Abstract
Using coimmunoprecipitation and glutathione S-transferase pulldown experiments, we found that polyomavirus large T antigen binds to p300 in vivo and in vitro. The N-terminal region of the viral protein, including the pRB binding motif, was dispensable for this interaction, which involved several regions within the C-terminal half of the large T antigen. Interestingly, anti-T antibody coimmunoprecipitated a subspecies of p300 which has high histone acetyltransferase activity.
DNA tumor viruses, the adenoviruses and the papoviruses, need cells in the S phase of the cell cycle for their replication. To cope with this requirement, they code for proteins which interact with regulators of growth and differentiation of the host cell and interfere with their normal function. Important cellular targets of viral proteins are the retinoblastoma-type proteins, pRB, p107, and p130. Adenovirus protein E1A, the large T antigens of simian virus 40 (SV40) and polyomavirus, and the E7 protein of human papillomaviruses share a protein sequence which includes the motif LXCXE, through which they bind to a characteristic region, called the pocket, of pRB-type proteins (reviewed in reference 23). The pocket proteins are cellular regulators of transcription which interact with members of the transcription factor family E2F. Binding sites for E2F are present in promoters of many genes coding for enzymes active in DNA replication and precursor production and in those of cell cycle-regulating proteins, such as G1/S- and S-phase specific cyclins (reviewed in reference 18). The viral proteins induce expression of such genes under conditions under which they normally are shut off.
The E1A protein was the first one for which another important cellular target, which again plays an important part in the regulation of growth and differentiation, was discovered (23). This protein, p300, is a member of a group of transcriptional coactivators which includes the CREB-binding protein CBP, known to play a substantial role in the regulation of cyclic-AMP-responsive genes (2, 3, 11, 21). p300/CBP has been found to be involved in diverse ways in the control of a large number of differentiation- and growth-specific genes. Alterations of the p300/CBP genes have recently been implicated in various diseases and malignancies (reviewed in references 12, 16, and 29). p300 interacts with a great variety of cellular transcription factors and was found to be associated with complexes containing TATA-binding protein (1, 8). It also interacts with P/CAF, a histone acetyltransferase (HAT). P/CAF was found to compete with the binding of the E1A protein to p300/CBP (38). A sequence located at the very N-terminal region of E1A and including conserved region 1, known to be important for its S phase-inducing ability, is required for p300 binding (7, 34, 35). Recently, p300 itself was found to exhibit HAT activity (6, 26). It is therefore speculated that p300/CBP is important for altering the chromatin structure of promoters. Interaction of p300 with E1A and other DNA tumor virus proteins likely results in a modification of this activity.
In contrast to the situation for the binding motifs for pocket proteins, which are present in highly homologous form in E1A, in the T antigens, and in the HPV E7 protein, the status is not as clear with respect to p300/CBP. While SV40 large T antigen (SV40LT) was recently found to bind p300 and its relatives (4, 10), amino acid sequences within the viral protein essential for this binding are not well defined. Whereas a sequence at the N terminus of SV40LT was first presumed to be important in analogy to the situation of the E1A protein (10, 37), sequences towards the C terminus were more recently found to be essential. These sequences at least in part overlap with the known binding site for the tumor suppressor protein p53 (19). SV40LT appears to form a ternary complex with p53 and p300/CBP (19), but the coactivator protein was also found to target p53 directly (5, 14, 20).
