Abstract
Amoebae of the genus Acanthamoeba can cause diseases such as amoebic keratitis and granulomatous amoebic encephalitis. Until now, treatment options for these diseases have not been fully effective and have several drawbacks. Therefore, research into new drugs is needed for more effective treatment of Acanthamoeba infections. Eugenol, a phenolic aromatic compound mainly derived from cloves, has a variety of pharmaceutical properties. In this study, nine eugenol derivatives (K1-K9), consisting of five new and four known compounds, were synthesized and screened for their antiamoebic properties against Acanthamoeba sp. The structure of these compounds was characterized spectroscopically by Fourier transform infrared (FTIR), Ultraviolet–Visible (UV–Vis), 1H and 13C Nuclear Magnetic Resonance (NMR) and mass spectrometer (MS). The derived molecules were screened for antiamoebic activity by determining IC50 values based on 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay and observation of amoeba morphological changes by light and fluorescence microscopy. Most of the tested compounds possessed strong to moderate cytotoxic effects against trophozoite cells with IC50 values ranging from 0.61 to 24.83 μg/mL. Observation of amoebae morphology by light microscopy showed that the compounds caused the transformed cells to be roundish and reduced in size. Furthermore, fluorescence microscopy observation using acridine orange (AO) and propidium iodide (PI) (AO/PI) staining showed that the cells have damaged membranes by displaying a green cytoplasm with orange-stained lysosomes. Acidification of the lysosomal structure indicated disruption of the internal structure of Acanthamoeba cells when treated with eugenol derivatives. The observed biological results were also confirmed by interaction simulations based on molecular docking between eugenol derivatives and Acanthamoeba profilin. These interactions could affect the actin-binding ability of the protein, disrupting the shape and mobility of Acanthamoeba. The overall results of this study demonstrate that eugenol derivatives can be considered as potential drugs against infections caused by Acanthamoeba.
Keywords: Synthesis, Anti-amoebic, Cytotoxicity, Eugenol derivatives, MTT assay
1. Introduction
The genus Acanthamoeba consists of a free-living amoeba (FLA). These unicellular protozoa were first discovered in 1930 by Castellani (Castellani, 1930). He reported their existence in a culture of Cryptococcus pararoseus yeast, and they were later named Acanthamoeba castellanii. Acanthamoeba can commonly be found in freshwater, dust and soil. In addition, they also possess the ability to live in diverse environments where they are isolated from mud, sediments, sewage, swimming pools, bottled mineral water and even lens cleansing solutions (Niyyati et al., 2009). Throughout their life cycles, Acanthamoeba can exist in two distinctive forms; motile trophozoite and resistant cyst (Anacarso et al., 2017).
Member of the Acanthamoeba is known to be facultative pathogens because they can cause disease when they come into contact with a susceptible host. Acanthamoeba keratitis (AK) and granulomatous amoebic encephalitis (GAE) are two severe human diseases caused by some species of Acanthamoeba. AK is a vision-threatening infection of the cornea, and it mostly affects contact lens wearers. This is mainly due to continued wearing of contact lenses, non-sterile contact lens rinsing and poor contact lens hygiene (Khan, 2006). Without proper treatment, AK can develop into blindness. GAE is a serious brain and spinal cord infection, occurring most often in individuals with weak immune systems (Lackner et al., 2010). GAE also affects those with existing underlying diseases, such as lupus erythematosus (Cha et al., 2006, Shirwadkar et al., 2006), diabetes (Mayer et al., 2011), HIV (Memari et al., 2017, Pietrucha-Dilanchian et al., 2012) and pneumonitis (Westmoreland et al., 2004). In recent years, AK cases have been more common compared to other complications from Acanthamoeba sp. due to the rising number of contact lens users (Marciano-Cabral and Cabral, 2003). In contrast with AK, GAE is a rare infection but almost all cases lead to death. Besides, Acanthamoeba can cause other diseases, for instance cutaneous ulcers, abscesses, and rhinosinusitis (Lorenzo-Morales et al., 2010).
To date, there have been no known drugs that are safe and effective to treat these diseases caused by the Acanthamoeba. Most drugs show human toxicity and cause unpleasant side effects such as nausea, dizziness and hypertension (Siddiqui et al., 2016). Moreover, diagnosis of these infections is more challenging as a result of the available treatments being lengthy and not fully effective against all strains (Lorenzo-Morales et al., 2008, Lorenzo-Morales et al., 2013). As such, the current treatments are rife with many problems, thus emphasizing a need to develop alternative antiamoebic agents.
Currently, compounds derived from natural products are receiving a growing interest, as possible antiamoebic agents. They have the potential to replace the existing drugs due to their interesting physicochemical features that show outstanding effectiveness in treating and preventing diseases caused by Acanthamoeba. Moreover, many natural products are more friendly to the environment. Generallly, natural products such as essential oils are generally recognized by the Food and Drug Administration as safe substances (Ait-Ouazzou et al., 2011). In this respect, natural products may be used to formulate drugs that will be more effective in treating Acanthamoeba-caused infection while minimizing unpleasant side effects.
One compound that has attracted the attention of many researchers is eugenol. Eugenol is a natural phenolic compound found in the essential oil of cloves. This compound has a chemically versatile structure and has a wide range of applications (Kaufman, 2015). Eugenol and its derived molecules have been used in medicinal practice for antibacterial (Devi et al., 2010, Teles et al., 2021, Bai et al., 2022), antifungal (de Souza et al., 2014, Ju et al., 2020, Zhao et al., 2021), anti-inflammatory (Daniel et al., 2009, Maurya et al., 2020, Anjum et al., 2022), antitumoral (Cui et al., 2019, Vladu et al., 2020), antiparasitic (Machado et al., 2011, ElGhannam et al., 2023) and antiviral properties (Lane et al., 2019, Gan et al., 2021). Despite its wide use, it is necessary to develop a better understanding of its mode on biological action for new applications in human health. For a while now, eugenol has received special attention for its antiamoebic activity, suggesting that it may be a promising alternative for treating infections brought on by Acanthamoeba (Anacarso et al., 2017). However, eugenol-derived molecules have not been fully explored in terms of their antiamoebic potential.
In our earlier research, the distinctive qualities of eugenol were exploited and optimized by synthetic means in order to take full advantage of their structural, chemical and biological diversity with regard to their therapeutic effects. (Rahim et al., 2017, Zamli et al., 2020, Zamli et al., 2021). Herein, we are interested to study the potential of eugenol derivatives as antiamoebic agents. To achieve this aim, we synthesized eugenol derivatives (K1-K9) by structurally modifying eugenol with different alkyl and acyl halides to improve their lipophilic character, which could enhance their antiamoebic activity. We then characterized and evaluated the synthesized eugenol derivatives against Acanthamoeba sp by investigating their IC50, changes in cellular morphology, as well as membrane integrity. Lastly, molecular docking simulation was performed to determine the potential biological target of the eugenol derivatives.
2. Experimental
2.1. Materials and methods
All chemicals were purchased from Merck, R&M, and Sigma-Aldrich and were used without purification unless otherwise stated. The reactions for the synthesis of derivatives were carried out in round bottom flasks. Thin-layer chromatography (TLC) coated with silica gel 60 F254 plastic-backed plates was used to monitor the progress of reactions. The spots were visualized under ultraviolet light of Shimadzu UV-1601PC at the wavelength 253 to 185 nm. The purification method by column chromatography was performed on a silica gel 60 column (230–400 mesh, Merck). The infrared (IR) spectra (KBr discs) were recorded in the 4000 to 400 cm−1 range on a Shimadzu IRTracer-100 Fourier Transform Infrared (FTIR) Spectrometer using single reflection ATR. The Ultraviolet–Visible (UV–Vis) spectrum of the derivatives was recorded on a Shimadzu UV-1601Pc Spectrophotometer using a 1 cm path length cuvette containing a solution of the synthesized compounds in methanol. The UV region scan was done in the range of 200 to 400 nm. The Nuclear Magnetic Resonance (NMR) spectra of 1H (400 MHz) and 13C (100 MHz) NMR were recorded on a Bruker Ultrashield 400 MHz spectrometer with deuterated chloroform (CDCl3) as a solvent, which was calibrated as internal standard at room temperature. The data were expressed as chemical shift values given in part per million (ppm) at a range between δH 0–9 ppm and δC 0–200 ppm. The mass spectrometers were recorded on LCMS Orbitrap Discovery (Thermo Scientific). The ionizing source used was electrospray ionization (ESI) and the samples were dissolved in methanol. For the biological investigations, IC50 values were recorded by an ELISA Microplate Reader (DynaTech, New York, USA) at the wavelength of 570 nm. The morphology of cells was observed under an inverted light microscope and fluorescence microscope (Olympus IX70, Louisiana, USA).
2.2. Chemistry
2.2.1. General procedure for preparation of compound K1
Compound K1 was prepared according to a published procedure (Rahim et al., 2017). Eugenol (1 equiv) was treated with potassium carbonate (1 equiv) and 4-isopropylbenzyl chloride (1.5 equiv) in anhydrous acetone (20 mL) under an inert condition. The mixture was refluxed at 60 °C with constant stirring for 24 h upon the adjustment of completion by TLC. After the consumption of the starting material, the reaction medium was diluted with 20 mL of distilled water and later extracted with dichloromethane (3 × 20 mL). Then, the residue was purified by using column chromatography (n-hexane: ethyl acetate) to produce K1 as a pure compound.
