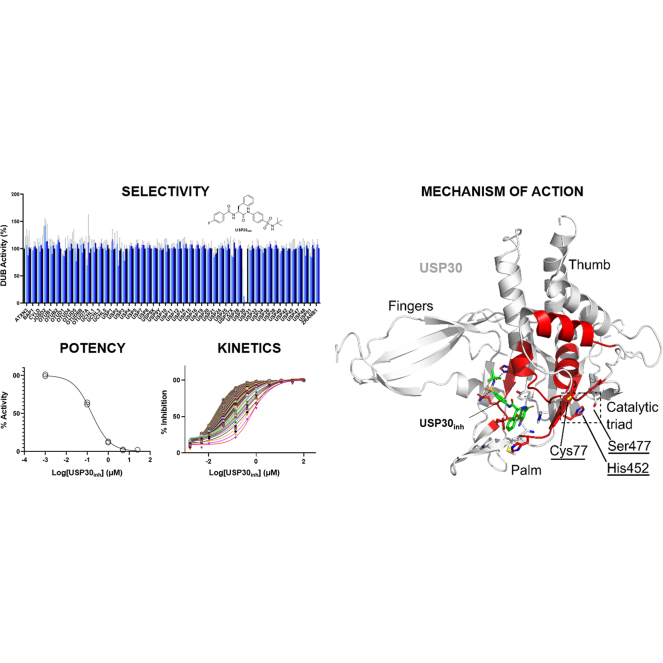
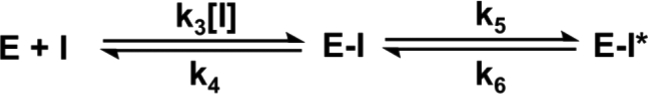Abstract
Dampening functional levels of the mitochondrial deubiquitylating enzyme Ubiquitin-specific protease 30 (USP30) has been suggested as an effective therapeutic strategy against neurodegenerative disorders such as Parkinson’s Disease. USP30 inhibition may counteract the deleterious effects of impaired turnover of damaged mitochondria, which is inherent to both familial and sporadic forms of the disease. Small-molecule inhibitors targeting USP30 are currently in development, but little is known about their precise nature of binding to the protein. We have integrated biochemical and structural approaches to gain novel mechanistic insights into USP30 inhibition by a small-molecule benzosulfonamide-containing compound, USP30inh. Activity-based protein profiling mass spectrometry confirmed target engagement, high selectivity, and potency of USP30inh for USP30 against 49 other deubiquitylating enzymes in a neuroblastoma cell line. In vitro characterization of USP30inh enzyme kinetics inferred slow and tight binding behavior, which is comparable with features of covalent modification of USP30. Finally, we blended hydrogen–deuterium exchange mass spectrometry and computational docking to elucidate the molecular architecture and geometry of USP30 complex formation with USP30inh, identifying structural rearrangements at the cleft of the USP30 thumb and palm subdomains. These studies suggest that USP30inh binds to this thumb–palm cleft, which guides the ubiquitin C terminus into the active site, thereby preventing ubiquitin binding and isopeptide bond cleavage, and confirming its importance in the inhibitory process. Our data will pave the way for the design and development of next-generation inhibitors targeting USP30 and associated deubiquitinylases.
Keywords: ubiquitin-specific protease 30, small-molecule benzosulfonamides, activity-based protein profiling, hydrogen–deuterium exchange mass spectrometry, deubiquitinylases, Parkinson's disease, mitophagy, drug discovery, structure-activity relationships, molecular docking simulations
Graphical Abstract
Highlights
-
•
The small-molecule benzosulfonamide USP30inh is highly potent and selective for neuronal USP30.
-
•
The inhibitor binds USP30 in a slow and tight manner and displays kinetic properties consistent with covalent attachment to USP30, despite its noncovalent design.
-
•
We identified regions within USP30 that undergo dramatic structural and conformational rearrangements by USP30inh, preventing Ub binding and decrease DUB activity.
-
•
Insights into USP30 inhibitory mechanistics will guide the development of next-generation inhibitors, which has relevance in neurodegeneration.
In Brief
We describe the dynamic structural interplay between USP30 and the small-molecule benzosulfonamide-containing compound, USP30inh. The inhibitor is highly potent and selective against endogenous USP30, binding in a slow and tight manner. Upon complex formation, significant perturbations occur in the conformational dynamics of the USP30 thumb–palm cleft region, proximal to its catalytic cysteine. Inhibitor binding here would sterically clash with Ub, preventing its binding and isopeptide bond cleavage. Our results help drive the design of the next generation of USP30 inhibitors.
Ubiquitination is essential to protein quality control, homeostasis, and life span (1). Damaged proteins are flagged for removal from cells with the covalent addition of ubiquitin (Ub), a small highly conserved 76-amino acid protein that is widely expressed across eukaryotic cell types (2, 3). This molecular “kiss of death” proceeds through the coordinated action of E1, E2, and E3 ligase enzymes, with targeted protein substrates conjugated by way of an isopeptide bond to either a single Ub molecule (monoubiquitination) or several repeating units (polyubiquitination) (4). Such modifications participate and control a vast array of cellular processes through proteolytic or nonproteolytic means, as seen in proteostasis, endocytic trafficking, NF-κB activation and inflammation, translation, DNA repair, and control of cell cycle processes (5). Besides the Ub–proteasome system subfamily, proteins selected for degradation can also be trafficked through the autophagy–lysosome pathway. In damaged mitochondria, such autophagic clearance of impaired proteins occurs through a highly selective and dedicated mechanism termed “mitophagy.” Several key players work tirelessly in the dysregulated mitochondrion to maintain overall cell integrity and survival, including the mitochondrial outer membrane (MOM)–associated Ub serine/threonine kinase PINK1 and the cytoplasmic E3 ligase Parkin (6). PINK1 phosphorylates Ub species on damaged proteins accumulating on the MOM, flagging them for elimination. This in turn recruits and activates endogenous Parkin (also by phosphorylation), initiating a hyperubiquitination cascade that gives the green light for mitophagy to proceed in a specialized autophagosome structure (7). Deubiquitinating enzymes (DUBs) counteract the actions of E3 ligases by removing Ub modifications, and several of these Ub-specific proteases (USPs) have been shown to oppose Parkin activity (8, 9). Of these, USP30 is the predominant active DUB to be directly implicated in mitophagy to-date, primarily due to its localization on the MOM, whilst also being linked to pexophagy and oxygen metabolism because of its widespread expression on peroxisomes (10). Interestingly, both Parkin and USP30 share an unusual preference for Lys6-linked Ub chains, the mitophagic importance of which has yet to be conclusively deciphered (11, 12).
Impaired mitophagy and oxidative stress have adverse roles in neurodegeneration, with both being linked to familial and sporadic forms of Parkinson’s Disease (PD) (13, 14); PINK1–Parkin-mediated mitophagy limits the build-up of toxic mitochondrial species in PD, and loss-of-function mutations occurring in the PINK1 and PRKN genes result in the progressive depletion of dopaminergic neurons of the basal ganglia and a rare hereditary form of juvenile Parkinsonism (15, 16). As USP30 antagonizes mitophagy through ubiquitination, its inhibition has been proposed as a novel therapeutic strategy to enhance mitochondrial turnover and clear damaged mitochondrial proteins, providing a much-needed strategy to improve outcomes in PD and other neurodegenerative disorders. Several small-molecule inhibitors targeting USP30 are in the pipeline, including phenylalanine derivatives, N-cyano pyrrolidines, and natural products (17, 18, 19). Perhaps those with the greatest potential, however, are a family of benzosulfonamides, most notably, the family member of CAS number 2242582-40-5 and hereon in referred to as “USP30inh”, which has been shown to boost mitophagy in dopaminergic neurons of PD patients by downregulating USP30 (20, 21). Little is currently known, however, regarding the intricacies of its noncovalent attachment to USP30 itself, and the structural basis of its inhibitory action. X-ray crystallography has recently provided structures of both human and zebrafish USP30, but these are solely in the context of attachment to Lys6-linked di-Ub moieties (11, 12). Hydrogen–deuterium exchange mass spectrometry (HDX-MS) is a complementary biophysical tool that can provide unique insights into protein structure, stability, dynamics, and function (22). In direct contrast to the static snapshot provided by the crystal structure, HDX-MS monitors the conformational dynamics of a system in solution, enabling the analysis of “proteins in motion.” The technique has shown particular utility for the rapid and reliable identification of small-molecule binding pockets on proteins (23). Whilst lacking the resolution of a crystal structure, the highly informative data obtained from HDX-MS can be combined with orthogonal structural, computational, biochemical, and/or biophysical techniques to define structure–activity relationships (SARs) and to direct drug discovery campaigns.