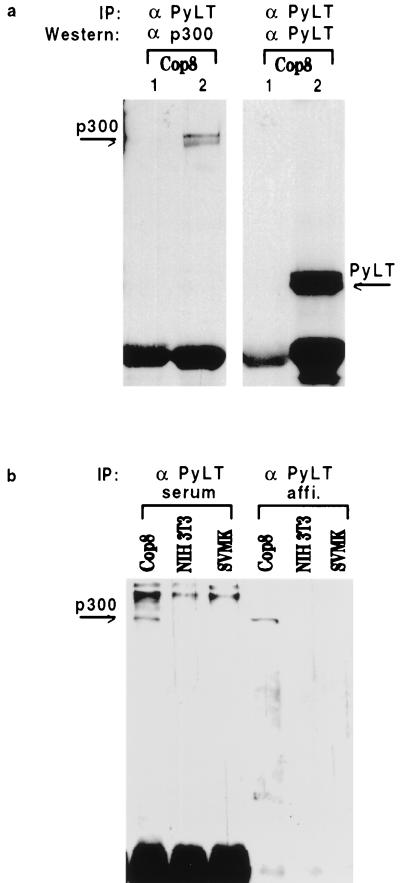
We are interested in functions of polyomavirus LT antigen (PyLT), a protein which shares properties with SV40LT but also differs in important attributes. A notable difference is that PyLT, in contrast to SV40LT, does not bind p53 (reviewed in reference 27). Previously we studied consequences of PyLT binding to pocket proteins (24, 25, 31). In this study we examined whether PyLT is able to interact with p300. As a first test for such potential interaction we carried out coimmunoprecipitation experiments using anti-p300 antibodies (RW 128 and RW 105, kindly provided by Richard Eckner) and anti-PyLT antibody (kindly provided by Egon Ogris). The immunoprecipitates were bound to protein A-Sepharose, and the beads were washed thoroughly. The eluted proteins were identified by immunoblotting with, respectively, antibody against p300 and anti-PyLT antibody, and the ECL system (Amersham) was used for detection. As shown in Fig. 1, anti-PyLT antibody precipitated p300 from extracts of COP8 cells. These are mouse C127 cells which are transformed by an origin-defective polyomavirus and express high levels of the T antigen. To rule out an unspecific binding of p300/CBP in immune complexes, anti-PyLT antibody was also tested in cells not expressing PyLT (Fig. 1b). No precipitation of p300 could be observed in this case. As expected, anti-PyLT antibody did not bring down p300 from extracts of cells devoid of PyLT, such as NIH 3T3 cells or SV40-transformed mouse kidney cells (SVMK cells). In order to verify that the protein precipitated by anti-PyLT antibody is in fact p300, proteins bound to protein A-Sepharose were eluted and reprecipitated with anti-p300 antibody. Western blotting of the second immunoprecipitate clearly identified the protein as p300 (not shown). In the converse experiment, anti-p300 only very poorly, if at all, precipitated PyLT from COP8 cell extracts (not shown). A similar observation was reported by Eckner et al. (10), who found that anti-SV40LT antibody did precipitate p300/CBP, but of 10 anti-p300 antibodies tested, only one precipitated a significant amount of SV40LT protein. Interestingly, in the same study, 9 of these 10 antibodies were found to efficiently precipitate E1A protein. This might point to differences in the interactions between p300 and E1A on the one hand and between p300 and the T antigens on the other hand, a possibility which is supported by data described below.
FIG. 1.
Coimmunoprecipitation of p300 and PyLT. (a) Cell extracts were prepared from COP8 cells by using earlier-described methods (19). The results were the same when buffers of even higher stringency (150 mM rather than 120 mM salt and 1% in place of 0.5% Nonidet P-40) were employed. Control serum (lane 1) or rabbit polyclonal anti-PyLT antibody (lane 2) was added to about 1 to 3 mg of cellular protein. After a 2-h incubation at 4°C, protein A-Sepharose beads were added and the mixture was left in the cold for at least another 2 h. The beads were then spun down and washed six times, and the bound proteins were eluted with sample buffer for gel electrophoresis. Anti-p300 antibody and anti-PyLT antibody were used for immunoblotting. The strong band at the bottom of the gels represents the immunoglobulin heavy chain. Experiments were repeated at least five times, and identical results were obtained. (b) Coimmunoprecipitation of p300 with anti-PyLT antibody requires the expression of PyLT in the cells. For immunoprecipitation, we used either the anti-PyLT antiserum, as in panel a, or an affinity purified antibody derived therefrom. The Western blotting was done with anti-p300 antibody.
