC45 (NM_003504.4), DONSON (NM_017613.3).
The use of hypothesis-free high throughput sequencing methods, alongside an increase in patient cohorts, continues to expand both the genetic and phenotypic spectrums of MGORS. This review presents an overview of the current MGORS-associated genes, focusing on molecular causes and clinical presentations for each gene.
Genes contributing to Meier-Gorlin syndrome
ORC1
The first individuals identified with biallelic variants in ORC1 have a generalised primordial dwarfism [4]. Based on the serendipitous finding of more severe ORC1 variants in a child with a complex congenital disorder with features reminiscent of MGORS, further sequencing efforts led to the discovery that variants in ORC1 are a common cause of MGORS, including the initial patient described by Gorlin [3, 18].
ORC1 individuals have characteristic MGORS features, though not all have the complete triad (Table 1) [19]. These individuals also show a spectrum of additional rarer phenotypes, including a range of neurological, respiratory, cardiac, gastrointestinal, urogenital, and musculoskeletal abnormalities [19]. This spectrum is relatively consistent with the MGORS presentations caused by other genes, however, ORC1 individuals have significantly more severe growth restrictions in height and head circumference [19, 20]. One individual has craniosynostosis [19], a feature typically seen in MGORS caused by variants in CDC45. Lethality is rare in MGORS but has been observed in a family segregating a frameshift variant together with the recurrent p.Arg105Gln variant [3], suggesting more severe variants could be linked to more severe outcomes in affected individuals. In considering the two originally described cohorts of individuals with variants in ORC1 (the generalised primordial dwarfism versus MGORS), individuals with MGORS tend to have at least one variant that is predicted to lower ORC1 protein levels, such as splicing, frameshift, or genomic deletions [3]. While not universal [7], this could point towards a pathological mechanism for why some patients have the additional specific features of MGORS that are absent from the primordial dwarfism cohort.
Many of the patients with ORC1-associated MGORS have substitutions in the N-terminal bromo adjacent homology (BAH) domain (Fig. 1), which is important for interactions between ORC and both chromatin-bound histones and DNA [21–23]. The most commonly reported MGORS ORC1 substitution, p.Arg105Gln, is located within this BAH domain and impairs the essential ORC function of chromatin binding [4, 23]. While there is variant clustering, missense variants affecting ORC1 are not solely confined to the BAH domain, for example, the p.Arg720Gln substitution is located within the ATPase domain and has a disruptive effect on ATP hydrolysis, which is essential for complex assembly on chromatin (Fig. 1) [24]. Other variants have splicing effects, decrease protein stability, or result in protein truncation (Fig. 1) [3]. The commonality amongst these variants appears to be the impaired rate of cell cycle progression [4].
Analysis of zebrafish models generated through morpholinos targeting orc1 showed significant decrease in overall growth, consistent with the MGORS phenotype [4]. Co-injection of morpholino and ORC1 mRNA demonstrated that this growth-restricted phenotype could be rescued by wildtype ORC1, but not ORC1 mRNA containing amino acid substitutions that impaired histone binding [21].
ORC4
Variants in ORC4 were identified independently by two groups in the first tranche of genes associated with MGORS [3, 7]. For ORC4, the most commonly reported causal variant is p.Tyr174Cys, which has been inherited in either a homozygous state or as a compound heterozygous variant inherited with nonsense variants [3, 7]. Tyr174 is located in the AAA + ATPase domain of ORC4 and is completely conserved throughout eukaryotes (Fig. 1) [3, 7]. Tyr174 is an important residue for ATPase activity, with activity decreased by 50% when the tyrosine is substituted to cysteine [24]. The p.Tyr174Cys change likely impairs the essential role of ORC4 ATP hydrolysis in facilitating ORC formation [7, 25]. Modelling of this variant in yeast caused reduced progression through S phase which impacted organism proliferation rates [7].
A Drosophila model studying the recurrent p.Tyr174Cys variant (p.Tyr162Cys in Drosophila) reported reduced viability in the homozygous state, along with tissue-specific effects in bristle spacing and length [26]. Similar bristle effects have also been observed in orc6 Drosophila mutants [27]. Females were sterile, likely due to decreased EdU incorporation affecting endoreplication, ultimately impacting chorion gene amplification.
Individuals with ORC4 variants generally have the clinical triad characteristic for MGORS and have the second most severe growth defects behind ORC1-associated individuals (Table 1) [20]. There also appears to be a genotype-phenotype correlation, as more severe features are present in individuals harbouring one loss-of-function variant in trans with a missense variant compared to those individuals with two missense variants [19]. Aligning with this, those cases with a predicted more severe genotype also commonly displayed additional clinical features, such as pulmonary emphysema (1/3 cases) or the need for tube feeding (2/3 cases) [19].
ORC6
Variants in ORC6 were also identified in the first tranche of genetic discoveries for MGORS [3]. ORC6 was screened as a candidate gene based on the role of the encoded protein as an ORC subunit, but this interaction appears to be more dynamic than other ORC6 subunits in different species, with human ORC6 being more loosely associated with the other ORC subunits [28]. Rather than being an integral subunit of ORC for binding to DNA, ORC6 appears to be more important in protein-protein interactions, such as with CDC6 [29]. Despite this, MGORS variants in ORC6 disrupt the ORC complex function in a similar fashion to other ORC variants.
Much like cases with ORC1 or ORC4 variants, not all individuals with ORC6-associated MGORS have all three of the characteristic features (10/11 cases with short stature, 7/8 cases with patellar hypo/aplasia) (Table 1) [19]. While most ORC6 MGORS patients have a relatively mild phenotype, one family is an exception, with a homozygous c.602_605delAGAA, p.Lys202Argfs*1, variant causing very severe growth and developmental defects in three terminated pregnancies [30]. Approximately one-third of reported individuals with causal variants in ORC6 have the biallelic missense variants c.2T>C and c.449+5G>A [19, 31]. While the c.2 T>C variant disrupts the translation start codon of ORC6 transcripts, the presence of nearby downstream in-frame methionine codons (Met20, Met50 and Met59) makes it theoretically possible that an N-terminal truncated protein could be translated, which could harbour some residual functionality. A minigene splicing assay confirmed that the intronic c.449+5G>A variant causes in-frame exon skipping of exon 4, which would result in a translated protein lacking 30 amino acids in the DNA binding domain (Fig. 1) [31].
Using Drosophila as a model, the introduction of the patient variant, p.Tyr232Ser (p.Tyr225Ser in Drosophila), caused impaired ORC formation by interrupting the interaction of orc6 with orc3, which was then further validated using human ORC6 in vitro [28]. Reducing orc6 levels using a knockdown model also showed that variants orthologous to p.Tyr232Ser and p.Lys23Glu were not able to rescue the mutant phenotype, the former impairing complex formation, and the latter impairing DNA binding. Both variants noticeably reduced pre-RC formation and DNA replication [27, 32].