As such, we have integrated several biophysical and structural approaches to help clarify the molecular and structural interplay of USP30inh binding to USP30. The endogenous cellular selectivity of USP30inh for USP30 inhibition was confirmed using activity-based protein profiling mass spectrometry (ABPP-MS) against a panel of 49 other endogenous DUBs in neuronal SH-SY5Y cells. Biolayer interferometry showed that USP30inh binds to USP30 in a slow and tight manner, which intriguingly, is consistent with the profile of covalent inhibition, despite it being a noncovalent binder. Finally, we combined HDX-MS and molecular docking simulations to elucidate the conformational dynamics and spatial preferences of USP30inh binding, identifying key residues in the inhibitory process itself. USP30inh induces conformational and structural rearrangements at the cleft of the USP30 thumb and palm subdomains, in a region encompassing its catalytic residues and the site of Ub binding. We postulate that these phenomena underlie the mechanism of inhibition of USP30inh. Dampening USP30 pharmacologically may represent a tractable treatment for PD and other mitophagy-related disorders. As no previous attempts to investigate the molecular basis of compound USP30inh binding to USP30 have been reported, our data will be instrumental in the development of rational next-generation inhibitors against USP30 and related DUBs.
Experimental Procedures
Purity
All compounds were >95% pure by HPLC analysis (supplemental Fig. S1). USP30inh refers to CAS number 2242582-40-5, which may be referred to as USP30 inhibitor compound 39 elsewhere.
Experimental Design and Statistical Rationale
The inhibition of USP30inh against endogenous USP30 was probed in SH-SY5Y cells using ABPP-MS. Cell extracts were treated with a range of inhibitor concentrations and quantified using label-free quantification and data-independent acquisition (DIA). All measurements were performed in biological duplicate. To decipher the conformational dynamic of inhibitor binding to a recombinant version of the protein, HDX-MS was performed. A USP30 sequence coverage map was generated following triplicate analysis of peptides acquired by MSe. Triplicate measurements were also obtained for both apo- and holo-USP30 across several deuterium-labeling time points. To estimate the statistical significance of deuterium uptake levels for apo- and holo-USP30, the average deuterium uptake value across triplicate measurements was first calculated for each USP30 peptide. We determined p values using a two-tailed one-sample Student’s t test calculated for all peptides across states, which allowed us to readily identify statistically significant changes in the deuterium uptake patterns of USP30 induced by inhibitor binding.
ABPP Assay
Hemagglutinin–Ub–Propargylamine Synthesis
Hemagglutinin (HA)–Ub–propargylamine (PA) synthesis was carried out as previously described (24, 25). Briefly, a pTYB construct was used to express Ub (Gly76del) in Escherichia coli. The Ub was tagged with a HA tag on the N terminus, and an intein–chitin binding domain on the C terminus. E. coli lysis was performed by sonication in 50 mM Hepes, 150 mM NaCl, 0.5 mM DTT. The protein was then bound to chitin bead slurry and incubated with 100 mM MesNa overnight (37 °C with agitation) to form HA–Ub–MesNa. The HA–Ub–MesNa was then incubated for 20 min with 250 mM PA (room temperature with agitation) and desalted to remove excess PA, resulting in the reactive activity-based probe HA–Ub–PA.
Cell Culture and Lysis
SH-SY5Y cells were cultured at 37 °C, 5% CO2 in Eagle’s minimum essential medium and Ham’s F12 nutrient mix (1:1), supplemented with 15% fetal bovine serum, 1% nonessential amino acids, and 2 mM Glutamax. Cells were collected by washing with PBS, followed by scraping in PBS and centrifugation at 200g. Cells were lysed in 50 mM Tris base, 5 mM MgCl2.6 H2O, 0.5 mM EDTA, 250 mM sucrose, 1 mM DTT by vortexing with acid washed beads (1:1 v/v) ten times (30 s vortexing, 1 min break on ice). Lysates were clarified at 600g for 10 min at 4 °C. Lysate protein concentration was then determined by bicinchoninic acid.
Inhibitor Selectivity With HA–Ub–PA Immunoprecipitation
HA–Ub–PA protein complexes were immunoprecipitated and analyzed using LC–MS/MS as previously described (26). USP30inh or dimethyl sulfoxide (DMSO) was incubated with 500 μg of SH-SH5Y protein lysate for 1 h at 37 °C. HA–Ub–PA was then incubated with the USP30inh-treated lysates for 45 min at 37 °C at a protein ratio of 1:200 (w/w). The reaction was quenched with the addition of 0.4% SDS and 0.5% NP-40 (IGEPAL CA-630) and diluted to 0.5 mg/ml with 50 mM Tris, 0.5% NP-40, 150 mM NaCl, and 20 mM MgCl2.6 H2O, pH 7.4. HA–Ub–PA protein complexes were immunoprecipitated using 150 μl of prewashed anti-HA agarose slurry overnight at 4 °C with end-over-end rotation. The agarose slurry was then washed four times, and the HA–Ub–PA protein complexes were eluted using 110 μl of 2× Laemmli buffer. To check for efficient immunoprecipitation, 10 μl of the eluates were run on a Western blot.
The remaining 100 μl of the eluates were reduced with 20 mM DTT for 10 min at 95 °C and alkylated with 40 mM of iodoacetamide for 30 min at room temperature in the dark. Proteins were then acidified to 1.2% phosphoric acid, diluted sixfold with 90% methanol/100 mM tetraethylammonium bromide, and captured/washed on an S-trap column according to the standard protocol (27). Proteins were digested on the S-trap column with 2 μg of trypsin overnight at 37 °C. Eluted peptides were then dried and resuspended in 2% acetonitrile (ACN) and 0.1% formic acid (FA).
LC–MS/MS
Peptides were analyzed using a Dionex Ultimate 3000 nano-ultra high pressure reversed-phase chromatography system coupled on-line to an Orbitrap Fusion Lumos mass spectrometer (Thermo Scientific). Samples were separated on an EASY-Spray PepMap RSLC C18 column (500 mm × 75 μm, 2 μm particle size; Thermo Scientific) over a 60 min gradient of 2 to 35% ACN in 5% DMSO, 0.1% FA, and at 250 nl/min. The column temperature was maintained at 50 °C with the aid of a column oven. The mass spectrometer was operated in positive polarity mode with a capillary temperature of 305 °C. DIA mode was utilized for automated switching between MS and MS/MS acquisition as described previously (28). Briefly, full scans (m/z 350–1650) were acquired in the Orbitrap with 120 k resolution and maximum injection time of 20 ms, followed by 40 DIA scan windows with variable widths (supplemental Table S1). MS/MS fragmentation was performed in the higher energy collisional dissociation cell with a collision energy set at 30%. MS2 scans were acquired in the Orbitrap between m/z 200 and 2000 at a resolution of 30 k. The minimum points-per-peak was enabled and set to 6 with a dynamic maximum injection. All data were acquired in profile mode.
DIA–MS Data Processing and Analysis
Data were analyzed using DIA-NN (version 1.8) with all settings as default (29). Specifically, we allowed for a maximum of one tryptic missed cleavage and fixed modifications of N-term M excision and carbamidomethylation of Cys residues. No variable modifications were selected. A Homo sapiens UniProt database (20,370 entries, retrieved on April 16, 2021) was used for the analysis. A default threshold of 1% false discovery rate was used at the peptide and protein levels. We used the library-free mode of DIA-NN to generate precursor and corresponding fragment ions in silico from the UniProt database (29). The software also generates a library of decoy precursors (negative controls). Retention time alignment was performed using endogenous peptides, and peak scores were calculated by comparison of peak properties between observed and reference spectra. Immunoprecipitations were carried out in duplicate, and any DUBs that were not present in both control samples and enriched greater than fivefold when compared with a no probe control were discarded from the analysis. Identifications that were assigned to multiple DUBs were not included in the analysis. MINDY3 was also removed from the dataset as it was of the lowest intensity, so may have been at the bottom of the instrument's dynamic range and did not produce stable values across the dataset. Duplicate measurements were acquired for all samples and used to determine experimental statistics. All MS raw files were deposited in PRIDE under the code PXD036574.
Enzyme Kinetics
In Vitro USP30 Activity Assay
Fluorescence intensity measurements were used to monitor the cleavage of a Ub–rhodamine (Ub-Rho110) substrate. All activity assays were performed in black 384-well plates in 20 mM Tris–HCl, pH 8.0, 150 mM potassium glutamate, 0.1 mM Tris(2-carboxyethyl)phosphine) (TCEP), and 0.03% bovine gamma globulin with a final assay volume of 20 μl. Compound IC50 values for DUB inhibition were determined as previously described (18). Briefly, an 11-point dilution series of compounds were dispensed into black 384-well plates using an Echo-550 Acoustic Liquid Handler (Beckman Coulter). USP30, 0.2 nM (residues 64–502Δ179–216 & 288–305; Viva Biotech [Shanghai] Ltd), was added, and the plates were preincubated for 30 min, 25 nM Ub-Rho110 (Ubiquigent) was added to initiate the reaction, and the fluorescence intensity was recorded for 30 min on a PHERAstar FSX (λEx = 485 nm, λEm = 520 nm) (BMG Labtech). Initial rates were plotted against compound concentration to determine IC50. Data were processed using analysis tools from Dotmatics (https://www.dotmatics.com/). RapidFire MS coupled to an Agilent QTof 6530 mass spectrometer (30) was used to confirm USP30 complex formation with USP30inh and assess cleavage of K6-linked di-Ub chains from USP30 in the presence and absence of the compound.