2.2.2. General procedure for preparation of compounds K2-K9
The synthesis of compounds K2-K9 was carried out following our previously reported protocol (Rahim et al., 2017, Zamli et al., 2021). Eugenol (1 equiv) was dissolved in anhydrous dichloromethane (20 mL) before trimethylamine (2.5 mL) was added. The mixture was magnetically stirred for 30 min at 0–5 °C. Then, an excess of acyl chloride (2 equiv) was slowly added. The reaction mixture was continued stirring for another 30 min at 0–5 °C before being allowed to reach room temperature and stirred for another 24 h. Monitoring of the reaction completion was done by TLC. Later, the mixture was diluted with distilled water and extracted with dichloromethane (3 × 20 mL). The organic layer was dried (MgSO4) and the solvent volume was removed under vacuum to give a crude product, which was purified via column chromatography. Hexane/ethyl acetate (3:1) was used as an eluent to afford the title product (K2-K9).
2.2.3. Characterization of eugenol derivatives
4-allyl-2-methoxyphenyl-4-isopropylbenzoate (K1) (Rahim et al., 2017): 69%; ATR 3074, 2958, 2873, 1639, 1508, 1226, 1029 cm−1; UV–Vis (MeOH): λmax = 220 nm, 279 nm; 1H NMR (400 MHz, CDCl3) δ 1.16 (d, J = 6.8 Hz, 6H, (CH3)2), 2.81 (sept, 1H, CH), 3.24 (d, J = 6.8 Hz, 2H, CH2), 3.78 (s, 3H, OCH3), 4.96–5.02 (m, 2H, CH = CH2), 5.00 (s, 2H, CH2), 5.82–5.92 (m, 1H, CH = CH2), 6.58 (dd, J = 8.0 Hz, J = 2.0 Hz, 1H, CHar), 6.65 (d, J = 2.0 Hz, 1H, CHar), 6.75 (d, J = 8.4 Hz, 1H, CHar), 7.13 (d, J = 8.0 Hz, 2H, CHar), 7.28 (d, J = 8.0 Hz, 2H, CHar) ppm; 13C NMR (100 MHz, CDCl3) δ 24.0 (2xCH3), 33.9 (CH), 39.8, (CH2), 55.9 (OCH3), 71.1 (CH2), 112.4 (CH ar), 114.1 (CH ar), 115.6 ( CH2), 120.4 (CH ar), 126.6 (CH ar), 127.5 (CH ar), 133.1 (Ar-C), 134.7 (Ar-C), 137.6 (CH ), 146.7 (Ar-C), 148.5 (Ar-C), 149.6 (Ar-C) ppm; Found (ES) [M−H]+ 295.17798 C20H23O2, requires 295.39542.
4-Allyl-2-methoxyphenyl-4-chlorobenzoate (K2) (Zamli et al., 2021): 90% yield); ATR 3078, 2931, 1735, 1597, 1504, 1257, 1064, 1010, 748 cm−1; UV–Vis (MeOH): λmax = 246 nm, 280 nm; 1H NMR (400 MHz, CDCl3) δ 3.43 (d, J = 6.4 Hz, 2H, CH2), 3.82 (s, 3H, OCH3), 5.11–5.18 (m, Hz, 2H, CH = CH2), 5.96–6.06 (m, 1H, CH CH2), 6.84 (dd, J = 8.0, J = 2.0 Hz, 1H, CHar), 6.86 (d, J = 1.6 Hz, 1H, CHar), 7.08 (d, J = 7.8 Hz, 1H, CHar), 7.50 (d, J = 8.8 Hz, 2H, CHar), 8.17 (d, J = 8.8 Hz, 2H, CHar) ppm; 13C NMR (100 MHz, CDCl3) δ 40.1 (CH2), 55.8 (OCH3), 112.8 (CH ar), 116.2 ( CH2), 120.7 (CH ar), 122.5 (CH ar), 127.9 (CH ar), 128.8 (CH ar), 131.7 (CH ar), 137.0 (CH ar),138.0 (HC ), 139.2 (Ar-C), 139.9 (Ar-C), 150.9 (Ar-C), 164.0 (C O) ppm; Found (ES) [M]+ 302.69186 C17H15ClO3, requires 302.75220.
4-Allyl-2-methoxyphenyl-4-methylbenzoate (K3) (Zamli et al., 2021): 81%; ATR 3070, 2931, 1724, 1604, 1508, 1261, 1033 cm−1; UV–Vis (MeOH): λmax = 240 nm, 273 nm 1H NMR (400 MHz, CDCl3) δ 2.47 (s, 3H, CH3), 3.44 (d, J = 6.8 Hz, 2H, CH2), 3.82 (s, 3H, OCH3), 5.12–5.18 (m, 2H, CH CH2), 5.97–6.07 (m, 1H, CH CH2), 6.84 (dd, J = 8.0, 2.0 Hz, 1H, CHar), 6.86 (d, J = 1.6 Hz, 1H, CHar), 7.09 (d, J = 7.6 Hz, 1H, CHar), 7.33 (d, J = 8.0 Hz, 2H, CHar), 8.13 (d, J = 8.4 Hz, 2H, CHar) ppm; 13C NMR (100 MHz, CDCl3) δ 21.7 (CH3), 40.1 (CH2), 55.9 (OCH3), 112.8 (CH ar), 116.1 ( CH2), 120.7 (CH ar), 122.7 (CH ar), 126.7 (Ar-C), 129.2 (CH ar), 130.3 (CH ar), 137.1 (Ar-C), 138.2 (HC ), 138.9 (Ar-C), 144.2 (Ar-C), 151.1 (Ar-C), 164.9 (C O) ppm; Found (ES) [M−H]+ 281.86893 C18H17O3, requires 281.32578.
4-allyl-2-methoxyphenyl-3,4-dichlorobenzoate (K4) Yield: 85%; ATR 3078, 2970, 1735, 1604, 1504, 1265, 1083, 1033, 748 cm−1; UV–Vis (MeOH): λmax = 243 nm, 274 nm, 280 nm; 1H NMR (400 MHz, CDCl3) δ 3.43 (d, J = 6.4 Hz, 2H, CH2), 3.82 (s, 3H, OCH3), 5.12–5.18 (m, 2H, CH CH2), 5.96–6.06 (m, 1H, CH = CH2), 6.84 (dd, J = 8.0 Hz, 1.6 Hz, 1H, CHar), 6.86 (d, J = 1.6 Hz, 1H, CHar), 7.07 (d, J = 8.0 Hz, 1H, CHar), 7.61 (d, J = 8.4 Hz, 1H, CHar), 8.05 (dd, J = 8.4 Hz, 2.0 Hz, 1H, CHar), 8.31 (d, J = 2 Hz, 1H, CHar) ppm; 13C NMR (100 MHz, CDCl3) δ 40.1 (CH2), 55.8 (OCH3), 112.8 (CH ar), 116.2 ( CH2), 120.7 (CH ar), 122.4 (CH ar), 129.3 (Ar-C), 129.4 (CH ar), 130.6 (CH ar), 132.1 (CH ar), 133.1 (Ar-C), 136.9 (Ar-C), 137.8 (HC ), 138.1 (Ar-C), 150.8 (Ar-C), 139.4 (Ar-C), 163.0 (C O) ppm; Found (ES) [M−H]+ 337.01820 C17H14Cl2O3, requires 337.19726.
4-Allyl-2-methoxyphenyl-3-methoxybenzoate (K5) Yield: 76%; ATR 3070, 2978, 1728, 1589, 1504, 1280, 1033 cm−1; UV–Vis (MeOH): λmax = 234 nm, 275 nm, 304 nm; 1H NMR (400 MHz, CDCl3) δ 3.44 (d, J = 6.8 Hz, 2H, CH2), 3.83 (s, 3H, OCH3), 3.90 (s, 3H, OCH3), 5.12–5.18 (m, 2H, CH CH2), 5.96–6.07 (m, 1H, CH = CH2), 6.84 (dd, J = 8.0 Hz, 1.6 Hz 1H, CHar), 6.86 (d, J = 1.6 Hz, 1H, CHar), 7.09 (d, J = 8.0 Hz, 1H, CHar), 7.19 (ddd, J = 8.0 Hz, 2.8 Hz, 0.8 Hz, 1H, CHar), 7.43 (t, J 8.0 Hz, 1H, CHar), 7.74 (dd, J = 2.8 Hz, 1.6 Hz, 1H, CHar), 7.85 (dt, J = 7.6 Hz, 1.2 Hz, 1H, CHar) ppm; 13C NMR (100 MHz, CDCl3) δ 40.1 (CH2), 55.5 (OCH3), 55.9 (OCH3), 112.8 (CH ar), 114.5 (CH ar), 116.1 ( CH2), 120.1 (CH ar), 120.7 (CH ar), 122.6 (CH ar), 122.7 (CH ar), 129.5 (CH ar), 130.8 (Ar-C), 137.1 (Ar-C), 138.2 (C C), 139.0 (Ar-C), 151.1 (Ar-C), 159.6 (Ar-C), 164.7 (C O) ppm; Found (ES) [M−H]+ 297.12039 C18H17O4, requires 297.32518.