CDT1
Variants in CDT1 appear to be the most common cause of MGORS, and much like ORC1 individuals, there is a spectrum of clinical presentation [19, 33]. Successful pregnancies have been noted in an adult case with MGORS but the female patient did experience complications requiring a hysterectomy [33]. While adult cases of MGORS known to the clinical genetics community are rare, several females have noted abnormalities of the reproductive tract including the uterus, suggesting this could be an under-reported issue for women with MGORS.
The most common variant seen in CDT1 is p.Arg462Gln, reported in 11 individuals in the literature so far. It is most often observed in combination with more disruptive variants such as premature stop variants [3, 7, 34]. A mouse model studying the orthologous variant (p.Arg474Gln) discovered that this variant partially impairs MCM binding [35], which may disrupt the essential role of CDT1 in MCM2-7 recruitment to origins of replication.
CDT1 patient substitutions including p.Arg462Gln were analysed in vitro [36]. Substitutions p.Arg462Gln and p.Glu468Lys disrupted the protein surface required for MCM6 binding, resulting in less efficient recruitment of MCM proteins to start sites. One of the more surprising results of this study was the discovery that the p.Ala66Thr variant acts in a hypermorphic manner, rather than hypomorphic as expected. This variant did not reduce stability of CDT1 and was consistently seen to induce increased re-replication compared to other variants and the wildtype control. Despite this, increased CHK1 phosphorylation in the presence of this variant strongly reduced proliferation in a colony-forming assay, suggesting the overall cellular consequence would still be reduced proliferation [36]. While this variant does not appear to impact MCM interactions, it is predicted to impair the binding of the inhibitor molecule cyclin A. This does not impair the nearby CDK-mediated phosphorylation and subsequent binding of SCF E3 ligase skp2 for degradation but nevertheless appears to act through a different, unidentified checkpoint in S or G2 phase to reduce overall proliferation [36].
CDC6
The first MGORS patient reported was homozygous for a missense variant located within the AAA+ ATPase domain in the encoded CDC6 protein [3]. Such a substitution would likely impact ATPase activity, hampering the ability of CDC6 to disengage from ORC as part of pre-RC assembly [37, 38]. The second patient reported was compound heterozygous for a nonsense and a missense variant [39]. In addition to the characteristic features of MGORS, this second individual was severely affected, with features of progeroid appearance at birth and lipodystrophy; features also observed in one individual with MCM7 variants [8], and in other replisome disorders affecting later stages of DNA replication initiation [13]. The patient had significant short stature and no microcephaly but did present with motor development delays. The residue affected by the substitution, Lys77, lies within a CDK2/cyclin A phosphorylation motif associated with cytoplasmic translocation to regulate CDC6 activity [39].
Both knockout and hypomorph cdc6 zebrafish models have been studied [40]. While the knockout was early embryonic lethal, the hypomorph model showed a similar growth profile to the individuals with CDC6 variants. Interestingly, male zebrafish had a shorter life span as well as reproductive defects, but such comparisons with affected individuals are not yet possible given the rarity and age of the CDC6 cohort.
GMNN
Pathogenic de novo variants in three individuals have been identified in GMNN [5], which encodes the negative regulator of DNA replication, GEMININ. Affected individuals had been previously clinically diagnosed with MGORS, with all possessing the classical triad of features. Microcephaly and genital anomalies were more variable (each recorded present in only one case), and there was some evidence of developmental delay (2/3 cases) (Table 1).
This is the only described example of an autosomal dominant inheritance pattern of MGORS, and the variants act through a specific mechanism to reduce DNA replication initiation. The N-terminal portion of GEMININ contains a destruction box (residues 23–31) similar to those found in mitotic cyclins [41]. APC ubiquitylation recognises this motif and signals for GEMININ degradation during late M/early G1 phase, thereby enabling the target of GEMININ, CDT1, to be active and assist the loading of MCM helicases on to chromatin [41, 42]. All three variants identified in GMNN lie near the 5′ end of the transcript. These disrupt translation of the full protein and either introduce a premature stop codon or are predicted to alter splicing of the first coding exon of GMNN. Despite the premature stop codons, the transcripts remained stable, with in vitro overexpression assays showing production of a smaller molecular weight product which is predicted to be translated from a downstream methionine (Met28). Given this protein translation starts within the destruction box motif, such a control mechanism for GEMININ degradation would not be active, ultimately causing less CDT1 activity and a similar effect as other MGORS genes on DNA replication initiation [5].
MCM3
Recently, a single individual from a consanguineous family has been described as homozygous for a missense variant in MCM3 [8]. The affected individual possessed some of the cardinal features of MGORS, including the facial gestalt and microtia, along with a proportionate reduction in growth, but patellae were reported as normal. She also had congenital lobar pulmonary emphysema. Despite functional investigations studying the variant in patient-derived cells, no effects on MCM3 transcript or protein levels, or on cell cycle progression could be demonstrated. The variant remains a strong candidate based on it’s location in the C-terminal domain, which is required in chromatin loading of the MCM complex [43], as well as supportive evidence through the identification of pathogenic variants in other MCM subunits in MGORS individuals.
MCM5
Only one family has been described thus far segregating variants in MCM5 [12]. The affected boy was biallelic for a frameshift and a missense variant in MCM5 and displayed typical MGORS features of short stature, microtia, patella agenesis, and bilateral cryptorchidism (Table 1). The individual had normocephaly, and unilateral kidney hypoplasia which likely caused recurrent intestinal infections. The authors studying this patient undertook a comprehensive suite of experiments, confirming a reduction of MCM5 protein in patient-derived cells in both whole-cell extracts and at chromatin, along with a concomitant reduction in MCM2 at chromatin [12]. These cells were slower to progress through S-phase but did not show an increase in replication stress nor any centrosome abnormalities. The missense variant, p.Thr466Ile, lies within the AAA+ ATPase domain of MCM5 (Fig. 1), and mutation of this residue in a yeast complementation assay reduced survival. Previously, zebrafish mcm5 mutants have been studied and showed a similar overall growth reduction in the absence of any other significant developmental anomalies [4, 44].
MCM7
MCM7 has recently been implicated in a broad range of phenotypes known to be associated with disorders of DNA replication [8, 45]. Three families have been described in the literature to date; an adult patient from one family has the classic MGORS features of short stature, microtia, and bilateral absent patella. In addition, she had proportional microcephaly and showed absence of breast tissue development (Table 1). She was compound heterozygous for nonsense and missense variants in MCM7. The second case showed a similar genotype, although the missense variant affected a residue in a different domain of MCM7. His set of clinical features was quite different, with normal growth parameters but a progeroid appearance at birth, as well as adrenal insufficiency and lipodystrophy – similar features have been observed in cases with CDC6 or POLD1 variants [13, 39]. In both cases, patient-derived cells showed a reduction of MCM7 protein, sufficient to cause a mild reduction in DNA replication in early S phase. In vitro studies of the missense variants confirmed that both impacted interactions within the MCM complex [8]. Most recently, a consanguineous family was described segregating a homozygous missense variant in MCM7 that causes mild microcephaly, short stature, and severe intellectual disability [45]. Interestingly, the missense variant in this third family was located close to the missense variant in the second case described, but the patients show quite different sets of clinical features. The MCM complex serves two roles in DNA replication: firstly, once activated it unwinds DNA and permits entry for the polymerases and other replisome components, and then secondly, the complex functions to continue this unwinding along the chromosome, to help ensure efficient DNA replication. Given the nonsense variants will elicit nonsense-mediated decay (causing null alleles), the position of the residues affected by the substitutions likely plays a role in causing different impacts to MCM7 functioning, and potentially leads to different phenotypes. Further investigation into the different consequences of these patient-informed substitutions could provide further insight.
