Abstract
Canonical splice site variants affecting the 5′ GT and 3′ AG nucleotides of introns result in severe missplicing and account for about 10% of disease-causing genomic alterations. Treatment of such variants has proven challenging due to the unstable mRNA or protein isoforms that typically result from disruption of these sites. Here, we investigate CRISPR-Cas9-mediated adenine base editing for such variants in the cystic fibrosis transmembrane conductance regulator (CFTR) gene. We validate a CFTR expression minigene (EMG) system for testing base editing designs for two different targets. We then use the EMG system to test non-standard single-guide RNAs with either shortened or lengthened protospacers to correct the most common cystic fibrosis-causing variant in individuals of African descent (c.2988+1G>A). Varying the spacer region length allowed placement of the editing window in a more efficient context and enabled use of alternate protospacer adjacent motifs. Using these modifications, we restored clinically significant levels of CFTR function to human airway epithelial cells from two donors bearing the c.2988+1G>A variant.
Keywords: MT: RNA/DNA Editing, canonical splice site variant, CSSV, CRISPR-Cas9-mediated adenine base editing, ABE, primary airway epithelial cells, CFTR function, cystic fibrosis, CF
Graphical abstract

Cutting and colleagues use adenine base adenine base editing to correct disease-causing canonical splice site variants, which are often difficult to treat, in the CFTR gene. Using modified RNA guides, the authors restore CFTR function in human airway epithelial cells for the most common disease-causing CFTR variant in individuals of African descent.
Introduction
Sequence variants that alter the canonical 5′ GT nucleotides of the splice donor or 3′ AG of the splice acceptor are estimated to account for ∼10% of disease-causing genetic variants. These canonical splice site variants (CSSVs) have severe effects as they invariably cause RNA missplicing, typically resulting in exon skipping and production of either no protein or aberrant protein.1,2,3,4 Loss of coding sequence caused by altered splicing generally results in a frameshift that introduces a premature termination codon, which, in turn, activates nonsense-mediated mRNA decay (NMD) leading to degradation of transcript. Even if NMD could be evaded,5,6,7,8,9,10 coding sequence lost to exon skipping and reading frame changes have to be recovered to generate a functional product. Consequently, gene delivery and genome editing are attractive strategies for treatment of individuals carrying CSSVs. While gene delivery is appealing as a “genotype agnostic” therapy, it has the limitation of requiring repeated dosing as therapeutic cargo and targeted cells are turned over.11,12 In contrast, genome editing has the potential to be a “one and done” precision therapy.
Genome editing technology is rapidly developing and encompasses a variety of approaches that target a nuclease or other enzyme to the genome in a sequence-specific manner.13,14,15,16,17 Many of these technologies utilize the bacterial clustered regularly interspersed palindromic repeats/CRISPR-associated protein (CRISPR-Cas) system.16,17 A modified form of CRISPR-Cas9 that performs targeted base editing has proven to be highly efficient with many advantages.18,19,20 Base editors typically use a Cas9 nuclease variant that no longer causes a double-strand DNA break, but only a nick in one strand.18,19,20 This greatly reduces the rate of unintended insertions/deletions, or genomic rearrangements. Adenine base editing is a particularly attractive approach for in vivo genome editing because of its high editing activity and low rate of undesired byproducts at the target site. However, deamination of nearby adenines (bystander editing) can create changes that introduce residue substitutions that may alter or severely limit function of the encoded protein. Furthermore, placement of the target site within the optimal editing window is limited by the distance downstream of the target where a Cas9-compatible protospacer adjacent motif (PAM) is present in the genomic sequence.19,21,22 Finally, testing and refinement of single-guide RNA (sgRNA) design for rare genetic variants is limited by cell line availability. We address the above issues by (1) correcting splice sites, which present a safer target as nearby nucleotide alterations in introns frequently have no functional consequences, (2) testing modifications in protospacer length that increase the range of potential splice site variants that can be efficiently corrected, and (3) using isogenic cell lines with an integrated splicing vector (expression minigene or EMG) bearing target splice site variants to allow for design optimization when primary cells are unavailable (i.e., for very rare variants). Using these approaches, we demonstrate the utility of adenine base editing as a treatment for CSSVs by correction of three different variants in the cystic fibrosis transmembrane conductance regulator (CFTR) gene,23 which cause cystic fibrosis (CF) and apply this approach to restoring CFTR function in primary human airway epithelial cells obtained from the upper and lower respiratory tract.
Results
CRISPR-Cas9-mediated adenine base editing of CFTR CSSVs integrated in an isogenic model of CF airway epithelial cells
Two disease-causing CFTR CSSVs, c.273+1G>A (405+1G>A in the legacy nomenclature) and c.3718-1G>A (legacy 3850-1G>A) (Table 1), which have a sequence context compatible with original versions of Cas9 base editors, were selected to assess genomic correction. Since cell lines bearing these rare variants are not available, we used CFTR EMGs which contain all of the exons of CFTR as well as select abridged or full-length CFTR intronic sequences and faithfully recapitulate splicing patterns observed in primary cells.1,24,25 As reported previously, both variants cause severe missplicing of EMGs when transiently expressed in HEK293 cells (Figure 1A).1 To evaluate genome editing, we integrated the EMGs bearing these splice site variants into the genome of the CF bronchial epithelial cell line, CFBE41o-, which has two non-expressing copies of the common CF-causing variant c.1521_1523delCTT (F508del) and thus produces no endogenous CFTR transcript or protein.26 CFBE14o- cells were previously modified to contain a single genomic Flp recombinase target site in chromosome 8,26 allowing for integration of CFTR EMGs into its genome and constitutive expression of CFTR bearing splice site variants.1,25,26 Lipofectamine-mediated transfection was used to introduce a DNA plasmid encoding NG-ABE8e (Addgene, no. 13849127,28) to CFBE cells stably expressing c.273+1G>A and VRQR-ABEmax (Addgene, no. 11981121,29,30,31) to c.3718-1G>A CFBE cells along with the sgRNA specific to each target (sequences shown in Table S1, gDNA results shown in Figure S1). These base editors were chosen based on the sequence context of both variants, which required Cas9 targeting to occur in the presence of non-NGG PAMs. CFTR channel function before and after editing was measured by short circuit current (Isc). Addition of forskolin facilitated CFTR channel opening leading to an increase in current as a result of ion transport. The CFTR-specific fraction of this increase was calculated by the change in current (ΔIsc) after addition of the CFTR-specific inhibitor inh-172. As expected, we observed negligible response to inh-172 in CFBE cells expressing each variant prior to editing (Figure 1B, unedited). After editing, we observed an ∼5-fold increase in CFTR function in cells expressing the c.273+1G>A variant (from 1.2 to 6.2 μA/cm2) as well as an ∼11-fold increase in CFTR function (from 1.3 to 14.5 μA/cm2) in cells expressing the c.3718-1G>A variant (Figure 1B, edited). These levels of function corresponded to ∼6% (c.273+1G>A) and ∼17% (c.3718-1G>A) of the CFTR current we observe in CFBE cells expressing the relevant wild-type (WT) EMG. Evading lung disease, the major cause of mortality in CF, requires at least 10% WT CFTR function although modest improvements are expected above 5% WT.23 These results illustrate that integrated EMGs provide a suitable platform for testing constructs for genome editing of CSSVs.
Table 1.
Canonical splice site variants of CFTR and possible gene editing approaches
| Legacy name | CFTR2 allele count | Intron in CFTR | HGVS | CFTR function | Modulator response | Gene editing approach |
|---|---|---|---|---|---|---|
| 621+1G>T | 1,323 | 4 | c.489+1G>T | none1 | non-responsive1 | prime editingAM32 |
| 1717-1G>A | 1,216 | 11 | c.1585-1G>A | – | – | adenine base editing19 |
| 3120+1G>A | 501 | 18 | c.2988+1G>A | none | non-responsive | adenine base editing |
| 1898+1G>A | 421 | 13 | c.1766+1G>A | – | – | adenine base editing |
| 711+1G>T | 274 | 5 | c.579+1G>T | none1 | non-responsive1 | prime editing |
| 2622+1G>A | 85 | 14 | c.2490+1G>A | – | – | adenine base editing |
| 1525-1G>A | 72 | 10 | c.1393-1G>A | – | – | adenine base editing |
| 712-1G>T | 42 | 5 | c.580-1G>T | – | – | prime editing |
| 406-1G>A | 40 | 3 | c.274-1G>A | – | – | adenine base editing |
| 405+1G>A | 39 | 3 | c.273+1G>A | none1 | non-responsive1 | adenine base editing |
| 1812-1G>A | 32 | 12 | c.1680-1G>A | – | – | adenine base editing |
| 1248+1G>A | 28 | 8 | c.1116+1G>A | – | – | adenine base editing |
| 4005+1G>A | 21 | 23 | c.3873+1G>A | – | – | adenine base editing |
| 3121-1G>A | 20 | 18 | c.2989-1G>A | – | – | adenine base editing |
| 1811+1G>C | 17 | 12 | c.1679+1G>C | – | – | C·G to G·C base editing20 |
| 4005+2T>C | 15 | 23 | c.3873+2T>C | residual function | – | cytosine base editing18 |
| 3500-2A>G | 13 | 20 | c.3368-2A>G | – | – | cytosine base editing |
| 3850-1G>A | 12 | 22 | c.3718-1G>A | none | responsive | adenine base editing |
| 1341+1G>A | 11 | 9 | c.1209+1G>A | – | – | adenine base editing |
| 4374+1G>T | 6 | 26 | c.4242+1G>T | – | – | prime editing |
Figure 1.