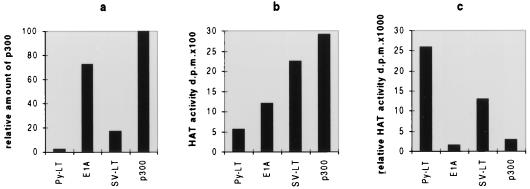
Considering the fact that p300 binds to HATs (38) and itself exhibits enzymatic activity (6, 26), we tested coimmunoprecipitates for HAT activity. Extracts from PyLT-expressing COP8 cells were precipitated with anti-PyLT antibody, those from E1A-expressing 293 cells were precipitated with anti-E1A antibody (Ad2-E1A; Santa Cruz), and those from SVMK cells were precipitated with anti-SV40LT antibody (pAB419). As a control, extracts from COP8 cells were precipitated with anti-p300 antibody. The immunoprecipitates were tested for HAT activity (9), and it was found that all those obtained with anti-viral-protein antibodies did exhibit this enzymatic activity. Interestingly, the HAT activity that precipitated with anti-PyLT antibody was considerably stronger than the activity that precipitated with anti-E1A antibody or with anti-SV40LT antibody (Fig. 2). This could be due to a greater affinity of PyLT to a subfraction of p300 which has high HAT activity. Since p300 not only has HAT activity by itself but also interacts with several other HATs like P/CAF, our data could also be interpreted as indicating that PyLT interacts with a subspecies of p300 capable of efficiently forming such HAT complexes. As p300 is a phosphoprotein which undergoes cell cycle-specific alteration (36), it is notable that SV40LT interacts preferentially with the underphosphorylated form of the protein and is capable of changing the phosphorylation status of p300 (4, 10). Interestingly, a similar characteristic of SV40LT was also noted with regard to the pRB analogs p130 and p107 (32). This contrasts with E1A, which appears to bind both unphosphorylated and phosphorylated p300 (4). It is not known whether there is any connection between the phosphorylation status of p300 and its HAT activity, but all these observations indicate that the viral proteins exhibit diversity and selectivity with regard to the subspecies of p300/CBP they bind to. This definitely influences the consequences of such binding and may explain why different viral proteins sometimes affect p300-regulated promoters in distinct ways.
FIG. 2.
HAT activity in coimmunoprecipitates. Extracts from PyLT-containing COP8 cells, E1A-containing 293 cells, and SV40LT-containing SVMK cells were utilized for coimmunoprecipitation with, respectively, anti-PyLT antibody, anti-E1A antibody, and anti-SV40LT antibody, as described for Fig. 1. In addition, p300 was directly precipitated from COP8 cell extract with anti-p300 antibody. Protein A beads carrying the respective immune complexes were split into two halves; one was used for immunoblotting of coprecipitated p300, and the other was used to determine HAT activity (9). (a) The p300 bands in immunoblots were quantitated, and the relative amounts of p300 were blotted with the signal obtained with anti-p300 antibody set at 100. (b) HAT activities as measured in the second set of aliquots of the precipitates. (c) HAT activities corrected for the amounts of p300 precipitated by the corresponding antibody. The experiments were repeated three times, and very similar results were obtained. As a control, all extracts were also precipitated with irrelevant serum which did not bring down p300 (see Fig. 1) and had only background activity in HAT assays (not shown).
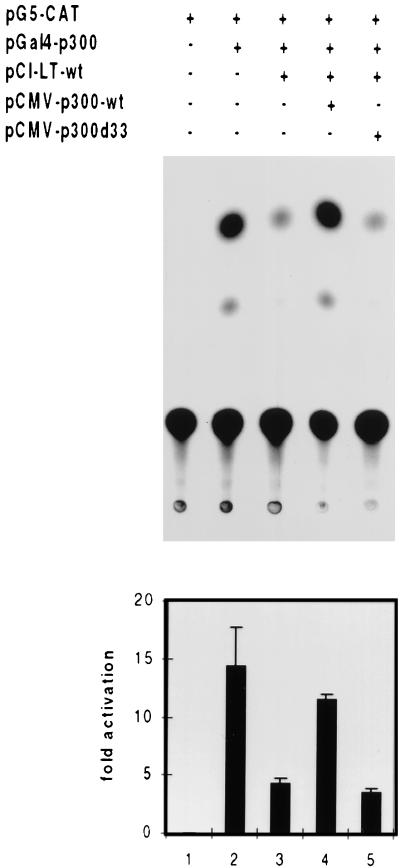
The experiments presented so far strongly suggest that PyLT and p300 bind to each other in vivo and that PyLT exhibits some preference for a subspecies of p300 which displays prominent HAT activity. Further support for such an in vivo interaction was obtained in the transfection experiment described in Fig. 3, in which all the transfected plasmids used (except for the chloramphenicol acetyltransferase [CAT] construct) carried the cytomegalovirus (CMV) promoter for expression. This experiment makes use of the intrinsic transactivating ability of p300. By using the calcium phosphate precipitation method, a GAL4-p300 construct (kindly provided by Antonio Giordano) was transfected into NIH 3T3 cells together with a promoter-CAT construct carrying GAL4 binding