CDC45
Variants in CDC45 underlie a MGORS subtype clinically characterised by the very common appearance of craniosynostosis [6, 46–48]. Outside of the CDC45 MGORS cohort, craniosynostosis has only been observed in two other MGORS individuals, one with ORC1 variants and another recently identified homozygous for a GINS2 variant (Table 1) [4, 11]. The strong association of craniosynostosis with CDC45 variants supports the use of this feature as a differential diagnosis in genetic testing. Identification of biallelic CDC45 variants in a MGORS cohort was the first evidence for the involvement of the pre-initiation and CMG helicase complexes in the pathogenesis of MGORS [6].
To date, biallelic CDC45 variants have been reported in 19 individuals across 15 families and underlie a broad spectrum of phenotypes that range from a classic MGORS presentation to syndromic craniosynostosis with short stature but no other MGORS features [6, 46–48]. The majority of clinically diagnosed MGORS individuals had the clinical triad of short stature, microtia, and patella hypo/aplasia [37–39]. MGORS-related genital anomalies (micropenis or clitoromegaly) were reported in at least two individuals and both females of suitable age were affected by mammary hypoplasia [37]. Craniosynostosis was present in many of the individuals with CDC45 variants but is an incompletely penetrant trait (13/14 individuals), as observed by the discordance of bicoronal craniosynostosis in two affected brothers (Table 1) [6]. Anorectal malformations are common and range from anal stenosis to an imperforate or anteriorly placed anus. Heart and palate malformations are occasionally present (7/13 and 2/13 cases respectively), and sparse eyebrows are invariant amongst all individuals with CDC45 variants. CDC45 is located on chromosome 22, within the genomic region associated with DiGeorge syndrome (MIM 188400). Sequencing of a cohort of individuals with DiGeorge syndrome revealed several hemizygous variants in CDC45 in those with atypical features similar to the MGORS cohort described [49]. Functional assays will be required to confirm the consequence of these variants on CDC45 levels and function and determine their involvement in the phenotypes presented, given some are found in a homozygous state in control populations, or lie within non-canonical or non-coding exons.
Across the CDC45 cohort there are a range of truncating, non-synonymous, and synonymous splice-altering variants but no biallelic truncating genotypes were reported. Missense variants do not cluster within a particular domain nor with a particular phenotype, but many are found in the conserved core region of CDC45 and are expected to destabilise the protein. In keeping with these predictions, cells derived from several patients demonstrated reduced CDC45 levels [6].
Interestingly, synonymous splice-altering variants appear to be more frequently found in the CDC45 cohort. These include variants c.318C>T and c.333C>T that are present in patients with syndromic craniosynostosis and the MGORS-subtype respectively. Both splicing variants cause the same effect of increased skipping of exon 4. The exclusion of this exon keeps the transcript in-frame and is observed at low levels in unaffected cells. However, since this removes part of the conserved DHH phosphoesterase domain, its presence is unexpected. A third synonymous splice-altering variant, c.630G>A, was found in the non-canonical exon 7 (numbering based on RefSeq: NM_001178010.2) in siblings with MGORS with craniosynostosis [46]. The splicing variant both reduced the level of canonical transcript and produced three alternative transcripts, all of which were expected to undergo nonsense-mediated decay. A fourth synonymous variant, c.1512C>T, has also been identified in MGORS, but functional analysis has been not undertaken to assess the impact on canonical splicing [47].
GINS2
Only one GINS2-associated case has been reported thus far, who is homozygous for a missense variant (p.Arg114Leu) [11]. In addition to the characteristic triad, this patient has many features in common with the CDC45-MGORS subtype, such as craniosynostosis, atrial septal defect, and an anteriorly placed anus (Table 1). The orthologous missense variant shows defects in a yeast assay of DNA replication, and the resolved CMG helicase structure places this residue at the interaction site between GINS2, CDC45, and MCM5 [11, 50].
GINS3
Recently, biallelic missense variants in GINS3 were also reported to underlie MGORS [10]. All affected individuals possessed at least one variant affecting p.Asp24, which was commonly substituted to glycine or less often, asparagine. Extensive analysis showed these variants impact on the protein stability of GINS3. Mutant GINS3 is reduced at chromatin, which has a broader effect on other members of the GINS complex and other protein components of the replisome. These effects then impact on replication speed, interorigin distance, and S phase length, ultimately slowing proliferation. Interestingly, unlike human counterparts homozygous for the same variant, the mouse model harbouring the p.Asp24Asn variant was embryonic lethal with fetal resorption starting from E16.5 [10]. Embryonic fibroblasts derived from E12.5 mutant embryos show premature senescence, which could potentially be due to reduced DNA replication.
The patient cohort demonstrated the characteristic features of MGORS, with a broad spectrum of other features (Table 1) [10]. All individuals with medical information demonstrated microcephaly (6/6 cases), but not all had short stature (6/7 cases). The growth restriction was not proportionate in many individuals, and while some had a more severe height restriction, others had a smaller head circumference. The facial gestalt common in MGORS was not so apparent in this cohort, although all had some degree of microtia or low-set ears. Patella agenesis was very common, but not universal (3/4 cases). Interestingly, many individuals showed neutropenia (4/5 cases), with one also showing B lymphopenia. Given the unique overlap of MGORS and neutropenia in GINS3 individuals, these features could be used diagnostically as differentials in candidate gene testing.
DONSON
Exome sequencing and phased genome sequencing identified DONSON as a novel disease gene in several MGORS patients [9, 16, 17]. DONSON is a replisome component and replication fork stabiliser that is involved in activating the ATR-dependent replication stress response [15]. DONSON is unique compared to all other MGORS genes as the encoded protein has no known role in either the DNA pre-RC or pre-IC [15].
All of the DONSON-MGORS cohort had short stature, microtia and patella agenesis (Table 1) [9, 16, 51]. Most had proportionate microcephaly, but one patient had an average head circumference for their age [9]. In vitro analysis of MGORS patient variants demonstrated that the substitutions reduced nuclear localisation of DONSON, and a deep intronic variant introduces a novel strong splice acceptor site causing a reading frameshift and premature stop codon, confirming the hypomorphic nature of these variants [9].