Isogenic CF bronchial epithelial cell lines with integrated expression minigenes provide a flexible platform for testing genome editing of CSSVs
(A) Left panel: schematic showing consequences of the CFTR variant c.273+1G>A (legacy 405+1G>A) when introduced to a CFTR expression minigene (EMG) containing abridged introns 1–4 and full-length intron 5 (EMG_i1-i5). c.273+1G>A caused complete skipping of exon three leading to a frameshift and introduction of a premature termination codon (PTC) that is predicted to lead to a severe reduction in CFTR transcript due to nonsense-mediated mRNA decay (NMD) resulting in no detectable CFTR function or modulator response.1 Right panel: schematic showing consequences of the CFTR variant c.3718-1G>A (legacy 3850-1G>A) when introduced to EMGi21-i23. Introduction of c.3718-1G>A to EMG_i21-i23, resulted in two misspliced RNA isoforms: one skipping 1 nt of exon 23 resulting in a frameshift and a second skipping exon 23 in its entirety resulting in deletion of 51 amino acids; both consequences cause loss of functional CFTR protein.1 (B) Adenine base editing restores CFTR channel function in CF bronchial epithelial (CFBE) cells stably expressing EMGs bearing canonical splice site variants. Left panel: representative tracings from short circuit current (Isc) measurements of CFTR channel function. Graphs on the left show unedited cell lines, right graphs show cell lines after adenine base editing. Top graphs are from CFBE cells expressing CFTR EMG_i1-i5 bearing c.273+1G>A. Bottom graphs are from CFBE cells expressing CFTR EMG_i21-i23 bearing c.3718-1G>A. Forskolin (10 μM) is applied to activate CFTR and CFTR-specific function is measured as the change in current (ΔIsc) after addition of the CFTR-specific inhibitor inh-172. Right panel: bar graph showing quantification of CFTR-specific function in CFBE cells bearing c.273+1G>A or c.3718-1G>A before and after editing shows successful recovery of CFTR function after adenine base editing. Data shown as mean ± SEM (n ≥ 3 technical replicates from one transfection). p value determined by t test. ∗∗∗p ≤ 0.001, ∗p ≤ 0.05.
Testing of protospacer-modified sgRNAs for correction of c.2988+1G>A in an isogenic cell line with integrated EMG
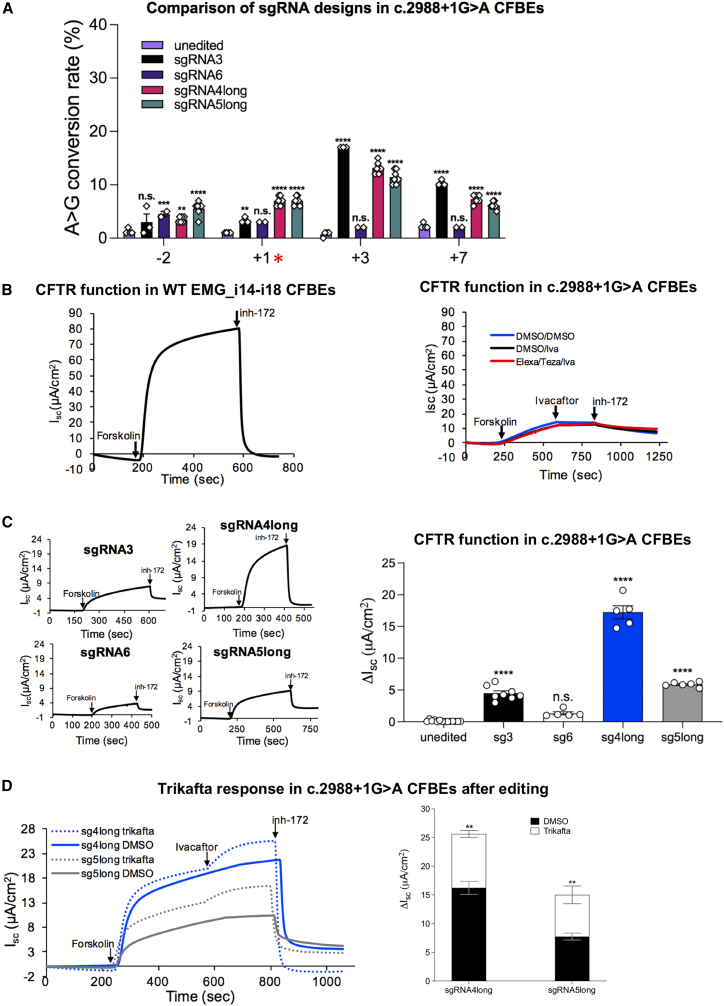
The evolution of a Cas9 base editor variant that can utilize an “NRCH” PAM33 allowed us to target the CSSV, c.2988+1G>A (legacy 3120+1G>A), which alters the donor splice site in intron 18 of CFTR. c.2988+1G>A is an important target as it is the most common CF-causing variant in individuals of African descent and cannot be treated with CFTR modulators (Table 1). Initial sgRNA designs using a 20 nt spacer (sgRNA3 and sgRNA6, Figure 2A) placed the target adenine at position A3 or position A6 of the sgRNA (relative to the 5′ end). Lipofectamine-mediated transfection was used to co-deliver NRCH-ABE8e plasmid DNA with plasmid DNA expressing either sgRNA3 or sgRNA6 to HEK293Flp cells with an integrated EMG_i14-i18 bearing c.2988+1G>A. Editing at the target site (+1) did not differ significantly from baseline for either guide RNA, while sgRNA3 incurred significant editing at nearby bystander sites at the +3 (c.2988+3A>G) and +7 (c.2988+7A>G) positions of the intron 18 splice donor (Figures 2B and 2C). In addition, we observed low levels of editing at the second to last nucleotide of the exon for sgRNA6 (c.2987A>G, “-2”); however, this did not reach significance when compared with unedited cells. We hypothesized that placing the target site in a different position may increase editing efficiency. To this end, we designed eight additional sgRNAs that would place c.2988+1 at A4, A5, A7, A8, or A9 (relative to the 5′ end, Figure 2A). To utilize alternate PAMs compatible with NRCH-ABE8e, we either shortened or lengthened the protospacer so that the 3′ end would be adjacent to a compatible PAM, while maintaining a perfect match between the sgRNA and genomic target (Figure 2A). In the first round of transfections, sgRNA4long and sgRNA5long produced the highest A>G conversion rates at the +1 nucleotide, while the remaining six designs did not edit at higher rates than sgRNA3 or sgRNA6 (Figure S2). We then performed additional transfections to assess editing efficiency across a greater number of technical replicates. After quantifying our results using EditR,34 we determined that sgRNA4long and sgRNA5long had A>G conversion rates at the +1 target of 10.1% ± 2.6% and 9.1% ± 1.8%, respectively (both p < 0.01 compared with unedited) (Figure 2C). Notably, sgRNA4long and sgRNA5long demonstrated significant editing at bystander sites, with sgRNA4long showing the highest level of A>G conversion at the +3 site (15.0% ± 3.8%).
Figure 2.
Modification of protospacer length increases the efficiency of adenine base editing of c.2988+1G>A in an isogenic cell line
(A) Schematic showing the target genomic sequence and 10 different sgRNA designs tested. Red nucleotides are exonic. Bold “A” indicates the target site (c.2988+1). Designs are named based on position of the target site within the sgRNA (relative to the 5′ end), number in parentheses indicates protospacer length (nt), and PAMs for each design correspond to those indicated in the gDNA sequence above. Underlined nucleotides denote positions 4–8 (theoretical optimal editing window for ABE8e) within each sgRNA. (B) Representative Sanger sequencing chromatograms showing editing of gDNA in HEK293Flp cells stably expressing EMG_i14-i18 bearing c.2988+1G>A using four sgRNA designs. Below each chromatogram is a schematic showing relevant sgRNA design. Nucleotides in red indicate exonic positions. Underlined nucleotides are positions where editing was observed. Red asterisk denotes target site. (C) Modified guide RNAs sgRNA4long and sgRNA5long improve on-target editing. Bar graph showing quantification of A>G conversion at each bystander site (−2, +3, +7) and the target site (+1, red asterisk). Values were determined using the Sanger sequencing deconvolution program EditR.34 Data shown as mean ± SEM (n ≥ 6; minimum two technical replicates from three transfections). p values were determined by two-way ANOVA followed by Dunnett’s multiple comparisons test. ∗∗∗∗p ≤ 0.0001, ∗∗p ≤ 0.01, ∗p ≤ 0.05, n.s. (not significant), p > 0.05.
To determine the functional consequences of the bystander edits, we introduced each variant to the EMG_i14-i18 plasmid DNA and assessed the effect on exon 18 inclusion. Only the +3 bystander edit (c.2988+3A>G) affected splicing, resulting in a 20% reduction in full-length transcript (S3AFig). Given that the −2 bystander would be predicted to produce an amino acid substitution (c.2987A>G; p.Gln996Arg), we also assessed its effect on CFTR protein processing when transiently expressed in HEK293 cells. Western blotting revealed that p.Gln996Arg generated full-length mature glycosylated CFTR (band C) and minimal amounts of incompletely glycosylated protein (band B) in proportions that did not differ from HEK293 cells expressing WT CFTR EMG_i14-i18 or WT CFTR cDNA (S3BFig). To determine if p.Gln996Arg decreased CFTR channel function, we introduced our EMG_i14-i18-bearing c.2987A>G into CFBE cells to achieve stable expression. Short circuit current measurements revealed CFTR-specific current in cells with p.Gln996Arg was not decreased from that observed in CFBE cells expressing WT EMG_i14-i18 (Figure S3C). One advantage of the EMG system is that the endogenous CFTR genes in the genome of the HEK293Flp cells remain intact, despite lack of expression.26 This allowed for assessment of targeting of bystander sites at a CFTR sequence without the c.2988+1G>A variant. Despite the 1 bp mismatch between the endogenous CFTR sequence and our c.2988+1G>A targeting sgRNAs, we observed editing at the +3 position of the endogenous CFTR genes (Figure S4). There was minimal editing of the −2 and +7 positions in the endogenous CFTR genes with any of the sgRNA designs (Figure S4). Global off-targets were assessed using the tool Cas-OFFinder,35 which predicted no off-targets with <3 mismatches for sgRNA5long and only one off-target with <3 mismatches for sgRNA4long, which was in an intergenic region (Table S2). All predicted off-targets with ≤3 mismatches were either intergenic or in non-coding regions.