4-allyl-2-methoxyphenyl benzo[d](Abid and Azam, 2006, Anjum et al., 2022)dioxole-5-carboxylate (K6) Yield: 84%; ATR 3062, 2970, 2908, 1735, 1597, 1504, 1265, 1033 cm−1; UV–Vis (MeOH): λmax = 223 nm, 268 nm, 303 nm; 1H NMR (400 MHz, CDCl3) δ 3.22 (d, J = 6.4 Hz, 2H, CH2), 3.72 (s, 3H, OCH3), 5.01–5.07 (m, 2H, CH CH2), 5.85–5.99 (m, 1H, CH CH2), 5.91 (s, 2H, CH2), 6.72 (dd, J = 8.0 Hz, 2 Hz, 1H, CHar), 6.75 (d, J = 1.6 Hz, 1H, CHar), 6.82 (d, J = 8.4 Hz, 1H, CHar), 6.97 (d, J = 8.0 Hz, 1H, CHar), 7.55 (d, J = 1.6 Hz, 1H, CHar), 7.75 (dd, J = 8.4 Hz, 1.6 Hz, 1H, CHar) ppm; 13C NMR (100 MHz, CDCl3) δ 40.1 (CH2), 55.9 (OCH3),101.9 (CH2), 108.1 (CH ar), 110.1 (CH ar), 112.8 (CH ar), 116.1 ( CH2), 120.7 (CH ar), 122.6 (CH ar), 123.4 (Ar-C), 126.3 (CH ar), 137.1 (Ar-C), 138.2 (HC ), 138.9 (Ar-C), 147.8 (Ar-C), 151.1 (Ar-C), 152.0 (Ar-C), 164.2 (C O) ppm; Found (ES) [M+H]+ 313.03870 C18H17O5 requires, 313.32458.
4-allyl-2-methoxyphenyl-3,5-dinitrobenzoate (K7) Yield: 50%; ATR 3086, 2931, 1743, 1627, 1543, 1504, 1342, 1265, 1033 cm−1; UV–Vis (MeOH): λmax = 226 nm, 270 nm; 1H NMR (400 MHz, CDCl3) δ 3.45 (d, J = 6.8 Hz, 2H, CH2), 3.84 (s, 3H, OCH3), 5.14–5.19 (m, 2H, CH CH2), 5.96–6.06 (m, 1H, CH CH2), 6.87 (d, J = 8.0 Hz, 1H, CHar), 6.89 (s, 1H, CHar), 7.12 (d, J = 8.0 Hz, 1H, CHar), 9.30 (s, 1H, CHar), 9.35 (s, 1H, CHar) ppm; 13C NMR (100 MHz, CDCl3) δ 40.1 (CH2), 55.8 (OCH3), 112.8 (CH ar), 116.5 ( CH2), 120.8 (CH ar), 122.0 (CH ar), 122.7 (CH ar), 130.0 (CH ar), 133.3 (Ar-C), 136.7 (Ar-C),137.3 (HC ), 140.1 (Ar-C), 150.5 (Ar-C), 148.7 (Ar-C), 160.8(C O) ppm; Found (ES) [M+H]+ 359.08914 C17H15N2O7, requires 359.31020.
4-allyl-2-methoxyphenyl-3-nitrobenzoate (K8) Yield: 86%; ATR 3093, 2916, 1739, 1612, 1527, 1508, 1346, 1288, 1056 cm−1; UV–Vis (MeOH): λmax = 221 nm, 270 nm 1H NMR (400 MHz, CDCl3) δ 3.44 (d, J = 6.8 Hz, 2H), 3.83 (s, 3H, OCH3), 5.12–5.19 (m, 2H, CH CH2), 5.98–6.04 (m, 1H, CH CH2), 6.86 (dd, J = 8.0 Hz, 2.0 Hz, 1H, CHar), 6.88 (d, J = 1.6 Hz, 1H, CHar), 7.10 (d, J = 8.0 Hz, 1H, CHar), 7.74 (t, J = 8.0 Hz, H, CHar), 8.50 (ddd, J = 8.0 Hz, 2.4 Hz, 1.2 Hz, H, CHar), 8.55 (dt, J = 7.6 Hz, 1.6 Hz, H, CHar), 9.07 (t, J = 2.0 Hz, H, CHar) ppm; 13C NMR (100 MHz, CDCl3) δ 40.1 (CH2), 55.8 (OCH3), 112.8 (CH ar), 116.3 ( CH2), 120.8 (CH ar), 122.3 (CH ar), 125.2 (CH ar), 127.8 (CH ar), 129.7 (CH ar), 131.3 (Ar-C), 135.9 (CH ar), 136.9 (Ar-C), 137.7 (HC ), 139.6 (Ar-C), 148.3 (Ar-C), 150.8 (Ar-C), 162.8 (C O) ppm; Found (ES) [M+H]+ 314.10376 C17H16NO5, requires 314.31264.
4-allyl-2-methoxyphenyl-4-butylbenzoate (K9) Yield: 79%; ATR 3062, 2931, 2862, 1735, 1604, 1512, 1265, 1064 cm−1; UV–Vis (MeOH): λmax = 241 nm, 277 nm 1H NMR (400 MHz, CDCl3) δ 0.97 (t, J = 7.2 Hz, 3H, CH3), 1.40 (sext, J = 7.6 Hz, 2H, CH2), 1.66 (quin, J = 7.6 Hz, 2H, CH2), 2.72 (t, J = 7.6 Hz, 2H, CH2), 3.43 (d, J = 6.8 Hz, 2H, CH2), 3.82 (s, 3H, OCH3), 5.11–5.18 (m, 2H, CH CH2), 5.96–6.06 (m, 1H, CH CH2), 6.83 (dd, J = 8.0 Hz, 2.0 Hz, 1H, CHar), 6.85 (d, J = 1.6 Hz, 1H, CHar), 7.08 (d, J = 8.0 Hz, 1H, CHar), 7.33 (d, J = 8.0 Hz, 2H, CHar), 8.14 (d, J = 8.4 Hz, 2H, CHar) ppm; 13C NMR (100 MHz, CDCl3) δ 13.9 (CH3), 22.3 (CH2), 33.3 (CH2), 35.8 (CH2), 40.1 (CH2), 55.9 (OCH3), 112.8 (CH ar), 116.1 ( CH2), 120.7 (CH ar), 122.7 (CH ar), 126.9 (Ar-C), 128.5 (CH ar), 130.3 (CH ar), 137.1 (Ar-C), 138.2 (HC ), 138.9 (Ar-C), 149.1 (Ar-C), 151.1 (Ar-C), 164.9 (C O) ppm; Found (ES) [M−H]+ 323.16705 C21H23O3, requires 323.40552.
2.3. Antiamoebic activity
The determination of antiamoebic activity of the synthesised eugenol derivatives was done on Acanthamoeba sp. following the method of Kusrini et al (2018). The 50% of inhibition concentration (IC50) of tested compounds was obtained by determining the nonlinear sigmoidal dose–response curve graph with various concentrations of eugenol derivatives compounds ranging from 0.47 to 30 μg/mL. Morphological and biochemical changes in Acanthamoeba cells after exposure to eugenol derivatives were also observed (Nakisah et al., 2012).
2.3.1. Media preparation
2.3.1.1. Page's amoeba saline (PAS) solution
Page's Amoeba Saline (PAS) solution was prepared by mixing of 5 mL of stock 1 (12.0 g of NaCl, 0.4 g of MgSO4·7H2O, 500 mL of distilled water) and 5 mL of stock 2 (14.0 g of Na2HPO4, 13.6 g of KH2PO4, 500 mL of distilled water). Afterward, distilled water was added until the volume reached 1000 mL.
2.3.1.2. Protease yeast glucose (PYG) media preparation
Protease yeast glucose (PYG) media was prepared by mixing 3.75 g of protease, 3.75 g of yeast extract and 7.5 g of D+ glucose in 5 mL of PAS solution. Distilled water was added to make up to 500 mL. Then, the media was autoclaved and stored at a low temperature of 4 °C prior to use.
2.3.1.3. Cell culture and maintenance
The Acanthamoeba sp., isolated from the corneal scrapings of a keratitis patient (supplied by Hospital Kuala Lumpur) was cultured in a T-25 tissue culture flask which contained 10 mL of PYG media. It was kept in an incubator at 30 °C. The cell's condition was examined for any signs of contamination under an inverted microscope. The media-changing process was done routinely when the cells confluence in order to maintain healthy cell cultures.
2.3.2. Sample stock solution
Eugenol derivatives stock solutions were prepared by dissolving 1 mg of each derivative in 40 μl of absolute DMSO, followed by adding 960 μl of sterile PYG media to make a 1 mg/mL solutions. Dissolution was facilitated by a vortex and the stock solutions were kept at 4 °C before use. Seven different concentrations of the samples were prepared from each stock solution through serial dilution to give the following final concentration of compounds: 30, 15, 7.50, 3.75, 1.88, 0.94, and 0.47 μg/mL. The concentration of DMSO in the most concentrated sample in treatment wells was 0.04% (w/v) and did not exceed 0.25% (w/v), which is the recommended threshold value of DMSO for anti-amoebic treatment (Wright et al., 1988). Negative effects against studied amoeba will occur if the concentration of DMSO exceeds this value.