Biallelic hypomorphic DONSON variants have previously been found to underlie a clinically distinct primordial dwarfism of severely disproportionate microcephaly and variable skeletal abnormalities mainly present in the upper limb (MISSLA; MIM 617604) [15, 51, 52]. A third associated phenotype is microcephalic-micromelia syndrome (MIMIS; MIM 251230), a severe form of primordial dwarfism associated with perinatal death [53, 54]. Microcephaly in MIMIS patients is severe but proportional to height restriction, with the identified MIMIS variants causing a severe or predicted complete loss of DONSON protein. Given the early embryonic lethality of a mouse knockout model [54], a rescue via an alternative transcript could be possible to explain the extended survival in MIMIS individuals.
The most pronounced phenotypic difference between DONSON-MGORS and DONSON-MISSLA individuals is the degree of severity of microcephaly versus short stature. DONSON-MISSLA exhibit a more severe microcephaly compared to height reduction, whereas these parameters are much more proportionate in DONSON-MGORS individuals [9]. Interestingly, like DONSON, pathogenic variants in multiple other genes encoding proteins also involved in replication fork stabilisation or the ATR-dependent stress response pathways cause a phenotypically similar primordial dwarfism, Seckel syndrome, that also features severely disproportionate microcephaly [13]. Some patients in the DONSON-MISSLA cohort exhibit phenotypic similarity to MGORS with patella hypoplasia and microtia present variably, for example, patient P19 from the original MISSLA cohort demonstrates a phenotype potentially better aligned with MGORS than with the rest of the MISSLA cohort [15]. This male patient is homozygous for p.Pro433Ser, a genotype independently reported in a different patient clinically diagnosed with MGORS [17]. Patient P19 had characteristic MGORS features of microtia, proportionate short stature, and microcephaly. While patella hypo/aplasia was not noted for this individual, mild genital hypoplasia was present, which is common in MGORS [20]. Three further patients from the initial MISSLA cohort (P9 and brothers P20-1 and P20-2) also show phenotypic overlap with MGORS. They each meet the diagnostic criteria for MGORS with microtia and patella hypo/aplasia, and brothers P20-1 and P20-2 also have cryptorchidism [15]. While the growth parameters of these cases are more in keeping with MISSLA, their skeletal anomalies are minor compared to the rest of the MISSLA cohort and restricted to fingers, toes, and metacarpals. Together, such cases highlight the complexity of reconciling a clinical and genetic diagnosis involving DONSON.
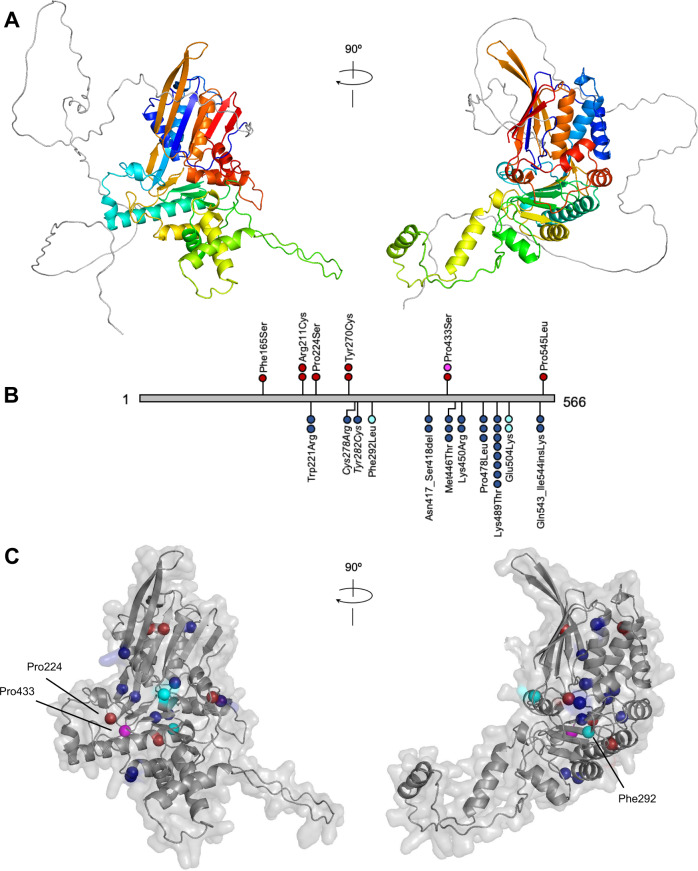
While the role of DONSON in DNA replication and cell cycle progression has become clearer [15, 55, 56], the protein itself is somewhat of an enigma. There are no predicted protein domains nor is it similar to any other known protein. With the recent release of AlphaFold [57, 58], we have been able to view a possible structure for DONSON for the first time. The AlphaFold prediction (AlphaFoldDB ID: HUMAN_DONS) (Fig. 2A) suggests an intrinsically disordered N-terminal region of approximately 150 amino acids, ahead of one interconnected domain that is joined together via a less ordered looped structure. This interconnected domain is predicted by AlphaFold to have a confident/very high model confidence with a pLDDT score of >70/>90.
Fig. 2. Predicted structure of human DONSON and the locations of DONSON-MGORS and MISSLA missense variants.
A Predicted structure of human DONSON (AlphaFoldDB ID: DONS_HUMAN). DONSON is composed of a 150-residue long intrinsically disordered N-terminal region (grey) followed by interfolded protein (residues 151-566) coloured rainbow from the N-terminal end (blue) to the C-terminal end (red). B Linear DONSON (grey) showing the location of missense variants found in MGORS (above) and MISSLA (below). Each sphere represents one individual: red - MGORS, blue - MISSLA, magenta - MISSLA individual with clear MGORS features, cyan - MISSLA patients with phenotypic overlap to MGORS. Variants Cys278Arg/Tyr282Cys are italicized as either one, or both, are pathogenic in a single individual. C Position of MGORS and MISSLA-affected residues within the predicted DONSON structure. Residues are coloured by patient phenotype as per B. Indicated residues are discussed further in the main text. No variants are in the 150-residue-long intrinsically disordered N-terminal region and so for clarity it was removed.
MGORS missense variants are spread throughout the DONSON protein sequence and do not cluster together in the linear protein sequence nor in the predicted 3D structure (Fig. 2B, C) [9]. MISSLA missense variants are also spread throughout the linear protein sequence, although they tend to lie towards the C-terminus. Most residues substituted in MISSLA are scattered throughout the 3D structure. Four of them are located within the core anti-parallel β-sheet, including Phe292, which is found in a patient with phenotypic overlap with MGORS. Although no MGORS variants are located within this core β-sheet structure, two proline residues that are substituted in MGORS, Pro224, and Pro433, are each found at the flank of a β-strand involved in this β-sheet. Further functional analysis will shed light on the role of this core in DONSON functioning.