Kinetic Assays–Determination of Kinetic Parameters for Slow-Tight Binding Inhibitors
Kinetic assays were performed in 384-well SensoPlate in 20 mM Tris–HCl, pH 8.0, 300 mM potassium glutamate, 0.1 mM TCEP, and 0.2% BGG with a final assay volume of 50 μl. An 11-point dilution series of compound was dispensed into assay plates, and 25 μl 2X Ub-Rho110 was added. The dispense function of the FLIPR Tetra (Molecular Devices) was used to add 25 μl 2X USP30 to give final assay concentrations of 5 and 180 nM for USP30 and Ub-Rho110, respectively. The fluorescence signal of the enzyme activity was monitored every 3 s for 1800 s (λEx = 470–495 nm, λEm = 515–575 nm, camera gain 70, exposure time 0.6 s, and excitation intensity 80%). Analysis was performed in GraphPad Prism, version 9.4.1 for Windows (GraphPad Software; www.graphpad.com). The time course data were normalized relative to enzyme in the absence of compound and used to generate inhibition curves at each time point.
As IC50 values are time dependent for USP30inh with no covalent labeling of USP30, shown by MS (supplemental Fig. S2), data were modeled to a slow-tight binding scheme (Fig. 1). Fitting of progress curves allows for calculation of relevant kinetic parameters (Supplemental Information).
Fig. 1.
Schematic showing slow-tight inhibitor (I) binding to enzyme (E). This is a two-step process with the fast formation of a less stable intermediate complex (E-I) defined by k3 and k4, followed by the formation of a more stable but still reversible complex (E-I∗) defined by k5 and k6. When k6 approaches zero, the complex is essentially irreversible.
Biolayer Interferometry
Biolayer interferometry was performed on an Octet RED384 system (Sartorius) at 25 °C in a buffer containing 20 mM Tris–HCl (pH 8), 100 mM NaCl, 2 mM TCEP, 0.05% Tween, and 1% DMSO. Biotinylated USP30 (residues 64–502Δ179–216 & 288–305; Viva Biotech [Shanghai] Ltd) was immobilized onto super streptavidin biosensors. After 60 s baseline detection, the association of defined concentrations of USP30inh (0–5 μM) was recorded over 300 s followed by dissociation in buffer over 600 s. Traces were normalized by double subtraction of baseline (no compound) and reference sensors (no USP30, association, and dissociation of compound) to correct for nonspecific binding to the sensors. Traces were analyzed using the Octet software (version 11.2; Sartorius).
HDX-MS
HDX Sample Preparation
USP30 was incubated in either the presence (holo-USP30) or absence (apo-USP30) of a twofold molar excess of USP30inh, ensuring that all complexes were fully formed and maintained over the course of the labeling reaction. Before the HDX-MS experiments, labelling (L), equilibration (E), and quench (Q) buffers were freshly prepared with D2O or H2O, respectively (buffers E and L: 50 mM Hepes, 400 mM NaCl, 2.0 mM TCEP, 10% glycerol [v/v] at pH 7.2; buffer Q: 50 mM potassium phosphate buffer, 2.0 M guanidine hydrochloride at pH 2.30). The USP30 protein sample was supplied at 66 μM and was diluted in buffer E to a final concentration of 11 μM, which equates to 16 pmol injected onto the pepsin column. Buffers E and L were equilibrated at 20 °C, whereas the protein samples and buffer Q were equilibrated at 0 °C.
HDX Cyclic Ion Mobility Mass Spectrometry
HDX-MS experiments were carried out on a fully automated HDX-2 system (supplied by Waters and previously described by Brown and Wilson (31). The exchange reaction was initiated by diluting 3.5 μl protein sample with a concentration of 11 μM into 56.5 μl buffer E for reference, or buffer L for D2O labeling, and incubated for several time points (0, 30, 60, 600, and 3600 s). A D2O/H2O ratio in excess of 90% guaranteed that the kinetics favored unidirectional exchange. Subsequently, the exchange reaction was stopped by mixing 50 μl of sample with 50 μl precooled buffer Q. Next, 50 μl of quenched sample was subjected to a temperature-controlled chromatography system (HDX M-Class UPLC; Waters). The protein was digested online by a pepsin column (Enzymate BEH pepsin column; 2.1 × 30 mm; Waters). Eluting peptides were trapped and washed on a C18 precolumn (C18 1.7 μM VanGuard 2.1 × 5 mm precolumn; Waters) at 100 μl/min for 3 min and separated on a reversed-phase column (C18 1.7 μM Acquity UPLC 1 × 100 mm reverse-phased column; Waters) with a linear gradient ranging from 5% ACN to 40% ACN plus 0.2% FA at 40 μl/min in 8 min, followed by a rapid rise to 99% ACN and holding for 0.3 min. ACN concentration was rapidly reduced to 5% and held there for 0.2 min, followed by a linear gradient back to 99% over 0.7 min, and holding that concentration for 0.1 min. Next, C18 columns were equilibrated with 95% H2O plus 0.2% FA for 4 min. The reversed-phase chromatographic system was kept at approximately 0 °C to reduce back-exchange. Peptides eluting from reversed-phase column were measured with a SELECT SERIES Cyclic IMS mass spectrometer (Waters) in HDMSE mode (m/z 50–2000). This mode utilizes ion mobility separation for orthogonal separation of the peptides (LC, ion mobility, m/z). The mass spectrometer was fitted with an electrospray source equipped with additional independent LockSpray probe (Leu–enkephalin lock mass solution was used, m/z 556.2771).
HDX-MS Data Processing and Analysis
All MS analyses were performed in triplicate for each time point and condition. Protein Lynx Global Server 3.0 (Waters Corporation) was used for peptide identifications. A peptic peptide sequence coverage map was generated in DynamX 3.0 HDX software (Waters Corporation). Peptide-level deuterium uptake data were also visualized in DynamX and reported as relative deuterium exchange levels expressed in either mass unit or fractional exchange. The latter was calculated by dividing the experimentally measured uptake by the theoretically maximum number of exchangeable backbone amide hydrogens that could be replaced within each peptide. This number corresponds to the number of amino acid residues present in the peptide minus the number of proline residues and minus one for the N terminus that back exchanges too rapidly to be measured by MS (32). A single charge state was considered per peptide. Data were also verified and visualized in MEMHDX (33), which uses a mixed-effects model for HDX-MS statistical validation, factoring in the time dependency of the HDX reaction and number of independent replicates. The software generates two adjusted p values for each peptide, the first for the “change in dynamics” and the second for the “magnitude of ΔD.” These p values were subsequently used to categorize the data by means of a “Logit” plot (not shown). HDX-MS raw files were deposited in PRIDE under the unique identifier PXD041582.
Molecular Docking
The crystal structure of human USP30 catalytic domain (residues K64-V502) in covalent complex with Ub–PA at 2.34 Å resolution represents the highest resolution human USP30 structure available in the Protein Data Bank ([PDB] code: 5OHK) (11) and was used as the target receptor for docking the selective USP30 benzosulfonamide inhibitor, compound USP30inh (20), using AutoDock Vina implemented in the program AMDock 1.5.2 (34). Coordinates for USP30inh were generated using ChemDraw Prime 16.0.1.4 and PRODRG implemented in the CCP4 software suite (35). The compound was docked using the simple docking mode and automatic defined search space in AMDock 1.5.2.
Results and Discussion
USP30inh is Highly Potent and Selective for Neuronal USP30
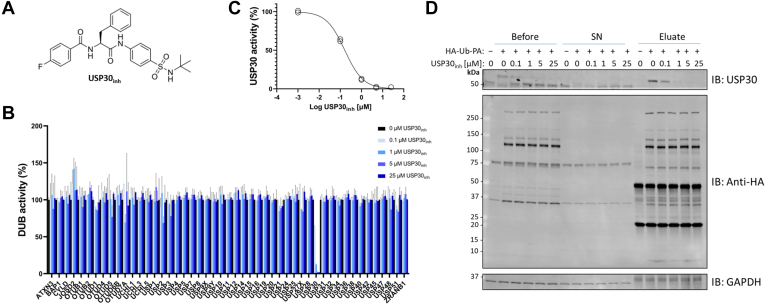
The efficacy and selectivity of USP30inh across a panel of cysteine-active DUBs was initially screened in SH-SY5Y neuroblastoma cell lysates by ABPP (Fig. 2A). SH-SY5Y cell extracts were treated with a range of inhibitor concentrations from 0.1 to 25 μM, followed by incubation with an HA-tagged Ub-based probe with a PA warhead (HA–Ub–PA). DUB-probe complexes were immunoprecipitated by way of their HA tag and quantified using label-free quantitation LC–MS/MS. We implemented a DIA MS regime to maximize the depth and reproducibility of the DUB profiling assay (supplemental Tables S1 and S2) (29, 36). The concentration-dependent competition between compound USP30inh and HA–Ub–PA for binding to USP30 confirmed target engagement and potency of the inhibitor in a cellular matrix, with an IC50 value in the nanomolar range (Fig. 2B). Moreover, USP30inh was found to be highly selective for USP30 as it had no significant activity against any of the other 49 endogenous DUBs detected in the experiment (Fig. 2B). The main cysteine-reactive DUB enzyme families were all represented, with proteins containing USP, ovarian tumor protease (OTU), Ub C-terminal hydrolase, and Josephin domains quantified (37, 38). The absence of USP30inh concentration-dependent inhibition for all other identified cysteine-active DUBs demonstrates the highly selective nature of the inhibitor. This selectivity is in line with previously published USP30 inhibitor selectivity data from both a recombinant DUB activity panel (21) and an ABPP-MS experiment on a smaller panel of endogenous DUBs identified from mouse brain tissue (25).