2.3.3. Determination of IC50 values by MTT assay
Experiments were carried out in 96-well plates at 30 °C under sterile conditions. Each well was seeded with a trophozoite suspension in three replicates (1 × 105 cells/well). After 24 h, the medium was removed after the cells have adhered to the bottom of the wells. Stock solutions of eugenol derivatives stock solution at concentrations ranging from 0.47 to 30 μg/mL were added to the wells and incubated for another 24 h. Then, the medium was removed and wells were washed with phosphate-buffered saline (PBS) to discard the remaining suspension. Next, 20 μl of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) solution (5 mg/mL) in PBS was added to each well and further incubated. After 4 h, 150 μl of DMSO was placed into the wells to dissolve the formed formazan crystal. The final solutions from all wells were read for their absorbance at 570 nm by an ELISA microplate reader. The readings obtained were plotted in GraphPad Prism software version 8.0.1 (GraphPad Inc.) to get a nonlinear sigmoidal dose–response curve. The cytotoxic activity was expressed as the IC50 value that corresponded to the concentration of the compounds which inhibited 50% of the Acanthamoeba population. The t-test was performed, where p < 0.005 was considered statistically significant.
2.3.4. Observation of Acanthamoeba morphological changes
Acanthamoeba (104 cells/mL) were treated with the eugenol-derived molecules at their IC50 concentration in 6-well plates. After 24 h incubation, the morphological changes of Acanthamoeba were evaluated and compared with those observed in control cultures. The negative control was healthy Acanthamoeba without any treatment while the positive control was chlorhexidine, a common agent used for treating AK infection. Observations were made directly from the culture flask under an inverted microscope (Leica Leitz, Wetzlar, Germany) which enabled the change in the acanthopodia structure as well as the shape and size of the cell to be viewed.
2.3.5. Evaluation of Acanthamoeba membrane integrity
Acridine orange (AO) and propidium iodide (PI) staining were used as fluorescence dyes to observe the integrity of the Acanthamoeba membrane after the cells (104 cells/mL) were treated with the compounds at their IC50 concentrations. The treatments including controls were carried out in 6-well plates at 30 °C for 24 h incubation time. Then, the medium was removed and wells were washed with PBS before undergoing a further AO/PI staining process. Stock solution for AO/PI was prepared by adding 2 μl AO (1 mg/ml) and 2 μl PI (1 mg/mL) in 996 μl PBS. The cell suspensions were then subjected to incubation for 15 min in the dark because the dyes were light-sensitive. Later, the Acanthamoeba cell suspensions were observed directly under an inverted fluorescence microscope (Leica Dmire, Wetzlar, Germany) in the dark.
2.3.6. Molecular docking simulation of eugenol derivatives to Acanthamoeba profilin (acPFN)
The atomic coordinates and the three-dimensional structure of Acanthamoeba castellanii profilin 1B (PDB ID: 1ACF), Acanthamoeba castellanii actin (PDB ID:4EFH) and Bos taurus profilin-beta-actin (PDB ID:2BTF) were retrieved from RCSB Protein Data Bank (https://www.rcsb.org/). To identify the actin binding site at the acPFN structure, molecular docking of 1ACF and 4EFH was performed using HADDOCK and superimposed with 2BTF. The RMSD of the superimposed structures was calculated. The SDF files of eugenol derivatives were converted into the mol2 format with 3D coordinates using Open Babel software (O’Boyle et al., 2011). To understand the intermolecular interaction between profilin and six eugenol derivatives, namely the 4-allyl-2-methoxyphenyl 4-isopropylbenzoate (K1), 4-allyl-2-methoxyphenyl 4-chlorobenzoate (K2), 4-allyl-2-methoxyphenyl 4-methylbenzoate (K3), 4-allyl-2-methoxyphenyl 3,4-dichlorobenzoate (K4), 4-allyl-2-methoxyphenyl 3-methoxybenzoate (K5) and 4-allyl-2-methoxyphenylbenzo[d] (Abid and Azam, 2006, Anjum et al., 2022) dioxole-5-carboxylate (KZ23), molecular docking simulations were performed using SwissDock server (Grosdidier et al., 2011). SwissDock employs VDW and electrostatic grids to determine the interaction energy between the protein and ligands, based on the EADock DSS (Dihedral Space Sampling) with CHARMM (Chemistry at HARvard Macromolecular Mechanics) forcefield (Kim et al., 2017). The predicted docking clusters were analyzed using UCSF-Chimera (Goddard et al., 2017, Razali and Shamsir, 2020). The 2D schematic diagram of the residues involved in the interactions was further analyzed using the LigPlot+ program (Laskowski and Swindells, 2011).
3. Result and discussion
3.1. Chemistry
Nine eugenol derived molecules were synthesized as outlined in Scheme 1, which comprised four reported (K1, K2, K3 and K5) and five new compounds (K4, K6, K7, K8 and K9). Compound K1 was prepared via the treatment of eugenol with 4-isopropylbenzyl chloride and potassium carbonate as according to a published method (Rahim et al., 2017). The compound yielded 69% of brownish-yellow solid. On the other hand, ester derivatives (K2-K9) were prepared by reacting the corresponding acyl halide with eugenol and triethylamine (Zamli et al., 2021). Compound K2-K9 was obtained in moderate to good percentage yield in the range of 50% to 86%.
Scheme 1.
Synthesis of eugenol derived molecules.
The structures of eugenol derivatives were established using spectral data from Fourier Transform Infrared Spectrometer (FTIR), 1H and 13C Nuclear Magnetic Resonance (NMR) and Mass Spectrometer (MS).
3.2. IC50 values by MTT assay
Various tests have been developed to study the proliferation and determination of the viability of Acanthamoeba sp. cells in vitro. MTT assay was used to measure the viability and proliferation as well as to determine the cytotoxic effect in Acanthamoeba cells since this organism also presented with mitochondria that produce the hydrogenase enzyme and are able to react with the MTT-producing formazan crystal (Kusrini et al., 2016). There is countless assays that have the same mechanisms as MTT, but most of them are only suitable for specific types of cells. MTT is the only assay that is suitable for any type of cell, including amoeba.
In this study, the optimization of MTT assay on Acanthamoeba sp. cells was performed in order to determine the number of viable cells that remained following the exposure to the eugenol derivatives. However, a slight modification to the MTT assay’s protocol was done, wherein each well of the 96-well plate was rinsed with PBS two times to ensure all the glucose content in the media was removed. This is due to MTT reagents being sensitive to glucose, which their presence in the medium can reduce the efficiency of the MTT reaction (Hashim et al., 2015). Then, various concentrations of eugenol-derived molecules ranging from 0.47 µg/mL to 30 µg/mL were exposed to Acanthamoeba trophozoite in order to determine the cytotoxicity potential of these synthesized molecules. The half maximal inhibitory concentration (IC50 value) for eugenol-derived molecules was acquired based on the absorbance reading by using the ELISA microplate reader at 570 nm. Simply put, the ELISA readings obtained were directly proportional to the number of viable cells present in the culture plate after 24 h of incubation time. Fig. 1 shows the curve derived from the GraphPad Prism software while the IC50 values are shown in Table 1.
Fig. 1.
Cytotoxicity curve of eugenol derived molecules after 24 h treatment, assessed based on cell viability percentage. The data represent as mean ± S.E.M from three replications.
Table 1.
The IC50 values of eugenol derivatives against Acanthamoeba sp.
| Compound | IC50 (μg/mL) |
|---|---|
| K1 | 24.83 ± 0.15 |
| K2 | 0.85 ± 0.16 |
| K3 | 19.57 ± 0.08 |
| K4 | 0.61 ± 0.30 |
| K5 | 6.10 ± 0.17 |
| K6 | 0.73 ± 0.26 |
| K7 | – |
| K8 | – |
| K9 | – |
-: not active up to concentration of 30 μg/mL. The values are expressed as mean ± S.E.M from triplicate. (t-test, p < 0.005). Reference drug: Chlorhexidine (IC50: 1.52 ± 0.45 μg/mL).
From the graph obtained, we found that EMD exhibited a wide range of antiamoebic activities with the IC50 values ranging from 0.61 to 24.83 ug/mL. The derivatives were synthesized with diverse modifications in the eugenol’s hydroxyl group, thus different derivatives comprised of different numbers of atoms and molecules. Modifications in the carbon chain nature (aliphatic or aromatic) causes stereo-electronic variations that help to determine the characteristics required to improve the activities of these molecules. A number of structure–activity relationships could be deduced from the results obtained. Among them, derivative K4 bearing the chloro moiety at the meta and para position on its phenyl ring, exhibited the most promising activity with an IC50 value of 0.61 mg/mL. Likewise, derivative K2 which also bears the chloro group, showed high activity with an IC50 value of 0.85 mg/mL. This structure-effect relationship revealed that the presence of chloro groups in K2 and K4 greatly enhanced the antiamoebic performance. A similar finding was reported by Ansari et al. (2015). Moreover, Martins et al. (2016) disclosed the vital role of the chloro group as a halogen atom bonded to the lateral chain of the compounds can provide additional lipophilicity and polarizability, which are important features to improve the antimicrobial activities. In addition, Yoon et al. (2022) emphasized that the incorporation of halogen atoms in a drug enhances its oral absorption and increases its cell membrane permeability, thus halogen is typically used to lead the development and optimization of the antimicrobial activity. The difference in IC50 values between derivatives K2 and K4 could be due to the difference in the total number of halogen atoms attached to the phenyl ring. According to Molchanova et al. (2020), the antimicrobial activity increases in parallel with the number of halogen atoms. On the other hand, the incorporation of the 1,3-dioxolane group in compound K6 exerted significant antiamoebic activity with an IC50 of 0.73 mg/mL. The result is supported by Schmidt et al. (2007), whereby the 1,3-dioxolane group contributes to the compounds’ antimicrobial activity. Compound K5 with the methoxy group showed a good toxicity effect against Acanthamoeba with an IC50 of 6.10 mg/mL. Bhutani et al., (1987) highlighted that in general, methoxy substituent groups of compounds can exhibit various biological activities, thus supporting the data acquired in the present study. In comparison, both compounds K1 and K3 only demonstrated moderate activity with an IC50 value of 24.83 mg/mL and 19.57 mg/mL, respectively. The results obtained could be attributed to the alkyl chain group attached to the phenyl ring. Raja et al. (2017) revealed that electron-donating groups on the aromatic compounds play an important role in raising the effectiveness of these in vitro activities.