sites (pG5-CAT; Clontech). This promoter was activated by GAL4-p300, and the activation was suppressed if PyLT was cotransfected. Furthermore, this suppression could be relieved by an additional transfection of p300 without GAL4 binding domains (kindly provided by Richard Eckner). As this p300 cannot by itself bind to the promoter, we have to assume that it functioned by binding to PyLT, thereby eliminating the inhibitory effect of the viral protein. In support of this assumption, we observed that a mutant form of p300 (p300del33; kindly donated by Richard Eckner) which is defective in interactions with several proteins in vivo (2, 11, 20) was unable to suppress the effect of PyLT in this assay (Fig. 3). In p300del33, amino acids 1737 to 1836 are deleted and the deletions affect the binding of proteins such as E1A, SV40LT, p53, and P/CAF. Our data allow us to include PyLT in the list of proteins of this type.
FIG. 3.
PyLT inhibits the transactivating activity of GAL4-p300 fusion protein from a Gal4-CAT promoter. This inhibition can be relieved by overexpression of p300 but not by p300del33, a mutant defective in the binding of a variety of proteins. By the calcium phosphate precipitation method, NIH 3T3 cells (2.5 × 105 cells/6-cm-diameter dish, plated 24 h before transfection) were transfected with the plasmids listed at the top of the figure, using 1 μg of pG5-CAT (this plasmid carries GAL4 binding sites), 2 μg of pGAL4-p300, 2 μg of PyLT, and 4 μg of either pCMV-p300 or pCMVp300del33. In all cases, the total amount of transfected DNA was brought to 10 μg by addition of empty plasmid. CAT activity was determined 48 h thereafter. Quantitation of the results is shown at the bottom of the figure. Each column displays the mean value of five independent experiments, and each vertical bar shows the standard deviation.
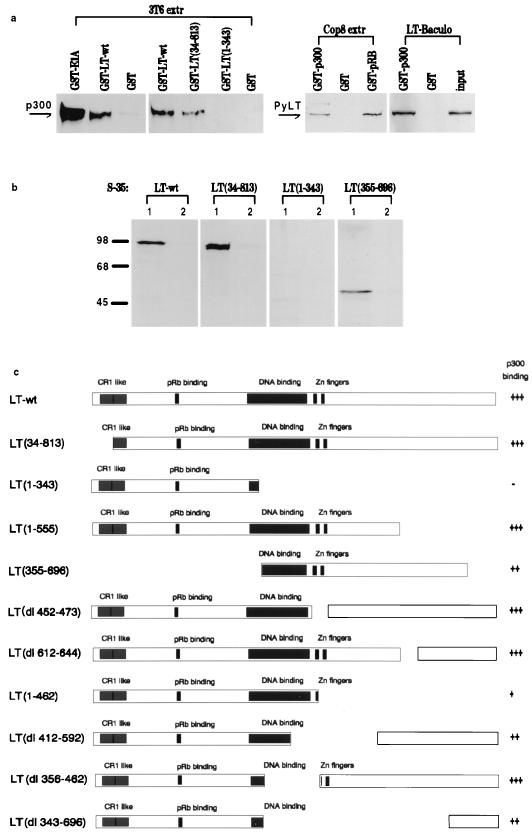
To verify the interaction between p300 and PyLT in vitro, glutathione S-transferase (GST) pulldown experiments were performed under conditions described previously (17). GST fusion proteins were constructed from PyLT and various mutated and truncated forms thereof as well as from the C-terminal part of p300 (amino acids 1570 to 2368), which is known to suffice for binding of E1A and SV40LT. The constructs were bound to glutathione-agarose beads and exposed to cell extracts containing p300 or PyLT or to in vitro-synthesized truncated versions of PyLT. As shown in Fig. 4a, GST-PyLT bound p300, albeit more weakly than GST-E1A, which was used as a positive control. Conversely, GST-p300 bound PyLT only slightly more weakly than the positive control, GST-pRB. Furthermore, GST-p300 bound not only PyLT present in cell extracts but also protein produced in and purified from baculovirus-infected f9 cells (kindly donated by Ingrid Mudrak), thus providing strong evidence for a direct interaction between p300 and PyLT. GST alone served as the negative control and did not bind any protein from the extracts. Also shown in this figure are some representative results obtained with GST fusion proteins containing fragments of PyLT. The fragment from amino acids 1 to 343, in contrast to fragments containing the C-terminal part of PyLT, did not bind p300 in this assay. A mutation of the RB binding site of PyLT also was without effect (not shown). For several reasons, it was difficult to express the truncated versions of PyLT in mouse cells after transfection for pulldown experiments with GSTp300, because it was impossible to ensure that all of the modified versions of PyLT are produced about equally in transfected cells and have similar stability, particularly as some of these proteins lack a nuclear localization signal. We therefore have chosen to synthesize the fragmented PyLT proteins in vitro and to use equal amounts of labelled proteins for binding to GST-p300. The results of this experiment are shown in Fig. 4b, which depicts the outcomes of representative pulldown experiments. The results are in good agreement with those shown in Fig. 4a. A summary of results obtained with mutated versions of PyLT is shown in Fig. 