Fig. 2.
USP30inhis highly potent and selective for USP30 inhibition in a cellular context.A, chemical structure of USP30inh (25). B, activity-based probe profiling of cysteine-reactive deubiquitinating enzymes (DUBs) with a hemagglutinin-tagged ubiquitin-based propargylamine warhead probe (HA–Ub–PA). SH-SY5Y lysates were incubated first with USP30inh at the indicated concentration for 1 h at 37 °C, followed by HA–Ub–PA for 45 min at 37 °C. DUB-probe complexes were immunoprecipitated by the HA tag and quantified by label-free LC–MS/MS. DUB intensity in the control with no USP30inh present is normalized as 100% activity. n = 2 for each condition. C, USP30 intensity values extracted from (B), demonstrating that USP30inh behaves in a concentration-dependent manner, with an IC50 of 0.16 μM. D, Western blot corresponding to data in (B and C). Immunoprecipitation of USP30 is reduced with increasing concentration of USP30inh. Anti-HA blot demonstrates efficient immunoprecipitation, and no USP30inh cross-reactivity with other HA–Ub–PA-labeled DUBS, in agreement with LC–MS/MS data in (B). USP30, ubiquitin-specific protease 30.
We have recently reported that USP30inh can be displaced by HA–Ub–PA over long incubations (25). Accordingly, a small amount of displacement during the 45 min HA–Ub–PA incubation was anticipated. It is therefore expected that the IC50 value of 0.16 μM that we obtained from our ABPP-MS assay is likely higher than the absolute IC50 inhibition concentration (Fig. 2, C and D).
USP30inh Binds USP30 in a Slow and Tight Manner
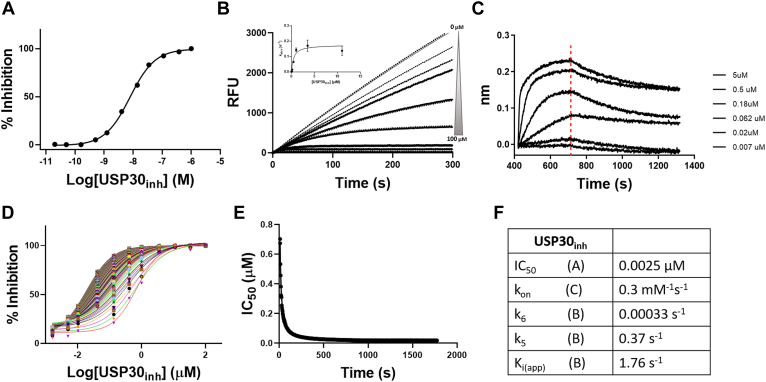
Once it was established that USP30inh downregulates endogenous USP30 activity in a highly selective fashion, we sought to rigorously profile its inhibitory properties using a recombinant version of the protein. Synthetic full-length USP30 is very unstable and difficult to solubilize (11). To circumvent this, we used a previously described truncated version of USP30 in our enzyme (and HDX-MS) assays, which readily went into solution and was determined to be stable over the time course of our experiments (supplemental Fig. S2). To assess enzyme deubiquitinating efficiency, the purified USP30 construct was incubated with a fluorogenic Ub–rhodamine substrate in both the presence and absence of USP30inh. This resulted in a calculated IC50 value for USP30inh of ∼2 nM in vitro, which was in-line (albeit 10-fold lower) with previous estimations (Fig. 3A) (20). Although both measurements remain in the nanomolar range, the 2 nM IC50 from the Ub–rhodamine assay is lower than the 162 nM IC50 from the ABPP. This could be attributable to differences in the endogenous and recombinant activity of USP30, nonspecific inhibitor occlusion in the cellular context of the ABPP, or displacement of USP30inh in the ABPP by HA–Ub–PA. Progress curves for Ub–rhodamine cleaved by USP30 were used to calculate the rate of inhibition. The kinetic constants k5, k6, and Ki gave values that were indicative of slow and tight binding behavior (Fig. 3, B, D, E, and F). The latter was visualized by a time-dependent shift of dose–response inhibition curves (Fig. 3D) as well as by plotting IC50 values against time (Fig. 3E). When considering the binding Figure 1 (Experimental Procedures section), the small value for k6 implies that it is behaving in an irreversible manner. The progress curves show typical features of an enzyme reaction in the presence of a slow binding inhibitor. Furthermore, two binding events are observed in the form of (a) an initial and (b) a steady-state velocity—both of which need to be considered during curve fitting and calculation of inhibitory rates (see Equations 2 and 3 in Supporting Information) (Fig. 3B).
Fig. 3.
Kinetic profiling of the noncovalent USP30 inhibitor USP30inh.Upper panel: A, dose-dependent inhibition of USP30 by USP30inh. B, reaction progress curves recorded on the FLIPR Tetra. Traditional method for determining kinetic constants associated with a two-step slow and tight binding inhibitor. kobs, determined by fitting the progress curves to Equation 2 (Supporting Information), is plotted versus [Compound] and fitted to Equation 3. (Supporting Information) to determine Ki, k5, and k6. C, biolayer interferometry showing binding of USP30inh to immobilized USP30 with no detectable dissociation. Lower panel: (D) Krippendorf method (49) (Supporting Information) was used as an alternative way of determining kinetic constants. Time-dependent IC50 curves. Each curve represents inhibition data at an individual incubation time from 3 to 1800 s. E, IC50 values versus incubation time fitted to Equation 1 (Supporting Information) to obtain Ki and kinact. As USP30inh is a noncovalent compound but has a k6 that is essentially 0, kinact in this case represents k5. F, data table of inhibition properties. USP30, ubiquitin-specific protease 30.
Biolayer interferometry experiments confirmed this slow and tight binding behavior (Fig. 3C). The compound had an association rate of 0.3 mM−1 s−1 and a very slow dissociation from USP30 that was comparable with features of covalent modification (Fig. 3F). This phenomenon is intriguing, as USP30inh is known to bind to USP30 by exclusively noncovalent means, as confirmed by our MS analysis in supplemental Fig. S2.
Finally, we explored the compound’s efficacy in the presence of more complex USP30–substrate interactions than Ub–rhodamine alone. We compared the cleavage of K6-linked di-Ub from USP30 in the presence and absence of USP30inh by RapidFire MS (supplemental Fig. S3). It was clearly demonstrated that USP30inh is able to inhibit the cleavage of K6-linked di-Ub from the substrate, confirming its activity in a more complex and physiologically relevant matrix.
HDX-MS Kinetics Identifies Key Residues at the USP30inh-Binding Interface of USP30
Knowledge is currently lacking on the precise location and mechanistics of USP30inh binding to USP30. HDX-MS experiments were consequently designed to pinpoint the key regions of USP30inh binding to USP30, whilst providing novel structural insights into the solution conformation and dynamics of complex formation. HDX relies on the natural isotopic exchange of the amide backbone hydrogens of a protein with deuterium when placed in a deuterated solution (22). This leads to protein mass increases that are directly measurable by MS, which can serve as direct probes of protein solvent accessibility and structure. Shielding of the deuterated solvent following introduction of a binding partner is indicative of a binding interface. We sought to identify such regions following USP30inh binding to USP30 by directly comparing the differences in HDX-MS uptake patterns of USP30 before (apo-USP30; in the presence of DMSO) and after (holo-USP30; in the presence of USP30inh) complex formation.