According to Lipinski’s rule of five (ROF), most “drug-like” molecules have a molecular weight ≤ 500, log P (partition coefficient) ≤ 5, number of hydrogen acceptors ≤ 10 and number of hydrogen donors ≤ 5 (Lipinski et al., 1997). These parameters were used as guidelines for oral absorption trends. All eugenol derived molecules were found in compliance with the Lipinski rule, except for compound K1, where the log P exceeded the threshold value. However, violation of one rule might not necessarily result in poor absorption, but the probability of poor absorption increases as a greater number of rules are violated. Violation of more than two rules is classified as “ALERT” (Choy and Prausnitz, 2011). Overall observation from the results of the cytotoxicity study revealed that compounds K2 and K4 containing the chloro group were more active than the other remaining compounds, which is in agreement with many works (Abid and Azam, 2006, Patel and Shaikh, 2010, Maurya et al., 2020). Moreover, it has been demonstrated that the chlorine in chlorhexidine contributes to its antiamoebic activity. Thus, it can be suggested that these compounds have good permeability across the cell membrane. Although the IC50 of chlorhexidine is lower than most of the synthesized compounds, chlorhexidine also induces side effects on the host cornea. It is hoped that eugenol derived molecules are less toxic to the host cells.
3.3. Acanthamoeba morphological changes
Light microscopy observation was performed to visualize the differences between untreated Acanthamoeba sp. cells and Acanthamoeba cells treated with eugenol derived molecules based on their morphological changes as presented in Fig. 2.
Fig. 2.
Light microscopy of Acanthamoeba sp.; (a) Untreated amoeba; (b) K1-treated amoeba; (c) K2-treated amoeba; (d) K3-treated amoeba; (e) K4-treated amoeba; (f) K5-treated amoeba; (g) K6-treated amoeba; (h) Chlorhexidine-treated amoeba (magnification 600×).
Based on the observation, the untreated Acanthamoeba sp. cells exhibited an irregular shape with a distinct structure of acanthopodia; a spiny surface projection, vacuoles and nucleus which indicated the active and vegetative forms as shown in Fig. 2a. As for the chlorhexidine-treated cells, the cells remained in irregular shape, however the vacuoles and acanthopodia almost disappeared. In contrast, eugenol derived molecules not only affected cell viability, but also induced morphological changes in the Acanthamoeba sp. cells. Eugenol derived-treated cells showed a decrease in cell size with the cells in a round shape and the absence of vacuoles (Fig. 2b), which indicated the encystment event has occurred. The encystment process occurs when Acanthamoeba sp. Cells experience stress or unfavourable conditions, which in this case was due to the presence of eugenol-derived molecules that inhibits their growth. This process is important for the survival of the Acanthamoeba sp. cells and it leads to the formation of resilient cysts from vegetative trophozoites. Consequently, the results obtained in this study indicate that eugenol derived molecules could induce the encystment event and create stress conditions towards Acanthamoeba sp. cells.
Furthermore, through this morphological assessment, one of the obvious changes detected was that exposure of eugenol-derived molecules to the cells caused the disruption of cell membrane that resulted in the loss of the acanthopodia structure. Acanthopodia are spike-like projections of Acanthamoeba sp. cells that help the cells in their locomotory, such as moving on a substrate and catching prey (Khan, 2006). Based on the result, the eugenol-derived molecules modified the structure of the acanthopodia at the surface of Acanthamoeba sp. cells. The treated cells became rounded and floated in the culture medium, perhaps as a result of the loss of acanthopodia. The loss of acanthopodia can result in a lesser pathogenicity effect to humans, as this structure plays an important role in the pathogenesis of Acanthamoeba sp. infections. The acanthopodia help Acanthamoeba sp. cells to make initial contact with the invaded host cells, by promoting the cells’ binding (Nakisah et al., 2012). Treatment with eugenol derivatives can thus inactivate Acanthamoeba to prevent it from affecting the host during pathogenesis. Likewise, the chlorhexidine-treated Acanthamoeba sp. cells (Fig. 2h) that acted as a positive control in this study gave comparable effects on the morphology of Acanthamoeba.
3.4. Acanthamoeba membrane integrity
Biochemical activities in Acanthamoeba cells were observed by using fluorescent microscopy based on AO and PI staining to confirm the mode of cell death induced by the eugenol-derived molecules. The distinction of the cell death type is very important and relevant in the determination of the cytotoxic effect of eugenol-derived molecules on Acanthamoeba sp. cells. The Acanthamoeba sp. cells may undergo three major types of cell death after being treated with the eugenol derivatives at IC50 value after incubation for 24 h. In general, these types of cell death are classified based on cell morphology features which are currently distinguished as apoptosis (known as type I cell death), autophagic (type II) and necrosis (type III) (Van Doorn, 2011). The fluorescence microscopy observation of Acanthamoeba sp. cells is presented in Fig. 3.
Fig. 3.
Fluorescence microscopy of Acanthamoeba sp. (a) Untreated amoeba; (b) K1-treated amoeba; (c) K2-treated amoeba; (d) K3-treated amoeba; (e) K4-treated amoeba; (f) K5-treated amoeba; (g)K6-treated amoeba; (h) Chlorhexidine-treated amoeba (magnification 600×).
In this study, AO and PI stains were used to differentiate the intact and non-intact membranes of the cells under the fluorescence microscope to determine the cell membrane’s integrity. The cell membrane plays an important role in maintaining the inner compartment of cells to ensure their survival. However, unfavourable conditions will cause the cell’s membrane integrity to be disrupted, which could induce apoptosis, autophagy, and necrosis of cells. According to Kusrini et al. (2016), the internal morphological changes in treated Acanthamoeba sp. cells cannot be confirmed and supported based on light microscopy observation alone, but need to be supported by using the fluorescence microscopy technique, so as to determine the mode of cell death induced by the cytotoxic compound used.
Based on the AO/PI staining results for the untreated Acanthamoeba sp. cells, the cells appeared as green fluorescence cells. The cells were fluorescent with green cytoplasm, green vacuoles and nuclei as can be seen in Fig. 3a. The green fluorescence cells with intact nucleus indicated that the cells are viable and healthy cells (Kusrini et al., 2018). According to Nakisah et al. (2012), AO is a membrane-permeable dye that is a nucleic acid selective dye that binds to the viable cells and thus produces green fluorescence in low concentration (Hashim et al., 2015).
However, for the Acanthamoeba sp. cells that were treated with eugenol derivatives, the result showed they were stained green with orange-yellow fluorescence organelles in green cytoplasm (Fig. 3b), indicating that the cells underwent the autophagic type of cell death. Autophagic cell death is a process that involves the formation of a double-membrane structure containing the sequestered cytoplasmic material, the autophagosome that subsequently fuses with the lysosome, forming an autolysosome, which later will end with the death of the autophagosome because of exposure to cytotoxic drugs (Hale et al., 2013). From the result obtained, the orange-yellow fluorescence precipitates observed were the active lysosomes that were stained by the AO dye. The lysosome is an organelle that is present in Acanthamoeba sp. cells that contain digestive enzymes (acid hydrolases) which will help in digesting excess or worn-out organelles, food particles and engulfed viruses or bacteria (Hashim and Amin, 2017). The high activity of hydrolytic enzymes in the lysosomes due to the reaction between the lysosomes and the treatment given is believed to have lowered the pH inside the lysosomes. The uptake of AO dye into the lysosomes was the result of an active proton pump in lysosomes where the high proton concentration (low pH) caused the AO, which could enter the lysosome in uncharged form, to become protonated and later be entrapped in the organelle and emit the orange fluorescence precipitates (Kusrini et al. 2016). Based on the fluorescence microscopy observation, it can be concluded that the AO dye became a cationic dye when entering the acidic compartment such as lysosome in low pH conditions, giving the orange fluorescence precipitates. The observation results for both untreated and treated Acanthamoeba sp. cells support the previous findings that were based on the light microscopy observation, thus proving that eugenol-derived molecules created pressure that caused morphological changes in Acanthamoeba sp. cells and induced the autophagic type of cell death after the cells were exposed to treatment at their IC50 value for 24 h of incubation.
