Abstract
T22, an analog of polyphemusin II (18 amino acid residues), was found to block T-tropic human immunodeficiency virus type 1 (HIV-1) entry into target cells as a CXCR4 inhibitor. We synthesized T134, a small analog (14 amino acid residues) of T22 with reduced positive charges. T134 exhibited highly potent activity and significantly less cytotoxicity in comparison to that of T22. T134 prevents the anti-CXCR4 monoclonal antibody from binding to peripheral blood mononuclear cells but has no effect on the binding of anti-CCR5 monoclonal antibodies. Since T134 inhibits the binding of stromal cell-derived factor-1 (SDF-1) to MT-4 cells, it seems that T134 prevents HIV-1 entry by binding to CXCR4. The bicyclam AMD3100 has also been shown to block HIV-1 entry via CXCR4 but not via CCR5. Both T134 and AMD3100 are CXCR4 antagonists and low-molecular-weight compounds but have different structures. Our results indicate that T134 is active against wild-type T-tropic HIV-1 strains and against AMD3100-resistant strains.
For anti-human immunodeficiency virus (anti-HIV) chemotherapy, the virus-cell fusion process is an attractive target. If specific drugs can inhibit the stage of virus-cell fusion, HIV type 1 (HIV-1) proviral DNA cannot be integrated into the cell genome, which prevents the spread of infection. For entry into target cells, HIV-1 requires a primary receptor, CD4, and coreceptors such as chemokine receptors. CXC chemokine receptor 4 (CXCR4) is a coreceptor for the entry of T-cell-line-tropic (T-tropic) strains of HIV-1 (15), and the CC chemokine receptor 5 (CCR5) serves as a coreceptor for macrophage tropic (M-tropic) strains of HIV-1 (1, 6, 11, 14). Therefore, compounds which interact with the chemokine receptors may be the ultimate hope for anti-HIV drugs. The ligands identified for these receptors, stromal cell-derived factor-1 (SDF-1) for CXCR4 (3, 22) and RANTES, macrophage inflammatory protein-1α (MIP-1α), and MIP-1β for CCR5 (7), were shown to be potent competitive inhibitors of HIV-1 entry into cells expressing the appropriate coreceptor.
We previously found that a synthetic peptide of T22 ([Tyr5,12, Lys7]-polyphemusin II), which consists of 18 amino acid residues and is an analog of polyphemusin II isolated from the hemocyte debris of American horseshoe crabs (Limulus polyphemus), showed strong anti-HIV activity in vitro (21, 28, 31). Recently, it was also found that T22 blocked T-tropic HIV-1 entry into target cells via CXCR4, not via CCR5 (19). In a series of investigations in the search for more active anti-HIV derivatives, we synthesized two smaller analogs, TW70 (des-[Cys8,13, Tyr9,12]-[d-Lys10, Pro11]-T22) and T134 (l-citrulline16-TW70 substituted for the C-terminal amide by a carboxylic acid), which have 14 amino acid residues and reduced basic amino acid residues less than T22 (27). TW70 and T134 showed more pronounced anti-HIV-1 activity than T22.
The bicyclam AMD3100 {octahydrochloride dihydrate of 1,1′ - [1,4 - phenylene - bis - (methylene)] - bis - 1,4,8,11 - tetra - azacyclotetradecane with a molecular weight of 830} has also been shown to block HIV-1 entry and membrane fusion via CXCR4 but not via CCR5 (13, 26). The structure of AMD3100 is different from that of T134 (10, 27), but both compounds function via CXCR4 to block HIV-1 entry into target cells. This investigation shows that T134 is a CXCR4 antagonist and attempts to clarify the cross-resistance observed between T134 and AMD3100, by using AMD3100-resistant HIV-1 prepared previously from a clinical HIV-1 isolate (2).