Correction of c.2988+1G>A and restoration of CFTR function in the isogenic CF airway cell model using protospacer-modified sgRNAs
To determine whether the on-target editing was sufficient to restore CFTR channel function, we transfected CFBE cells expressing c.2988+1G>A with NRCH-ABE8e plasmid DNA and each of the top 4 sgRNA designs. Editing at the target and bystander sites was assessed to verify that the sgRNA designs used in HEK293Flps functioned similarly in CFBE cells. We observed minimal editing using sgRNA3 (mean 3.3% ± 0.3%, p < 0.01) or sgRNA6 (3.0% ± 0.0%, p = 0.06) compared with unedited cells. As in HEKs, sgRNA4long and sgRNA5long had greater levels of editing at the +1 site (7%; p < 0.0001 for both) compared with unedited cells (Figure 3A). The CFBE data are based on technical replicates from a single transfection compared with multiple independent transfections performed in HEKs, which likely explains the reduced variability observed in CFBE cell experiments.
Figure 3.
Modified sgRNAs correct c.2988+1G>A and restore CFTR function in isogenic CFBE cells
(A) Bar graph showing quantification of A>G conversion (red asterisk denotes target site) after delivery of base editor and sgRNA to CFBE cells stably expressing EMG_i14-i18 bearing c.2988+1G>A. Values were determined using the Sanger sequencing deconvolution program EditR.34 Data shown as mean ± SEM (n ≥ 2 technical replicates from one transfection). p values were determined by two-way ANOVA followed by Dunnett’s multiple comparisons test. ∗∗∗∗p ≤ 0.0001, ∗∗∗p ≤ 0.001, ∗∗p ≤ 0.01, ∗p ≤ 0.05, n.s., p > 0.05. (B) Left panel: representative tracing from short circuit current (Isc) measurements of baseline CFTR channel function in WT EMG_i14-i18 CFBE cells. Right panel: representative tracings from short circuit current (Isc) measurements of CFTR channel function showing lack of baseline function and response to modulator therapies in CFBE cells stably expressing CFTR EMG_i14-i18 bearing c.2988+1G>A. (C). Left panel: representative short circuit current (Isc) tracings from c.2988+1G>A CFBE cells after editing with four sgRNA designs. Right panel: quantification of CFTR function in c.2988+1G>A CFBE cells (change in current after addition of inh-172, ΔIsc) after editing with four sgRNA designs. Data shown as mean ± SEM (n ≥ 5 technical replicates from one transfection). p values were determined by one-way ANOVA followed by Dunnett’s test for multiple comparisons. ∗∗∗∗p ≤ 0.0001, n.s., p > 0.05, when compared with unedited control. (D) CFTR function recovered by adenine base editing is augmented by treatment with CFTR modulators. Left panel: representative short circuit current (Isc) tracings on CFBE cells stably expressing EMG_i14-i18 bearing c.2988+1G>A after editing with either sgRNA4long or sgRNA5long at baseline (DMSO vehicle control treated, solid lines) or after 24 h treatment with elexacaftor/tezacaftor followed by acute addition of ivacaftor (dotted lines). Right panel: quantification of CFTR function (ΔIsc) in edited cells at baseline (DMSO vehicle control treated) or after treatment with triple combination therapy (elexacaftor/tezacaftor/ivacaftor). Data shown as mean ± SEM (n ≥ 3 technical replicates from one transfection). p values were determined by t test. ∗∗p ≤ 0.01.
Previous studies have shown that c.2988+1G>A causes complete skipping of exon 18, which is predicted to result in a frameshift leading to a premature termination codon that engages NMD, and ultimately severe reduction in CFTR RNA and loss of CFTR function.1,24 While cells expressing the WT EMG generate robust CFTR currents (Figure 3B, left), cells bearing c.2988+1G>A exhibited minimal CFTR-specific current (change in current after application of inh-172, i.e., ΔIsc) even after the addition of CFTR modulators (acute addition of ivacaftor or 24 h incubation with elexacaftor/tezacaftor followed by acute addition of ivacaftor; Figure 3B, right). Editing of c.2988+1G>A CFBE cells with sgRNA3 (4.5 ± 0.4 μA/cm2 [Figure 3C, top left tracing and black bar]) and sgRNA5long (5.9 ± 0.2 μA/cm2 [Figure 3C, bottom right tracing and gray bar]) resulted in modest increases in CFTR function, while sgRNA4long achieved the higher CFTR function of 17.2 ± 1.0 μA/cm2 (Figure 3C, top right tracing and blue bar). This corresponds to ∼12% of what we observe in CFBE cells expressing WT EMG_i14-i18. Consistent with the lack of significant on-target editing, sgRNA6 showed insignificant functional restoration (Figure 3C, p = 0.08). WT CFTR channel function can be augmented by addition of modulators.1,36 To assess whether modest levels of recovered CFTR function after editing could be boosted by modulators, we treated with either a combination of elexacaftor and tezacaftor or a DMSO vehicle control. Elexacaftor/tezacaftor-treated cells received ivacaftor acutely during Isc measurements to evaluate the effect of triple-combination therapy (Figure 3D, left dotted lines), while vehicle control received DMSO (Figure 3D, left solid lines). Compared with baseline function (DMSO control; Figure 3D, right black bars), both sgRNA4long and sgRNA5long corrected cells showed a significant increase in CFTR channel activity with the addition of elexacaftor/tezacaftor/ivacaftor (Figure 3D, right white bars). Notably, with the addition of modulators, cells edited with sgRNA4long achieved a total ΔIsc of 25.6 ± 0.6 μA/cm2, which corresponds to ∼17% WT.
Base editing with modified sgRNA corrects mRNA splicing and recovers CFTR function in primary human bronchial epithelial cells
To assess clinical applicability, we evaluated editing of c.2988+1G>A in primary human bronchial epithelial (HBE) cells obtained from an individual with CF using the two most efficient sgRNA designs (sgRNA4long and sgRNA5long). Electroporation of cells in suspension allowed for co-delivery of either NRCH-ABE8e plasmid DNA with sgRNA expressing plasmid DNA or NRCH-ABE8e mRNA with modified synthetic sgRNA (Table S1). Delivery of GFP mRNA was more efficient than delivery of GFP plasmid DNA as determined by fluorescence microscopy 24 or 48 h post-electroporation (Figure S5). Comparison of GFP-expressing cells to the total number of cells electroporated (∼3.0 × 105) indicated relatively low levels of delivery (i.e., <2% for plasmid DNA and <25% for mRNA), which we attribute in part to loss of cell viability after electroporation. Once a confluent monolayer of cells was formed on the filters (∼5 days), cultures were transitioned to air-liquid interface (ALI) culture to allow for differentiation. Delivery of base editor and sgRNA in RNA form achieved 74.7% correction of the c.2988+1G>A allele with sgRNA4long and 89.5% correction with sgRNA5long. Editing efficiency was calculated by subtracting the contribution from the in trans allele (c.1521_1523delCTT, p.Phe508del; legacy F508del) as described previously37 (dashed line, Figure 4A). Editing of the +1 site was less efficient when plasmid DNA encoded editor and guide RNA were used. Similarly, bystander editing of the +3 and +7 sites was lower when DNA was used (Figure 4B). The higher editing efficiency of the mRNA editor and guides at the target site and bystander sites likely reflect higher transfection rates and improved cell viability, as noted with the GFP controls (Figure S5). The two sgRNAs generated similar editing rates at the +3 and +7 sites, while only sgRNA5long caused significant A>G conversion at the −2 site, the same pattern observed in the isogenic cell lines. Finally, both sgRNA4long and sgRNA5long delivered in mRNA form edited the c.2988+3 and c.2988+7 positions at rates well over 50%, indicating that a fraction of this editing also occurred at the +3 and +7 sites in CFTR bearing F508del.
Figure 4.
Adenine base editing corrects c.2988+1G>A, recovers normally spliced transcript, and restores CFTR channel function in primary HBE cells
(A) Top: quantification of correct nucleotide (%G) at the c.2988+1 target position. Values were determined using the Sanger sequencing deconvolution program EditR.34 Dashed line at 50% indicates the contribution of G sequence from the in trans c.1521_1523delCTT (F508del) allele. Data shown as mean ± SEM (n ≥ 2; minimum one technical replicate each from two transfections). p values were determined by two-way ANOVA followed by Dunnett’s multiple comparisons test. ∗∗∗∗p ≤ 0.0001, ∗∗p ≤ 0.01, n.s., p > 0.05. Bottom: table showing allelic editing efficiency under each condition, which was calculated as follows: Allelic editing % = ((%Gedited – %Gunedited)/%Aunedited) × 100.37 (B) Quantification of A>G conversion at each bystander site (−2, +3, +7) with different sgRNA designs and delivery approaches. Values were determined using the Sanger sequencing deconvolution program EditR.34 Data shown as mean ± SEM (n ≥ 2; minimum one technical replicate each from two transfections). p values were determined by two-way ANOVA followed by Dunnett’s multiple comparisons test. ∗∗∗∗p ≤ 0.0001, ∗∗∗p ≤ 0.001, ∗∗p ≤ 0.01, n.s., p > 0.05. (C) Left panel: mean percent of total stable CFTR mRNA derived from the corrected c.2988+1G>A allele. Values were determined by fragment analysis assessing relative abundance of F508del allele (F508del) and transcript from the c.2988+1G>A allele (non-F508del) in unedited and edited cells. Error bars indicate SEM derived from three technical replicates (data in Table S3). Right panel: scatterplot comparing the rate of A>G conversion at the target site (DNA editing %) to the relative amount of non-F508del transcript. Trend line was generated by simple linear regression. (D) Left and middle panel: representative short circuit current (Isc) tracings on primary c.2988+1G>A/F508del primary HBE cells unedited (red), edited with sgRNA4long (blue), and edited with sgRNA5long (gray). Base editor and sgRNAs were delivered either as plasmid DNA (left) or RNA (middle). “Unedited” cells received base editor, but no sgRNA. To measure CFTR function in primary airway cells, amiloride (100 μM) is added to inhibit epithelial sodium channels and CFTR is activated using a combination of forskolin (10 μM) and IBMX (100 μM). As with isogenic CFBE cells, CFTR-specific function is measured as the change in current (ΔIsc) after addition of the CFTR-specific inhibitor inh-172. Right panel: quantification of CFTR-specific chloride transport before and after editing. Gray shading indicates range of values observed in WT/WT cells. Dashed line indicates average ΔIsc observed in WT/WT HBE cells (Table S4). Data shown as mean ± SEM (n = 4; two technical replicates from two transfections). p values were determined by two-way ANOVA followed by Dunnett’s multiple comparisons test. ∗∗∗∗p ≤ 0.0001, ∗∗∗p ≤ 0.001, n.s., p > 0.05. Recovery of normal splicing of the c.2988+1G>A allele correlates with recovery of CFTR function. Scatterplot comparing estimated abundance of non-F508del transcript with % WT CFTR channel function, which was calculated by comparing average ΔIsc values graphed in (D) to determine WT/WT primary HBE ΔIsc values (Table S4). Trend line was generated by simple linear regression.