From the evidence currently available, there is no obvious genotypic/phenotypic relationship between MGORS and MISSLA patients. While both syndromes are caused by reduced function, specific variants or genotypes have not been compared to understand the relative severity on protein activity or function, such as their effect on protein-protein interactions.
The involvement of a replication fork stabilisation factor in MGORS potentially challenges MGORS as being the consequence of disrupted early-replication organisation. MGORS could therefore be considered to be a disorder of broader DNA replication processes including late-stage replisome stability, or alternatively DONSON may have additional unknown roles in early replication processes. Given that new complexities of DNA replication initiation are still being uncovered [59], and the recent confirmation of DONSON as a DNA replication protein, the latter remains a possibility. Further investigation into DONSON domains and function could help to clarify whether there are genotypic/phenotypic relationships between the clinically distinct syndromes.
Non-canonical functions of MGORS-associated proteins
A number of other cellular functions have been described for proteins linked to MGORS. For example, ORC6 acts to promote cell abscission during cytokinesis and also has a role in mismatch repair during DNA synthesis [60, 61], while CDT1 enables stable kinetochore-microtubule interactions during mitosis [62, 63]. Non-canonical functions of some of the encoded proteins include the presence at the centrosome or cilium (reviewed in ref. [64]). There is accumulating evidence supporting ORC and MCM complexes at the centrosome [65], with this organelle serving as a possible hub for cell cycle progression interactions, such as ORC1 with cyclin E [66]. ORC1 assists in regulating centriole and centrosome duplication via interaction of two separate domains. The cyclin-CDK2 binding domain contains MGORS variants; p.Arg105Gln entirely abolishes inhibition of cyclin E-CDK2, while p.Phe89Ser and p.Glu127Gly partially impede inhibition of both cyclin A-CDK2 and cyclin E-CDK2. The result of abolished Cyclin-CDK inhibition is centrosome reduplication and loss of centrosome copy number control [67].
Cilia defects have been observed at a low level in MGORS patient-derived cells, or when MGORS proteins are subjected to siRNA in reporter cell models [64]. Zebrafish models, using morpholinos targeting some of the MGORS proteins, show some phenotypes that overlap with those observed in zebrafish models of ciliopathies. In these models, rescuing with a patient-informed mutant mRNA did not rescue the ciliopathic phenotypes, but wildtype mRNA also only resulted in a partial phenotypic rescue. Based on these observations, a hypothesis has been put forward that MGORS could be considered a ciliopathy [64, 68, 69]. Such a suggestion needs to be considered with care, for both clinical and biological reasons. Clinically, ciliopathies are distinguished by characteristic features such as retinal dystrophy, polydactyly, cystic kidneys, or obesity [70] requiring specific clinical management, which have never been observed in MGORS individuals [2, 19]. Furthermore, while many of the ORC and MCM subunits have been found localised at the centrosome, not all MGORS-associated proteins have, such as CDC45, or proteins linked to MGORS-like clinical features, such as RECQL4 or POLE. The presence of these proteins at the centrosome is intriguing, and suggests complex biology linking DNA replication, cell cycle progression and centrosome duplication and ciliogenesis, but the pathognomonic relevance to MGORS is less certain. Given the long list of MGORS associated genes identified as highlighted in this review, which account for a majority of affected individuals, the most parsimonious explanation remains that MGORS is a disorder of DNA replication.
The expanding spectrum of disorders linked to DNA replication
While the MGORS variants in the genes encoding the pre-RC were amongst the first germline variants to be described involving DNA replication, since then, many components in this complex process have become linked to genetic disorders [13]. It is common for these syndromes to be characterised by growth restriction, microcephaly and/or short stature. As further individuals and more disease-causing genes are identified, some interesting patterns are appearing which give tantalising clues to the underlying developmental biology disrupted by such variants. A key example is effects on haematopoiesis. Neutropenia has not been described as a common feature associated with any other MGORS gene before, with GINS3 being the first description of such an overlap. However, neutropenia is commonly associated with variants in other GINS subunits, GINS1 and GINS4 [71, 72], and in at least some individuals with a biallelic variant in the functionally related subunit of the MCM complex, MCM4, but not GINS2, GINS3 or MCM3, MCM5 or MCM7 [73]. These clinical findings in GINS3 adds to the accumulating evidence for the role of a sub-section of replication initiation factors in haematopoiesis and immune cell functioning [13].
Microtia and patella hypo/aplasia have been considered differential features of MGORS [74], but clinical genetic findings reveals a more complicated picture. These features have also been reported in individuals with variants in genes such as RECQL4 and POLE. Such patients do not have the facial gestalt that is commonly observed in MGORS but these overlaps do indicate a broader link between skeletal and/or cartilage development and efficient organisation and initiation of DNA replication. There is emerging evidence linking DNA replication initiation to cell fate [75, 76], and the wealth of human genetics summarised here adds to this. It will be fascinating to learn how tissue-specific development and the ubiquitous requirement of DNA replication initiation are so intimately linked.
Conclusion
High-throughput sequencing approaches in clinical genetics have expanded the molecular and clinical understanding of MGORS. As the number of genes associated with MGORS increases, we see the appearance of subtypes, notably the prevalence of craniosynostosis in CDC45-associated MGORS and neutropenia in GINS3-associated MGORS. The classical triad of short stature, microtia, and patella hypo/aplasia that historically defined MGORS is more variable than previously thought. Conversely, mammary hypoplasia is the most consistent feature observed in MGORS, and external genital abnormalities and skeletal anomalies are also common. While gene discoveries continue to provide insights into MGORS and the link between DNA replication and human development, unique individuals illustrate the clinical complexities of a multi-gene disorder such as MGORS.
Acknowledgements
Thank you to Dr. Karen Knapp for assistance with DONSON protein modelling.
Author contributions
LSB conceived the idea. LSB wrote the manuscript with the help of END and MR.
Funding
END and MR are each supported by a University of Otago Masters Scholarship. Open Access funding enabled and organized by CAUL and its Member Institutions.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Emily Nielsen-Dandoroff, Mischa S. G Ruegg.