4c. This indicates that the N terminus of PyLT, including the binding site for pRB, is not required for the interaction with p300. In the case of SV40LT, a mutation of the pRB binding site was found to affect the interaction with p300/CBP; such mutations also failed to alter the phosphorylation status of the coactivator protein (10). On the other hand, the information obtained with fragments of the C-terminal part of PyLT argues that the binding site is complex and that more than one region of the protein is involved. For better comparison of the efficiencies of binding of different LT fragments, the amount of radioactive fragment bound to GST-p300 relative to the input amount was calculated. As indicated, different truncated versions of PyLT bind to p300 with different efficiencies. In particular, there seem to be two regions which contribute to the interaction: one of these is close to the Zn fingers, and the other one is located at the C terminus. The N-terminal and the C-terminal halves of PyLT seem to form domains which can function independently (13, 15). Our results thus imply that it is the C-terminal domain of PyLT which is involved in binding p300 and that two subdomains therein appear to play a role. This indicates that the three-dimensional structure is important for this interaction, a conclusion also evident from observations made for the binding of Smad proteins to CBP/p300 (16a). Furthermore, the complexity of the p300 binding region of PyLT corresponds to similar findings for the LT from SV40. It points to a difference in the binding specificities for p300 between these two viral proteins and the adenovirus protein E1A. SV40LT was shown to form complexes in which both p300 and p53 are present, and the major site of interaction between p300 and SV40LT coincides with the albeit broadly defined binding region for p53 in the C-terminal half of the protein. In this context it is worth noting that PyLT, in contrast to SV40LT, does not bind to the p53 protein; PyLT therefore associates with p300 despite its inability to interact with p53.
FIG. 4.
GST pulldown experiments. (a) Various GST fusion proteins, as outlined at the top of the figure, were bound to glutathione-agarose (the binding buffer was the same as the extraction buffer described in reference 10, except that the amount of Nonidet P-40 was reduced to 0.2%) and mixed with extracts of 3T6 cells (3T6 extr) or COP8 cells (Cop8 extr). The beads were washed, bound proteins were then eluted, and the amount of p300 or PyLT was determined by immunoblotting. GST-E1A was used as a positive control for the binding of p300, and GST-pRB was used as a positive control for the binding of PyLT. Also shown are the results of an experiment in which PyLT produced in and purified from insect cells as a baculovirus construct (LT-Baculo) was employed instead of a cell extract. The amount of purified PyLT applied onto the gel for straight immunoblotting was 5% of the amount used in the GST binding experiment. wt, wild type. (b) Examples of experiments demonstrating the interaction between GST-p300 and in vitro-synthesized, 35S-labelled PyLT or fragments thereof (produced essentially as described in reference 17). Using suitable restriction enzymes, fragments and deletions of PyLT were produced and cloned into pCIneo (Promega) for in vitro transcription and translation. Labelled fragments were incubated with GST-p300 (lanes 1) or GST alone (lanes 2) immobilized on glutathione-agarose beads. Bound protein was eluted from the washed beads, separated in sodium dodecyl sulfate gels, and visualized by autoradiography. Five percent of the amount of radioactive fragment added to beads was subjected to electrophoresis in parallel to obtain the signal of the input (not shown). (c) Summary of attempts to define regions of PyLT involved in binding to p300. The efficiency of binding was calculated relative to the amount of input radioactive fragment. The gels were autoradiographed, and the bands were scanned. +++ corresponds to 8 to 11%, ++ corresponds to 5 to 6%, and + corresponds to 2 to 3% binding of input fragment.