We first studied the anti-HIV-1 activity of T22 and its derivatives, TW70 and T134. Table 1 summarizes the amino acid sequence of each test compound and their anti-HIV activities and cytotoxicities in MT-4 cells by the MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide] method (21, 23). We have already investigated the structure-activity relationship of more than 140 analogs of T22 (27, 30). These investigations showed that the analogs that were small in size and contained reduced basic amino acids exhibited highly potent activity and low cytotoxicity. Table 1 shows that T134 exhibited much higher potency and lower cytotoxicity than T22 and TW70, and therefore it was more suitable for further analysis in the investigation of the mechanism of these types of compounds.
TABLE 1.
Anti-HIV activities of T22, TW70, and T134 in MT-4 cellsa
| Compound | Sequenceb | CC50c (μg/ml) | EC50 (μg/ml) | SId |
|---|---|---|---|---|
| T22 | R-R-W-C-Y-R-K-C-Y-K-G-Y-C-Y-R-K-C-R-CONH2 | 49 | 0.076 | 645 |
| TW70 | R-R-W-C-Y-R-K-dK-P-Y-R-K-C-R-CONH2 | 40 | 0.030 | 1333 |
| T134 | R-R-W-C-Y-R-K-dK-P-Y-R-Ci-C-R-COOH | 305 | 0.009 | 33889 |
MT-4 cells were infected with HIV-1IIIB (multiplicity of infection, 0.01).
The substituted amino acids are underlined; deleted amino acids are double-underlined. Ci, l-citrulline.
CC50, 50% cytotoxic concentration.
SI, selectivity index based on the ratio of CC50 to EC50.
The possibility that T134 inhibits T-tropic HIV-1 but not M-tropic HIV-1 infection as exhibited by T22 was investigated with the multinuclear activation of the galactosidase indicator (MAGI) assay in MAGI-CCR5 cells (17). Table 2 demonstrates that T134 and its derivatives inhibited only T-tropic HIV-1 strains such as HIV-1IIIB and not the M-tropic HIV-1 strains such as HIV-1JR-FL. This data suggests that T134 and other derivatives of T22 inhibit the T-tropic HIV-1 infection mediated by CXCR4 but not the M-tropic HIV-1 infection mediated by CCR5.
TABLE 2.
Effects of T22, TW70, and T134 on T-tropic or M-tropic HIV-1-infected MAGI-CCR5 cells
| Compound and concn (μg/ml) | No. of blue cells (mean ± SD)a
|
|
|---|---|---|
| T-tropic HIV-1b | M-tropic HIV-1c | |
| None | 296 ± 5 | 430 ± 8 |
| T22 | ||
| 10 | 0 ± 0 | 377 ± 33 |
| 0.1 | 13 ± 3 | 347 ± 82 |
| TW70 | ||
| 10 | 4 ± 1 | 337 ± 7 |
| 0.1 | 6 ± 2 | 372 ± 60 |
| T134 | ||
| 10 | 5 ± 2 | 372 ± 75 |
| 0.1 | 3 ± 1 | 328 ± 68 |
The MAGI assay was performed as described by Kimpton and Emerman (17), except that DEAE-dextran was not used.
HIV-1IIIB.
HIV-1JR-FL.