To verify that correction of c.2988+1G>A restored normal CFTR splicing, and thus stability of transcript, the relative abundance of CFTR mRNA from each allele was determined. We amplified the region encompassing the 3 bp deletion in CFTR with F508del using 5′ 6FAM-tagged primers and quantified transcripts generated from each allele based on the length of the product generated (253 bp for F508del and 256 bp for non-F508del; Figure S6). As noted previously, c.2988+1G>A causes a reduction in CFTR RNA and loss of CFTR function.1,24 Therefore, we expected that the transcript from CFTR with c.2988+1G>A (i.e., a transcript that has WT sequence at the F508 location) in unedited cells would be reduced in quantity compared with transcript from CFTR with F508del. Indeed, <20% of the total CFTR transcript amplified by RT-PCR was generated from CFTR with c.2988+1G>A (Table S3). We observed a modest increase in the relative abundance of non-F508del CFTR transcript after editing with base editor plasmid DNA. However, cells edited with base editor mRNA and either sgRNA4long or sgRNA5long produced substantially more non-F508del transcript (Figure 4C, left), consistent with editing restoring normal splicing and mRNA stability of transcript generated by the corrected c.2988+1G>A allele. One limit of this approach is that bystander edits on both alleles likely alter transcript stability, which may also affect the relative abundance of the two transcripts. Despite this possibility, we observed a linear correlation (r = 0.87) between allelic conversion and edited full-length transcript abundance (Figure 4C, right).
CFTR function was assessed by short circuit current measurement of fully differentiated HBE cells (∼14–21 days on ALI culture). Following application of amiloride to inhibit the epithelial Na+ channel (ENaC), CFTR activity was stimulated using a combination of forskolin and IBMX (3-isobutyl-1-methylxanthine). As noted previously, CFTR chloride transport was measured as the change in current in response to CFTR-specific inhibitor, inh-172 (ΔIsc). Prior to editing, cells showed minimal current (1.9 ± 0.4 μA/cm2). Transfection with plasmid DNA-encoded base editor and sgRNAs generated a modest response of 5.7 ± 0.4 and 4.3 ± 0.9 μA/cm2 after editing with sgRNA4long and sgRNA5long, respectively (Figure 4D, left). CFTR function was markedly higher after delivery of base editors and sgRNAs in RNA form (Figure 4D, middle). Notably, cells electroporated with NRCH-ABE8e mRNA and sgRNA4long showed a ΔIsc of 16.7 ± 2.4 μA/cm2, which equates to ∼50% of the function observed in WT/WT primary HBE cells (Figure 4D, right). The % WT CFTR function demonstrated robust correlation with abundance of edited full-length mRNA (Figure 4E, r = 0.91), indicating that restoration of normal splicing was critical to recovery of channel function. To assess potential off-targets in a primary cell context whole-genome sequencing (WGS) was performed for unedited cells and cells edited with either sgRNA4long or sgRNA5long. Only one SNP (rs2076277) was identified in an edited sample that was not present in the unedited sample. However, this was located within an intergenic region in the highly variable MHC locus38 and could not be explained by either homology to sgRNA sequence or off-target deaminase activity.
Base editing with modified sgRNAs restores CFTR function in primary human nasal epithelial cells
Primary human nasal epithelial (HNE) cells are a useful system for study of CF airway cells, in addition to primary HBE cells.39 We therefore evaluated editing efficiency in freshly acquired HNE cells from an individual bearing c.2988+1G>A in trans with c.1438 G>A (p.Gly480Ser; legacy G480S). ABE8e and sgRNA4long or sgRNA5long were delivered in RNA form via electroporation. Editing effects were assessed by gDNA sequencing, single-cell RNA sequencing (scRNA-seq) and CFTR channel function measurement. The pattern of on-target and bystander editing observed for each sgRNA design in HNE cells corroborated the HBE results (Figure 5A). Note that unedited has overlapping A nucleotide from CFTR gene bearing c.2988+1G>A (green) and G nucleotide from CFTR gene bearing c.1438 G>A (black) peaks. After correction of c.2988+1G>A, the G nucleotide peak increased and A nucleotide peak decreased in abundance. Editing efficiency in HNE cells was comparable with that observed in HBE cells with 81.3% A>G conversion from sgRNA4long and 89.6% with sgRNA5long (Figure 5A). To assess editing effects on CFTR RNA quantity directly and to determine which cell types are contributing to CFTR expression and function in the primary airway model, we performed scRNA-seq on c.2988+1G>A/c.1438 G>A HNE cells edited (sgRNA4long) and unedited (no sgRNA) after differentiation. We observed an increase in the percent of cells expressing CFTR across all subtypes, except deuterosomal cells, after editing (Figure S7A). Notably the number of cells expressing CFTR RNA transcript and the average CFTR expression level increased by ∼0.6-fold (Figure S7B). As with HBE cells, no CFTR channel function was observed prior to editing (0.9 μA/cm2; Figure 5B). CFTR function was restored in edited cells with both guide RNAs to ranges within or above values observed in WT/WT HNE cells (Figure 5B, right40).
Figure 5.
Base editing restores CFTR function in primary HNE cells bearing c.2988+1G>A
(A) Two sgRNA designs delivered in RNA form result in A>G conversion at both the target site (+1) and bystander sites (−2, +3, +7). Representative Sanger sequencing chromatograms showing CFTR exon 18/intron 18 region in unedited cells (top left) and A>G conversion in the region surrounding c.2988+1 after editing with sgRNA4long (top right) or sgRNA5long (bottom right). Note that A and G nucleotide peaks overlap in unedited and only the A peak is read by the software. Below each chromatogram is schematic showing relevant sgRNA design. Nucleotides in red indicate exonic positions. Underlined nucleotides are positions where editing was observed. Red asterisk denotes target site (c.2988+1 position). Table shows allelic editing efficiency under each condition, which was calculated as follows: Allelic editing % = ((%Gedited – %Gunedited)/%Aunedited) × 100.37 (B) Left panel: representative short circuit current (Isc) tracings of c.2988+1G>A/c.1438G>A primary HNE cells unedited (red), edited with sgRNA4long (blue), and edited with sgRNA5long (gray). “Unedited” cells received base editor, but no sgRNA. Right panel: quantification of CFTR-specific chloride transport (change after addition of inh-172, ΔIsc) in edited and unedited primary cells. Gray shading indicates range of values observed in WT/WT HNE cells as reported previously.40 Data shown as mean ± SD (two technical replicates from one transfection). p values were determined by one-way ANOVA followed by Dunnett’s multiple comparisons test. ∗∗∗∗p ≤ 0.0001, ∗∗∗p ≤ 0.001.
Discussion
Genome editing offers a viable therapeutic solution for correction of CSSVs, and base editing is particularly attractive as the targeted and potential bystander nucleotides are in non-coding (i.e., intronic) sequence. Primary cells are not available for many rare variants and are in limited quantity for more common variants necessitating the use of model cell lines. Previous work has established EMGs as a reliable model system for assessing CFTR splicing, protein processing, and channel function.1,24,25 Isogenic cell lines stably expressing EMGs bearing rare CSSVs, c.273+1G>A and c.3718 – 1G>A, enabled us to efficiently optimize an ABE strategy, validating the use of this approach. Although only low levels of DNA editing were achieved, recovery of CFTR chloride transport was clearly evident. The recently evolved NRCHCas933 with a broader targeting scope permitted the editing of new targets including c.2988+1G>A (legacy 3120+1G>A). As c.2988+1G>A is the 20th most common variant in the CF population and the most common CF-causing variant in individuals with CF of African descent,41,42,43 we were able to obtain primary cells for this variant.