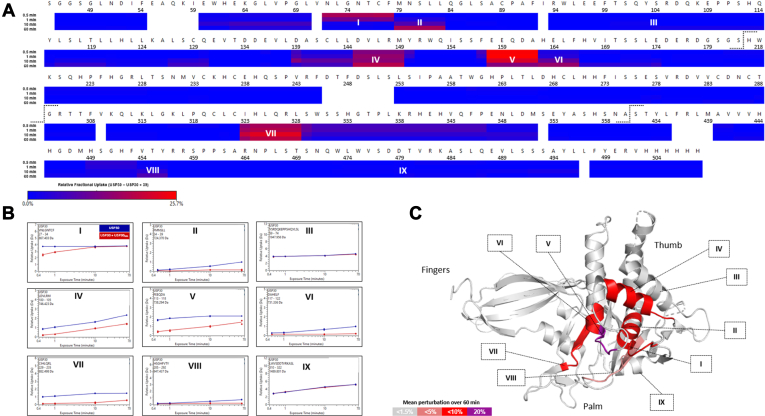
Following digestion of unlabeled USP30 with pepsin, a total of 723 peptides were generated for the protein, 133 of which were shortlisted for downstream data analysis (supplemental Figs. S4 and S5). Selected peptides covered 96.2% of the USP30 sequence, with an average of 4.19 peptides covering each amino acid. The kinetics of deuterium uptake was analyzed for all regions of USP30, which included USP domains 1 to 6 and the catalytic triad at Cys77, His452, and Ser477 (supplemental Fig. S6A). From three independent replicates, the relative fractional exchange was calculated for all peptides at each of the four time points 30, 60, 600, and 3600 s, and plotted as a function of peptide position (Fig. 4A and supplemental Fig. S6B).
Fig. 4.
HDX-MS characterizes the conformational dynamics of USP30inhbinding to USP30.A, residue-level heat map indicating that USP30inh induces solvent protection in several regions of USP30. The plot displays the difference in relative fractional uptake between the holo- and apo-form of the protein over 1 h. Regions of that have the greatest perturbation following USP30inh binding are labeled regions I–IX. B, comparative uptake plots of regions I–IX for apo- and holo-USP30 states. C, integrated HDX-MS and X-ray crystal structure of USP30 in complex with di-Ub. Regions of perturbation between apo- and holo-USP30 states of HDX-MS data are colored according to magnitude of change. The data indicate that the USP30inh-binding interface is located between the USP30 thumb and palm domains of the protein. Numbering is in accordance with the crystal structure of 5OHK. Dotted lines indicate the site of cleavage and removal of unstable disordered sequences from the full-length USP30 protein. HDX-MS, Hydrogen–deuterium exchange mass spectrometry; Ub, ubiquitin; USP30, ubiquitin-specific protease 30.
Following incubation with compound USP30inh in conditions conducive to binary complex formation, our comparative HDX-MS data indicate that the majority of USP30 is unaffected by inhibitor binding, with no differences in deuterium uptake kinetics between apo and holo states (Fig. 4A). This suggests that USP30inh binding is confined to smaller subsections of the protein. Indeed, several short regions of USP30 had significant shielding from the solvent in the presence of the inhibitor as compared with the DMSO control, indicative of regions involved in compound binding (Fig. 4, A and B). The areas of most significant perturbation included at two peptides spanning the USP30 catalytic Cys77 residue, N72–L83 (labeled regions I and II in Fig. 4 and highlighted in representative uptake plots in Fig. 4B), in addition to peptides mapping to D145–M149 (region IV), E158–F166 (regions V and VI), I323–L328 (region VII), and finally, H449–Y456 (region VIII), which encompasses the catalytic His452. These were in direct contrast to regions III and IX, which gave identical HDX-MS behaviors in both the apo and holo forms, reflecting the majority of the USP30 protein sequence (Fig. 4B). The shielding induced by inhibitor was greatest at the 1 h time point, where a difference of >25% was observed in the relative fractional uptake between states. Results were confirmed by the presence of multiple overlapping peptides displaying equivalent HDX-MS activity. Interestingly, no difference in conformational dynamics was observed between states for the region covering the catalytic Ser477 (region IX), suggesting that this site is not vital to the USP30inh inhibitory process, but rather, may have a greater influence in determining USP30 Ub linkage preferences (11, 12, 39). Nevertheless, targeting this site to improve USP30 inhibition efficacies may prove fruitful in future design regimes.
No X-ray crystal structure of apo-USP30 or USP30 in complex with USP30inh currently exists. We therefore mapped our solution HDX-MS data to PDB code 5OHK, which at a resolution of 2.34 Å, represents the highest resolution 3D structure of the protein currently available in the PDB (11). It is worth noting that this structure corresponds to USP30 in covalent complex with Lys6-linked di-Ub, and as such, may give rise to subtle discrepancies when comparing across results. Nevertheless, because of the lack of more suitable alternatives, we felt it a worthwhile pursuit to map our apo- and holo-USP30 HDX-MS data to this 3D model. To facilitate a facile overview of the entire dataset, HDX-MS results were collapsed into a single datapoint by calculating the mean perturbation between apo and holo states across the four labeling time points. USP30 constitutes three subdomains designated “thumb, palm, and fingers,” which is in line with related USP family members of elucidated structure (8, 40). Strikingly, the shielded USP30 peptides in the presence of USP30inh all cluster to the same spatially adjacent region of the protein, which lies at the interface of its palm and thumb subdomains (Fig. 4C). Moreover, the regions with the greatest perturbation as highlighted previously, I323–L328 (region VII) and H449–Y456 (region VIII), cover areas of the protein that lie opposite to each other on the 3D structure. They may represent an entrance vector to the USP30-binding pocket, which is anticipated to be closer to the site of greatest perturbation at E158–F166 (regions V and VI) and the nearby catalytic Cys77. The importance of these residues to the inhibitory process is further strengthened by their correlation with the proposed region of USP30 binding to the Ub C-terminal tail in the crystal structure (11).
Binding of USP30inh Alters USP30 Conformation and Induces Rigidification in Several Regions
Although likely to be in good agreement with the majority of 5OHK, the structural makeup of apo-USP30 has yet to be experimentally confirmed. As yet, no high-resolution crystal structure exists, which is of a direct consequence of the poor stability of the full-length protein itself (11). As stated earlier, a highly truncated USP30 construct was used in this study, which was devoid of its N-terminal mitochondrial intermembrane domain and adjacent transmembrane domain. Furthermore, several long disordered regions were cleaved, and multiple hydrophobic residues mutated out, resulting in substantially improved protein stability and solubility. As HDX-MS is not reliant on successful crystallization trials, we saw this as an opportunity to describe the solution structural integrity of apo-USP30, which would allow us to elucidate its mode of binding to USP30inh.
The conformational landscape of apo-USP30 generally follows the arrangement of its USP domains; some of the most solvent-exposed regions of the protein are found at the linker regions connecting individual domains, most noticeably between USP domains 1 and 2, 4 and 5, and 5 and 6 (supplemental Fig. S6, A and B). Conversely, USP domains 1 and 5 and the N-terminal end of USP domain 6 are largely protected from the solvent and inaccessible (supplemental Fig. S6B). This is in good agreement with HDX-MS data recently acquired on the full-length apo-USP30 protein, where the USP domains were shown to be in a conformation that was generally hidden from the solvent and connected by several exposed linkers (11). However, because of the instability of the full-length species, HDX-MS was performed on a much shorter timescale compared with our own study, with only a sole 3 s labeling time point measured. Furthermore, an appreciation of USP30 dynamics could not be extracted from this single time point. Looking at apo-USP30 dynamics across the several labeling time points described herein, an increase in the rate of deuteration over the time course of the experiment was observed across the majority of the protein (supplemental Fig. S6B). This dynamic HDX-MS behavior is indicative of the presence of secondary structural elements, thereby confirming the highly structured nature of the protein. The regions with the greatest dynamic HDX-MS behavior were found within USP domains 2, 3, 4, and 6 (supplemental Fig. S6B). Conversely, no dynamic HDX-MS events were observed in several regions of apo-USP30, and the maximum level of deuteration was reached immediately, indicating structural disorder. These unstructured regions map to the N- and C-terminal extremities of the protein and within USP domains 1 (residues 71–78), 2 (residues 130–136), and 5 (residues 439–453). There is a high level of overlap when mapping the structural data inferred from the HDX-MS to the crystal structure of 5OHK for both apo- and holo-USP30 (supplemental Fig. S7).
Several regions of USP30 undergo structural transitions in the presence of USP30inh, which are potentially significant in terms of inhibitory mechanistics. First, multiple segments of USP30 become completely blocked and inaccessible to the solvent following inhibitor binding. These include a region directly adjacent to the catalytic Cys77 at F78–L83 and an area of USP domain 2 covering E158–F166, as highlighted in purple in supplemental Fig. S7. This suggests that USP30inh induces a conformation of USP30 that not only blocks off the catalytic region and its surroundings from the solvent, but importantly, also prevents access and binding of Ub itself. A second structural phenomenon is also evident, specifically the conversion of intrinsically disordered loops in the absence of USP30inh, to rigid structural elements in the presence of the compound (supplemental Fig. S7). These disorder-to-order transitions likely embody functional significance (32), and in USP30, these are found at the catalytic Cys77, represented by peptide V71–F78, a section of USP domain 2 at R148–F154, and a long chain of residues spanning Q326–L349.
Tracking these structural rearrangements across individual labeling time points allows us to propose a general timeline of inhibition (supplemental Fig. S8). Taking Q326–L349 as an example, peptides mapping to this region of USP30 undergo significant structural transitions at the earliest time points monitored (30 and 60 s), which are completed in the later stages of our experimental time course. Conversely, peptides proximal to the catalytic Cys77 become blocked and solvent inaccessible primarily in the latter half of our experiment (600 and 3600 s). The fact that the residue (and adjacent regions) most crucial to USP30 catalysis, Cys77, is most significantly perturbed in the latter stages of our experiment could go some way to explain the slow and tight binding behavior observed for USP30inh in our enzyme kinetics analyses (Fig. 3).