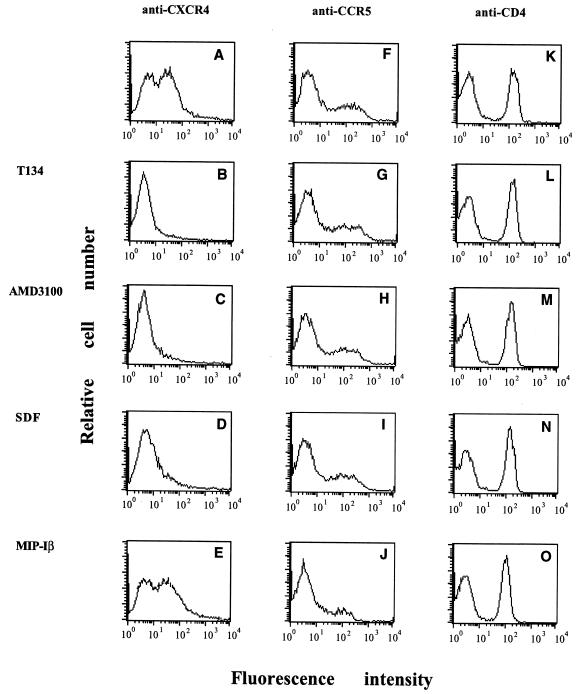
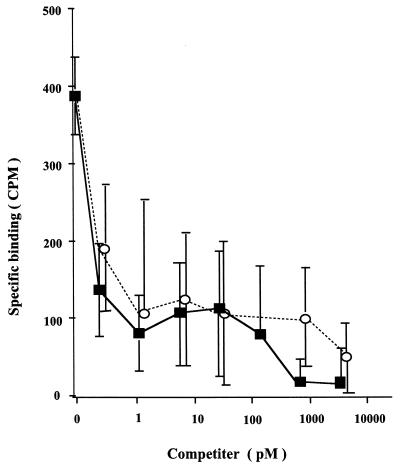
We used anti-CXCR4 and -CCR5 monoclonal antibodies (MAbs) to examine whether T134 interacted with these chemokine receptors. Preincubation of peripheral blood mononuclear cells (PBMC) with T134 caused binding inhibition of 12G5 (R & D Systems Inc., Minneapolis, Minn.). This binding inhibition was also observed for our synthesized SDF-1[1-67] (29) and AMD3100 (4). As shown in Fig. 1, T134, AMD3100, and SDF-1 inhibited the binding activity of 12G5 to PBMC; however, none of these compounds affected the binding of anti-CCR5 MAb (2D7; NIH AIDS Research and Reference Reagent Program) to PBMC and only MIP-1α inhibited the binding of 2D7 to PBMC. T134 and AMD3100 had no effect on the binding of anti-CD4 MAbs to PBMC. These findings show that T134 interacts specifically with CXCR4 to prevent both 12G5 binding and coreceptor activity. The specific interaction of T134 with CXCR4 was also confirmed in other human T-cell lines such as MT-4 (data not shown). To investigate the interaction of T134 with the binding of CXCR4 and its ligand SDF-1α, a binding inhibition experiment was performed. Figure 2 demonstrates that T134 inhibits the binding of 125I-labeled SDF-1α (0.02 nM; DuPont-NEN, Boston, Mass.) to MT-4 cells and complete inhibition was observed at a concentration of 0.8 nM; AMD3100 inhibition of SDF-1α binding was observed at the same concentration. These results reveal that T134 directly blocks the interaction of virus, anti-CXCR4 MAb, or SDF-1 with CXCR4. Our results suggest that the structure of T134 mimics the region of SDF that is involved in binding to CXCR4. In fact, the CXC chemokines have a core structure consisting of three antiparallel β-sheets (16) and T134 is also comprised of an antiparallel β-sheet structure similar to that of T22 and TW70 (27). Our results were also supported by Murakami and colleagues, who demonstrated that cellular signal transduction induced by SDF-1 binding to CXCR4, detected by using Ca2+ mobilization, was inhibited by T22 (19).
FIG. 1.
Effect on binding to PBMC of an anti-CXCR4 MAb (12G5), an anti-CCR5 MAb (2D7), or an anti-CD4 MAb in the presence of T134, SDF, AMD3100, or MIP-α. PBMC were preincubated with T134 (5 μg/ml) (B, G, and L), AMD3100 (5 μg/ml) (C, H, and M), SDF-1 (5 μg/ml) (D, I, and N), or MIP-1α (5 μg/ml) (E, J, and O) at 4°C for 30 min. 12G5 (5 μg/ml) (A to E), 2D7 (5 μg/ml) (F to J), or the anti-CD4 MAb (5 μg/ml) (K to O) was then added to the PBMC, and they were incubated at 4°C for 30 min and labeled with fluorescein isothiocyanate-conjugated anti-mouse immunoglobulin G. After the cells were stained, CXCR4, CCR5, and CD4 expression was analyzed with a FACScan flow cytometer. The results are representative of one of three independent experiments.