References
- 1.Klingseisen A, Jackson AP. Mechanisms and pathways of growth failure in primordial dwarfism. Genes Dev. 2011;25:2011–24. doi: 10.1101/gad.169037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Munnik SA, Hoefsloot EH, Roukema J, Schoots J, Knoers NV, Brunner HG, et al. Meier-Gorlin syndrome. Orphanet J Rare Dis. 2015;10:114. doi: 10.1186/s13023-015-0322-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bicknell LS, Bongers EM, Leitch A, Brown S, Schoots J, Harley ME, et al. Mutations in the pre-replication complex cause Meier–Gorlin syndrome. Nat Genet. 2011;43:356–9. doi: 10.1038/ng.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bicknell LS, Walker S, Klingseisen A, Stiff T, Leitch A, Kerzendorfer C, et al. Mutations in ORC1, encoding the largest subunit of the origin recognition complex, cause microcephalic primordial dwarfism resembling Meier–Gorlin syndrome. Nat Genet. 2011;43:350–5. doi: 10.1038/ng.776. [DOI] [PubMed] [Google Scholar]
- 5.Burrage LC, Charng WL, Eldomery MK, Willer JR, Davis EE, Lugtenberg D, et al. De Novo GMNN mutations cause autosomal-dominant primordial dwarfism associated with Meier-Gorlin Syndrome. Am J Hum Genet. 2015;97:904–13. doi: 10.1016/j.ajhg.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fenwick AL, Kliszczak M, Cooper F, Murray J, Sanchez-Pulido L, Twigg SR, et al. Mutations in CDC45, encoding an essential component of the pre-initiation complex, cause Meier-Gorlin Syndrome and Craniosynostosis. Am J Hum Genet. 2016;99:125–38. doi: 10.1016/j.ajhg.2016.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guernsey DL, Matsuoka M, Jiang H, Evans S, Macgillivray C, Nightingale M, et al. Mutations in origin recognition complex gene ORC4 cause Meier-Gorlin syndrome. Nat Genet. 2011;43:360–4. doi: 10.1038/ng.777. [DOI] [PubMed] [Google Scholar]
- 8.Knapp KM, Jenkins DE, Sullivan R, Harms FL, von Elsner L, Ockeloen CW, et al. MCM complex members MCM3 and MCM7 are associated with a phenotypic spectrum from Meier-Gorlin syndrome to lipodystrophy and adrenal insufficiency. Eur J Hum Genet. 2021;29:1110–20. doi: 10.1038/s41431-021-00839-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knapp KM, Sullivan R, Murray J, Gimenez G, Arn P, D’Souza P, et al. Linked-read genome sequencing identifies biallelic pathogenic variants in DONSON as a novel cause of Meier-Gorlin syndrome. J Med Genet. 2020;57:195–202. doi: 10.1136/jmedgenet-2019-106396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McQuaid ME, Ahmed K, Tran S, Rousseau J, Shaheen R, Kernohan KD, et al. Hypomorphic GINS3 variants alter DNA replication and cause Meier-Gorlin syndrome. JCI Insight. 2022;7:e155648. doi: 10.1172/jci.insight.155648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nabais SaMJ, Miller KA, McQuaid M, Koelling N, Wilkie AOM, Wurtele H, et al. Biallelic GINS2 variant p.(Arg114Leu) causes Meier-Gorlin syndrome with craniosynostosis. J Med Genet. 2022;59:776–80. doi: 10.1136/jmedgenet-2020-107572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vetro A, Savasta S, Russo Raucci A, Cerqua C, Sartori G, Limongelli I, et al. MCM5: a new actor in the link between DNA replication and Meier-Gorlin syndrome. Eur J Hum Genet. 2017;25:646–50. doi: 10.1038/ejhg.2017.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bellelli R, Boulton SJ. Spotlight on the Replisome: Aetiology of DNA replication-associated genetic diseases. Trends Genet. 2021;37:317–36. doi: 10.1016/j.tig.2020.09.008. [DOI] [PubMed] [Google Scholar]
- 14.Fragkos M, Ganier O, Coulombe P, Mechali M. DNA replication origin activation in space and time. Nat Rev Mol Cell Biol. 2015;16:360–74. doi: 10.1038/nrm4002. [DOI] [PubMed] [Google Scholar]
- 15.Reynolds JJ, Bicknell LS, Carroll P, Higgs MR, Shaheen R, Murray JE, et al. Mutations in DONSON disrupt replication fork stability and cause microcephalic dwarfism. Nat Genet. 2017;49:537–49. doi: 10.1038/ng.3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karaca E, Posey JE, Bostwick B, Liu P, Gezdirici A, Yesil G, et al. Biallelic and De Novo variants in DONSON reveal a clinical spectrum of cell cycle-opathies with microcephaly, dwarfism, and skeletal abnormalities. Am J Med Genet A. 2019;179:2056–66. doi: 10.1002/ajmg.a.61315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nerakh G, Vineeth VS, Tallapaka K, Nair L, Dalal A, Aggarwal S. Microcephalic primordial dwarfism with predominant Meier-Gorlin phenotype, ichthyosis, and multiple joint deformities-Further expansion of DONSON Cell Cycle-opathy phenotypic spectrum. Am J Med Genet A. 2022;188:2139–46. doi: 10.1002/ajmg.a.62725. [DOI] [PubMed] [Google Scholar]
- 18.Gorlin RJ, Cervenka J, Moller K, Horrobin M, Witkop CJ., Jr Malformation syndromes. A selected miscellany. Birth Defects Orig Artic Ser. 1975;11:39–50. [PubMed] [Google Scholar]
- 19.de Munnik SA, Bicknell LS, Aftimos S, Al-Aama JY, van Bever Y, Bober MB, et al. Meier-Gorlin syndrome genotype-phenotype studies: 35 individuals with pre-replication complex gene mutations and 10 without molecular diagnosis. Eur J Hum Genet. 2012;20:598–606. doi: 10.1038/ejhg.2011.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Munnik SA, Otten BJ, Schoots J, Bicknell LS, Aftimos S, Al-Aama JY, et al. Meier-Gorlin syndrome: growth and secondary sexual development of a microcephalic primordial dwarfism disorder. Am J Med Genet A. 2012;158A:2733–42. doi: 10.1002/ajmg.a.35681. [DOI] [PubMed] [Google Scholar]
- 21.Kuo AJ, Song J, Cheung P, Ishibe-Murakami S, Yamazoe S, Chen JK, et al. The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome. Nature. 2012;484:115–9. doi: 10.1038/nature10956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Onishi M, Liou GG, Buchberger JR, Walz T, Moazed D. Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Mol Cell. 2007;28:1015–28. doi: 10.1016/j.molcel.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 23.Zhang W, Sankaran S, Gozani O, Song J. A Meier-Gorlin syndrome mutation impairs the ORC1-nucleosome association. ACS Chem Biol. 2015;10:1176–80. doi: 10.1021/cb5009684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tocilj A, On KF, Yuan Z, Sun J, Elkayam E, Li H, et al. Structure of the active form of human origin recognition complex and its ATPase motor module. Elife. 2017;6:e20818. doi: 10.7554/eLife.20818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siddiqui K, Stillman B. ATP-dependent assembly of the human origin recognition complex. J Biol Chem. 2007;282:32370–83. doi: 10.1074/jbc.M705905200. [DOI] [PubMed] [Google Scholar]