The difference between the E1A protein, which has at the N terminus a well defined region that is involved in p300 binding, and the T antigens from SV40 and polyomavirus, in which ill-defined domains within the C-terminal parts of the proteins play a role in interactions with p300, points once more to interesting distinctions between proteins encoded by different DNA tumor viruses. It is to be expected that this difference has functional consequences. On the other hand, in contrast to the well-defined sites of interaction of the T proteins with pRB and its relatives, p107 and p130, or with the DnaJ-type chaperones (28, 30, 33), which allow production of point mutants for the study of the biological effects of such mutations, the situation for sites of interaction with p300 does not allow such investigations at the present time, and such investigations may turn out to be altogether difficult to achieve. Furthermore, while the importance of the site of interaction of the E1A protein with p300/CBP for S-phase inducing and immortalizing functions of the viral protein is well documented, experiments with SV40LT and PyLT suggest that in these cases the binding sites for pRB (and its relatives) and the J domain, both situated at the N termini of the proteins, are sufficient for S-phase induction (13, 15, 22, 39). What then is the role of the p300/CBP binding capacity of these viral proteins? In this context it has to be pointed out that the vast majority of experiments with the viral proteins were conducted with fibroblast cell lines from rodents, most frequently murine 3T3 cells. Results obtained under these conditions may represent specialized cases which do not require the full repertoire of virus functions and, therefore, cannot necessarily be extrapolated to other cell types, in particular to differentiated cells. It is likely that interference of the T antigens with p300/CBP-regulated processes is mandatory for efficient virus replication in various differentiated cells.
Acknowledgments
We are grateful to Richard Eckner, Antonio Giordano, Ingrid Mudrak, and Egon Ogris for materials and to Egon Ogris, Hans Rotheneder, and Christian Seiser for help and discussion.
This work was supported by the Fonds zur Förderung der wissenschaftlichen Forschung.
REFERENCES
- 1.Abraham S E, Lobo S, Yaciuk P, Wang H G H, Moran E. p300 and p300-associated proteins are components of TATA-binding protein (TBP) complexes. Oncogene. 1993;8:1639–1647. [PubMed] [Google Scholar]
- 2.Arany Z, Sellers W R, Livingston D M, Eckner R. E1A-associated p300 and CREB-associated CBP belong to a conserved family of coactivators. Cell. 1994;77:799–800. doi: 10.1016/0092-8674(94)90127-9. [DOI] [PubMed] [Google Scholar]
- 3.Arany, Z., D. Newsome, E. Olread, D. M. Livingston, and R. Eckner. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature (London) 374:81–84. [DOI] [PubMed]
- 4.Avantaggiati M L, Carbone M, Graessmann A, Nakatani Y, Howard B, Levine A S. The SV40 large T antigen and adenovirus E1A oncoproteins interact with distinct isoforms of the transcriptional co-activator, p300. EMBO J. 1996;25:2236–2248. [PMC free article] [PubMed] [Google Scholar]
- 5.Avantaggiati M L, Ogryzko V, Gardner K, Giordano A, Levine A S, Kelly K. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- 6.Bannister A J, Kouzarides T. The CBP coactivator is a histone acetyltransferase. Nature (London) 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 7.Barbeau, D., R. Charboneau, S. G. Whalen, S. T. Bayley, and P. E. Branton. Functional interactions within adenovirus E1A protein complexes. Oncogene 9:359–373. [PubMed]