FIG. 2.
Competitive inhibition of 125I-SDF-1α binding to MT-4 cells by T134 and AMD3100. The counts per minute of 125I-SDF-1α specifically bound to MT-4 cells in the presence of the indicated T134 (■) or AMD3100 (○) concentrations were determined. Specific binding was estimated by subtracting from each data point the amount of nonspecific binding observed in the presence of excess unlabeled SDF. T134, AMD3100, or unlabeled SDF immediately followed by 0.02 nM 125I-SDF-1α was added to MT-4 cells in suspension in cold binding buffer (RPMI 1640 medium containing 10% fetal calf serum and 25 mM HEPES [pH 7.5]). The resulting sample were gently rocked for 1 h at 4°C, washed, solubilized with NaOH, and counted in a gamma counter. The data are the means ± standard deviations from three replicate determinations.
Several variants isolated from patients with one class of protease inhibitor exhibited cross-resistance to other structurally diverse protease inhibitors (8, 9, 24). This type of cross-resistance is frequently observed in non-nucleoside reverse transcriptase inhibitors (5, 18), and it creates a disadvantage for clinical use of the same class antiviral drugs in combination therapy. AMD3100 was found to function in the same manner as T134. However, the chemical structures of these compounds were extremely different (10, 27). Therefore, we were interested in investigating whether cross-resistance like that found with protease inhibitors or non-nucleoside reverse transcriptase inhibitors would occur in these two anti-HIV-1 compounds. AMD3100-resistant HIV-1 was generated from a clinical HIV-1 isolate (A018A) after several passages of the virus in cell culture in the presence of AMD3100 (2). The anti-HIV-1 activity of AMD3100 was assessed by using the MAGI-CCR5 assay for wild-type clinically isolated HIV-1 and AMD3100-resistant HIV-1 strains. Table 3 demonstrates that the concentration of AMD3100 required to achieve 50% antiviral activity in AMD3100-resistant strains was 20- to 27-fold higher than that needed for the wild type (strain A018A). However, approximately equal concentrations of T134 were needed to achieve 50% effective concentrations (EC50s) in both the wild-type and AMD3100-resistant strains. This suggests that both T134 and AMD3100 inhibit HIV-1 infection via CXCR4, but these two compounds have no cross-resistance to each other. Cross-resistance was also not exhibited when zidovudine, 1-ethoxymethyl-5-ethyl-6-(phenylthio)-uracil (E-EPU), or dextran sulfate was tested against AMD3100-resistant strains (data not shown).
TABLE 3.
Insensitivity of T134 to AMD3100-resistant virus
| Compound | EC50 (nM)a
|
||
|---|---|---|---|
| A018A | A031-1 | A031-2 | |
| T134 | 18.5 | 19.1 | 28.6 |
| AMD3100 | 49.0 | 1,000.0 | 1,330.0 |
| Zidovudine | 50.6 | 24.0 | 16.2 |
The EC50 was calculated on the basis of the reduction in the number of blue cells induced by wild-type HIV-1 (A018A) and AMD3100-resistant HIV-1 isolates (A031-1 and A031-2) in the MAGI assay.