Interestingly, we found that ABE performed more efficiently in primary airway epithelial cells than in model cell lines. We observed the highest editing efficiency in primary cells that underwent electroporation-mediated mRNA delivery, consistent with previous studies.44 Of note, plasmid DNA-encoded reagents had greater editing efficiency in primary cells than observed in the immortalized cell lines with integrated EMG targets. While differences in transfection approach (i.e., electroporation in primaries vs. Lipofectamine in cell lines) could in part explain this discrepancy, it is also possible that some aspect of the EMG prevented efficient editing, such as altered chromatin context. In support of this concept, the endogenous CFTR genes in HEK293Flp cells were found to undergo bystander editing at a rate greater than or equal to that observed at the integrated site. In addition, chromosomal instability within the immortalized cell lines26 could increase the number of Flp recombinase target sites present within the genome of each HEK or CFBE cell, thus resulting in multiple EMG integrations and a greater number of potential editing targets. Despite these shortcomings, EMGs and model cell lines provided a flexible and abundant resource, as demonstrated by the editing of the “leaky” CFTR splice variant c.2657+5G>A (legacy 2789+5G>A), published while this article was in revision.45
Extending the spacer sequence beyond the typical 20 nucleotides for S. pyogenes sgRNAs improved the editing efficiency of the c.2988+1G>A EMG in HEK293 cells, and the recovery of CFTR function in c.2988+1G>A EMG expressing CFBE cells, which translated faithfully to successful editing and recovery of CFTR function in primary airway cells. Placing the target in position A4 or A5 (relative to the 5′ end) of the sgRNA yielded the greatest efficiency, consistent with patterns of editing seen previously for ABE8e.28 To place the target in these positions we had to alter the protospacer length to reach a compatible PAM for the NRCHCas9.33 This approach allowed for testing of a total of seven different placements of the target site (as opposed to three placements using only standard sgRNAs), while maintaining a perfect match between the target site and protospacer. We found that shortening the protospacer (sgRNA4short and sgRNA5short) yielded little to no editing, while lengthening (sgRNA4long and sgRNA5long) resulted in higher levels of editing and recovery of function as compared with the 20 nt protospacer designs (sgRNA3 and sgRNA6). While previous studies altering protospacer length for base editing are limited,46,47 our results support the hypothesis that a lengthened spacer could be used to alter the editing window thereby allowing for more optimal placement of an intended target when PAM availability is limited. In addition, our three best designs (sgRNA3, sgRNA4long, and sgRNA5long) utilized the same PAM (CGTT), consistent with previously reported NRCHCas9 sequence preferences.33
While altering sgRNA design did increase efficiency, it did not eliminate bystander edits. This is likely due to the close proximity of the target site to the most efficiently edited bystander (c.2988+3A>G). However, one advantage of targeting CSSVs is that bystanders have a greater chance of being benign, as the majority of the editing window will fall within the intron. While intronic bystanders can be detrimental, the effect is often modest (e.g., partial missplicing of c.2988+3A>G). Notably, the misspliced CFTR isoform induced by bystander editing at c.2988+3 is the same isoform generated by c.2988+1G>A, thus there is no new protein product with this unintended change. In addition to the EMG system allowing for verification that none of the bystander edits resulted in severe missplicing, it also allowed for assessment of CFTR protein processing and function in the case of the exonic bystander (c.2987A>G; p.Gln996Arg). Since this is not a known naturally occurring CFTR variant (www.genet.sickkids.on.ca), it was particularly important to experimentally verify that c.2987A>G caused no functional defect. The EMG also permitted assessment of editing at the endogenous, non-targeted allele, akin to what may be observed in a compound heterozygote. While correcting the targeted mutation restored full-length CFTR expression and CFTR function, we did observe c.2988+3 editing in the endogenous allele. This propensity should not prevent correction of a targeted patient allele, but is important to note that the other CFTR gene may be subject to bystander editing. Notably, bystander editing complicates interpretation of the degree of functional recovery that can be achieved by a given rate of editing. We hypothesize that differences in bystander editing profiles may explain discrepancies between similar on-target editing rates, but differing degrees of recovery of CFTR function when comparing our candidate sgRNA designs (i.e., sgRNA3 vs. sgRNA6 or sgRNA4long vs. sgRNA5long). While individual bystanders did not show evidence of severe functional consequences, it is possible that combinations of these edits at higher rates and editing at the in trans allele, where relevant, mitigate some of the increase in CFTR mRNA levels that we would expect for a given level of editing. Although we do report a linear correlation between DNA editing and relative abundance of stable transcript from the c.2988+1G>A allele, we acknowledge technical limitations in to our assessment of total RNA (i.e., amplification of intermediate products of mRNA processing). However, RNA sequencing performed at the single-cell level corroborates our findings that correction of c.2988+1G>A results in restored expression of stable full-length CFTR RNA. Thus, we conclude that, in the case of CSSVs, the critical factor in restoring CFTR channel function is generation of full-length WT transcript. This is supported by the strong linear correlation observed between relative abundance of non-F508del transcript and restoration of CFTR function.
We observed functional restoration in both primary HBE cells (c.2988+1G>A/c.1521_1523delCTT) and primary HNE cells (c.2988+1G>A/c.1438G>A). Variation in the % WT function between the two cell types can be attributed to interindividual variation in expression,48 differences in passage number (which was greater in HBE cells as compared with HNE cells), and the underlying variability in CFTR function observed in WT/WT primary cells. Nevertheless, both cell types showed levels of CFTR function after editing that were equivalent to >50% WT, well above the established threshold for a clinically significant improvement (∼10% WT49). Thus, our results support the findings of Krishnamurthy et al.,37 showing that clinically significant recovery of function can be achieved without clonal selection of edited cells. Ex vivo editing approaches such as electroporation have proven highly successful in blood disorders such as sickle cell disease and β-thalassemia50 and have been considered for CF.51 scRNA-seq data reported here demonstrate that genomic correction of non-differentiated primary cells can increase the population of progenitor cells (basal and suprabasal cells) expressing CFTR. However, treatment of the multiorgan manifestations of CF23 will likely require an in vivo editing approach. Successful in vivo genome editing for CF will require efficient delivery to epithelial stem cells in affected tissues. Thus, systemic delivery approaches such as nanoparticles52,53,54 or viral vectors46,55 are promising for future development.
As treatment of CF has progressed from symptom mitigation to targeting of the precise molecular defect, clinical outcomes have significantly improved.56,57,58 However, this improvement is driven by successful treatment of ∼90% of individuals using protein-targeted modulator therapies, while ∼10% remain without a viable precision therapy.57,58 Of the 3,475 individuals in the CFTR2 database (cftr2.org) reported to have at least one CSSV, 804 (23%) do not have a CFTR modulator eligible variant in their other CFTR gene. Modification of base editing designs for correction of the most common CF-causing variant among individuals of African descent (c.2988+1G>A41,42,43) allowed for successful translation to two different types of primary airway cells. Notably, individuals with this variant are statistically more likely to have a second c.2988+1G>A allele or an in trans variant that is rare and/or modulator non-responsive. Importantly, about 50% of the 450 individuals with CF reported to harbor this variant are not currently eligible for modulator therapy (cftr2.org). Thus, there is a particular unmet need for a therapeutic approach for c.2988+1G>A, which warrants continued exploration.
Beyond CF, splice variants in general are estimated to account for 10%–50% of all disease-causing genetic variants.59,60 Given their critical nature, variants in the canonical splice dinucleotides are likely to cause severe clinical manifestations with limited options for RNA and protein targeted therapies.60,61 While base editing therapeutics have tended to focus on the more common variants within a particular disease of interest (e.g., W1282X in the case of CF37,62), we believe that these findings emphasize the advantage of considering such an approach for any CSSV for which the sequence context allows. In addition, considering length-modified protospacers may allow for efficient adenine base editing of otherwise non-targetable or low efficiency sites. This could facilitate expansion of precision therapies to more individuals, especially when such variants are rare or otherwise challenging to treat.
Materials and methods
Generation of EMGs and stable expression cell lines
EMGs were constructed as described previously,24,63 and variants to be studied were introduced by site-directed mutagenesis as described previously1,24,25,63 (Table S1). EMGs were stably integrated into a single genomic locus using a Flp recombinase targeting system in either HEK293Flps (Flp-In-293 cells, Invitrogen, catalog no. R75007) or CFBE cells (CF8Flps26) as described previously.1,25,26
Cloning of crRNA sequences into sgRNA vector
Target-specific sequences were cloned into Cas9 sgRNA vector plasmid, which was a gift from Su-Chun Zhang (Addgene, no. 6846364). Oligos were designed with crRNA sequence and overhangs (Table S1). Cloning was performed by restriction digest and ligation.65 Ligation reactions were transformed into XL10 Gold ultracompetent cells (Agilent), which were plated on agar with 50 μg/mL kanamycin (Quality Biologics). Kanamycin-resistant colonies were picked, grown in liquid culture, and plasmid DNA was purified and sequence verified.
Cell culture
All cells were maintained at 37°C with 5% CO2. HEK293Flp cells were maintained in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (PS) with 50 μg/mL Zeocin. After stable integration, Zeocin was replaced with 100 μg/mL Hygromycin B. CF8Flp cells were maintained in Gibco’s minimal essential medium with 10% FBS and 1% PS. Zeocin (50 μg/mL) was added for maintenance prior to stable transfection, and switched to 200 μg/mL Hygromycin B to maintain stable integration.
Primary HBE cells (c.2988+1G>A/c.1521_1523delCTT) were obtained through the CFF therapeutics lab biobank. Primary HNE cells (c.2988+1G>A/c.1438G>A) were obtained by nasal brushing of an individual bearing one copy of c.2988+1G>A. Brushing was performed by Dr. Christian Merlo at Johns Hopkins Hospital under IRB no. 00116966. Cells were propagated on a feeder layer of 3T3 mouse fibroblasts irradiated with 30 Gy. To allow for expansion, cells were maintained in the presence of 10 μM reagent Y-27632 2HCl (ROCK inhibitor, Selleckchem) with the addition of 1.25 μg/mL amphotericin B, 2 μg/mL fluconazole, and 10 μg/mL nystatin for HBE cells.
Adenine base editing
Immortalized cells
Adenine base editors and sgRNAs were delivered in plasmid DNA form with Lipofectamine/LTX (Invitrogen, catalog no. A12621). For HEK293Flps, transfection was performed in either a 6- or 24-well plate. For CF8Flps, transfection was performed in a 6-well plate. Plasmid DNA (3 or 0.6 μg) was delivered per well of each 6- or 24-well plate, respectively. A 2:1 weight ratio of base editor:sgRNA yielded a 1:1 M ratio. Seventy-two hours post-transfection, cells were scaled up or gDNA or RNA was collected.