In contrast, after being treated with chlorhexidine as a positive control treatment, the cells underwent a necrotic type of cell death. After being exposed to the chlorhexidine at IC50 value in 24 h of incubation time, all the Acanthamoeba sp. cells were stained with bright orange cytoplasm (Fig. 3h) which indicated the necrotic type of cell death. According to Hashim et al. (2015), necrotic cell death occurs when the cells lose their membrane and lyse, which allows the uptake of the PI staining dye and produces red or orange colour fluorescence. Therefore, from this observation, chlorhexidine could induce necrotic type of cell death as it caused the rupture of the Acanthamoeba sp. cell membrane and thus allowed the diffusion of the PI staining dye.
3.5. Molecular docking simulation of Acanthamoeba profilin with eugenol derivatives
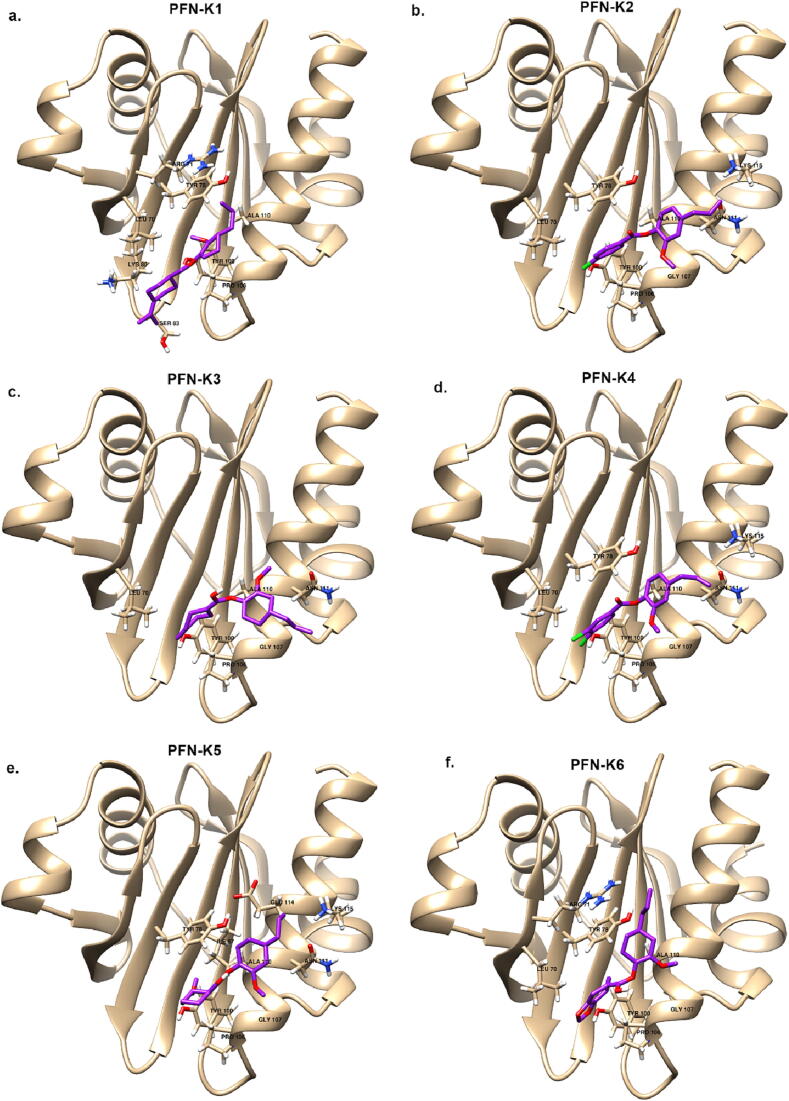
Molecular docking simulations were performed to investigate the binding affinities of the eugenol derivatives on an Acanthamoeba protein, the profilin 1B (PDB ID: 1ACF). PFN from Acanthamoeba castellanii (acPFN) demonstrates a canonical profilin fold, which is composed of seven anti-parallel β-strands sandwiched by four helices (Fig. 4). Profilin is an important actin cytoskeleton regulator that is involved in a variety of actin-based cellular processes. This protein plays a vital role in filament assembly which determines the shape of the Acanthamoeba cell. Thus, we docked eugenol derivatives with profilin 1B (PDB ID: 1ACF) at the actin binding site (Fig. 5). However, to date, there is no crystal structure of profilin-beta-actin from Acanthamoeba castellanii. To validate our docking results, we performed molecular docking of profilin (PDB ID: 1ACF) with actin (PDB ID: 4EFH) from Acanthamoeba castellanii and superimposed the docked structure with the crystal structure of profilin-beta-actin (PDB ID: 2BTF) (supplementary file, Fig. 37 and Fig. 38). The docking simulations revealed the interactions of all six eugenol derivatives with acPFN at the actin binding area, thus supporting our experimental data on the potential of eugenols as anti-amoebic drugs against Acanthamoeba sp. Three eugenol derivatives have the lowest binding affinity based on the ΔG value, which gives the estimated free energy of binding: 4-allyl-2-methoxyphenyl 3,4-dichlorobenzoate (K4) with −7.01 kcal/mol, 4-allyl-2-methoxyphenyl 4-chlorobenzoate (K2) with −6.69 kcal/mol, and 4-allyl-2-methoxyphenylbenzo[d] (Abid and Azam, 2006, Anjum et al., 2022) dioxole-5-carboxylate (K6) with −6.51 kcal/mol. This can be directly attributed to the hydrogen bond interaction with the Tyr 100 residue. The average binding energies of other derivatives (K5, K3 and K1) were between −6.40 and −6.22 kcal/mol (Table 2). The stability of eugenol derivatives is associated with the number of hydrophobic interactions with surrounding residues, including Leu 70, Tyr 78, Pro 106, Gly 107, and Ala 110 indicating that these residues play an important role in acPFN activity. All the eugenol derivatives were found to bind at the actin-binding site pocket with strong hydrophobic interactions with profilin. These interactions could affect the protein's actin-binding capabilities, causing the Acanthamoeba's shape and mobility to be disrupted, which is consistent with the observed morphological changes and loss of acanthopodia structure after exposure to the eugenol derivatives.
Fig. 4.
Overview of Acanthamoeba castellanii profilin 1B structure (PFN). The PFN architecture contains four α-helices at the outer layer (shown in blue) and seven β-strands at the inner core region (shown in red). The figure was drawn in ribbon representation using UCSF Chimera.
Fig. 5.
The binding mode of six eugenol derivatives (purple stick models) a) 4-allyl-2-methoxyphenyl 4-isopropylbenzoate (K1) b) 4-allyl-2-methoxyphenyl 4-chlorobenzoate (K2) c) 4-allyl-2-methoxyphenyl 4-methylbenzoate (K3) d) 4-allyl-2-methoxyphenyl 3,4-dichlorobenzoate (K4) e) 4-allyl-2-methoxyphenyl 3-methoxybenzoate (K5) and f) 4-allyl-2-methoxyphenylbenzo[d] (Abid and Azam, 2006, Anjum et al., 2022) dioxole-5-carboxylate (K6).
Table 2.
Molecular docking simulation of Acanthamoeba profilin with eugenol derivatives.
| Code | Molecular summary | Compound structure | Binding energy (kcal/mol) | Interacting amino acid residues |
|---|---|---|---|---|
| K1 | 4-allyl-2-methoxyphenyl 4-isopropylbenzoate |  |
−6.22 | Leu 70, Arg 71, Tyr 78, Lys 80, Ser 83, Tyr 100, Pro 106, Ala 110 |
| K2 | 4-allyl-2-methoxyphenyl 4-chlorobenzoate |  |
−6.69 | Leu 70, Tyr 78, Tyr 100, Pro 106, Gly 107, Ala 110, Asn 111, Lys 115 |
| K3 | 4-allyl-2-methoxyphenyl 4-methylbenzoate |  |
−6.28 | Leu 70, Tyr 100, Pro 106, Gly 107, Ala 110, Asn 111 |
| K4 | 4-allyl-2-methoxyphenyl 3,4-dichlorobenzoate | 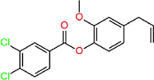 |
−7.01 | Leu 70, Tyr 78, Tyr 100, Pro 106, Gly 107, Ala 110, Asn 111, Lys 115 |
| K5 | 4-allyl-2-methoxyphenyl 3-methoxybenzoate |  |
−6.40 | Tyr 78, Ille 87, Tyr 100, Pro 106, Gly 107, Ala 110, Asn 111, Glu 114, Lys 115 |
| K6 | 4-allyl-2 methoxyphenyl benzo[d][1,3] dioxole-5-carboxylate |  |
−6.51 | Leu 70, Arg 71, Tyr 78, Tyr 100, Pro 106, Gly 107, Ala 110 |
4. Conclusion
In conclusion, nine compounds were successfully synthesized by structural modification of eugenol. The derivatives were spectroscopically characterized by FTIR, 1H NMR and 13C NMR. Most of the derivatives used in this study demonstrated promising antiamoebic activity against Acanthamoeba sp. with IC50 values ranging from 0.61 to 24.83 mg/mL. Among them, compound K4 exhibited the strongest activity, and this finding correlated with the presence of chloro groups in the molecular structure. Through microscopic observation, the tested compounds were found to cause Acanthamoeba cells to become inactive and disrupted their membrane integrity. The loss of the acanthopodia structure is also consistent with molecular docking simulations that show strong interactions between eugenol derivatives and profilin, that can disrupt the protein's actin-binding capacity. Considering these compounds are very cytotoxic to the Acanthamoeba cells and have high potential for use as antiamoebic agents, the effects of eugenol derivatives against host cells should be further investigated.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research is partly funded by the Ministry of Higher Education, Malaysia under Research Acculturation Collaborative Effort RACE/F1/ST3/UMT/5 (Vote RACE-50068). The authors also would like to thank the Faculty of Science and Marine Environment, Universiti Malaysia Terengganu, Terengganu for providing the space and facility to carry out the work.