Molecular Docking Proposes Key Residues Important for USP30inh Binding to USP30
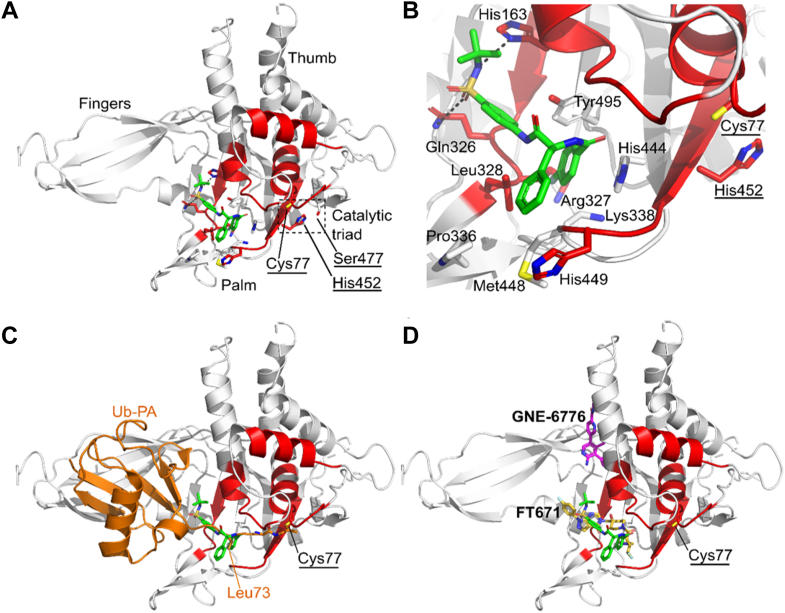
To further refine our HDX-MS findings, we explored the binding mode of USP30inh to USP30 computationally. We performed molecular docking simulations using the simple docking mode in AMDock software, with the human USP30 catalytic domain from the crystal structure of USP30 in complex with Ub–PA (PDB code: 5OHK) acting as the target receptor for the compound (11, 20, 34). The in silico binding pose of USP30inh with the highest-ranking docking score has estimated affinity and Ki values of −7.9 kcal/mol and 1.62 μM, respectively, compared with a reported experimental IC50 value of approximately 20 nM (20). USP30inh is predicted to bind to the thumb–palm cleft that guides the Ub C terminus into the active site, residing approximately 7.4 Å away at its closest point from the thiol side chain of catalytic Cys77 (Fig. 5A). The benzyl moiety of USP30inh is flanked by Pro336, Met448, and the side chains of Leu328 and His449 (Fig. 5B). The fluorophenyl moiety is flanked by the side chains of Leu328, Arg327, Lys338, and Tyr495, with a π-stacking interaction with His444. The N-tert-butyl sulfonamide moiety is anchored by hydrogen-bonding interactions with the side chains of Gln326 and His163. Compared with the structure of USP30 in complex with Ub–PA (PDB code: 5OHK), the modeled position of USP30inh would sterically clash with the C-terminal tail of the Ub substrate with the fluorophenyl moiety sitting in an equivalent position to the side chain of Ub Leu73, thereby preventing Ub binding and isopeptide bond cleavage (Fig. 5C).
Fig. 5.
Modeled structure of human USP30 in complex with USP30inh.A, structure of human USP30 catalytic domain highlighting the modeled position of USP30inh shown as a stick representation and colored green. The thumb, palm, and finger's subdomains of the catalytic domain and catalytic triad (Cys77, Ser477, and His452; underlined) are highlighted. Regions identified in the HDX-MS analysis of USP30 in the presence of USP30inh are colored red. B, close-up view of the putative USP30inh binding site highlighting key residues and hydrogen-bonding interactions represented as dotted lines. C, superposition of Ub–PA (orange; PDB code: 5OHK) on the docked structure. USP30inh sterically clashes with the C-terminal tail of the Ub substrate, thereby preventing Ub binding and isopeptide bond cleavage. D, superposition of the USP7 inhibitors, FT671 (yellow carbon atoms) and GNE-6776 (magenta carbon atoms) in complex with USP7, onto the docked structure. USP30inh putatively binds to an equivalent site in the thumb–palm cleft compared with FT671. Figure was prepared using PyMOL (The PyMOL Molecular Graphics System, version 2.4.1; Schrödinger, LLC). PA, propargylamine; PDB, Protein Data Bank; Ub, ubiquitin; USP30, ubiquitin-specific protease 30.
Crystal structures of USP7 in complex with the small-molecule inhibitors, FT761 (PDB code: 5NGE) (40) and GNE-6776 (PDB code: 5UQX) (41), reveal two distinct inhibitor binding modes that attenuate Ub binding and inhibit the DUB activity. FT671 binds to the thumb–palm cleft and resides approximately 5 Å away from the catalytic cysteine, whereas GNE-6776 interacts with acidic residues in the USP7 catalytic domain that mediate hydrogen-bonding interactions with the Ub Lys48 side chain and binds 12 Å from the catalytic cysteine. A comparison of the docked structure of USP30inh with the USP7 inhibitor complexes suggests that USP30inh is most likely to bind to an equivalent site to FT671 (Fig. 5D). In addition, the docked pose of USP30inh correlates well with the HDX-MS data, with the predicted binding site of USP30inh being flanked by residues residing in peptides E158–F166, I323–L328, and H449–Y456, which become solvent protected in HDX-MS (Fig. 5, A and B). The HDX-MS analysis also implicates peptides N72–L83 (which contains the catalytic Cys77) and D139–M149 in structural rearrangements upon compound binding. These regions reside further from the predicted binding site of USP30inh. However, compound binding may potentially cause conformational rearrangements of the catalytic domain remote from the binding site resulting in these regions becoming protected upon compound binding. Similar conformational rearrangements are seen in human USP7, in which the inhibitors bind to the catalytically incompetent apoform state with the switching loop “in”, as compared with the catalytically competent Ub-bound state with the switching loop “out”. It is possible that USP30 may exhibit similar dynamic conformational flexibility.
SAR of USP30 and USP30inh
The inhibitory activity of USP30inh and a series of analogs were reported by Mitobridge Therapeutics (20). The SAR of this series indicates that the sulfonamide N–H hydrogen bond donor is essential for inhibitory activity since replacement of this with N–Me resulted in a complete loss of inhibition. This finding is consistent with the hydrogen-bonding interaction of the sulfonamide N–H with His163 shown by our predicted binding pose.
The reported SAR also showed that the two amide N–H groups are important for potency against USP30 indicating that they may form hydrogen-bonding interactions; however, the docking simulation did not find hydrogen-bonding interactions between these N–H groups and USP30. It may be that the predicted binding mode needs further refinement or that the methylated amide groups lose inhibitory activity against USP30 for other reasons such as a steric clash between the methyl groups and USP30.
The docking simulation also indicated that the benzyl group of USP30inh contributes to binding by occupying a lipophilic pocket rather than forming π-stacking interactions. This is also consistent with the reported SAR, which indicates that the benzyl group can be replaced by other lipophilic groups, that is, a cyclohexylmethyl group. The 4-fluorophenyl group also occupies a lipophilic group in the predicted binding pose, and the docking simulation indicates that it can form a π-stacking interaction with His444. The reported SAR indicates that the 4-fluorophenyl group can be replaced both by other aromatic rings, which could maintain the π-stacking interaction (where the fluorine substituent is moved around the phenyl ring or where the phenyl ring is replaced with a pyridyl ring) or by a cyclohexyl ring that would not be able to form a π-bond indicating. This would suggest that any π-stacking interactions formed by the 4-fluorphenyl group are not essential for potent inhibition on USP30.
Comparison of USP30inh to Other Small-Molecule Compounds
Recently, the structures of human USP30 in complex with covalent inhibitors, 552 (PDB code: 8D1T; supplemental Fig. S9A) and 829 (PDB code: 8D0A; supplemental Fig. S10A), have been deposited in the PDB (https://www.lens.org/images/patent/WO/2020036940/A1/WO_2020_036940_A1.pdf). Comparisons with the structure of USP30 in complex with Ub–PA (supplemental Fig. S9, B and C; supplemental Fig. S10, B and C) reveal distinct differences in the conformation in blocking loops 1 and 2 and a switching loop region (residues 150–162), indicating that these regions are likely to be highly flexible and adopt different conformations upon Ub or compound binding. The covalent-inhibitor binding sites are flanked by regions implicated in noncovalent inhibitor USP30inh binding from the HDX-MS analysis (supplemental Figs. S9A and S10A). Comparison with the docked structure of USP30 in complex with USP30inh reveals that USP30inh is likely to bind to a similar region in the thumb–palm cleft compared with 552 and 829 (supplemental Figs. S9D and S10D), with the benzyl moiety of USP30inh overlaying on the cyclopropyl nitrile and cyclopropyl pyrazine moieties of 552 and 829, respectively (supplemental Figs. S9E and S10E). Compared with 552 and 829, the docked pose of USP30inh extends toward the finger’s subdomain. It is conceivable that conformational flexibility in blocking loops 1 and 2 and the switching loop region, not accounted for in the modeling, may facilitate USP30inh to bind closer to the catalytic cysteine, Cys77, compared with the docked pose.