We had also intended to prepare HIV-1 strains resistant to T22 or T134 by the cell culture methods used to generate AMD3100-resistant HIV-1. However, we were unable to prepare T134-resistant strains for more than 25 passages. T22 and T134 may not induce resistant viruses easily. De Clercq and colleagues reported that a mutant of HIV-1NL4-3 with decreased sensitivity to AMD3100 has multiple amino acid substitutions in env glycoprotein 120 (gp120), most of them within, or in proximity to, the V3 loop (12). But they also reported that it was difficult to obtain a completely resistant AMD3100 virus (12, 25). This information may show the potential of these CXCR4 antagonists as therapeutic drugs. It was reported that SDF-1α-resistant HIV-1NL4-3, which was more easily prepared than AMD3100-resistant HIV-1NL4-3, had been produced. Of the nine mutations detected in gp120 of the SDF-1α-resistant virus, four were located in the V3 domain and all four were also detected in the AMD3100-resistant virus (25). The SDF-1α-resistant virus became resistant to SDF-1β and to anti-CXCR4 MAbs. However, AMD3100 was still active against the SDF-1α-resistant virus. Although HIV-1NL4-3 with complete resistance to AMD3100 was not obtained, it was shown that a larger number of mutations were present in the gp120 of the AMD3100-resistant virus than in the gp120 of SDF-1α-resistant virus. AMD3100 and T134 may not induce resistant virus easily in comparison with SDF-1α. These results may show that these compounds have a much stronger interaction with CXCR4 than the natural ligand SDF-1 itself, a finding which is also reflected by the fact that T134 and AMD3100 compete with SDF-1 at much lower concentrations (under 1 pM) than that for 125I-SDF-1α (20 pM). Since T134 and AMD3100 are much smaller in size than SDF-1, these compounds may be able to interact with CXCR4 at a higher affinity than SDF-1. It was reported that knocking out the SDF-1 gene in mice creates a lethal phenotype. Thus, SDF-1 might be a necessary chemokine for prenatal viability, B lymphopoiesis, bone marrow myelopoiesis, and cardiac ventricular septal formation (20). However, the biological importance of CXCR4 for T lymphocyte function and whether blocking of the function of CXCR4 is detrimental to the adult host are not clear.
In our present study, we can conclude that T134 has anti-HIV-1 activity against not only the wild type but also AMD3100-resistant strains. We produced two different CXCR4 antagonists, which, should HIV-1 acquire resistance to one of the inhibitors, would allow for the use of another inhibitor to suppress the resistant strain. This observation indicates the potential for using these inhibitors as preventive and/or therapeutic drugs for HIV infections.
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science and Culture of Japan and a Research Grant from the Human Science Foundation. M.P. is grateful to the Japanese Foundation for AIDS Prevention, Tokyo, Japan, for a fellowship.
Anti-CCR5 MAb (2D7) and MAGI-CCR5 cells were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, with 2D7 from LeukoSite, Inc., and MAGI-CCR5 from Julie Overbaugh.
REFERENCES
- 1.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 2.Baba, M., N. Yamamoto, R. Pauwels, Z. Debyser, S. Shigeta, E. De Clercq, G. Bridger, G. Henson, and M. Abrams. 1993. In vitro isolation of bicyclam-resistant variants of human immunodeficiency virus type 1. Antiviral Res. 20:(Suppl. 1.1):93.