Footnotes
Peer review under responsibility of King Saud University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jsps.2023.101703.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- Abid M., Azam A. Synthesis, characterization and antiamoebic activity of 1-(thiazolo [4, 5-b] quinoxaline-2-yl)-3-phenyl-2-pyrazoline derivatives. Bioorg. Med. Chem. Lett. 2006;16(10):2812–2816. doi: 10.1016/j.bmcl.2006.01.116. [DOI] [PubMed] [Google Scholar]
- Ait-Ouazzou A., Cherrat L., Espina L., Lorán S., Rota C., Pagán R. The antimicrobial activity of hydrophobic essential oil constituents acting alone or in combined processes of food preservation. Innov. Food Sci. Emerg. Technol. 2011;12(3):320–329. [Google Scholar]
- Anjum N.F., Shanmugarajan D., Shivaraju V.K., Faizan S., Naishima N.L., Kumar B.R.P., Javidac S., Purohit M.N. Novel derivatives of eugenol as potent antiinflammatory agents via PPARg agonism: rational design, synthesis, analysis, PPARg protein binding assay and computational studies. RSC Adv. 2022;12:16966–16978. doi: 10.1039/d2ra02116a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anacarso, I., Sabia, C., de Niederhäusern, S., Iseppi, R., Condò, C., Bondi, M. and Messi, P. (2017) In vitro evaluation of the amoebicidal activity of rosemary (Rosmarinus officinalis L.) and cloves (Syzygium aromaticum L. Merr. & Perry) essential oils against Acanthamoeba polyphaga trophozoites. Natural Product Research, 33(4), 606–611. [DOI] [PubMed]
- Ansari M.F., Siddiqui S.M., Agarwal S.M., Vikramdeo K.S., Mondal N., Azam A. Metronidazole hydrazone conjugates: design, synthesis, antiamoebic and molecular docking studies. Bioorg. Med. Chem. Lett. 2015;25(17):3545–3549. doi: 10.1016/j.bmcl.2015.06.091. [DOI] [PubMed] [Google Scholar]
- Bai X., Li X., Liu X., Xing Z., Su R., Wang Y., Xia X., Shi C. Antibacterial effect of eugenol on Shigella flexneri and its mechanism. Foods. 2022;11:2565. doi: 10.3390/foods11172565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutani K.K., Sharma G.L., Ali M. Plant based antiamoebic drugs; Part I. Antiamoebic activity of phenanthroindolizidine alkaloids; common structural determinants of activity with emetine. Planta Med. 1987;53(06):532–536. doi: 10.1055/s-2006-962803. [DOI] [PubMed] [Google Scholar]
- Castellani A. An amoeba found in culture of yeast: preliminary note. Am. J. Trop. Med. Hyg. 1930;33:160. [Google Scholar]
- Cha J.H.J., Furie K., Kay J., Walensky R.P., Mullins M.E., Hedley-Whyte E.T. Case 39–2006: A 24-year-old woman with systemic lupus erythematosus, seizures, and right arm weakness. N. Engl. J. Med. 2006;355(25):2678–2689. doi: 10.1056/NEJMcpc069030. [DOI] [PubMed] [Google Scholar]
- Choy Y.B., Prausnitz M.R. The rule of five for non-oral routes of drug delivery: ophthalmic, inhalation and transdermal. Pharm. Res. 2011;28(5):943–948. doi: 10.1007/s11095-010-0292-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Z., Liu Z., Zeng J., Chen L., Wu Q., Mo J., Zhang G., Song Y., Xu W., Zhang S., Guo X. Eugenol inhibits non-small cell lung cancer by repressing expression of NF-κB-regulated TRIM59. Phytother. Res. 2019;33(5):1562–1569. doi: 10.1002/ptr.6352. [DOI] [PubMed] [Google Scholar]
- Daniel A.N., Sartoretto S.M., Schmidt G., Caparroz-Assef S.M., Bersani-Amado C.A., Cuman R.K.N. Anti-inflammatory and antinociceptive activities A of eugenol essential oil in experimental animal models. Rev. Bras. 2009;19(1B):212–217. [Google Scholar]
- de Souza T.B., Orlandi M., Coelho L.F.L., Malaquias L.C.C., Dias A.L.T., de Carvalho R.R., Silva N.C., Carvalho D.T. Synthesis and in vitro evaluation of antifungal and cytotoxic activities of eugenol glycosides. Med. Chem. Res. 2014;23(1):496–502. [Google Scholar]
- Devi K.P., Nisha S.A., Sakthivel R., Pandian S.K. Eugenol (an essential oil of clove) acts as an antibacterial agent against Salmonella typhi by disrupting the cellular membrane. J. Ethnopharmacol. 2010;130(1):107–115. doi: 10.1016/j.jep.2010.04.025. [DOI] [PubMed] [Google Scholar]
- ElGhannam M., Dar Y., ElMehlawy M.H., Mokhtar F.A., Bakr L. Eugenol; effective anthelmintic compound against foodborne parasite Trichinella spiralis muscle larvae and adult. Pathogens. 2023;12:127. doi: 10.3390/pathogens12010127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan X., Wang Z., Hu D. Synthesis of novel antiviral ferulic acid-eugenol and isoeugenol hybrids using various link reactions. J. Agric. Food Chem. 2021;69(46):13724–13733. doi: 10.1021/acs.jafc.1c05521. [DOI] [PubMed] [Google Scholar]
- Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., Ferrin T.E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2017 doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosdidier A., Zoete V., Michielin O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011;39 doi: 10.1093/nar/gkr366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale A.N., Ledbetter D.J., Gawriluk T.R., Rucker E.B., III Autophagy: regulation and role in development. Autophagy. 2013;9(7):951–972. doi: 10.4161/auto.24273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashim, F., Amin, N.M., 2017. Insights into the prominent effect of mahanimbine on Acanthamoeba castellanii: Cell profiling analysis based on microscopy techniques. In: AIP Conference Proceedings. vol. 1817, no. 11, pp. 030002.