Conclusions
Mitochondrial pathway disruption has been linked to a spectrum of pathophysiological conditions, from neurodegeneration and acute, chronic kidney, and cardiovascular diseases, through to hepatocellular carcinoma and peroxisome biogenesis disorders (7, 42, 43, 44). USP30 represents an actionable drug target of these conditions through its participation in PINK1/Parkin-mediated mitophagy, BAX/BAK-dependent apoptosis, oncogenesis, and pexophagy (19, 45, 46, 47). USP30 regulates mitophagy by antagonizing Parkin-mediated ubiquitination, and its inhibition has been shown to have significant therapeutic potential against PD and similar neurodegenerative disorders. Drug discovery efforts targeting USP30 have yielded the highly potent and selective small-molecule benzosulfonamide inhibitor, compound USP30inh (18, 20, 48). Combining state-of-the-art proteomics, HDX-MS and molecular docking, we have described the dynamic structural interplay between USP30 and USP30inh. The inhibitor binds to USP30 in a slow and tight manner, and displays kinetic properties consistent with covalent attachment to USP30, despite its noncovalent design. Collectively, our integrative structural biology lens successfully identified regions within USP30 that undergo dramatic structural and conformational rearrangements in the presence of USP30inh, which prevent Ub binding and decrease DUB activity. X-ray data for USP30 in complex with USP30inh will undoubtedly complement these observations and will, combined with molecular dynamics studies, drive the development of next-generation inhibitors.
Data Availability
The ABPP proteomics data files have been deposited to the ProteomeXchange Consortium and can be located under the PRIDE dataset identifier PXD036574. Annotated spectra ABPP data can be viewed following deposition to PanoramaWeb, which can be accessed using the ProteomeXchange ID code of PXD042782 and permanent link https://panoramaweb.org/LDvSvO.url. Similarly, the HDX-MS dataset can be located using the code PXD041582. PDB ID code 5OHK represents the crystal structure of human USP30 catalytic domain (residues K64–V502) in covalent complex with Ub–PA at 2.34 Å resolution. This is the highest resolution structure of human USP30 currently available in the PDB.
Supplemental Data
This article contains supplemental data.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Drs Roman Fischer and Simon Davis from the Oxford Translational Proteomics Hub for their help with ABPP-MS data acquisition and deposition. We also thank Daryl S. Walter from Evotec and Jeff Schkeryantz from Bristol Myers Squibb for the highly useful project discussions. Our gratitude goes to Alzheimer’s Research UK for their support and funding for the ARUK-Oxford Drug Discovery Institute (grant no.: ARUK-2021DDI-OX). We also thank our other funders, including the late Mr and Mrs James Hardwick for funding the ODDI Medicinal Chemistry Team, and the G & K Boyes Charitable Trust.
Funding and additional information
We thank our other funders, including the Chinese Academy of Medical Sciences Innovation Fund for Medical Science, China (grant number: 2018-I2M-2-002) (to A. P.-F. and B. M. K.) and Bayer (to B. M. K.).
Author contributions
J. B. D. and B. M. K. conceptualization; D. P. O. B., H. B. L. J., F. G., E. J. M., I. V., M. A., A. P.-F., and A. P. T. methodology; M. A. and A. P. T. software; D. P. O. B., H. B. L. J., F. G., E. J. M., A. P.-F., and A. P. T. validation; D. P. O. B., H. B. L. J., F. G., E. J. M., K. S. E., A. P.-F., and A. P. T. formal analysis; D. P. O. B., H. B. L. J., F. G., E. J. M., K. S. E., M. A., A. P.-F., and A. P. T. investigation; M. A., P. B., J. B. D., and B. M. K. resources; D. P. O. B., H. B. L. J., and F. G. data curation; D. P. O. B., H. B. L. J., F. G., E. J. M., I. V., M. A., A. P.-F., and A. P. T. writing–original draft; D. P. O. B., H. B. L. J., F. G., E. J. M., K. S. E., I. V., M. A., P. E. B., J. B. D., A. P.-F., A. P. T., and B. M. K. writing–review & editing; D. P. O. B., H. B. L. J., F. G., E. J. M., A. P.-F., and A. P. T. visualization; P. E. B., J. B. D., and B. M. K. supervision; P. E. B., J. B. D., and B. M. K. project administration; P. E. B., J. B. D., and B. M. K. funding acquisition.
Contributor Information
Darragh P. O'Brien, Email: darragh.obrien@ndm.ox.ac.uk.
Benedikt M. Kessler, Email: benedikt.kessler@ndm.ox.ac.uk.
Supplemental Data
References
- 1.Popovic D., Vucic D., Dikic I. Ubiquitination in disease pathogenesis and treatment. Nat. Med. 2014;20:1242–1253. doi: 10.1038/nm.3739. [DOI] [PubMed] [Google Scholar]
- 2.Ciechanover A. The ubiquitin proteolytic system and pathogenesis of human diseases: a novel platform for mechanism-based drug targeting. Biochem. Soc. Trans. 2003;31:474–481. doi: 10.1042/bst0310474. [DOI] [PubMed] [Google Scholar]
- 3.Pickart C.M., Eddins M.J. Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 4.Mulder M.P., Witting K., Berlin I., Pruneda J.N., Wu K.P., Chang J.G., et al. A cascading activity-based probe sequentially targets E1-E2-E3 ubiquitin enzymes. Nat. Chem. Biol. 2016;12:523–530. doi: 10.1038/nchembio.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Damgaard R.B. The ubiquitin system: from cell signalling to disease biology and new therapeutic opportunities. Cell Death Differ. 2021;28:423–426. doi: 10.1038/s41418-020-00703-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pollock L., Jardine J., Urbe S., Clague M.J. The PINK1 repertoire: not just a one trick pony. Bioessays. 2021;43 doi: 10.1002/bies.202100168. [DOI] [PubMed] [Google Scholar]
- 7.Bingol B., Sheng M. Mechanisms of mitophagy: PINK1, Parkin, USP30 and beyond. Free Radic. Biol. Med. 2016;100:210–222. doi: 10.1016/j.freeradbiomed.2016.04.015. [DOI] [PubMed] [Google Scholar]
- 8.Komander D., Clague M.J., Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 9.Nguyen T.N., Padman B.S., Lazarou M. Deciphering the molecular signals of PINK1/Parkin mitophagy. Trends Cell Biol. 2016;26:733–744. doi: 10.1016/j.tcb.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 10.Marcassa E., Kallinos A., Jardine J., Rusilowicz-Jones E.V., Clague M.J., Urbe S. New aspects of USP30 biology in the regulation of pexophagy. Autophagy. 2019;15:1634–1637. doi: 10.1080/15548627.2019.1615304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gersch M., Gladkova C., Schubert A.F., Michel M.A., Maslen S., Komander D. Mechanism and regulation of the Lys6-selective deubiquitinase USP30. Nat. Struct. Mol. Biol. 2017;24:920–930. doi: 10.1038/nsmb.3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sato Y., Okatsu K., Saeki Y., Yamano K., Matsuda N., Kaiho A., et al. Structural basis for specific cleavage of Lys6-linked polyubiquitin chains by USP30. Nat. Struct. Mol. Biol. 2017;24:911–919. doi: 10.1038/nsmb.3469. [DOI] [PubMed] [Google Scholar]
- 13.Pickrell A.M., Youle R.J. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson's disease. Neuron. 2015;85:257–273. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agarwal S., Muqit M.M.K. PTEN-induced kinase 1 (PINK1) and Parkin: unlocking a mitochondrial quality control pathway linked to Parkinson's disease. Curr. Opin. Neurobiol. 2022;72:111–119. doi: 10.1016/j.conb.2021.09.005. [DOI] [PubMed] [Google Scholar]
- 15.Kitada T., Asakawa S., Hattori N., Matsumine H., Yamamura Y., Minoshima S., et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 16.Valente E.M., Abou-Sleiman P.M., Caputo V., Muqit M.M., Harvey K., Gispert S., et al. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 17.Yue W., Chen Z., Liu H., Yan C., Chen M., Feng D., et al. A small natural molecule promotes mitochondrial fusion through inhibition of the deubiquitinase USP30. Cell Res. 2014;24:482–496. doi: 10.1038/cr.2014.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rusilowicz-Jones E.V., Jardine J., Kallinos A., Pinto-Fernandez A., Guenther F., Giurrandino M., et al. USP30 sets a trigger threshold for PINK1-PARKIN amplification of mitochondrial ubiquitylation. Life Sci. Alliance. 2020;3 doi: 10.26508/lsa.202000768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang F., Gao Y., Zhou L., Chen J., Xie Z., Ye Z., et al. USP30: structure, emerging physiological role, and target inhibition. Front. Pharmacol. 2022;13 doi: 10.3389/fphar.2022.851654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kluge A.F., Lagu B.R., Maiti P., Jaleel M., Webb M., Malhotra J., et al. Novel highly selective inhibitors of ubiquitin specific protease 30 (USP30) accelerate mitophagy. Bioorg. Med. Chem. Lett. 2018;28:2655–2659. doi: 10.1016/j.bmcl.2018.05.013. [DOI] [PubMed] [Google Scholar]