- 3.Bleul C C, Farzan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J, Springer T A. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature. 1996;382:829–833. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 4.Bridger G J, Skerlj R T, Thornton D, Padmanabhan S, Martellucci S A, Henson G W, Abrams M J, Yamamoto N, De Vreese K, Pauwels R, De Clercq E. Synthesis and structure-activity relationships of phenylenebis-(methylene)-linked bis-tetraazamacrocycles that inhibit HIV replication. Effects of macrocyclic ring size and substituents on the aromatic linker. J Med Chem. 1995;38:366–378. doi: 10.1021/jm00002a019. [DOI] [PubMed] [Google Scholar]
- 5.Bymes V W, Emini E A, Schleif W A, Condra J H, Schneider C L, Long W J, Wolfgang J A, Graham D J, Gotlib L, Schlabach A J. Susceptibilities of human immunodeficiency virus type 1 enzyme and viral variants expressing multiple resistance-engendering amino acid substitutions to reserve transcriptase inhibitors. Antimicrob Agents Chemother. 1994;38:1404–1407. doi: 10.1128/aac.38.6.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 7.Cocchi F, DeVico A L, Demo A G, Arya S K, Gallo R C, Lusso P. Identification of RANTES, MIP-1α, and MIP-1β as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 8.Condra J H, Holder D J, Schleif W A, Blahy O M, Danovich R M, Gabryelski L J, Graham D J, Laird D, Quintero J C, Rhodes A, Robbins H L, Roth E, Shivaprakash M, Yang T, Chodakewitz J A, Deutsch P J, Leavitt R Y, Massari F E, Mellors J W, Squires K E, Steigbigel R T, Teppler H, Emini E A. Genetic correlates of in vivo viral resistance to indinavir, a human immunodeficiency virus type 1 protease inhibitor. J Virol. 1996;70:8270–8276. doi: 10.1128/jvi.70.12.8270-8276.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Condra J H, Schleif W A, Blahy O M, Gabryelski L J, Graham D J, Quintero J C, Rhodes A, Robbins H L, Roth E, Shivaprakash M, Titus D, Yang T, Teppler H, Squires K E, Deutsch P J, Emini E A. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature. 1995;374:569–571. doi: 10.1038/374569a0. [DOI] [PubMed] [Google Scholar]
- 10.De Clercq E, Yamamoto N, Pauwels R, Balzarini J, Witvrouw M, De Vreese K, Debyser Z, Rosenwirth B, Peichl P, Datema R, Thornton D, Skerlj R, Gaul F, Padmanabhan S, Bridger G, Henson G, Abrams M. Highly potent and selective inhibition of human immunodeficiency virus by the bicyclam derivative JM3100. Antimicrob Agents Chemother. 1994;38:668–674. doi: 10.1128/aac.38.4.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng H, Liu R, Ellmeiner W, Choe S, Unutmaz D, Burkhart M, Di.Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–667. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 12.De Vreese K, Kofler-Mongold V, Leutgeb C, Weber V, Vermeire K, Schacht S, Anne J, De Clercq E. The molecular target of bicyclams, potent inhibitors of human immunodeficiency virus replication. J Virol. 1996;70:689–696. doi: 10.1128/jvi.70.2.689-696.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donzella G A, Schols D, Lin S W, Este J A, Nagashima K A, Maddon P J, Allaway G P, Sakmar T P, Henson G, De.Clercq E, Moore J P. AMD3100, a small molecule inhibitor of HIV-1 entry via the CXCR4 co-receptor. Nat Med. 1998;4:72–77. doi: 10.1038/nm0198-072. [DOI] [PubMed] [Google Scholar]
- 14.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 15.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 16.Harrison P M, Sternberg M J. The disulphide beta-cross: from cystine geometry and clustering to classification of small disulphide-rich protein folds. J Mol Biol. 1996;264:603–623. doi: 10.1006/jmbi.1996.0664. [DOI] [PubMed] [Google Scholar]
- 17.Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larder B A, Kellam P, Kemp S D. Convergent combination therapy can select viable multidrug-resistant HIV-1 in vitro. Nature. 1993;365:451–453. doi: 10.1038/365451a0. [DOI] [PubMed] [Google Scholar]