Primary cells
Base editor and sgRNA were delivered to non-differentiated primary cells via electroporation. For DNA delivery experiments, base editor plasmid DNA and sgRNA plasmid were combined in a 2:1 weight ratio and a total of 1 μg of DNA was used per reaction. In the no sgRNA control, sgRNA plasmid DNA was substituted with equivalent volume of H2O. pcDNA3-EGFP plasmid DNA was a gift from Doug Golenbock (Addgene, no. 13031). Base editor mRNA was obtained as a custom product from Trilink Biotechnologies using their mammalian-optimized UTR sequences. mRNAs used full substitution of uracil for N1-methylpseudouridine, co-transcriptional 5′ capping with the CleanCap AG analog resulting in a 5′ Cap1 structure, and included a 120 nt poly(A) tail. For RNA delivery experiments, base editor mRNA was at a concentration of 2 μg/μL and synthetic sgRNA (Table S1, IDT) was at a concentration of 100 μM. Editor and sgRNA were combined in a 3:1 volume ratio and a total of 1 μL of RNA was used for each electroporation reaction. In the no sgRNA control, synthetic sgRNA was substituted with equivalent volume of DEP-C H2O and for GFP control eGFP mRNA with unmodified bases and CleanCap was used (TriLink Biotechnologies, catalog no. L-7601). Electroporation was performed using the Neon Transfection System (Invitrogen) with 10 μL Neon tips (Invitrogen, catalog no. MPK1025). Each electroporation reaction consisted of 1.5 × 105 cells resuspended in 9 μL of buffer R (Invitrogen, catalog no. MPK1025) and combined with 1 μL of nucleic acid mix. Electroporation conditions were as follows: 1,400 V, 20 ms, 2 pulses.66 After electroporation, cells were plated onto human collagen type IV (Sigma, no. C6745-1ML) coated snapwell filters (Costar, catalog no. 3801) in a 6-well plate seeded with irradiated mouse fibroblast cells (3T3). Two electroporation reactions were added to each filter. Fluorescence microscopy was performed 24 or 48 h post-electroporation (for RNA and DNA delivery, respectively) to validate successful transfection of GFP control. Transfection efficiency was estimated by comparing the number of GFP+ cells (counted in ImageJ) to the total number of cells plated per filter (3.0 × 105). Cells were allowed to recover from electroporation in medium containing ROCK inhibitor until confluency was reached (∼5–10 days), at which point cells were moved to differentiation medium (no ROCK inhibitor). The next day, apical medium was removed, starting ALI culture. Cells were maintained on ALI culture for 14–21 days before short circuit measurements and collection of gDNA and RNA.
Assessment of editing efficiency
For HEKs, genomic DNA (gDNA) was collected 72 h post-transfection. For CFBE cells and primary airway cells, gDNA was collected directly from snapwell filters after short circuit measurements were taken. Genomic DNA (gDNA) extraction was performed using the DNeasy Blood & Tissue Extraction Kit (QIAGEN, catalog no. 69504). Following extraction, editing was assessed by PCR amplification of the relevant CFTR exon (Table S1). For HEK and CFBE experiments PCR was performed with KOD hot start master mix (Millipore Sigma). For primary cells, PCR was performed using HotStarTaq DNA polymerase (QIAGEN, catalog no. 203203). For both cell lines and primary cells PCR products were purified, Sanger sequenced, and editing rate at the target site and bystander sites were quantified using EditR.34
RNA analysis
cDNA synthesis
An input of 500 ng of RNA was used with the iScript cDNA synthesis kit (Bio-Rad). For immortalized cells, cDNA was diluted 1:10.
Fragment analysis
cDNA was used as a template for RT-PCR performed with KOD hot start master mix (Millipore Sigma) and a forward primer bearing a 5′ 6FAM tag (Table S1). PCR products were separated on an Applied Biosystems 3730 DNA Analyzer capillary electrophoresis system at the Johns Hopkins Genetics Resources Core Facility (GRCF). GeneScan 500 Rox was used as an internal size standard. To calculate relative RNA isoform quantity, the area under the curve (AUC) for each isoform was compared with the total AUC for that sample. For assessment of missplicing after bystander editing (Figure S3) amplification from exon 16 to exon 19 allowed for determination of the relative abundance of normally spliced and exon 18 skipped transcript. For primary cells, amplification from exon 11 to exon 13 allowed for quantification of the relative abundance of transcript from the F508del allele (F508del) and transcript from the c.2988+1G>A allele (non-F508del) (Figure S6; Table S3).
Immunoblotting
Immunoblotting was performed in HEK293 cells transiently expressing CFTR plasmid DNAs as described previously.1
Evaluation of CFTR channel function
CFBE cells
Cells were grown on filters until a transepithelial resistance of >200 Ω was reached (6–8 days). Filters were then mounted on Ussing chambers and a chloride gradient was established using asymmetrical buffers as described previously.67 Short circuit current measurements were taken using a multi-channel voltage-current clamp amplifier (Physiologic Instruments) and the data acquisition program Acquire and Analyze. After equilibration, forskolin (10 μM) was added to the basolateral chamber to activate channel opening. The CFTR-specific inhibitor, inh-172 was used (10 μM, apical chamber) to inhibit the channel. The drop in current after addition of inhibitor allowed for quantification of CFTR function (ΔIsc).
Primary cells
CFTR function was assessed in primary cells by mounting differentiated filters on Ussing chambers using a symmetrical buffer (126 mM NaCl, 25 mM NaHCO3, 5 mM KCl, 2.5 mM Na2HPO4, 1.8 mM CaCl2, 1 mM MgSO4, 10 mM dextrose). Buffer pH was maintained at ∼7.3–7.4 by continuous circulation with carbogen gas (95% O2/5% CO2) and temperature was maintained at 37°C. The apically located sodium epithelial channel (ENaC) was inactivated by addition of amiloride. A combination of forskolin and IBMX was added to the basolateral chamber to stimulate channel opening. Inh-172 was used to inhibit CFTR channel function and quantify ΔIsc.
Drug testing
To test CFTR channel response to Trikafta (elexacaftor/tezacaftor/ivacaftor), cells were treated with either correctors (tezacaftor + elexacaftor, 3 μM each) or vehicle control (DMSO, equivalent volume) for 24 h. The potentiator (ivacaftor, 10 μM) or additional DMSO was delivered acutely to the apical chamber.
Calculation of % WT
CFBE cells
Average WT ΔIsc was determined independently for each EMG. Experimental values were then compared with respective WT EMG average to calculate approximate % WT.
Primaries
Percent of WT function was calculated by comparing ΔIsc values obtained in experiments to average observed in WT/WT primary control cells. For HBE cells, this average value came from six technical replicates each from two independent HBE cell lines obtained from different individuals (Table S4). For HNE cells, this average came from previously reported data.40
Off-target analysis
Computational prediction of off-targets
gRNA-dependent off-targets for top two sgRNAs were assessed using Cas-OFFinder35 run in command line using GRCh37/hg19 with length set to accommodate the longer designs and mismatch number set to five. Potential off-targeting of both NGN and NRCH PAM-containing sequences was evaluated. Predicted off-target loci with ≤3 mismatches are reported in Table S2.
WGS to assess off-targets in a primary cell context
Library preparation and sequencing was performed by the Johns Hopkins GRCF on the Illumina NovaSeq 6000 with 30× genomic coverage. PLINK files were converted to VCF format using --recode.68 VCF files were then directly compared as data frames using base R functions (R 4.1.2). Only SNPs where both samples had a call were considered.
scRNA-seq
Library preparation was performed in the Cutting lab using 10X Genomics v.3.1 3′ Library Preparation chemistry with a dual indexing system. Sequencing was performed by the Johns Hopkins GRCF via paired-end sequencing 2 × 100 cycles using a NovaSeq 6000 Illumina sequencing machine. Analysis of scRNA-seq results was performed using 10X Genomics Cell Ranger 3.1.0 followed by the Seurat package (v.4.1) in R created by the Satija lab.69 Subsetting was determined based on comparisons of the number of genes expressed, UMI count, and percentage of reads mapped to mitochondrial RNA. A log normalization with a scale factor of 10,000 was used. Cell type was assigned based on known transcriptional markers (basal KRT5+ TP63+; cycling basal KRT5+ TOP2A+; secretory SCGB1A1+; secretory [goblet] MUC5AC+; deuterosomal DEUP1+ FOXJ1+; ciliated DEUP1– FOXJ1+).70
Statistical analysis
GraphPad Prism v.9.3.0 (GraphPad Software), was used to perform all statistical analysis.
Data and code availability
RNA-seq and whole-genome sequencing data reported in this article are available from the corresponding author upon reasonable request.
Acknowledgments
The authors thank the individuals with CF for the donation of primary airway cells and Priyanka Bhatt, CFF Therapeutics Lab, for assistance with shipping and methodology for culture of primary HBE cells. We thank Dr. Dimitri Avramopoulos (Johns Hopkins University) for the use of the Neon electroporation system. The graphical abstract was created in BioRender.com. This work was supported by the Cystic Fibrosis Foundation (CUTTIN20G0 to G.R.C and SHARMA19I0 to N.S.) and a Research Innovation award from Vertex Pharmaceuticals (to N.S.), by US National Institutes of Health awards (U01 AI142756, RM1 HG009490, and R35 GM118062 to D.R.L.), the Howard Hughes Medical Institute (to D.R.L.), and the Helen Hay Whitney Fellowship (to G.A.N.).
Author contributions
Conceptualization, G.R.C., A.T.J., and N.S.; data curation, K.S.R.; formal analysis, A.T.J., A.C.E., and E.W.K.; project administration, G.R.C.; supervision, G.R.C., N.S., and D.R.L.; funding acquisition, G.R.C., N.S., D.R.L., and G.A.N.; methodology, A.T.J., G.A.N., and N.S.; investigation, A.T.J., E.W.K., S.M., A.D.B., K.C.P., and D.L.O.; resources, G.R.C., N.S., G.A.N., D.R.L., C.A.M., and S.U.P.; software, A.C.E.; validation, K.C.P.; visualization, A.T.J. and E.W.K.; writing – original draft, A.T.J. and G.R.C.; writing – review & editing, A.T.J., G.R.C., N.S., G.A.N., K.C.P., and E.W.K.
Declaration of interests
D.R.L. is a consultant and equity owner of Prime Medicine, Beam Therapeutics, Pairwise Plants, Nvelop Therapeutics, and Chroma Medicine, companies that use or deliver genome editing or epigenome-modulating agents. G.R.C. and G.A.N. are consultants of the U.S. Cystic Fibrosis Foundation.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2023.06.020.