- 21.Rusilowicz-Jones E.V., Barone F.G., Lopes F.M., Stephen E., Mortiboys H., Urbe S., et al. Benchmarking a highly selective USP30 inhibitor for enhancement of mitophagy and pexophagy. Life Sci. Alliance. 2022;5 doi: 10.26508/lsa.202101287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Engen J.R., Wales T.E. Analytical aspects of hydrogen exchange mass spectrometry. Annu. Rev. Anal. Chem. (Palo Alto Calif.) 2015;8:127–148. doi: 10.1146/annurev-anchem-062011-143113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marciano D.P., Dharmarajan V., Griffin P.R. HDX-MS guided drug discovery: small molecules and biopharmaceuticals. Curr. Opin. Struct. Biol. 2014;28:105–111. doi: 10.1016/j.sbi.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borodovsky A., Ovaa H., Kolli N., Gan-Erdene T., Wilkinson K.D., Ploegh H.L., et al. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem. Biol. 2002;9:1149–1159. doi: 10.1016/s1074-5521(02)00248-x. [DOI] [PubMed] [Google Scholar]
- 25.Jones H.B.L., Heilig R., Fischer R., Kessler B.M., Pinto-Fernandez A. ABPP-HT - high-throughput activity-based profiling of deubiquitylating enzyme inhibitors in a cellular context. Front. Chem. 2021;9 doi: 10.3389/fchem.2021.640105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pinto-Fernandez A., Davis S., Schofield A.B., Scott H.C., Zhang P., Salah E., et al. Comprehensive landscape of active deubiquitinating enzymes profiled by advanced chemoproteomics. Front. Chem. 2019;7:592. doi: 10.3389/fchem.2019.00592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.HaileMariam M., Eguez R.V., Singh H., Bekele S., Ameni G., Pieper R., et al. S-trap, an ultrafast sample-preparation approach for shotgun proteomics. J. Proteome Res. 2018;17:2917–2924. doi: 10.1021/acs.jproteome.8b00505. [DOI] [PubMed] [Google Scholar]
- 28.Muntel J., Kirkpatrick J., Bruderer R., Huang T., Vitek O., Ori A., et al. Comparison of protein quantification in a complex background by DIA and TMT workflows with fixed instrument time. J. Proteome Res. 2019;18:1340–1351. doi: 10.1021/acs.jproteome.8b00898. [DOI] [PubMed] [Google Scholar]
- 29.Demichev V., Messner C.B., Vernardis S.I., Lilley K.S., Ralser M. DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods. 2020;17:41–44. doi: 10.1038/s41592-019-0638-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerken P.A., Wolstenhulme J.R., Tumber A., Hatch S.B., Zhang Y., Muller S., et al. Discovery of a highly selective cell-active inhibitor of the histone lysine demethylases KDM2/7. Angew. Chem. Int. Ed. Engl. 2017;56:15555–15559. doi: 10.1002/anie.201706788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown K.A., Wilson D.J. Bottom-up hydrogen deuterium exchange mass spectrometry: data analysis and interpretation. Analyst. 2017;142:2874–2886. doi: 10.1039/c7an00662d. [DOI] [PubMed] [Google Scholar]
- 32.O'Brien D.P., Durand D., Voegele A., Hourdel V., Davi M., Chamot-Rooke J., et al. Calmodulin fishing with a structurally disordered bait triggers CyaA catalysis. PLoS Biol. 2017;15 doi: 10.1371/journal.pbio.2004486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hourdel V., Volant S., O'Brien D.P., Chenal A., Chamot-Rooke J., Dillies M.A., et al. MEMHDX: an interactive tool to expedite the statistical validation and visualization of large HDX-MS datasets. Bioinformatics. 2016;32:3413–3419. doi: 10.1093/bioinformatics/btw420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valdes-Tresanco M.S., Valdes-Tresanco M.E., Valiente P.A., Moreno E. AMDock: a versatile graphical tool for assisting molecular docking with Autodock Vina and Autodock4. Biol. Direct. 2020;15:12. doi: 10.1186/s13062-020-00267-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones H.B.L., Heilig R., Davis S., Fischer R., Kessler B.M., Pinto-Fernandez A. ABPP-HT∗-deep meets fast for activity-based profiling of deubiquitylating enzymes using advanced DIA mass spectrometry methods. Int. J. Mol. Sci. 2022;23:3263. doi: 10.3390/ijms23063263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Komander D. Mechanism, specificity and structure of the deubiquitinases. Subcell. Biochem. 2010;54:69–87. doi: 10.1007/978-1-4419-6676-6_6. [DOI] [PubMed] [Google Scholar]
- 38.Clague M.J., Urbe S., Komander D. Breaking the chains: deubiquitylating enzyme specificity begets function. Nat. Rev. Mol. Cell Biol. 2019;20:338–352. doi: 10.1038/s41580-019-0099-1. [DOI] [PubMed] [Google Scholar]
- 39.Cunningham C.N., Baughman J.M., Phu L., Tea J.S., Yu C., Coons M., et al. USP30 and parkin homeostatically regulate atypical ubiquitin chains on mitochondria. Nat. Cell Biol. 2015;17:160–169. doi: 10.1038/ncb3097. [DOI] [PubMed] [Google Scholar]
- 40.Turnbull A.P., Ioannidis S., Krajewski W.W., Pinto-Fernandez A., Heride C., Martin A.C.L., et al. Molecular basis of USP7 inhibition by selective small-molecule inhibitors. Nature. 2017;550:481–486. doi: 10.1038/nature24451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kategaya L., Di Lello P., Rouge L., Pastor R., Clark K.R., Drummond J., et al. USP7 small-molecule inhibitors interfere with ubiquitin binding. Nature. 2017;550:534–538. doi: 10.1038/nature24006. [DOI] [PubMed] [Google Scholar]
- 42.Honsho M., Okumoto K., Tamura S., Fujiki Y. Peroxisome biogenesis disorders. Adv. Exp. Med. Biol. 2020;1299:45–54. doi: 10.1007/978-3-030-60204-8_4. [DOI] [PubMed] [Google Scholar]
- 43.Schmidt M.F., Gan Z.Y., Komander D., Dewson G. Ubiquitin signalling in neurodegeneration: mechanisms and therapeutic opportunities. Cell Death Differ. 2021;28:570–590. doi: 10.1038/s41418-020-00706-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pan W., Wang Y., Bai X., Yin Y., Dai L., Zhou H., et al. Deubiquitinating enzyme USP30 negatively regulates mitophagy and accelerates myocardial cell senescence through antagonism of Parkin. Cell Death Discov. 2021;7:187. doi: 10.1038/s41420-021-00546-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bingol B., Tea J.S., Phu L., Reichelt M., Bakalarski C.E., Song Q., et al. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature. 2014;510:370–375. doi: 10.1038/nature13418. [DOI] [PubMed] [Google Scholar]
- 46.Liang J.R., Martinez A., Lane J.D., Mayor U., Clague M.J., Urbe S. USP30 deubiquitylates mitochondrial Parkin substrates and restricts apoptotic cell death. EMBO Rep. 2015;16:618–627. doi: 10.15252/embr.201439820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Riccio V., Demers N., Hua R., Vissa M., Cheng D.T., Strilchuk A.W., et al. Deubiquitinating enzyme USP30 maintains basal peroxisome abundance by regulating pexophagy. J. Cell Biol. 2019;218:798–807. doi: 10.1083/jcb.201804172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harrigan J.A., Jacq X., Martin N.M., Jackson S.P. Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat. Rev. Drug Discov. 2018;17:57–78. doi: 10.1038/nrd.2017.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krippendorff B.F., Neuhaus R., Lienau P., Reichel A., Huisinga W. Mechanism-based inhibition: deriving K(I) and k(inact) directly from time-dependent IC(50) values. J. Biomol. Screen. 2009;14:913–923. doi: 10.1177/1087057109336751. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The ABPP proteomics data files have been deposited to the ProteomeXchange Consortium and can be located under the PRIDE dataset identifier PXD036574. Annotated spectra ABPP data can be viewed following deposition to PanoramaWeb, which can be accessed using the ProteomeXchange ID code of PXD042782 and permanent link https://panoramaweb.org/LDvSvO.url. Similarly, the HDX-MS dataset can be located using the code PXD041582. PDB ID code 5OHK represents the crystal structure of human USP30 catalytic domain (residues K64–V502) in covalent complex with Ub–PA at 2.34 Å resolution. This is the highest resolution structure of human USP30 currently available in the PDB.