- 19.Murakami T, Nakajima T, Koyanagi Y, Tachibana K, Fujii N, Tamamura H, Yoshida N, Waki M, Matsumoto A, Yoshie O, Kishimoto T, Yamamoto N, Nagasawa T. A small molecule CXCR4 inhibitor that blocks T cell line-tropic HIV-1 infection. J Exp Med. 1997;186:1389–1393. doi: 10.1084/jem.186.8.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagasawa T, Hirota S, Tachibana K, Takakura N, Nishikawa S, Kitamura Y, Yoshida N, Kikutani H, Kishimoto T. Defect of B-cell lymphopoiesis and bone-marrow myelopoiesis in mice lacking the CXC chemokine PBSF/SDF-1. Nature. 1996;382:635–638. doi: 10.1038/382635a0. [DOI] [PubMed] [Google Scholar]
- 21.Nakashima H, Masuda M, Murakami T, Koyanagi Y, Matsumoto A, Fujii N, Yamamoto N. Anti-human immunodeficiency virus activity of a novel synthetic peptide, T22([Tyr-5,12, Lys-7]polyphemusin II): a possible inhibitor of virus-cell fusion. Antimicrob Agents Chemother. 1992;36:1249–1255. doi: 10.1128/aac.36.6.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier J L, Arenzana-Seisdedos F, Schwarts O, Heard J-M, Clark-Lewis I, Legler D F, Loetscher M, Baggiolini M, Moser B. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature. 1996;382:833–835. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- 23.Pauwels R, Balzarini J, Baba M, Snoeck R, Schols D, Herdewijn P, Desmyter J, De Clercq E. Rapid and automated tetrazolium-based colorimetric assay for the detection of anti-HIV compounds. J Virol Methods. 1988;20:309–321. doi: 10.1016/0166-0934(88)90134-6. [DOI] [PubMed] [Google Scholar]
- 24.Schock H B, Garsky V M, Kuo L C. Mutational anatomy of an HIV-1 protease variant conferring cross-resistance to protease inhibitors in clinical trials. J Biol Chem. 1996;271:31957–31963. doi: 10.1074/jbc.271.50.31957. [DOI] [PubMed] [Google Scholar]
- 25.Schols D, Este J A, Cabrera C, De Clercq E. T-cell-line-tropic human immunodeficiency virus type 1 that is made resistant to stromal cell-derived factor 1α contains mutations in the envelope gp120 but does not show a switch in coreceptor use. J Virol. 1998;72:4032–4037. doi: 10.1128/jvi.72.5.4032-4037.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schols D, Struyf S, Van Damme J, Este J A, Henson G, De Clercq E. Inhibition of T-tropic HIV strains by selective antagonization of the chemokine receptor CXCR4. J Exp Med. 1997;186:1383–1388. doi: 10.1084/jem.186.8.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamamura H, Arakaki R, Funakoshi H, Imai M, Otaka A, Ibuka T, Nakashima H, Murakami T, Waki M, Matsumoto A, Yamamoto N, Fujii N. Effective lowly cytotoxic analogs of an HIV-cell fusion inhibitor, T22([Tyr5,12, Lys7]-polyphemusin II) Bioorg Med Chem. 1998;6:231–238. doi: 10.1016/s0968-0896(97)10037-2. [DOI] [PubMed] [Google Scholar]
- 28.Tamamura H, Kuroda M, Masuda M, Otaka A, Funakoshi S, Nakashima H, Yamamoto N, Waki M, Matsumoto A, Lancelin J M, Kohda D, Tate S, Inagaki F, Fujii N. A comparative study of the solution structures of tachyplesin I and a novel anti-HIV synthetic peptide, T22([Tyr5,12,Lys7]-polyphemusin II), determined by nuclear magnetic resonance. Biochim Biophys Acta. 1993;1163:209–216. doi: 10.1016/0167-4838(93)90183-r. [DOI] [PubMed] [Google Scholar]
- 29.Tamamura H, Matsumoto F, Sakano K, Otaka A, Ibuka T, Fujii N. Unambiguous synthesis of stromal cell-derived factor-1 by regioselective disulfide bond formation using a DMSO-aqueous HCl system. Chem Commun. 1998;1998:151–152. [Google Scholar]
- 30.Tamamura H, Murakami T, Masuda M, Otaka A, Takada W, Ibuka T, Nakashima H, Waki M, Matsumoto A, Yamamoto N, Fujii N. Structure-activity relationships of an anti-HIV peptide, T22. Biochem Biophys Res Commun. 1994;205:1729–1735. doi: 10.1006/bbrc.1994.2868. [DOI] [PubMed] [Google Scholar]
- 31.Tamamura H, Otaka A, Murakami T, Ibuka T, Sakano K, Waki M, Matsumoto A, Yamamoto N, Fujii N. An anti-HIV peptide, T22, forms a highly active complex with Zn(II) Biochem Biophys Res Commun. 1996;229:648–652. doi: 10.1006/bbrc.1996.1858. [DOI] [PubMed] [Google Scholar]