- 8.Dallas P B, Yaciuk P, Moran E. Characterization of monoclonal antibodies raised against p300: both p300 and CBP are present in intracellular TBP complexes. J Virol. 1997;71:1726–1731. doi: 10.1128/jvi.71.2.1726-1731.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eberharter A, Lechner T, Goralik-Schremel M, Loidl P. Purification and characterization of the cytoplasmic histone acetyltransferase B of maize embryos. FEBS Lett. 1996;386:75–81. doi: 10.1016/0014-5793(96)00401-2. [DOI] [PubMed] [Google Scholar]
- 10.Eckner R, Ludlow J W, Lill N L, Oldread E, Arany Z, Modjtahedi N, DeCaprio J A, Livingston D M, Morgan J A. Association of p300 and CBP with simian virus 40 large T antigen. Mol Cell Biol. 1996;16:3454–3464. doi: 10.1128/mcb.16.7.3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckner R, Ewen M E, Newsome D, Gerdes M, DeCaprio J A, Lawrence J B, Livingston D. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 12.Giles, R. H., D. J. M. Peters, and M. H. Breuning. Conjunction dysfunction: CBP/p300 in human disease. Trends Genet. 14:178–183. [DOI] [PubMed]
- 13.Gjorup O V, Rose P E, Holman P S, Bockus B J, Schaffhausen B S. Protein domains connect cell cycle stimulation directly to initiation of DNA replication. Proc Natl Acad Sci USA. 1994;91:12125–12129. doi: 10.1073/pnas.91.25.12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu W, Shi X-L, Roeder E G. Synergistic activation of transcription by CBP and p53. Nature (London) 1997;387:819–822. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 15.Holman P S, Gjoerup O V, Davin T, Schaffhausen B S. Characterization of an immortalizing N-terminal domain of polyomavirus large T antigen. J Virol. 1994;68:668–673. doi: 10.1128/jvi.68.2.668-673.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janknecht R, Hunter T. Transcriptional control: versatile molecular glue. Curr Biol. 1996;6:951–954. doi: 10.1016/s0960-9822(02)00636-x. [DOI] [PubMed] [Google Scholar]
- 16a.Janknecht R, Wells N J, Hunter T. TGF-β-stimulated cooperation of Smad proteins with the coactivators CBP/p300. Genes Dev. 1998;12:2114–2119. doi: 10.1101/gad.12.14.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karlseder J, Rotheneder H, Wintersberger E. Interaction of Sp1 with the growth- and cell cycle-regulated transcription factor E2F. Mol Cell Biol. 1996;16:1659–1667. doi: 10.1128/mcb.16.4.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LaThangue N B. DRTF1/E2F: an expanding family of heterodimeric transcription factors implicated in cell-cycle control. Trends Biochem Sci. 1994;19:108–114. doi: 10.1016/0968-0004(94)90202-x. [DOI] [PubMed] [Google Scholar]
- 19.Lill N L, Tevethia M J, Eckner R, Livingston D M, Modjtahedi N. p300 family members associate with the carboxyl terminus of simian virus 40 large tumor antigen. J Virol. 1997;71:129–137. doi: 10.1128/jvi.71.1.129-137.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lill N L, Grossman S R, Ginsberg D, DeCaprio J, Livingston D M. Binding and modulation of p53 by p300/CBP coactivators. Nature (London) 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 21.Lundblad J R, Kwok R P S, Laurance M E, Harter M L, Goodman R H. Adenoviral E1A-associated protein p300 as a functional homologue of the transcriptional co-activator CBP. Nature (London) 1995;374:85–88. doi: 10.1038/374085a0. [DOI] [PubMed] [Google Scholar]
- 22.Marsilio E, Cheng S H, Schaffhausen B, Paucha E, Livingston D M. The T/t common region of simian virus 40 large T antigen contains a distinct transformation-governing sequence. J Virol. 1991;65:5647–5652. doi: 10.1128/jvi.65.10.5647-5652.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moran E. DNA tumor virus transforming proteins and the cell cycle. Curr Opin Genet Dev. 1993;3:63–70. doi: 10.1016/s0959-437x(05)80342-9. [DOI] [PubMed] [Google Scholar]