- 26.McDaniel SL, Hollatz AJ, Branstad AM, Gaskill MM, Fox CA, Harrison MM. Tissue-specific DNA replication defects in drosophila melanogaster caused by a Meier-Gorlin Syndrome Mutation in Orc4. Genetics. 2020;214:355–67. doi: 10.1534/genetics.119.302938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balasov M, Akhmetova K, Chesnokov I. Humanized Drosophila Model of the Meier-Gorlin Syndrome reveals conserved and divergent features of the Orc6 Protein. Genetics. 2020;216:995–1007. doi: 10.1534/genetics.120.303698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bleichert F, Balasov M, Chesnokov I, Nogales E, Botchan MR, Berger JM. A Meier-Gorlin syndrome mutation in a conserved C-terminal helix of Orc6 impedes origin recognition complex formation. Elife. 2013;2:e00882. doi: 10.7554/eLife.00882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomae AW, Baltin J, Pich D, Deutsch MJ, Ravasz M, Zeller K, et al. Different roles of the human Orc6 protein in the replication initiation process. Cell Mol Life Sci. 2011;68:3741–56. doi: 10.1007/s00018-011-0675-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shalev SA, Khayat M, Etty DS, Elpeleg O. Further insight into the phenotype associated with a mutation in the ORC6 gene, causing Meier-Gorlin syndrome 3. Am J Med Genet A. 2015;167A:607–11. doi: 10.1002/ajmg.a.36906. [DOI] [PubMed] [Google Scholar]
- 31.Nazarenko MS, Viakhireva IV, Skoblov MY, Soloveva EV, Sleptcov AA, Nazarenko LP. Meier-Gorlin Syndrome: Clinical misdiagnosis, genetic testing and functional analysis of ORC6 mutations and the development of a prenatal test. Int J Mol Sci. 2022;23:9234. doi: 10.3390/ijms23169234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balasov M, Akhmetova K, Chesnokov I. Drosophila model of Meier-Gorlin syndrome based on the mutation in a conserved C-Terminal domain of Orc6. Am J Med Genet A. 2015;167A:2533–40. doi: 10.1002/ajmg.a.37214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knapp KM, Murray J, Temple IK, Bicknell LS. Successful pregnancies in an adult with Meier-Gorlin syndrome harboring biallelic CDT1 variants. Am J Med Genet A. 2021;185:871–6. doi: 10.1002/ajmg.a.62016. [DOI] [PubMed] [Google Scholar]
- 34.Kim YM, Lee YJ, Park JH, Lee HD, Cheon CK, Kim SY, et al. High diagnostic yield of clinically unidentifiable syndromic growth disorders by targeted exome sequencing. Clin Genet. 2017;92:594–605. doi: 10.1111/cge.13038. [DOI] [PubMed] [Google Scholar]
- 35.Jee J, Mizuno T, Kamada K, Tochio H, Chiba Y, Yanagi K, et al. Structure and mutagenesis studies of the C-terminal region of licensing factor Cdt1 enable the identification of key residues for binding to replicative helicase Mcm proteins. J Biol Chem. 2010;285:15931–40. doi: 10.1074/jbc.M109.075333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pozo PN, Matson JP, Cole Y, Kedziora KM, Grant GD, Temple B, et al. Cdt1 variants reveal unanticipated aspects of interactions with cyclin/CDK and MCM important for normal genome replication. Mol Biol Cell. 2018;29:2989–3002. doi: 10.1091/mbc.E18-04-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang F, Riera A, Evrin C, Sun J, Li H, Speck C, et al. Cdc6 ATPase activity disengages Cdc6 from the pre-replicative complex to promote DNA replication. Elife. 2015;4:e05795. doi: 10.7554/eLife.05795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herbig U, Marlar CA, Fanning E. The Cdc6 nucleotide-binding site regulates its activity in DNA replication in human cells. Mol Biol Cell. 1999;10:2631–45. doi: 10.1091/mbc.10.8.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zabnenkova V, Shchagina O, Makienko O, Matyushchenko G, Ryzhkova O. Novel compound heterozygous variants in the CDC6 gene in a Russian patient with Meier-Gorlin Syndrome. Appl Clin Genet. 2022;15:1–10. doi: 10.2147/TACG.S342804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yao L, Chen J, Wu X, Jia S, Meng A. Zebrafish cdc6 hypomorphic mutation causes Meier-Gorlin syndrome-like phenotype. Hum Mol Genet. 2017;26:4168–80. doi: 10.1093/hmg/ddx305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McGarry TJ, Kirschner MW. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–53. doi: 10.1016/S0092-8674(00)81209-X. [DOI] [PubMed] [Google Scholar]
- 42.Yoshida K, Oyaizu N, Dutta A, Inoue I. The destruction box of human Geminin is critical for proliferation and tumor growth in human colon cancer cells. Oncogene. 2004;23:58–70. doi: 10.1038/sj.onc.1206987. [DOI] [PubMed] [Google Scholar]
- 43.Frigola J, Remus D, Mehanna A, Diffley JF. ATPase-dependent quality control of DNA replication origin licensing. Nature. 2013;495:339–43. doi: 10.1038/nature11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ryu S, Holzschuh J, Erhardt S, Ettl AK, Driever W. Depletion of minichromosome maintenance protein 5 in the zebrafish retina causes cell-cycle defect and apoptosis. Proc Natl Acad Sci USA. 2005;102:18467–72. doi: 10.1073/pnas.0506187102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ravindran E, Gutierrez de Velazco C, Ghazanfar A, Kraemer N, Zaqout S, Waheed A, et al. Homozygous mutation in MCM7 causes autosomal recessive primary microcephaly and intellectual disability. J Med Genet. 2022;59:453–61. doi: 10.1136/jmedgenet-2020-107518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Knapp KM, Fellows B, Aggarwal S, Dalal A, Bicknell LS. A synonymous variant in a non-canonical exon of CDC45 disrupts splicing in two affected sibs with Meier-Gorlin syndrome with craniosynostosis. Eur J Med Genet. 2021;64:104182. doi: 10.1016/j.ejmg.2021.104182. [DOI] [PubMed] [Google Scholar]
- 47.Li X, Zhang LZ, Yu L, Long ZL, Lin AY, Gou CY. Prenatal diagnosis of Meier-Gorlin syndrome 7: a case presentation. BMC Pregnancy Childbirth. 2021;21:381. doi: 10.1186/s12884-021-03868-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ting CY, Bhatia NS, Lim JY, Goh CJ, Vasanwala RF, Ong CC, et al. Further delineation of CDC45-related Meier-Gorlin syndrome with craniosynostosis and review of literature. Eur J Med Genet. 2020;63:103652. doi: 10.1016/j.ejmg.2019.04.009. [DOI] [PubMed] [Google Scholar]