Supplemental information
References
- 1.Joynt A.T., Evans T.A., Pellicore M.J., Davis-Marcisak E.F., Aksit M.A., Eastman A.C., Patel S.U., Paul K.C., Osorio D.L., Bowling A.D., et al. Evaluation of both exonic and intronic variants for effects on RNA splicing allows for accurate assessment of the effectiveness of precision therapies. PLoS Genet. 2020;16:e1009100. doi: 10.1371/journal.pgen.1009100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lampe A.K., Zou Y., Sudano D., O’Brien K.K., Hicks D., Laval S.H., Charlton R., Jimenez-Mallebrera C., Zhang R.-Z., Finkel R.S., et al. Exon skipping mutations in collagen VI are common and are predictive for severity and inheritance. Hum. Mutat. 2008;29:809–822. doi: 10.1002/humu.20704. [DOI] [PubMed] [Google Scholar]
- 3.Tang P.H., Velez G., Tsang S.H., Bassuk A.G., Mahajan V.B. VCAN Canonical Splice Site Mutation is Associated With Vitreoretinal Degeneration and Disrupts an MMP Proteolytic Site. Invest. Ophthalmol. Vis. Sci. 2019;60:282–293. doi: 10.1167/iovs.18-25624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Varela P., Carvalho G., Martin R.P., Pesquero J.B. Fabry disease: GLA deletion alters a canonical splice site in a family with neuropsychiatric manifestations. Metab. Brain Dis. 2021;36:265–272. doi: 10.1007/s11011-020-00640-0. [DOI] [PubMed] [Google Scholar]
- 5.Keenan M.M., Huang L., Jordan N.J., Wong E., Cheng Y., Valley H.C., Mahiou J., Liang F., Bihler H., Mense M., et al. Nonsense-mediated RNA Decay Pathway Inhibition Restores Expression and Function of W1282X CFTR. Am. J. Respir. Cell Mol. Biol. 2019;61:290–300. doi: 10.1165/rcmb.2018-0316OC. [DOI] [PubMed] [Google Scholar]
- 6.Sharma J., Du M., Wong E., Mutyam V., Li Y., Chen J., Wangen J., Thrasher K., Fu L., Peng N., et al. A small molecule that induces translational readthrough of CFTR nonsense mutations by eRF1 depletion. Nat. Commun. 2021;12:4358. doi: 10.1038/s41467-021-24575-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crawford D.K., Mullenders J., Pott J., Boj S.F., Landskroner-Eiger S., Goddeeris M.M. Targeting G542X CFTR nonsense alleles with ELX-02 restores CFTR function in human-derived intestinal organoids. J. Cyst. Fibros. 2021;20:436–442. doi: 10.1016/j.jcf.2021.01.009. [DOI] [PubMed] [Google Scholar]
- 8.Lueck J.D., Yoon J.S., Perales-Puchalt A., Mackey A.L., Infield D.T., Behlke M.A., Pope M.R., Weiner D.B., Skach W.R., McCray P.B., Ahern C.A. Engineered transfer RNAs for suppression of premature termination codons. Nat. Commun. 2019;10:822. doi: 10.1038/s41467-019-08329-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J., Zhang Y., Mendonca C.A., Yukselen O., Muneeruddin K., Ren L., Liang J., Zhou C., Xie J., Li J., et al. AAV-delivered suppressor tRNA overcomes a nonsense mutation in mice. Nature. 2022;604:343–348. doi: 10.1038/s41586-022-04533-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Welch E.M., Barton E.R., Zhuo J., Tomizawa Y., Friesen W.J., Trifillis P., Paushkin S., Patel M., Trotta C.R., Hwang S., et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007;447:87–91. doi: 10.1038/nature05756. [DOI] [PubMed] [Google Scholar]
- 11.Choi S.H., Engelhardt J.F. Gene Therapy for Cystic Fibrosis: Lessons Learned and Paths Forward. Mol. Ther. 2021;29:428–430. doi: 10.1016/j.ymthe.2021.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.George L.A., Ragni M.V., Rasko J.E.J., Raffini L.J., Samelson-Jones B.J., Ozelo M., Hazbon M., Runowski A.R., Wellman J.A., Wachtel K., et al. Long-Term Follow-Up of the First in Human Intravascular Delivery of AAV for Gene Transfer: AAV2-hFIX16 for Severe Hemophilia B. Mol. Ther. 2020;28:2073–2082. doi: 10.1016/j.ymthe.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kandavelou K., Mani M., Durai S., Chandrasegaran S. Magic” scissors for genome surgery. Nat. Biotechnol. 2005;23:686–687. doi: 10.1038/nbt0605-686. [DOI] [PubMed] [Google Scholar]
- 14.Li T., Huang S., Jiang W.Z., Wright D., Spalding M.H., Weeks D.P., Yang B. TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res. 2011;39:359–372. doi: 10.1093/nar/gkq704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller J.C., Tan S., Qiao G., Barlow K.A., Wang J., Xia D.F., Meng X., Paschon D.E., Leung E., Hinkley S.J., et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 16.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaudelli N.M., Komor A.C., Rees H.A., Packer M.S., Badran A.H., Bryson D.I., Liu D.R. Programmable base editing of A·T to G·C in genomic DNA without DNA cleavage. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koblan L.W., Arbab M., Shen M.W., Hussmann J.A., Anzalone A.V., Doman J.L., Newby G.A., Yang D., Mok B., Replogle J.M., et al. Efficient C·G-to-G·C base editors developed using CRISPRi screens, target-library analysis, and machine learning. Nat. Biotechnol. 2021;39:1414–1425. doi: 10.1038/s41587-021-00938-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang T.P., Zhao K.T., Miller S.M., Gaudelli N.M., Oakes B.L., Fellmann C., Savage D.F., Liu D.R. Circularly permuted and PAM-modified Cas9 variants broaden the targeting scope of base editors. Nat. Biotechnol. 2019;37:626–631. doi: 10.1038/s41587-019-0134-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang L., Zhang X., Wang L., Yin S., Zhu B., Xie L., Duan Q., Hu H., Zheng R., Wei Y., et al. Increasing targeting scope of adenosine base editors in mouse and rat embryos through fusion of TadA deaminase with Cas9 variants. Protein Cell. 2018;9:814–819. doi: 10.1007/s13238-018-0568-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cutting G.R. Cystic fibrosis genetics: from molecular understanding to clinical application. Nat. Rev. Genet. 2015;16:45–56. doi: 10.1038/nrg3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma N., Sosnay P.R., Ramalho A.S., Douville C., Franca A., Gottschalk L.B., Park J., Lee M., Vecchio-Pagan B., Raraigh K.S., et al. Experimental assessment of splicing variants using expression minigenes and comparison with in silico predictions. Hum. Mutat. 2014;35:1249–1259. doi: 10.1002/humu.22624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharma N., Evans T.A., Pellicore M.J., Davis E., Aksit M.A., McCague A.F., Joynt A.T., Lu Z., Han S.T., Anzmann A.F., et al. Capitalizing on the heterogeneous effects of CFTR nonsense and frameshift variants to inform therapeutic strategy for cystic fibrosis. PLoS Genet. 2018;14:e1007723. doi: 10.1371/journal.pgen.1007723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gottschalk L.B., Vecchio-Pagan B., Sharma N., Han S.T., Franca A., Wohler E.S., Batista D.A.S., Goff L.A., Cutting G.R. Creation and characterization of an airway epithelial cell line for stable expression of CFTR variants. J. Cyst. Fibros. 2016;15:285–294. doi: 10.1016/j.jcf.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishimasu H., Shi X., Ishiguro S., Gao L., Hirano S., Okazaki S., Noda T., Abudayyeh O.O., Gootenberg J.S., Mori H., et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science. 2018;361:1259–1262. doi: 10.1126/science.aas9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richter M.F., Zhao K.T., Eton E., Lapinaite A., Newby G.A., Thuronyi B.W., Wilson C., Koblan L.W., Zeng J., Bauer D.E., et al. Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat. Biotechnol. 2020;38:883–891. doi: 10.1038/s41587-020-0453-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Zheng Z., Joung J.K. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kleinstiver B.P., Prew M.S., Tsai S.Q., Topkar V.V., Nguyen N.T., Zheng Z., Gonzales A.P.W., Li Z., Peterson R.T., Yeh J.-R.J., et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koblan L.W., Doman J.L., Wilson C., Levy J.M., Tay T., Newby G.A., Maianti J.P., Raguram A., Liu D.R. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat. Biotechnol. 2018;36:843–846. doi: 10.1038/nbt.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anzalone A.V., Randolph P.B., Davis J.R., Sousa A.A., Koblan L.W., Levy J.M., Chen P.J., Wilson C., Newby G.A., Raguram A., Liu D.R. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576:149–157. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller S.M., Wang T., Randolph P.B., Arbab M., Shen M.W., Huang T.P., Matuszek Z., Newby G.A., Rees H.A., Liu D.R. Continuous evolution of SpCas9 variants compatible with non-G PAMs. Nat. Biotechnol. 2020;38:471–481. doi: 10.1038/s41587-020-0412-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kluesner M.G., Nedveck D.A., Lahr W.S., Garbe J.R., Abrahante J.E., Webber B.R., Moriarity B.S. EditR: A Method to Quantify Base Editing from Sanger Sequencing. CRISPR J. 2018;1:239–250. doi: 10.1089/crispr.2018.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bae S., Park J., Kim J.-S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shaughnessy C.A., Zeitlin P.L., Bratcher P.E. Elexacaftor is a CFTR potentiator and acts synergistically with ivacaftor during acute and chronic treatment. Sci. Rep. 2021;11:19810. doi: 10.1038/s41598-021-99184-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krishnamurthy S., Traore S., Cooney A.L., Brommel C.M., Kulhankova K., Sinn P.L., Newby G.A., Liu D.R., McCray P.B. Functional correction of CFTR mutations in human airway epithelial cells using adenine base. Nucleic Acids Res. 2021;49:10558–10572. doi: 10.1093/nar/gkab788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gulko B., Hubisz M.J., Gronau I., Siepel A. A method for calculating probabilities of fitness consequences for point mutations across the human genome. Nat. Genet. 2015;47:276–283. doi: 10.1038/ng.3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brewington J.J., Filbrandt E.T., LaRosa F.J., Moncivaiz J.D., Ostmann A.J., Strecker L.M., Clancy J.P. Brushed nasal epithelial cells are a surrogate for bronchial epithelial CFTR studies. JCI Insight. 2018;3:e99385. doi: 10.1172/jci.insight.99385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raraigh K.S., Paul K.C., Goralski J.L., Worthington E.N., Faino A.V., Sciortino S., Wang Y., Aksit M.A., Ling H., Osorio D.L., et al. CFTR bearing variant p.Phe312del exhibits function inconsistent with phenotype and negligible response to ivacaftor. JCI Insight. 2022;7:e148841. doi: 10.1172/jci.insight.148841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Macek M., Mackova A., Hamosh A., Hilman B.C., Selden R.F., Lucotte G., Friedman K.J., Knowles M.R., Rosenstein B.J., Cutting G.R. Identification of common cystic fibrosis mutations in African-Americans with cystic fibrosis increases the detection rate to 75. Am. J. Hum. Genet. 1997;60:1122–1127. [PMC free article] [PubMed] [Google Scholar]