- 24.Mudrak I, Ogris E, Rotheneder H, Wintersberger E. Coordinated trans activation of DNA synthesis- and precursor-producing enzymes by polyomavirus large T antigen through interaction with the retinoblastoma protein. Mol Cell Biol. 1994;14:1886–1892. doi: 10.1128/mcb.14.3.1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogris E, Rotheneder H, Mudrak I, Pichler A, Wintersberger E. A binding site for transcription factor E2F is a target for trans activation of murine thymidine kinase by polyomavirus large T antigen and plays an important role in growth regulation of the gene. J Virol. 1993;67:1765–1771. doi: 10.1128/jvi.67.4.1765-1771.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 27.Pipas J M. Common and unique features of T antigens encoded by the polyomavirus group. J Virol. 1992;66:3979–3985. doi: 10.1128/jvi.66.7.3979-3985.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheng Q, Denis D, Ratnofsky M, Roberts T M, DeCaprio J A, Schaffhausen B. The DnaJ domain of polyomavirus large T antigen is required to regulate Rb family tumor suppressor function. J Virol. 1997;71:9410–9416. doi: 10.1128/jvi.71.12.9410-9416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shikama N, Lyon J, LaThangue N B. The p300/CBP family: integrating signals with transcription factors and chromatin. Trends Cell Biol. 1997;7:230–236. doi: 10.1016/S0962-8924(97)01048-9. [DOI] [PubMed] [Google Scholar]
- 30.Srinivasan A, McClellan A J, Vartikar J, Marks I, Cantalupo P, Li Y, Whyte P, Rundell K, Brodsky J L, Pipas J M. The amino-terminal transforming region of simian virus 40 large T and small t antigens functions as a J domain. Mol Cell Biol. 1997;17:4761–4773. doi: 10.1128/mcb.17.8.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stiegler P, Schüchner S, Lestou V, Wintersberger E. Polyomavirus large T antigen-dependent DNA amplification. Oncogene. 1997;14:987–995. doi: 10.1038/sj.onc.1200904. [DOI] [PubMed] [Google Scholar]
- 32.Stubdal H, Zalvide J, DeCaprio J A. Simian virus 40 large T antigen alters the phosphorylation state of the RB-related proteins p130 and p107. J Virol. 1996;70:2781–2788. doi: 10.1128/jvi.70.5.2781-2788.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stubdal H, Zalvide J, Campbell K S, Schweitzer C, Roberts T M, DeCaprio J. Inactivation of pRB-related proteins p130 and p107 mediated by the J domain of simian virus 40 large T antigen. Mol Cell Biol. 1997;17:4979–4990. doi: 10.1128/mcb.17.9.4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang H G H, Moran E, Yaciuk P. E1A promotes association between p300 and pRB in multimeric complexes required for normal biological activity. J Virol. 1995;69:7917–7924. doi: 10.1128/jvi.69.12.7917-7924.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang H G H, Rikitake Y, Carter M C, Yaciuk P, Abraham S E, Zerler B, Moran E. Identification of specific adenovirus E1A N-terminal residues critical to the binding of cellular proteins and to the control of cell growth. J Virol. 1993;67:476–488. doi: 10.1128/jvi.67.1.476-488.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yaciuk P, Moran E. Analysis with specific polyclonal antiserum indicates that the E1A-associated 300-kDa product is a stable nuclear phosphoprotein that undergoes cell cycle phase-specific modification. Mol Cell Biol. 1991;11:5389–5397. doi: 10.1128/mcb.11.11.5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yaciuk P, Carter M C, Pipas J M, Moran E. Simian virus 40 large-T antigen expresses a biological activity complementary to the p300-associated transforming function of the adenovirus E1A gene products. Mol Cell Biol. 1991;11:2116–2124. doi: 10.1128/mcb.11.4.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang X-J, Ogryzko V V, Nishikawa J, Howard B, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature (London) 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 39.Zalvide J, DeCaprio J A. Role of pRb-related proteins in simian virus 40 large-T-antigen-mediated transformation. Mol Cell Biol. 1995;15:5800–5810. doi: 10.1128/mcb.15.10.5800. [DOI] [PMC free article] [PubMed] [Google Scholar]