- 49.Unolt M, Kammoun M, Nowakowska B, Graham GE, Crowley TB, Hestand MS, et al. Pathogenic variants in CDC45 on the remaining allele in patients with a chromosome 22q11.2 deletion result in a novel autosomal recessive condition. Genet Med. 2020;22:326–35. doi: 10.1038/s41436-019-0645-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simon AC, Sannino V, Costanzo V, Pellegrini L. Structure of human Cdc45 and implications for CMG helicase function. Nat Commun. 2016;7:11638. doi: 10.1038/ncomms11638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schulz S, Mensah MA, de Vries H, Frober R, Romeike B, Schneider U, et al. Microcephaly, short stature, and limb abnormality disorder due to novel autosomal biallelic DONSON mutations in two German siblings. Eur J Hum Genet. 2018;26:1282–7. doi: 10.1038/s41431-018-0128-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Danyel M, Cheng Z, Jung C, Boschann F, Pantel JT, Hajjir N, et al. Differentiation of MISSLA and Fanconi anaemia by computer-aided image analysis and presentation of two novel MISSLA siblings. Eur J Hum Genet. 2019;27:1827–35. doi: 10.1038/s41431-019-0469-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abdelrahman HA, John A, Ali BR, Al-Gazali L. Further delineation of the Microcephaly-Micromelia Syndrome associated with loss-of-function variants in DONSON. Mol Syndromol. 2019;10:171–6. doi: 10.1159/000497337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Evrony GD, Cordero DR, Shen J, Partlow JN, Yu TW, Rodin RE, et al. Integrated genome and transcriptome sequencing identifies a noncoding mutation in the genome replication factor DONSON as the cause of microcephaly-micromelia syndrome. Genome Res. 2017;27:1323–35. doi: 10.1101/gr.219899.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bandura JL, Beall EL, Bell M, Silver HR, Botchan MR, Calvi BR. Humpty dumpty is required for developmental DNA amplification and cell proliferation in Drosophila. Curr Biol. 2005;15:755–9. doi: 10.1016/j.cub.2005.02.063. [DOI] [PubMed] [Google Scholar]
- 56.Fuchs F, Pau G, Kranz D, Sklyar O, Budjan C, Steinbrink S, et al. Clustering phenotype populations by genome-wide RNAi and multiparametric imaging. Mol Syst Biol. 2010;6:370. doi: 10.1038/msb.2010.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bockholt M, Mehnert-Theuerkauf A, Vehling S. [Association of illness perceptions with demoralization and psychological distress in cancer: a longitudinal study. Psychother Psychosom Med Psychol. 2021;71:464–72. doi: 10.1055/a-1522-8500. [DOI] [PubMed] [Google Scholar]
- 58.Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022;50:D439–D44. doi: 10.1093/nar/gkab1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ratnayeke N, Baris Y, Chung M, Yeeles JTP, Meyer T. CDT1 inhibits CMG helicase in early S phase to separate origin licensing from DNA synthesis. Mol Cell. 2023;83:26–42. doi: 10.1016/j.molcel.2022.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bernal JA, Venkitaraman AR. A vertebrate N-end rule degron reveals that Orc6 is required in mitosis for daughter cell abscission. J Cell Biol. 2011;192:969–78. doi: 10.1083/jcb.201008125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin YC, Liu D, Chakraborty A, Kadyrova LY, Song YJ, Hao Q, et al. Orc6 is a component of the replication fork and enables efficient mismatch repair. Proc Natl Acad Sci USA. 2022;119:e2121406119. doi: 10.1073/pnas.2121406119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Agarwal S, Smith KP, Zhou Y, Suzuki A, McKenney RJ, Varma D. Cdt1 stabilizes kinetochore-microtubule attachments via an Aurora B kinase-dependent mechanism. J Cell Biol. 2018;217:3446–63. doi: 10.1083/jcb.201705127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Varma D, Chandrasekaran S, Sundin LJ, Reidy KT, Wan X, Chasse DA, et al. Recruitment of the human Cdt1 replication licensing protein by the loop domain of Hec1 is required for stable kinetochore-microtubule attachment. Nat Cell Biol. 2012;14:593–603. doi: 10.1038/ncb2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tingler M, Philipp M, Burkhalter MD. DNA replication proteins in primary microcephaly syndromes. Biol Cell. 2022;114:143–59. doi: 10.1111/boc.202100061. [DOI] [PubMed] [Google Scholar]
- 65.Knockleby J, Lee H. Same partners, different dance: involvement of DNA replication proteins in centrosome regulation. Cell Cycle. 2010;9:4487–91. doi: 10.4161/cc.9.22.14047. [DOI] [PubMed] [Google Scholar]
- 66.Hossain M, Stillman B. Opposing roles for DNA replication initiator proteins ORC1 and CDC6 in control of Cyclin E gene transcription. Elife. 2016;5:e12785. doi: 10.7554/eLife.12785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hossain M, Stillman B. Meier-Gorlin syndrome mutations disrupt an Orc1 CDK inhibitory domain and cause centrosome reduplication. Genes Dev. 2012;26:1797–810. doi: 10.1101/gad.197178.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maerz LD, Casar Tena T, Gerhards J, Donow C, Jeggo PA, Philipp M. Analysis of cilia dysfunction phenotypes in zebrafish embryos depleted of Origin recognition complex factors. Eur J Hum Genet. 2019;27:772–82. doi: 10.1038/s41431-019-0338-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stiff T, Alagoz M, Alcantara D, Outwin E, Brunner HG, Bongers EM, et al. Deficiency in origin licensing proteins impairs cilia formation: implications for the aetiology of Meier-Gorlin syndrome. PLoS Genet. 2013;9:e1003360. doi: 10.1371/journal.pgen.1003360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Avasthi P, Marshall WF. Stages of ciliogenesis and regulation of ciliary length. Differentiation. 2012;83:S30–42. doi: 10.1016/j.diff.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Conte MI, Poli MC, Taglialatela A, Leuzzi G, Chinn IK, Salinas SA, et al. Partial loss-of-function mutations in GINS4 lead to NK cell deficiency with neutropenia. JCI Insight. 2022;7:e154948. doi: 10.1172/jci.insight.154948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cottineau J, Kottemann MC, Lach FP, Kang YH, Vely F, Deenick EK, et al. Inherited GINS1 deficiency underlies growth retardation along with neutropenia and NK cell deficiency. J Clin Invest. 2017;127:1991–2006. doi: 10.1172/JCI90727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gineau L, Cognet C, Kara N, Lach FP, Dunne J, Veturi U, et al. Partial MCM4 deficiency in patients with growth retardation, adrenal insufficiency, and natural killer cell deficiency. J Clin Invest. 2012;122:821–32. doi: 10.1172/JCI61014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bongers EM, Opitz JM, Fryer A, Sarda P, Hennekam RC, Hall BD, et al. Meier-Gorlin syndrome: report of eight additional cases and review. Am J Med Genet. 2001;102:115–24. doi: 10.1002/ajmg.1452. [DOI] [PubMed] [Google Scholar]
- 75.Carroll TD, Newton IP, Chen Y, Blow JJ, Nathke I. Lgr5(+) intestinal stem cells reside in an unlicensed G(1) phase. J Cell Biol. 2018;217:1667–85. doi: 10.1083/jcb.201708023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Matson JP, Dumitru R, Coryell P, Baxley RM, Chen W, Twaroski K, et al. Rapid DNA replication origin licensing protects stem cell pluripotency. Elife. 2017;6:e30473. doi: 10.7554/eLife.30473. [DOI] [PMC free article] [PubMed] [Google Scholar]