- Hashim F., Rahman N.A.A., Amin N.M. Morphological analysis on the toxic effect of manganese on Acanthamoeba sp. isolated from Setiu Wetland, Terengganu: An in vitro study. Procedia Environ. Sci. 2015;30:15–20. [Google Scholar]
- Ju J., Xie Y., Yu H., Guo Y., Cheng Y., Qian H., Yao W. Analysis of the synergistic antifungal mechanism of eugenol and citral. LWT. 2020;123 [Google Scholar]
- Kaufman T.S. The multiple faces of eugenol. A versatile starting material and building block for organic and bio-organic synthesis and a convenient precursor toward bio-based fine chemicals. J. Braz. Chem. Soc. 2015;26(6):1055–1085. [Google Scholar]
- Khan N.A. Acanthamoeba: biology and increasing importance in human health. FEMS Microbiol. Rev. 2006;30(4):564–595. doi: 10.1111/j.1574-6976.2006.00023.x. [DOI] [PubMed] [Google Scholar]
- Kim S., Lee J., Jo S., Brooks C.L., Lee H.S., Im W. CHARMM-GUI ligand reader and modeler for CHARMM force field generation of small molecules. J. Comput. Chem. 2017;38(21):1879–1886. doi: 10.1002/jcc.24829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusrini E., Hashim F., Azmi W.N.N.W.N., Amin N.M., Estuningtyas A. A novel antiamoebic agent against Acanthamoeba sp.—A causative agent for eye keratitis infection. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2016;153:714–721. doi: 10.1016/j.saa.2015.09.021. [DOI] [PubMed] [Google Scholar]
- Kusrini E., Hashim F., Gunawan C., Mann R., Azmi W.N.N.W.N., Amin N.M. Anti-amoebic activity of acyclic and cyclic-samarium complexes on Acanthamoeba. Parasitol. Res. 2018;117(5):1409–1417. doi: 10.1007/s00436-018-5814-x. [DOI] [PubMed] [Google Scholar]
- Lackner P., Beer R., Broessner G., Helbok R., Pfausler B., Brenneis C., Auer H., Walochnik J., Schmutzhard E. Acute granulomatous Acanthamoeba encephalitis in an immunocompetent patient. Neurocrit. Care. 2010;12(1):91–94. doi: 10.1007/s12028-009-9291-z. [DOI] [PubMed] [Google Scholar]
- Lane T., Anantpadma M., Freundlich J.S., Davey R.A., Madrid P.B., Ekins S. The natural product eugenol is an inhibitor of the ebola virus in vitro. Pharm. Res. 2019;36(7):1–6. doi: 10.1007/s11095-019-2629-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R.A., Swindells M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011;51(10):2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997;23(1–3):3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- Lorenzo-Morales J., Kliescikova J., Martinez-Carretero E., De Pablos L.M., Profotova B., Nohynkova E., Osuna A., Valladares B. Glycogen phosphorylase in Acanthamoeba spp.: determining the role of the enzyme during the encystment process using RNA interference. Eukaryot. Cell. 2008;7(3):509–517. doi: 10.1128/EC.00316-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo-Morales J., Marciano-Cabral F., Lindo J.F., Visvesvara G.S., Maciver S.K. Pathogenicity of amoebae. Exp. Parasitol. 2010;126:2–3. doi: 10.1016/j.exppara.2010.05.003. [DOI] [PubMed] [Google Scholar]
- Lorenzo-Morales J., Martín-Navarro C.M., López-Arencibia A., Arnalich-Montiel F., Piñero J.E., Valladares B. Acanthamoeba keratitis: an emerging disease gathering importance worldwide? Trends Parasitol. 2013;29(4):181–187. doi: 10.1016/j.pt.2013.01.006. [DOI] [PubMed] [Google Scholar]
- Machado M., Dinis A.M., Salgueiro L., Custódio J.B., Cavaleiro C., Sousa M.C. Anti-Giardia activity of Syzygium aromaticum essential oil and eugenol: effects on growth, viability, adherence and ultrastructure. Exp. Parasitol. 2011;127(4):732–739. doi: 10.1016/j.exppara.2011.01.011. [DOI] [PubMed] [Google Scholar]
- Marciano-Cabral F., Cabral G. Acanthamoeba spp. as agents of disease in humans. Clin. Microbiol. Rev. 2003;16(2):273–307. doi: 10.1128/CMR.16.2.273-307.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins R.M., Farias M.D.A., Nedel F., de Pereira C.M., Lencina C., Lund R.G. Antimicrobial and cytotoxic evaluation of eugenol derivatives. Med. Chem. Res. 2016;25:2360–2367. [Google Scholar]
- Maurya A.K., Agarwal K., Gupta A.C., Saxena A., Nooreen Z., Tandon S., Ahmad A., Bawankule D.U. Synthesis of eugenol derivatives and its anti-inflammatory activity against skin inflammation. Nat. Prod. Res. 2020;34(2):251–260. doi: 10.1080/14786419.2018.1528585. [DOI] [PubMed] [Google Scholar]
- Mayer P.L., Larkin J.A., Hennessy J.M. Amebic encephalitis. Surg. Neurol. Int. 2011;2 doi: 10.4103/2152-7806.80115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memari F., Niyyati M., Joneidi Z. Pathogenic Acanthamoeba T4 genotype isolated from mucosal tissue of a patient with HIV infection: a case report. Iran. J. Parasitol. 2017;12(1):143. [PMC free article] [PubMed] [Google Scholar]
- Molchanova N., Nielsen J.E., Sørensen K.B., Prabhala B.K., Hansen P.R., Lund R., Barron A.E., Jenssen H. Halogenation as a tool to tune antimicrobial activity of peptoids. Sci. Rep. 2020;10(1):14805. doi: 10.1038/s41598-020-71771-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakisah M.A., Muryany M.I., Fatimah H., Fadilah R.N., Zalilawati M.R., Khamsah S., Habsah M. Anti-amoebic properties of a Malaysian marine sponge Aaptos sp. on Acanthamoeba castellanii. World J. Microbiol. Biotechnol. 2012;28(3):1237–1244. doi: 10.1007/s11274-011-0927-8. [DOI] [PubMed] [Google Scholar]
- Niyyati M., Lorenzo-Morales J., Rezaie S., Rahimi F., Mohebali M., Maghsood A.H., Motevalli-Haghi A., Martin-Navarro C.M., Farnia S., Valladares B., Rezaeian M. Genotyping of Acanthamoeba isolates from clinical and environmental specimens in Iran. Exp. Parasitol. 2009;121(3):242–245. doi: 10.1016/j.exppara.2008.11.003. [DOI] [PubMed] [Google Scholar]
- O’Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R. Open Babel: An Open chemical toolbox. J. Cheminf. 2011;3(10):1–14. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel N.B., Shaikh F.M. Synthesis and antimicrobial activity of new 4-thiazolidinone derivatives containing 2-amino-6-methoxybenzothiazole. Saudi Pharmaceut. J. 2010;18(3):129–136. doi: 10.1016/j.jsps.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrucha-Dilanchian P., Chan J.C., Castellano-Sanchez A., Hirzel A., Laowansiri P., Tuda C., Visvesvara G.C., Qvarnstorm Y., Ratzan K.R. Balamuthia mandrillaris and Acanthamoeba amebic encephalitis with neurotoxoplasmosis coinfection in a patient with advanced HIV infection. J. Clin. Microbiol. 2012;50(3):1128–1131. doi: 10.1128/JCM.06252-11. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Rahim N.H.C.A., Asari A., Ismail N., Osman H. Synthesis and antibacterial study of eugenol derivatives. Asian J. Chem. 2017;29(1):22. [Google Scholar]
- Raja M.R.C., Velappan A.B., Chellappan D., Debnath J., Mahapatra S.K. Eugenol derived immunomodulatory molecules against visceral leishmaniasis. Eur. J. Med. Chem. 2017;139:503–518. doi: 10.1016/j.ejmech.2017.08.030. [DOI] [PubMed] [Google Scholar]
- Razali S.A., Shamsir M.S. Characterisation of a catalytic triad and reaction selectivity in the dual mechanism of the catalyse hydride transfer in xylitol phosphate dehydrogenase. J. Mol. Graph. Model. 2020;97 doi: 10.1016/j.jmgm.2020.107548. [DOI] [PubMed] [Google Scholar]
- Schmidt M., Ungvári J., Glöde J., Dobner B., Langner A. New 1, 3-dioxolane and 1, 3-dioxane derivatives as effective modulators to overcome multidrug resistance. Bioorg. Med. Chem. 2007;15(6):2283–2297. doi: 10.1016/j.bmc.2007.01.024. [DOI] [PubMed] [Google Scholar]
- Shirwadkar C.G., Samant R., Sankhe M., Deshpande R., Yagi S., Schuster F.L., Sriram R., Visvesvara G.S. Acanthamoeba encephalitis in patient with systemic lupus, India. Emerg. Infect. Dis. 2006;12(6):984. doi: 10.3201/eid1206.060087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui R., Aqeel Y., Khan N.A. The development of drugs against Acanthamoeba infections. Antimicrob. Agents Chemother. 2016;60(11):6441–6450. doi: 10.1128/AAC.00686-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teles A.M., Silva-Silva J.V., Fernandes J.M.P., Abreu-Silva A.L., Calabrese K.D.S., Mendes Filho N.E., Mouchrek A.D., Almeida-Souza F. GC-MS characterization of antibacterial, antioxidant, and antitrypanosomal activity of syzygium aromaticum essential oil and eugenol. Evid.-Based Complem. Alternat. Med. 2021;2021 doi: 10.1155/2021/6663255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doorn W.G. Classes of programmed cell death in plants, compared to those in animals. J. Exp. Bot. 2011;62(14):4749–4761. doi: 10.1093/jxb/err196. [DOI] [PubMed] [Google Scholar]
- Vladu A.F., Marin S., Neacsu I., Truşcă R., Kaya M., Kaya D., Popa A., Popiană C., Cristescu I., Orlov C., Ficau D., Ficau A., Udeanu D.I., Valescu B.S., Nikolouzakis T.K., Gurevich L., Kuskov A.N., Nitipar C. Spongious fillers based on collagen-hydroxyapatite-eugenol acetate with therapeutic potential in bone cancer. Farmacia. 2020;68(2):313–321. [Google Scholar]
- Westmoreland S.V., Rosen J., MacKey J., Romsey C., Xia D.L., Visvesvera G.S., Mansfield K.G. Necrotizing meningoencephalitis and pneumonitis in a simian immunodeficiency virus-infected rhesus macaque due to Acanthamoeba. Vet. Pathol. 2004;41(4):398–404. doi: 10.1354/vp.41-4-398. [DOI] [PubMed] [Google Scholar]
- Yoon H.R., Chai C.C., Kim C.H., Kang N.S. A Study on the Effect of the Substituent against PAK4 Inhibition Using In Silico Methods. Int. J. Mol. Sci. 2022;23(6):3337. doi: 10.3390/ijms23063337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamli K.M., Asari A., Hashim F., Yusoff H.M., Mohamad H., Abdullah F. Anti-Amoebic Activity of Eugenol Derivatives against Acanthamoeba castellanii. Malaysian J. Chem. 2020;22(3):100–110. doi: 10.1016/j.jsps.2023.101703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamli K.M., Asari A., Khaw K.Y., Murugaiyah V., Al-Rashida H.M., Yusoff H.M., Hasnah N.H.A.W. Cholinesterase inhibition activity and molecular docking study of eugenol derivatives. Sains Malaysiana. 2021;50(4):1037–1045. [Google Scholar]
- Zhao Y., Wang Q., Wu X., Jiang M., Jin H., Tao K., Hou T. Unraveling the polypharmacology of a natural antifungal product, eugenol, against Rhizoctonia solani. Pest Manag. Sci. 2021;77(7):3469–3483. doi: 10.1002/ps.6400. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.