- 42.Padoa C., Goldman A., Jenkins T., Ramsay M. Cystic fibrosis carrier frequencies in populations of African origin. J. Med. Genet. 1999;36:41–44. doi: 10.1136/jmg.36.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stewart C., Pepper M.S. Cystic Fibrosis in the African Diaspora. Ann. Am. Thorac. Soc. 2017;14:1–7. doi: 10.1513/AnnalsATS.201606-481FR. [DOI] [PubMed] [Google Scholar]
- 44.Newby G.A., Yen J.S., Woodard K.J., Mayuranathan T., Lazzarotto C.R., Li Y., Sheppard-Tillman H., Porter S.N., Yao Y., Mayberry K., et al. Base editing of haematopoietic stem cells rescues sickle cell disease in mice. Nature. 2021;595:295–302. doi: 10.1038/s41586-021-03609-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Amistadi S., Maule G., Ciciani M., Ensinck M.M., De Keersmaecker L., Ramalho A.S., Guidone D., Buccirossi M., Galietta L.J.V., Carlon M.S., Cereseto A. Functional restoration of a CFTR splicing mutation through RNA delivery of CRISPR adenine base editor. Mol. Ther. 2023;31:1647–1660. doi: 10.1016/j.ymthe.2023.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ryu S.-M., Koo T., Kim K., Lim K., Baek G., Kim S.-T., Kim H.S., Kim D.E., Lee H., Chung E., Kim J.S. Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat. Biotechnol. 2018;36:536–539. doi: 10.1038/nbt.4148. [DOI] [PubMed] [Google Scholar]
- 47.Kim Y.B., Komor A.C., Levy J.M., Packer M.S., Zhao K.T., Liu D.R. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat. Biotechnol. 2017;35:371–376. doi: 10.1038/nbt.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aksit M.A., Bowling A.D., Evans T.A., Joynt A.T., Osorio D., Patel S., West N., Merlo C., Sosnay P.R., Cutting G.R., Sharma N. Decreased mRNA and protein stability of W1282X limits response to modulator therapy. J. Cyst. Fibros. 2019;18:606–613. doi: 10.1016/j.jcf.2019.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCague A.F., Raraigh K.S., Pellicore M.J., Davis-Marcisak E.F., Evans T.A., Han S.T., Lu Z., Joynt A.T., Sharma N., Castellani C., et al. Correlating Cystic Fibrosis Transmembrane Conductance Regulator Function with Clinical Features to Inform Precision Treatment of Cystic Fibrosis. Am. J. Respir. Crit. Care Med. 2019;199:1116–1126. doi: 10.1164/rccm.201901-0145OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frangoul H., Altshuler D., Cappellini M.D., Chen Y.-S., Domm J., Eustace B.K., Foell J., de la Fuente J., Grupp S., Handgretinger R., et al. CRISPR-Cas9 Gene Editing for Sickle Cell Disease and β-Thalassemia. N. Engl. J. Med. 2021;384:252–260. doi: 10.1056/NEJMoa2031054. [DOI] [PubMed] [Google Scholar]
- 51.Suzuki S., Crane A.M., Anirudhan V., Barillà C., Matthias N., Randell S.H., Rab A., Sorscher E.J., Kerschner J.L., Yin S., et al. Highly Efficient Gene Editing of Cystic Fibrosis Patient-Derived Airway Basal Cells Results in Functional CFTR Correction. Mol. Ther. 2020;28:1684–1695. doi: 10.1016/j.ymthe.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robinson E., MacDonald K.D., Slaughter K., McKinney M., Patel S., Sun C., Sahay G. Lipid Nanoparticle-Delivered Chemically Modified mRNA Restores Chloride Secretion in Cystic Fibrosis. Mol. Ther. 2018;26:2034–2046. doi: 10.1016/j.ymthe.2018.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Musunuru K., Chadwick A.C., Mizoguchi T., Garcia S.P., DeNizio J.E., Reiss C.W., Wang K., Iyer S., Dutta C., Clendaniel V., et al. In vivo CRISPR base editing of PCSK9 durably lowers cholesterol in primates. Nature. 2021;593:429–434. doi: 10.1038/s41586-021-03534-y. [DOI] [PubMed] [Google Scholar]
- 54.Song C.-Q., Jiang T., Richter M., Rhym L.H., Koblan L.W., Zafra M.P., Schatoff E.M., Doman J.L., Cao Y., Dow L.E., et al. Adenine base editing in an adult mouse model of tyrosinaemia. Nat. Biomed. Eng. 2020;4:125–130. doi: 10.1038/s41551-019-0357-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yeh W.-H., Shubina-Oleinik O., Levy J.M., Pan B., Newby G.A., Wornow M., Burt R., Chen J.C., Holt J.R., Liu D.R. In vivo base editing restores sensory transduction and transiently improves auditory function in a mouse model of recessive deafness. Sci. Transl. Med. 2020;12:eaay9101. doi: 10.1126/scitranslmed.aay9101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ramsey B.W., Davies J., McElvaney N.G., Tullis E., Bell S.C., Dřevínek P., Griese M., McKone E.F., Wainwright C.E., Konstan M.W., et al. A CFTR potentiator in patients with cystic fibrosis and the G551D mutation. N. Engl. J. Med. 2011;365:1663–1672. doi: 10.1056/NEJMoa1105185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Keating D., Marigowda G., Burr L., Daines C., Mall M.A., McKone E.F., Ramsey B.W., Rowe S.M., Sass L.A., Tullis E., et al. VX-445-Tezacaftor-Ivacaftor in Patients with Cystic Fibrosis and One or Two Phe508del Alleles. N. Engl. J. Med. 2018;379:1612–1620. doi: 10.1056/NEJMoa1807120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Joshi D., Ehrhardt A., Hong J.S., Sorscher E.J. Cystic fibrosis precision therapeutics: Emerging considerations. Pediatr. Pulmonol. 2019;54:S13–S17. doi: 10.1002/ppul.24547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stenson P.D., Ball E.V., Mort M., Phillips A.D., Shiel J.A., Thomas N.S.T., Abeysinghe S., Krawczak M., Cooper D.N. Human Gene Mutation Database (HGMD): 2003 update. Hum. Mutat. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- 60.Wang G.-S., Cooper T.A. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat. Rev. Genet. 2007;8:749–761. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- 61.Krawczak M., Thomas N.S.T., Hundrieser B., Mort M., Wittig M., Hampe J., Cooper D.N. Single base-pair substitutions in exon-intron junctions of human genes: nature, distribution, and consequences for mRNA splicing. Hum. Mutat. 2007;28:150–158. doi: 10.1002/humu.20400. [DOI] [PubMed] [Google Scholar]
- 62.Geurts M.H., de Poel E., Amatngalim G.D., Oka R., Meijers F.M., Kruisselbrink E., van Mourik P., Berkers G., de Winter-de Groot K.M., Michel S., et al. CRISPR-Based Adenine Editors Correct Nonsense Mutations in a Cystic Fibrosis Organoid Biobank. Cell Stem Cell. 2020;26:503–510.e7. doi: 10.1016/j.stem.2020.01.019. [DOI] [PubMed] [Google Scholar]
- 63.Lee M., Roos P., Sharma N., Atalar M., Evans T.A., Pellicore M.J., Davis E., Lam A.-T.N., Stanley S.E., Khalil S.E., et al. Systematic Computational Identification of Variants That Activate Exonic and Intronic Cryptic Splice Sites. Am. J. Hum. Genet. 2017;100:751–765. doi: 10.1016/j.ajhg.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen Y., Cao J., Xiong M., Petersen A.J., Dong Y., Tao Y., Huang C.T.-L., Du Z., Zhang S.-C. Engineering Human Stem Cell Lines with Inducible Gene Knockout using CRISPR/Cas9. Cell Stem Cell. 2015;17:233–244. doi: 10.1016/j.stem.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moyer T.C., Holland A.J. Generation of a conditional analog-sensitive kinase in human cells using CRISPR/Cas9-mediated genome engineering. Methods Cell Biol. 2015;129:19–36. doi: 10.1016/bs.mcb.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Koh K.D., Siddiqui S., Cheng D., Bonser L.R., Sun D.I., Zlock L.T., Finkbeiner W.E., Woodruff P.G., Erle D.J. Efficient RNP-directed Human Gene Targeting Reveals SPDEF Is Required for IL-13-induced Mucostasis. Am. J. Respir. Cell Mol. Biol. 2020;62:373–381. doi: 10.1165/rcmb.2019-0266OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Raraigh K.S., Han S.T., Davis E., Evans T.A., Pellicore M.J., McCague A.F., Joynt A.T., Lu Z., Atalar M., Sharma N., et al. Functional Assays Are Essential for Interpretation of Missense Variants Associated with Variable Expressivity. Am. J. Hum. Genet. 2018;102:1062–1077. doi: 10.1016/j.ajhg.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Second-generation PLINK: rising to the challenge of larger and richer datasets | GigaScience | Oxford Academic. https://academic.oup.com/gigascience/article/4/1/s13742-015-0047-8/2707533?login=true [DOI] [PMC free article] [PubMed]
- 69.Hao Y., Hao S., Andersen-Nissen E., Mauck W.M., Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zaragosi L.E., Deprez M., Barbry P. Using single-cell RNA sequencing to unravel cell lineage relationships in the respiratory tract. Biochem. Soc. Trans. 2020;48:327–336. doi: 10.1042/BST20191010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq and whole-genome sequencing data reported in this article are available from the corresponding author upon reasonable request.